Abstract
In a broad range of species—including humans—it has been demonstrated that telomere length declines throughout life and that it may be involved in cell and organismal senescence. This potential link to ageing and thus to fitness has triggered recent interest in understanding how variation in telomere length is inherited and maintained. However, previous studies suffer from two main drawbacks that limit the possibility of understanding the relative importance of genetic, parental and environmental influences on telomere length variation. These studies have been based on (i) telomere lengths measured at different time points in different individuals, despite the fact that telomere length changes over life, and (ii) parent–offspring regression techniques, which do not enable differentiation between genetic and parental components of inheritance. To overcome these drawbacks, in our study of a songbird, the great reed warbler, we have analysed telomere length measured early in life in both parents and offspring and applied statistical models (so-called ‘animal models') that are based on long-term pedigree data. Our results showed a significant heritability of telomere length on the maternal but not on the paternal side, and that the mother's age was positively correlated with their offspring's telomere length. Furthermore, the pedigree-based analyses revealed a significant heritability and an equally large maternal effect. Our study demonstrates strong maternal influence on telomere length and future studies now need to elucidate possible underlying factors, including which types of maternal effects are involved.
Keywords: Acrocephalus arundinaceus, great reed warbler, telomere inheritance, ageing, maternal effects, animal model
1. Introduction
Telomeres are repetitive sequences located at the ends of chromosomes. They are highly conserved between lineages and are important because they protect chromosomes from degradation and fusion, thereby maintaining chromosome integrity [1,2]. The telomeres decrease in length throughout life of an organism owing to cell division and through the effects of oxidative stress; this eventually leads to cell death, which may play a role in organismal ageing [3–7]. Several studies have shown a link between telomere length (or rate of telomere loss) and individual lifespan or physiological condition [5,7–11], implying that telomere length is an important fitness proxy involved in senescence. Despite its apparent association with senescence and fitness, it has been found that telomere length is highly variable between individuals of the same age class and appears to have a relatively large heritable component [7–12]. However, evidence is accumulating that telomere length is also influenced by environmental factors [7,13,14]. Central questions in telomere research are currently how variation in telomere length is maintained over time in the population and to what degree telomere length is under genetic and non-genetic (parental or other environmental) influence.
Telomere inheritance has been very actively researched in humans, and several studies have found a significant association between the fathers' telomere length and that of their offspring, but not between mothers' telomere length and that of their offspring [15–17]. However, in the only study where the telomere length was measured at childbirth, a highly significant mother–offspring telomere length regression was found [18]. Thus, the inheritance pattern of telomeres in humans is presently unresolved, which is further emphasized by two recent meta-analyses that reached different conclusions. One of these studies concluded a stronger maternal than paternal inheritance and significant paternal age effect [12], while the other study found no significant difference between mother–offspring and father–offspring telomere length regressions [19]. In vertebrates beyond humans, the inheritance pattern of telomere length has seldom been investigated. A study in the sand lizard (Lacerta agilis) found a significant correlation between parents and offspring, with a stronger correlation between fathers and sons than between mothers and daughters [20]. Three bird species have been investigated so far: the kakapo (Strigops habroptilus), where a significant regression was found only between mothers and offspring (but not between fathers and offspring) [21], and the collared flycatcher (Ficedula albicollis), where a cross-fostering study revealed heritability and/or a parental effect, but the effect of parental sex on telomere inheritance could not be investigated [22]. Finally, a recent study in king penguin (Aptenodytes patagonicus) showed maternal inheritance of telomere length [23].
Most previous studies suffer from two drawbacks that limit the potential to understand the relative importance of genetic and environmental influence of telomere length variation. First, most studies have been based on telomere length measurements taken from the offspring and parents at different times in their lives. This is problematic because telomeres shorten with age and potentially at different rates depending on exposure to environmental factors [7,13,14]. Indeed, evidence is accumulating that telomere length can change over time even in the germ cells and that this can influence offspring telomere length [24–26]. It is, therefore, possible that paternal or maternal age at the time of fertilization can affect offspring telomere length. This could result in a carry-over effect of parental age to the next generation [7]. In humans, there is evidence that offspring telomere length is positively correlated with their father's age [15,25,27], but it shows no or a negative correlation with their mother's age [28]. However, there are to our knowledge no such studies conducted on any other species. With analyses of data on mixed-age individuals it is not possible to control for potential environmental effects, which can seriously bias the heritability estimates [24] and hamper our understanding of how genetic and non-genetic factors contribute to early life telomere length. The inability to control for environmental effects can potentially explain the large range of estimates of heritability of telomere length in different studies, and e.g. in humans, the heritability estimates range from 36 to 90% [15,18,29,30]. Studies measuring the telomere length early in life in both parents and offspring will be decisive for our understanding of telomere inheritance, because they take account of the variation in telomere length caused by individual differences in life experiences.
Second, previous studies of the inheritance pattern of telomere length have used parent–offspring regressions, which is unfortunate as these techniques do not allow the separation of the genetic and non-genetic components. However, to tease apart the relative contributions of genetic and non-genetic factors in determining variation in telomere length, a so-called ‘animal model’ approach (a linear mixed model (LMM) estimated with restricted maximum likelihood) can be applied by using reliable extensive pedigrees from long-term study of (wild) populations [31]. So far, however, no study has used this approach. This type of study is of general interest, because if we can understand how and why non-genetic factors impact early life telomere length, this may reveal general biological patterns that may emerge, e.g. identifying factors that slow-down early life telomere shortening (which later in life may postpone senescence).
In this study, we evaluate the influence of genetic and parental factors on offspring telomere length in a wild population of a songbird, the great reed warbler (Acrocephalus arundinaceus), studied over the past 30 years [32,33]. To overcome the drawbacks of previous studies, we measured telomere length very early in life in both parents and offspring and apply ‘animal models' based on resolved pedigree data [34,35]. This approach enabled us to analyse the relative importance of genetic, environmental, parental age and maternal effects on telomere length in a way that has previously not been possible in vertebrates.
2. Material and methods
(a). Great reed warbler study population
This study is based on life-history data collected from great reed warblers breeding at Lake Kvismaren (59°10′ N, 15°25′ E), in southern Central Sweden, between 1983 and 2013. Daily visits were performed throughout the breeding season and the majority of the breeding males and females were colour-ringed with individual-unique combinations, allowing identification of individuals from a distance. Details of fieldwork and morphology, life-history and reproduction data are described elsewhere [36,37]. A blood sample of 20–100 μl was collected from each captured adult and stored in SET-buffer at −80°C until DNA extraction.
