Abstract
The competition-relatedness hypothesis (CRH) predicts that the strength of competition is the strongest among closely related species and decreases as species become less related. This hypothesis is based on the assumption that common ancestry causes close relatives to share biological traits that lead to greater ecological similarity. Although intuitively appealing, the extent to which phylogeny can predict competition and co-occurrence among species has only recently been rigorously tested, with mixed results. When studies have failed to support the CRH, critics have pointed out at least three limitations: (i) the use of data poor phylogenies that provide inaccurate estimates of species relatedness, (ii) the use of inappropriate statistical models that fail to detect relationships between relatedness and species interactions amidst nonlinearities and heteroskedastic variances, and (iii) overly simplified laboratory conditions that fail to allow eco-evolutionary relationships to emerge. Here, we address these limitations and find they do not explain why evolutionary relatedness fails to predict the strength of species interactions or probabilities of coexistence among freshwater green algae. First, we construct a new data-rich, transcriptome-based phylogeny of common freshwater green algae that are commonly cultured and used for laboratory experiments. Using this new phylogeny, we re-analyse ecological data from three previously published laboratory experiments. After accounting for the possibility of nonlinearities and heterogeneity of variances across levels of relatedness, we find no relationship between phylogenetic distance and ecological traits. In addition, we show that communities of North American green algae are randomly composed with respect to their evolutionary relationships in 99% of 1077 lakes spanning the continental United States. Together, these analyses result in one of the most comprehensive case studies of how evolutionary history influences species interactions and community assembly in both natural and experimental systems. Our results challenge the generality of the CRH and suggest it may be time to re-evaluate the validity and assumptions of this hypothesis.
Keywords: evolutionary ecology, molecular phylogenetics, community ecology
1. Introduction
A long-standing aim of ecological research is to understand how evolutionary relatedness influences the strength of species interactions and the ability of species to coexist [1–4]. Darwin [5] proposed that competition should be strongest between close relatives because they share with each other more traits that influence species interactions, such as habitat use, the types of resources consumed and potential shared predators. As such, he argued that closely related species should be the least likely to coexist. This idea is now commonly called the competition-relatedness hypothesis (CRH) [1], which is sometimes extended to suggest that species must evolve to differ by some minimum amount in order to stably coexist—an idea known as the phylogenetic limiting similarity [6]. Comprehensive tests of this hypothesis are critical because strong support for the CRH could motivate biologists to use phylogenetic distances between species to make important management decisions. For example, it has already been argued that phylogenetic distances could help prioritize species for conservation that have the greatest ecological uniqueness [7], help identify species that pose the greatest risk of being invasive [8] and help maximize the restoration of ecological function in degraded ecosystems [9].
Despite important potential applications of the CRH, there is far from consistent support for it. While select studies have found support for the CRH for herbaceous plants [8], arbuscular mycorrhizal fungi [10] and microbes [11–13], an increasing number of studies, performed in a wide variety of systems, have shown that evolutionary relationships do not predict the nature of interactions among species [1,14–21]. Venail et al. [17] recently summarized 20 experimental tests (see their table 1) and found that only six studies to date provided clear support for the CRH, and most of those varied across experimental treatments or conditions. Although the summary was informal, it did suggest that support for the CRH is less common than often presumed, and results are often inconsistent, even within individual studies. This, along with emerging evidence that ecological traits often lack phylogenetic signal [22], suggests we need a more critical evaluation of the CRH.
A failure to detect evidence in support of the CRH could be due to at least three limitations. First, there may be systematic biases in estimates of evolutionary relationships caused by the use of data-poor phylogenies. Evolutionary relatedness is often measured using metrics of phylogenetic diversity (PD) generated from taxonomy or incomplete phylogenies estimated using a small number of publicly available genes. More data-rich phylogenetic analyses commonly alter previously published results owing to major shifts in branch topology [23–30], such as the rearrangement of deep nodes or entire clades within the tree that lead to systematic biases when calculating PD. Second, the statistical methods may be inadequate to detect relationships between PD and ecological parameters. Recent work based on data simulations suggest that, under a Brownian model of trait evolution, trait dissimilarity may increase nonlinearly as species diverge [31]. Divergence may also cause variances in species traits to increase as a function of PD [32]. Studies that use general linear models to relate ecological variables to PD may fail to identify nonlinearities or account for heteroskedasticity of variances that mask relationships. Third, most tests of the CRH stem from laboratory experiments or experimental field settings where conditions do not reflect ecological interactions in natural systems that have changed over evolutionary time periods [33]. In fact, niche differences that allow species to be unique, and thus minimize their interactions, may be diminished and impossible to detect in short-term experiments [34]. The challenges of testing the CRH motivate the examination of select model systems in depth.
In a recent sequence of papers using freshwater green algae as a model system, we have failed to find support for the CRH [15–17]. All three papers were based on laboratory experiments that: (i) used publicly available gene sequences to estimate molecular phylogenies, (ii) manipulated the phylogenetic distance separating experimental species pairs, and then (ii) grew the species in monocultures and bicultures to measure how species interact. First, Fritschie et al. [15] used a relatively crude measure of species interactions calculated from relative densities (RD, cell density polyculture : monoculture), but analysed a large number of combinations of different species. Second, Venail et al. [17] had less breadth (28 combinations of eight species), but used a more refined metric of competition, measuring growth rates of a focal species introduced into a steady-state population of a potential competitor. Third, Narwani et al. [16], who also analysed 28 species combinations, provided the most refined and generalizable [35,36] estimates of competition and coexistence by using the mutual invisibility criterion to quantify the strength of niche as well as fitness differences among species pairs. Despite differences in experimental details, all three studies led to the same conclusion that evolutionary relatedness does not predict the strength of species interactions, or the probability of coexistence among common species of freshwater green algae.
Most studies examining mechanisms of coexistence use null community phylogenetic models or rely on experimental data that can be correlated with PD, while few if any combine these two approaches. Here, we extend in multiple ways our previous work on algal phylogenetic ecology and show—counter to the CRH—that phylogenetic distance does not predict ecological traits in the laboratory or natural systems. We first present a new, state of the art, data-rich, transcriptome-based phylogeny for 53 species of common and culturable freshwater green algae that have been used in laboratory studies. In addition, we find no support for statistical explanations for a lack of fit between PD and laboratory-measured ecological variables. Finally, we expand beyond experimental inferences by examining biogeographic patterns of species co-occurrence across 1077 natural lake communities in North America. Consistent with laboratory results, we show that species compositions of an overwhelming majority of North American lakes (more than 99%) are randomly composed with respect to phylogeny. Taken together, these results provide one of the most comprehensive examinations of the relationship between evolutionary history and species interactions and community assembly in both natural and experimental systems.
