Abstract
In this study ultra performance liquid chromatography (UPLC) coupled to time-of-flight mass spectrometry in the MSE mode was used for rapid and comprehensive analysis of metabolites in the serum of mice exposed to internal exposure by Cesium-137 (137Cs). The effects of exposure to 137Cs were studied at several time points after injection of 137CsCl in mice. Over 1800 spectral features were detected in the serum of mice in positive and negative electrospray ionization modes combined. Detailed statistical analysis revealed that several metabolites associated with amino acid metabolism, fatty acid metabolism, and the TCA cycle were significantly perturbed in the serum of 137Cs-exposed mice compared with that of control mice. While metabolites associated with the TCA cycle and glycolysis increased in their serum abundances, fatty acids such as linoleic acid and palmitic acid were detected at lower levels in serum after 137Cs exposure. Furthermore, phosphatidylcholines (PCs) were among the most perturbed ions in the serum of 137Cs-exposed mice. This is the first study on the effects of exposure by an internal emitter in serum using a UPLC–MSE approach. The results have put forth a panel of metabolites, which may serve as potential serum markers to 137Cs exposure.
Keywords: mass spectrometry, UPLC−MSE, metabolomics, lipidomics, radiation, internal emitter, Cesium-137
Introduction
Exposure to internal emitters such as cesium-137 is an inevitable consequence of nuclear accidents, such as Chernobyl and Fukushima Daiichi.1 In addition to such large-scale events, occupational and industrial exposure, as in the case of the Goiania scrap metal incident or as in the case of a terrorist-plotted radiological dispersal device, raise serious concerns. Because of its environmental persistence and ease of dispersal, 137Cs poses a great health risk to the general public. Thus, our team set out to develop early and robust markers for 137Cs exposure in the easily obtainable biofluids, urine, and serum. Such exposure markers can be used to screen and assess the risk of exposure in a given population, which, in turn, may help triage the affected population in a faster and more efficient manner.
Many of the previous ionizing radiation (IR) studies focused on the DNA damage/repair machinery and its markers in biological samples.2−4 Although such genomic and transcriptomic studies have helped bring to light many facts about IR-induced injury and inflammation responses, they are limited in scope and have failed to capture the molecular and structural effects of IR in cells. Proteomics and more recently metabolomics have filled this gap and contributed to the field of biodosimetry by revealing the molecular targets of IR and their associated signaling networks. Advances in technology, particularly in the field of mass spectrometry, have enabled our laboratory to make significant progress in determining even the slight and subtle changes in the molecular composition, namely, the metabolome, of biofluids as a result of exposure to external beam and internal emitters5,6 using mouse models.
This study is complementary to our previous work on the metabolic perturbations in urine of C57/Bl6 mice over a 30-day period after exposure to 137Cs, through employing the sensitivity of time-of-flight mass spectrometry (TOFMS) coupled to high resolving power of ultra performance liquid chromatography (UPLC).6 Herein we utilized the same platform to determine the unique and robust responses of serum metabolites to 137Cs exposure but in a new approach entailing a fast and simple lipid profiling method with relative quantitation called MSE. By using this method, we were able to not only identify several different classes of lipids along with their relative abundances but also gain structural information on these chemical species. Furthermore, we utilized a comprehensive statistical analysis workflow we have specifically developed for metabolomics, called MetaboLyzer, to determine 137Cs-specific perturbations in metabolites/lipids and their respective pathways.7 The statistically significant metabolites and lipids were then compared with known external-beam γ-irradiation markers to determine similarities and differences in terms of responses to these two types of exposure.
To date, this is the first time that mass spectrometry has been utilized to study the effects of exposure to an internal emitter in serum of mice. The results of this study and other radiation-induced signaling studies in easily accessible biofluids may help uncover the mechanism behind IR-induced inflammation and injury, which will ultimately lead to the discovery of novel biomarkers of IR exposure.
Materials and Methods
Materials
The following fatty acids and lipid standards were purchased from Avanti Polar lipids (Alabaster, AL): sphingolipid mix (SM) II, phosphatidylethanolamine PE (14:0/14:0), phosphatidylcholine PC (14:0/14:0), phosphatidic acid PA (14:0/14:0), phosphatidylserine PS (14:0/14:0), phosphatidylinositol PI (17:0/20:4), and lysophosphatidylcholine LPC (17:1). Prostaglandin standard and leukotriene were purchased from Cayman Chemical (Ann Arbor, MI), and fatty acid standard FA(17:1) was purchased from Nu-Chek Prep (Elysian, MN). Debrisoquine sulfate, 4-nitrobenzoic acid (4-NBA), and UPLC-grade solvents such as acetonitrile, water, and isopropanol were purchased from Fisher Scientific (Hanover Park, IL). Glucose, riboflavin, arachidonic acid, linoleic acid, oleic acid, palmitic acid, hippuric acid, nicotinic acid, lactic acid, uridine, taurine, α-ketobutyric acid, uric acid, hydroxyphenylpyruvic acid, acetylcarnitine, and carnitine were purchased from Sigma-Aldrich (Seelze, Germany). The tandem MS spectrum of dityrosine provided by Hanft et al. was used as a validation reference spectrum.8 In addition, the MS/MS spectra provided by Scripps Center for Metabolomics, METLIN, (La Jolla, CA) were used as reference spectra for hydroquinone, inositol, gentisic acid, and dihydrolipoamide. METLIN was also used to putatively identify the ion at m/z of 337.1048 as the sodium adduct of 7,8-dihydropteroic acid.
Animal Irradiation and Sample Collection
All animal experiments were conducted in accordance with applicable federal and state guidelines and were approved by the Institutional Animal Care and Use Committee of the Lovelace Biomedical and Environmental Research Institute (LBERI). C57Bl/6 mice (approximately 10–12 weeks old, 25–30 g) were received from Charles River Laboratories (Frederick, MD) and were quarantined for 14 days prior to group assignment by body-weight stratification for randomization into the study.
Animals were injected intraperitoneally with 8.0 ± 0.3 MBq 137CsCl solution in a volume of 50 μL. Nearly identical biokinetics have been found following inhalation, intraperitoneal, or intravenous administration of 137Cs, which was confirmed in a pilot study comparison of the biokinetics between intravenous and intraperitoneal administration in C57Bl/6 mice (unpublished results). On the basis of these results, intraperitoneal administration was chosen for the current study. After dose administration, mice were housed individually in microisolator cages with lead shielding used to minimize cross-irradiation from adjacent mice. All animals had unlimited access to Teklad Certified Global Rodent Diet 2016 (Harlan Teklad, Madison, WI) and water except during dose administration and whole-body in vivo counting. Control mice gained weight steadily throughout the study. Mice injected with 137CsCl initially lost weight, then resumed weight gain at approximately the same rate as the unexposed controls from day 3 after injection. No adverse effects were noted on the animals during the course of the study. The absorbed doses were calculated as previously described by Stabin et al.9 using the RATDOSE software.10 Serum was collected at necropsy by cardiac stick at 2, 3, 5, 20, and 30 days postinjection. Mice at each time point were sacrificed as previously described.6 Each time point consists of eight mice per group with the exception of day 2 and day 30 control mice with seven mice per group (Table 1).
