Abstract

Ionizing radiation is widely used in cancer therapy; however, cancer cells often develop radioresistance, which compromises the efficacy of cancer radiation therapy. Quantitative assessment of the alteration of the entire kinome in radioresistant cancer cells relative to their radiosensitive counterparts may provide important knowledge to define the mechanism(s) underlying tumor adaptive radioresistance and uncover novel target(s) for effective prevention and treatment of tumor radioresistance. By employing a scheduled multiple-reaction monitoring analysis in conjunction with isotope-coded ATP affinity probes, we assessed the global kinome of radioresistant MCF-7/C6 cells and their parental MCF-7 human breast cancer cells. We rigorously quantified 120 kinases, of which 1/3 exhibited significant differences in expression levels or ATP binding affinities. Several kinases involved in cell cycle progression and DNA damage response were found to be overexpressed or hyperactivated, including checkpoint kinase 1 (CHK1), cyclin-dependent kinases 1 and 2 (CDK1 and CDK2), and the catalytic subunit of DNA-dependent protein kinase. The elevated expression of CHK1, CDK1, and CDK2 in MCF-7/C6 cells was further validated by Western blot analysis. Thus, the altered kinome profile of radioresistant MCF-7/C6 cells suggests the involvement of kinases on cell cycle progression and DNA repair in tumor adaptive radioresistance. The unique kinome profiling results also afforded potential effective targets for resensitizing radioresistant cancer cells and counteracting deleterious effects of ionizing radiation exposure.
Keywords: breast cancer, radioresistance, protein kinases, MRM, isotope-coded ATP affinity probe, ionizing radiation
Introduction
There are increasing public concerns about the safety of environmental ionizing radiation as well as industrial and medical applications of ionizing radiation. Ionizing radiation, which arises from both natural and anthropogenic sources, may cause damage to exposed individuals depending on radiation dose and duration.1 In this way, ionizing radiation may emanate from secondary particles from cosmic rays or decay of naturally occurring radioisotopes and arise from nuclear reactors or during high-energy physics experiments.2,3 Inadvertent exposure to ionizing radiation may result in DNA damage, cell death, and ultimately lead to human diseases including cancer.2 On the other hand, the ionizing radiation’s capability to damage DNA also forms the basis for cancer radiotherapy.4 However, cellular response toward ionizing radiation varies among different cell types; the toxic effects on normal cells and tissues may elicit adverse human health consequences, whereas resistance of tumor cells toward ionizing radiation may render cancer radiation therapy less effective.5 Accumulating evidence suggests that mammalian cells including many types of tumor cells are able to develop adaptive radioresistance against ionizing radiation by activating a prosurvival signaling network.6,7 The so-called tumor adaptive radioresistance creates a barrier for further improvement of cancer patient survival by ionizing radiation-based anticancer modalities.7
Tumor aggressiveness in metastatic lesions is the lethal cause of cancer patients and is associated with the tumor-initiating cells, also known as cancer stem cells, which display enhanced self-renewal, elevated DNA repair capacity and radioresistance.5,8,9 Radioresistant cells are capable of surviving under many genotoxic stress conditions including the therapeutic ionizing radiation, and this defective response in radiation therapy may be innate or acquired.10 Along this line, radioresistance may arise from self-repair mechanisms in cells, mainly DNA damage repair,11 or repopulation of radioresistant cancer stem cells.7,12 Tumor heterogeneity was linked to different levels of radioresistance, and a group of clones isolated from the MCF-7 breast cancer cells after long-term fractionated radiation were found to be more resistant to radiation than were the parental MCF-7 tumor cells,13 which supports the concept of the presence of cancer stem cells.11,14 One of the radioresistant clones, that is, MCF-7/C6,15−17 was found to be enriched in breast cancer stem cells (BCSCs; ALDH+/CD44+/CD24–/low) and exhibit an enhanced prosurvival network of NF-κB and HER-2 expression,7,18 which suggests that the radioresistant MCF-7/C6 cells are present as the most aggressive breast cancer cells. However, the precise mechanisms underlying this radioresistant phenotype remain elusive.
Kinases are an important superfamily of enzymes that catalyze the phosphorylation of small intracellular molecules and proteins that are critical in the maintenance of a homeostatic cellular environment.19 Aberrant regulation of kinases affects a myriad of cellular processes including cell signaling, proliferation, and apoptosis. As mentioned above, HER-2 is implicated in the development of radioresistance.7,18 In addition, several other kinase-mediated cell signaling pathways were found to play an important role in cancer radioresistance.20 Thus, a thorough interrogation of the kinome reprogramming in cells with tumor radioresistance will not only provide practical perspectives about increasing the efficacy of cancer radiotherapy, but also afford important knowledge for developing potential effective molecules that can significantly prevent or treat tumors that are resistant to cancer radiation therapy.
With the advances in mass spectrometry instrumentation and the availability of bioinformatic tools, studies of global proteome and kinome become feasible.21,22 We developed a multiple-reaction monitoring (MRM)-based approach, together with the use of isotope-coded ATP-affinity probes, for global kinome profiling, which enabled the simultaneous assessment of the expression/activity of more than 300 kinases in human cells and tissues.23 In this approach, the binding moiety of the ATP affinity probe interacts specifically with ATP-binding proteins, including kinases, and subsequently forms a covalent bond with lysine near the ATP binding sites, which enables efficient labeling, enrichment, and identification of ATP-binding proteins from complex protein mixtures.24,25 The stable isotope-coded linker of the ATP affinity probe further facilitates quantitative comparison of kinase expression/activity between different samples.22 Moreover, with a preconstructed MRM kinome library, scheduled MRM analysis can be achieved with precisely predicted retention time window, which renders high-throughput and simultaneous quantification of protein kinases feasible.23 This MRM-based targeted proteomic analysis exhibited better sensitivity, accuracy, and reproducibility than discovery-based shotgun proteomic method.23 In the current study, we employed the same approach and conducted differential kinome analysis of MCF-7/WT human breast-cancer cells and the corresponding radioresistant MCF-7/C6 cells.