2. Material and methods
(a). Taxon selection
To estimate a phylogeny for use in ecological studies of North American freshwater green algae, we relied on two criteria to select target species for sequencing: (i) we chose species that are common and abundant across North America, and (ii) we chose species that were culturable in laboratory conditions and readily available in public culture collections (electronic supplementary material, table S1). We used the 2007 Environmental Protection Agency (EPA) National Lakes Assessment (NLA) database of cell density for the dominant plankton of representative lakes across North America (http://www.water.epa.gov/type/lakes/NLA_data.cfm) as a guide, and cross-referenced this list with international culture collections of algae. The EPA survey gives exhaustive taxonomic lists and estimates of cell density for 1077 lakes across North America—all sampled in 2007 using consistent methodology. Although this dataset represents a ‘snapshot-in-time’, it is presently the most comprehensive dataset of algal biogeography in the United States that we are aware of. Although phylogenetically nested within green algae [37], we chose not to include embryophytes (land plants) in our analyses because multiple gene duplication events within the group make overall orthology assignment and subsequent phylogenetic analysis more difficult and because land plants are outside of the ecological scope of our study. Except for land plants, our sampling includes the four primary taxonomic classes within Charophyta and five within Chlorophyta, but excludes Ulvophyceae (which are rare in freshwater) and some early diverging lineages. The electronic supplementary material gives details of algal culture conditions.
(b). RNA extraction and transcriptome sequencing
Algal cells were removed from culture media using serial centrifugation and either preserved in RNAlater at −20°C or processed immediately. We used the Macherey-Nagel NucleoSpin Plant extraction kit to isolate total RNA. First and second strand cDNA synthesis was performed using Clontech SMARTer cDNA and Clontech Advantage 2 PCR kits. We purified double-stranded cDNA using Agencourt Ampure XP beads and checked quality with a Bioanalyzer 2000. Libraries were constructed using the Illumina TruSeq kits and 100 bp paired-end sequencing was performed on an Illumina HiSeq 2000 platform using a strategy of eight barcoded, multiplexed samples per lane.
(c). Quality control and de novo assembly
We used a workflow in Galaxy [38] to process raw FASTQ output files for quality control (QC) and perform de novo assembly. For QC, files were initially groomed with FASTQ Groomer [39], followed by removal of sequencing artefacts, quality filtering and finally filtered short read data were plotted for visual inspection [39,40]. Following QC, we generated an initial de novo assembly of filtered reads with Trinity, using a minimum contig length of 150 bps [41]. We then re-assembled Trinity output using iAssembler [42] because we found Trinity often outputs multiple contigs with 100% pairwise identity that might be discarded as paralogues in downstream analyses.
(d). Orthologue determination
One of the major methodological challenges to using transcriptomic information in phylogenetics is determination of orthologues, because evaluating paralogues as orthologues leads to misleading conclusions about species relationships. One popular approach for determining orthology is the HaMStR algorithm [43]. This approach begins by defining a set of ‘primer-taxa’ whose genomes are fully sequenced and whose species relationships can be determined. Because full genomes are known from the primer-taxa, they provide a set of ‘core-orthologues’, gene families with one and only one representative gene in each of the primer-taxa. Each aligned core-orthologue gene family then provides the data for constructing a profile Hidden Markov Model (pHMM). Next, each pHMM provides a search image to find an orthologue in sequence data from each of the species to be added to the phylogenetic analysis (electronic supplementary material, figure S1). Further details are provided in the electronic supplementary material.
(e). Phylogenomics
After defining all orthologues in our dataset, we conducted multi-step phylogenomic analyses (electronic supplementary material, table S2 and figure S2) with the principle purpose of removing poorly translated and/or artefactual sequences and estimating a phylogenetic tree from a concatenated dataset. We removed problematic sequences by identifying genes on long terminal branches, which often results from problems in translation from DNA to protein. We used the WAG model because it was the most common for the nuclear genes in our dataset based on PROTTEST and the Aikake information criterion [44], while CPREV for most common for chloroplast genes. To do so, we first aligned each gene family with Muscle v. 3.8 [45], then estimated the maximum likelihood (ML) gene tree under WAG and CPREV models, respectively, for each orthologue, then we removed genes on branches that were 3 s.d. longer than that tree's median branch length. We realigned the remaining genes and conducted a second round of removing genes on long terminal branches, this time removing those that were 5 s.d. from the mean. For more detail please refer to the electronic supplementary material.
(f). Phylogenetic diversity
In experimental ecology, PD is defined as the total phylogenetic distance among species and is used to examine the correlation between evolutionary relatedness and numerous ecological variables [4,7,46]. We were interested in testing whether conclusions of these studies would change using PDs generated from larger, and far more robust pools of genetic data, and whether the results of these experiments predict a pattern of community structure. We also chose to reanalyse these data in order to demonstrate the use and applicability of our new phylogenetic framework for experimental studies investigating the effects of evolutionary history on community dynamics. We calculated PD with our phylogenomic data for algae used in three recent studies [15–17] using Osiris phylogenetic tools, implemented in Galaxy [47]. The data we re-analysed had previously relied on PD-values generated from publicly available data. Each study used at least one taxon for which data were not publicly available. In each of those cases, the phylogenetic position of the taxon with missing data had to be inferred using congeneric representatives for which data were available, assuming that those genera were monophyletic. Based on our current tree, our assumption of monophyly and the use of congeneric substitute taxa were both justified.
(g). Experimental ecology analyses
In Venail et al. [17], we performed a mesocosm laboratory experiment in which we studied the influence of evolutionary relatedness on the prevalence and strength of competitive and facilitative interactions among 28 pairs of freshwater green algal species. For each of eight species, we first measured the invasion success of the focal species when introduced into steady-state populations of all other resident species. Then, we compared the growth rate of the focal species when grown alone in monoculture to its growth rate when introduced as an invader into a steady-state population. The change in the focal species' population growth rate as an invader was used as a measure of the strength of its interaction with the resident species (what is called ‘sensitivity’ to competition). We observed no significant relationship between the phylogenetic distance separating two interacting species and the success of invasion, nor the prevalence or strength of either competition or facilitation. Thus, our results rejected the hypothesis that close relatives compete strongly and contesting recent evidence that facilitation is likely to occur more frequently between distant relatives. For this study, we re-analysed the data by replacing previous PD values with the ones originated using our new molecular phylogeny described above. Statistical tests conducted identically to those described in the original paper [17].