The great reed warbler breeds in Europe and Western Asia and winters in Africa [38]. It has a facultatively polygynous social mating system, where a male can have two to five breeding females in his territory simultaneously, although 38% of the territorial males form pair bonds with only one female and 21% of territorial males are unmated [39–41]. We visited all breeding territories almost daily (every 1–3 days) throughout the breeding season and located almost all of the nesting attempts. The parentage of chicks has been confirmed by molecular methods [39,42,43]. The frequency of extra-pair paternity is low in this population, involving only about 3% of the chicks [42,43], and the pedigrees have been corrected for these events. Still, we excluded all extra-pair young from the analyses in this study. The dataset used is based on early life telomere length of individuals hatched between 1988 and 2002, because the genetic parentage has been established for this subsample. We included only nestlings that had at least one parent whose telomere length was measured early in life. Early life telomere lengths of both parents and offspring were measured from the blood samples taken when these birds were 8–10-day-old nestlings (in most cases 9 days old) [39–41]. A total of 193 individuals (139 offspring from 46 broods, which were produced by 25 females and 29 males) were measured. For 18 of the parents (8 females and 10 males), we could not determine early life telomere length because DNA samples were not available from their nestling period. These individuals were excluded from the heritability analysis (although we kept the relatedness links of these individuals in the pedigree, but without information on the phenotype), but they were included in the analysis of the effects of parental age at fertilization on offspring early life telomere length. Numbers of included individuals varied between years from 2 to 20 (mean = 12.6 per year). At our study site, great reed warblers show remarkably high breeding site fidelity [33,44]. Among the previous year's breeders that survive the winter, 92% return to breed at Lake Kvismaren [44]. When considering only birds hatched in Kvismaren that returned to breed at this site (philopatric individuals), breeding site fidelity in consecutive years was even higher (almost 100%). Therefore, we included only philopatric birds in the study to maximize the number of years an individual was sampled.
(b). Pedigree
The great reed warbler pedigree spanning the years 1985–2004 contains 735 individuals [34,35]. We pruned this pedigree prior to the present analyses to remove uninformative links [45]. The pruned pedigree contained 203 unique individuals with the following links: 154 maternities, 154 paternities, 254 full sibs, 467 maternal sibs, 470 paternal sibs, 213 maternal half sibs, 216 paternal half sibs, 118 maternal grandmothers, 118 maternal grandfathers, 123 paternal grandmothers and 123 paternal grandfathers.
(c). Molecular analyses
DNA was extracted using standard phenol/chloroform methods and diluted to a concentration of 1 ng μl−1. Real-time PCRs (qPCRs) were performed in a Mx3000P QPCR system (Stratagene) for quantification of telomeres by using the primer set tel (tel 1b 5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ and tel 2b 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′) [46]. To obtain an accurate measurement of the total DNA content in a qPCR reaction, a second reaction was carried out with host-specific primer set sfsr/3 (sfsr/3-Fb 5′-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3′ and sfsr/3-Rb 5′-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3′) [47,48], which amplify a single-copy nuclear sequence (114 bp) that is ultra-conserved across vertebrates [49]. Each reaction of 25 μl included 5 μl (1 ng μl−1) DNA, 12.5 μl Supermix (Platinum SYBR-green q-PCR SuperMix-UDG, Invitrogen), 0.1 μl ROX, 0.5 μl (10 μM) of each tel primer or 1 μl (10 μM) of each sfsr/3 primer and ddH2O. We ran 30 thermal cycles for telomere measurements and 40 thermal cycles for total DNA contents. Samples were incubated at 50°C for 2 min and 95°C for 10 min before running thermal cycling [95°C for 15 s, (57°C for sfsr/3 and 58°C for tel for 30 s) and 72°C for 30 s]. Owing to different annealing temperatures, the sfsr/3 and tel primer sets were run on different plates.
Each sample was run in duplicates for both the tel and the sfsr/3 primer sets and we used the mean of the Ct-values of these duplicates in all calculations. Every 96-well plate included a ‘golden sample’ (one great reed warbler sample of 1 ng μl−1) [46], two negative controls and a serial dilution of a carefully quantified sample (using a NanoDrop 2000, Thermo Scientific) from another great reed warbler (25, 5, 1, 0.2 and 0.04 ng of DNA per well) to produce standard curves. This serially diluted standard was used on all plates and for both sets of primers. We discarded and then re-ran samples on plates producing standard curves where the qPCR efficiency was outside 85–115%.
From the Ct values of qPCR reactions (mean Ct ± s.d.; telomere = 14.5 ± 1.1, single-copy gene = 24.4 ± 1.0), we calculated the amounts of the telomere sequence and the single-copy nuclear sequence relative to their respective standard curves. Because the standard curves are expressed in ng μl−1 this is the unit reported from the qPCR. These values were standardized across the plates by dividing them by the plate value of the ‘golden sample’ in order to obtain plate-adjusted amounts of telomere sequence (T) and single-copy nuclear sequence (S). Finally, we calculated the relative telomere length by the ratio of T and S (T/S), which is the standard used in several recent qPCR-based telomere studies [9,46]. In qPCR studies, differences in gene copy numbers are often expressed as ΔΔCt-values; however, the two approaches can easily be compared because the −log2 of T/S is equal to ΔΔCt [50]. We have used the T/S ratio as a relative measure of ‘telomere length’ in this study.
Next, we evaluated the within-plate repeatability (i.e. between technical replicates of the same sample) for non-normalized values obtained by the primer sets sfsr/3 and tel. We used six plates (three plates for each primer set sfsr/3 and tel) and each plate contained 42 samples that were run in duplicates. We analysed the data using linear mixed model (LMM) fitted with restricted maximum likelihood (REML, lme4 package in R v. 2.15.1 [51]) and included the sample ID as a random factor. The within-plate repeatability was very high for both primers (tel, mean ICC = 0.99; sfsr/3, mean ICC = 0.98; see electronic supplementary material, table S1). To calculate between-plate repeatability we used 15 random samples run three times in duplicates on different plates. Also for these analyses, we used LMM fitted with REML (lme4 package in R v. 2.15.1 [51]). The response values were either the T/S ratios or the standard curve-adjusted values from the qPCR output fitting the sample ID as a random factor. The method we have used also shows very high between-plate repeatability (tel, mean intra class correlation (ICC) = 0.96; sfsr/3, mean ICC = 0.94). Finally, we calculated the repeatability of the T/S ratio (i.e. our estimate of ‘telomere length’ after normalization for both the ‘golden sample’ and the total DNA content in the sample) using the same statistical method as above. Our method also shows high repeatability of the T/S ratio (ICC = 0.93; see electronic supplementary material, table S1).