Table 1. Cumulative Doses and Average Body Weights of Mice in Each Study Group.
| study group | collection time (days)a | avg cumulative dose (Gy) | SDb | avg body weight pre ± SD (kg)c | avg body weight post ± SD (kg)d |
|---|---|---|---|---|---|
| control | 2 | NA | NA | 26.01 ± 1.39 | 26.40 ± 1.44 |
| control | 3 | NA | NA | 26.44 ± 1.18 | 27.18 ± 1.08 |
| control | 5 | NA | NA | 27.06 ± 1.63 | 28.43 ± 1.69 |
| control | 20 | NA | NA | 26.62 ± 1.02 | 30.32 ± 1.29 |
| control | 30 | NA | NA | 27.13 ± 0.33 | 32.59 ± 1.77 |
| 137Cs | 2 | 1.95 | 0.11 | 26.02 ± 1.04 | 25.56 ± 1.06 |
| 137Cs | 3 | 2.70 | 0.37 | 26.09 ± 1.30 | 24.90 ± 1.07 |
| 137Cs | 5 | 4.14 | 0.35 | 26.41 ± 1.27 | 25.87 ± 1.20 |
| 137Cs | 20 | 9.46 | 0.41 | 25.74 ± 0.89 | 27.06 ± 1.16 |
| 137Cs | 30 | 9.91 | 1.20 | 25.65 ± 0.82 | 27.55 ± 0.88 |
Serum collection time is indicated as the number of days after treatment.
SD stands for standard deviation.
Average body weight of mice per study group before treatment (pre) in kilograms.
Average body weight of mice per study group after treatment (post) in kilograms.
Sample Preparation for Mass Spectrometry Analysis
Serum samples were prepared by adding one part serum to four parts of a chilled chloroform and methanol mixture (2:1 v/v) containing nonendogenous metabolite and lipid standards in a sterile siliconized tube. The list of nonendogenous internal standards along with their concentrations used in this experiment can be found in Supplementary Table 1 in the Supporting Information. Each sample was then vortexed vigorously for 30 s at room temperature followed by centrifugation at 13 000g for 5 min to separate the polar and nonpolar species into two phases. The upper aqueous phase containing primarily polar metabolites was collected, dried in a vacuum to <10 μL, and resuspended in 50 μL of 50% acetonitrile. The lower phase was also collected, dried under a gentle stream of nitrogen, and resuspended in 50 μL of a isopropanol and 50% acetonitrile mixture (1:1 v/v). To establish appropriate standard curves for the lipid internal standards, we carried out a series of two-fold dilutions at the initial concentration of 350 μg/mL to the final concentration of 2 μg/mL for phosphatidylcholine 14:0/14:0 (PC 14:0/14:0), phosphatidylethanolamine 14:0/14:0 (PE 14:0/14:0), phosphatidylglycerol d4PGE2 (PG d4PGE2), and sphingolipid mix II (SM mix II). The concentration range for lysophosphatidylcholine 17:1 (LPC 17:1) and fatty acid 17:1 (FA 17:1) internal standards was at 128 to 1 μg/mL. The internal standards were also spiked into pooled control serum samples and processed via UPLC–MS at every seven injection intervals. These nonendogenous internal standards were quickly identified based on their unique mass, retention time, and fragmentation profiles. The calculated standard curve for each internal standard was then used to determine the relative abundance of different classes of lipids in each ionization mode.
Mass Spectrometry Analysis
The metabolomic analysis was performed by injecting 2 μL of aliquot of each sample into a reverse-phase 50 × 2.1 mm H-class UPLC Acquity 1.7-μM BEH C18 column (Waters Corp, Milford, MA) coupled to a time-of-flight mass spectrometry (TOFMS). The mobile phase consisted of water and 0.1% formic acid (solvent A), 100% acetonitrile (solvent B). The gradient for the metabolomic analysis switched from 98% aqueous solvent A to 40% solvent A and 60% solvent B after 4 min and to 98% solvent B at 8 min for 1 min and back to 98% solvent A for the last 2 min of the 11 min gradient at a flow rate of 0.5 mL/min. The Xevo G2-S mass spectrometer (Waters) was operated in both MS and MSE modes scanning a 50–1200 m/z range with low collision energy of 10.0 eV for the precursor ions and collision energy ramp of 10–50 eV for the product ions. The lipidomic samples were injected into a CSH C18 column 150 μm × 100 mm (Waters) with the H-class UPLC Acquity. The solvents used for the lipidomic analysis were 50% acetonitrile with 0.1% formic acid and 10 mM ammonium formate (solvent C) and isopropanol/acetonitrile (90:10 v:v) with 10 mM ammonium formate (solvent D).The gradient started with 60% solvent C at 0.45 mL/min for the initial 8 min, then switched to 100% solvent D for 1 min, and back to 60% solvent C for the remaining 2 min of the 11 min long gradient. The Xevo G2-S QTOF mass spectrometer was operated in positive (ESI+) and negative (ESI–) electrospray ionization (ESI) modes over a mass range of 50 to 1200 Da in two channels, MS and MSE. The low energy MS channel was operated at 10.0 eV of collision energy while the MSE channel included an energy ramp of 10–50 eV. The lock-spray consisted of leucine–enkephalin (556.2771 [M + H]+ and 554.2615 [M-H]−). The MS data were acquired in centroid mode and processed using MassLynx software (Waters) as described later.
Statistical Analysis
As described previously5 MarkerLynx software (Waters) was used to construct a data matrix consisting of the retention time, m/z, and abundance value (via the normalized peak area) for each ion using the raw MS chromatograms. To determine the peak areas of internal standards, QuanLynx (Waters) was used. For analyzing the MSE data, the high energy scans (fragments) were aligned with low energy scans (precursors) in MarkerLynx. Our in-house statistical analysis program, MetaboLyzer, was used to analyze the data and identify statistically significant ions.7 MetaboLyzer allowed for extraction of the ions with nonzero abundance values, which were detected in at least 70% of samples in each study group, called complete-presence ions. Data were then log-transformed and analyzed for statistical significance via the nonparametric Mann–Whitney U statistical hypothesis test (p value <0.05). Statistical significance testing for ions with nonzero abundance values in at least 70% of the samples in only one group (partial-presence ions) were analyzed as categorical variables for presence status (i.e., nonzero abundance) via Fisher’s exact test (p value <0.05). The log-transformed data for statistically significant complete-presence ions were then utilized for principal component analysis (PCA) via singular value decomposition for the purpose of data visualization
Metabolic Pathway Analysis
Statistically significant ions were putatively identified via MetaboLyzer, which utilizes the Human Metabolome Database (HMDB), LipidMaps, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database.11 The m/z values were used to putatively assign IDs to the ions by neutral mass elucidation, which was accomplished by considering the possible adducts, H+, Na+, or NH4+ in the ESI+ mode and H– and Cl– in the ESI– mode. The masses were then compared with the exact mass of small molecules in the databases, from which putative metabolites were identified with a mass error of 20 ppm (ppm) or less. KEGG-annotated pathways associated with these putative metabolites were also identified. To extract structural information on the putative identities of the metabolite, we explored MSE data via QuanLynx for alignment of the low energy scans with high energy scans. The fragmentation pattern of each metabolite and lipid of interest was compared against that of its pure chemical form either in online databases or the in-house database.
