Abstract
Evolving data suggests that marrow hematopoietic stem cells show reversible changes in homing, engraftment and differentiation phenotype with cell cycle progression. Furthermore, marrow stem cells are a cycling population. Traditional concepts hold that the system is hierarchical, but the information on the lability of phenotype with cycle progression suggests a model in which stem cells are on a reversible continuum. Here we have investigated mRNA expression in murine lineage negative Stem Cell Antigen-1 positive stem cells of a variety of cell surface epitopes and transcription regulators associated with stem cell identity or regulation. At isolation these stem cells expressed almost all cell surface markers, and transcription factors studied, including receptors for G-CSF, GM-CSF and IL-7. When these stem cells were induced to transit cell cycle in vitro by exposure to interleukin-3 (IL-3), Il-6, IL-11 and steel factor some (CD34,CD45R c-kit, Gata-1, Gata-2, Ikaros and Fog) showed stable expression over time, despite previously documented alterations in phenotype, while others showed variation of expression between and within experiments. These included Sca-1, Mac-1, c-fms and c-mpl. Tal-1,endoglin and CD4 showed variation between experiments. These studies indicate that defined marrow stem cells express a wide variety of genes at isolation and with cytokine induced cell cycle transit show marked and reversible phenotype lability. Altogether, the phenotypic plasticity of gene expression for murine stem cells indicates a continuum model of stem cell regulation and extends the model to reversible expression with cell cycle transit of mRNA for cytokine receptors and stem cell markers.
Keywords: Stem Cell, Phenotype, Hematopoiesis, Cell Cycle, Gene Expression, Plasticity
Introduction
A tenet of experimental marrow stem cell biology is that the process of producing hematopoietic cells involves “the sequential commitment of multipotential hematopoietic stem cells to gradually more restricted progenitor cells, and finally to the functionally distinct cells of mature blood, including red blood cells, platelets, neutrophils, eosinophils, basophils, monocytes, macrophages and lymphocytes” (1). An alternative model of hematopoiesis, suggests that at the stem progenitor cell level, rather than sequential commitment, there exists continually changing windows of transcriptional opportunity, related to stem cell cycle phase and chromatin alterations associated with cell cycle transit (2-6). In this model the stem cell represents different reversible functional states of the same cell.
The traditional hierarchical model is based on a significant body of experimental work (7-9). Early kinetic studies indicated sequential differentiation with loss of proliferative potential in the differentiated erythroid and granulocytic pathways. Myeloblasts differentiated into promyelocytes, which in turn differentiated into myelocytes. All these cells were shown to proliferate by tritiated thymidine labeling. There then followed a nonproliferative maturation sequence with metamyelocytes, bands and polymorphonuclear granulocytes (10). The erythroid pathway showed a similar sequence from proerythroblasts to basophilic erythroblast to polychromatic erythroblast to orthochromatic erythroblast and reticulocytes.
The definition of granulocyte-macrophage progenitors by in vitro clonal culture indicated the existence of a more primitive cell committed to these two lineages (11,12). It was natural to assume that this granulocyte-macrophage progenitor had differentiated from the multipotent colony forming unit spleen (CFU-S) (13). The progenitor was more rapidly cycling than the multipotent cell and was relatively lineage restricted (14,15). However, attempts to physically isolate these two cell types were unsuccessful. Further support for a hierarchical stem cell model derived from characterization of in vivo stem cells with short and long-term repopulation capacity separated on the basis of rhodamine staining (16). However, elegant experiments by Ogawa and coworkers indicated that daughter cells of a single early progenitor stem cell could pursue totally different lineage choices (17-20). Thus, different pathways such as erythrocyte/megakaryocyte versus granulocytes/macrophages could be chosen during one cell cycle transit. This was evidence against an ordered hierarchy. Furthermore, Snodgrass and Keller (21) transplanted retrovirally labeled marrow cells and showed that individual clones could change their differentiation phenotypes over time post- transplantation. In a similar vein, when single lineage-defined progenitors selected from early murine colony starts were grown under permissive conditions and analyzed by single cell RT-PCR, heterogeneous expression of the basic helix-loop-helix (bHLH) transcription factors, SCL, Lyl-1, E12/E47 and Id-1 were seen (22). Some progenitors for a particular lineage expressed the factors, while others did not. In 9 instances, two sister cells from the same developing colony were sampled; discordance in gene expression was seen for Id (56%), E12 (22%) and SCL (44%). These data did not indicate an ordered transcriptional hierarchy. These data also ruled against a straightforward hierarchical model of hematopoiesis.
The most primitive murine marrow stem cells are all either continually progressing through cell cycle at a relatively slow rate or intermittently exiting and entering cycle. The cycling nature of the marrow stem cell was definitively shown by Bradford and colleagues (23) with continuous oral administration of BrdU to mice. They isolated primitive lineage negative, rhodamine-low and Hoechst-low (LRH) stem cells over the time of the feeding schedule and demonstrated 60% BrdU labeling by week 4 and estimated a T1/2 for labeling of 19 days. This work was confirmed by two other groups (24,25). In addition, primitive stem cells are easily induced into a high cycling rate in vitro or in vivo (26,27). Further work has indicated that the functional phenotype of the stem cell changes reversibly with cell cycle transit (28-30). Studies have shown that 8 week or 6 month engraftment of unseparated marrow or purified marrow stem cells reversibly change with IL-3, IL-6, IL-11 and steel factor cytokine-induced cycle passage, and that marked alterations can occur at 2−4 hour intervals. The engraftment defect appears to be related to a homing defect (31) which in turn may be related to alterations in various adhesion proteins, particularly VLA4 (32,33). Global gene expression of LRH stem cells exposed to IL-3, IL-6, IL-11 and steel factor in vitro, as determined by differential display of 3’ end restriction fragments of cDNA was reversed as to intensity at time points in culture when homing and engraftment were decreased (34).
An alternative cytokine cocktail, TPO, Flt3 and steel factor, while giving somewhat different kinetics of LRH cycle transit, showed similar reversible cycle related changes in engraftment. In addition reversible increases in progenitor number were demonstrated and in general increases in progenitors were tied to decreases in engraftable stem cells, a phenomenon termed stem/progenitor cell inversion (35).
LRH cells stimulated to transit cell cycle and then subcultured in a set cytokine cocktail (G-SCF, GM-CSF and steel factor) showed impressive reversible differentiation hotspots; megakaryocyte differentiation was induced at early S phase and granulocyte differentiation later (36). Other studies have indicated that marrow cells, cycling in response to IL-3, IL-6, IL-11 and steel factor demonstrate cycle related points of increased homing to lung and differentiation to lung cells (37). Thus the functional phenotype of unseparated marrow or highly purified murine marrow stem cells shows marked variability as the cells transit cell cycle. These results have laid the basis for a continuum model of hematopoiesis in which the phenotypic potential of stem cells is constantly changing over time in relations to cell cycle phase or even circadian rhythm. A key to this model is that these changes are not unidirectional but rather reversible. The model proposes that at the stem/progenitor cell level there is a continuum of potential, until the cells are exposed to specific inducers at the appropriate window of transcriptional potential. This model indicates that there is not a hierarchy at the stem/progenitor cell level (Figures 2-6).
