Abstract
Within protozoa or human macrophages Legionella pneumophila evades the endosomal pathway and replicates within an ER-derived vacuole termed the Legionella-containing vacuole (LCV). The LCV membrane-localized AnkB effector of L. pneumophila is an F-box protein that mediates decoration of the LCV with lysine48-linked polyubiquitinated proteins, which is essential for intravacuolar replication. Using high-throughput LC–MS analysis, we have identified the total and ubiquitinated host-derived proteome of LCVs purified from human U937 macrophages. The LCVs harboring the AA100/130b WT strain contain 1193 proteins including 24 ubiquitinated proteins, while the ankB mutant LCVs contain 1546 proteins with 29 ubiquitinated proteins. Pathway analyses reveal the enrichment of proteins involved in signaling, protein transport, phosphatidylinositol, and carbohydrate metabolism on both WT and ankB mutant LCVs. The ankB mutant LCVs are preferentially enriched for proteins involved in transcription/translation and immune responses. Ubiquitinated proteins on the WT strain LCVs are enriched for immune response, signaling, regulation, intracellular trafficking, and amino acid transport pathways, while ubiquitinated proteins on the ankB mutant LCVs are enriched for vesicle trafficking, signaling, and ubiquitination pathways. The complete and ubiquitinated LCV proteome within human macrophages illustrates complex and dynamic biogenesis of the LCV and provides a rich resource for future studies.
Keywords: Legionnaires’ disease, AnkB, Dot/Icm, polyubiquitin, LCV, proteome, signaling
Introduction
Legionella pneumophila is a facultative intracellular pathogen that causes a severe form of pneumonia termed Legionnaires disease.1L. pneumophila is primarily found in freshwater environments where it infects a wide array of amoeba host.2,3 Through adaptation to the amoeba host, L. pneumophila has evolved elaborate mechanisms to replicate within amoeba and human macrophages.2,4 Upon inhalation of L. pneumophila-containing aerosols, L. pneumophila enters alveolar macrophages, where it evades the endocytic pathway and resides in a rough endoplasmic reticulum-derived vacuole referred to as the Legionella-containing vacuole (LCV).1,5,6
Biogenesis of the LCV is governed by the Dot/Icm Type IV secretion system7,8 that delivers ∼300 effector proteins into the host cell.9−11 This repertoire of effector proteins manipulates a myriad of host cellular processes including evasion of lysosomal fusion, modulation of host cell pro-apoptotic and antiapoptotic pathways, and acquisition of nutrients through manipulation of host cell proteasome machinery.1,6,11−14 Loss of single translocated Dot/Icm substrates rarely causes a detectable defect in intravacuolar proliferation of L. pneumophila.11,15 However, intracellular replication in amoeba and human macrophages is dependent on the eukaryotic-like F-box effector protein AnkB in the AA100/130b and the Paris strain16,17 but not in the Philadelphia-derived Lp02 strain.18 Despite the presence of at least four additional F-box effector proteins in L. pneumophila, the AnkB effector is essential for polyubiquitination of the LCV.17−19 AnkB is Dot/Icm-translocated and is anchored into the LCV membrane through host-mediated farnesylation,20,21 where the F-box domain of AnkB directly interacts with the host SCF1 (SKP1/CUL1/F-box) E3 ubiquitin ligase complex17−19 on the LCV membrane,22 to promote lysine48-linked polyubiquitination of the LCV.13 In human cells and amoeba, the lysine48-linked polyubiquitinated proteins on the LCV are degraded through ubiquitin-dependent proteolysis by the proteasome, generating an increased level of free cellular amino acids above the threshold required as a source of carbon and energy for intracellular replication of L. pneumophila.13 The identity of the ubiquitinated proteins on the LCV remains unknown.
Many intracellular pathogens form a replicative niche within specialized vacuoles in the host cell.23,24 The proteomes of Yersinia pseudotubercuolosis and Salmonella typhimurium-containing vacuoles have been partially characterized from J774A.1 mouse macrophages.25 The LCV proteome of the JR32 Philadelphia-derived strain of L. pneumophila has been generated using the mouse macrophage cell line Raw 264.7.26 In addition, the proteome of the LCV of the Corby strain of L. pneumophila in the amoeba host Dictyostelium discoideum has been profiled.26−28 The Raw 264.7 mouse macrophage cell line is permissive for L. pneumophila despite originating from the BALB/C mouse, whose primary macrophages are nonpermissive to L. pneumophila infection.29,30 Importantly, the molecular bases of permissiveness of human and mouse macrophages to infection by L. pneumophila are distinct.29,31−34 Therefore, it is important to determine the LCV proteome within human macrophages, which are the only known mammalian host for L. pneumophila. Because of the essential requirement of the polyubiquitinated proteins on the LCV for intracellular replication, their identification can be valuable to decipher biogenesis and dynamic remodeling of the LCV.
We determined the complete and the ubiquitinated proteome of the LCV in U937 human macrophage cell line for the WT AA100/130b strain and the isogenic ankB mutant. The intravacuolar proliferation defect of the ankB mutant is due to its inability to decorate the LCV with lysine48-linked polyubiquitinated proteins, but the mutant is localized within an ER-derived LCV that evades lysosomal fusion similar to the wild-type strain.16,19 Therefore, the ankB mutant is a unique and useful genetic tool to analyze the total and ubiquitinated proteome of nonreplicative LCV that is not decorated with polyubiquitinated proteins and contrasts that with the replicative polyubiquitinated LCV of the WT strain. We identified 1193 host proteins localized to the WT strain LCV and 1546 on the ankB mutant LCV with metabolism proteins, in particular, phophatidylinositol and carbohydrate, cellular signaling, and protein transport are significantly represented. The ankB mutant LCV proteome contained >80% of the WT strain LCV proteome. The additional 354 proteins on the ankB mutant LCV are primarily involved in transcription/translation and the immune response. Of the 24 ubiquitinated proteins on the WT strain LCV, a large portion (25%) are involved in immune response signaling and regulation (interferon regulatory factor 7 and interleukin-1 receptor-associated kinase 1), while proteins involved in transport and intracellular trafficking were also identified. In contrast, the ankB mutant LCV contained 29 ubiquitinated proteins primarily involved in signaling (integrin beta-1, beta-2, and alpha-5) and vesicle trafficking (Rab1A, Rab14). The WT and ankB mutant ubiquitinated proteomes each contained p97, tubulin, and the neutral amino acid transporter SLC3A2. Further analysis of the identified nature of native ubiquitinated host proteins on the LCV within human macrophages could prove valuable to determine how L. pneumophila exploits human macrophages.
Methods
Bacterial Strains and Cell Culture
L. pneumophila strain AA100/130b (ATCC BAA-74) and the isogenic ankB mutant16 were grown on BCYE agar plates for 3 days at 37 °C prior to use in infections as described previously.16 U937 cells were cultured using RPMI1640 media as we described previously.19
Legionella-Containing Vacuole Purification
A total of 6 × 108 U937 macrophages were plated in T175 cm flask in 60 mL of RPMI media supplemented with 10% FBS. At a multiplicity of infection of 50 bacteria per cell, Legionella-containing vacuoles were formed by internalization of bacteria diluted in 20 mL of RPMI media. Internalization of the bacteria was performed for 30 min at 37 °C under 5% CO2. Cells were washed three times in PBS and then incubated in growth media for 4 h at 37 °C under 5% CO2. After 4 h, cells were washed three times in PBS (4 °C) and scraped into a 50 mL screw cap centrifuge tube and pelleted at 4 °C for 5 min at 450g. Cells were resuspended in homogenization buffer (250 mM sucrose, 20 mM Hepes/KOH (pH 7.2) + 0.5 mM EGTA (pH 8.0) and pelleted by centrifugation at 675g for 6 min at 4 °C. Cells were then resuspended in homogenization buffer with protease inhibitors (Roche cocktail) at 2 × 108/mL. Cells were lysed with a dounce homogenizer on ice and visualized under light and confocal microscopy to ensure effective cell lysing and Legionella vacuole integrity. Whole cells and nuclei were then pelleted in an1.5 mL tube for 3.5 min at 344g. The supernatant was placed in a new 1.5 mL tube and centrifuged for 3.5 min at 344g, resulting in the postnuclear supernatant (PNS). The PNS was brought to a final concentration of 39% sucrose. The sucrose solutions for the gradient were made in w/v in 20 mM Hepes/KOH (pH 7.2). The sucrose gradient was made by layering the PNS (39% sucrose) onto 2 mL of 55% sucrose layered onto 1 mL of 65% sucrose in a 14 mm × 89 mm Beckman ultracentrifuge tube. We then layered 2 mL of 10% sucrose onto 2 mL of 25% sucrose solution onto the PNS. The sucrose gradient was centrifuged for 1 h at 100 000g at 4 °C in a swinging bucket rotor (Beckman SW41). The LCVs were isolated from the 55–65% interface using a 16g needle and not disturbing any other fraction. LCVs were placed into 10 mL of PBS (4C) and centrifuged at 40 000g (SW41) for 30 min at 4 °C. Pelleted LCVs were solubilized in 1% Triton X-100 in PBS for 30 min on ice. Following centrifugation at 10 000g for 5 min to pellet bacteria, the supernatant containing eukaryotic proteins associated with the LCV was stored at −80 °C.