Results
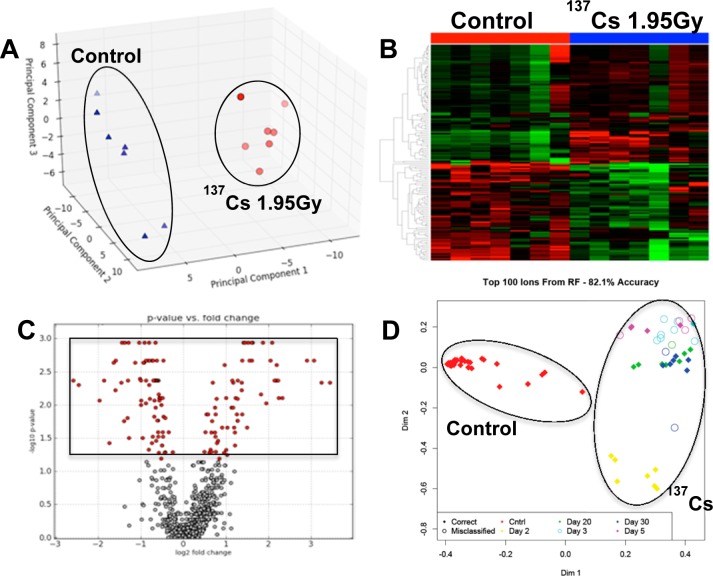
In this study we utilized the sensitivity of mass spectrometry (Xevo-G2, Waters) to detect changes in the sera of mice exposed to 137Cs over the course of 30 days. The comprehensive analysis of serum metabolites and lipids along with their relative abundances were made possible by a feature of the Xevo-G2 called MSE, where many precursor, neutral loss, and product ion scans are acquired for every injection. The acquired raw chromatograms were preprocessed and subsequently analyzed via MetaboLyzer. We initially focused on the overall metabolomic profiles of sera. More than 1800 spectral features were detected in both ESI modes combined. These spectral features were used to determine the changes in the serum metabolome of mice after exposure to 137Cs at different time points/doses (Table 1). For example, the overall serum metabolomic profile of mice 2 days after 137CsCl injection at a cumulative dose of 1.95 Gy showed significant changes when compared with the serum metabolomic profile of matched control mice, as expected. This is evident from the clear separation in PCA of Figure 1A, where 137Cs-exposed mice (red circles) are tightly clustered and clearly dichotomized from the control mice (blue triangles). The heatmap in Figure 1B shows a panel of putative metabolites on the vertical axis, whose serum abundances change significantly (p < 0.05) 2 days post-137Cs-exposure and contributed the most to the separation seen in the PCA of Figure 1A. Each circle in the volcano plot of panel C represents a putative metabolite, with the boxed red circles on the top half representing those, which were statistically significantly perturbed in the serum of 137Cs-exposed mice 2 days after exposure. This initial statistical analysis using traditional tests was extended to other time points/doses with similar results. To see how the overall metabolomic profiles of all time points/doses compared, we used Random Forests ranking on the entire metabolomic data set. The resulting proximity-matrix-based MDS plot in Figure 1D shows that the overall metabolomic profile of serum from control mice is clearly separated from that of 137Cs-exposed mice at all time point/doses based on the top 100 ranked putative metabolite. These 100 putative metabolite can be used to assign with 82% accuracy any serum sample to its correct dose group based on its relative serum abundance (data from ESI+ mode).
Figure 1.
Comparative analysis of serum metabolomic profiles of control mice and those exposed to 137Cs at a cumulative dose of 1.95 Gy at 2 days post-exposure. Panel A is a principle component analysis (PCA) plot showing clear separation of metabolomic signatures of sera from control (blue triangles) and 137Cs-exposed mice (red circles). Panel B is a heatmap of metabolites whose serum levels change significantly 2 days post-137Cs-exposure. The top half of this heatmap displays metabolites whose levels in serum dropped post-exposure and those of metabolites on the bottom half increased post-exposure after 2 days. Panel C is a volcano plot, which highlights many statistically significant metabolites post-exposure. Statistical significance was determined via Mann–Whitney U test (p value <0.05). Panel D is an MDS plot generated in Random Forests showing the spatial separation between the overall metabolomic profiles of serum samples from control mice and those of serum samples from 137Cs-exposed mice at 2, 3, 5, 20, and 30 days post-exposure. All of the figures were created using ESI+ data. Panels A–C were created in MetaboLyzer, while panel D was generated in Random Forests.
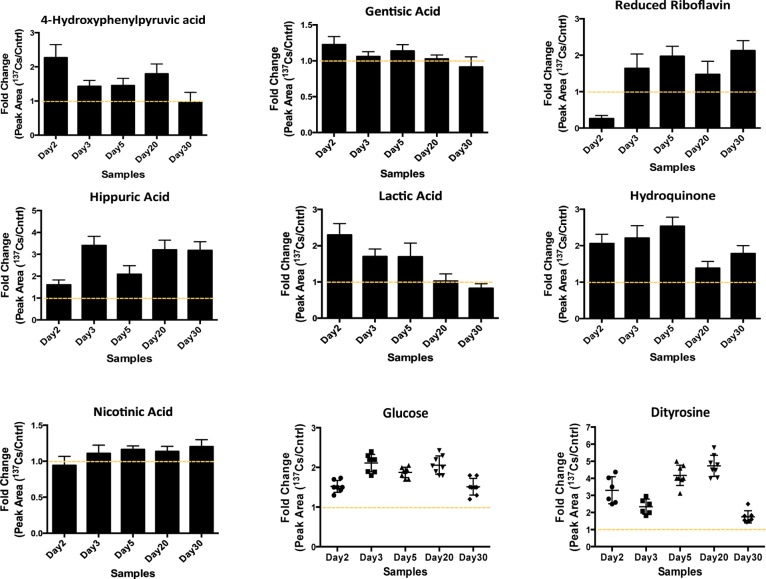
After thorough analysis of the statistically significant metabolites from all of the time-points/doses we were able to group the metabolites into several key pathways, such as riboflavin metabolism and linoleic acid metabolism in ESI+ mode (Supplemental Figure 1 in the Supporting Information ) and glycolysis, TCA cycle, tyrosine, and phenylalanine metabolism in ESI– mode. The results of this analysis suggested that exposure to 137Cs caused an increase in the serum levels of metabolites from these pathways particularly at earlier time points (day 3 and day 5 post-exposure). Some metabolites’ levels dropped to their pre-exposure levels after the initial increase at days 3 and 5, but most metabolites showed a persistent increase in their response to 137Cs during the entire course of the study, as shown in Figure 2. For instance, two metabolites associated with riboflavin pathway, hydroquinone, and riboflavin (reduced form), showed increased serum levels post-exposure, particularly after 3 and 5 days (cumulative doses of 2.70 and 4.14 Gy). The levels of these metabolites remained elevated until 30 days post-exposure (the last time-point in the study).