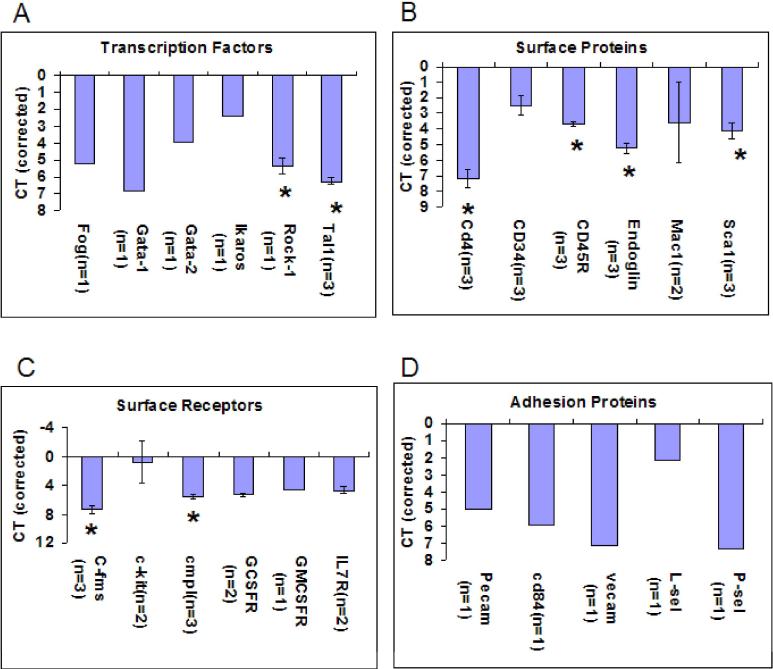
Figure 2. Gene Expression at Isolation (0Hours).
Cycle threshold (CT) values at 0hour Lin-Sca-1+ isolation. CT values are corrected for endogenous control (Beta-2 microglobulin). The X-axis is inverted to demonstrate the relationship to expression. The higher the number of cycles to detect mRNA, the lower the mRNA expression which is directly related to the CT value. Thus points near zero represent the highest mRNA expression. Higher CT values indicate less expression. Expression of mRNA in this stem cell population is analyzed by looking at (A) Transcription Factors (B) Surface Proteins (C) Surface Receptors and (D) Adhesion Proteins. Error bars indicate standard error between the experiments with the n value indicating the number of experiments analyzed. Significant difference (*) was analyzed with Student's t-test ( 2-tailed) and p<.005 is indicated with these comparisons: Rock1 to Tal1 (Figure 2A); CD4 to CD3, CD45R, Endoglin & Sca1 (Figure 2B); CD45R to Endoglin & Sca1 (Figure 2B); Endoglin to Sca1 (Figure 2B); C-fms to cmpl (Figure 2C).
Figure 6. Stem Cell Continuum Model.
This model shows promoter regions, interactions with chromatin, transcriptional regulation and proposed alterations and interactions with cell cycle progression and inducer exposure. In order for expression to occur, the complementing transcription complex must form with the promoter region. External factors such as chromatin, correlating to the specific phase of cell cycle can block this alignment. This is both a stochastic and deterministic model of gene expression.
The stem cell has been placed in an ordered hierarchy by its cell surface markers, cytokine receptors and transcription factors (38-44). This is not consistent with a continuum as outlined above. Accordingly, in the present studies we have examined mRNA expression for selected transcription factors, cytokine receptors, and cell surface markers which have been implicated as defining stem cells. Our hypothesis is that the genetic profile of early stem cells will show cycle related reversible fluctuations.
Materials and Methods
Animals
C57BL/6 for lineage negative, stem cell antigen-1 positive (Lin-Sca-1+) population studies were obtained from Taconic Farms (Germantown, NY). All animals were housed in micro-insulator cages, in a conventional clean facility for at least one week prior to experimental use. The animals in this Animal Care Committee approved study were maintained in accordance with the guidelines of the Institutional Animal Care and Use Committee of Roger Williams Medical Center and recommendations in the Guide for the care and use of laboratory animals (45). All animals were 6−8 weeks of age at the time of bone marrow harvest.
Isolation of Whole Bone Marrow
Six to 8-week old male mice were sacrificed. Bone marrow was collected from femurs, tibiae, iliac crests and spines for Lin-Sca-1+ isolation by grinding bones in phosphate-buffered saline (PBS) supplemented with 5% heat inactivated fetal calf serum (HI FCS); Hyclone, Logan, VT) and 1% penicillin/streptomycin (P/S) (Life /Technologies/Gibco/BRL) using a mortar and pestle. The bone fragments were washed multiple times and the supernatant cell suspension and wash fractions filtered through a 40-μm filter (Becton Dickinson, Franklin Lakes, NJ) to remove large bone particles. High lipid concentrations were reduced by centrifugation and re-suspension of the cells in fresh buffer. The cells were incubated at 4 degrees Celsius for 5 minutes, so that small bone particles could settle out. The cell supernatant, depleted of these fragments, was then diluted to 107 cells/ml PBS with 5% HI FCS and 1% Penicillin/Streptomycin (PBS buffer).