In a series of small-scale laboratory experiments in tissue culture well-plates, Narwani et al. [16] used a measure of sensitivity to competition to estimate the size of niche differences (ND) and competitive inequalities among species (relative fitness differences (RFD)). They also determined whether two species could coexist based on the criterion of mutual invasibility [48]. They measured species' sensitivities to competition by comparing a given species' population-level growth rate when invading an established population of another species to its growth rate when alone. They measured sensitivity ND, RFD and coexistence for 28 species pairs and compared these measures to the phylogenetic distance among species based on a phylogeny that was constructed using 18 s and rbcL sequences available on GenBank. For one species, Cosmarium turpinii, sequence data were not available, and thus, sequences from two taxa of the same genus were used to calculate PD as the closest available estimate. The original analysis suggested that phylogenetic distance was not a significant predictor of ND, RFD or coexistence. These analyses were performed again using phylogenetic distances from the new phylogenomic tree presented here, and all analyses were performed as reported in the original publication [16].
To test for evidence of the CRH among 23 freshwater algae species, Fritschie et al. [15] related PD to a metric of species interaction strength (RD: the ratio of a species' monoculture density to its density when grown with one other species) for 216 pairwise species combinations grown for 40 days in 1 ml microcosms. The authors found no significant effect of PD on the magnitude, variance or type of interaction among species, concluding that the CRH did not operate among this experimental group of algae. We reproduced the authors' analyses (simple linear regression and ANOVA) using their original interaction strength data and our newly calculated PD values.
In addition to repeating the analyses of the original three publications using our more resolved and data-rich phylogeny, we also performed additional analyses that addressed recent concerns about statistical methods that fail to detect nonlinearities or account for heterscedasticity in variances. Specifically, Letten & Cornwell [31] recently showed that under Brownian motion (a common null model of trait evolution), trait dissimilarity among species increases nonlinearly as species diverge. These authors suggested that values of PD be square-root transformed prior to analyses to linearize the response. We not only analysed transformed PD values, we went a step further and fitted data from all three studies to power functions (y = a × PDb), which have the flexibility to quantify any nonlinear relationship that is monotonically increasing or decreasing. Any values of the scaling parameter b that significantly differ from zero (no relationship) and are not equal to 1 (a linear relationship), provide evidence of a nonlinear relationship between PD and the ecological response variable. In addition to specific tests for nonlinearity, we also tested whether increased variance was positively correlated with PD [32] using Bruesch–Pagan (BP) tests of variance (heteroscedasticity) in the response variables. Please refer to the electronic supplementary material for more detailed methods of transformation.
(h). Phylogenetic structure of natural communities
As the CRH states that competition should be strongest between close relatives, it has been used to predict a pattern of phylogenetic overdispersion where evolutionary divergence between co-occurring species in natural communities is greater than expected from a null model that calculates values of PD for any set of species chosen from the phylogeny at random [3]. In order to test the hypothesis that green algae co-occurring in North American lakes are non-random assemblages with respect to their evolutionary history, we combined our newly developed phylogenomic dataset with publicly available data from GenBank. We used the NLA algal phytoplankton count data to compile a list of all genera surveyed. Algae in the NLA were mostly identified to the taxonomic level of genus owing to inherent uncertainty algal identification and the lack of distinguishing morphological characteristics among species. This necessitated the use of single taxonomic representatives per genus. We identified 99 genera in the NLA survey within the Chlorophyta and Charophyta. Of the 99 genera, our original phylogenomic dataset covered 43 genera, and an additional 45 genera were added to our phylogeny using 18 s and rbcL sequences from NCBI GenBank, while 10 were excluded from subsequent analyses due to the lack of data (electronic supplementary material, table S4). Data from NCBI GenBank (18 s and rbcL sequences) were aligned using Muscle v. 3.8 [45]. The GenBank alignment was then concatenated with the phylogenomic data. We conducted a partitioned analysis of these data, using RaxML v. 7.4.8 [49]. We applied a GTR model to the GenBank data (based on results from jModelTest), a WAG model for the nuclear amino acid data and a CPREV model for the chloroplast data. We used 100 bootstrap replicates to assess robustness of the topology, and then used Picante v. 1.6 in R v. 3.1 [50] to test whether communities of genera inhabiting 1077 lakes surveyed in the NLA are more genetically diverse than predicted by chance.
Within Picante, we used the ses.mpd (standardized effect size of mean pairwise distances) and ses.mntd (standardized effect size of mean nearest taxon distances (MNTDs)) functions to calculate community structure [50]. Both methods compare phylogenetic relatedness to the pattern expected under a null model of phylogeny or community randomization. We used null models that randomly shuffle the tip labels of the phylogeny and randomized the community data matrix with both trial-swap and independent swap algorithms. Positive values and high quantiles indicate phylogenetic overdispersion, while negative values and low quantiles indicate phylogenetic clustering. In order to assess the significance of phylogenetic community structure across the lakes, we used a minimum of 10 000 randomizations, which was increased to 100 000 simulations for samples with low p-values (Bonferroni corrected).
3. Results
(a). New transcriptomic resources for green algae
We sequenced 53 new green algae transcriptomes (electronic supplementary material, table S1), using Illumina HiSeq technology. RNA extractions from algal cultures averaged 14.5 μg per extraction of RNA and double-stranded cDNA libraries averaged 7.8 μg per library. We found no correlation between total RNA and/or total cDNA with the amount or quality of the resulting Illumina sequence data. The average number of reads per species after filtering the data for low-quality sequences (using default cut-off values in Filter by Quality from the FASTX toolbox) were 13 693 372 left-hand reads and 12 987 520 right-hand reads. This amounted to approximately 1.3 billion base pairs per species. Assemblies using Trinity [41] resulted in an average of 25 279 contigs (totalling an average of 6 201 293 bps) with a mean contig length of 241 bps and a mean GC content of 46.3%. Not only do these data provide a solid foundation for the phylogenetic focus of our current work, they can also be used in gene expression analyses and serve as valuable pilot data for future whole genome sequencing projects.
(b). Novel orthologues for green algae
The use of EvolMap in our Galaxy workflow allowed us to customize our search for orthologues within the taxonomic range of our choice, resulting in the identification of a vast quantity of orthologous genes. In fact, our results constitute the largest available collection of orthologous genes for green algae to date, making them a valuable resource for any researcher wanting to reconstruct the evolutionary relationships of green algae. Using EvolMap, we identified a total of 1846 nuclear genomic core-orthologues and 38 plastid core-orthologues shared across the six full genome sequences available for green algae. Average unaligned sequence length of the orthologues was 442 amino acids, with a minimum of 52 amino acids and a maximum of 3991 amino acids. All sequence descriptions, lengths, eValues, similarities and GO annotations from Blast2GO have been made available (electronic supplementary material, table S5).