Figure 2.
Changes in the serum abundances of selected metabolites post 137Cs exposure are represented as the ratio of their responses in 137Cs-exposed mice to those in control mice. The responses were calculated as the area under the peak for each metabolite in 137Cs-treated serum divided by that in control serum (Y axis). These metabolites were selected based on their statistical significance as determined by Mann–Whitney U test (p value <0.05) and biological importance. The identities of these ions were validated via MS/MS against pure standards or through MS/MS spectra published in online databases (METLIN20 and HMDB). Tight clustering of fold-change values for glucose and dityrosine is shown at each time point.
Furthermore, the serum abundance of a few metabolites associated with energy metabolism was determined to be significantly attenuated as a result of exposure to 137Cs. Lactic acid is an important metabolite of energy metabolism, and changes in its levels may be indicative of a change in energy supply and glycolytic flux.12 The serum levels of this metabolite increased as a result of exposure to 137Cs in the earlier time points and returned to its pre-exposure levels by the end of the experiment (day 30). Increased level of this metabolite is associated with increased glycolysis and ROS production.13 Interestingly, we determined a similar increase in the levels of dityrosine, which is a cross-linked species formed upon protein modification due to ROS-induced damage. The rise in dityrosine levels during earlier time points matched the increase in serum lactic acid during these time points, which may suggest a link between these two metabolites and internal emitter exposure-induced ROS damage. Additionally, glucose level was determined to be elevated by almost two-fold in mice exposed to 137Cs starting as early as 2 days post-exposure. Together with an increase in serum levels of nicotinic acid, members of the TCA cycle such as citrate, malate, and metabolites associated with tyrosine and phenylalanine metabolism (Figure 2 and Table 2) may suggest an increase in the rate of glycolysis. Taurine and uric acid listed in Table 2 have been reported in gamma irradiation studies as potential markers of external beam exposure.14,15 These metabolites were also found to be significantly perturbed in the urine of 137Cs-exposed mice in this study.6 Taurine shows a significant decrease in its serum concentration post-137Cs-exposure, while its urinary abundance was determined to increase in the same mice post-exposure. Uric acid in the urine of these mice showed a late increased response, while its serum levels suggest an early increase in response. Inositol is yet another metabolite reported in literature as a potential serum marker for gamma irradiation, whose increased response to radiation in a dose-specific manner may indicate a perturbation in hepatic lipid metabolism.16 We determined that the serum levels of this metabolite increased post 137Cs-exposure; however, this persistent increase was not dose-specific and may suggest a more systemic perturbation in lipid metabolism as a result of exposure to an internal emitter.
Table 2. Examples of Statistically Significant Serum Metabolites along with the Fold Changes in Their Responses to 137Cs Exposure after 2, 3, 5, 20, and 30 Days (D) of Exposure.
| fold changea |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| ESI mode | ID | name | p value (Mann–Whitney U test) | D2 | D3 | D5 | D20 | D30 | |
| 1 | POS | 203.0523_0.2908 | inositol | 4.40 × 10–3 | 1.30 | 1.19 | 1.29 | 1.70 | 1.26 |
| 2 | POS | 208.0821_2.1842 | dihydrolipoamide | 1.17 × 10–3 | 2.99 | 2.63 | 1.92 | 3.50 | 2.45 |
| 3 | POS | 337.1048_4.0617 | 7,8-dihydropteroic acidb | 8.66 × 10–3 | 0.67 | 0.51 | 0.48 | 0.41 | 0.38 |
| 4 | POS | 245.0782_3.4928 | uridine | 4.33 × 10–3 | 0.53 | 0.42 | 0.38 | 0.28 | 0.70 |
| 5 | NEG | 124.0065_0.2973 | taurine | 2.33 × 10–3 | 0.55 | 0.46 | 0.67 | 0.75 | 0.87 |
| 6 | NEG | 101.0234_0.2805 | 2-ketobutyric acid | 9.40 × 10–3 | 1.70 | 1.33 | 1.37 | 1.86 | 1.21 |
| 7 | NEG | 133.0134_0.3209 | malic acid | 2.53 × 10–3 | 0.88 | 0.98 | 1.53 | 1.87 | 1.25 |
| 8 | NEG | 167.0196_0.3154 | uric acid | 5.83 × 10–4 | 1.60 | 1.45 | 1.63 | 1.52 | 0.98 |
Fold change was calculated by dividing the relative concentration of a metabolite in post 137Cs exposure serum samples by that in respective control samples.
Putative name was assigned based on accurate mass matched to METLIN database (mass error <10 ppm).