One potential problem of using qPCR to study telomeres in birds is that it measures telomere sequences at the end of the chromosomes together with any interstitial telomeric repeats. Interstitial telomeric repeats probably occur in most birds and can vary among individuals of the same species, however, the interstitial repeats do not change in length over an individual's life [52,53]. To evaluate whether our qPCR measurement captures variation in telomere length without being seriously affected by any interstitial telomeric repeats, we investigated the annual qPCR measurements of telomere repeat numbers in 65 individuals that survived to 1–6 years of age.
(d). Statistics
To investigate the narrow sense heritability (h2) of telomere length, we first ran parent–offspring regressions. Some of the individuals produced offspring in more than one year, in most cases with different partners. We therefore pooled all offspring belonging to the same mother or father, respectively, to calculate mean offspring telomere length for each parent. For the sex-specific analyses, we conducted regression analyses of early life telomere length of parents and their offspring: mother versus mid-son, mother versus mid-daughter, father versus mid-son and father versus mid-daughter. We also ran regression analyses to investigate the parental age effect on offspring early life telomere length.
To investigate the factors that may influence offspring telomere length, we LMM estimated with REML (lme4 package in R v. 2.15.1). We included the following variables as fixed factors in the models: offspring sex; mother's age; mother early life telomere length; and father's age and father's early life telomere length, with all two-way interactions. As both females and males produced broods in different years, we added mother identity and father identity as random effects, with broods nested inside mother and father. We used the package LMERConvenienceFunction to forward-fit random effects (using log-likelihood ratio testing) and to back-fit fixed factors (on F-values, t-values and log-likelihood ratio testing). We used the function pamer.fnc to compute ANOVAs with upper- and lower-bound p-values and percentage of deviance explained [54] (table 1).
Table 1.
Results of a linear mixed model (LMM) that investigated factors that could be associated with offspring early life telomere length in great reed warblers. All factors were included simultaneously in the model. As both females and males produced broods in different years, we added mother identity and father identity as random effects with broods nested inside mother and father. d.f., nominator degree of freedom; upper.den.d.f., upper-boundary denominator degree of freedom; upper.p.val, upper-boundary significant value; lower.den.d.f., lower boundary denominator degree of freedom; lower.p.val, lower boundary significant value; expl.dev.(%), proportion of deviance explained. Significant p values are given in bold.
| Model | estimates ± s.e. | d.f. | F-value | upper.den.d.f. | upper.p.val | lower.den.d.f. | lower.p.val | expl.dev. (%) |
|---|---|---|---|---|---|---|---|---|
| offspring sex | −0.043 ± 0.034 | 1 | 1.089 | 79 | 0.297 | 17 | 0.309 | 0.226 |
| mother age | 0.071 ± 0.022 | 1 | 15.098 | 79 | 0.0002 | 17 | 0.001 | 3.107 |
| father age | 0.015 ± 0.028 | 1 | 0.141 | 79 | 0.708 | 17 | 0.712 | 0.029 |
| mother telomere length | 0.647 ± 0.148 | 1 | 19.004 | 79 | <0.0001 | 17 | <0.001 | 4.003 |
| father telomere length | −0.130 ± 0.235 | 1 | 0.307 | 79 | 0.581 | 17 | 60.58 | 0.063 |
Finally, to evaluate genetic and non-genetic effects that may affect early life telomere length, we analysed our pedigree data [34,35] using an ‘animal model’ approach. The animal model is a LMM estimated with restricted maximum likelihood (REML), that takes all relationships from the pedigree into account, adjusts for fixed effects, and simultaneously divides the remaining phenotypic variance into environmental and genetic components [31,55]. We used the software ASReml v. 3.0 [56] to fit our model. Based on the results from the LMM (table 1), we included ‘mother age’ as a fixed factor (because it significantly affected telomere length, see table 1). As random factors we included mother identity (to estimate maternal effects), brood identity (to estimate brood-specific environmental effects, other than maternal effects) and the individual identity linked to the pedigree (to estimate additive genetic effects). To obtain significance levels (see table 2), we compared models with a log-likelihood ratio test [55]. As our dataset is rather small for employing ‘animal models', it should be stated that it is likely to be underpowered for more complex models.
Table 2.
Additive genetic and maternal effects (estimate ± s.e.) on offspring early life telomere length (TL) estimated by ‘animal model’ analyses. Six models (1–6) are presented, where the first one is the null model. The next models (2–4) show how variance is partitioned when factors are added to the null model, starting with brood (model 2), followed by maternal effect (model 3) and then genetic effect (model 4). The last two models (5–6) show how variance is partitioned when excluding brood but including first maternal effect (model 5) and then adding genetic effect (model 6). In all these models, ‘mother age’ is included as a fixed factor. Significance levels of the factors are estimated with log-likelihood (logL) ratio test and factors with p < 0.001 are indicated in bold. Degrees of freedom are 167 for all models. VP, phenotypic variance; VE, environmental (residual) variance; e2, environmental (residual) effect; VB, brood variance; b2, brood effect; VM, maternal variance; m2, maternal effect; VA, additive genetic variance; h2, heritability; logL, log-likelihood estimate; *, model did not fully converge; n.e., not estimated.
