Abstract
Proteomics of human milk has been used to identify the comprehensive cargo of proteins involved in immune and cellular function. Very little is known about the effects of gestational diabetes mellitus (GDM) on lactation and breast milk components. The objective of the current study was to examine the effect of GDM on the expression of proteins in the whey fraction of human colostrum. Colostrum was collected from women who were diagnosed with (n = 6) or without (n = 12) GDM at weeks 24–28 in pregnancy. Colostral whey was analyzed for protein abundances using high-resolution, high-mass accuracy liquid chromatography tandem mass spectrometry. A total of 601 proteins were identified, of which 260 were quantified using label free spectral counting. Orthogonal partial least-squares discriminant analysis identified 27 proteins that best predict GDM. The power law global error model corrected for multiple testing was used to confirm that 10 of the 27 proteins were also statistically significantly different between women with versus without GDM. The identified changes in protein expression suggest that diabetes mellitus during pregnancy has consequences on human colostral proteins involved in immunity and nutrition.
Keywords: gestational diabetes mellitus, human colostrum, lactation, LC−MS/MS, multivariate analysis, whey, proteome
Introduction
In addition to nutrients, breast milk delivers bioactive proteins that possess a wide range of biological activities that promote the normal development and maturation of the innate immune system.1 Bioactive proteins in human milk exert protection against infectious diseases via antimicrobial and immunomodulatory activities that confer passive immunity to the breastfed infant.1,2 For example, secretory immunoglobulin A (sIgA), the predominant immunoglobulin in human milk,3 provides protection to neonates by coating microorganisms, inhibiting colonization, and neutralizing viral and bacterial endotoxins.1 Interestingly, the metabolic state of the mother during pregnancy has been found to influence the expression of protective proteins in breast milk. Concentrations of immunoglobulins IgA and IgG4,5 and complement C3 protein5 were lower in colostrum of women with hyperglycemia versus normoglycemic women, which suggests a link between maternal insulin sensitivity and the immune functions of milk. Gestational diabetes mellitus (GDM), a complex disease characterized by elevated blood glucose,6 has immediate and lasting metabolic and immune consequences in infants exposed to maternal diabetes in-utero.7−9 For example, the concentration of sIgA and its glycosylation in breast milk collected at 14 days postpartum was significantly lower in women with versus without GDM.10
Proteomic profiling of milk using high-sensitivity, label-free, semiquantitative mass spectrometry is a potentially useful diagnostic tool to reveal the multifunctional properties of milk,11−14 the health state of the lactating mother,15 and effects in the breastfed infant.16 For example, changes in 75 low abundance proteins in milk were identified as potential biomarkers of mastitis in dairy cows.15 Recently, Molinari et al. identified 55 differentially expressed proteins between pooled skim milk from mothers delivering at term versus preterm, which suggests that mammary development during pregnancy influences protein synthetic and transport pathways in lactation;17 however, the proteomic analysis of breast milk from women with metabolic dysregulation during pregnancy such as GDM has not been reported.
The objective of this study was to determine the effects of GDM on the proteome of human whey colostrum. We hypothesized that proteins involved in immune function found in colostral whey would be different between women with GDM compared to women without GDM. In the current study, whey was isolated from human colostrum collected from women with and without GDM and analyzed by high-resolution, high-mass accuracy liquid chromatography tandem mass spectrometry (LC–MS/MS). The findings reported herein document for the first time differences in colostral whey due to GDM in mothers and illustrate the potential for the characterization of milk for immune, metabolic, and developmental functions.
Materials and Methods
Subject Enrollment
Colostrum samples from primiparous and multiparous women who gave birth to term infants and were diagnosed with (n = 6) or without GDM (n = 12) were prepared and analyzed for this study. Primiparous women have only given birth to one infant, and multiparous women have given birth to two or more infants. GDM was screened for during each subjects’ routine 24–28 week prepartum clinical visit using a 50 g 1 h oral glucose tolerance test (OGTT). Subjects whose 1 h OGTT was less than 7.8 mmol/L comprised the normal glycemic subject pool (non-GDM). Subjects whose 1 h OGTT exceeded 7.8 mmol/L completed a 3 h 100 g OGTT for a final GDM diagnosis. During pregnancy, two of the subjects with GDM controlled their blood sugar with diet, two with diet and oral insulin sensitizing medications, and two with insulin.
Milk Sample Collection and Processing
Colostrum samples were collected between the first and third day of life postpartum by hand expression from one breast by the subject with assistance by an International Board Certified Lactation Consultant and frozen immediately in subjects’ homes or transported to the lab on ice and stored at −80 °C. Samples from each subject were analyzed independently without pooling. Anthropometric and health history and status upon collection were obtained from self-reported health history questionnaires. Samples were deidentified to protect patient privacy and ensure blinding during proteomic analysis. The West Virginia University and University of California Davis Institutional Review Boards approved all aspects of the study, and informed consent was obtained from all subjects. This trial was registered on clinicaltrials.gov (ClinicalTrials.gov, identifier: NCT01817127).
Protein Extraction
When thawed, 5 μL of protease inhibitor (Roche, complete mini EDTA-free, 50× stock) was added to each individual 250 μL colostrum sample. Colostrum sample volumes ranged from 70–250 μL. Casein was depleted with the addition of CaCl2 to adjust the final calcium concentration to 0.06 M. The pH was adjusted to 4.6, and samples were incubated at room temperature for 1 h.18 The samples were centrifuged at 13 000 × g at 4 °C for 30 min twice, and 100 μL of aqueous middle whey fraction was collected. The proteins were precipitated, and lipids in the whey were removed using the Wessel and Flügge method.19 In brief, methanol–chloroform–water was added to the whey samples prior to several centrifugation steps. The supernatant containing the lipids was then discarded, and the protein pellet was resuspended in 50 mM ammonium bicarbonate up to 1:4 (v/v) and sonicated in 15 min intervals until the pellet was fully solubilized in solution. Protein concentrations were determined using NanoDrop spectrophotometer (ND-1000 Spectrophotometer, NanoDrop, Wilmington, DE), and 5 μL of the same protease inhibitor was added to each sample prior to freezing overnight at −80 °C. On the following day, thawed samples (at room temperature) were measured for their protein concentrations using the NanoDrop spectrophotometer. Sample volumes were adjusted to contain 100 μg of whey protein.