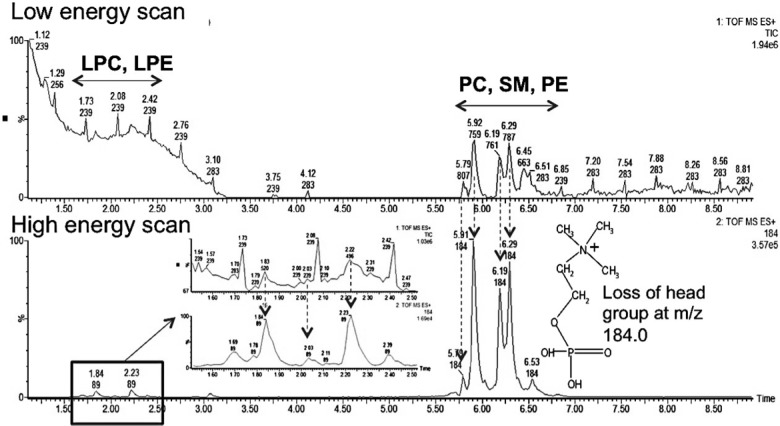
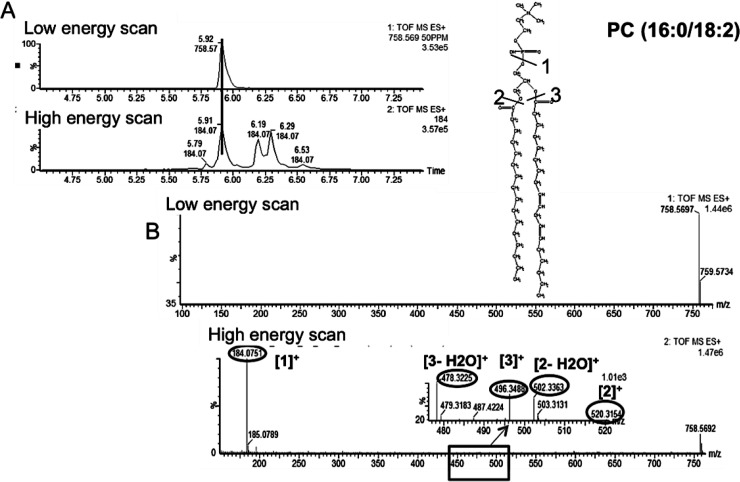
Our initial metabolomic analysis of the data was followed by LC–MSE lipidomics. This involved assigning the detected lipids to their respective classes. Figure 3 demonstrates how retention time window and specific fragments of lipid head groups can be used to do just that. The use of lipid and fatty acid standards in both ESI modes further facilitated their identification due to the differences in preferential ionization of polar head groups. For instance, in ESI+ mode, the phosphatidylcholine (PC) and lysophosphatidylcholine (LPC) standards were used for identification and relative quantification of lipids belonging to these classes, while in ESI– mode (phosphatidylethanolamine (PE), lysophosphatidylethanolamine (LPE), phosphatidylglycerol (PG), and (fatty acid) FA were used. The two-fold serial dilutions established linearity between peak area and concentration for the mentioned standards (Supplemental Table 1 in the Supporting Information). More than 800 spectral features were detected in both ESI modes combined, from which 48 were determined to be PCs, 12 SMs, 7 PEs, 15 LPCs, 2 LPEs, 3 FAs, and 1 PG. We were able to detect more PCs than the other lipid species in this study. Among the PCs, the ion at m/z of 758.5685 and retention time of 5.95 min was determined to be the most statistically significant PC. Figure 4 shows the low and high energy scans for this PC with calculated fatty acid chains of 16 carbons long and 18 carbons long with two double bonds. The cleavage of the phospho-headgroup of PCs gives rise to a fragment at m/z of 184.0752, which can be used to search for all PC and LPC classes of phospholipid along with SMs. To gain further insight into the chemical structure of these lipids, we explored the MSE data at high collision energy for identification of other fragments of the precursor ion at the desired retention time. For instance, for the precursor ion at m/z of 758.5685 and retention time of 5.95 min, the low and high energy scans were aligned, and the fragments of the precursor ion were identified. This lead to the identification of fragments at m/z of 478.3211, which corresponds to the loss of a 16 carbon long fatty acyl chain and a water molecule from the precursor ion. The proposed sn2 reaction that gave rise to the peak at m/z of 478.3211 is depicted in Figure 4 (ESI+ mode) along with a similar mechanism for the fragment at m/z of 520.3643. On the basis of these fragments we identified the precursor ion at m/z of 758.5685 as PC(16:0/18:2), as shown in Figure 4. This demonstrates how LC–MSE data can be carefully mined to gain more structural information on lipids in addition to assigning them to their respective classes. A similar approach was taken to identify FAs and lipid species such PEs and LPCs within their specified retention time window and mass error window (<10 ppm). A few examples of lipids identified via this approach are provided in Table 3.
Figure 3.
UPLC–MSE low and high energy scans of various classes of lipids in sera of mice. The low energy scan highlights the specific retention time window for each class. The high energy scan highlights a specific fragment at m/z of 184.0 that can be used to identify lipids with a phosphate headgroup.
Figure 4.
Base peak of a PC and its chemical structure (top) along with its MS/MS spectrum and identified fragments (bottom). The MS/MS spectrum was obtained using described MSE method at high collision energy in ESI+ mode. The low and high energy scans were first aligned in the expected retention time window (A). The individual fragments of the precursor ion were determined by mining the high energy scan spectrum (B).
Table 3. Examples of Statistically Significant Serum Lipids along with the Fold Changes in Their Responses to 137Cs Exposure after 2, 3, 5, 20, and 30 days (D) of Exposure.
| fold changeq |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| ESI mode | ID | lipid class | p value (Mann–Whitney U test) | D2 | D3 | D5 | D20 | D30 | |
| 1 | POS | 758.5736_6.5062 | PC | 2.00 × 10–2 | 0.88 | 0.89 | 0.78 | 0.74 | 0.81 |
| 2 | POS | 780.5493_5.6732 | PC | 1.40 × 10–4 | 0.65 | 0.69 | 0.68 | 0.67 | 0.43 |
| 3 | POS | 518.3218_1.9790 | LysoPC | 5.89 × 10–3 | 1.03 | 3.96 | 3.69 | 4.03 | 4.12 |
| 4 | POS | 542.3215_1.6057 | LysoPC | 3.74 × 10–2 | 1.78 | 4.56 | 4.74 | 3.90 | 3.51 |
| 5 | POS | 496.3401_2.2871 | LysoPC | 2.60 × 10–2 | 1.80 | 2.31 | 2.16 | 2.07 | 2.66 |
| 6 | POS | 703.5717_5.5076 | SM | 4.12 × 10–2 | 1.66 | 2.00 | 1.94 | 1.79 | 1.15 |
| 7 | POS | 725.5578_6.1051 | SM | 1.75 × 10–02 | 3.21 | 4.74 | 5.12 | 4.77 | 5.02 |
| 8 | NEG | 722.5070_5.5520 | PE | 1.89 × 10–7 | 5.27 | 3.78 | 3.24 | 2.05 | 1.57 |
| 9 | NEG | 776.5450_6.4326 | PE | 7.04 × 10–5 | 3.12 | 4.28 | 2.46 | 1.59 | 1.23 |
| 10 | NEG | 476.2776_2.5595 | LysoPE | 8.29 × 10–5 | 0.36 | 0.69 | 0.73 | 0.74 | 0.89 |
| 11 | NEG | 387.2142_4.7629 | PG | 3.55 × 10–3 | 0.30 | 0.36 | 0.57 | 0.63 | 0.48 |
Fold change was calculated by dividing the relative concentration of a metabolite in post 137Cs exposure serum samples by that in respective control samples.