(c). Phylogenomic resolution of green algal relationships
The phylogenomic framework we provide for green algae is well supported and extremely data-rich compared with previous phylogenies of green algae (see the electronic supplementary material for a systematic discussion). The density of genetic data present within a phylogenetic matrix can have notable effects on the resulting phylogeny [51–53]. We used Phylocatenator [47] to generate five alternative matrices of concatenated alignments with different parameters and chose the one with the highest average bootstrap support. Results from these tests showed that the density of the matrix and the minimum length of aligned amino acids both affected average bootstrap score of the resulting tree. Increasing the density of the matrix improved bootstrap support values (from an average of 25.2% to an average of 79.2% for all nodes) and the overall topology, resulting in the monophyly of major groups. The highest average bootstrap score across all nodes was obtained by increasing density of the matrix to 10 species per gene and 20 genes per species as well as setting a minimum aligned gene length of 100 amino acids. This phylogeny resulted in the monophyly of major groups of green algae (electronic supplementary material, figure S3).
We find that sequentially removing genes contributing to long branches yields increased bootstrap values. We tested the use of a variety of parameters when using the Long Branch removal tool, including the average bootstrap score prior and post long-branch removal and found an improvement of 6.4% across all nodes. We then calculated the average bootstrap score of resulting topologies after one iteration of long-branch removal, and after two iterations of the same process. Our tests concluded that two iterations of long-branch removal improved average bootstrap scores by 3.6% compared with a single iteration of the process. Prior to concatenating nuclear and chloroplast data, we calculated the average bootstrap score for a nuclear dataset, a chloroplast dataset and a combined dataset. Our tests support the concatenation of nuclear and chloroplast data with an average bootstrap score 9% higher than the chloroplast data and 6% higher than the nuclear data on its own. Our final dataset totalled 59 taxa and 19 949 amino acids across 25 chloroplast genes and 94 nuclear genes. Some of the orthologues were not retained due to the parameters set in Phylocatenator, specifically by setting a minimum of 10 genes per species and 10 species per gene.
(d). Phylogenetic diversity and the strength of species interactions
Values of PD generated from our dataset and those generated by a couple of publicly available genetic markers are different, primarily owing to the larger number of substitutions in our dataset (our study relied on over 100 genes, whereas the previous studies were only able to use a couple). We found that the difference in PD values increased with the number of taxa used in each experiment. Furthermore, compared to the tree generated from publicly available data (GenBank), our phylogenomic-based tree resulted in significantly higher mean bootstrap scores (65.2% compared with 79.5%), indicating that our phylogenomic data offer a significant statistical improvement over the phylogeny generated from GenBank.
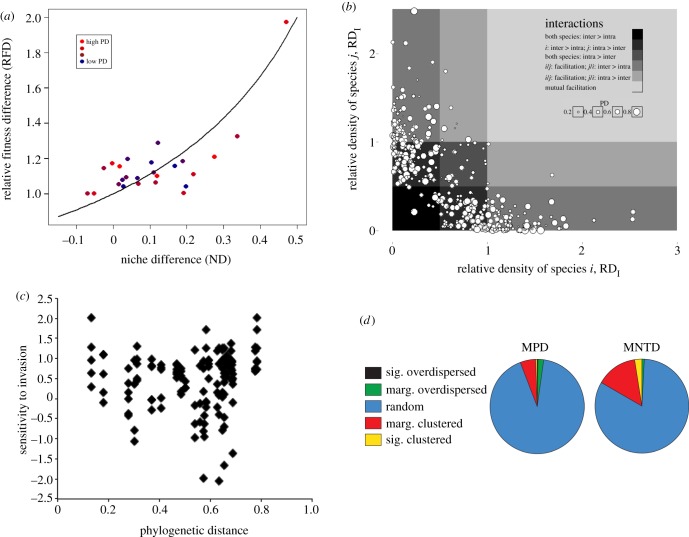
Using our phylogenomic dataset, we found that PD generally does not predict the strength of species interactions for green algae, as measured by RFD, ND, cell densities and sensitivity to invasion (figure 1).
Figure 1.
A reanalysis of the results of three published studies using PD values from the generated herein. (a) Relative fitness difference against niche difference with PDs colour coded from high to low; (b) The distribution of algae communities across an interaction gradient. Each data point is a single biculture and its size is scaled to PD between its constituent species. The background colour scales along a gradient of competition strength; the dark grey indicates that both species experienced stronger inter- versus intraspecific competition (RDx < 0.5: n = 12), while the lightest grey indicates that species were mutually facilitated. (c) Average sensitivity to invasion. (d) Pie charts depicting phylogenetic community structure across lakes in the EPA dataset. MPD and MNTD are presented as alternative metrics of community structure. Sig. denotes statistically significant structure, whereas Marg. denotes marginally significant structure.
The original analysis of effects of phylogenetic distance on the mechanisms and outcome of competition in Narwani et al. [16] using the 18 s and rbcL-based phylogeny indicated there were no significant impacts of phylogenetic distance on ND (F1,23 = 0.56, p = 0.46), RFD (F1,23 = 2.23, p = 0.15) or coexistence (χ2 = 0.11, p = 0.74). These results were confirmed using phylogenetic distances calculated based on the significantly improved phylogenomic tree presented here (ND: F1,23 = 0.31, p = 0.59, RFD: F1,23 = 1.80, p = 0.19 and coexistence: χ2 = 0.04, p = 0.84; figure 1a). We also tested whether PD influenced the strength of species interactions by calculating species' RD: their performance with a competitor relative to their performance when grown alone. Consistent with results of the original experiment [15], PD was not related to several ecologically meaningful RD metrics, including its average values across 216 pairwise combinations (original analysis: slope = −0.12, r2 < 0.01, p = 0.19; new analysis: slope = 0.06, r2 < 0.01, p = 0.21; figure 1b). Moreover, the forms of interspecific interactions experienced in biculture (estimated from the joint distribution of species' RDs) were not related to the interacting species' PD (original: F4,422 = 1.63, p = 0.15; new: F4,422 = 0.81, p = 0.52; figure 1b). We then tested whether PD influenced species' ability to successfully invade an established culture and compared the growth rate of the invader when rare to the growth rate of same species in monoculture (sensitivity), and found that evolutionary relatedness did not determine the strength of species interactions (linear regression, r2 = 0.0036, p = 0.469, n = 135; figure 1c). This confirms our previous study using GenBank based PD values [17] that found the evolutionary relatedness of species had no influence on the sensitivity of growth rates to species interactions (linear regression, r2 = 0.0004, p = 0.82, n = 135). Thus, despite a major advance in rigour of the phylogeny used to generate estimates of PD, none of the conclusions from our prior studies were altered.