In-Solution Digestion
Whey protein samples were digested in-solution using a modified standard trypsin digestion protocol.13 Briefly, acetonitrile (100%) (ThermoFisher Scientific, Waltham, MA) was added to each sample for a final concentration of 6% in 50 mM ammonium bicarbonate prior to protein reduction with tris(2-carboxyethyl)-phosphine (TCEP) (Pierce, Rockford, IL) and alkylation with iodoacetamide (Sigma Life Sciences, St. Louis, MO), followed by treatment with dithiothreitol (Acros Organics, Fair Lawn, NJ). Lactose was removed from the samples using Amicon dialysis membrane with a molecular cutoff of 3000 (Millipore, Billerica, MA) in ammonium bicarbonate (50 mM). The samples were treated with reductively methylated trypsin20 at a 1:30 enzyme-to-protein ratio (w/w) overnight at room temperature. The digested samples were centrifuged at 13 000 × g at room temperature for 2 min, and the supernatant was dried in a speed vacuum and resuspended in Milli-Q water (18.2 MΩ; Millipore, Billerica, MA) prior to being desalted with Aspire RP30 desalting tips (Thermo Fisher Scientific, Inc., Waltham, MA). The desalted peptides were dried under speed vacuum and dissolved in 2% ACN/0.1% TFA for LC–MS/MS analysis.
LC–MS/MS
LC separation was done on a Proxeon Easy-nLC II HPLC (Thermo Scientific) with a Proxeon nanospray source. The digested peptides were reconstituted in 2% acetonitrile/0.1% trifluoroacetic acid, and duplicates of 3 μg of each sample were loaded onto a 100 μm × 25 mm Magic C18 100 Å 5U reverse phase trap where they were desalted online before they were separated using a 75 μm × 150 mm Magic C18 200 Å 3U reverse phase column. Peptides were eluted using a gradient of 0.1% formic acid (A) and 100% acetonitrile (B) with a flow rate of 300 nL/min. A 90 min gradient ran with 5–35% B over 70 min, 35–80% B over 8 min, 80% B for 1 min, 80–5% B over 1 min, and finally held at 5% B for 10 min. Each of the gradients was followed by a 1 h column wash.
Mass spectra were collected on an Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific) in a data-dependent mode with one MS precursor scan followed by 15 MS/MS scans. A dynamic exclusion of 60 s was used. MS spectra were acquired with a resolution of 70 000, and a target of 1 × 106 ions or a maximum injection time of 2 ms. MS/MS spectra were acquired with a resolution of 17 500 and a target of 5 × 104 ions or a maximum injection time of 60 ms. Peptide fragmentation was performed using higher-energy collision dissociation (HCD) with a normalized collision energy (NCE) value of 27. Unassigned charge states as well as +1 and ions > +5 were excluded from MS/MS fragmentation.
Database Searching
Tandem mass spectra were extracted, and charge states were deconvoluted and deisotoped. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version CYCLONE (2013.02.01.1)). X! Tandem was set up to search the Uniprot Human reference database (May 2013, 20 252 entries) with an equal number of reverse sequences and 47 nonhuman common laboratory contaminant proteins from the common Repository of Adventitious Proteins database (http://www.thegpm.org/crap/), assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 PPM and a parent ion tolerance of 20 PPM. Carbamidomethyl of cysteine was specified in X! Tandem as a fixed modification. Dehydration of the N-terminus, glu → pyro-Glu of the N-terminus, ammonia loss of the N-terminus, gln → pyro-Glu of the N-terminus, deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, dioxidation of methionine and tryptophan, and acetylation of the N-terminus were specified in X! Tandem as variable modifications.
Criteria for Protein Identification
Scaffold (version Scaffold 4.0.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 87.0% probability by the Scaffold local false discovery rate (lFDR) Naïve Bayes classifier algorithm, with a minimum of two identified peptides per protein. Protein probabilities were assigned by the Protein Prophet algorithm.21 Proteins containing similar peptides that could not be resolved by MS/MS were grouped into clusters to satisfy the principles of parsimony. By using these parameters, a peptide decoy false discovery rate was calculated as 0.27% on the peptide level and 4.9% on the protein level.
Spectral Counting and Shared Peptide Refinement
Scaffold 4.0.0, was used to sum and normalize the spectral counts and group peptides into proteins. Label-free quantitation was performed on normalized total spectral counts for each protein. Scaffold 4.0.0 normalizes total spectral counts for each protein by multiplying the unweighted spectral counts for each protein in each sample by the ratio of the average total unweighted spectral counts in all samples and the total unweighted spectral counts in each sample. Proteins were retained in the final data analysis if >0 normalized total spectral counts were found in ≥ 50% of the samples collected from women with GDM or without GDM.
Statistical Analysis of Subject Characteristics
Anthropometric measurements were checked for normality using SPSS version 22.0 for Windows (SPSS, Chicago, IL) and transformed appropriately. To determine differences for anthropometric measurements between women with versus without GDM, normalized data were analyzed by independent samples t test, and data that could not be normalized were analyzed by Mann–Whitney U test (α = 0.05).
Orthogonal Partial Least Squared Discriminant Analysis
An orthogonal signal corrected partial least-squares discriminant (O-PLS-DA)22,23 model was developed to identify the top 10% of all differentially expressed proteins between women with and without GDM. Modeling was conducted on protein abundance expressed as normalized spectral abundance factors (NSAF), adjusted for the time of colostrum collection, mean centered, and scaled to unit variance. Leave-one-out cross-validation was used to fit a preliminary two latent variable (LV) O-PLS-DA model to discriminate between women with and without GDM, and model scores and loadings were used for feature selection. Features (proteins) were selected based on their fulfillment of two criteria: (1) NSAF spectral counts were significantly correlated with the O-PLS-DA model scores24 (Spearman’s rho, P ≤ 0.05); and (2) the absolute value of the model loadings on the first latent variable 1 (LV1) ≥ 90th quantile,25 where LV1 is the model component that captures the maximum difference between colostral whey proteins from women with and without GDM.