Materials and Methods
Cell Culture
All reagents unless otherwise stated were obtained from Sigma-Aldrich (St. Louis, MO). MCF-7/WT human breast cancer cells were purchased from ATCC (Manassas, VA). MCF-7+FIR30 (MCF-7/C6) radioresistant clone is a single clone (#6) that survives from 15 fractionated ionizing radiations at 2 Gy each.13,15 MCF-7/WT and MCF-7/C6 cells were cultured in Eagle’s minimum essential medium (EMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, Life Technologies, Grand Island, NY) and 1% antibiotic-antimycotic solution (Life Technologies). Cells were maintained in a humidified atmosphere with 5% CO2 at 37 °C with media renewal every 2–3 days.
Sample Preparation and Isotope-Coded ATP Affinity Probe Labeling
MCF-7/WT and MCF-7/C6 cells were harvested at 80% confluency, at a density of ∼5 × 105 cells/mL. Cells were collected and lysed as previously described.23 Briefly, after they were washed with ice-cold phosphate-buffered saline (PBS) three times, the cells were lysed with 0.7% CHAPS lysis buffer and further homogenized with a cell homogenizer. The cell lysates were passed through Illustra NAP-25 columns (GM Healthcare Bio-Sciences, Pittsburgh, PA) for the removal of endogenous nucleotides, and protein concentrations in the resultant lysates were subsequently determined by Bicinchoninic Acid Kit for protein determination.
The isotope-coded ATP affinity probes were synthesized previously.22,26 Cell lysates (1.5 mg) from MCF-7/WT and MCF-7/C6 cells were incubated with heavy or light isotope-coded ATP affinity probe until its final concentration reached 100 μM. As described elsewhere,23 after incubation at room temperature for 2.5 h, heavy and light ATP probe-labeled cell lysates were mixed at a 1:1 ratio (w/w). Subsequently, 10 kDa Amicon Ultra-15 centrifugal filter units (EMD Millipore, Billerica, MA) and sequencing-grade trypsin (Roche Applied Science, Indianapolis, IN) were employed for sample purification and tryptic digestion with the use of the filter-aided sample preparation method.27 After enrichment using avidin-agarose resin, the eluted peptides were desalted using OMIX C18 pipet tips (Agilent, Santa Clara, CA).
Scheduled LC–MRM Analysis and Data Processing
Scheduled liquid chromatography coupled with multiple-reaction monitoring (LC–MRM) analyses were performed on a TSQ Vantage triple-quadrupole mass spectrometer (Thermo Fisher, San Jose, CA) coupled with a nanoelectrospray ionization source and an EASY n-LCII HPLC system (Thermo Fisher). Detailed experimental conditions were described previously.23 Briefly, the HPLC separation was conducted automatically with a 40 mm trapping column (150 μm i.d.) and a 200 mm analytical column (75 μm i.d., PicoTip Emitter, New Objective, Woburn, MA) packed with 5 and 3 μm reversed-phase ReproSil-Pur C18 material (120 Å in pore size, Dr. Maisch, Germany), respectively. The peptides were separated using a 130 min linear gradient of 2–35% acetonitrile in 0.1% formic acid at a flow rate of 230 nL/min. The spray voltage was 1.9 kV, and Q1 and Q3 resolutions were 0.7 Da. According to the maximum number of transitions per cycle, the cycle time was set at 4–5 s. Data were then processed against the MRM-based kinome library23 on Skyline, version 1.4.0.4421.28 Subsequent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene ontology (GO) analyses were conducted using DAVID Bioinformatics Resources, version 6.7.29
Western Blot
Western blot was performed as described elsewhere.23 Briefly, equal amounts of lysates from MCF-7/WT and MCF-7/C6 cells were electrophoresed on a 12% sodium-dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel with a 4% stacking gel. After protein transfer, the nitrocellulose membrane was blocked and incubated with the corresponding primary and then secondary antibodies. Checkpoint kinase 1 (CHK1) (2G1D5) mouse mAb (Cell Signaling, Beverly, MA), cyclin-dependent kinase 1 (CDK1) (E161) rabbit mAb (Abcam, Cambridge, MA), and cyclin-dependent kinase 2 (CDK2) (78B2) rabbit mAb (Cell Signaling) were employed as the primary antibodies, and actin was used as the loading control.
Results and Discussion
Strategy for Global Kinome Profiling
Despite being extensively studied, the exact mechanisms that underly tumor adaptive radioresistance to therapeutic ionizing radiation remain unresolved.7,30 We found that tumor heterogeneity plays a key role in cell repopulation in breast cancer cells and the radioresistant clones.13 The radioresistant clones including MCF-7/C615−17 were found to be enriched with BCSCs (ALDH+/CD44+/CD24–/low),7,18 which suggests that the radioresistant MCF-7/C6 cells represent the most aggressive breast cancer cells.31 Therefore, revealing active factors signaling the radioresistant phenotype of MCF-7/C6 cells may allow for the discovery of specific targets or development of small-molecule inhibitors for the prevention and treatment of radioresistant cancer. To exploit the kinome reprogramming accompanied by the development of radioresistance, we conducted an in-depth comparative analysis of the global kinome of MCF-7/WT and MCF-7/C6 human breast cancer cells. To this end, we adopted a previously established global kinome profiling method that relies on LC–MRM and isotope-coded ATP affinity probes.22,23 The probe consists of a binding moiety of ATP, an enrichment moiety of desthiobiotin, and an isotope-coded linker with six hydrogens or deuterons for light or heavy isotope labeling.