| model | VP | VE | e2 | VB | b2 | VM | m2 | VA | h2 | logL |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. TL = mother age | 0.114 ± 0.012 | 0.114 ± 0.012 | 1 | — | — | — | — | — | — | 91.68 |
| 2. TL = mother age + brood | 0.106 ± 0.016 | 0.015 ± 0.002 | 0.141 ± 0.029 | 0.091 ± 0.017 | 0.848 ± 0.029 | — | — | — | — | 172.5 |
| 3. TL = mother age + brood + maternal effect | 0.106 ± 0.018 | 0.015 ± 0.002 | 0.141 ± 0.031 | 0.043 ± 0.020 | 0.405 ± 0.146 | 0.048 ± 0.014 | 0.454 ± 0.137 | — | — | 176.3 |
| 4. TL = mother age + brood + genetic effect + maternal effect* | 0.091 ± 0.013 | 0 ± 0 (n.e.) | 0 ± 0 (n.e.) | 0.016 ± 0.015 | 0.184 ± 0.151 | 0.042 ± 0.013 | 0.467 ± 0.039 | 0.032 ± 0.004 | 0.349 ± 0.068 | 179.5 |
| 5. TL = Telomere length = mother age + maternal effect | 0.112 ± 0.019 | 0.039 ± 0.005 | 0.351 ± 0.071 | — | — | 0.072 ± 0.020 | 0.650 ± 0.071 | — | — | 142.98 |
| 6. TL = mother age + genetic effect + maternal effect | 0.110 ± 0.019 | 0.006 ± 0.008 | 0.051 ± 0.077 | — | — | 0.051 ± 0.017 | 0.470 ± 0.093 | 0.051 ± 0.015 | 0.480 ± 0.120 | 155.5 |
3. Results
We found that telomere repeat number in great reed warblers decreases significantly with age (F1,108 = 98.772, p < 0.0001; figure 1) with no effect of sex (F1,63 = 0.11, p = 0.71). This relationship strongly implies that the chromosome-end telomere repeats is the main component measured when using the qPCR method in great reed warblers, thus providing a good estimate of relative telomere length (without being seriously affected by interstitial telomeric repeats).
Figure 1.
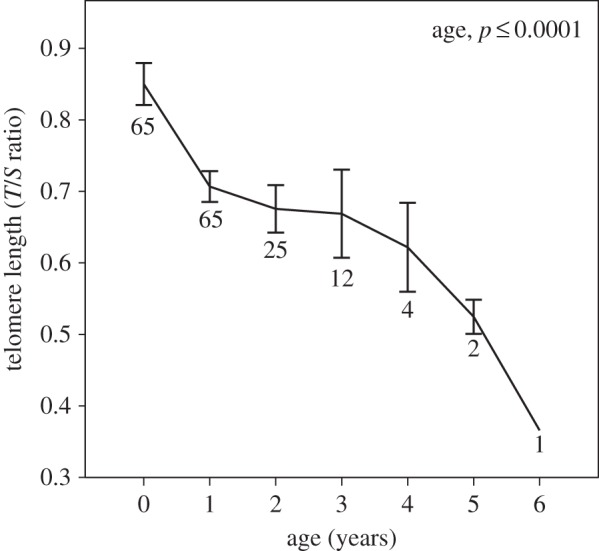
Relationship between age and telomere length in great reed warblers. The sample size (N) for each age group is given below the error bar. T/S ratio is a relative telomere length estimated by dividing the sample's telomere copy number (T) by its copy number of an ultra-conserved single-copy nuclear sequence (S) (see §2).
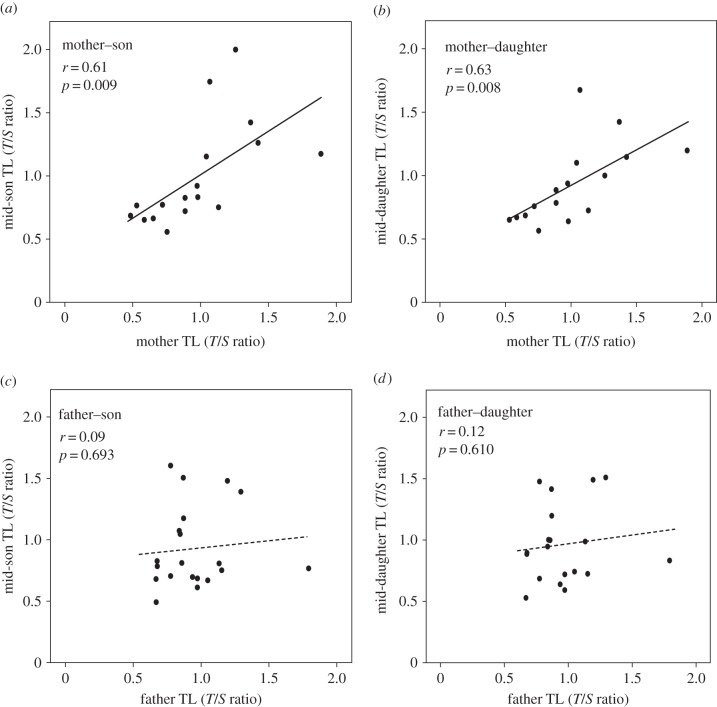
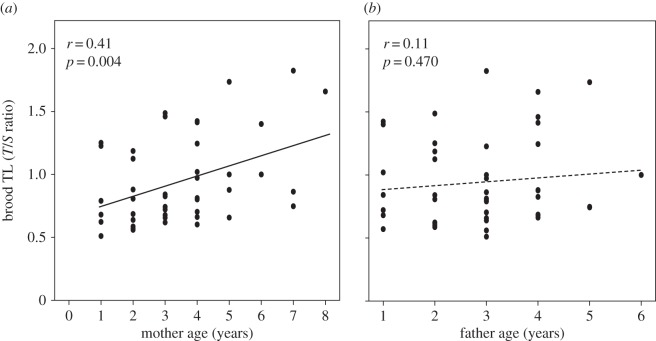
Parent–offspring regression analyses revealed a significant relationship between mother and offspring early life telomere length with a similar magnitude for both sons (b = 0.69 (95% CI = 0.20–1.18), r = 0.61, p = 0.009; figure 2a) and daughters (b = 0.56 (95% CI = 0.17–0.95), r = 0.63, p = 0.008; figure 2b). However, there was no significant relationship between father and offspring early life telomere length, either for sons (b = 0.12 (95% CI = −0.50–0.72), r = 0.09, p = 0.69; figure 2c) or daughters (b = 0.14 (95% CI = −0.4–0.71), r = 0.12, p = 0.61; figure 2d). The heritability estimates (b × 2) for the significant mother–mid-offspring regression was h2 = 1.08 (0.54 × 2) and for the non-significant father–mid-offspring regression h2 = 0.28 (0.14 × 2) (electronic supplementary material, figure S1a,b). The parent–offspring regression estimates of the narrow sense heritability h2 can, however, be confounded by environmental effects. This is emphasized by our finding that mother age was significantly positively correlated with offspring early life telomere length (r = 0.41, p = 0.004, N = 139 offspring from 46 broods; figure 3a), whereas there was no effect of father age on offspring early life telomere length (r = 0.11, p = 0.47, N = 139 offspring from 46 broods; figure 3b).