Lin−Sca-1+ Hematopoietic Stem Cell (HSC) Purification
Isolation of lineage negative, stem cell antigen-1 positive (Lin-Sca-1+) purified stem cells was obtained using bone marrow isolated from the iliacs, femurs, tibiae and spine of C57BL/6 mice 6 to 8 weeks of age. A low-density fraction (1.320±.001 g/ml) was isolated on Optiprep (Accurate Chemical and Scientific Corporation, Westbury, NY). The cells were lineage depleted with the following primary rat antibodies: anti-B220, anti-MAC-1, anti-GR1, anti-Lyt-2, anti-L3T4, and Ter119 (BD PharMingen, San Diego, CA). Each batch of antibody was evaluated by flow cytometry analysis for the concentration, which resulted in the greatest shift in mean channel fluorescence and/or the greatest percentage of positive cells detected. The optimal dilution for each antibody was at a final concentration of 0.1ug/106 cells ( 0.5ug/106 cells for GR-1). After a 15-minute incubation on ice, the labeled cells were washed in 1× Dulbecco's Phosphate-buffered saline, without calcium or magnesium chloride (PBS) (Invitrogen Corp., Carlsbad, CA), 5% heat-inactivated fetal calf serum (HI FCS) (Hyclone, Logan, UT) and resuspended in PBS buffer. The cells were incubated with washed sheep anti-rat IgG conjugated immunomagnetic polystyrene spheres (M-450 Dynabeads; Dynal, Lake Success, NY) at 4°C for 20 minutes by adding beads in a drop-like fashion to obtain a 1:5 bead to cell ratio. The beads were suspended in PBS buffer and, when added to the cells, resulted in 1.5 times the original cell volume. After the 20 minute incubation, immunomagnetic bead-rosetted cells were removed using a magnetic particle concentrator (Dynal, MPC-6), and the unrosetted cells remaining in suspension were harvested by pipette (46). Immunomagnetically lineage depleted cell suspensions were washed and resuspended at cell concentrations of 3 to 5 × 106 cells per ml in PBS buffer and incubated with biotin conjugated rat anti-mouse Ly6A/E (stem cell antigen-1) monoclonal antibody (Pharmingen, SanJose, CA) at a concentration of 1 ug/106 cells on ice for 20 minutes. The cells were washed twice in ice-cold PBS buffer, resuspended at a cell concentration of 3 to 5 × 106 per ml in PBS buffer and then incubated at 1μg streptavidin, allophycocyanin crosslinked conjugate (Molecular Probes, Carlsbad, CA) per million cells for 20 minutes on ice. Aliquots of the bead-free cells were also stained with isotype-matched immunoglobulin. The cells were then washed with ice-cold PBS buffer and resuspended in PBS containing 1 μg/ml propidium iodide (PI; Sigma Chemical Co., St. Louis, MO). The sample was filtered through a 35 μm small blue cap filter (Falcon Cat # 2235). Fluorescence-activated cell sorting was performed on a MoFlo cell sorter (Cytomation, Fort Collins, CO). Cells were sequentially selected for sorting as PI negative, FITC negative and Sca-1 positive (Figure 1B).
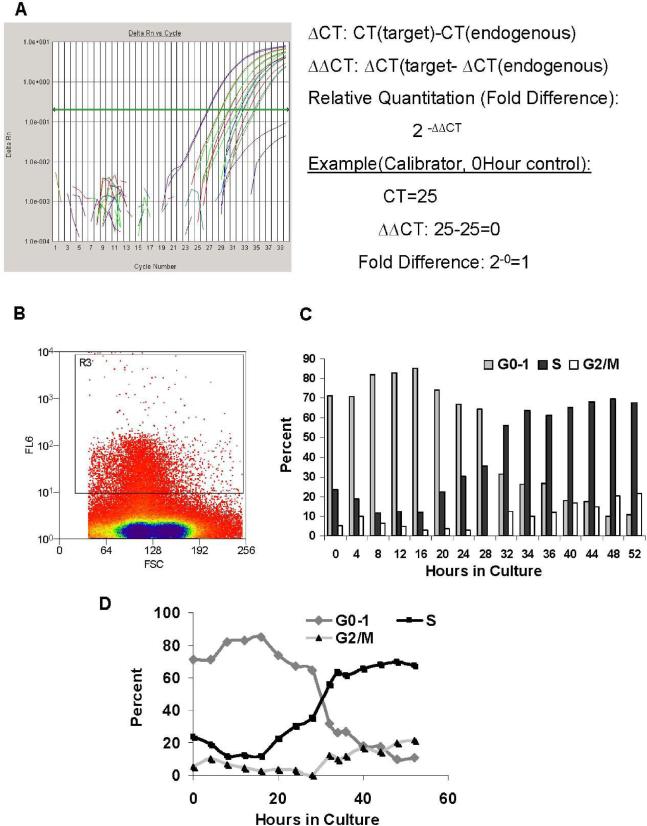
Figure 1. Methods. (A) Quantitation of Gene Expression.
Gene expression was calculated, using Applied Biosystem's Delta, Delta CT (ΔΔCT) method from results obtained on the ABI 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). Relative quantitation (RQ) or “fold difference” is represented in relation to the 0Hour, non-cultured control. (B) Lin−Sca-1+ Histogram (sort). Cells were sorted based on Sca-1+ characteristics. FL6= Allophycocyanin; FSC=Forward Scatter. (C) Cell cycle Kinetics: Lin−Sca-1+ Propidium Iodide Treatment Through Cell Cycle in IL-3, IL-6, IL-11, SCF (Bar Graph). Cycling status of the cells was analyzed at different times in culture by flow cytometry of propidium iodide labeled cells. (D) Cell cycle Kinetics: Lin−Sca-1+ Propidium Iodide Treatment Through Cell Cycle in IL-3, IL-6, IL-11, SCF (Line Graph). Cycling status of the cells was analyzed at different times in culture by flow cytometry of propidium iodide labeled cells
Lin-Sca-1+ cultures
Lin− Sca-1+ marrow cells were established in Teflon (non-adherent) bottle cultures with DMEM + 15% HI FCS+ 1% P/S + 1% L-glutamine and cytokines at the following concentrations: IL-3, 50U/ml, IL-11, 50ng/ml, IL-6, 50ng/ml and steel factor 50ng/ml. Cells were cultured at 1×106cells/ml in Teflon bottles at 37°C, 5% CO2 in a humidified water-jacketed incubator (Forma Scientific, Marietta, OH). Gene expression was evaluated immediately (time 0, before culture), and in different experiments at 2 ,3, 4, 8, 16, 22, 23, 24, 25, 32, 40 and 48 hours.
Cell Cycle Evaluation with Propidium Iodide Staining
Lin− Sca-1+ populations were analyzed for cell cycle status at different time points in culture under the same conditions previously stated. Cellular DNA was labeled with propidium iodide (Stain DNA Assay PI (RPI-2), Nova Century Scientific) and percent of cells stained was analyzed on a MoFlo cell sorter (Cytomation, Fort Collins, CO) to assess the cycling status of the cells at different times in culture (Figure 1C).