Figure 1 depicts the general workflow for the scheduled LC–MRM analysis. After 80% confluency was reached, MCF-7/WT and the corresponding γ-radiation-selected radioresistant MCF-7/C6 cells were harvested and lysed separately. In the forward labeling experiment, lysates of MCF-7/WT and MCF-7/C6 cells were incubated with the heavy and light isotope-coded ATP affinity probes, respectively. The labeling was reversed in the reverse labeling experiment. To achieve a reliable differential analysis of the global kinome of these two lines of cells, we conducted one forward and two reverse labeling experiments. As noted previously,26 the ATP component of the affinity probe binds to the ATP-binding pocket of kinases during incubation. Upon binding, the acyl phosphate group in the probe reacts with the side chain amino group of the nearby conserved lysine residue, which gives rise to the labeling of kinases. After the incubation, the heavy and light ATP probe-labeled cell lysates were mixed together in equal mass. Subsequently, the filter-aided sample preparation method was utilized for protein purification and tryptic digestion. The resulting peptides were subsequently enriched using avidin agarose. After peptide elution and desalting, the samples were subjected to scheduled LC–MRM analyses (Figure 1). The MRM transitions and the corresponding collision energies for all monitored kinase peptides are listed in Table S1 of the Supporting Information. For kinase quantification, integration of peak areas for the monitored transitions of kinase peptides was processed using Skyline.28
Figure 1.

General workflow for the quantitative scheduled MRM analysis of the entire kinome using isotope-coded ATP affinity probes. Flowcharts of the forward labeling and LC–MRM experiment.
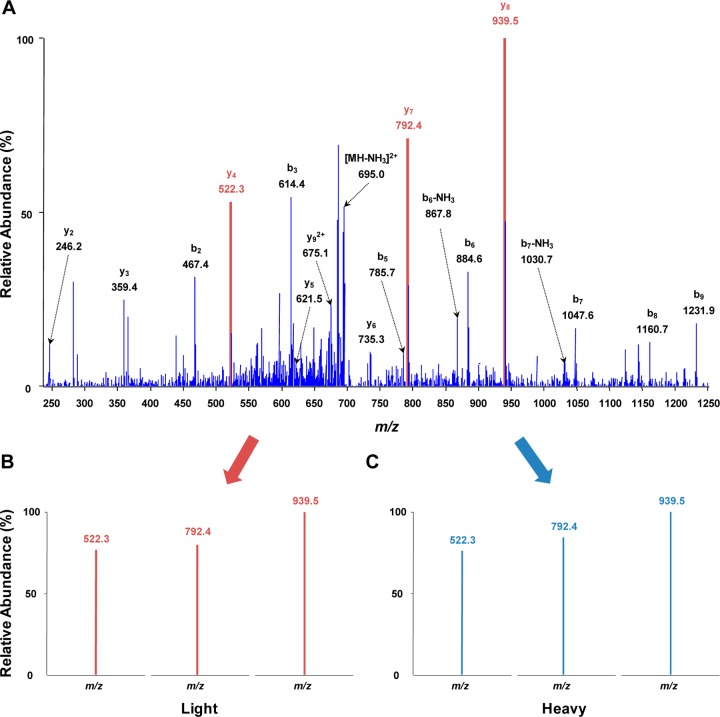
To achieve global kinome quantification in high throughput, we utilized our previously constructed kinome library for scheduled MRM analysis.23 For the kinome library construction, large-scale data-dependent analyses of isotope-coded desthiobiotinylated kinome were performed, from which the tandem mass spectra and retention times for kinase peptides were extracted for scheduled MRM analysis using Skyline. With this method, we were able to monitor a total of ∼2000 transitions, which corresponded to approximately 400 peptides derived from more than 300 kinases, in two LC–MRM analyses. As illustrated in Figure 2, panel A, the tandem mass spectrum (MS/MS) of GK*FGNVYLAR (K* designates isotope-labeled desthiobiotin-conjugated lysine residue) derived from aurora kinase A was acquired from previous data-dependent analysis for the kinome library construction. On the basis of the MS/MS, transitions for the formation of the three most abundant y-ions (y4, y7, and y8) were chosen for the scheduled MRM analysis of this kinase peptide. Transitions monitored in the current LC–MRM analysis for the corresponding light- and heavy-labeled peptides are shown in Figure 2, panels B and C, respectively. The distribution patterns of the three y-ions derived from the light- and heavy-labeled peptides were very similar to that in the MS/MS acquired from previous data-dependent analysis, which supports the reliable identification and quantification of the corresponding peptides.
Figure 2.
Representative MS/MS depicted the reliable quantification of aurora kinase A. (A) The MS/MS of aurora kinase A peptide GK*FGNVYLAR (K* designates isotope-labeled desthiobiotin-conjugated lysine) in the kinome library from previous data-dependent analysis. (B,C) The monitored transitions from the current LC–MRM analysis for the same light and heavy-labeled desthiobiotinylated peptides, respectively.