Antibodies and Confocal Microscopy
Isolated LCVs were plated onto 24-well coverslips pretreated with poly-l-lysine and allowed to adhere for 1 h. Extravacuolar L. pneumophila were labeled with a rabbit anti-L. pneumophila antibody prior to permeabilization for 1 h. LCVs were then permeabilized with methanol (−20 °C) for 5 min, blocked with 3% BSA for 1 h, and then labeled with a mouse anti-L. pneumophila antibody for 1 h. Secondary antibodies used were Alexa Flour goat antimouse 488 and Alexa Flour goat antirabbit 555. Polyubiquitinated proteins present on the LCVs were labeled with a mouse antipolyubiquitin antibody (Enzo Life Sciences) for 1 h prior to vacuole membrane permeabilization and visualized with Alexa Flour goat antimouse 488.
Protein Digestion
For mass spectrometry analysis, TCA-precipitated proteins were resuspended in 8 M urea and 50 mM Tris pH 8.5. Resuspended peptides were diluted 1:1 with 50 mM Tris pH 8.5 to lower the urea concentration to 4 M prior to digestion with endoproteinase Lys-C (10 ng/uL) for 8 h. Digestions were further diluted 4:1 with 50 mM Tris pH 8.5 to lower the urea concentration to 1 M urea prior to digestion with trypsin (5 ng/uL) for 8 h. Peptides were desalted using a 50 mg tC18 Sep-Pak (Waters).
Ubiquitinated Protein Immunoprecipitation
Ubiquitinated peptides were immunoprecipitated using ubiquitin motif immunoaffinity beads (Cell Signaling Technology), as recommended by the manufacturer with the following modifications.35 Desalted peptides were dissolved in 1.0–1.4 mL of IAP buffer (50 mM MOPS-NaOH, pH 7.2, 10 mM Na2HPO4, 50 mM NaCl). The pH was adjusted to 7.2 with 1 M NaOH. The peptide solution was cleared by centrifugation for 5–10 min at 13 200g. The supernatant was transferred to a new tube and cooled on ice for 10 min. The cooled peptide solution was transferred to the tube with the antibody beads and incubated on a rotator at 4 °C for 60 min. The beads were washed four times with 1 mL of IAP buffer, followed by one wash with 1 mL water. The peptides were eluted from the beads by adding 55 μL of 5% formic acid (FA), mixed, and incubated at room temperature for 10 min. The beads and elution buffer were transferred to a Teflon spin column, and the eluate was collected by centrifugation. The beads were washed with 45 μL of 0.1% TFA and combined with the first eluate.35 The peptides were desalted using StageTips,36 and nanoLC–MS2 data were collected.
nanoLC–MS2 Data Collection
In collaboration with Dr. Steve Gygi, Dr. Ryan Kunz, and Ross Tomaino at the Harvard University Taplin Mass Spectometry Facility, mass spectrometry data were collected on an Orbitrap Fusion mass spectrometer equipped with an easy nano-LC 1000 for sample handling and liquid chromatography. Peptides were separated on a 75 μm × 30 cm hand-pulled fused silica microcapillary column with a needle tip diameter less than 10 μm and packed with 1.8 μm 120 Å GP-C18 beads from Sepax Technologies. The column was equilibrated with buffer A (3% ACN + 0.125% FA). An equal amount of proteins for all samples with ∼2 μg/each was loaded onto the column at 100% buffer A. Separation and elution from the column were achieved using a 90 min 3–25% gradient of buffer B (100% ACN + 0.125% FA). Survey scans of peptide precursors from 400 to 1400 m/z were performed at 120 K resolution (at 200 m/z); AGC, 50k; max injection time, 100 ms; monoisotopic precursor selection turned on; charge state, 2–6; dynamic exclusion, 45s with a 10 ppm tolerance. Tandem MS was performed in top speed mode (2 s cycles) starting with the most intense precursor having an intensity greater than 5k. Parent ions were isolated in the quadrupole (0.7 m/z isolation window). Collision-induced dissociation was performed in the ion trap with a rapid scan rate; 35% collision energy; AGC, 10k; max injection time, 35 ms; parallelizable time was turned on.
Mass Spectrometry Data Analysis
A suite of in-house software tools were used for .RAW file processing, controlling peptide and protein false discovery rates, and assembling peptide level data into protein level data.35,37 The “in house software” refers to software used in the Gygi laboratory. The MS/MS spectra were searched using the SEQUEST algorithm38 against a composite protein database consisting of all protein sequences from the Uniprot human database (88 501 proteins) along with nonsense and common contaminating proteins (111 proteins) in both the forward and reverse direction. Sequest parameters used to search the MS data were: precursor tolerance, 50 ppm; fragment ion tolerance, 1 Da; fully tryptic; 2 missed cleavages; variable modifications of oxidized methionine (15.9949 Da), alkylation of cysteine (57.0214), and diGlycine motif on lysine (114.0429). A target-decoy strategy was used to determine false discovery rates.39 The peptide-level false discovery rate was restricted to <1% by using linear discriminate analysis based on several different SEQUEST parameters including Xcorr ≥1.0, deltaXcorr, charge state, and a minimum peptide length of seven amino acids.37 An algorithm similar to Ascore was used for diGlycine localization and site quantification.35,40 The localization score is based on the Ascore algorithm, where a localization score ≥19 indicated >99% certainty in site localization.40 Protein identifications for the complete proteome of the WT strain and ankB mutant strain are based on identification of at least two unique peptides. The proteome list was obtained from analyses of two LCV samples of the WT strain and the ankB mutant, and only the proteins that were reproducible in both samples were included in our analyses.
MetaCore Software Analysis
MetaCore by Thomson Reuters is an integrated software suite that is available online for functional analysis of various aspects of screening data.41,42 MetaCore software determines the most significant relationships among the proteins analyzed such as Pathway Maps, GO Processes, Process Networks, and Metabolomic Networks. For our analysis, the software determined the most significant relationships shared among the WT strain LCV proteome and a separate analysis for the ankB mutant strain. The excel spreadsheet containing the complete WT strain LCV proteome and the ankB mutant stain LCV proteome were separately uploaded into the start page, and an enrichment analysis workflow was generated.
Results
Purification of the LCV from Human Macrophages
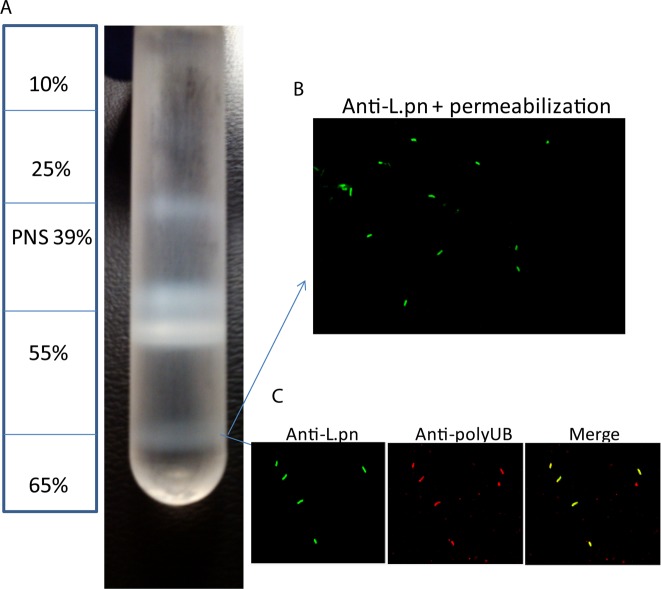
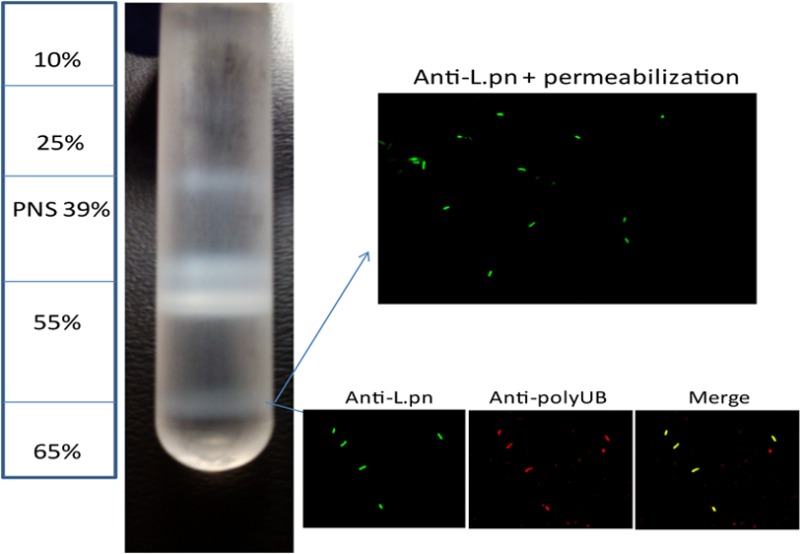
To determine the identity of host proteins on the LCV, we infected U937 human macrophages with L. pneumophila WT strain AA100/130b or its isogenic ankB mutant strain. The ankB mutant strain evades lysosomal fusion and is localized within an ER-derived LCV, similar to the WT strain.16 However, the ankB mutant strain fails to replicate within human macrophages and amoeba due to the levels of cellular amino acids being below the threshold needed as the major source of carbon and energy to support intravacuolar proliferation of L. pneumophila.13,14,43 The U937 human macrophage cell line is widely used in studies on Legionella-human macrophage interaction and was used in this study instead of primary human monocytes due to the need of a large number of cells to isolate LCVs, because only ∼20% of the cells become infected. Following a 4 h infection of 0.6 × 109 U937 macrophages by each of the strains, the LCVs were purified according to previously reported protocols with minor modifications.25,44 Cells were lysed through dounce homogenization, and the PNS was used to isolate LCVs through density ultracentrifugation on a discontinuous sucrose gradient, which resulted in purified LCVs at the 55–65% interface (Figure 1A). To ensure vacuole integrity following purification, isolated LCVs were evaluated using confocal microscopy after differential membrane permeabilization as well as vacuole marker staining to ensure the LCV membranes were intact. Vacuoles were labeled with a polyclonal anti-Legionella antibody prior to vacuolar permeabilization, which resulted in ∼20% of bacteria being labeled, while 100% of bacteria were labeled after vacuolar membrane permeabilization, indicating that the LCV membrane is intact on ∼80% of the isolated LCVs (Figure 1B). We next evaluated the presence of polyubiquitinated proteins, which showed that ∼70% of isolated LCVs were decorated with polyubiqutinated proteins (Figure 1C). Isolated LCVs were solubilized in 1% Triton X-100, and the eukaryotic proteins associated with the LCV were identified by high-throughput liquid chromatography coupled to tandem mass spectrometry (LC–MS). The MS was loaded with 2 μg of protein for each LCV sample. A positive protein identification for the proteome was based on at least two unique peptides. Although numerous proteins were identified, it is likely that some proteins present with scarce quantities on the LCV are not detectable by our analyses.