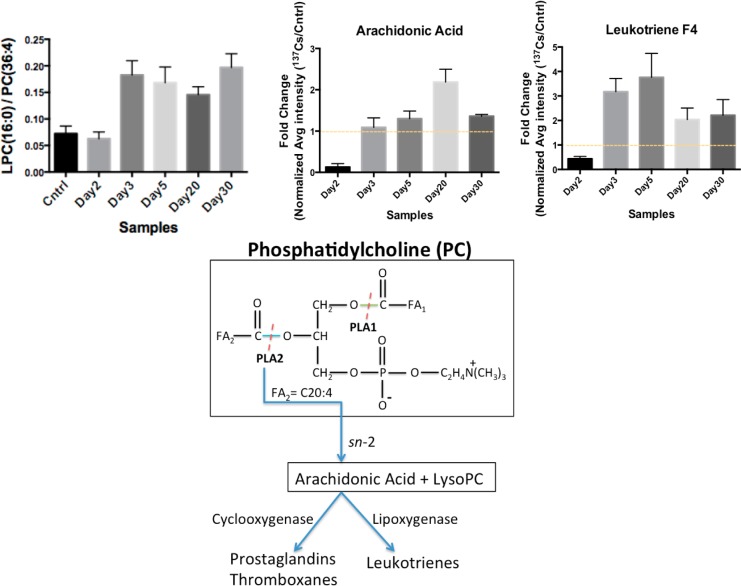
Statistical analysis on the identified lipid species revealed that the serum levels of PCs and LPCs were highly affected post-137Cs-exposure. PCs undergo hydrolysis of their acyl FA chains by the actions of phospholipases (PL) A1 and A2 to form LPCs. For instance, PC(36:4) with m/z of 782.5681 undergoes hydrolysis of its C20:4 acyl chain with m/z of 305.3305 by the actions of PLA2, resulting in the release of arachidonic acid (C20) and LPC(16:0) with m/z of 478.3457, as depicted in Figure 5A. Arachidonic acid is a known inflammation marker and was determined to have elevated serum levels post-Cs-exposure (Figure 5B). The calculated ratio of LPC(16:0) peak area to the precursor PC(36:4) peak area in the serum of Cs exposed mice showed an almost four-fold increase when compared with the calculated ratio in the serum of control mice (Figure 5B) as early as 3 days post-exposure and continued to be elevated throughout the course of the study (Figure 5C). This along with elevated levels of arachidonic acid in the serum of Cs-exposed mice suggests an increase in the activity of PLA2 as a result of inflammatory response to 137Cs exposure. Arachidonic is further acted on by lipoxygenases to form leukotrienes or by cyclogenases to form PGs and thromboxanes. We identified leukotriene F4 to be elevated in the sera of 137Cs-exposed mice as expected (Figure 5D).
Figure 5.
Ratio of LPC(16:0) to PC(36:4) and the subsequent formation of arachidonic acid and synthesis of leukotrienes are collectively a measure of phospholipase 2 (LPA2) activity and are used to gauge the level of inflammation. The data suggest an increase in the ratio of LPC/PC, which corresponds to an increase seen in the post-Cs-exposure serum levels of arachidonic acid and leukotrienes. An increase in the formation of arachidonic acid via an sn-2 reaction can serve as an inflammation marker. The error bars represent the standard deviation values for each time point.
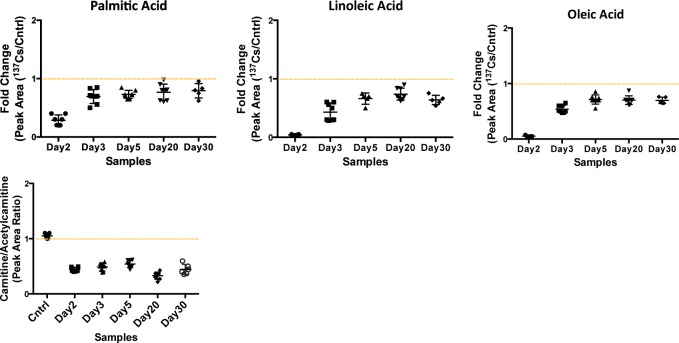
In addition to the above phospholipids, we studied the serum levels of three fatty acids, linoleic, oleic, and palmitic acids, along with acyl-carnitine and acetylcarnitine species. Fatty acids are important sources of energy, and they are converted into acyl-CoA during β-oxidation to be used in the TCA cycle. Our data suggest a slight decrease in the serum levels of these three fatty acids post-137Cs-exposure, accompanied by a decrease in the relative concentration of free carnitine (Figure 6). However, the serum levels of acetylcarnitine increase by 35% post 137Cs exposure, which may suggest a decrease in β-oxidation. Together with our metabolomics data, this indicates a shift in energy production from fatty acids oxidation to glycolysis as a result of 137Cs exposure. It is important to note that the urine metabolomics analysis on these mice revealed opposite changes in the levels of TCA cycle metabolites and carnitine species to what we observed in the serum.
Figure 6.
Significant decrease is observed in the serum abundances of fatty acids: palmitic, linoleic, and oleic acid. As a crude measure of fatty acid β-oxidation, the ratio of free carnitine to acetylcarnitine was calculated (peak area ratio shown on y axis), which also suggests a decrease in serum-free carnitine levels post-137Cs-exposure.
Discussion
In this study, we focused on the changes in serum metabolites and lipids from mice exposed to an internal emitter, 137Cs, which follows our previously published work in the urine of these mice. Here we took advantage of an untargeted, data-independent, and hybrid technology called MSE to rapidly yet comprehensively analyze mouse serum samples after 2, 3, 5, 20, and 30 days of exposure to 137Cs and gain insight into robust responses to 137Cs exposure. Recent metabolomic studies of serum from rats and mice exposed to external gamma irradiation by gas chromatography (GC)–TOFMS have indicated important changes in the serum levels of amino acids such as serine and lysine, isocitrate, glycerol, stearic acid, steroid hormones such as progesterone, and cholesterol.16 These studies focused in particular on the volatile small metabolites and steroid hormones via GC–TOFMS providing a specific yet narrow window into metabolic changes in serum of the animals post gamma irradiation. Studying the complex network of metabolites and lipids and their exposure-specific responses is a monumental task and cannot be covered in any single study; however, our LC–MSE approach can provide insight into a wider range of molecules, from small polar metabolites to lipids, with relative quantification in serum.
As with previous gamma irradiation studies from external sources, we initially focused on the metabolomic data and the overall metabolomic signature of serum from mice exposed to 137Cs compared with that of serum from control mice. As expected, 137Cs exposure significantly perturbed the levels of many serum metabolites as determined by statistical testing (MetaboLyzer, Mann–Whitney U-test p < 0.05) as early as 2 days post-exposure at a cumulative dose of 1.95 Gy. The magnitude of change in the levels of these metabolites was large enough to affect the differential metabolomic profile of serum in the exposed mice. This is seen from the clear separation of profiles in the PCA plot of Figure 1A. The heatmap and the volcano plot in Figure 1 highlight the specific 137Cs exposure responses of selected metabolites at day 2 compared with their respective controls. The metabolomic profile of serum from 137Cs-exposed mice at all time-points/doses was clearly distinguishable from that of their respective controls (Figure 1D). More thorough exploration of the data indicates changes in the metabolites associated with tyrosine metabolism, riboflavin metabolism, glycolysis, and TCA cycle. Two of the TCA cycle metabolites were also found to be significantly perturbed in the urine of these mice,6 but in the opposite direction. For instance, urinary levels of citrate and malate were found to have decreased, while in serum their levels appeared elevated post-137Cs-exposure. A key metabolite of glycine and serine metabolism, 2-ketobutyric acid, which feeds into acetyl-CoA production, and TCA cycle was also found at higher concentration post-137Cs-exposure throughout the course of the study. This suggests an up-regulation of TCA cycle and its feeder pathways in the serum of mice exposed to 137Cs. We additionally found that glucose serum levels in mice post 137Cs exposure were elevated while the average body weight of mice in all of the study groups remained steady (Table 1). Thus, the increase in serum levels of glucose and TCA cycle associate metabolites is a result of exposure to 137Cs and not food intake. The serum levels of uric acid appeared elevated post exposure. This change is similar to what had been seen in the urine of these mice and those exposed to gamma irradiation.6,15 However, taurine was detected at lower concentration in the serum of 137Cs-exposed mice, unlike what had been seen in the urine of these mice and in the urine of mice exposed to gamma irradiation at similar doses.14
Inositol was recently shown by Liu et al. to increase in the serum of mice exposed to gamma irradiation in a dose–response manner.16 We also found the levels of this metabolite to be elevated in the serum of 137Cs-exposed mice. Inositol serum levels remained elevated throughout the course of the study, with the response being greatest at day 20 and decreasing to levels at earlier time-points by day 30 (Table 2). An increase in serum concentration of inositol is associated with perturbation of lipid metabolism.