Global Kinome of Radioresistant MCF-7/C6 Breast Cancer Cells versus Parental MCF-7 Cells
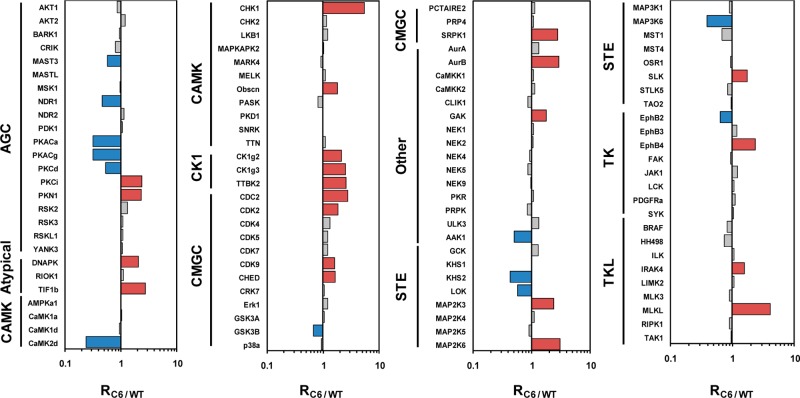
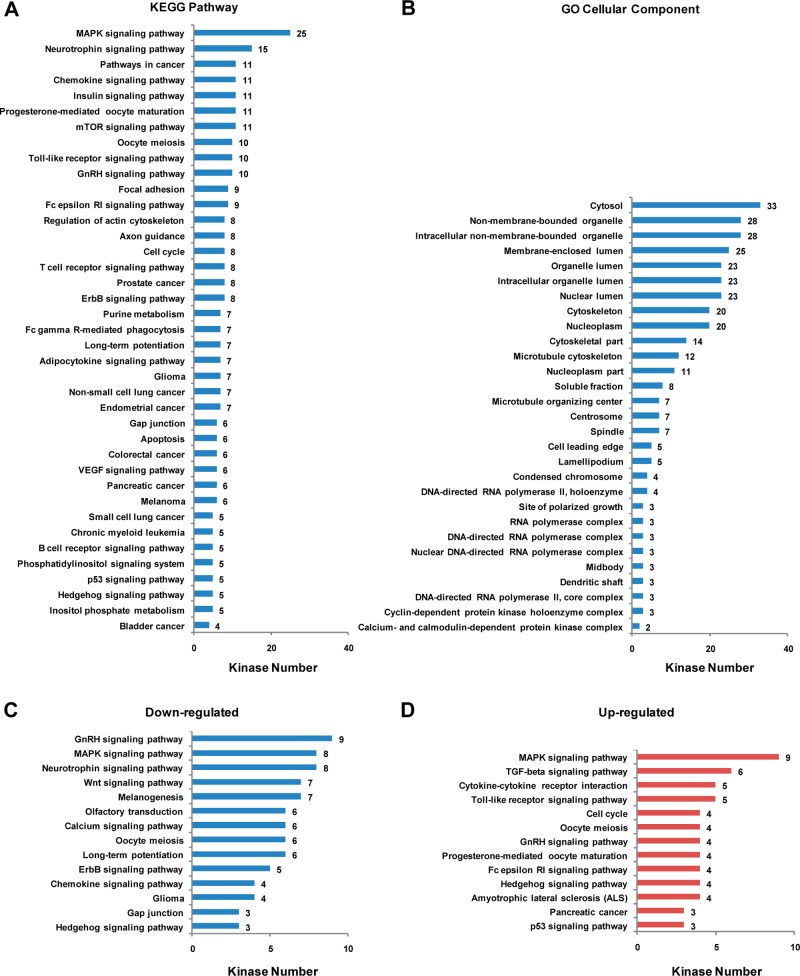
In the current study, we were able to rigorously quantify 120 kinases from one forward and two reverse labeling experiments (Table S2, Supporting Information). The average relative standard deviation for all the quantified kinases is 14%. Thus, we employed cutoff ratios of >1.5 and <0.67 to define the significantly up- and down-regulated kinases, respectively. With the use of this criterion, 24 and 13 of the quantified kinases were significantly up- and down-regulated in MCF-7/C6 relative to MCF-7/WT cells, respectively (Table S3, Supporting Information and Figure 3). To the best of our knowledge, this represents the first in-depth analysis of the alteration of the entire kinome after breast cancer cells develop radioresistance. Kinases from all kinase families except that the RGC and PKL families were successfully detected. These kinases cover a major part of cellular components and are involved in a large collection of cellular pathways, as revealed by GO and KEGG pathway analysis (Figure 4). Figure 4, panels A and B list the GO cellular components and KEGG pathways as well as the number of kinases in each component or pathway with a p-value <0.05. For example, 25, 10, and 8 key kinase modulators involved in MAPK, Toll-like receptor (TLR), and ErbB signaling pathways were successfully quantified with our method. Among them, TLR signaling pathway is well-known for its critical role in innate immune response.32 Some members of TLRs such as TLR5 and TLR9 were found to trigger cancer recurrence after radiotherapy.33,34
Figure 3.
Global kinome comparison of MCF-7/WT and radioresistant MCF-7/C6 human breast cancer cells. Shown in the histogram are the quantification results of the entire kinome in the two cell lines. Blue and red bars represent those kinases that are significantly down- and up-regulated in MCF-7/C6 relative to MCF-7/WT cells, respectively.
Figure 4.