Figure 2.
Parent–offspring regressions of early life telomere length (TL) in great reed warblers; (a) between mother and sons (17 mothers, 31 broods and 56 sons), (b) between mother and daughters (17 mothers, 31 broods and 43 daughters), (c) between father and sons (19 fathers, 36 broods and 65 sons) and (d) between father and daughters (19 fathers, 36 broods and 47 daughters). Some of the individuals produced offspring in more than 1 year (almost always with different partners), and we therefore pooled data for all the offspring that belonged to the same mother or father, respectively, to calculate mean offspring telomere length for each parent.
Figure 3.
Relationship between a parent's age at breeding and the brood-mean early life telomere length (TL) of the offspring, for (a) mothers (46 broods, 139 offspring) and (b) fathers (46 broods, 139 offspring). Statistics in the figures are from regression analyses.
The LMM analysis showed a significant positive relationship between the mother's early life telomere length and the early life telomere length of their offspring (F1,79 = 19.01, p < 0.001; table 1). Moreover, there was a significant positive relationship between the mother's age at the time of the specific breeding event and the early life telomere length of the offspring resulting from that breeding event (F1,79 = 15.098, p < 0.001; table 1, see also figure 3a). There was no relationship between offspring early life telomere length and either the father's early life telomere length or the father's age at the time of a breeding event (p > 0.50; table 1). Offspring sex did not explain any of the variation in offspring early life telomere length (p > 0.20; table 1), and all two-way interactions were non-significant (all p > 0.05).
The differences in the slope values of the mother–offspring (figure 2a,b) and father–offspring (figure 2c,d) regressions suggest a strong maternal effect on the inheritance of telomere length, an effect that potentially can mask the presence of an additive effect. The presence of a strong maternal, but no paternal effect on the inheritance pattern of telomere length is further supported by the LMM analyses (table 1).
To tease apart the additive genetic and environmental components of the phenotypic variance in early life telomere length, we used the extensive pedigree and the ‘animal model’ approach. Based on the LMM (table 1), maternal age was included as a covariate fixed effect to account for variation in offspring telomere length caused by the age of the mother. As random effects, we fitted additive genetic effects, maternal effects (this time, the general maternal effects based on consistent differences between individual mothers) and brood effect. Both brood effect and maternal effect are significant (model 3, table 2). When including the additive genetic effects into the model, the model does not fully converge (the parameters can still change 1–2%) and the residuals cannot be estimated. In this model, the heritability was estimated to be 0.349 (±0.068 s.e., p < 0.001). We also ran a model that did fully converge, only including maternal effect and additive genetic effect, where the heritability was estimated to be 0.480 (±0.120 s.e., p < 0.0001). The maternal effect did not change considerably between these models (models 4 and 6 in table 2) varying between 0.467 (±0.039 s.e., p < 0.0001) and 0.470 (±0.093 s.e., p < 0.0001; table 2).
4. Discussion
Our analyses showed strong heritability and pronounced maternal effects on early life telomere length in great reed warblers. Previous studies in a number of different organisms and species have reported significant heritability of telomere length. However, since these studies are based on parent–offspring regressions [15,17,20,21,29,30], none of them has been able to disentangle the relative contribution of the additive genetic and parental effects to telomere length. The strength of our study is that it is based on a dataset (193 individuals including mothers, fathers and young from 46 broods) embedded in a resolved pedigree [34,35], which enabled us to use ‘animal models' [31] to disentangle the additive genetic and maternal effects. When applying these tools to the great reed warbler dataset where we had measured early life telomere length in both parents and their offspring, we could demonstrate that both additive genetic effects (h2 = 35–48%) and maternal effects (47%) contributed to offspring early life telomere length. The strong genetic and maternal effects on offspring early life telomere length are likely to be important for offspring performance and fitness, because in our study species an individual's early life telomere length is positively associated with its longevity [57].
Our study shows that offspring early life telomere length can be strongly influenced by maternal effects. This has important implications and raises new questions in telomere research. A first step would be to find out to what extent maternal effects also influence offspring early life telomere length also in other species of birds and mammals, including in humans. Second, we can begin to analyse which maternal factors contribute to early life telomere length in the offspring. For example, ecological studies have shown that maternal (or experimental) modulation of egg yolk composition (e.g. concentrations of hormones and antibodies) can impact behaviour, growth, immune function and offspring survival [58–60]. Moreover, patterns of growth and exposure to stress early in life have been associated with reduced telomere length in humans and rats [61,62]. Hence, maternal influence on growth, immune response and stress in their offspring might also affect offspring telomere length during the prenatal and early postnatal developmental stages. Future studies should try to identify which types of maternal effects are decisive for the control of offspring early life telomere length and during which stage (prenatal or early postnatal) of development such factors have the strongest effects.
Another remarkable finding in our study was that offspring telomere length was positively related to maternal age but not to paternal age. By contrast, in humans, offspring telomere length is positively correlated with paternal but not with maternal age [15,25,27]. One proposed explanation for this pattern has been that older men have sperm with longer telomeres [25,63,64], possibly as a consequence of the high activity of telomerase found in testes [25,65]. This higher telomerase activity is thought to be owing to the need for continued production of male gametes throughout life [66]. The argument explaining why maternal age does not affect offspring telomere length in humans has been that the entire population of ova is developed at a very early age, after which it is retained throughout life without further cell divisions, preventing any ageing effects of the mother on offspring telomere length [25,67]. However, these mechanisms can obviously not explain the sex-specific age-dependent pattern found in our songbird study, because we found a positive association between maternal age and the telomere length of their offspring, but no such relationship with father age. The association between mother age and offspring telomere length could be explained by a higher telomerase activity in the ovaries of female birds, a process that is age- and/or condition-dependent. However, this is speculative and remains to be investigated. Another possible explanation for longer telomere length in offspring produced by older females could be that these young experience a slower rate of telomere loss before ca 9 days of age (our measurement point), and that this slower telomere loss rate is caused by some type of maternal effect that is enhanced as the female becomes older (see above). Previous studies have shown that parental age at breeding can influence offspring phenotype via variation in nest attendance, incubation temperature or other maternal effects [60,68,69]. In great reed warblers, older females arrive earlier in spring, which may allow them to occupy better territories for breeding and gain more help from the males in feeding nestlings. By contrast, younger females arrive later and, therefore, more often settle as secondary females (with socially polygynous males) and thereby get limited or no help from the male with nestling feeding, or they settle as monogamous females in low quality territories with young or low quality males, situations that may induce costs and reduce phenotypic condition in young females [40,41,70]. Such social, temporal and spatial variation may affect female condition and hence play a role in the telomere degradation of their offspring.