Figure 1.
LCV purification using a discontinuous sucrose gradient. U937 macrophages were infected with WT L. pneumophila or the isogenic ankB strain at an MOI of 50 for 30 min washed and the infection proceeded for 4 h. Cells were lysed through dounce homogenization, and the postnuclear supernatant was used to isolate LCVs through density ultracentrifugation on a discontinuous sucrose gradient. (A) Diagram of the sucrose gradient showing the isolated LCVs at 55–65% interface and (B) confocal microscopy of isolated LCVs labeled with mouse anti-L. pneumophila following vacuole membrane permeabilization. (C) Confocal microscopy of isolated LCVs labeled with rabbit anti-L. pneumophila antiserum and mouse antipolyubiquitin antibodies.
Total LCV Proteome
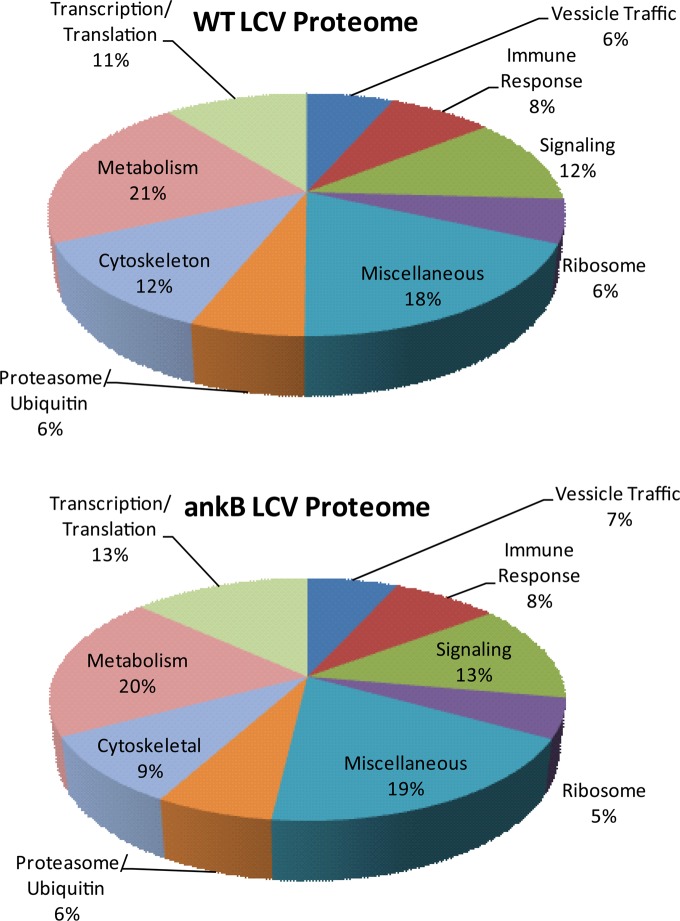
The proteome of WT strain AA100/130b LCV contained 1193 eukaryotic proteins, while the LCV of the ankB mutant strain contained 1546 eukaryotic proteins (Suppl. Tables 1 and 2 in the Supporting Information). We profiled the proteins according to various cellular functions, including transcription/translation, vesicle trafficking, immune response, ribosomal proteins, ubiquitination, proteasome machinery components, signaling, cytoskeleton arrangement, and metabolism (Figure 2). Using these categories we identified the largest proportion of WT strain LCV proteins to be involved in metabolism (21%) while cytoskeleton arrangement (12%), signaling (12%), and transcription/translation (11%) were highly represented (Figure 2). Proteins involved in immune response (8%), ribosome machinery (6%), vesicle trafficking (6%), and ubiquitin-dependent proteolysis (6%) were significantly represented among the WT strain LCV proteome (Figure 2). Despite the presence of additional proteins on the ankB mutant LCV, the overall distribution of the proteins based on cellular function was very similar to the WT strain LCV proteome (Figure 2). The large numbers of shared proteins on isolated LCVs of WT strain and the ankB mutant were consistent with the findings that the ankB mutant strain is localized in an ER-derived LCV that evades lysosomal fusion16 but lacks sufficient levels of amino acids for intravacuolar proliferation.13 The ankB mutant LCV contained 80% of the proteins on the WT strain LCV with an additional 354 proteins primarily involved in regulation and initiation of translation, transcription, apoptosis, and immune response signaling. It is likely that degradation of lysine48-linked polyubiquitinated proteins on the WT strain LCV renders them undetectable by proteomic analysis. The degradation of lysine48-linked polyubiquitinated proteins on the WT strain LCV also alters the relative abundance of identifiable proteins in the proteome of the two strains.
Figure 2.
Functional classification of eukaryotic proteins localized to the LCV. The 1193 eukaryotic proteins localized to the WT strain LCV and the 1546 localized to the ankB mutant LCV identified by high-throughput LC–MS were grouped according to their cellular function according to the UniProt and GeneCards database sets.
Proteome data of the WT strain LCV regarding metabolism identified proteins involved in neutral amino acid transport (SLC1A5, SLC38A2, SLC3A2), cationic amino acid transport (SLC7A1), and monocarboxylate transport (SLC16A1, SLC16A3) as well as a variety of ATPases involved in calcium, sodium, and potassium transport. A large proportion of metabolism-related proteins are involved in carbohydrate metabolism, especially glycolysis or glucose transport (7%), while 5% are involved in lipid metabolism. Both the ankB mutant and WT strain LCV proteomes harbor phosphatidylinositol phosphatases (INPP5D, INPPL1), kinases (PI4KA), phosphatidylinositol 3,4,5-trisphosphate-dependent GTPase-activating proteins (ARAP1, ASAP1), and guanine exchange factors (SWAP70, PREX1, DEF6) (Suppl. Tables 1 and 2 in the Supporting Information). There are also proteins that bind to membranes or vesicles enriched in phosphatidylinositol that function in early endosomal trafficking (CLINT1, RUFY1) and receptor-mediated endocytosis (EPN1).
Consistent with the findings that the LCV of the WT strain and the ankB mutant strain is RER-derived, the proteome data revealed 70–80 (6%) 40S and 60S ribosomal associated proteins present on WT and ankB mutant LCVs (Figure 2). Our data identified 6 to 7% of the WT, and ankB mutant LCV proteomes play a role in vesicular trafficking, which included eight small Rab GTPases (Rab1, Rab2, Rab5, Rab6, Rab7, Rab10, Rab11, Rab14) as well as four sorting nexins and multiple ADP ribosylation factors involved in endocytic recycling, vesicle transport, and vesicle budding (Suppl. Table 3 in the Supporting Information). The ankB mutant LCV had four additional Rab GTPases (Rab8, Rab13, Rab27, and Rab35) (Suppl. Tables 1 and 2 in the Supporting Information). The high similarity in ribosomal and vesicular trafficking proteins in WT and ankB mutant LCV proteomes is very consistent with the findings that biogenesis of the ankB mutant LCV is very similar to that of the WT strain.16
Proteins involved in apoptosis and immune responses, which comprise 8% of the WT strain LCV proteome, include multiple major histocompatibility complex proteins, ligands, and receptors involved in T-cell adhesion (intercellular adhesion molecule 1, 3) and signaling (leukocyte-associated immunoglobulin-like receptor 1, HCLS1 binding protein 3), promoters of cell apoptosis (BCL2-associated athanogene 6, caspase recruitment domain family member 6), inhibitors of apoptosis (defender against cell death 1, FAM129B), and TNF-α regulators and proteins, which promote LPS-induced TNF-α production (thymocyte selection associated family member 2) (Suppl. Tables 1 and 2 in the Supporting Information). The proteome of the ankB mutant LCV contained 84 of the 92 immune response proteins on the WT strain LCV with an additional 42 proteins. These include seven clusters of differentiation molecules (CD109, CD47, CD58, CD63, CD82, CD83, CD84), two BCL2-associated proteins (BAG3, BAX), and two negative regulators of apoptosis (TNFAIP8 and AVEN) (Suppl. Tables 1 and 2 in the Supporting Information).