GO and KEGG pathway analysis of kinases detected in the LC–MRM analysis. The number of kinases in the (A) KEGG pathways and (B) GO cellular components with p-values <0.05 are listed. Displayed are also the significantly (C) down- and (D) up-regulated kinases involved KEGG pathways in MCF-7/C6 relative to MCF-7/WT cells.
It is worth noting that, in the current labeling strategy, the ATP-affinity probe forms a covalent amide bond with the side chain of lysine residue at the ATP-binding site and may share conserved sequence.22,35 To increase the specificity and reliability of this approach, we excluded those labelings that emanate from nonspecific interactions. In this vein, only peptides that contain the predetermined ATP-binding motifs (HRDxKxxN, VAxK, or GxxxxGK) or those without a binding motif but were frequently identified in previous shotgun proteomic studies were chosen for the scheduled MRM monitoring.23 Thus, some targeted peptides may be attributed to multiple protein kinases; however, the aforementioned quantification results for the 120 kinases were all based on unique peptides that can only be attributed to the specific targeted kinases. Aside from the 120 rigorously quantified kinases, we also determined the ratios of peptides derived from 75 other kinases; nevertheless, these peptides may arise from multiple kinases, and further experiments are needed to determine the accurate levels of their expression/activity (Table S4, Supporting Information).
The quantification results of the 120 rigorously quantified kinases are depicted in the histogram in Figure 3. Among these kinases, 69% of quantified kinases exhibited very similar levels of expression/activity in the MCF-7/WT and MCF-7/C6 cells. The remaining 20% and 11% of kinases were significantly increased and decreased, respectively. Among the different kinase families, the cell kinase 1 family displayed the most distinct alterations in expression level/activity, with all the kinases quantified in this family being elevated in the radioresistant cells. Similarly, for tyrosine kinase-like and atypical families of kinases, all significantly altered kinases were up-regulated in MCF-7/C6 cells. On the other hand, considerably more kinases in the protein kinase A/G/C family were significantly decreased in MCF-7/C6 cells than those that were increased. From KEGG pathway analysis of these kinases, we discovered that a variety of pathways were altered in the radioresistant cells compared to the parental MCF-7 cells. As depicted in Figure 4, panels C and D, most pathways are unambiguously activated or inhibited in the radioresistant MCF-7/C6 cells. However, for certain pathways, such as MAPK signaling pathway, the regulation of kinase expression/activity displayed the opposite trend, which may suggest the roles of some of these kinases in other cellular pathways or that the aberrant regulation of some kinases is compensated by others in the same pathway.
Ionizing Radiation Perturbed the Expression/Activity of Kinases in Cell Cycle Progression as Well as DNA Damage Response and Repair
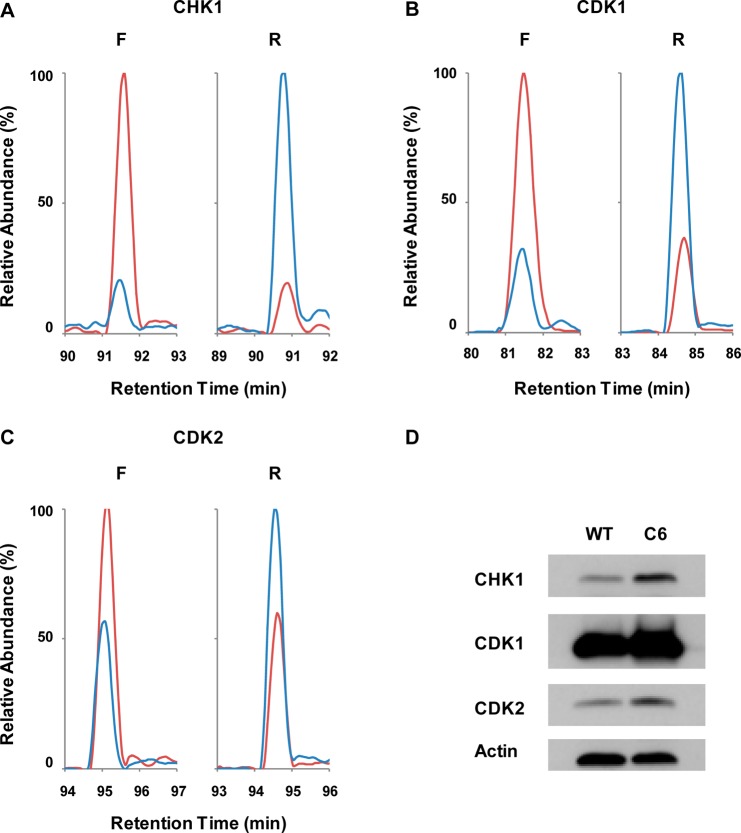
Four kinases involved in cell cycle regulation and DNA damage response/repair were found to be overexpressed/activated in the radioresistant MCF-7/C6 relative to MCF-7/WT cells, which include CHK1, CDK1, CDK2, and the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs). Figure 5, panels A–C show the extracted ion-chromatograms of DIK*PENLLLDER from CHK1 (ratio of 5.46), DLK*PQNLLIDDK from CDK1 (ratio of 2.76), and DLK*PQNLLINTEGAIK from CDK2 (ratio of 1.82), where K* denotes isotope-labeled desthiobiotin-conjugated lysine residue. To further confirm these quantification results, we monitored the expression levels of CHK1, CDK1, and CDK2 in these two lines of cells by Western blot analysis. Our results showed that all three kinases displayed elevated expression in MCF-7/C6 relative to MCF-7/WT cells, which is in agreement with our MRM quantification results (Figure 5D). Therefore, enabled by this global kinome analysis, we demonstrated unambiguously the elevated expression/activation of kinases involved in cell cycle regulation in the radioresistant cells, which may better facilitate cell cycle progression in the presence of genotoxic stress introduced by ionizing radiation.