To the best of our knowledge, this is the first study to show the relative effect of both the genetic and parental factors that determine telomere length in a vertebrate. The results of this study clearly show that there is a significant additive genetic as well as an equally large maternal component on early life telomere length in great reed warblers. Our study of a songbird, as well as parent–offspring regression studies of other bird species [21,23], supports the view that telomere length is under strong maternal influence in birds. This contrasts the situation in humans, where telomere length appears to be under paternal influence [15–17], although results of human studies are presently equivocal [12,19]. Furthermore, we found an association between offspring telomere length and their mother's age, which is in contrast to studies of humans where offspring telomere length instead is linked to father's age [15,25,27]. Taken together, our study provides several novel insights into the relative importance of genetic and non-genetic factors that determine early life telomere length in vertebrates. These results now call for experimental studies to further elucidate the factors that may underlie the maternal effects on offspring early life telomere length. Moreover, it also highlights the need for a broader taxonomic survey regarding which parental sex determines telomere inheritance patterns, as well as the relative contribution of genetic and non-genetic factors to early life telomere length.
Acknowledgements
We thank J-Å. Nilsson, M. Strandh and A. Marzal for critical discussions and comments on the manuscript.
Funding statement
This work was supported by grants from the Swedish Research Council (to S.B., B.H. and D.H.), from the Higher Education Commission of Pakistan (M.A.), by the Centre for Animal Movement Research (CAnMove) financed by a Linnaeus excellence research grant (349–2007–8690) from the Swedish Research Council and Lund University, and by support from Kvismare Bird Observatory (report 173).
References
- 1.de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110. ( 10.1101/gad.1346005) [DOI] [PubMed] [Google Scholar]
- 2.Blasco MA. 2007. Telomere length, stem cells and aging. Nat. Chem. Biol. 3, 640–649. ( 10.1038/nchembio.2007.38) [DOI] [PubMed] [Google Scholar]
- 3.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. 1992. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA 89, 10 114–10 118. ( 10.1073/pnas.89.21.10114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA. 2003. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet 361, 393–395. ( 10.1016/S0140-6736(03)12384-7) [DOI] [PubMed] [Google Scholar]
- 5.Haussmann MF, Winkler DW, Vleck CM. 2005. Longer telomeres associated with higher survival in birds. Biol. Lett. 1, 212–214. ( 10.1098/rsbl.2005.0301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbig U, Jobling WA, Chen BPC, Chen DJ, Sedivy JM. 2004. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a). Mol. Cell 14, 501–513. ( 10.1016/S1097-2765(04)00256-4) [DOI] [PubMed] [Google Scholar]
- 7.Monaghan P. 2010. Telomeres and life histories: the long and the short of it. Ann. NY Acad. Sci. 1206, 130–142. ( 10.1111/j.1749-6632.2010.05705.x) [DOI] [PubMed] [Google Scholar]
- 8.Barrett EL, Burke TA, Hammers M, Komdeur J, Richardson DS. 2013. Telomere length and dynamics predict mortality in a wild longitudinal study. Mol. Ecol. 22, 249–259. ( 10.1111/mec.12110) [DOI] [PubMed] [Google Scholar]
- 9.Heidinger BJ, Blount JD, Boner W, Griffiths K, Metcalfe NB, Monaghan P. 2012. Telomere length in early life predicts lifespan. Proc. Natl Acad. Sci. USA 109, 1743–1748. ( 10.1073/pnas.1113306109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn T, Robertson BC, Gemmell NJ. 2010. The use of telomere length in ecology and evolutionary biology. Heredity (Edinb.) 105, 497–506. ( 10.1038/hdy.2010.113) [DOI] [PubMed] [Google Scholar]
- 11.Joeng KS, Song EJ, Lee KJ, Lee J. 2004. Long lifespan in worms with long telomeric DNA. Nat. Genet. 36, 607–611. ( 10.1038/ng1356) [DOI] [PubMed] [Google Scholar]
- 12.Broer L, et al. 2013. Meta-analysis of telomere length in 19 713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet. 21, 1163–1168. ( 10.1038/ejhg.2012.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattan V, et al. 2008. Chronic oxidative stress induces a tissue-specific reduction in telomere length in CAST/Ei mice. Free Radic. Biol. Med. 44, 1592–1598. ( 10.1016/j.freeradbiomed.2008.01.007) [DOI] [PubMed] [Google Scholar]
- 14.Shlush LI, et al. 2011. Telomere elongation followed by telomere length reduction, in leukocytes from divers exposed to intense oxidative stress—implications for tissue and organismal aging. Mech. Ageing Dev. 132, 123–130. ( 10.1016/j.mad.2011.01.005) [DOI] [PubMed] [Google Scholar]
- 15.Njajou OT, et al. 2007. Telornere length is paternally inherited and is associated with parental lifespan. Proc. Natl Acad. Sci. USA 104, 12 135–12 139. ( 10.1073/pnas.0702703104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nordfäll K, Larefalk A, Lindgren P, Holmberg D, Roos G. 2005. Telomere length and heredity: indications of paternal inheritance. Proc. Natl Acad. Sci. USA 102, 16 374–16 378. ( 10.1073/pnas.0501724102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordfäll K, Svenson U, Norrback KF, Adolfsson R, Roos G. 2010. Large-scale parent–child comparison confirms a strong paternal influence on telomere length. Eur. J. Hum. Genet. 18, 385–389. ( 10.1038/ejhg.2009.178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akkad A, Hastings R, Konje JC, Bell SC, Thurston H, Williams B. 2006. Telomere length in small-for-gestational-age babies. BJOG 113, 318–323. ( 10.1111/j.1471-0528.2005.00839.x) [DOI] [PubMed] [Google Scholar]
- 19.Eisenberg DT. 2014. Inconsistent inheritance of telomere length (TL): is offspring TL more strongly correlated with maternal or paternal TL? Eur. J. Hum. Genet. 22, 8–9. ( 10.1038/ejhg.2013.202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsson M, Pauliny A, Wapstra E, Uller T, Schwartz T, Blomqvist D. 2011. Sex differences in sand lizard telomere inheritance: paternal epigenetic effects increases telomere heritability and offspring survival. PLoS ONE 6, e17473 ( 10.1371/journal.pone.0017473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horn T, Robertson BC, Will M, Eason DK, Elliott GP, Gemmell NJ. 2011. Inheritance of telomere length in a bird. PLoS ONE 6, e17199 ( 10.1371/journal.pone.0017199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voillemot M, Hine K, Zahn S, Criscuolo F, Gustafsson L, Doligez B, Bize P. 2012. Effects of brood size manipulation and common origin on phenotype and telomere length in nestling collared flycatchers. BMC Ecol. 12, 17 ( 10.1186/1472-6785-12-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichert S, et al. 2014. Maternal telomere length inheritance in the king penguin. Heredity. ( 10.1038/hdy.2014.60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilley D, Herbert BS, Huda N, Tanaka H, Reed T. 2008. Factors impacting human telomere homeostasis and age-related disease. Mech. Ageing Dev. 129, 27–34. ( 10.1016/j.mad.2007.10.010) [DOI] [PubMed] [Google Scholar]