Real Time RT-PCR
Total RNA was isolated from cultured cells, using PicoPure RNA Isolation Kit (Arcturus Mountain View, CA). Real Time reverse transcription-polymerase chain reaction (Real Time RT-PCR) was performed in a two-step reaction. Reverse transcription was achieved with the TaqMan® RT Reagents kit (Applied Biosystems, cat.no. N8080234, Foster City, CA) and the PCR reaction was performed, using samplings from the same cDNA aliquot for multiple genes. Reactions for each specific gene were performed in triplicates to ensure reliability of data. Applied Biosystem's pre-developed, TaqMan® Gene Expression Assays were used in the PCR reaction for gene expression analysis. The TaqMan® assay is a 20× concentrated nucleotide mix which includes 2 unlabeled PCR primers (900nM each final concentration) and 1 FAM™ dye-labeled TaqMan® MGB probe (250nM final concentration). The PCR reaction was carried out with the 2× TaqMan® PCR mastermix (Applied Biosystems, cat.no. 4304437, Foster City, CA) and 20× assay mix in 25μl reactions in the ABI7000 Sequence Detection System (Applied Biosystems, Foster City, CA) on the same thermal profile of 50°C×2min., 95°C×10min., 40cycles (95°C×15sec., 60°C×1min.). Beta-2-microglobulin (Applied Biosystems assay no. Mm00437762_m1) was used as an endogenous control in conjunction with genes of interest. RNA was analyzed for surface expression of FOG (Applied Biosystems assay no. Mm00494336_m1), Gata-1 (Applied Biosystems assay no. Mm00484678_m1), Gata-2 (Applied Biosystems assay no. Mm00492300_m1), Ikaros (Applied Biosystems assay no. Mm00456421_m1), Rhodamine-associated coiled-coil forming kinase 1 (Rock-1) (Applied Biosystems assay no. Mm00485745_m1), CD4 (Applied Biosystems assay no. Mm00442754_m1), Pu-1 (Applied Biosystems assay no. Mm00488140_m1), cfms (Applied Biosystems assay no. Mm00432689_m1), Pecam (Applied Biosystems assay no. Mm00476702_m1), P-selectin (Applied Biosystems assay no. Mm00441295_m1), CD84 (Applied Biosystems assay no. Mm00488934_m1), c-kit (Applied Biosystems assay no. Mm00445212_m1), Vcam1 (Applied Biosystems assay no. Mm00449197_m1), L-selectin (Applied Biosystems assay no. Mm00441291_m1), Endoglin (Applied Biosystems assay no. Mm00468256_m1), Tal-1 (Applied Biosystems assay no. Mm00441665_m1), CD34 (Applied Biosystems assay no. Mm00519283_m1), CD45R (Applied Biosystems assay no. Mm00448463_m1), c-mpl (Applied Biosystems assay no. Mm00440310_m1), Flt3 (Applied Biosystems assay no. Mm00438996_m1), G-CSFR (Applied Biosystems assay no. Mm00432735_m1), GM-CSFR (Applied Biosystems assay no. Mm00438331_g1), Mac-1 (Applied Biosystems assay no. Mm00434455_m1), Sca-1 (Applied Biosystems assay no. Mm00485928_m1) and IL7R (Applied Biosystems assay no. Mm00434295_m1).
Quantitation of Gene Expression
Gene expression was calculated in relation to a calibrator or non-treated sample (0 hour control), using the cycle threshold (CT) values and calculating relative quantitation (RQ) or “fold difference” with Applied Biosystems's “delta, delta CT” method (ΔΔCT). Each sample was corrected with beta-2-microglobulin as an endogenous control and then normalized against the calibrator sample. Samples with endogenous CT values 35 or greater in a 40 cycle reaction were discarded due to poor quantity of RNA. Fold difference is represented as 2−ΔΔCT, with the calibrator sample being equal to one (Figure 1A).
Statistical Analysis
Statistical analysis was performed on data when three or more experiments were averaged together. Significant difference of gene expression was analyzed with Student's t-test (2-tailed analysis) and p values less than .005 were noted with an asterisk (Figure 2).
Results
The quantitation of gene expression using Applied Biosystem's ΔΔCT method is shown in Figure 1A.
Beta-2 microglobulin was an endogenous housekeeping correction. The levels of expression are presented as CT minus endogenous control. The more positive the value the lower the expression. The Lin-Sca-1+ histogram from the FACS separation is presented in Figure 1B and the cell cycle characteristics as determined by propidium staining and flow cytometry is seen in Figure 1C. In Figure 2 we present baseline values at time 0 of selected transcription factors, adhesion proteins ,surface proteins and surface receptors.
Not surprisingly c-kit and CD34 were the most highly expressed, but a striking feature was the wide spread expression of many selected transcription factors, surface proteins and surface receptors. Of particular interest were the expression of MAC-1 and c-fms, markers felt to characterize differentiated cell populations. mRNA for the IL-7R was also seen along with expression of mRNA for G-CSF and GM-CSF receptors.
In evaluating the changes in gene expression over time correlated with time after cytokine exposure and cell cycle phase we studied transcription factors, cell surface receptors, surface proteins and adhesion proteins.
Studies at 2, 3, 4, 8, 16, 22, 24, 25, 32, 40 and 48 hours of culture in IL-3, IL-6, IL-11 and steel factor showed stability of expression of CD34, CD45R, c-kit, Gata-1, Gata-2, Ikaros, Fog and several cell surface receptors and adhesion proteins. (Table 1)
Changes Through Cycle
| Gene | Changes Through Cycle | Number of Experiments |
|---|---|---|
| CD34 | no change | 3 |
| CD45R | no change | 3 |
| ckit | no change | 3 |
| IL7R | no change | 3 |
| GMCSFR | no change | 1 |
| GSCFR | no change | 2 |
| Fog | no change at 24H, 48H | 2 |
| Gata-1 | no change at 24H, 48H | 2 |
| Gata-2 | no change at 24H, 48H | 2 |
| IKAROS | no change at 24H, 48H | 2 |
| Psel | no change | 2 |
| Pecam | no change | 2 |
| Vecam | no change | 1 |
| Lsel | no change | 1 |
| CD84 | no change | 1 |
| Rock-1 | no change at 24H, 48H | 1 |
In different experiments, the following times were assessed: Hours (2,3,4,8,16,22,24,25,32,40,48)
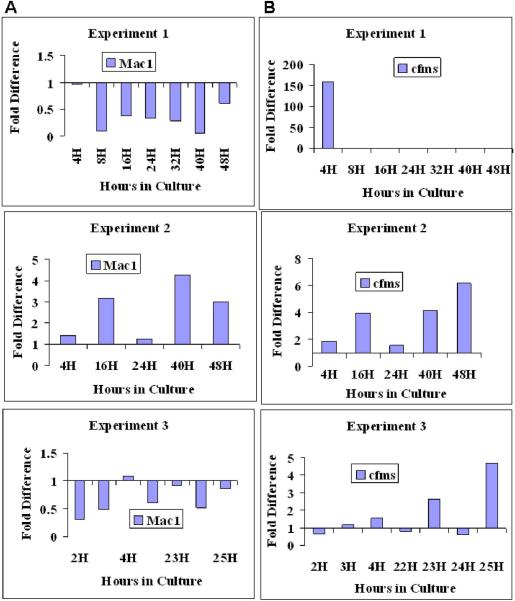
We were interested in gene expression which varied both within and between experiments. In Figure 3 we see marked variances of Mac-1 mRNA expression between experiments at 16, 40 and 48 hours. Also noted is that in experiment 2 Mac-1 shows increased expression at 16 hours with decreased expression at 24 hours followed by an increase in expression at 40 hours (Figure 3A). Thus there is lability of mRNA expression at certain time intervals. A putative differentiation marker for macrophages is c-fms, and there is variation between experiments at 4, 16, 40 and 48 hours and in experiment 2 we see increases in gene expression at 16 and 40 hours, but not at 24 hours representing reversible fluctuation in expression (Figure 3B).