The classification performance of the selected and excluded feature models were validated and compared using Monte Carlo cross-validation (MCCV)26 and permutation testing. MCCV was carried out by randomly selecting 2/3 of the subjects as a training set (to build models) and using 1/3 of the subjects to test the models, whil the proportion of women with and without GDM was maintained in the full data set. This procedure was repeated 100 times and used to estimate distributions for the model performance statistics, fit to training data (Q2), area under the receiver operator characteristic curve (AUC), sensitivity (true positive rate), and specificity (true negative rate). Permutation testing (prediction of randomly assigned phenotype labels) was combined with the described MCCV model cross-validation and used to estimate the probability of achieving the model’s predictive performance by chance, through comparison of the actual model Q2, AUC, sensitivity, and specificity to those of the null hypothesis as defined by the permuted models. Independent sample t tests were used to asses O-PLS-DA model significance through comparison of actual to permuted model and between selected and excluded feature model’s performance statistics.
Power Law Global Error Model
The power law global error model (PLGEM), a parametric test successfully applied to protein spectral counts27 and transcriptomic data,28 was used to test for significant differences (FDR adjusted P ≤ 0.05) in proteins identified by O-PLS-DA. PLGEM was carried out on time (h) of postpartum colostrum collection-adjusted NSAF protein abundances. All women were classified into early (<20 h, n = 6) or late (>20 h, n = 12) postpartum colostrum collection groups. A linear model was fit to NSAF protein abundances from early and late colostrum collection groups, and the residuals were then tested for significant differences between GDM and non-GDM using PLGEM. The significance level for the PLGEM test statistic (i.e., P-values) was adjusted for the false discovery rate associated with the multiple hypothesis testing according to Benjamini and Hochberg29 and is reported as Padj.
Gene Ontology Enrichment Analysis
Gene ontology (GO) enrichment analysis was conducted using the AmiGO v1.8,30 the GO consortium’s annotation and ontology toolkit (GO database release 2014–01–04), using the GO Term Enrichment tool.31 All differentially expressed proteins between colostral whey from women with versus without GDM (PLGEM, Padj ≤ 0.05) were evaluated for enrichment in GO terms for biological processes, molecular functions, and cellular components. All identified proteins were used as a background set. Significantly overrepresented terms were identified using the hypergeometric test (P ≤ 0.05).
Protein–Protein Interaction Network
A Gaussian graphical Markov network was calculated to model empirical protein–protein interactions among all O-PLS-DA selected colostral whey proteins that were differentially expressed between women with and without GDM. To limit the scope of the partial correlation analysis to the strongest protein–protein empirical relationships, q-order partial correlations32 were first calculated to identify direct from indirect protein associations. Q-order partial correlations (q = 1, 5, 8, 12) were calculated using 1000 replications each and used to estimate the average nonrejection rate (β) for all pairwise relationships. Analysis of the relationship between vertex number, edge degree (connections), and β was used to select β = 0.6 for edge acceptance. Coefficients of partial correlation and P-values were calculated for all q-value identified connections. Cytoscape33 was used to visualize all conditionally independent empirical protein–protein interactions (P ≤ 0.05). All multivariate and statistical analyses on proteomics data were conducted in R v3.0.1.34
Results
Subjects
All women in the study gave birth to healthy singleton term infants. Upon colostrum collection, women did not report illness or mastitis. Subject characteristics are reported in Table 1. Of the anthropometric measurements reported in Table 1, only maternal prepregnancy weight and BMI were significantly higher in women with versus without GDM (P < 0.01). Additionally, colostrum was collected significantly sooner after birth in women with versus without GDM (P < 0.05) (Table 1).
Table 1. Subject Characteristics of Women with and without Gestational Diabetes Mellitus.
| Characteristics | non-GDM (n = 12) |
GDM (n = 6) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| mean | ± | SD | min | max | mean | ± | SD | min | max | |
| maternal age (y) | 34.7 | ± | 2.1 | 32.0 | 39.0 | 34.5 | ± | 3.4 | 31.0 | 40.0 |
| maternal height (m) | 1.6 | ± | 0.1 | 1.5 | 1.8 | 1.6 | ± | 0.1 | 1.5 | 1.7 |
| maternal prepregnancy weight (kg)a | 59.2 | ± | 5.6 | 51.4 | 68.0 | 83.1 | ± | 19.3 | 63.5 | 113.6 |
| maternal prepregnancy BMI (kg/m2)a | 22.0 | ± | 1.4 | 20.1 | 25.0 | 31.6 | ± | 7.6 | 22.6 | 43.0 |
| gestational age of infant (wk) | 39.4 | ± | 1.3 | 37.0 | 41.5 | 38.5 | ± | 0.8 | 37.0 | 39.0 |
| infant birth weight (g) | 3.4 | ± | 0.3 | 2.9 | 3.9 | 3.5 | ± | 0.4 | 3.1 | 4.0 |
| collection of colostrum postdelivery (h)b | 57.8 | 22.9 | 9.0 | 77.0 | 28.2 | 30.4 | 4.6 | 74.0 | ||
| Frequencies | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Parity | ||||||||||
| primiparous | 6 | 1 | ||||||||
| multiparous | 6 | 5 | ||||||||
| Delivery Mode | ||||||||||
| c-section | 1 | 1 | ||||||||
| vaginal | 11 | 5 | ||||||||
| Infant Sex | ||||||||||
| female | 6 | 2 | ||||||||
| male | 6 | 4 | ||||||||
Different between non-GDM and GDM; log-transformed, independent samples t test, P < 0.01.
Different between non-GDM and GDM; independent samples t test, P < 0.05.
Characterization of Whey Proteins in Human Colostrum
Isolation of the aqueous whey fraction of colostrum collected between the first and third day of life postpartum was conducted by a series of centrifugation, casein precipitation, and fat removal. Peptide identifications were accepted if they could be established at greater than 87.0% probability by the Scaffold Local FDR algorithm, with a minimum of two identified peptides per protein and 4.9% protein FDR, which corresponded to a 0.27% peptide decoy FDR for peptide identification. After decoy peptides and proteins with nonhuman accessions were excluded, 601 proteins were identified in colostral whey samples isolated from women with and without GDM (Table 1, Supporting Information). These 601 identified proteins were filtered to only include peptide species with >0 normalized spectral counts in ≥ 50% of the samples collected from women with GDM or without GDM. With this procedure, 60% of the originally measured 601 proteins were removed, and 260 final proteins were retained for further statistical and multivariate data analysis.