- 25.Kimura M, et al. 2008. Offspring's leukocyte telomere length, paternal age, and telomere elongation in sperm. PLoS Genet. 4, e37 ( 10.1371/journal.pgen.0040037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L, et al. 2007. Telomere lengthening early in development. Nat. Cell Biol. 9, 1436–1441. ( 10.1038/ncb1664) [DOI] [PubMed] [Google Scholar]
- 27.Unryn BM, Cook LS, Riabowol KT. 2005. Paternal age is positively linked to telomere length of children. Aging Cell 4, 97–101. ( 10.1111/j.1474-9728.2005.00144.x) [DOI] [PubMed] [Google Scholar]
- 28.Keefe DL. 2007. Telomeres and meiosis in health and disease. Cell Mol. Life Sci. 64, 115–116. ( 10.1007/s00018-006-6462-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bischoff C, et al. 2005. The heritability of telomere length among the elderly and oldest-old. Twin Res. Hum. Genet. 8, 433–439. ( 10.1375/twin.8.5.433) [DOI] [PubMed] [Google Scholar]
- 30.Vasa-Nicotera M. 2005. Mapping of a major locus that determines telomere length in humans. Am. J. Hum. Genet. 76, 373 ( 10.1086/426734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruuk LE. 2004. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. Lond. B 359, 873–890. ( 10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bensch S, Waldenström J, Jonzén N, Westerdahl H, Hansson B, Sejberg D, Hasselquist D. 2007. Temporal dynamics and diversity of avian malaria parasites in a single host species. J. Anim. Ecol. 76, 112–122. ( 10.1111/j.1365-2656.2006.01176.x) [DOI] [PubMed] [Google Scholar]
- 33.Hansson B, Jack L, Christians JK, Pemberton JM, Åkesson M, Westerdahl H, Bensch S, Hasselquist D. 2007. No evidence for inbreeding avoidance in a great reed warbler population. Behav. Ecol. 18, 157–164. ( 10.1093/beheco/arl062) [DOI] [Google Scholar]
- 34.Åkesson M, Bensch S, Hasselquist D, Tarka M, Hansson B. 2008. Estimating heritabilities and genetic correlations: comparing the ‘animal model’ with parent–offspring regression using data from a natural population. PLoS ONE 3, e1739 ( 10.1371/journal.pone.0001739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarka M, Åkesson M, Beraldi D, Hernández-Sánchez J, Hasselquist D, Bensch S, Hansson B. 2010. A strong quantitative trait locus for wing length on chromosome 2 in a wild population of great reed warblers. Proc. R. Soc. B 277, 2361–2369. ( 10.1098/rspb.2010.0033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bensch S, Hasselquist D, Nielsen B, Hansson B. 1998. Higher fitness for philopatric than for immigrant males in a semi-isolated population of great reed warblers. Evolution 52, 877–883. ( 10.2307/2411282) [DOI] [PubMed] [Google Scholar]
- 37.Westerdahl H, Asghar M, Hasselquist D, Bensch S. 2012. Quantitative disease resistance: to better understand parasite-mediated selection on major histocompatibility complex. Proc. R. Soc. B 279, 577–584. ( 10.1098/rspb.2011.0917) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cramp S. 1993. Hand book of the birds of Europe, the Middle East and North Africa: the birds of the Westren Palearctic volume VI: warblers. Oxford, UK: Oxford University Press. [Google Scholar]
- 39.Hansson B, Bensch S, Hasselquist D, Lillandt BG, Wennerberg L, Von Schantz T. 2000. Increase of genetic variation over time in a recently founded population of great reed warblers (Acrocephalus arundinaceus) revealed by microsatellites and DNA fingerprinting. Mol. Ecol. 9, 1529–1538. ( 10.1046/j.1365-294x.2000.01028.x) [DOI] [PubMed] [Google Scholar]
- 40.Hasselquist D. 1998. Polygyny in great reed warblers: a long-term study of factors contributing to male fitness. Ecology 79, 2376–2390. ( 10.1890/0012-9658(1998)079[2376:PIGRWA]2.0.CO;2) [DOI] [Google Scholar]
- 41.Bensch S. 1996. Female mating status and reproductive success in the great reed warbler: is there a potential cost of polygyny that requires compensation? J. Anim. Ecol. 65, 283–296. ( 10.2307/5875) [DOI] [Google Scholar]
- 42.Arlt D, Hansson B, Bensch S, Von Schantz T, Hasselquist D. 2004. Breeding synchrony does not affect extra-pair paternity in great reed warblers. Behaviour 141, 863–880. ( 10.1163/1568539042265699) [DOI] [Google Scholar]
- 43.Hasselquist D, Bensch S, von Schantz T. 1996. Correlation between male song repertoire, extra-pair paternity and offspring survival in the great reed warbler. Nature 381, 229–232. ( 10.1038/381229a0) [DOI] [Google Scholar]
- 44.Hansson B, Bensch S, Hasselquist D, Nielsen B. 2002. Restricted dispersal in a long-distance migrant bird with patchy distribution, the great reed warbler. Oecologia 130, 536–542. ( 10.1007/s00442-001-0831-2) [DOI] [PubMed] [Google Scholar]
- 45.Hadfield JD. 2010. MCMC methods for multi–response generalised linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22.20808728 [Google Scholar]
- 46.Criscuolo F, Bize P, Nasir L, Metcalfe NB, Foote CG, Griffiths K, Gault EA, Monaghan P. 2009. Real-time quantitative PCR assay for measurement of avian telomeres. J. Avian Biol. 40, 342–347. ( 10.1111/j.1600-048X.2008.04623.x) [DOI] [Google Scholar]
- 47.Asghar M, Hasselquist D, Bensch S. 2011. Are chronic avian haemosporidian infections costly in wild birds? J. Avian Biol. 42, 530–537. ( 10.1111/j.1600-048X.2011.05281.x) [DOI] [Google Scholar]
- 48.Asghar M, Westerdahl H, Zehtindjiev P, Ilieva M, Hasselquist D, Bensch S. 2012. Primary peak and chronic malaria infection levels are correlated in experimentally infected great reed warblers. Parasitology 139, 1246–1252. ( 10.1017/S0031182012000510) [DOI] [PubMed] [Google Scholar]
- 49.Bejerano G, et al. 2004. Ultraconserved elements in the human genome. Science 304, 1321–1325. ( 10.1126/science.1098119) [DOI] [PubMed] [Google Scholar]
- 50.Cawthon RM. 2002. Telomere measurement by quantitative PCR. Nucl. Acid Res. 30, 10e47 ( 10.1093/nar/30.10.e47) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bates D, Maechler M, Bolker B, Walker S, Christensen RHB, Singmann H. 2014. lme4: linear mixed-effects models using Eigen and S4. R package version 1.1–6 See https://CRAN.R-project.org/package=lme4.
