Abstract

Biomarkers capable of detecting and targeting epithelial ovarian cancer cells for diagnostics and therapeutics would be extremely valuable. Ovarian cancer is the deadliest reproductive malignancy among women in the U.S., killing over 14 000 women each year. Both the lack of presenting symptoms and high mortality rates illustrate the need for earlier diagnosis and improved treatment of this disease. The glycosyltransferase enzyme GnT-III encoded by the Mgat3 gene is responsible for the addition of GlcNAc (N-acetylglucosamine) to form bisecting N-linked glycan structures. GnT-III mRNA expression is amplified in ovarian cancer tissues compared with normal ovarian tissue. We use a lectin capture strategy coupled to nano-ESI–RPLC–MS/MS to isolate and identify the membrane glycoproteins and unique glycan structures associated with GnT-III amplification in human ovarian cancer tissues. Our data illustrate that the majority of membrane glycoproteins with bisecting glycosylation are common to both serous and endometrioid histological subtypes of ovarian cancer, and several have been reported to participate in signaling pathways such as Notch, Wnt, and TGFβ.
Keywords: biomarkers, ovarian cancer, mass spectrometry, glycoproteins, bisecting N-linked glycans, human tissue
Introduction
The discovery of biomarkers useful for the early detection of epithelial ovarian cancer (EOC) has been complicated by several factors: (i) there are different histological subtypes of EOC that have very unique genetic alterations, (ii) the tissues and serum samples available for biomarker discovery are frequently collected from patients in later stages of the disease, and (iii) the expected sensitivity and specificity required for a biomarker to detect EOC is high due to the lower disease incidence and high risk surgical interventions. Despite these obstacles, proteomic studies profiling potential biomarkers for EOC have the potential to increase our knowledge about this disease, leading to improvements in clinical disease management.
Glycoproteins account for ∼80% of the proteins located at the cell surface and in the extracellular environment. Therefore, strategies to identify glycoproteins that have tumor-specific glycosylation changes could lead to the discovery of novel diagnostic and therapeutic markers. In our present study, we are targeting the neutral N-linked glycan structure known as bisecting glycosylation that is amplified in ovarian cancer tissues, focusing on membrane proteins. Membrane proteins play pivotal roles in regulating signaling pathways, and these proteins can frequently be found both on the cell surface and in circulation due to enzyme release making them ideal potential biomarkers.
Bisecting glycosylation, added by the glycosyltransferase known as GnT-III, is normally expressed on glycoproteins in the brain and kidney.1 The functional effects of amplified bisecting glycosylation on the growth and spread of ovarian cancer have not been determined previously. In this manuscript, we have identified membrane proteins with bisecting glycosylation from human primary endometrioid and serous ovarian cancer tissues and determined the glycan structures from these proteins using mass spectrometry methods. Functional annotation of this data demonstrates that membrane proteins with bisecting glycosylation in ovarian cancer have significant functions including: cell adhesion, control of immune responses, regulation of protein transport, and participation in signaling pathways such as Notch, Wnt, and TGFβ. Also, our glycan structural analysis from primary ovarian tumor tissues demonstrates that GnT-III overexpression in human ovarian cancer produces bisected N-linked glycans that have dramatic reductions in branching and extensions and may be useful as a biomarker for the development of therapeutic and diagnostic advances.
Materials and Methods
Materials
Biotinylated E-PHA (Phaseolus vulgaris erythroagglutinin) and streptavidin-HRP were obtained from Vector Laboratories (Burlingame, CA). PNGaseF was from New England Biolabs (Ipswich, MA). Chymotrypsin and sequencing grade trypsin were obtained from Promega. Vydac C18 silica columns were obtained from The Nest Group. The EGFR receptor antibody, HA antibody, 4G10 antibody, and all secondary antibodies were obtained from Santa Cruz Biotechnologies. The streptavidin-FITC, EGF-FITC, and EEA1 antibody were obtained from Invitrogen. The mouse HA-tagged GnT-III expression construct was a gift from Dr. Pamela Stanely (Albert Einstein College of Medicine). The puromycin resistance shRNA lentivirus construct was obtained from SBI System Biosciences. Fresh frozen human endometrioid ovarian cancer (EOC) tissues (n = 3), human serous ovarian cancer tissues (n = 3), normal human ovary tissues (n = 2), and isolated ovarian surface epithelial cells (100 mg each) were obtained from the Ovarian Cancer Institute (Atlanta, GA). Institutional review board approval for this research was obtained from the Georgia Institute of Technology, The University of Georgia, Northside Hospital (Atlanta, GA) and The University of Arkansas for Medical Sciences.