Identification of Differentially Expressed Colostral Whey Proteins between Women with versus without Maternal Gestational Diabetes Mellitus
O-PLS-DA, a multivariate classification model, was used to identify the top 10% (n = 27) of all protein discriminants between women with and without GDM (Table 1, Supporting Information). The limited study sample size precluded the use of a single hold-out test set for model validation, and instead, 100 rounds of MCCV were used to generate robust model performance statistics (Table 2, Supporting Information). O-PLS-DA scores from a model based on the 27 selected variables demonstrate clear visual separation between the two phenotypes (Figure 1). Of these 27 proteins, three were correlated with maternal prepregnancy BMI: alpha-2-HS-glycoprotein (Spearman’s rho = −0.61, P < 0.01), complement C1r subcomponent-like protein (Spearman’s rho = −0.49, P < 0.05), and transmembrane protein 201 (Spearman’s rho = 0.48, P < 0.05).
Figure 1.
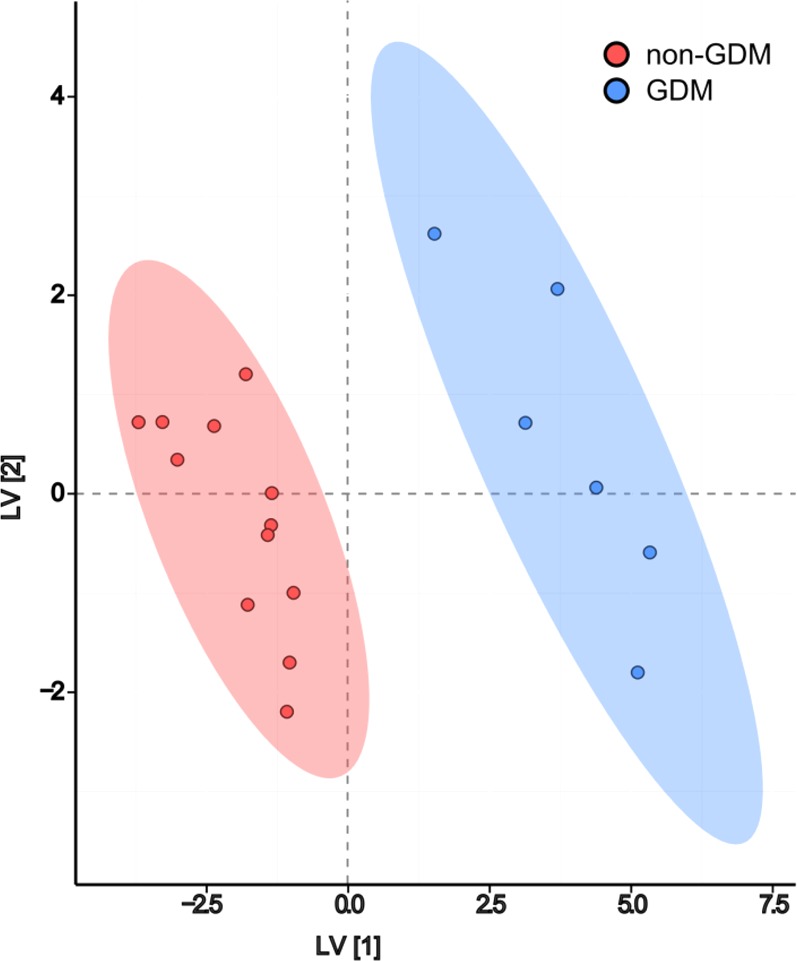
Scores plot displaying discrimination between women with and without GDM based on 27 selected colostral whey proteins using orthogonal signal correction partial least-squares discriminant analysis. Edge width and color encode the magnitude and direction of partial correlations (P < 0.05) among all selected proteins based on O-PLS-DA. Vertex size and shape display the magnitude and direction of the fold-difference in protein expression in colostral whey from women with GDM relative to women without GDM (mean GDM/mean non-GDM). Significantly differentially expressed proteins (Power Law Global Error Model, Padj ≤ 0.05) are identified with thick black borders.
The prediction of maternal phenotype using the 27 selected proteins was compared to the classification performance of a model based on all excluded proteins (n = 233) and validated using MCCV and permutation testing. Comparison of model classification performance metrics (Q2, AUC, sensitivity, and specificity) were used to asses model significance. On the basis of test/training and permutation testing, the selected O-PLS-DA model displayed significantly higher classification performance (P < 0.05) for the test set (AUC = 0.791 ± 0.14) than did both the permuted model (random chance) (AUC = 0.475 ± 0.21) and a model using all 233 excluded proteins (AUC = 0.370 ± 0.19) (Table 2, Supporting Information). On the basis of PLGEM statistical analysis, with adjustment for time of collection and multiple hypothesis testing, 10 of the 27 O-PLS-DA selected proteins were significantly different between women with versus without GDM (Table 2). Colostral whey from women with GDM contained higher abundances of apolipoprotein D, Ig heavy chain V–II region ARH-77, and prostasin and lower abundances of alpha-2-HS-glycoprotein, apolipoprotein A1 and E, 14–3–3 protein zeta/delta, protein disulfide-isomerase, protein DJ-1, and protein FAM3D (Table 2).