Proteins involved in cellular signaling pathways on the WT strain and the ankB mutant LCV include tyrosine-protein kinases that play a role in the response to environmental stress and cytokines such as TNF-α (MAP4), regulation of cell growth, differentiation, migration and the immune response (CSK), cytoskeleton remodeling in response to extracellular stimuli, cell motility and receptor endocytosis (ABL2, ARAP1). Additional signaling proteins included Rho GTPase activating proteins (ARHGAP1, ARHGAP17, ARHGAP18), Rab GTPase activating proteins (TBC1D10B, TBC1D15, TBC1D5), and guanine nucleotide-binding proteins (G proteins).
The WT strain and ankB mutant LCV proteomes contained proteins involved in cytoskeletal membrane integrity and organization (actin, coronin) and proteins that regulate actin and microtubule polymerization (ARP2/3 complex, KANK1, LASP1). Interestingly, molecular chaperones that play a role in the folding of actin and tubulin (CCT2-CCT8) were present on the LCV, possibly indicating that following initial LCV formation various cytoskeleton proteins are constantly recruited to the LCV membrane.31
Validation of many of the proteins detected in the proteome of the LCV in our analyses comes from many published studies that showed that at least 17 of the proteins identified in our proteome have been already shown to be localized to the LCV using different strategies, such as confocal microscopy (Suppl. Table 3 in the Supporting Information). The WT strain and ankB mutant LCV contain a significant portion (6%) of proteins involved in ubiquitination and proteosomal degradation (Figure 2). In agreement with findings of colocalization of SKP122 and p9745 with the LCV by confocal microscopy, our proteome data identified the SKP1 component of the SCF E3 ubiquitin ligase complex and the ATPase p97 on the LCV (Suppl. Table 3 in the Supporting Information). In addition, both proteomes contained RAD23A and RAD23B, which serve as multiubiquitin chain receptors that bind to the 26S proteasome and deliver lysine48-linked polyubiquitintated proteins for proteasomal degradation. Four E2 ubiquitin conjugating enzymes and eight E3 ubiquitin ligases were identified on the WT strain LCV (Table 1). The E3 ubiquitin ligases identified on the WT strain LCV regulate apoptosis, NF-κβ activation, and IFN-β production,46,47 while the E2 ubiquitin conjugation enzymes catalyze the synthesis of lysine48 and lysine63-linked polyubiquitin chains48,49 (Table 1). The eight ubiquitin-specific peptidases on the WT strain LCV can act as deubiquitinases (VCPIP1(p97), USP14, USP15, USP9X), which are able to remove ubiquitin moieties from polyubiquitin chains, which prevent proteasomal degradation, or are involved in lysine48-linked polyubiquitination disassembly (USP5) (Table 1). The ankB mutant LCV contained 9 E3 ubiquitin ligases of which three were found on the WT strain LCV (Table 1). The E3 ubiquitin ligases on the ankB mutant LCV are involved in regulation of DNA, p53 activation, and mTORC1 signaling pathway.50,51 The ankB mutant LCV contained four additional ubiquitin-conjugating enzymes including two atypical ubiquitin-conjugating enzymes (UBE2H, UBE2O) and UBE2M, which catalyze the attachment of the ubiquitin like protein NEDD8 onto other proteins (Table 1).
Table 1. Ubiquitin Conjugation and Degradation Enzymes.
| WT strain LCV | |
|---|---|
| E3 Ubiquitin Ligase | |
| ARIH1 | ariadne RBR E3 ubiquitin protein ligase 1 |
| BIRC6 | baculoviral IAP repeat containing 6 |
| HERC1 | E3 ubiquitin protein ligase HERC1 |
| HUWE1 | E3 ubiquitin protein ligase HUWEI |
| NEDD4 | E3 ubiquitin-protein ligase NEDD4 |
| RNF31 | ring finger protein 31 |
| TRIM21 | tripartite motif containing 21 |
| UBR4 | ubiquitin protein ligase E3 component n-recognin 4 |
| E2 Ubiqutin-Conjugating Enzymes | |
| UBE2K | ubiquitin-conjugating enzyme E2K |
| UBE2L3 | ubiquitin-conjugating enzyme E2L3 |
| UBE2N | ubiquitin-conjugating enzyme E2N |
| UBE2V1 | ubiquitin-conjugating enzyme E2 variant 1 |
| Ubiquitin Specific Peptidase | |
| USP10 | ubiquitin specific peptidase 10 |
| USP14 | ubiquitin specific peptidase 14 |
| USP15 | ubiquitin specific peptidase 15 |
| USP47 | ubiquitin specific peptidase 47 |
| USP5 | ubiquitin specific peptidase 5 |
| USP8 | ubiquitin specific peptidase 8 |
| USP9X | ubiquitin specific peptidase 9 |
| VCP | valosin containing protein (p97) |
|
ankB LCV | |
| E3 Ubiquitin Ligase | |
| ARIH2 | ariadne RBR E3 ubiquitin protein ligase 2 |
| BIRC6 | baculoviral IAP repeat containing 6 |
| HUWE1 | E3 ubiquitin protein ligase HUWEI |
| HECTD3 | HECT domain containing E3 ubiquitin protein ligase 3 |
| MYCBP2 | MYC binding protein 2, E3 ubiquitin protein ligase |
| RNF130 | ring finger protein 130 |
| TRIP12 | E3 ubiquitin protein ligase TRIP12 |
| UBR4 | ubiquitin protein ligase E3 component n-recognin 4 |
| UBR5 | ubiquitin protein ligase E3 component n-recognin 5 |
| E2 Ubiqutin-Conjugating Enzymes | |
| UBE2H | ubiquitin-conjugating enzyme E2H |
| UBE2J1 | ubiquitin-conjugating enzyme E2J1 |
| UBE2K | ubiquitin-conjugating enzyme E2K |
| UBE2L3 | ubiquitin-conjugating enzyme E2L3 |
| UBE2M | ubiquitin-conjugating enzyme E2M |
| UBE2N | ubiquitin-conjugating enzyme E2N |
| UBE2O | ubiquitin-conjugating enzyme E2O |
| UBE2V1 | ubiquitin-conjugating enzyme E2 variant 1 |
| Ubiquitin Specific Peptidase | |
| USP10 | ubiquitin specific peptidase 10 |
| USP14 | ubiquitin specific peptidase 14 |
| USP15 | ubiquitin specific peptidase 15 |
| USP19 | ubiquitin specific peptidase 19 |
| USP24 | ubiquitin specific peptidase 24 |
| USP34 | ubiquitin specific peptidase 34 |
| USP47 | ubiquitin specific peptidase 47 |
| USP5 | ubiquitin specific peptidase 5 |
| USP8 | ubiquitin specific peptidase 8 |
| USP9X | ubiquitin specific peptidase 9 |
| VCP | valosin containing protein (p97) |
MetaCore Analysis of the Total LCV Proteome
Bioinformatic analysis of the LCV proteome was performed using MetaCore by Thomson Reuters software.41,42 MetaCore software determined the most significant relationships among pathway maps, process networks, go processes, and metabolomic pathways from eukaryotic proteins localized to the LCV. The top pathway maps identified in WT and ankB mutant LCV proteomes include regulation of the actin cytoskeleton by Rho GTPases, the role of PKA in cytoskeleton remodeling, and integrin-mediated cell adhesion. Signaling pathways including G-protein signaling - RhoA regulation pathway and G protein-α 12 signaling pathways were also identified in the proteome pathway enrichment analysis. The generated pathway maps correlate with the proteome data that reveal a large proportion (12%) of LCV proteins are involved in cytoskeletal formation of the LCV.
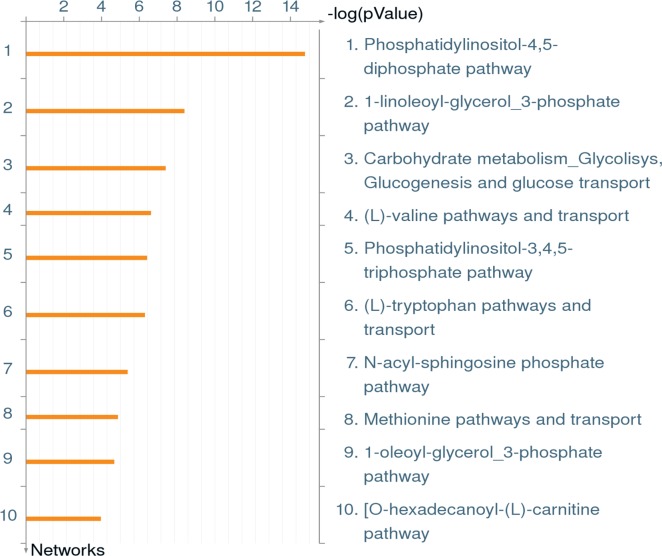
Bioinformatic analysis of metabolic networks in the WT strain LCV proteome revealed the phosphatidylinositol-4,5-diphosphate pathway and phosphatidylinositol-3,4,5-triphosphate pathway as two of the top five (Figure 3). These data reflect the aforementioned results of a wide array of phosphatidylinositol-dependent proteins in the WT strain and the ankB mutant LCV proteomes. This is also consistent with findings that have shown the LCV is decorated with phospatidylinositol-4 phosphate (PI(4)P) to which multiple effector proteins (SidC, SidM, SdcA, RidL) bind on the LCV membrane.26,52 Carbohydrate metabolism (glycolysis, gluconeogenesis, and glucose transport) was the third most abundant metabolomic network identified, while valine, tryptophan, and methionine synthesis and transport were identified in the top eight metabolic pathways represented in the LCV proteome (Figure 3).