- 52.Nakagawa S, Gemmell NJ, Burke T. 2004. Measuring vertebrate telomeres: applications and limitations. Mol. Ecol. 13, 2523–2533. ( 10.1111/j.1365-294X.2004.02291.x) [DOI] [PubMed] [Google Scholar]
- 53.Foote CG, Vleck D, Vleck CM. 2013. Extent and variability of interstitial telomeric sequences and their effects on estimates of telomere length. Mol. Ecol. Res. 13, 417–428. ( 10.1111/1755-0998.12079) [DOI] [PubMed] [Google Scholar]
- 54.Tremblay A. 2011. LMERConvenienceFunctions: a suite of functions to back-fit fixed effects and forward-fit random effects, as well as other miscellaneous functions. R package v. 1.6.3. package v. 1.6.3.
- 55.Wilson AJ, et al. 2010. An ecologist's guide to the animal model. J. Anim. Ecol. 79, 13–26. ( 10.1111/j.1365-2656.2009.01639.x) [DOI] [PubMed] [Google Scholar]
- 56.Gilmour AR, Gogel BJ, Cullis BR, Thompson R. 2009. ASReml user guide release 3.0 VSN International Ltd, Hemel Hempstead, HP1 1ES, UK www.vsni.co.uk.
- 57.Asghar M. 2012. Living with parasites: avian malaria, telomere length and life history trade-offs, p. 120 Lund, Sweden: Media Tryck, Lund University press. [Google Scholar]
- 58.Hasselquist D, Nilsson JA. 2009. Maternal transfer of antibodies in vertebrates: trans-generational effects on offspring immunity. Phil. Trans. R. Soc. B 364, 51–60. ( 10.1098/rstb.2008.0137) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasselquist D, Tobler M, Nilsson J-Å. 2012. Maternal modulation of offspring immune function in vertebrates. In Eco-Immunology (eds Nelson RM, Demas G.), pp. 165–224. Oxford, UK: Oxford University Press. [Google Scholar]
- 60.Krist M. 2011. Egg size and offspring quality: a meta-analysis in birds. Biol. Rev. Camb. Phil. Soc. 86, 692–716. ( 10.1111/j.1469-185X.2010.00166.x) [DOI] [PubMed] [Google Scholar]
- 61.Entringer S, Epel ES, Kumsta R, Lin J, Hellhammer DH, Blackburn EH, Wust S, Wadhwa PD. 2011. Stress exposure in intrauterine life is associated with shorter telomere length in young adulthood. Proc. Natl Acad. Sci. USA 108, E513–E518. ( 10.1073/pnas.1107759108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarry-Adkins JL, Chen JH, Smith NS, Jones RH, Cherif H, Ozanne SE. 2009. Poor maternal nutrition followed by accelerated postnatal growth leads to telomere shortening and increased markers of cell senescence in rat islets. FASEB J. 23, 1521–1528. ( 10.1096/fj.08-122796) [DOI] [PubMed] [Google Scholar]
- 63.Baird DM, et al. 2006. Telomere instability in the male germline. Hum. Mol. Genet. 15, 45–51. ( 10.1093/hmg/ddi424) [DOI] [PubMed] [Google Scholar]
- 64.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. 1996. Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet. 18, 173–179. () [DOI] [PubMed] [Google Scholar]
- 65.Zalenskaya IA, Zalensky AO. 2002. Telomeres in mammalian male germline cells. Int. Rev. Cytol. 218, 37–67. ( 10.1016/S0074-7696(02)18011-9) [DOI] [PubMed] [Google Scholar]
- 66.Eisenberg DT. 2011. An evolutionary review of human telomere biology: the thrifty telomere hypothesis and notes on potential adaptive paternal effects. Am. J. Hum. Biol. 23, 149–167. ( 10.1002/ajhb.21127) [DOI] [PubMed] [Google Scholar]
- 67.Arbeev KG, Hunt SC, Kimura M, Aviv A, Yashin AI. 2011. Leukocyte telomere length, breast cancer risk in the offspring: the relations with father's age at birth. Mech. Ageing Dev. 132, 149–153. ( 10.1016/j.mad.2011.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bogdanova MI, Nager RG, Monaghan P. 2007. Age of the incubating parents affects nestling survival: an experimental study of the herring gull Larus argentatus. J. Avian Biol. 38, 83–93. ( 10.1111/j.2007.0908-8857.03701.x) [DOI] [Google Scholar]
- 69.Nilsson JF, Stjernman M, Nilsson JA. 2008. Experimental reduction of incubation temperature affects both nestling and adult blue tits Cyanistes caeruleus. J. Avian Biol. 39, 553–559. ( 10.1111/j.0908-8857.2008.04199.x) [DOI] [Google Scholar]
- 70.Sejberg D, Bensch S, Hasselquist D. 2000. Nestling provisioning in polygynous great reed warblers (Acrocephalus arundinaceus): do males bring larger prey to compensate for fewer nest visits? Behav. Ecol. Sociobiol. 47, 213–219. ( 10.1007/s002650050658) [DOI] [Google Scholar]