Table 2. Differentially Expressed Proteins in Human Colostral Whey between Women with and without Gestational Diabetes Mellitus.
| protein | accessiona | non-GDMb | GDMb | FDc | Padjd | ranke |
|---|---|---|---|---|---|---|
| Higher in GDM | ||||||
| apolipoprotein D | P05090 | 17.1 ± 12 | 48.1 ± 20 | 2.8 | 0.002 | 8 |
| Ig heavy chain V–II region ARH-77 | P06331 | 1.15 ± 1.7 | 2.04 ± 1.6 | 1.8 | 0.015 | 2 |
| prostasin | Q16651 | 4.59 ± 2 | 7.95 ± 4.2 | 1.7 | 0.041 | 5 |
| Lower in GDM | ||||||
| alpha-2-HS-glycoprotein | P02765 | 20.1 ± 8.6 | 9.36 ± 4.4 | 0.5 | 0.048 | 7 |
| apolipoprotein A1 | P02647 | 38.2 ± 13 | 16.8 ± 14 | 0.4 | 0.012 | 1 |
| apolipoprotein E | P02649 | 9.81 ± 5.3 | 3.21 ± 3 | 0.3 | 0.017 | 6 |
| 14–3–3 protein zeta/delta | P63104 | 6.23 ± 3.3 | 2.11 ± 1.8 | 0.3 | 0.028 | 10 |
| protein disulfide-isomerase | P07237 | 1.36 ± 1.7 | 0.275 ± 0.51 | 0.2 | 0.050 | 9 |
| protein DJ-1 | Q99497 | 1.08 ± 0.79 | 0.097 ± 0.24 | 0.1 | 0.007 | 3 |
| protein FAM3B | P58499 | 2.77 ± 1.6 | 0.876 ± 1.3 | 0.3 | 0.032 | 4 |
Uniprot database identifier (http://www.uniprot.org/).
Values are reported as the mean ± standard deviation of normalized total spectra.
Fold difference, FD; of the means of normalized total spectra for GDM relative to non-GDM.
False discovery rate adjusted (q = 0.05) P-value for power law global error model.
Rankings are based on the absolute values of the O-PLS-DA model loadings on the LV1 for shown proteins.
Gene Ontology Enrichment Analysis
The 10 expressed colostral whey proteins that were significantly different between women with and without GDM were enriched (P < 0.05) for the biological process associated with the “regulation of transport” (GO:0051049). No other GO terms were statistically enriched.
Protein–Protein Dependency Network
The colostral whey proteins selected by O-PLS-DA were used in a protein–protein interaction network to identify direct empirical relationships (partial correlations, P ≤ 0.05) among all 27 proteins (Figure 2). Nineteen of the 27 proteins were directly correlated, of which 17 were positively and two were negatively associated with one another. Sixteen proteins were lower and three were higher in colostral whey from women with GDM compared to women without GDM (Figure 2 and Table 1, Supporting Information). Of the 19 associated proteins, only seven were significantly different (PLGEM, P < 0.05) between colostral whey from women with versus without GDM (14–3–3 protein zeta/delta, alpha-2-HS-glycoprotein, apolipoprotein A1, apolipoprotein D, Ig heavy chain V–II region ARH-77, protein disulfide-isomerase, and protein FAM3D).
Figure 2.
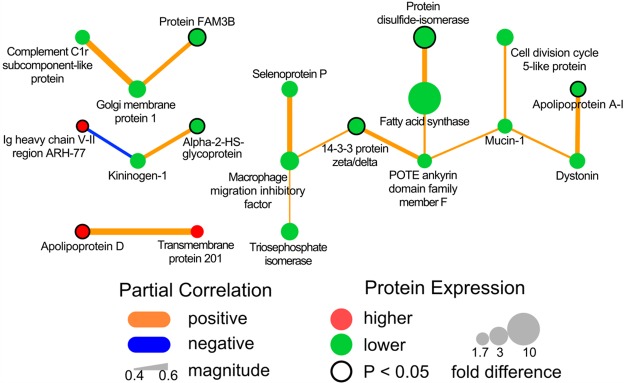
Empirical protein–protein interaction network for differentially expressed colostral whey proteins between women with and without GDM.
Discussion
LC–MS/MS Approach
Proteomic profiling of milk using high-sensitivity, label-free, semiquantitative mass spectrometry has revealed multifunctional properties of milk and its dynamic nature to meet the immunologic, metabolic, and developmental demands of the neonate.11−14 Recently, a focus on the proteins of least abundance in breast milk has received attention for their unique bioactive roles in infant immunity, development, and growth. By using ProteoMiner to enrich low abundance proteins coupled with LC–MS/MS, Liao et al. identified a total of 115 proteins in human whey, of which 35% were involved in the immune response, and several proteins were differentially regulated between early and mature milk.13 By using ion-exchange and SDS-PAGE based protein fractionation methods followed by LC–MS/MS, Gao et al. identified 976 whey proteins, of which 152 were significantly regulated between transitional and mature milk.11 In contrast to the study reported by Gao et al., we did not use fractionation methods prior to LC–MS/MS. However, because of the high mass accuracy, high-resolution, and scan speed of the Q Exactive orbitrap mass spectrometer, we were able to confidently identify 601 proteins from the whey fraction of human milk with a low false discovery rate at the protein and peptide level. Additionally, we used a low peptide decoy FDR (0.27%) compared to the protein FDR (4.9%) because there were over 470 000 peptides in this data set compared to only 601 proteins. It is common in data sets with large numbers of identified spectra that condense to a much smaller number of proteins to have a highly inflated protein decoy FDR compared to the spectra or peptide decoy FDR.35
Multivariate and Statistical Analyses
In this study, we used multivariate modeling and statistical analyses to identify differentially expressed colostral whey proteins (i.e., proteins that were up- or down-regulated) between women with versus without GDM. Identification of differentially expressed proteins between these two groups posed two major challenges: (1) limitation of the sample size and test power precluding the identification of diagnostic markers for GDM; and (2) overcoming the relationship between protein expression and the time of sample collection postpartum. By using power analysis, we have previously shown that GDM had a large effect on the human milk glycome36 and that the current experimental design is sufficiently powered to detect differences in colostral whey proteins between the two groups. However, because of the large number of proteins measured, even proteins that display moderate effect sizes may not yield significant results in statistical tests due to the large penalty for multiple hypotheses tested. To reduce the FDR we used O-PLS-DA based feature selection37 to identify the top 10% of all multivariate protein discriminants between women with and without GDM, which were then tested using the univariate statistical test, PLGEM. Human milk protein composition and abundance are known to vary based on the postpartum period.11−13 To account for this issue and ensure that changes in colostral protein abundances were not associated with differences in postpartum time of sample collection, both O-PLS-DA modeling and PLGEM analyses were conducted on NSAF protein abundances adjusted for the time of collection postpartum using a linear model. With the aforementioned strategy, O-PLS-DA modeling was used to identify 27 colostral whey proteins that best discriminated women with versus without GDM, of which 10 were identified to be significantly differentially expressed (PLGEM Padj < 0.05) after an adjustment was made for multiple hypothesis testing. Gene ontology enrichment analysis and protein–protein empirical interaction networks were used to help identify the biological context for the noted differences in colostral whey proteins between women with versus without GDM.