Figure 3.
MetaCore enrichment analysis of metabolic networks in the WT STRAIN LCV proteome. The complete WT strain LCV proteome was analyzed through MetaCore software, using the enrichment analysis function, to identify the most significant metabolic networks.
When proteins found only on the WT strain LCV but not on the ankB mutant LCV were analyzed through MetaCore many of the pathways were consistent with the total WT strain LCV proteome. The finding that the ankB mutant proteome contained 80% of the proteins present in the WT strain LCV proteome is consistent with the MetaCore analysis of very similar pathways in both of the proteomes. Interestingly, Ubiquitin-proteosomal proteolysis was identified as the second most abundant process network involved in WT strain LCV-specific proteins despite the findings that the ankB mutant LCV proteome contains a majority of the proteins found in the WT strain LCV proteome. These results are consistent with the requirement of polyubiquitination of the WT strain LCV for intracellular replication.
Ubiquitinated Proteome of the LCV
To identify ubiquitinated proteins on the WT and ankB mutant LCV, the solubilized LCV proteins were immunoprecipitated with antiubiquitin antibodies and identified by LC–MS. We identified 24 ubiquitinated proteins on the WT strain LCV such as Annexin A2, plasminogen activator inhibitor, Rab5 GDP/GTP exchange factor, and transitional endoplasmic reticulum ATPase (p97) (Table 2). The polyubiquitinated proteins identified on the LCV were lysine6-, lysine11-, lysine27-, and lysine48-linked as evident from the findings that ubiquitin was ubiquitinated on the four lysine residues K6, K11, K27, and K48. Interestingly, 6 (25%) ubiquitinated proteins play a role in the immune response such as interleukin-1 receptor-associated kinase 1 and Interferon regulatory factor 7, which play a key role in the signaling and regulation of the immune system against pathogens. Furthermore, we identified two amino acid transporters (SLC3A2 and SLC1A4) and a sodium bicarbonate transporter (SLC4A7) to be ubiquitinated on the WT strain LCV. We analyzed these proteins through the Mammalian Ubiquitination Site Database,53 which contains a comprehensive list of all ubiquitination sites on mammalian proteins. The search revealed that 17 of the identified ubiquitinated proteins have been shown to be ubiquitinated on the same lysine residues, as identified in our LC–MS analyses. However, three of the ubiquitinated proteins identified (plasminogen activator inhibitor 2, mast cell-expressed membrane protein 1, isoform 13 of sodium bicarbonate cotransporter 3) were not in the database, while four (vimentin, target of Myb protein 1, receptor-type tyrosine-protein phosphatase C, transient receptor potential cation channel subfamily V member 2) were in the database but have been previously shown to be ubiquitinated on other lysine residues than the ones we identified in our analysis. The ankB mutant LCV contained 29 ubiquitinated proteins primarily involved in signaling (Integrin beta-1, beta-2 and alpha-5, Rho GDP-dissociation inhibitor 2) and ubiquitination (cullin-5, the E3 ubiquitin ligase LAPTM5, and the E2 ubiquitin-conjugating enzyme E2N) (Table 3). The ankB mutant LCV had multiple ubiqutinated transporters, including the monocarboxylate transporter (SLC16A3), the glucose transporter (SLC2A3), and neutral amino acid transporters (SLC3A2, SLC1A5) as well as 2 ATPases (ATP2B4 and ATP13A3) (Table 3).
Table 2. Ubiquitinated Proteome of the WT Strain LCVa.
| protein | ubiquitination site | localization score |
|---|---|---|
| ubiquitin | 6, 11, 27, 48 | all 1000 |
| target of Myb protein 1b | 452 | 1000 |
| plasminogen activator inhibitor 2b | 240 | 1000 |
| annexin A2b | 115 | 1000 |
| 4F2 cell-surface antigen heavy chain (SLC3A2) | 21, 46 | 1000, 49.5 |
| receptor-type tyrosine-protein phosphatase Cb | 715 | 1000 |
| vimentinb | 223 | 1000 |
| elongation factor 2b | 239 | 1000 |
| DNA-directed RNA polymerase II subunit RPB2b | 847 | 1000 |
| glycine-tRNA ligaseb | 197 | 1000 |
| neutral amino acid transporter A (SLC1A4)b | 528 | 1000 |
| interleukin-1 receptor-associated kinase 1b | 397 | 1000 |
| transitional endoplasmic reticulum ATPase (p97) | 668 | 1000 |
| myosin light polypeptide 6b | 81 | 1000 |
| tubulin beta chain | 324 | 1000 |
| mast cell-expressed membrane protein 1b | 46 | 1000 |
| interferon regulatory factor 7b | 341 | 1000 |
| histone H2A type 1-Cb | 119 | 1000 |
| peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidaseb | 419 | 1000 |
| phosphatidylinositol 4-phosphate 5-kinase type-1 alphab | 103 | 1000 |
| transmembrane protein 59b | 315 | 78.19 |
| toll-interacting proteinb | 63 | 1000 |
| Rab5 GDP/GTP exchange factorb | 263 | 1000 |
| transient receptor potential cation channel subfamily V member 2b | 103 | 1000 |
| isoform 13 of Sodium bicarbonate cotransporter 3 (SLC4A7)b | 1071 | 1000 |
Localization score≥19 is>99% certainty
WT strain LCV-specific ubiquitinated proteins.
Table 3. Ubiquitinated Proteome of the ankB Mutant LCVa.
| protein | ubiquitination site | localization score |
|---|---|---|
| ubiquitin | 6, 11, 27, 48 | all 1000 |
| membrane-associated progesterone receptor component 1 | 105 | 1000 |
| carboxy-terminal domain small phosphatase 2 (CTDSP2) | 20 | 1000 |
| monocarboxylate transporter 4 (SLC16A3) | 431 | 1000 |
| fructose-bisphosphate aldolase A | 28 | 1000 |
| integrin beta-2 | 755, 765 | 1000 |
| plasminogen activator inhibitor 2 | 335 | 1000 |
| integrin beta-1 | 774, 784 | 1000 |
| protein disulfide-isomerase | 81 | 1000 |
| transitional endoplasmic reticulum ATPase (p97) | 668 | 1000 |
| isoform 2 of 4F2 cell-surface antigen heavy chain (SLC3A2) | 39 | 1000 |
| integrin alpha-5 | 1042 | 1000 |
| solute carrier family 2, glucose transporter member 3 (SLC2A3) | 490 | 1000 |
| CD44 antigen | 715 | 1000 |
| 40S ribosomal protein S3 | 214 | 48.53 |
| plasma membrane calcium-transporting ATPase 4 | 1194 | 1000 |
| proteasome subunit alpha type-2 | 64 | 26.04 |
| Rho GDP-dissociation inhibitor 2 | 63 | 1000 |
| tubulin beta chain | 324 | 1000 |
| ubiquitin-conjugating enzyme E2N | 82 | 1000 |
| Ras-related protein Rab-14 | 193 | 1000 |
| Ras-related protein Rab-1A | 61 | 1000 |
| Isoform B of Ras-related C3 botulinum toxin substrate 1 | 166, 185 | 1000, 69.81 |
| Rho-related GTP-binding protein RhoG | 130 | 1000 |
| lysosomal-associated transmembrane protein 5 | 233, 248 | 1000 |
| nascent polypeptide-associated complex subunit alpha | 142 | 1000 |
| isoform 2 of neutral amino acid transporter Member 5 (SLC1A5) | 335 | 1000 |
| cullin-5 | 724 | 1000 |
| probable cation-transporting ATPase 13A3 | 128 | 1000 |
| transmembrane protein 9B | 171 | 1000 |
Localization score ≥19 is >99% certainty.
While the WT strain and ankB mutant LCV had some common ubiquitinated proteins (p97, SLC3A2, tubulin), there was a significant difference in the ubiquitinated proteome among the two strains, as can be seen by the bolded proteins in the WT strain ubiquitinated proteome table. The ankB mutant LCV had multiple ubiquitinated proteins involved in intracellular trafficking (Rab1A, Rab14) and two GTPases (Rac1, RhoG), while the WT strain contained only one ubiquitinated protein involved in intracellular trafficking (Rab5 GDP/GTP exchange factor). Proteasomal degradation of lysine48-linked polyubiquitinated proteins on the WT strain LCV may explain the major differences in the modified host proteins between the two strains.