Figure 3. Gene Expression Through Cell Cycle: Lin-Sca-1+ in Culture with IL-3, IL-6, IL-11, SCF.
Expression patterns in individual experiments are represented using quantitation methods shown in Figure 1. Fold difference is expressed in relation to 0 hour sample (calibrated to a value of 1). The 0 hour control is represented as a baseline of 1 at the X axis. (A) Mac-1 expression through cycle. (B) cfms expression through cycle.
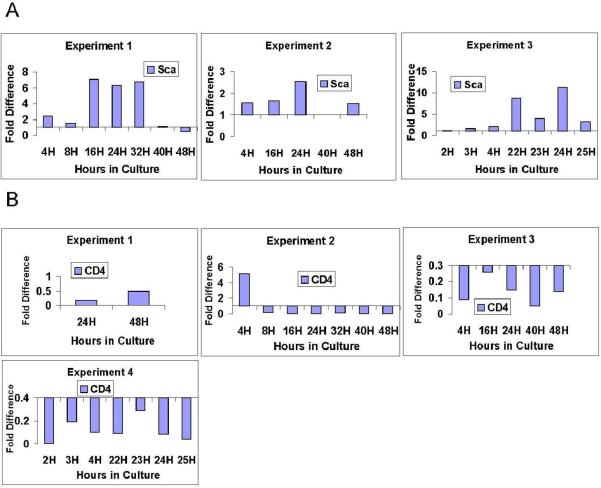
CD4 marks both T cells and early stem/progenitor cells. CD4 expression varies between experiments at 4 hours (Figure 4). The stem cell marker Sca-1, used for separation in these experiments, varied between experiments at 16 hours and showed reversible fluctuations in experiment 1 and 3 (Figure 4).
Figure 4. Gene Expression Through Cell Cycle: Lin-Sca-1+ in Culture with IL-3, IL-6, IL-11, SCF.
Expression patterns in individual experiments are represented using quantitation methods shown in Figure 1. Fold difference is expressed in relation to 0 hour sample (calibrated to a value of 1). The 0 hour control is represented as a baseline of 1 at the X axis. (A) Sca1 expression through cycle. (B) CD4 expression through cycle.
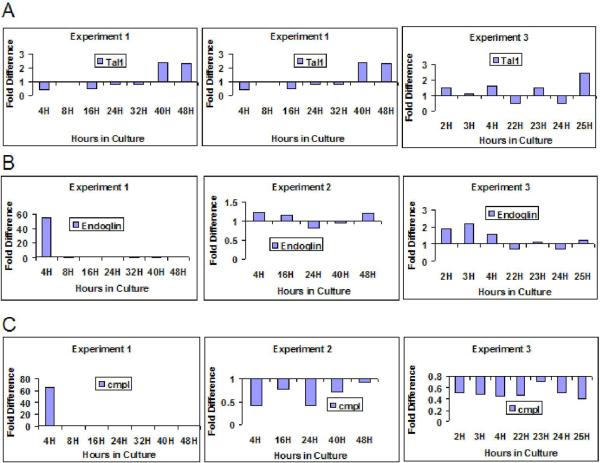
In Figure 5 we present the stem cell surface markers cmpl and endoglin along with the putative stem cell transcription factor Tal-1. Expression of cmpl varied between experiments at 4 hours and showed reversible fluctuations in experiments 2 and 3. Endoglin expression also varied between experiments at 4 hours, but otherwise showed minimal fluctuations at the different time points assessed. The putative stem cell transcription factor Tal 1 showed a variation between experiments at 16 hours.
Figure 5. Gene Expression Through Cell Cycle: Lin-Sca-1+ in Culture with IL-3, IL-6, IL-11, SCF.
Expression patterns in individual experiments are represented using quantitation methods shown in Figure 1. Fold difference is expressed in relation to 0 hour sample (calibrated to a value of 1). The 0 hour control is represented as a baseline of 1 at the X axis. (A) Tal1 expression through cycle. (B) Endoglin expression through cycle. (C) cmpl expression through cycle.
These data indicate that expression of a variety of stem or differentiation specific genes shows variation between experiments and reversible fluctuations within experiments tied to time after cytokine exposure and cell cycle phase.
Discussion
The definition of hematopoietic stem cells and the regulation of their differentiation has been extensively investigated and many defining molecular characteristics have been identified.
Surface markers and transcription regulators have been highlighted. Gata-1, Gata-2 and Fog have been implicated in erythroid and megakaryocyte lineage determination, PU.1 and Ikaros in myeloid and lymphoid regulation, respectively (38). Hierarchical models have been constructed in which transcriptional regulators sequentially determine lineage differentiation. The regulatory picture is, of course, more complicated. While elegant knockout and knock-in studies have indicated relative dependence for different lineage pathways on these factors, the results have not been totally concordant. In addition, it is clear now that most transcription factors cooperatively interact with multiple other factors to exert their biologic effects. Recently increasing note has been made of cross-antagonism of lineage-specific transcription factors (1). Gata-1 blocks the interaction of PU.1 with its cofactor c-JUN antagonizing the action of PU.1 (39-41), while PU.1 represses Gata-1 by disrupting its capacity to bind to DNA (42). Similarly, Fog expression is downregulated at the transcriptional level with C/EBP-β mediated differentiation of avian multipotent hematopoietic progenitors to eosinophils and Fog represses E12/E47 expression. Thus transcriptional regulation of hematopoiesis is characterized by involvement of multiple transcription factors which can act either cooperatively or antagonistically by a variety of mechanisms.
In a similar fashion different cell surface determinants have been felt to characterize the “stemness” of cell populations (47-52). These have included CD34, cmpl, endoglin, and Sca-1. Recent work has also indicated that CD4 and Mac-1 while prominently expressed on T cells and macrophages/granulocytes also mark early stem cell populations.
We first analyzed the relative expression of different genes in Lin- Sca-1+ cells at isolation (time 0). We have utilized B2-microglobulin as an endogenous standard. Lin-Sca-1+ murine stem cells are a “cycling” population of stem cells, as are most other defined stem cells classes. Perhaps only the LRH murine marrow stem cells are isolated as a noncycling population, although as noted, above, they then proceed over time to transit cell cycle. The cycle characteristics of Lin-Sca-1+ cells are shown in Figure 1, in a separative histogram. Also shown is the progression of these cells through cell cycle under stimulation with IL-3, IL-6, IL-11, and steel factor. At isolation (0 hour), approximately 70% are in G0/G1 with over 20% in S and about 5 % in G2/M. Virtually every factor analyzed was found present in one or many experiments, consistent with other work indicating that early stem cells show expression of a wide variety of genes (41).