We also found no evidence that nonlinear relationship or heteroskedasticity of variances masked our ability to detect significant relationships. Square-root and power function transformations of PD values for all three studies confirmed that neither linear or nonlinear relationships exist between PD and the ecological response variables measured for green algae. We found no relationships between square-root transformed PD and coexistence (χ2 = 0.08, p = 0.77, n = 27), ND (F1,23 = 0.32; p = 0.58, adj-r2 = 0.03), RFD (F1,23 = 1.96; p = 0.17, adj-r2 = 0.04), sensitivity to invasion (F1,133 = 0.12, p = 0.73, adj-r2 = 0.01) or relative yield (RY) (F1,425 = 1.51; p = 0.22, adj-r2 < 0.01). Power functions also yielded no significant relationship between PD and ND (b = 0.30, t = 0.52, p = 0.61, r2 = 0.63), RFD (b = 0.10, t = 1.37, p = 0.18, r2 = 0.21) or sensitivity to competition (b = −0.03, t = −0.09, p = 0.93). Furthermore, there was also no relationship between PD and the variances of NDs (BP = 0.19, p = 0.66), RFDs (BP = 0.56, p = 0.45), sensitivities (BP = 0.49, p = 0.49) or RYs (BP = 1.91, p = 0.17). These analyses lead us to conclude that our previous lack of support for the CRH is not a limitation of the statistical methodology used to summarize relationships.
(e). Random phylogenetic community structure
Our analyses of phylogenetic community structure revealed that the species composition of over 99% of North American lake communities is random with respect to phylogenetic relatedness. The GenBank alignment resulted in 2722 nucleotides, which were concatenated with the 19 949 amino acids from the phylogenomic data matrix. The final phylogenetic dataset consisted of 92 taxa and 22 761 bps. The resulting tree from the ML analysis was well supported and in agreement with results from previous studies (electronic supplementary material, figure S4). The results from all metrics and null models used to assess community structure revealed random phylogenetic distribution of taxa across North American lakes. Specifically, standardized effect size analyses using mean phylogenetic distance (MPD) and MNTD metrics found that 99.3 and 97.6% (respectively) of the lakes were composed of communities with no significant phylogenetic structure (figure 1d). Results supported a slight tendency towards phylogenetic clustering in a select subset of lakes (MPD = 0.5%; MNTD = 2.3%). We differentiate our results between ‘marginal’ (p = 0.005–0.05; p = 0.95–0.99) and significant phylogenetic signal to highlight that even under a more ‘relaxed’ statistical scenario without Bonferroni correction (p < 0.005; p > 0.99), the lack of phylogenetic community structure remains. Our results highlight the lack of any relationship between phylogenetic distance and community composition, contrary to the predictions of the CRH.
4. Discussion
Taken together, our results refute the CRH by confirming that close relatives are no more likely than distant relatives to be in competition, and no more or less likely to co-occur or coexist in the same system. Rather than indirectly inferring coexistence mechanisms from phylogenetic patterns, we test the relationship between phylogeny and coexistence mechanisms using experimental data and null community phylogenetic models. Our new data-rich phylogenetic framework provides considerably improved estimates of PD than used in previous studies, yet still shows that the strength and the outcome of competition are not a function of relatedness. Our results also confirm that determinants of species coexistence (RFD and ND), relative yield and sensitivity to invasion are not correlated with evolutionary relatedness, contrary to the predictions of CRH. The lack of relationships was not altered after we accounted for statistical limitations that have been proposed by some to explain the lack of support for the CRH. We also used the new phylogeny to examine whether patterns of co-occurrence among 99 genera in 1077 lakes throughout North America were random, or non-random with respect to phylogeny. We found a pervasive lack of phylogenetic signal in algal community structure across North America, indicating that evolutionary relatedness does not predict patterns of co-occurrence in the overwhelming majority of algal lake communities. To our knowledge, no other studies have reported random phylogenetic community structure at the continental scale shown in this paper.
Despite the large spatial scale of these analyses, and the pervasive absence of phylogenetic signal, it is important to be aware of the limitations of the EPA's NLA dataset. This dataset provides only a snapshot of community structure in lakes (sampled only in the summer months of 2007), as the data were based on a single sampling effort rather than a temporal series that could provide data on fluctuations in community structure through time. This snapshot may or may not be representative of overall community structure. If species in these lakes coexist primarily by temporal niche differences—for example, by taking advantage of seasonal fluctuations in temperature, light or nutrient availability—then a single summer sampling effort might not reflect variation in the species pools of lakes, or their ecologically unique characteristics. While we know of no dataset that has highly resolved timeseries at the continental scale, it would be a valuable exercise to supplement the analyses we have presented here with analyses of smaller subsets of lakes that do have resolved timeseries, or to repeat our analyses when the EPA publishes data from its second, 2012 sampling effort for the NLA. The phylogeny we report here can be used as a framework for such future studies.
Furthermore, the EPA relied primarily on genus-level taxonomic identification since species-level identification of microscopic green algae can be time consuming, and diagnosis often requires both morphological and molecular data in order to determine the correct taxonomic placement. Therefore, in order to test community structure, we had to use a phylogeny with genera (electronic supplementary material, figure S4) as our operational taxonomic unit. Several recent studies have examined the effect of taxonomic scale on testing phylogenetic community structure and concluded that as taxonomic scale becomes finer, patterns of overdispersion tend to increase, while broader taxonomic scale leads to more phylogenetic clustering [54,55]. Although we cannot test this hypothesis directly (as we are restricted by the EPAs taxonomic scale), we do acknowledge that sampling at a finer taxonomic scale could result in more phylogenetic structure than observed herein. We would, however, point out that all of our experimental studies have clearly documented widespread evidence of competition among focal species, as well as variation in the degree of niche differences, and trait variation among species. Therefore, we have studied the ecological phenomenon of interest (e.g. competition) at the appropriate phylogenetic scale in our laboratory-based studies, and it is clear that evolutionary relatedness does not predict competitive interactions across interaction strengths that span from total competitive exclusion to very weak competition.
Finally, green algae are a particularly ancient group with fossils dating back well over 500 Myr [56] and molecular clock estimates placing their origin at more than a billion years [57]. This makes green algae much older than typical groups used for phylogenetic community studies (namely vertebrates and angiosperms). At this timescale, it is possible that phylogenetic signal becomes obscured owing to convergent evolution and/or horizontal gene transfer. While we cannot address these possibilities directly, we do note that our sampling strategy covers both very closely and distantly related species, and there is no empirical evidence to suggest that increased phylogenetic distance between species obscures the signal of ecologically relevant traits. In fact, recent meta-analyses of empirical data suggest that beyond a certain threshold, increasingly more distant relatives are not more divergent in phenotype [58].
