Abstract
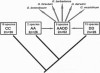
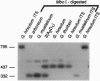
Polyploidy is a prominent process in plant evolution; yet few data address the question of whether homeologous sequences evolve independently subsequent to polyploidization. We report on ribosomal DNA (rDNA) evolution in five allopolyploid (AD genome) species of cotton (Gossypium) and species representing their diploid progenitors (A genome, D genome). Sequence data from the internal transcribed spacer regions (ITS1 and ITS2) and the 5.8S gene indicate that rDNA arrays are homogeneous, or nearly so, in all diploids and allopolyploids examined. Because these arrays occur at four chromosomal loci in allopolyploid cotton, two in each subgenome, repeats from different arrays must have become homogenized by interlocus concerted evolution. Southern hybridization analysis combined with copy-number estimation demonstrate that this process has gone to completion in the diploids and to completion or near-completion in all allopolyploid species and that it most likely involves the entire rDNA repeat. Phylogenetic analysis demonstrates that interlocus concerted evolution has been bidirectional in allopolyploid species--i.e., rDNA from four polyploid lineages has been homogenized to a D genome repeat type, whereas sequences from Gossypium mustelinum have concerted to an A genome repeat type. Although little is known regarding the functional significance of interlocus concerted evolution of homeologous sequences, this study demonstrates that the process occurs for tandemly repeated sequences in diploid and polyploid plants. That interlocus concerted evolution can occur bidirectionally subsequent to hybidization and polyploidization has significant implications for phylogeny reconstruction, especially when based on rDNA sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnheim N., Krystal M., Schmickel R., Wilson G., Ryder O., Zimmer E. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin B. G. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the compositae. Mol Phylogenet Evol. 1992 Mar;1(1):3–16. doi: 10.1016/1055-7903(92)90030-k. [DOI] [PubMed] [Google Scholar]
- Enea V., Corredor V. The evolution of plasmodial stage-specific rRNA genes is dominated by gene conversion. J Mol Evol. 1991 Feb;32(2):183–186. doi: 10.1007/BF02515391. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Dixon M. T. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991 Dec;66(4):411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Hillis D. M., Moritz C., Porter C. A., Baker R. J. Evidence for biased gene conversion in concerted evolution of ribosomal DNA. Science. 1991 Jan 18;251(4991):308–310. doi: 10.1126/science.1987647. [DOI] [PubMed] [Google Scholar]
- Hsiao C., Chatterton N. J., Asay K. H., Jensen K. B. Phylogenetic relationships of 10 grass species: an assessment of phylogenetic utility of the internal transcribed spacer region in nuclear ribosomal DNA in monocots. Genome. 1994 Feb;37(1):112–120. doi: 10.1139/g94-014. [DOI] [PubMed] [Google Scholar]
- Masterson J. Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science. 1994 Apr 15;264(5157):421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- Nagylaki T. Evolution of multigene families under interchromosomal gene conversion. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3796–3800. doi: 10.1073/pnas.81.12.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritland C., Straus N. A. High evolutionary divergence of the 5.8S ribosomal DNA in Mimulus glaucescens (Scrophulariaceae). Plant Mol Biol. 1993 Jul;22(4):691–696. doi: 10.1007/BF00047409. [DOI] [PubMed] [Google Scholar]
- Savard L., Michaud M., Bousquet J. Genetic diversity and phylogenetic relationships between birches and alders using ITS, 18S rRNA and rbcL gene sequences. Mol Phylogenet Evol. 1993 Jun;2(2):112–118. doi: 10.1006/mpev.1993.1011. [DOI] [PubMed] [Google Scholar]
- Seperack P., Slatkin M., Arnheim N. Linkage disequilibrium in human ribosomal genes: implications for multigene family evolution. Genetics. 1988 Aug;119(4):943–949. doi: 10.1093/genetics/119.4.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWiel P. L., Voytas D. F., Wendel J. F. Copia-like retrotransposable element evolution in diploid and polyploid cotton (Gossypium L.). J Mol Evol. 1993 May;36(5):429–447. doi: 10.1007/BF02406720. [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K., Nazar R. A conserved core structure in the 18-25S rRNA intergenic region from tobacco, Nicotiana rustica. Plant Mol Biol. 1991 Aug;17(2):189–194. doi: 10.1007/BF00039493. [DOI] [PubMed] [Google Scholar]
- Wendel J. F. New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Kawata T., Iida Y., Kato A., Tanifuji S. Nucleotide sequences of the 5.8S rRNA gene and internal transcribed spacer regions in carrot and broad bean ribosomal DNA. J Mol Evol. 1989 Oct;29(4):294–301. doi: 10.1007/BF02103617. [DOI] [PubMed] [Google Scholar]
- Zimmer E. A., Jupe E. R., Walbot V. Ribosomal gene structure, variation and inheritance in maize and its ancestors. Genetics. 1988 Dec;120(4):1125–1136. doi: 10.1093/genetics/120.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer E. A., Martin S. L., Beverley S. M., Kan Y. W., Wilson A. C. Rapid duplication and loss of genes coding for the alpha chains of hemoglobin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2158–2162. doi: 10.1073/pnas.77.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]