Figure 5.
Development of radioresistance is correlated with elevation of kinases involved in cell cycle progression and checkpoint control. Shown are the extracted-ion chromatograms for transitions monitored for the following peptides: (A) DIK*PENLLLDER (K* designates isotope-labeled desthiobiotin-conjugated lysine residue) from CHK1; (B) DLK*PQNLLIDDK from CDK1; and (C) DLK*PQNLLINTEGAIK from CDK2 in the LC–MRM experiments. (red) Light- and (blue) heavy- labeled peptides in (left) forward and (right) reverse labeling experiments. (D) Western blot analysis reveals the increased expression of CHK1, CDK1, and CDK2 in MCF-7/C6 relative to MCF-7/WT cells.
Cancer radiation therapy relies primarily on the radiation-induced DNA lesions, particularly DNA strand breaks.4,36 Differences in DNA repair capacities may contribute directly to variations in the efficacies of cancer radiation therapy.37 In this regard, radioresistant cancer may possess a more robust DNA damage repair machinery through inherited or acquired mechanisms.5 Most DNA repair events require the initiation of cell cycle arrest, which provides time for genome repair before cell cycle progression continues.38 Along this line, CHK1 is activated in response to DNA damage, which facilitates transduction of checkpoint signal for the induction of cell cycle arrest.39 Thus, activation of CHK1 could be a critical contributor to cancer radioresistance. In this vein, the connection between CHK1 activation and induction of radioresistance was reported previously.40 CHK1’s role in radioresistance may stem from the zinc-finger E-box binding homeobox 1-enhanced ubiquitin-specific-processing protease 7 activity, which controls the activation and deubiquitination of CHK1 through an ATM-dependent mechanism.41 To overcome the CHK1-induced radioresistance, inhibitors have been utilized for the improvement of radiotherapy.42 Likewise, as a cell cycle regulator, CDK1 was found to be up-regulated in radioresistant cells in our study, which was believed to contribute to radioresistance through phosphorylations of checkpoint proteins and modulators of DNA repair.38 In this context, it was found recently that CDK1 can relocate to mitochondria, which leads to the phosphorylation of a cluster of mitochondrial targets including complex I and enhances mitochondrial ATP generation at G2/M transition in fast-growing cells.43 The results from the present study further support the notion that CDK1 may exert two layers of function in enhancing cell survival, that is, by increasing the DNA repair capacity and augmenting cellular energy supply through modulation of mitochondrial metabolism. Nevertheless, the interplay of these two functions of cell survival, especially in tumor adaptive radioresistance, awaits further investigation.
Nonhomologous end-joining constitutes the major mechanism for repairing ionizing radiation-induced DNA double strand breaks.44 We found that DNA-PKcs, a key component of this repair pathway, was overexpressed/activated, which is consistent with its documented role in cancer radioresistance.45 It was reported that DNA-PKcs participates in acquired radioresistance through the activation of protein kinase B and inactivation of glycogen synthase kinase-3β in a positive feedback loop, which thereby leads to cyclin D1 overproduction and triggers the DNA damage response pathway in cancer cells upon exposure to ionizing radiation.46 In keeping with this notion, inhibition of DNA-PKcs was shown to sensitize MCF-7, MDA-MB-231, and T47D human breast cancer cells toward ionizing radiation.47 Aside from the aforementioned kinases whose overexpression is known to be important in radioresistance, we also observed the overexpression/activation of a number of other kinases, including MLKL, EphB4, etc. These kinases may constitute novel targets to ameliorate radioresistance in cancer therapy, and further studies are needed to determine whether this is the case.
Conclusions
Although extensively studied, the mechanisms that underly tumor adaptive radioresistance remain unclear. The innate or acquired radioresistance compromises the effectiveness of radiation therapy for cancer patients, an urgent clinical issue that must be addressed. To unveil the molecular mechanisms of radioresistance, we performed quantitative comparison of the global kinome of MCF-7 human breast cancer cells and the corresponding ionizing radiation-selected radioresistant clone, that is, MCF-7/C6 cells. In particular, we employed scheduled LC–MRM coupled with isotope-coded desthiobiotin-conjugated ATP-affinity probes for the quantification of the entire kinome of these two cell lines. We were able to rigorously quantify a total of 120 kinases, which covered around 25% of the entire human kinome. Approximately one-third of these quantified kinases displayed significant alterations in expression or activity, which consists of a majority of cellular components and regulates multiple cellular processes.
Notably, several key regulators of cell cycle progression and maintenance of genomic stability were found to be overexpressed/hyperactivated in the radioresistant MCF-7/C6 cells, including CHK1, CDK1, CDK2, and DNA-PKcs. The elevated expression of CHK1, CDK1, and CDK2 was further confirmed by Western blot analysis. Given that DNA lesion induction is one of the major deleterious effects elicited by ionizing radiation,4 efficiency in DNA repair is a deciding factor for the cellular sensitivity toward ionizing radiation. Cell cycle arrest is indispensable for providing cells enough time to complete DNA repair.48 The above-mentioned kinases play pivotal roles in triggering cell cycle checkpoint and regulating cell cycle progression. In addition, some of these kinases are also actively engaged in the phosphorylation of mediators in DNA repair pathways. Thus, the above findings of global kinome alterations associated with radioresistance provide new knowledge to understand tumor adaptive radioresistance and offer potential targets to sensitize cancer cells toward radiation therapy and to achieve better remission of cancer.