Rapid evolution of ecological traits in green algae has been demonstrated previously [59–62] and could explain why we find no evidence supporting the CRH, no correlations between PD and ecology and no phylogenetic signal in traits. If ecologically relevant traits were conserved across algal lineages, we would expect to find a significant pattern of phylogenetic community structure across North American lakes. Testing the phylogenetic signal of ecological traits is beyond the scope of our work, however, our companion study quantified 17 morphological and physiological traits that are thought to regulate competition among green algae exhibit and found little to no phylogenetic signal of any of these traits [63]. The absence of phylogenetic signal in traits and communities could be explained by a propensity for rapid trait evolution among green algae. Green algae have short generation times, are subject to high levels of predation and may therefore be more likely to experience rapid trait divergence than other well-studied groups [59–62]. Although traits that have been shown to evolve rapidly are typically involved in predator defence (cell morphology, spines, etc.), it is also possible that traits involved in interspecific competitive interactions evolve rapidly. Phenotypic plasticity in algae could also lead to rapid trait evolution [64–67], further obscuring phylogenetic signal of such traits. Different abiotic variables across lakes could lead to local adaptation, driving rapid evolution at a local scale.
Our study suggests that we may need to question the assumptions and predictions of the CRH [1,2,15,16,22]. These assumptions have provided a useful foundation for our understanding of species interactions in a community context, yet our results show that the mechanisms of competition are not always evolutionarily conserved. Some studies have found evidence of phylogenetic overdispersion in natural communities [68–73], while others have not [18,20,21], suggesting that the pattern is not as widespread as predicted by the CRH. Contrary to those predictions, we show that close relatives of green algae do not compete strongly in bicultures and natural algal communities are not structured based on phylogenetic distance. Our results challenge the assumption that close relatives inherit more similar ecological traits than distant relatives, calling into question whether the CRH can explain mechanisms of community assembly for green algae.
In addition to prompting reconsideration of the CRH, the new phylogeny presented here has much potential to enhance future work in phylogenetics, experimental ecology and gene expression. The resulting tree can be used to guide manipulations of PD in future experimental ecological studies and further evaluate the assumptions of the CRH. The phylogenomic framework we provide for green algae is well supported and extremely data-rich compared with previous phylogenies of green algae. Furthermore, the fact that it has been created specifically for culturable species that can be used in future experimental studies makes it a particularly valuable tool. Such studies are important and timely as they help inform conservation policy by predicting the consequences of biodiversity loss and the role of evolutionary history plays in community assembly. This work can also inform future experimental studies focusing on the evolutionary history of ecologically important traits that affect ecosystem function, as well as investigating the genetic basis of those traits. Our results suggest that phylogenetic patterns do not necessarily predict ecological similarity among species, and we urge future studies to use caution when interpreting such patterns, particularly as they apply to decisions in species conservation and management.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
Thanks to Nana He for culture establishment, Paul Weakliem for extensive technical assistance, Karl Loepker and Roger Ngo for developing some tool wrappers, Lewis Louise for advice on the phylogeny and Nicole Leung for assisting with laboratory protocols.
Data accessibility
All phylogenetic datasets and orthologs are available on Dryad (doi:10.5061/dryad.c574h). Raw Illumina files can be accessed using the NCBI SRA archive under BioProject Accession PRJNA237822 (see table S1 for species specific accessions). Bioinformatics tools can be accessed at http://galaxy-dev.cnsi.ucsb.edu/osiris, downloaded from https://bitbucket.org/osiris_phylogenetics/osiris_phylogenetics, and read about at http://osiris-phylogenetics.blogspot.com.
Funding statement
We acknowledge support from the Center for Scientific Computing at the CNSI and MRL: an NSF MRSEC (DMR-1121053) and NSF CNS-0960316. This work was supported by a US National Science Foundation DIMENSIONS of Biodiversity grant to T.H.O. (DEB-1046307), C.F.D. (DEB-1046075) and B.J.C. (DEB-1046121).
References
- 1.Cahill JF, et al. 2008. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspect. Plant Ecol. Evol. Syst. 10, 41–50. ( 10.1016/j.ppees.2007.10.001) [DOI] [Google Scholar]
- 2.Mayfield MM, Levine JM. 2010. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13, 1085–1093. ( 10.1111/j.1461-0248.2010.01509.x) [DOI] [PubMed] [Google Scholar]
- 3.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. ( 10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 4.Cavender-Bares J, Kozak KH, Fine PVA, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715. ( 10.1111/j.1461-0248.2009.01314.x) [DOI] [PubMed] [Google Scholar]
- 5.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: J. Murray. [PMC free article] [PubMed] [Google Scholar]
- 6.MacArthur RH, Levins R. 1967. Limiting similarity convergence and divergence of coexisting species. Am. Nat. 101, 377–385. ( 10.1086/282505) [DOI] [Google Scholar]
- 7.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 8.Strauss SY, Webb CO, Salamin N. 2006. Exotic taxa less related to native species are more invasive. Proc. Natl Acad. Sci. USA 103, 5841–5845. ( 10.1073/pnas.0508073103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kettenring KM, Mercer KL, Reinhardt Adams C, Hines J. 2014. Application of genetic diversity- ecosystem function research to ecological restoration. J. Appl. Ecol. 51, 339–348. ( 10.1111/1365-2664.12202) [DOI] [Google Scholar]
- 10.Maherali H, Klironomos JN. 2007. Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748. ( 10.1126/science.1143082) [DOI] [PubMed] [Google Scholar]