Colostral Whey Proteins Higher in GDM
One of the 10 proteins that were differentially expressed in colostral whey from women with GDM compared to women without GDM is the variable region of the heavy chain of an immunoglobulin (Ig heavy chain V–II region ARH-77). Immunoglobulins are secreted by plasma cells in the mammary gland and throughout the body. During mature lactation, immunoglobulins enter milk via transcytosis across mammary epithelial cells; however, during the production of colostrum, the tight junctions that join mammary epithelial cells are leaky, which allows passage of immunoglobulins and white blood cells via the paracellular pathway. That is, the immunoglobulins pass from interstitial spaces, between the mammary epithelial cells, and into the colostrum. Thus, in the context of the transition from colostrum to mature milk, immunoglobulins are a marker of the tightness of the junctions and the stage of milk production.38 In our study, without correction for time of postpartum milk collection, 29 of the differentially expressed proteins were regions of immunoglobulins, which suggests that women at the earliest and latest times of collection were in different stages of milk production (colostrum vs transitional milk production); however, after the adjustment for the time of milk collection was made, only Ig heavy chain V–II region ARH-77 was significantly higher in colostral whey from women with GDM (Table 2). This suggests that women with GDM may have a delayed progression from colostrum to transitional milk production, otherwise known as a delayed lactogenesis. It is known that obese women have delayed lactogenesis;38 yet this protein was not significantly correlated with prepregnancy BMI, which suggests that independent of obesity, GDM may delay the onset of copious milk production. Furthermore, even though women with GDM were disproportionally multiparous (Table 1), have a faster onset of milk production,39 the effect of GDM was strong enough to overcome that bias. However, these data should be taken with caution because unlike other immunoglobulins, this protein was in low abundance in colostral whey and thereby susceptible to low reproducibility and high analytical variance. Orthogonal analysis is warranted to confirm if this protein contributes to the biological variation in predicting maternal phenotype.
Apolipoprotein D is a lipocalin involved in lipid transport and fluctuates during gestation and fetal development. It has previously been shown that plasma apolipoprotein D levels decrease during pregnancy and further decrease in women with excessive gestational weight gain. Interestingly, plasma apolipoprotein returned to baseline after birth more quickly in lactating women compared to those who were not breastfeeding.40 In our study, colostral apolipoprotein D abundance was higher in women with GDM versus in women without GDM. Liao et al. showed that apolipoprotein D levels were higher in colostrum relative to mature milk.13 Interestingly, two other apolipoproteins found in human milk (apolipoprotein A1 and apolipoprotein E) were significantly lower in colostral whey from women with versus without GDM. These data are supported by human trials that reported lower plasma levels of LDL and HDL cholesterol in women with GDM compared to healthy controls.41 Thus, it is possible that the higher apolipoprotein D level in colostral whey from women with GDM is due to delayed lactogenesis. However, future human studies are warranted to test this hypothesis.
Colostral Whey Proteins Lower in GDM
Whey colostral proteins involved in de novo lipid synthesis were uniformly lower in GDM. Differences in these proteins were positively correlated with one another and constituted the largest connected cluster within the empirical protein–protein interaction network (Figure 2). Although the lipid content in colostrum was not measured in this study, these results suggest that women with GDM produce lipid-poor milk compared to that of women without GDM. Morceli et al. reported 1.8-fold lower total lipid content in colostrum from diabetic women with hyperglycemia than from normoglycemic women.5 In mice fed a high-fat (HF) diet, obese HF-fed mice produce lipid-poor milk compared to that of lean HF-fed mice.42 Importantly, in our study, none of the lipid synthesis related proteins reduced by GDM were observed to be significantly correlated with prepregnancy BMI, which suggests that GDM (or insulin resistance), and not obesity per se, is associated with low-fat milk production. Thus, we hypothesize that women with GDM produce lipid-poor milk and that this effect is independent of obesity.
Circulating alpha-2-HS-glycoprotein (also known as fetuin-A), a liver-derived protein that down-regulates insulin signaling in peripheral tissues and was recently identified as a marker of insulin resistance, inflammation, and adiposity,43−45 was lower in women with GDM. Serum alpha-2-HS-glycoprotein concentrations are elevated in humans with metabolic disorders but can also be elevated during negative energy balance (NEB), which serves to preserve glucose utilization by peripheral tissues. In dairy cows, alpha-2-HS-glycoprotein increases during early lactation and causes a switch toward using fatty acids as an energy source instead of glucose.46 Unlike cows, humans do not heavily rely on body stores for milk synthesis but perhaps experience mild NEB during the first days of lactation given the energetic needs of birthing, recovery, and initiation of lactation. In a study of the human whey proteome across lactation, alpha-2-HS-glycoprotein did not appear to be differentially abundant among colostrum and later stages of lactation, but subjects were not excluded based on prepregnancy BMI or metabolic disease, and the data was not analyzed in the context of metabolic status.13 In our study, we found colostral whey alpha-2-HS-glycoprotein to be lower in women with GDM relative to those without GDM; however, colostral whey alpha-2-HS-glycoprotein was also negatively correlated with prepregnancy BMI; thus, women with higher BMI also had lower levels of colostral whey alpha-2-HS-glycoprotein. While it is not possible to determine whether high BMI or GDM status is the driver of lower colostrum alpha-2-HS-glycoprotein, future research is necessary to confirm if individuals with high BMI or GDM have a blunted response to the metabolic changes of early lactation.
Protein FAM3B, also known as PANDER (pancreatic derived factor), a cytokine likely to be cosecreted with insulin47 by the pancreatic alpha- and beta-cells, was significantly lower in the colostral whey from women with GDM and was not correlated with prepregnancy BMI. While the physiological role of FAM3B is incompletely described, its role in insulin signaling and glucose homeostasis supports our hypothesis that GDM impacts milk production and composition.