Acknowledgments
This work was partly supported by the National Institutes of Health (R01 ES019873 to Y.W. and R01 CA152313 to J.L.).
Supporting Information Available
A list of MRM transitions and the corresponding collision energies for all monitored peptides; expression ratios of all the peptides and kinases quantified; a list of all significantly changed kinases; a list of kinases that were quantified based on unique peptides only or based on conserved peptides; and a zipped Skyline file for the MRM library. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Little M. Risks associated with ionizing radiation: Environmental pollution and health. Br. Med. Bull. 2003, 68, 259–275. [DOI] [PubMed] [Google Scholar]
- Doll R. Hazards of ionising radiation: 100 years of observations on man. Br. J. Cancer 1995, 72, 1339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melott A. L.; Thomas B. C. Astrophysical ionizing radiation and Earth: A brief review and census of intermittent intense sources. Astrobiology 2011, 11, 343–61. [DOI] [PubMed] [Google Scholar]
- Prise K. M.; Schettino G.; Folkard M.; Held K. D. New insights on cell death from radiation exposure. Lancet Oncol. 2005, 6, 520–8. [DOI] [PubMed] [Google Scholar]
- Baumann M.; Krause M.; Hill R. Exploring the role of cancer stem cells in radioresistance. Nat. Rev. Cancer 2008, 8, 545–554. [DOI] [PubMed] [Google Scholar]
- Ahmed K. M.; Li J. J. NF-kappa B-mediated adaptive resistance to ionizing radiation. Free Radical Biol. Med. 2008, 44, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duru N.; Candas D.; Jiang G.; Li J. J. Breast cancer adaptive resistance: HER2 and cancer stem cell repopulation in a heterogeneous tumor society. J. Cancer Res. Clin. Oncol. 2014, 140, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S.; Wu Q.; McLendon R. E.; Hao Y.; Shi Q.; Hjelmeland A. B.; Dewhirst M. W.; Bigner D. D.; Rich J. N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [DOI] [PubMed] [Google Scholar]
- Rich J. N. Cancer stem cells in radiation resistance. Cancer Res. 2007, 67, 8980–4. [DOI] [PubMed] [Google Scholar]
- Conger A. D.; Luippold H. J. Studies on the mechanism of acquired radioresistance in cancer. Cancer Res. 1957, 17, 897–903. [PubMed] [Google Scholar]
- Bao S.; Wu Q.; McLendon R. E.; Hao Y.; Shi Q.; Hjelmeland A. B.; Dewhirst M. W.; Bigner D. D.; Rich J. N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–60. [DOI] [PubMed] [Google Scholar]
- Reya T.; Morrison S. J.; Clarke M. F.; Weissman I. L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–11. [DOI] [PubMed] [Google Scholar]
- Li Z.; Xia L.; Lee M. L.; Khaletskiy A.; Wang J.; Wong J. Y. C.; Li J. J. Effector genes altered in MCF-7 human breast cancer cells after exposure to fractionated ionizing radiation. Radiat. Res. 2001, 155, 543–553. [DOI] [PubMed] [Google Scholar]
- Pajonk F.; Vlashi E.; McBride W. H. Radiation resistance of cancer stem cells: The 4 R’s of radiobiology revisited. Stem Cells 2010, 28, 639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K. M.; Dong S.; Fan M.; Li J. J. Nuclear factor-kappaB p65 inhibits mitogen-activated protein kinase signaling pathway in radioresistant breast cancer cells. Mol. Cancer Res. 2006, 4, 945–55. [DOI] [PubMed] [Google Scholar]
- Guo G.; Wang T.; Gao Q.; Tamae D.; Wong P.; Chen T.; Chen W. C.; Shively J. E.; Wong J. Y.; Li J. J. Expression of ErbB2 enhances radiation-induced NF-kappaB activation. Oncogene 2004, 23, 535–45. [DOI] [PubMed] [Google Scholar]
- Cao N.; Li S.; Wang Z.; Ahmed K. M.; Degnan M. E.; Fan M.; Dynlacht J. R.; Li J. J. NF-kappaB-mediated HER2 overexpression in radiation-adaptive resistance. Radiat. Res. 2009, 171, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duru N.; Fan M.; Candas D.; Menaa C.; Liu H. C.; Nantajit D.; Wen Y.; Xiao K.; Eldridge A.; Chromy B. A.; Li S.; Spitz D. R.; Lam K. S.; Wicha M. S.; Li J. J. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clin. Cancer Res. 2012, 18, 6634–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. C.; Qi R. Z.; Paudel H.; Zhu H. J. Regulation and function of protein kinases and phosphatases. Enzyme Res. 2011, 2011, 794089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsova I.; Skvortsov S.; Stasyk T.; Raju U.; Popper B. A.; Schiestl B.; von Guggenberg E.; Neher A.; Bonn G. K.; Huber L. A.; Lukas P. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics 2008, 8, 4521–33. [DOI] [PubMed] [Google Scholar]
- Mann M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 2006, 7, 952–958. [DOI] [PubMed] [Google Scholar]
- Xiao Y.; Guo L.; Wang Y. Isotope-coded ATP probe for quantitative affinity profiling of ATP-binding proteins. Anal. Chem. 2013, 85, 7478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Guo L.; Wang Y. A targeted quantitative proteomics strategy for global kinome profiling of cancer cells and tissues. Mol. Cell. Proteomics 2014, 13, 1065–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patricelli M. P.; Szardenings A. K.; Liyanage M.; Nomanbhoy T. K.; Wu M.; Weissig H.; Aban A.; Chun D.; Tanner S.; Kozarich J. W. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 2007, 46, 350–8. [DOI] [PubMed] [Google Scholar]