We also have characterized gene expression in Lin-Sca-1+ murine stem cells driven through cell cycle by exposure to IL-3, IL-6, IL-11, and steel factor There are particular points of interest in the situations in which gene expression changes over time and also when there are no changes over time. In Table 1 we show that the transcription factors Fog, Gata-1, Gata-2, Ikaros were stable over a limited number of time points but points at which clear changes in stem cell engraftment and differentiation phenotype have been previously reported. In other studies we have shown changes in Fog, nfe2 and Fli1 at a point of megakaryocyte differentiation potential (36) and here we show variation in the expression of Tal 1 with cycle transit (Figure 5). Thus expression of particular transcription factors may or may not correspond to the functional phenotype of a marrow stem cell. The complexity of the transcriptional regulation of hematopoiesis has recently been emphasized. In addition it is clear that definition of the action of transcription factors by developmental or knockout experiments can be misleading as in the case of Tal-1/SCL which was critical during development but appeared to play a minor role in stem cell biology in adult animals (53). A number of other studies have suggested caution in assigning specific roles to various transcription factors. The present data is consistent with a continually changing gene expression with cycle passage of stem cells probably tied to cell cycle. In this context, specific transcription factors would be acting cooperatively with a number of other factors in the context of cell cycle phase. Only if the proper inducer and chromatin configuration were present at a time of intrinsic or induced transcriptional factor expression would a specific differentiation event ensue (Figure 6).
The schemas of a well ordered differentiation cascade characterized by an ordered sequence of transcription factor (or cell surface marker expression, vide infra) is probably not correct. Rather a highly plastic interactive continuum is envisioned with extraordinary differentiation potential when exposed to the right inductive influence.
The surface markers mac-1, c-fms, Sca and CD4 all showed marked inter-experiment variability. They also show time periods in which reversible alterations of expression were seen. These changes suggest that specific stem cell surface markers are also labile and only define stem cells under certain specific circumstances. This concept is presented in Figure 7.
Figure 7. Fluctuating Surface Phenotype of Stem Progenitor Cells with Cell Cycle Progression.
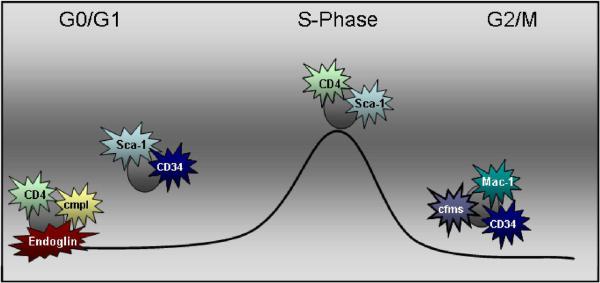
This model demonstrates fluctuations in stem cell phenotype through cell cycle.
Ogawa and colleagues have previously shown that CD34 varies with the activation state of the cell, and others have raised question of whether CD34 positive or negative cells are the true stem cell 54−56). Our present data indicate that some the stem cell surface epitopes vary reversibly with cell cycle. This may explain the wide variety of reported stem cell phenotypes. Work by Spangrude and colleagues (57) clearly showed that after transplantation, the Lin− c-kit+ Sca-1+ phenotype no longer identified the true stem cell. Thus, while helpful, under many circumstances, the nature of the stem cell is not defined by its surface epitopes, but rather only by its specific functional state.
Altogether, these findings are consistent with the continuum model of stem cell regulation described above (Figures 2-6)and indicate that transcription factors, cell surface markers and stem cell markers vary on a cell cycle/cytokine related continuum, but that this variability may not be consistent between experiments carried out at different time points.
Striking features of the above data are the generality of expression of virtually any genes tested, the heterogeneity of expression in freshly isolated stem cells and the varying modulation seen over time in cytokine stimulated cultures.
Reproducibility of Results
These discrepant results can be viewed as reflecting intrinsic and temporal heterogeneity with differences expressed over short time intervals or differences seen at a larger time scale of days, weeks or months possibly relating to circadian rhythm. Each experiment was internally checked by using a housekeeping gene (beta-2 microglobulin) as an endogenous correction control and running samples on triplicates. Furthermore, genes with CT values of endogenous control at 35 or greater in a 40 cycle reaction were discarded due to poor RNA quality. CT values 35 or greater on target genes were regarded as not expressed. Thus the resultant data is reproducible within each experiment. We propose that the variability between experiments represents the true state of the biology of marrow stem cells and the influence of many experimental variables some of which are known such as circadian variations occurring over time frames of hours or over longer and varied time frames. Cell cycle variations possibly at relatively short time intervals could also explain the variability, but it must be acknowledged that there are probably many more variables of which we are at present unaware.
Lastly, the variable expression of c-fms, a macrophage marker with reversible elevations at particular points in culture is intriguing. One possibility is that this so-called promiscuous gene expression is unrelated to biological outcomes. Alternatively, these points could represent points in time where a stem cell is open to a particular fate or even approaches a specific differentiated phenotype.
Conclusion
Altogether, the phenotypic plasticity of gene expression for purified murine stem cells indicate a continuum model of stem progenitor cell regulation and extends the stem cell continuum model to reversible expression with time in cytokines and cycle transit to cytokine receptors and stem cell markers.
Acknowledgements
We would like to thank Sandra Bibby for her administrative support.
Funding Sources:
Contract Grant Sponsor: National Institute of Health
Contract Grant Numbers: P20 RR018757, R01 HL73749
Contract Grant Sponsor: National Institute of Diabetes and Digestive and Kidney Diseases
Contract Grant Numbers: DK61858, K08 DK064980, PO-1 DK50222, PO-1 HL56920.