- 11.Jiang L, Tan J, Pu Z. 2010. An experimental test of Darwin's naturalization hypothesis. Am. Nat. 175, 415–423. ( 10.1086/650720) [DOI] [PubMed] [Google Scholar]
- 12.Violle C, Nemergut DR, Pu Z, Jiang L. 2011. Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett. 14, 782–787. ( 10.1111/j.1461-0248.2011.01644.x) [DOI] [PubMed] [Google Scholar]
- 13.Peay KG, Belisle M, Fukami T. 2012. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc. R. Soc. B 279, 749–758. ( 10.1098/rspb.2011.1230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoustra SE, Dench J, Dali R, Aaron SD, Kassen R. 2012. Antagonistic interactions peak at intermediate genetic distance in clinical and laboratory strains of Pseudomonas aeruginosa. BMC Microbiol. 12, 40 ( 10.1186/1471-2180-12-40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritschie KJ, Cardinale BJ, Alexandrou MA, Oakley TH. 2013. Evolutionary history and the strength of species interactions: testing the phylogenetic limiting similarity hypothesis. Ecology 95, 1407–1417. ( 10.1890/13-0986.1) [DOI] [PubMed] [Google Scholar]
- 16.Narwani A, Alexandrou MA, Oakley TH, Carroll IT, Cardinale BJ. 2013. Experimental evidence that evolutionary relatedness does not affect the ecological mechanisms of coexistence in freshwater green algae. Ecol. Lett. 16, 1373–1381. ( 10.1111/ele.12182) [DOI] [PubMed] [Google Scholar]
- 17.Venail PA, Narwani A, Fritschie K, Alexandrou MA, Oakley TH, Cardinale BJ. 2014. The influence of phylogenetic relatedness on species interactions among freshwater green algae in a mesocosm experiment. J. Ecol. 102, 1288–1299. ( 10.1111/1365-2745.12271) [DOI] [Google Scholar]
- 18.Bennett JA, Lamb EG, Hall JC, Cardinal-McTeague WM, Cahill JF. 2013. Increased competition does not lead to increased phylogenetic overdispersion in a native grassland. Ecol. Lett. 16, 1168–1176. ( 10.1111/ele.12153) [DOI] [PubMed] [Google Scholar]
- 19.Dostal P. 2011. Plant competitive interactions and invasiveness: searching for the effects of phylogenetic relatedness and origin on competition intensity. Am. Nat. 177, 655–667. ( 10.1086/659060) [DOI] [PubMed] [Google Scholar]
- 20.Kunstler G, Lavergne S, Courbaud B, Thuiller W, Vieilledent G, Zimmermann NE, Kattge J, Coomes DA. 2012. Competitive interactions between forest trees are driven by species’ trait hierarchy, not phylogenetic or functional similarity: implications for forest community assembly. Ecol. Lett. 15, 831–840. ( 10.1111/j.1461-0248.2012.01803.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Best RJ, Caulk NC, Stachowicz JJ. 2013. Trait vs. phylogenetic diversity as predictors of competition and community composition in herbivorous marine amphipods. Ecol. Lett. 16, 72–80. ( 10.1111/ele.12016) [DOI] [PubMed] [Google Scholar]
- 22.Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003. ( 10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 23.Zhang ZG, Ye Z-Q, Shi P. 2011. Phylogenomic reconstruction of lactic acid bacteria: an update. BMC Evol. Biol. 11, 1 ( 10.1186/1471-2148-11-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Philippe H, et al. 2009. Phylogenomics revives traditional views on deep animal relationships. Curr. Biol. 19, 706–712. ( 10.1016/j.cub.2009.02.052) [DOI] [PubMed] [Google Scholar]
- 25.Xi ZX, et al. 2012. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl Acad. Sci. USA 109, 17 519–17 524. ( 10.1073/pnas.1205818109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pick KS, et al. 2010. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol. Biol. Evol. 27, 1983–1987. ( 10.1093/molbev/msq089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lartillot N, Philippe H. 2008. Improvement of molecular phylogenetic inference and the phylogeny of Bilateria. Phil. Trans. R. Soc. B 363, 1463–1472. ( 10.1098/rstb.2007.2236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou XM, Xu S, Yang Y, Zhou K, Yang G. 2011. Phylogenomic analyses and improved resolution of Cetartiodactyla. Mol. Phylogeneti. Evol. 61, 255–264. ( 10.1016/j.ympev.2011.02.009) [DOI] [PubMed] [Google Scholar]
- 29.Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463, 1079–1U98. ( 10.1038/nature08742) [DOI] [PubMed] [Google Scholar]
- 30.Dunn CW, et al. 2008. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452, 745–749. ( 10.1038/nature06614) [DOI] [PubMed] [Google Scholar]
- 31.Letten AD, Cornwell WK. In press. Trees, branches and (square) roots: why evolutionary relatedness is not linearly related to functional distance. Methods Ecol. Evol. ( 10.1111/2041-210X.12237) [DOI] [Google Scholar]
- 32.Godoy O, Kraft NJ, Levine JM. 2014. Phylogenetic relatedness and the determinants of competitive outcomes. Ecol. Lett. 17, 836–844. ( 10.1111/ele.12289) [DOI] [PubMed] [Google Scholar]
- 33.Srivastava DS, et al. 2004. Are natural microcosms useful model systems for ecology? Trends Ecol. Evol. 19, 379–384. ( 10.1016/j.tree.2004.04.010) [DOI] [PubMed] [Google Scholar]
- 34.Zimmerman E, Cardinale BJ. 2013. Does biodiversity affect the productivity of natural lake ecosystems? Oikos 123, 267–278. ( 10.1111/j.1600-0706.2013.00777.x) [DOI] [Google Scholar]
- 35.Chesson P. 2000. General theory of competitive coexistence in spatially-varying environments. Theor. Popul. Biol. 58, 211–237. ( 10.1006/tpbi.2000.1486) [DOI] [PubMed] [Google Scholar]
- 36.Adler PB, HilleRisLambers J, Levine JM. 2007. A niche for neutrality. Ecol. Lett. 10, 95–104. ( 10.1111/j.1461-0248.2006.00996.x) [DOI] [PubMed] [Google Scholar]
- 37.Timme RE, Bachvaroff TR, Delwiche CF. 2011. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 7, e29696 ( 10.1371/journal.pone.0029696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giardine B, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. ( 10.1101/gr.4086505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blankenberg D, Gordon A, Von Kuster G, Coraor N, Taylor J, Nekrutenko A. 2010. Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785. ( 10.1093/bioinformatics/btq281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7, e30619 ( 10.1371/journal.pone.0030619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, Zhao L, Gao J, Fei Z. 2011. iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinform. 12, 453 ( 10.1186/1471-2105-12-453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebersberger I, Strauss S, von Haeseler A. 2009. HaMStR: profile hidden Markov model based search for orthologs in ESTs. BMC Evol. Biol. 9, 157 ( 10.1186/1471-2148-9-157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. ( 10.1093/bioinformatics/bti263) [DOI] [PubMed] [Google Scholar]