- Qiu H.; Wang Y. Probing adenosine nucleotide-binding proteins with an affinity-labeled nucleotide probe and mass spectrometry. Anal. Chem. 2007, 79, 5547–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.; Guo L.; Jiang X.; Wang Y. Proteome-wide discovery and characterizations of nucleotide-binding proteins with affinity-labeled chemical probes. Anal. Chem. 2013, 85, 3198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski J. R.; Zougman A.; Nagaraj N.; Mann M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–62. [DOI] [PubMed] [Google Scholar]
- MacLean B.; Tomazela D. M.; Shulman N.; Chambers M.; Finney G. L.; Frewen B.; Kern R.; Tabb D. L.; Liebler D. C.; MacCoss M. J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourguignon L. Y.; Shiina M.; Li J. J. Hyaluronan–CD44 interaction promotes oncogenic signaling, microRNA functions, chemoresistance, and radiation resistance in cancer stem cells leading to tumor progression. Adv. Cancer Res. 2014, 123, 255–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips T. M.; McBride W. H.; Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. J. Natl. Cancer Inst. 2006, 98, 1777–85. [DOI] [PubMed] [Google Scholar]
- Akira S.; Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [DOI] [PubMed] [Google Scholar]
- Kauppila J. H.; Mattila A. E.; Karttunen T. J.; Salo T. Toll-like receptor 5 (TLR5) expression is a novel predictive marker for recurrence and survival in squamous cell carcinoma of the tongue. Br. J. Cancer 2013, 108, 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C.; Kozlowska A.; Nechaev S.; Li H.; Zhang Q.; Hossain D. M.; Kowolik C. M.; Chu P.; Swiderski P.; Diamond D. J.; Pal S. K.; Raubitschek A.; Kortylewski M. TLR9 signaling in the tumor microenvironment initiates cancer recurrence after radiotherapy. Cancer Res. 2013, 73, 7211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G.; Whyte D. B.; Martinez R.; Hunter T.; Sudarsanam S. The protein kinase complement of the human genome. Science 2002, 298, 1912–34. [DOI] [PubMed] [Google Scholar]
- Jackson S. P. Sensing and repairing DNA double-strand breaks. Carcinogenesis 2002, 23, 687–96. [DOI] [PubMed] [Google Scholar]
- Buchholz T. A. Radiation therapy for early-stage breast cancer after breast-conserving surgery. N. Engl. J. Med. 2009, 360, 63–70. [DOI] [PubMed] [Google Scholar]
- Branzei D.; Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Hunter T. Roles of CHK1 in cell biology and cancer therapy. Int. J. Cancer 2014, 134, 1013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. J.; Wu S. P.; Liu J. B.; Shi Y. S.; Huang X.; Zhang Q. B.; Yao K. T. MYC regulation of CHK1 and CHK2 promotes radioresistance in a stem cell-like population of nasopharyngeal carcinoma cells. Cancer Res. 2013, 73, 1219–31. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Wei Y.; Wang L.; Debeb B. G.; Yuan Y.; Zhang J.; Yuan J.; Wang M.; Chen D.; Sun Y.; Woodward W. A.; Liu Y.; Dean D. C.; Liang H.; Hu Y.; Ang K. K.; Hung M. C.; Chen J.; Ma L. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 2014, 16, 864–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolderson E.; Richard D. J.; Zhou B. B.; Khanna K. K. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin. Cancer Res. 2009, 15, 6314–20. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Fan M.; Candas D.; Zhang T. Q.; Qin L.; Eldridge A.; Wachsmann-Hogiu S.; Ahmed K. M.; Chromy B. A.; Nantajit D.; Duru N.; He F.; Chen M.; Finkel T.; Weinstein L. S.; Li J. J. Cyclin B1/CDK1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev. Cell 2014, 29, 217–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R.; Ma Y.; Pannicke U.; Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell. Biol. 2003, 4, 712–20. [DOI] [PubMed] [Google Scholar]
- Kienker L. J.; Shin E. K.; Meek K. Both V(D)J recombination and radioresistance require DNA–PK kinase activity, though minimal levels suffice for V(D)J recombination. Nucleic Acids Res. 2000, 28, 2752–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura T.; Kakuda S.; Ochiai Y.; Nakagawa H.; Kuwahara Y.; Takai Y.; Kobayashi J.; Komatsu K.; Fukumoto M. Acquired radioresistance of human tumor cells by DNA–PK/AKT/GSK3beta-mediated cyclin D1 overexpression. Oncogene 2010, 29, 4826–37. [DOI] [PubMed] [Google Scholar]
- Ciszewski W. M.; Tavecchio M.; Dastych J.; Curtin N. J. DNA–PK inhibition by NU7441 sensitizes breast cancer cells to ionizing radiation and doxorubicin. Breast Cancer Res. Treat. 2014, 143, 47–55. [DOI] [PubMed] [Google Scholar]
- Zhou B. B.; Elledge S. J. The DNA damage response: Putting checkpoints in perspective. Nature 2000, 408, 433–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.