Lipid and fatty acid metabolism were further investigated in the second phase of this study. The statistically significant lipids were assigned to their respective classes using a shotgun approach LC–MSE method as previously described. We used the elution window and alignment of low and high energy scans as shown in Figure 2 to do this. A few examples of lipids in each identified class are shown in Table 3. The PCs were detected at lower concentrations in the serum of 137Cs exposed mice, while LPCs were found at elevated levels. We further investigated the fragmentation pattern of a PC species at m/z of 782.5681. The phospho- headgroup of all PCs is cleaved at high collision energy scan to give rise to a peak at m/z of 184. Furthermore, the sn1 and sn2 reactions on the two fatty acyl chains of PCs give rise to distinct peaks corresponding to the loss of each acyl chain. As for the high energy scan of PC at 782.5681, two fragments were aligned with the low energy scan, which corresponded to the loss of an acyl chain with 16 carbons and no double bonds and a 20 carbon long chain with 4 double bonds. Thus, the identity assigned to the peak at m/z of 782.5681 was PC(36:4). PCs are converted into LPCs by the action of phospholipase A2 (PLA2) and a fatty acid. Therefore, we checked for the presence of a LPC(16:0) species and a fatty acyl chain of C20:4, which corresponds to arachidonic acid. Both of these molecules were found in the serum of Cs-exposed mice at higher levels than PC(36:4). The ratio of concentration of PC(36:4) to the formed LPC(16:0) thus may be indicative of the activity of PLA2. This ratio is shown in Figure 4 along with a proposed pathway for action of this enzyme. The ratio suggests an increased activity for PLA2. Furthermore, arachidonic acid is an inflammation marker and its increased levels in the 137Cs-exposed mice may suggest radiation-induced inflammation in these mice and subsequently an increase in the activity of PLA2. Arachidonic is further converted to leukotrienes by lipogenases. We detected leukotriene F4 in the serum at higher concentration in 137Cs-exposed mice (Figure 4).
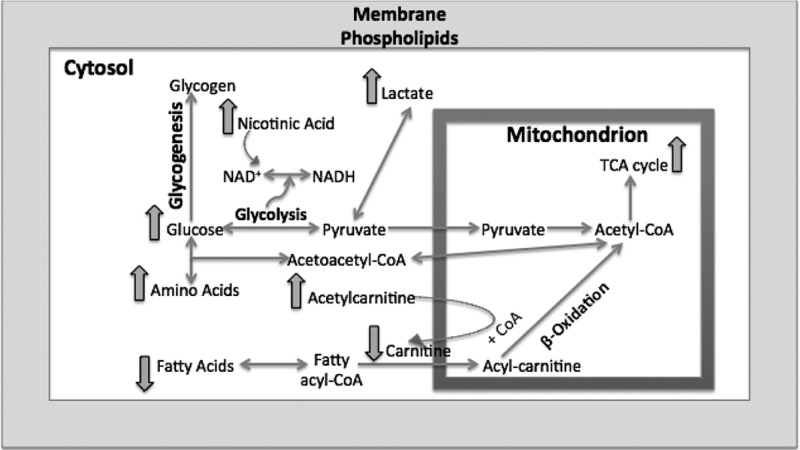
Furthermore, we analyzed the fatty acid elution window in the ESI– mode. As suggested by our previous pathway analysis, we expected to see perturbations in the levels of linoleic acid. In addition, we studied the serum levels of three fatty acids, linoleic, oleic, and palmitic acids, along with acyl-carnitine and acetylcarnitine species. Fatty acids are important sources of energy, and they are converted into acyl-CoA during β-oxidation in the cytosol and transported into mitochondria via carnitines to ultimately form acetyl-CoA to be used in the TCA cycle. The carnitine is then deposited back into cytosol.17 Our data suggest a slight decrease in the serum levels of the three fatty acids post-137Cs exposure, accompanied by a decrease in the relative concentration of free carnitine. However, the serum levels of acetylcarnitine increased by almost 40% post 137Cs exposure, which may suggest a decrease in β-oxidation. Together with our metabolomics data, this indicates a shift in energy production from fatty acids oxidation to glycolysis as a result of 137Cs exposure. Figure 7 depicts the pathways indicated as significantly perturbed in this study and how they are interconnected in energy production. Amino acids such as tyrosine and phenylalanine enter the energy production pathway by forming acetyl-CoA. Fatty acids also ultimately form acetyl-CoA through β-oxidation. In addition to fatty acids and amino acids, glucose also is converted into acetyl-CoA through pyruvate production. All of these pathways help provide a steady flow of acetyl-CoA into mitochondria to be used in the TCA cycle. It is known that in the presence of environmental stimuli and stress, such as radiation exposures, these pathways become perturbed, which will ultimately affect the function of mitochondria and TCA cycle energy production.18 Furthermore, a recent gene profiling study found significant changes in the expression of genes associated with mitochondrial processes such as the electron transport chain, and cellular respiration in white blood cells from the same samples used in our study.19 A change in mitochondria function can also throw the balance between ROS production and scavenging off. Our data suggest that exposure to 137Cs may perturb fatty acid oxidation and trigger an increase in TCA cycle and its feeder pathways. This may lead to higher levels of ROS production and further damage to structural proteins and enzymes. An increase in the serum abundance of dityrosine, a marker of protein oxidation, may further support the proposed mechanism in Figure 7. Further exploration of enzymatic activity and chemical assays along with more in-depth and targeted metabolomics and lipidomics is necessary to independently validate these results and paint a more complete biosignature for the effects of 137Cs exposure in serum.
Figure 7.
Pathway illustration of the overall metabolomic and lipidomic changes in mouse serum post 137Cs exposure. At least two metabolites in each noted pathway were found to be significantly perturbed (p value <0.05). The overall pathway analysis indicates a shift in energy metabolism from fatty acid oxidation to glycolysis.