- 45.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5, 1–19. ( 10.1186/1471-2105-5-113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cadotte MW, Jonathan Davies T, Regetz J, Kembel SW, Cleland E, Oakley TH. 2010. Phylogenetic diversity metrics for ecological communities: integrating species richness, abundance and evolutionary history. Ecol. Lett. 13, 96–105. ( 10.1111/j.1461-0248.2009.01405.x) [DOI] [PubMed] [Google Scholar]
- 47.Oakley THAlexandrou MA, Ngo R, Pankey MS, Churchill CK, Chen W, Lopker KB. 2014. Osiris: accessible and reproducible phylogenetic and phylogenomic analyses within the Galaxy workflow management system. BMC Bioinform. 15, 230 ( 10.1186/1471-2105-15-230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 49.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. ( 10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 50.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464. ( 10.1093/bioinformatics/btq166) [DOI] [PubMed] [Google Scholar]
- 51.Oakley TH, Wolfe JM, Lindgren AR, Zaharoff AK. 2013. Phylotranscriptomics to bring the understudied into the fold: monophyletic Ostracoda, fossil placement, and Pancrustacean phylogeny. Mol. Biol. Evol. 30, 215–233. ( 10.1093/molbev/mss216) [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Ezpeleta N, Brinkmann H, Roure B, Lartillot N, Lang BF, Philippe H. 2007. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 56, 389–399. ( 10.1080/10635150701397643) [DOI] [PubMed] [Google Scholar]
- 53.Philippe H, Brinkmann H, Lavrov DV, Littlewood DTJ, Manuel M, Wörheide G, Baurain D. 2011. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 9, e1000602 ( 10.1371/journal.pbio.1000602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cavender-Bares J, Keen A, Miles B. 2006. Phylogenetic structure of floridian plant communities depends on taxonomic and spatial scale. Ecology 87, S109–S122. ( 10.1890/0012-9658(2006)87[109:PSOFPC]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 55.Swenson NG, Enquist BJ, Pither J, Thompson J, Zimmerman JK. 2006. The problem and promise of scale dependency in community phylogenetics. Ecology 87, 2418–2424. ( 10.1890/0012-9658(2006)87[2418:TPAPOS]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 56.Wellman CH, Osterloff PL, Mohiuddin U. 2003. Fragments of the earliest land plants. Nature 425, 282–285. ( 10.1038/nature01884) [DOI] [PubMed] [Google Scholar]
- 57.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. 2004. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818. ( 10.1093/molbev/msh075) [DOI] [PubMed] [Google Scholar]
- 58.Kelly S, Grenyer R, Scotland RW. 2014. Phylogenetic trees do not reliably predict feature diversity. Div. Distrib. 20, 600–612. [Google Scholar]
- 59.Yoshida T, Ellner SP, Jones LE, Bohannan BJM, Lenski RE, Hairston NG. 2007. Cryptic population dynamics: rapid evolution masks trophic interactions. PLoS Biol. 5, 1868–1879. ( 10.1371/journal.pbio.0050235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG. 2003. Rapid evolution drives ecological dynamics in a predator-prey system. Nature 424, 303–306. ( 10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 61.Becks L, Ellner SP, Jones LE, Hairston NG. 2012. The functional genomics of an eco-evolutionary feedback loop: linking gene expression, trait evolution, and community dynamics. Ecol. Lett. 15, 492–501. ( 10.1111/j.1461-0248.2012.01763.x) [DOI] [PubMed] [Google Scholar]
- 62.Jones LE, Becks L, Ellner SP, Hairston NG, Yoshida T, Fussmann GF. 2009. Rapid contemporary evolution and clonal food web dynamics. Phil. Trans. R. Soc. B 364, 1579–1591. ( 10.1098/rstb.2009.0004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Narwani A, Vouaux A, Zhou C, Miller C, Alexandrou MA, Oakley TH, Cardinale BJ. In press Phylogeny is a poor predictor of ecological traits of freshwater green algae. New Phytol.
- 64.Scoville AG, Pfrender ME. 2010. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc. Natl Acad. Sci. USA 107, 4260–4263. ( 10.1073/pnas.0912748107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lurling M. 2003. Phenotypic plasticity in the green algae Desmodesmus and Scenedesmus with special reference to the induction of defensive morphology. Ann. Limnol. 39, 85–101. ( 10.1051/limn/2003014) [DOI] [Google Scholar]
- 66.Stoyneva MP, Descy JP, Vyverman W. 2007. Green algae in Lake Tanganyika: is morphological variation a response to seasonal changes? Hydrobiologia 578, 7–16. ( 10.1007/s10750-006-0428-1) [DOI] [Google Scholar]
- 67.Neustupa J, Stastny J, Hodac L. 2008. Temperature-related phenotypic plasticity in the green microalga Micrasterias rotata. Aquat. Microb. Ecol. 51, 77–86. ( 10.3354/ame01184) [DOI] [Google Scholar]
- 68.Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. 2004. Phylogenetic overdispersion in Floridian oak communities. Am. Nat. 163, 823–843. ( 10.1086/386375) [DOI] [PubMed] [Google Scholar]
- 69.Letcher SG. 2010. Phylogenetic structure of angiosperm communities during tropical forest succession. Proc. R. Soc. B 277, 97–104. ( 10.1098/rspb.2009.0865) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slingsby JA, Verboom GA. 2006. Phylogenetic relatedness limits co-occurrence at fine spatial scales: evidence from the schoenoid sedges (Cyperaceae: Schoeneae) of the Cape Floristic Region, South Africa. Am. Nat. 168, 14–27. ( 10.1086/505158) [DOI] [PubMed] [Google Scholar]
- 71.Davies TJ, Meiri S, Barraclough TG, Gittleman JL. 2007. Species co-existence and character divergence across carnivores. Ecol. Lett. 10, 146–152. ( 10.1111/j.1461-0248.2006.01005.x) [DOI] [PubMed] [Google Scholar]
- 72.Cooper N, Rodriguez J, Purvis A. 2008. A common tendency for phylogenetic overdispersion in mammalian assemblages. Proc. R. Soc. B 275, 2031–2037. ( 10.1098/rspb.2008.0420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kozak KH, Larson A, Bonett RM, Harmon LJ. 2005. Phylogenetic analysis of ecomorphological divergence, community structure, and diversification rates in dusky salamanders (Plethodontidae: Desmognathus). Evolution 59, 2000–2016. ( 10.1111/j.0014-3820.2005.tb01069.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All phylogenetic datasets and orthologs are available on Dryad (doi:10.5061/dryad.c574h). Raw Illumina files can be accessed using the NCBI SRA archive under BioProject Accession PRJNA237822 (see table S1 for species specific accessions). Bioinformatics tools can be accessed at http://galaxy-dev.cnsi.ucsb.edu/osiris, downloaded from https://bitbucket.org/osiris_phylogenetics/osiris_phylogenetics, and read about at http://osiris-phylogenetics.blogspot.com.