Discussion
During the past several years characterization of the LCV, through cellular and biochemical studies, has provided some insights into how L. pneumophila form a replicative niche in the host cell. However, mechanisms of LCV biogenesis remain elusive. In this study, we sought to identify the total LCV proteome as well as the ubiquitinated proteins present on the LCV within human macrophages. The cellular functions of the identified proteins were widely distributed with cellular metabolism proteins representing the largest proportion of the WT strain and ankB mutant LCV proteome at ∼20%. Signaling and cytoskeletal proteins were also highly represented. The ankB mutant LCV contained 80% of the proteins on the WT strain LCV with 569 unique proteins primarily involved in immune response, transcription and translation. The additional proteins on the ankB mutant but not the WT strain LCV could be due to proteasomal degradation of lysine48-linked polyubiquitinated proteins in the WT strain LCV.13 This may also explain the difference in the relative abundance of proteins in the two proteomes because the ankB mutant LCV contained 14 additional ribosomal proteins and 8 additional clusters of differentiation molecules compared with the WT strain LCV. However, this proteome analysis revealed proteins present on the WT strain and the ankB mutant strain LCV and not the relative abundance of the identified proteins. In addition, scarce proteins may not be detectable in our proteomic analyses of the LCV.
The LCV proteome contains a unique set of antiapoptotic proteins that could enhance host-cell survival to enable intracellular bacterial growth and may explain the inhibition of apoptosis in L. pneumophila infected cells.12,54,55 We identified the Inhibitor of Apoptosis (IAP) protein Birc6, which binds and inhibits effector caspases 3 and 7.46 Because caspase 3 has been shown to be triggered by L. pneumophila,55−58 the role of Birc6 should be evaluated to determine the effect Birc6 has during infection. This is of particular importance because activation of caspase 3 does not lead to apoptosis of L. pneumophila-infected macrophages,55 which exhibit a strong antiapoptotic phenotype.12,54,55 Birc6 also possesses E3 ubiquitin ligase activity that has been shown to ubiquitinate apoptotic proteins resulting in their proteosomal degradation.46,59 In addition to Birc6, we identified the protein defender against cell death (DAD1), which has been implicated in downstream effects of BCL2 to prevent cell death.60 Because the LCV also harbors eukaryotic pro-apoptotic proteins (BCL2-associated athanogene 6, apoptosis-inducing factor) further investigation into the dynamics, balance, and functions of these antagonistic proteins could prove valuable to understand how L. pneumophila modulates a fine balance between pro- and antiapoptotic events in the infected cells.12,33,43,44 Further investigation into the function and recruitment of antiapoptotic proteins on the LCV could explain why permissive macrophages are particularly resistant to exogenous apoptotic stimuli during L. pneumophila infection.12,54,55,61,62
The WT strain and ankB mutant LCV contains a significant portion (6%) of proteins involved in post-translational modifications such as ubiquitination and farnesylation. It has been shown that multiple L. pneumophila effector proteins undergo host-mediated farnesylation, which enable their anchoring into host membranes.20,63,64 There are no reports of L. pneumophila effector proteins undergoing ubiquitination. However, further characterization of the E3 ubiquitin ligases identified on the LCV could prove valuable to determine if they catalyze ubiquitination of L. pneumophila effector proteins through lysine48 or non-lysine48-linked polyubiquitination.65
Lysine48-linked polyubiquitination of the LCV through the AnkB effector is required for intracellular replication.13 Previous studies have shown the valosin-containing protein (p97) participates in the removal of polyubiquitinated proteins from the LCV.45 Proteome data identified the SKP1 component of the SCF1 E3 ubiquitin ligase complex on the LCV as well as RAD23A and RAD23B, which serve as multiubiquitin chain receptors that bind to the 26S proteasome and deliver lysine48-linked polyubiquitintated proteins for protesomal degradation.66,67 Four E2 ubiquitin-conjugating enzymes (UBE2K, UBE2N, UBE2L3, UBEV1) were identified on the WT strain LCV; however, only UBE2K and possibly UBE2L3 catalyze the synthesis of lysine48-linked polyubiquitinated proteins.48 Therefore, it could be speculated that the E2 ubiquitin-conjugating enzyme UBE2K or UBE2L3 delivers ubiquitin moieties to the SCF1 E3 ubiquitin ligase complex for lysine48-linked polyubiquitination of the AnkB substrates and RAD23A or RAD23B along with p97 delivers the polyubiquitinated proteins to the proteasome to generate an increase in free cellular amino acids.
L. pneumophila is auxotrophic for seven amino acids; therefore, retrieval of host cell amino acids is essential for intracellular replication.14,68 It is not known how L. pneumophila transports amino acids through the LCV membrane to provide the carbon and energy source for intracellular replication.13 We have identified five amino acid transporters (SLC7A5, SLC3A2, SLC38A2, SLC1A5, SLC1A4)69 in the LCV proteome. It has been shown that the amino acid transporter SLC1A5 is required for L. pneumophila replication in human macrophages.70 Because of the essential need for the acquisition of host cell amino acids, further investigation into the localization and function of the host SLC amino acid transporters and the specific amino acid transporter they import into the LCV lumen could provide insightful information on how intravacuolar L. pneumophila import amino acids from the host cell cytosol through the vacuolar membrane.
Although amino acids are the major sources of carbon and energy for L.pneumophila, the organism utilizes the Entner–Doudoroff pathway to convert exogenous glucose to pyruvate.71,72L. pneumophila mutants deficient in the Entner–Doudoroff pathway have a significant defect in intracellular growth in A549 human epithelial cells, A/J mouse macrophages, and Acanthamoeba culbertstoni.71 The presence of the glucose transporters (SLC2A1, SLC2A14, SLC2A3) on the LCV is consistent with the import and utilization of glucose by intravacuolar L. pneumophila as a potential source of carbon and energy during intravacuolar growth.71,72
The LCV proteome contained two monocarboxylate transporters (SLC16A1, SLC16A3) that transport pyruvate across eukaryotic membranes.73 Interestingly, pyruvate supplementation rescues the ankB mutant for intracellular replication in human macrophages and amoeba.13 Therefore, host-derived pyruvate may constitute an important source of carbon and energy for intravacuolar L. pneumophila, and import of pyruvate by the LCV is likely mediated by SLC16A1 and SLC16A3 identified in the LCV proteome.
We identified 24 ubiquitinated proteins on the WT strain LCV and 29 on the ankB mutant LCV. Ubiquitination of these proteins could have a wide range of effects due to the complexity and altered location or function of proteins modified by various ubiquitin polymer linkages. Of the 24 ubiquitinated proteins on the WT strain LCV, 6 are involved in the immune response (T cell activation, IL-1, and Toll-like receptor signaling). Ubiquitin was found to be ubiquitinated on four lysine residues (K6, K11, K27, and K48), which indicates polyubiquitin chains are formed on some of these ubiquitinated proteins through linkages other than lysine48.13 Interestingly, the transitional endoplasmic reticulum ATPase (p97) that participates in the removal of lysine48-linked polyubiquitinated proteins from the LCV45 was found to be ubiqutinated on the LCV. Some of the 21 ubiquitinated proteins specific to the WT strain are likely to be substrates for AnkB. In addition, the ubiquitinated proteins on the LCV are likely ubiquitinated by various L. pneumophila ubiquitin ligases (F-box, U-box) and host E3 ligases identified in the proteome. A previous study analyzed ubiquitinated proteins from Raw 264.7 macrophage whole cell lysates during L. pneumophila infection.74 Importantly, our ubiquitinated proteome is of the LCV; however, there were some common ubiquitinated proteins (vimentin, SLC4A7, elongation factor 2).74 Further studies are needed to determine the lysine48-linked polyubiquitinated proteins as well as the lysine6-, lysine11-, and lysine27-linked polyubiquitinated proteins localized to the LCV to assess their role in the intracellular infection.
The small GTPase Rab5 is a crucial regulator of early endosomes.75 Rab5 is activated by the guanine nucleotide exchange factor (Rabex-5), which was found to be ubiquitinated on the WT strain LCV. The L. pneumophila effector VipD interacts with activated endosomal Rab5 and is required for endolysosomal evasion.76,77 Because of the importance of Rab5 during L. pneumophila infection, the ubiquitination of Rabex-5 could pose another strategy for endolysosomal evasion by L. pneumophila.
The LCV proteome has been generated for the JR32 Philadelphia-derived strain of L. pneumophila in the mouse macrophage cell line Raw 264.7,26,27 which identified 1156 proteins.26 Proteins involved in cellular metabolism are the most abundant in our proteome as well as the previously characterized JR32 strain proteome.26 However, in our LCV proteome within human macrophages, proteins involved in metabolism (∼20%) were less abundant compared with previous reports (∼50%).26 Solute carriers, 40S and 60S ribosomal proteins and proteins involved in vesicular trafficking (sorting nexins, Rab GTPases), were very similar in the LCV proteomes of the JR32 and AA100/130b strains within mice and human macrophages, respectively.26 The LCV proteome of the JR32 strain in mouse macrophages had a larger number of 28S and 39S mitochondrial ribosomal proteins as well as NADH dehydrogenase proteins26 compared with the AA100/130b strain in human macrophages. While phophatidylinositol-dependent proteins have been identified (Sac1, Ship1) in the JR32 strain LCV;26 we identified phophatidylinositol-dependent GTPase activating proteins and guanine exchange factors that were not detectable in the LCV proteome of the JR32 strain within mouse macrophages. Interestingly, the E3 ubiquitin ligases and E2 ubiquitin-conjugating enzymes we identified have not been detected in the LCV of the JR32 proteome within Raw 264.7 mouse macrophages.26 The differences between the LCV proteome of the JR32 and AA100/130b strains could be attributed to the different L. pneumophila strains that exhibit plasticity in ∼30% of the genome78,79 or more importantly the difference in the LCV between mouse and human macrophages or both.