References
- 1.Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–519. doi: 10.1016/s0959-437x(00)00226-4. [DOI] [PubMed] [Google Scholar]
- 2.Quesenberry PJ, Colvin GA, Lambert JF. The chiaroscuro stem cell: A unified stem cell theory. Perspective, Blood. 2002;100:4266–4271. doi: 10.1182/blood-2002-04-1246. [DOI] [PubMed] [Google Scholar]
- 3.Quesenberry PJ, Colvin GA, Abedi M, Cerny J, Dooner M, Moore B, McAuliffe C, Demers D, Greer D, Parent A, Badiavas E, Lum L, Falanga V. The Marrow Stem Cell: The Continuum. Bone Marrow Transplantation. 2003;32:S19–22. doi: 10.1038/sj.bmt.1703938. [DOI] [PubMed] [Google Scholar]
- 4.Quesenberry PJ, Dooner G, Dooner M, Colvin G. The Stem Cell Continuum: Considerations on the Heterogeneity and Plasticity of Marrow Stem Cells. Stem Cell. 2004 doi: 10.1385/SCR:1:1:029. Review. [DOI] [PubMed] [Google Scholar]
- 5.Quesenberry PJ. The continuum model of stem cell regulation. Curr Opin Hematol. 2006;13:216–221. doi: 10.1097/01.moh.0000231417.08031.ac. [DOI] [PubMed] [Google Scholar]
- 6.Quesenberry PJ, Colvin G, Dooner G, Dooner M, Aliotta JM, Johnson K. The Stem Cell Continuum: Cell Cycle, Injury and Phenotype Lability. Ann N Y Acad Sci. 2007 doi: 10.1196/annals.1392.016. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 7.Quesenberry PJ, Levitt L. Hematopoietic stem cells. New Engl J Med. 1979;301(14):755–761. doi: 10.1056/NEJM197910043011404. Review. [DOI] [PubMed] [Google Scholar]
- 8.Quesenberry PJ, Levitt L. Hematopoietic stem cells (second of three parts). New Engl J Med. 1979;301(15):819–23. doi: 10.1056/NEJM197910113011505. 1979. Review. [DOI] [PubMed] [Google Scholar]
- 9.Quesenberry PJ, Levitt L. Hematopoietic stem cells (third of three parts). New Engl J Med. 1979;301(16):868–72. doi: 10.1056/NEJM197910183011605. Review. [DOI] [PubMed] [Google Scholar]
- 10.Cronkite EP. Hemopoietic stem cells: An analytic review of hemopoiesis. Pathobiol Annu. 1975;5:35–69. Review. [PubMed] [Google Scholar]
- 11.Bradley TR, Metcalf D. The growth of mouse bone marrow cells in vitro. Aust J Exp Biol Med Sci. 1966;44(3):287–299. doi: 10.1038/icb.1966.28. [DOI] [PubMed] [Google Scholar]
- 12.Pluznik DH, Sachs L. The cloning or normal “mast” cells in tissue culture. J Cell Physiol. 1965;66(3):319–324. doi: 10.1002/jcp.1030660309. [DOI] [PubMed] [Google Scholar]
- 13.Till JE, McCulloch EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–222. [PubMed] [Google Scholar]
- 14.Rickard KA, Shadduck RK, Howard DE, Stohlman F., Jr A differential effect of hydroxyurea on hemopoietic stem cell colonies in vitro and in vivo. Proc Soc Exp Biol Med. 1970;134(1):152–156. doi: 10.3181/00379727-134-34749. [DOI] [PubMed] [Google Scholar]
- 15.Iscove NN, Till JE, McCulloch EA. The proliferative states of mouse granulopoietic progenitor cells. Proc Soc Exp Biol Med. 1970;134(1):33–36. doi: 10.3181/00379727-134-34721. [DOI] [PubMed] [Google Scholar]
- 16.Bertoncello I, Hodgson GS, Bradley TR. Multiparameter analysis of transplantable hemopoietic stem cells: I. The separation and enrichment of stem cells homing to marrow and spleen on the basis of rhodamine-123 fluorescence. Exp Hematol. 1985;13(10):999–1006. [PubMed] [Google Scholar]
- 17.Ogawa M, Pharr PN, Suda T. Stochastic nature of stem cell functions in culture. Prog Clin Biol Res. 1985;184:11–19. [PubMed] [Google Scholar]
- 18.Nakahata T, Ogawa M. Hemopoietic colony-forming cells in umbilical cord blood with extensive capability to generate mono- and multipotential hemopoietic progenitors. J Clin Invest. 1982;70(6):1324–1328. doi: 10.1172/JCI110734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suda T, Suda J, Ogawa M. Single-cell origin of mouse hemopoietic colonies expressing multiple lineages in variable combinations. Proc Natl Acad Sci USA. 1983;80(21):6689–6693. doi: 10.1073/pnas.80.21.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suda T, Suda J, Ogawa M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci USA. 1984;81(8):2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snodgrass R, Keller G. Clonal fluctuation within the haematopoietic system of mice reconstituted with retrovirus-infected stem cells. EMBO J. 1987;6(13):3955–3960. doi: 10.1002/j.1460-2075.1987.tb02737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quesenberry PJ, Iscove NN, Cooper C, Brady G, Newberger PE, Stein GS, Stein JS, Reddy GP, Pearson-White S. Expression of Basic Helix-Loop-Helix Transcription Factors in Explant Hematopoietic Progenitors. J. Cell. Biochem. 1996;61:478–488. doi: 10.1002/(SICI)1097-4644(19960601)61:3%3C478::AID-JCB15%3E3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Bradford GB, Williams B, Rossi R, Bertoncello I. Quiescence, cycling, and turnover in the primitive hematopoietic stem cell compartment. Exp Hematol. 1997;25(5):445–453. [PubMed] [Google Scholar]
- 24.Cheshier SH, Morrison SJ, Liao X, Weissman IL. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96(6):3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang L, Reddy PV, McAuliffe CI, Colvin G, Quesenberry PJ. Studies on BrdU labeling of hematopoietic cells: stem cells and cell lines. J Cell Physiol. 2003;197(2):251–260. doi: 10.1002/jcp.10357. [DOI] [PubMed] [Google Scholar]
- 26.Reddy GP, Tiarks CY, Pang L, Wuu J, Hsieh CC, Quesenberry PJ. Cell cycle analysis and synchronization of pluripotenet hematopoietic progenitor stem cells. Blood. 1997;90:2293–2299. [PubMed] [Google Scholar]
- 27.Nilsson SK, Dooner MS, Quesenberry PJ. Synchronized cell-cycle induction of engrafting long-term repopulating stem cells. Blood. 1997;90(11):4646–4650. [PubMed] [Google Scholar]
- 28.Peters SO, Kittler EL, Ramshaw HS, Quesenberry PJ. Murine marrow cells expanded in culture with IL-3, IL-6, IL-11, and SCF acquire an engraftment defect in normal hosts. Exp Hematol. 1995;23(5):461–469. [PubMed] [Google Scholar]
- 29.Peters SO, Kittler EL, Ramshaw HS, Quesenberry PJ. Ex vivo expansion of murine marrow cells with interleukin-3 (IL-3), IL-6, IL-11, and stem cell factor leads to impaired engraftment in irradiated hosts. Blood. 1996;87(1):30–37. [PubMed] [Google Scholar]