Conclusion
In this study we used a fast and robust LC–MSE technique, which allows one to use the elution profile of precursor masses and the fragmentation profiles obtained during high collision energy scans to elucidate structural information on the detected spectral features. By taking advantage of this technique we were able to comprehensively study the effects of exposure to 137Cs, an internal emitter, in the serum of mice. The findings suggest an up-regulation of TCA cycle and down-regulation of fatty acid β-oxidation as a result of exposure to 137Cs.
Acknowledgments
We thank Georgetown University’s Radiation Safety Office and the Proteomic and Metabolomics Shared Resources, NIH P30 CA51008, for making the analysis of the radioactive specimens possible. We also acknowledge Drs. Michael Stabin and Luiz Bertelli for calculation of the murine dose coefficients. This study was supported by the National Institute of Health (National Institute of Allergy and Infectious Diseases) grant U19 A1067773 awarded to Dr. David J. Brenner and the Proteomic and Metabolomics Shared Resources, NIH P30 CA51008.
Glossary
Abbreviations
- 137Cs
Cesium-137
- IR
ionizing radiation
- TOF-MS
time-of-flight mass spectrometry
- UPLC
ultra performance liquid chromatography
- PC
phosphatidylcholine
- ESI
electrospray ionization
Supporting Information Available
Metabolic pathway enrichment plot is shown in Supplemental Figure 1 to highlight the most perturbed pathways post 137Cs exposure. Information on the internal standards used in this study is presented in Supplemental Table 1. A brief discussion and appropriate references on the changes in the gene expression profile of blood post 137Cs exposure in relation with the metabolomic data is offered. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Fushiki S. Radiation hazards in children - lessons from Chernobyl, Three Mile Island and Fukushima. Brain Dev. 2013, 35, 220–227. [DOI] [PubMed] [Google Scholar]
- Ahmed M. M.; Sells S. F.; Venkatasubbarao K.; Fruitwala S. M.; Muthukkumar S.; Harp C.; Mohiuddin M.; Rangnekar V. M. Ionizing radiation-inducible apoptosis in the absence of p53 linked to transcription factor EGR-1. J. Biol. Chem. 1997, 272, 33056–33061. [DOI] [PubMed] [Google Scholar]
- Amundson S. A.; Do K. T.; Shahab S.; Bittner M.; Meltzer P.; Trent J.; Fornace A. J. Jr. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat. Res. 2000, 154, 342–346. [DOI] [PubMed] [Google Scholar]
- Artuso M.; Esteve A.; Bresil H.; Vuillaume M.; Hall J. The role of the ataxia telangiectasia gene in the p53, WAF1/CIP1(p21)- and GADD45-mediated response to DNA damage produced by ionising radiation. Oncogene 1995, 11, 1427–1435. [PubMed] [Google Scholar]
- Laiakis E. C.; Hyduke D. R.; Fornace A. J. Jr. Comparison of mouse urinary metabolic profiles after exposure to the inflammatory stressors γ radiation and lipopolysaccharide. Radiat. Res. 2012, 177, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudarzi M.; Weber W.; Mak T. D.; Chung J.; Doyle-Eisele M.; Melo D.; Brenner D. J.; Guilmette R. A.; Fornace A. J. Jr. Development of urinary biomarkers for internal exposure by cesium-137 using a metabolomics approach in mice. Radiat. Res. 2014, 181, 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak T. D.; Laiakis E. C.; Goudarzi M.; Fornace A. J. Jr. MetaboLyzer: a novel statistical workflow for analyzing Postprocessed LC-MS metabolomics data. Anal. Chem. 2014, 86, 506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanft F.; Koehler P. Quantitation of dityrosine in wheat flour and dough by liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 2005, 53, 2418–2423. [DOI] [PubMed] [Google Scholar]
- Stabin M. G.; Peterson T. E.; Holburn G. E.; Emmons M. A. Voxel-based mouse and rat models for internal dose calculations. J. Nucl. Med. 2006, 47, 655–659. [PubMed] [Google Scholar]
- Miller G.; Bertelli L.; Klare K.; Weber W.; Doyle-Eisele M.; Guilmette R. Softwre for empirical building of biokinetic models for normal and decorporation-affected data. Health Phys. 2012, 103, 484–494. [DOI] [PubMed] [Google Scholar]
- Kanehisa M.; Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhup S.; Dadhich R. K.; Porporato P. E.; Sonveaux P. Multiple Biological Activities of Lactic Acid in Cancer: Influences on Tumor Growth,Angiogenesis and Metastasis. Curr. Pharm. Des. 2012, 18, 1319–1330. [DOI] [PubMed] [Google Scholar]
- Guido C.; Whitaker-Menezes D.; Lin Z.; Pestell R. G.; Howell A.; Zimmers T. A.; Casimiro M. C.; Aquila S.; Ando’ S.; Martinez-Outschoorn U. E.; Sotgia F.; Lisanti M. P. Mitochondrial Fission Induces Glycolytic Reprogramming in Cancer-Associated Myofibroblasts, Driving Stromal Lactate Production, and Early Tumor Growth. Oncotarget 2012, 3, 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyburski J. B.; Patterson A. D.; Krausz K. W.; Slavik J.; Fornace A. J. Jr.; Gonzalez F. J.; Idle J. R. Radiation metabolomics. 1. Identification of minimally invasive urine biomarkers for gamma-radiation exposure in mice. Radiat. Res. 2008, 170, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. H.; Patterson A. D.; Krausz K. W.; Kalinich J. F.; Tyburski J. B.; Kang D. W.; Luecke H.; Gonzalez F. J.; Blakely W. F.; Idle J. R. Radiation metabolomics. 5. Identification of urinary biomarkers of ionizing radiation exposure in nonhuman primates by mass spectrometry-based metabolomics. Radiat. Res. 2012, 178, 328–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Wang Z.; Zhang X.; Qiao Y.; Wu S.; Dong F.; Chen Y. Selection of candidate radiation biomarkers in the serum of rats exposed to gamma-rays by GC/TOFMS-based metabolomics. Radiat. Prot. Dosim. 2013, 154, 9–17. [DOI] [PubMed] [Google Scholar]
- Chegary M.; Brinke H. T.; Ruiter J. P.; Wijburg F. A.; Stoll M. S. K.; Minkler P. E.; van Weeghel M.; Schulz H.; Hoppel C. L.; Wanders R. J. A.; Houten S. M. Mitochondrial long chain fatty acid β-oxidation in man and mouse. Biochim. Biophys. Acta 2009, 1797, 806–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S.; Gorman A. M.; Hori O.; Samali A. Cellular Stress Responses: Cell Survival and Cell Death. Int. J. Cell Biol. 2010, 2010, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S.; Ghandhi S. A.; Weber W.; Doyle-Eisele M.; Melo D.; Guilmette R.; Amundson S. A. Gene expression responses of mice after a single dose of 137Cs as an internal emitter. Radiat. Res. 2014, 182, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R.; Cho K.; Uritboonthai W.; Zhu Z.; Patti G. J.; Siuzdak G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.