This report outlines the total and novel ubiquitinated proteome of the LCV that include E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases. The phosphatidylinositol-dependent proteins in our LCV proteome and the MetaCore bioinformatic analysis of metabolomic networks in the WT strain LCV proteome clearly show the phosphatidylinositol-4,5-diphosphate pathway as the most abundant, while amino acid synthesis and transport are also highly represented in the LCV proteome. The amino acids, pyruvate, and glucose transporters in the LCV proteome show a unique mechanism of import of host sources of carbon and energy for intravacuolar nutrition and metabolism of L. pneumophila. Further investigation into the ubiquitinated proteins and the various nutrient transporters localized to the LCV could prove valuable to understanding how L. pneumophila manipulates host cell machinery to acquire nutrients and circumvent a starvation response during infection.13,22 Our findings should provide a rich resource for future investigations of numerous cellular processes and pathways involved in biogenesis and dynamic remodeling of the LCV within human macrophages.
Acknowledgments
We gratefully acknowledge Dr. Steve Gygi, Dr. Ryan Kunz, and Ross Tomaino at the Harvard University Taplin Mass Spectometry Facility for their proteomic analyses and for their helpful discussion. Y.A.K. is supported by Public Health Service Awards R01AI069321 and R21AI107978 from NIAID and by the commonwealth of Kentucky Research Challenge Trust Fund. We thank all members of the Abu Kwaik laboratory for reading of the manuscript and insightful discussion.
Supporting Information Available
Supplementary Table 1. Complete WT strain LCV proteome from infected U937 human macrophages. Supplementary Table 2. Complete ankB mutant LCV proteome from infected U937 human macrophages. Supplementary Table 3. Validated LCV associated eukaryotic proteins. Proteins identified in the Mass Spectometry screen that have previously been shown to localize at the LCV during infection. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Isberg R. R.; O’Connor T. J.; Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat. Rev. Microbiol. 2009, 7113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A. M.; Von Dwingelo J. E.; Price C. T.; Abu Kwaik Y. Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 2013, 44307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M.; Horn M.; Wagner M.; Santic M.; Abu Kwaik Y. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 2005, 71120–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Quadan T.; Price C. T.; Abu Kwaik Y. Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 2012, 206299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J. C.; Roy C. R. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 2002, 412945–954. [DOI] [PubMed] [Google Scholar]
- Shin S.; Roy C. R. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell. Microbiol. 2008, 1061209–1920. [DOI] [PubMed] [Google Scholar]
- Vogel J. P.; Andrews H. L.; Wong S. K.; Isberg R. R. Conjugative transfer by the virulence system of Legionella pneumophila. Science (New York, N.Y.) 1998, 2795352873–876. [DOI] [PubMed] [Google Scholar]
- Segal G.; Purcell M.; Shuman H. A. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. U.S.A. 1998, 9541669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Banga S.; Tan Y.; Zheng C.; Stephenson R.; Gately J.; Luo Z. Q. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PloS One 2011, 63e17638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z. Q. Targeting One of its Own: Expanding Roles of Substrates of the Legionella Pneumophila Dot/Icm Type IV Secretion System. Front. Microbiol. 2011, 2, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac D. T.; Isberg R. Master manipulators: an update on Legionella pneumophila Icm/Dot translocated substrates and their host targets. Future Microbiol. 2014, 93343–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Zant A.; Jones S.; Asare R.; Suttles J.; Price C.; Graham J.; Kwaik Y. A. Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell. Microbiol. 2007, 91246–264. [DOI] [PubMed] [Google Scholar]
- Price C. T.; Al-Quadan T.; Santic M.; Rosenshine I.; Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science (New York, N.Y.) 2011, 33460621553–1557. [DOI] [PubMed] [Google Scholar]
- Price C. T.; Richards A. M.; Von Dwingelo J. E.; Samara H. A.; Abu Kwaik Y. Amoeba host-Legionella synchronization of amino acid auxotrophy and its role in bacterial adaptation and pathogenic evolution. Environ. Microbiol. 2014, 162350–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger A. W.; Isberg R. R. Legionella pneumophila Dot/Icm translocated substrates: a sum of parts. Curr. Opin. Microbiol. 2009, 12167–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodor S.; Price C. T.; Habyarimana F.; Kalia A.; Abu Kwaik Y. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 2008, 704908–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomma M.; Dervins-Ravault D.; Rolando M.; Nora T.; Newton H. J.; Sansom F. M.; Sahr T.; Gomez-Valero L.; Jules M.; Hartland E. L.; Buchrieser C. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell. Microbiol. 2010, 1291272–1291. [DOI] [PubMed] [Google Scholar]
- Ensminger A. W.; Isberg R. R. E3 ubiquitin ligase activity and targeting of BAT3 by multiple Legionella pneumophila translocated substrates. Infect. Immun. 2010, 7893905–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T.; Al-Khodor S.; Al-Quadan T.; Santic M.; Habyarimana F.; Kalia A.; Kwaik Y. A. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog. 2009, 512e1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T.; Al-Quadan T.; Santic M.; Jones S. C.; Abu Kwaik Y. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J. Exp. Med. 2010, 20781713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Quadan T.; Price C. T.; London N.; Schueler-Furman O.; AbuKwaik Y. Anchoring of bacterial effectors to host membranes through host-mediated lipidation by prenylation: a common paradigm. Trends Microbiol. 2011, 1912573–579. [DOI] [PubMed] [Google Scholar]
- Bruckert W. M.; Price C. T.; Abu Kwaik Y. Rapid nutritional remodeling of the host cell upon attachment of Legionella pneumophila. Infect. Immun. 2014, 82172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth D. E.; Heinzen R. A. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell. Microbiol. 2007, 94829–840. [DOI] [PubMed] [Google Scholar]
- Cossart P.; Roy C. R. Manipulation of host membrane machinery by bacterial pathogens. Curr. Opin. Cell Biol. 2010, 224547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. D.; Finlay B. B. Isolation and characterization of Salmonella typhimurium and Yersinia pseudotuberculosis-containing phagosomes from infected mouse macrophages: Y. pseudotuberculosis traffics to terminal lysosomes where they are degraded. Eur. J. Cell Biol. 1998, 77135–47. [DOI] [PubMed] [Google Scholar]
- Hoffmann C.; Finsel I.; Otto A.; Pfaffinger G.; Rothmeier E.; Hecker M.; Becher D.; Hilbi H. Functional analysis of novel Rab GTPases identified in the proteome of purified Legionella-containing vacuoles from macrophages. Cell. Microbiol. 2014, 1671034–1052. [DOI] [PubMed] [Google Scholar]
- Urwyler S.; Nyfeler Y.; Ragaz C.; Lee H.; Mueller L. N.; Aebersold R.; Hilbi H. Proteome analysis of Legionella vacuoles purified by magnetic immunoseparation reveals secretory and endosomal GTPases. Traffic (Copenhagen, Den.) 2009, 10176–87. [DOI] [PubMed] [Google Scholar]
- Shevchuk O.; Batzilla C.; Hagele S.; Kusch H.; Engelmann S.; Hecker M.; Haas A.; Heuner K.; Glockner G.; Steinert M. Proteomic analysis of Legionella-containing phagosomes isolated from Dictyostelium. Int. J. Med. Microbiol. 2009, 2997489–508. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y.; Klein T. W.; Newton C. A.; Widen R.; Friedman H. Growth of Legionella pneumophila in thioglycolate-elicited peritoneal macrophages from A/J mice. Infect. Immun. 1988, 562370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salins S.; Newton C.; Widen R.; Klein T. W.; Friedman H. Differential induction of gamma interferon in Legionella pneumophila-infected macrophages from BALB/c and A/J mice. Infect. Immun. 2001, 6963605–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter A.; Caution K.; Abu Khweek A.; Tazi M.; Abdulrahman B. A.; Abdelaziz D. H.; Voss O. H.; Doseff A. I.; Hassan H.; Azad A. K.; Schlesinger L. S.; Wewers M. D.; Gavrilin M. A.; Amer A. O. Caspase-11 promotes the fusion of phagosomes harboring pathogenic bacteria with lysosomes by modulating actin polymerization. Immunity 2012, 37135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelaziz D. H.; Amr K.; Amer A. O. Nlrc4/Ipaf/CLAN/CARD12: more than a flagellin sensor. Int. J. Biochem. Cell Biol. 2010, 426789–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amer A. O. Modulation of caspases and their non-apoptotic functions by Legionella pneumophila. Cell. Microbiol. 2010, 122140–147. [DOI] [PubMed] [Google Scholar]