Herein we report that GDM had an impact on the human colostral whey proteome. This was an observational study that demonstrated proof of concept that alterations in whey proteins in GDM have short-term implications for the transport of infant nutrition. These findings should be further explored in large prospective cohort studies that include analysis of milk over the course of lactation and associated effects of maternal gestational diabetes mellitus on infant nutrient status.
Acknowledgments
This project was made possible in part by support from the University of California Discovery Program (05GEB01NHB), California Dairy Research Foundation, West Virginia University, Sigma Theta Tau International Pi Lambda Phi, the National Institute of Health (HD059127 and HD061923), and the West Coast Metabolomics Center (NIH 1 U24 DK097154).
Glossary
Abbreviations
- gestational diabetes mellitus
GDM
- liquid chromatography tandem mass spectrometry
LC–MS/MS
- orthogonal signal corrected partial least-squares discriminant analysis
O-PLS-DA
- power law global error model
PLGEM
Supporting Information Available
Subject characteristics of women with and without gestational diabetes mellitus; differentially expressed proteins in human colostral whey between women with and without gestational diabetes mellitus; all 601 measured human colostral whey proteins from women with and without gestational diabetes mellitus; validation of orthogonal signal corrected partial least-squares discriminant analysis model; and selected proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
J.T.S. and J.B.G. conceived the project; J.T.S. and I.C. designed and executed the clinical study; D.S.G. prepared the samples for analysis; D.W. and B.S.P. analyzed the samples and generated the data; D.G., D.G.L. and J.T.S. analyzed the data; D.G., D.G.L. and J.T.S. wrote the paper; all authors reviewed and revised the manuscript. D.G. and J.T.S. contributed equally to this project.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Brandtzaeg P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 1562S8–S15. [DOI] [PubMed] [Google Scholar]
- Lönnerdal B. Nutritional roles of lactoferrin. Curr. Opin. Clin. Nutr. Metab. Care 2009, 123293–297. [DOI] [PubMed] [Google Scholar]
- Chirico G.; Marzollo R.; Cortinovis S.; Fonte C.; Gasparoni A. Antiinfective properties of human milk. J. Nutr. 2008, 13891801S–1806S. [DOI] [PubMed] [Google Scholar]
- França E. L.; Calderon I. M. P.; Vieira E. L.; Morceli G.; Honorio-França A. C. Transfer of maternal immunity to newborns of diabetic mothers. Clin. Dev. Immunol. 2012, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morceli G.; França E.; Magalhães V.; Damasceno D.; Calderon I.; Honorio-França A. Diabetes induced immunological and biochemical changes in human colostrum. Acta Paediatr. 2011, 1004550–556. [DOI] [PubMed] [Google Scholar]
- Diagnosis and classification of diabetes mellitus. Diabetes Care 2011, 34Suppl. 1S62–S69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy L.; Casas J. P.; Hingorani A. D.; Williams D. Type 2 diabetes mellitus after gestational diabetes: A systematic review and meta-analysis. Lancet 2009, 37396771773–1779. [DOI] [PubMed] [Google Scholar]
- Clausen T. D.; Mathiesen E. R.; Hansen T.; Pedersen O.; Jensen D. M.; Lauenborg J.; Damm P. High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes. Diabetes Care 2008, 312340–346. [DOI] [PubMed] [Google Scholar]
- See e4 in:Kumar R.; Ouyang F.; Story R. E.; Pongracic J. A.; Hong X.; Wang G.; Pearson C.; Ortiz K.; Bauchner H.; Wang X. Gestational diabetes, atopic dermatitis, and allergen sensitization in early childhood. J. Allergy Clin. Immunol. 2009, 12451031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz J. T.; Totten S. M.; Huang J.; Grapov D.; Durham H. A.; Lammi-Keefe C. J.; Lebrilla C.; German J. B. Human milk secretory immunoglobulin A and lactoferrin N-glycans are altered in women with gestational diabetes mellitus. J. Nutr. 2013, 143121906–1912jn. 113.180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.; McMahon R. J.; Woo J. G.; Davidson B. S.; Morrow A. L.; Zhang Q. Temporal changes in milk proteomes reveal developing milk functions. J. Proteome Res. 2012, 1173897–3907. [DOI] [PubMed] [Google Scholar]
- Liao Y.; Alvarado R.; Phinney B.; Lonnerdal B. Proteomic characterization of human milk fat globule membrane proteins during a 12 month lactation period. J. Proteome Res. 2011, 1083530–41. [DOI] [PubMed] [Google Scholar]
- Liao Y.; Alvarado R.; Phinney B.; Lönnerdal B. Proteomic characterization of human milk whey proteins during a twelve-month lactation period. J. Proteome Res. 2011, 1041746–1754. [DOI] [PubMed] [Google Scholar]
- Reinhardt T. A.; Lippolis J. D. Developmental changes in the milk fat globule membrane proteome during the transition from colostrum to milk. J. Dairy Sci. 2008, 9162307–18. [DOI] [PubMed] [Google Scholar]
- Boehmer J. L.; DeGrasse J. A.; McFarland M. A.; Tall E. A.; Shefcheck K. J.; Ward J. L.; Bannerman D. D. The proteomic advantage: Label-free quantification of proteins expressed in bovine milk during experimentally induced coliform mastitis. Vet. Immunol. Immunopathol. 2010, 1384252–266. [DOI] [PubMed] [Google Scholar]
- Mangé A.; Tuaillon E.; Viljoen J.; Nagot N.; Bendriss S.; Bland R. M.; Newell M.-L.; Van de Perre P.; Solassol J. Elevated concentrations of milk β2-microglobulin are associated with increased risk of breastfeeding transmission of HIV-1 (vertical transmission study). J. Proteome Res. 2013, 12125616–5625. [DOI] [PubMed] [Google Scholar]
- Molinari C. E.; Casadio Y. S.; Hartmann B. T.; Livk A.; Bringans S.; Arthur P. G.; Hartmann P. E. Proteome mapping of human skim milk proteins in term and preterm milk. J. Proteome Res. 2012, 1131696–1714. [DOI] [PubMed] [Google Scholar]