- 30.Habibian HK, Peter SO, Hsie CC, Wuu J, Vergilis K, Grimaldi CI, Reilly J, Carlson J, Frimberger AE, Stewart FM. The fluctuating phenotype of the lymphohematopoietic stem cell with cell cycle transit. J Exp Med. 1998;188(2):393–398. doi: 10.1084/jem.188.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerny J, Dooner MS, McAuliffe CI, Habibian H, Stencil K, Berrios V, Reilly J, Carlson J, Cerny AM, d'Hondt L, Benoit B, Lambert JF, Colvin G, Nilsson S, Becker P, Quesenberry P. Homing of purified murine lymphohematopoietic stem cells: A cytokine-induced defect. J Hematother Stem Cell Res. 2002;11:913–922. doi: 10.1089/152581602321080574. [DOI] [PubMed] [Google Scholar]
- 32.Becker PS, Nilsson SK, Li Z, Berrios VM, Dooner MS, Cooper CI, Hsieh CC, Quesenberry PJ. Adhesion receptor expression by hematopoietic cell lines and murine progenitors: Modulation by cytokines and cell cycle status. Exp Hematol. 1999;27:533–541. doi: 10.1016/s0301-472x(98)00037-x. [DOI] [PubMed] [Google Scholar]
- 33.Berrios VM, Dooner GJ, Nowakowski G, Frimberger A, Valinski H, Quesenberry PJ, Becker PS. The molecular basis for the cytokine-induced defect in homing and engraftment of hematopoietic stem cells. Exp Hematol. 2001;29(11):1326–1335. doi: 10.1016/s0301-472x(01)00734-2. [DOI] [PubMed] [Google Scholar]
- 34.Lambert JF, Liu M, Colvin GA, Dooner M, McAuliffe CI, Becker PS, Forget BG, Weissman SM, Quesenberry PJ. Marrow stem cells shift gene expression and engraftment phenotype with cell cycle transit. J Exp Med. 2003;197(11):1563–1572. doi: 10.1084/jem.20030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colvin GA, Lambert JF, Moore BE, Carlson JE, Dooner MS, Abedi M, Cerny J, Quesenberry PJ. Intrinsic hematopoietic stem cell/progenitor plasticity: Inversions. J Cell Physiol. 2004;199:20–31. doi: 10.1002/jcp.10436. [DOI] [PubMed] [Google Scholar]
- 36.Colvin GA, Dooner MS, Dooner GJ, Sanchez-Guijo FM, Demers DA, Abedi M, Ramanathan M, Chung S, Pascual S, Quesenberry PJ. Stem cell continuum: directed differentiation hotspots. Exp Hematol. 2007;35:96–107. doi: 10.1016/j.exphem.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Dooner M, Cerny J, Colvin G, Demers D, Pimentel J, Greer D, Abedi M, McAuliffe C, Quesenberry P. Homing and conversion of murine hematopoietic stem cells to lung. Blood Cells Mol Dis. 2004;32:47–51. doi: 10.1016/j.bcmd.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Orkin SH. Transcription Factors and Hematopoietic Development. J Biol Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 39.Rekhtman N, Raparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription cofactors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang P, Behre G, Pan J, Iwama A, Wara-Aswapati N, Radonska HS, Auron PE, Tenen DG, Sun Z. Negative cross-talk between hematopoietic regulators: GATA-1 proteins repress PU.1. Proc Natl Acad Sci USA. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 42.Zhang P, Zhang X, Iwama A, Yu C, Smith KA, Mueller BU, Narravula S, Torbett BE, Orkin SH, Tenen DG. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. [PubMed] [Google Scholar]
- 43.Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002;298(5593):601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 44.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Research Council . Laboratory Animal Management: Rodents. National Academy Press; Washington, D.C.: 1996. pp. 141–146. [Google Scholar]
- 46.Boswell HS, Wade PM, Jr, Quesenberry PJ. Thy-1 antigen expression by murine high-proliferative capacity hematopoietic progenitor cells: Relation between sensitivity to depletion by Thy-1 antibody and stem cell generation potential. J Immunol. 1984;133(6):2940–2949. [PubMed] [Google Scholar]
- 47.Zhang CC, Lodish HF. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood. 2005;105:4314–4320. doi: 10.1182/blood-2004-11-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen CZ, Li L, Li M, Lodish HF. The endoglin (positive) sca-1 (positive) rhodamine (low) phenotype defines a near-homogeneous population of long-term repopulating hematopoietic stem cells. Immunitiy. 2003;19:525–533. doi: 10.1016/s1074-7613(03)00265-6. [DOI] [PubMed] [Google Scholar]
- 49.Broxmeyer HE, Cooper S, Lasky LA, De Sauvage F. Identificatrionof a massive reserve of hematopoietic progenitors in mice. Stem Cells Dev. 2005;14:105–110. doi: 10.1089/scd.2005.14.105. [DOI] [PubMed] [Google Scholar]
- 50.Holmes C, Stanford WL. Stem Cell Antigen-1: Expression, function and Enigma. Stem Cells. 2007:22. doi: 10.1634/stemcells.2006-0644. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 51.Ishida A, Zeng H, Ogawa M. Expression of lineage markers by CD34+ hematopoietic stem cells of adult mice. Exp Hematol. 2002;30:361–365. doi: 10.1016/s0301-472x(01)00795-0. [DOI] [PubMed] [Google Scholar]
- 52.Wineman JP, Gilmore GL, Gritzmacher C, Torbett BE, Muller-Sieburg CE. CD4 is expressed on murine pluripotent hematopoietic stem cells. Blood. 1992;80:1717–1724. [PubMed] [Google Scholar]
- 53.Mikkola HK, Klintman J, Yank H, Hock H, Schlaeger TM, Fujiwara Y, Orkin SH. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-leukaemia SCL/tal-1 gene. Nature. 2003;421(6922):547–551. doi: 10.1038/nature01345. [DOI] [PubMed] [Google Scholar]
- 54.Ogawa M, Tajima F, Ito T, Sato T, Laver JH, Deguchi T. CD34 expression by murine hematopoietic stem cells. Ann N Y Acad Sci. 2001;938:139–145. doi: 10.1111/j.1749-6632.2001.tb03583.x. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa M. Changing phenotypes of hematopoietic stem cells. Exp Hematol. 2002;30(1):3–6. doi: 10.1016/s0301-472x(01)00770-6. [DOI] [PubMed] [Google Scholar]
- 56.Zanjani ED, Almeida-Porada G, Livingston AG, Zeng H, Ogawa M. Reversible expression of CD34 by adult human bone marrow long-term engrafting hematopoietic stem cells. Exp Hematol. 2003;31(5):406–412. doi: 10.1016/s0301-472x(03)00051-1. [DOI] [PubMed] [Google Scholar]
- 57.Spangrude GJ, Brooks DM, Tumas DB. Long-term repopulation of irradiated mice with limiting numbers of purified hematopoietic stem cells: in vivo expansion of stem cell phenotype but not function. Blood. 1995;85(4):1006–1016. [PubMed] [Google Scholar]