- Abdelaziz D. H.; Gavrilin M. A.; Akhter A.; Caution K.; Kotrange S.; Khweek A. A.; Abdulrahman B. A.; Grandhi J.; Hassan Z. A.; Marsh C.; Wewers M. D.; Amer A. O. Apoptosis-associated speck-like protein (ASC) controls Legionella pneumophila infection in human monocytes. J. Biol. Chem. 2011, 28653203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.; Bennett E. J.; Huttlin E. L.; Guo A.; Li J.; Possemato A.; Sowa M. E.; Rad R.; Rush J.; Comb M. J.; Harper J. W.; Gygi S. P. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 442325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J.; Mann M.; Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 281896–1906. [DOI] [PubMed] [Google Scholar]
- Huttlin E. L.; Jedrychowski M. P.; Elias J. E.; Goswami T.; Rad R.; Beausoleil S. A.; Villen J.; Haas W.; Sowa M. E.; Gygi S. P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 14371174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J. K.; McCormack A. L.; Yates J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994, 511976–989. [DOI] [PubMed] [Google Scholar]
- Elias J. E.; Gygi S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 43207–214. [DOI] [PubMed] [Google Scholar]
- Beausoleil S. A.; Villen J.; Gerber S. A.; Rush J.; Gygi S. P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006, 24101285–1292. [DOI] [PubMed] [Google Scholar]
- Ekins S.; Nikolsky Y.; Bugrim A.; Kirillov E.; Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol. Biol. 2007, 356, 319–350. [DOI] [PubMed] [Google Scholar]
- van Leeuwen D. M.; Pedersen M.; Knudsen L. E.; Bonassi S.; Fenech M.; Kleinjans J. C.; Jennen D. G. Transcriptomic network analysis of micronuclei-related genes: a case study. Mutagenesis 2011, 26127–32. [DOI] [PubMed] [Google Scholar]
- Abu Kwaik Y.; Bumann D. Microbial quest for food in vivo: ’nutritional virulence’ as an emerging paradigm. Cell. Microbiol. 2013, 156882–890. [DOI] [PubMed] [Google Scholar]
- Derre I.; Isberg R. R. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 2004, 7253048–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer M. S.; Kirton D.; Bader J. S.; Isberg R. R. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006, 24e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T.; Pohl C.; Pyrowolakis G.; Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol. Cell 2004, 146801–811. [DOI] [PubMed] [Google Scholar]
- Zhong Q.; Gao W.; Du F.; Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 2005, 12171085–1095. [DOI] [PubMed] [Google Scholar]
- Christensen D. E.; Brzovic P. S.; Klevit R. E. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat. Struct. Mol. Biol. 2007, 1410941–948. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Pickart C. M. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J. Biol. Chem. 1990, 2653521835–21842. [PubMed] [Google Scholar]
- Gudjonsson T.; Altmeyer M.; Savic V.; Toledo L.; Dinant C.; Grofte M.; Bartkova J.; Poulsen M.; Oka Y.; Bekker-Jensen S.; Mailand N.; Neumann B.; Heriche J. K.; Shearer R.; Saunders D.; Bartek J.; Lukas J.; Lukas C. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 2012, 1504697–709. [DOI] [PubMed] [Google Scholar]
- Han S.; Kim S.; Bahl S.; Li L.; Burande C. F.; Smith N.; James M.; Beauchamp R. L.; Bhide P.; DiAntonio A.; Ramesh V. The E3 ubiquitin ligase protein associated with Myc (Pam) regulates mammalian/mechanistic target of rapamycin complex 1 (mTORC1) signaling in vivo through N- and C-terminal domains. J. Biol. Chem. 2012, 2873630063–30072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S. S.; Ragaz C.; Reus K.; Nyfeler Y.; Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006, 25e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.; Zhou T.; He B.; Yu H.; Guo X.; Song X.; Sha J. mUbiSiDa: a comprehensive database for protein ubiquitination sites in mammals. PloS One 2014, 91e85744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V. P.; Isberg R. R. NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J. Exp. Med. 2006, 20392177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Zant A.; Santic M.; Molmeret M.; Jones S.; Helbig J.; Abu Kwaik Y. Incomplete activation of macrophage apoptosis during intracellular replication of Legionella pneumophila. Infect. Immun. 2005, 7395339–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Hammad L. A.; Hsu F.; Mao Y.; Luo Z. Q. Induction of caspase 3 activation by multiple Legionella pneumophila Dot/Icm substrates. Cell. Microbiol. 2013, 15111783–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L. Y.; Abu Kwaik Y. Activation of caspase 3 during Legionella pneumophila-induced apoptosis. Infect. Immun. 1999, 6794886–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M.; Zink S. D.; Han L.; Abu-Zant A.; Asari R.; Bitar D. M.; Abu Kwaik Y. Activation of caspase-3 by the Dot/Icm virulence system is essential for arrested biogenesis of the Legionella-containing phagosome. Cell. Microbiol. 2004, 6133–48. [DOI] [PubMed] [Google Scholar]
- Dubrez-Daloz L.; Dupoux A.; Cartier J. IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle 2008, 781036–1046. [DOI] [PubMed] [Google Scholar]
- Nakashima T.; Sekiguchi T.; Kuraoka A.; Fukushima K.; Shibata Y.; Komiyama S.; Nishimoto T. Molecular cloning of a human cDNA encoding a novel protein, DAD1, whose defect causes apoptotic cell death in hamster BHK21 cells. Mol. Cell. Biol. 1993, 13106367–6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S.; Gao P.; Shen X.; Fiscus V.; Zong W. X.; Chen L.; Luo Z. Q. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc. Natl. Acad. Sci. U.S.A. 2007, 104125121–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M. F.; Banga S.; Barry K. C.; Shen X.; Tan Y.; Luo Z. Q.; Vance R. E. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011, 72e1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C. T.; Jones S. C.; Amundson K. E.; Kwaik Y. A. Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of legionella pneumophila. Front. Microbiol. 2010, 1, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S. S.; Charron G.; Hang H. C.; Roy C. R. Lipidation by the host prenyltransferase machinery facilitates membrane localization of Legionella pneumophila effector proteins. J. Biol. Chem. 2010, 2854534686–34698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y.; Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 138508–523. [DOI] [PubMed] [Google Scholar]
- Raasi S.; Orlov I.; Fleming K. G.; Pickart C. M. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J. Mol. Biol. 2004, 34151367–1379. [DOI] [PubMed] [Google Scholar]
- Goh A. M.; Walters K. J.; Elsasser S.; Verma R.; Deshaies R. J.; Finley D.; Howley P. M. Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins. BMC Biochem. 2008, 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunder E.; Gillmaier N.; Kutzner E.; Herrmann V.; Lautner M.; Heuner K.; Eisenreich W. Amino Acid Uptake and Metabolism of Legionella pneumophila Hosted by Acanthamoeba castellanii. J. Biol. Chem. 2014, 2893021040–21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweikhard E. S.; Ziegler C. M. Amino acid secondary transporters: toward a common transport mechanism. Curr. Top. Membr. 2012, 70, 1–28. [DOI] [PubMed] [Google Scholar]
- Wieland H.; Ullrich S.; Lang F.; Neumeister B. Intracellular multiplication of Legionella pneumophila depends on host cell amino acid transporter SLC1A5. Mol. microbiol. 2005, 5551528–1537. [DOI] [PubMed] [Google Scholar]
- Harada E.; Iida K.; Shiota S.; Nakayama H.; Yoshida S. Glucose metabolism in Legionella pneumophila: dependence on the Entner-Doudoroff pathway and connection with intracellular bacterial growth. J. Bacteriol. 2010, 192112892–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eylert E.; Herrmann V.; Jules M.; Gillmaier N.; Lautner M.; Buchrieser C.; Eisenreich W.; Heuner K. Isotopologue profiling of Legionella pneumophila: role of serine and glucose as carbon substrates. J. Biol. Chem. 2010, 2852922232–22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A. P. The SLC16 gene family - structure, role and regulation in health and disease. Mol. Aspects Med. 2013, 342–3337–349. [DOI] [PubMed] [Google Scholar]
- Ivanov S. S.; Roy C. R. Pathogen signatures activate a ubiquitination pathway that modulates the function of the metabolic checkpoint kinase mTOR. Nat. Immunol. 2013, 14121219–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer A.; Gilleron J.; Bogorad R. L.; Marsico G.; Nonaka H.; Seifert S.; Epstein-Barash H.; Kuchimanchi S.; Peng C. G.; Ruda V. M.; Del Conte-Zerial P.; Hengstler J. G.; Kalaidzidis Y.; Koteliansky V.; Zerial M. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 2012, 4857399465–470. [DOI] [PubMed] [Google Scholar]
- Gaspar A. H.; Machner M. P. VipD is a Rab5-activated phospholipase A1 that protects Legionella pneumophila from endosomal fusion. Proc. Natl. Acad. Sci. U.S.A. 2014, 111124560–4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku B.; Lee K. H.; Park W. S.; Yang C. S.; Ge J.; Lee S. G.; Cha S. S.; Shao F.; Heo W. D.; Jung J. U.; Oh B. H. VipD of Legionella pneumophila targets activated Rab5 and Rab22 to interfere with endosomal trafficking in macrophages. PLoS Pathog. 2012, 812e1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C.; Rusniok C.; Bruggemann H.; Zidane N.; Magnier A.; Ma L.; Tichit M.; Jarraud S.; Bouchier C.; Vandenesch F.; Kunst F.; Etienne J.; Glaser P.; Buchrieser C. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004, 36111165–1173. [DOI] [PubMed] [Google Scholar]
- Flynn K. J.; Swanson M. S. Integrative conjugative element ICE-betaox confers oxidative stress resistance to Legionella pneumophila in vitro and in macrophages. mBio 2014, 53e01091-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.