- Kunz C. L. B. Human milk proteins: Separation of whey proteins and their analysis by polyacrylamide gel electrophoresis, fast protein liquid chromatography (FPLC) gel filtration, and anion-exchange chromatography. Am. J. Clin. Nutr. 1989, 49, 464–470. [DOI] [PubMed] [Google Scholar]
- Wessel D. F.; Flugge U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 1381141–143. [DOI] [PubMed] [Google Scholar]
- Rice R. H.; Rocke D. M.; Tsai H.-S.; Silva K. A.; Lee Y. J.; Sundberg J. P. Distinguishing mouse strains by proteomic analysis of pelage hair. J. Invest. Dermatol. 2009, 12992120–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii A. I.; Keller A.; Kolker E.; Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003, 75174646–4658. [DOI] [PubMed] [Google Scholar]
- Trygg J.; Wold S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 163119–128. [Google Scholar]
- Svensson O.; Kourti T.; MacGregor J. F. An investigation of orthogonal signal correction algorithms and their characteristics. J. Chemom. 2002, 164176–188. [Google Scholar]
- Wiklund S.; Johansson E.; Sjostrom L.; Mellerowicz E. J.; Edlund U.; Shockcor J. P.; Gottfries J.; Moritz T.; Trygg J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models. Anal. Chem. 2008, 801115–22. [DOI] [PubMed] [Google Scholar]
- Palermo G.; Piraino P.; Zucht H. D. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics-type data. Adv. Appl. Bioinf. Chem. 2009, 2, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q.-S.; Liang Y.-Z. Monte Carlo cross validation. Chemom. Intell. Lab. Syst. 2001, 5611–11. [Google Scholar]
- Pavelka N.; Pelizzola M.; Vizzardelli C.; Capozzoli M.; Splendiani A.; Granucci F.; Ricciardi-Castagnoli P. A power law global error model for the identification of differentially expressed genes in microarray data. BMC Bioinf. 2004, 5, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N.; Fournier M. L.; Swanson S. K.; Pelizzola M.; Ricciardi-Castagnoli P.; Florens L.; Washburn M. P. Statistical similarities between transcriptomics and quantitative shotgun proteomics data. Mol. Cell. Proteomics 2008, 74631–44. [DOI] [PubMed] [Google Scholar]
- Benjamini Y.; Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 571289–300. [Google Scholar]
- Carbon S.; Ireland A.; Mungall C. J.; Shu S.; Marshall B.; Lewis S.; Ami G. O. H. Web Presence Working, G., AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 252288–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle E. I.; Weng S.; Gollub J.; Jin H.; Botstein D.; Cherry J. M.; Sherlock G. GO::TermFinder—Open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 2004, 20183710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo R.; Roverato A. Reverse engineering molecular regulatory networks from microarray data with qp-graphs. J. Comput. Biol. 2009, 162213–27. [DOI] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13112498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011.
- Zhang B.; Wang J.; Wang X.; Zhu J.; Liu Q.; Shi Z.; Chambers M. C.; Zimmerman L. J.; Shaddox K. F.; Kim S.; Davies S. R.; Wang S.; Wang P.; Kinsinger C. R.; Rivers R. C.; Rodriguez H.; Townsend R. R.; Ellis M. J. C.; Carr S. A.; Tabb D. L.; Coffey R. J.; Slebos R. J. C.; Liebler D. C.; N. C. I. C. P. T. A. C. Proteogenomic characterization of human colon and rectal cancer. Nature 2014, 5137518382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz J. T.; Totten S. M.; Huang J.; Grapov D.; Durham H. A.; Lammi-Keefe C. J.; Lebrilla C.; German J. B. Human milk secretory immunoglobulin a and lactoferrin N-glycans are altered in women with gestational diabetes mellitus. J. Nutr. 2013, 143121906–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grapov D.; Fahrmann J.; Hwang J.; Poudel A.; Jo J.; Periwal V.; Fiehn O.; Hara M. Diabetes associated metabolomic perturbations in NOD mice. Metabolomics 2014, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M. C.Milk secretion: An overview: UCHSC: Denver, CO, 1998. http://mammary.nih.gov/reviews/lactation/Neville001/.
- Dewey K. G.; Nommsen-Rivers L. A.; Heinig M. J.; Cohen R. J. Risk factors for suboptimal infant breastfeeding behavior, delayed onset of lactation, and excess neonatal weight loss. Pediatrics 2003, 1123607–619. [DOI] [PubMed] [Google Scholar]
- Do Carmo S.; Forest J.-C.; Giguère Y.; Masse A.; Lafond J.; Rassart E. Modulation of apolipoprotein D levels in human pregnancy and association with gestational weight gain. Reprod. Biol. Endocrinol. 2009, 7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaran A. Pregnancy-induced hyperlipoproteinemia: Review of the literature. Reprod. Sci. 2009, 165431–437. [DOI] [PubMed] [Google Scholar]
- Wahlig J. L.; Bales E. S.; Jackman M. R.; Johnson G. C.; McManaman J. L.; MacLean P. S. Impact of high-fat diet and obesity on energy balance and fuel utilization during the metabolic challenge of lactation. Obesity 2012, 20165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan N.; Hennige A. M.; Staiger H.; Machann J.; Schick F.; Kröber S. M.; Machicao F.; Fritsche A.; Häring H.-U. α2-Heremans–Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 2006, 294853–857. [DOI] [PubMed] [Google Scholar]
- Pal D.; Dasgupta S.; Kundu R.; Maitra S.; Das G.; Mukhopadhyay S.; Ray S.; Majumdar S. S.; Bhattacharya S. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat. Med. 2012, 1881279–1285. [DOI] [PubMed] [Google Scholar]
- Stefan N.; Häring H.-U. Circulating fetuin-A and free fatty acids interact to predict insulin resistance in humans. Nat. Med. 2013, 194394–395. [DOI] [PubMed] [Google Scholar]
- Wathes D. C.; Clempson A. M.; Pollott G. E. Associations between lipid metabolism and fertility in the dairy cow. Reprod., Fertil. Dev. 2012, 25148–61. [DOI] [PubMed] [Google Scholar]
- Yang J.; Robert C. E.; Burkhardt B. R.; Young R. A.; Wu J.; Gao Z.; Wolf B. A. Mechanisms of glucose-induced secretion of pancreatic-derived factor (PANDER or FAM3B) in pancreatic β-cells. Diabetes 2005, 54113217–3228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