Membrane Protein Extraction, E-PHA Binding, and Peptide Preparation
Tissue or cells (100 mg) were homogenized in 10 mM Hepes pH 7.5. The slurry was placed in a Dounce glass homogenizer, and cells were lysed with 10 strokes of the large and fine pestle. The solution was incubated on ice for 1 h to continue the hypotonic cell lysis. Nuclei were removed by multiple centrifugations at 3000 rpm at 4 °C. The supernatant was placed in a Beckman ultracentrifuge tube, and the solution was centrifuged at 100 000g for 1 h at 4 °C to isolate a total membrane pellet. The pellet was rinsed and sonicated into 40 mM ammonium bicarbonate/10 mM DTT at room temperature for 2 h to reduce proteins. An equal volume of iodoacetamide (10 mg/mL) in 40 mM ammonium bicarbonate was added, and the solution was mixed by vortexing and placed in the dark for 45 min. The solution was dialyzed into 10 mM ammonium bicarbonate using Tube-O–Dialyzer membranes overnight at 4 °C. Protein concentration was measured using BCA assay, and 500 μg of total proteins was used for E-PHA capture, while 60 μg was digested with sequencing-grade trypsin overnight for prelectin nano-ESI–RPLC–MS/MS analysis. Sodium chloride was added to each E-PHA capture sample (final 150 mM), 5 mM calcium chloride, 5 mM magnesium chloride, and 10 μg biotinylated E-PHA lectin prior to overnight incubation at 4 °C. Lectin-bound proteins were captured using 100 μL of paramagnetic streptavidin particles (Promega) at 4 °C for 2 h. Beads were washed three times with 10 mM ammonium bicarbonate supplemented with salts. Proteins were eluted from beads using 4 M urea/4 mM DTT/40 mM ammonium bicarbonate at 50 °C for 1 h. Proteins were digested with 5 μg of sequencing-grade trypsin overnight at 37 °C. Peptides were acidified to a final of 0.1% trifluoroacetic acid and purified using C18 spin columns. Peptides were dried in a speedVac prior to resuspension in 19.5 μL of buffer A (0.1% formic acid) and 0.5 μL of buffer B (75% acetonitrile, 0.1% formic acid). Tryptic peptides were analyzed by nanoflow LC–MS/MS with a Thermo Orbitrap Velos mass spectrometer equipped with a Waters nanoACQUITY LC system. Tryptic peptides were separated by reverse-phase Jupiter Proteo resin (Phenomenex) on a 100 × 0.1 mm column using a nanoAcquity UPLC system (Waters). Peptides were eluted using a 120 min gradient from 98:2 to 40:60 buffer A/B ratio. Buffer A = 0.1% formic acid, 0.05% acetonitrile, buffer B = 0.1% formic acid, 75% acetonitrile. Eluted peptides were ionized by electrospray (2.0 kV), followed by MS/MS analysis using collision-induced dissociation on a Thermo LTQ Orbitrap Velos mass spectrometer. MS data were acquired using the FTMS analyzer in a profile mode at a resolution of 60 000 over a mass range of 375 to 1500 m/z. MS/MS data were collected from the top 15 peaks from each MS scan using the ion trap analyzer in centroid mode and normal mass range with normalized collision energy of 35.0 with a dynamic exclusion of two repeat counts at 30 s duration.
Permethylation and Glycan Analysis
To facilitate the analysis of oligosaccharides by MS, N-linked glycans from E-PHA captured glycoproteins were released by N-glycanase and permethylated as described previously.2 Briefly, E-PHA captured glycoproteins were digested with trypsin and chymotrypsin. The resulting digests were enriched for glycopeptides prior to PNGaseF (Prozyme) treatment to release the N-linked glycans. Contaminating peptides and salts were removed using Sep-Pak C18 chromatography, and the resulting glycans were permethylated prior to analysis using a linear ion trap mass spectrometer (LTQ Orbitrap; Thermo Scientific). Detection and relative quantification of bisecting N-linked glycan structures were carried out by using the total ion mapping (TIM) function of the Xcalibur software package version 2.0 (Thermo Fisher Scientific) as described previously.3 The TIM profiles were initially filtered with neutral loss of terminal HexNAc (Δ mass, 260 (1+), 130 (2+), and 86.7 (3+)) to assess the presence of bisecting N-linked glycan structures. N-glycan structures carrying terminal HexNAc were further subjected to manual MSn analysis to determine bisecting N-linked glycan structures.
Proteomic Data Analysis
Raw peptide data were converted to mzXML, and MS/MS spectra were searched against the International Protein Index (IPI) human database (IPI.HUMAN.v.3.71) using MyriMatch.4 All unambiquous identifications matched to multiple peptide sequences were excluded. Proteins identified required 2+ peptides and were grouped using IDPicker software.5 IDPicker incorporates searches against a separate reverse database, probability match from MyriMatch, and DeltCN scores to achieve a false-discovery rate of <5%. Information on MyriMatch and IDPicker can be found at http://fenchurch.mc.vanderbilt.edu/software.php. Variance for membrane protein extractions and E-PHA capture between normal and tumor tissues were calculated by measuring the number of peptides identified for proteins that adhere to E-PHA from membrane extractions in a non-glycan-dependent manner such as HSPA2 and RPL9. Values observed for tumor tissues for HSPA2 (CV 0.63/8 peptides) and RPL9 (CV 1.36/4 peptides). Values observed for normal tissues HSPA2 (CV 0.71/7 peptides) and RPL9 (CV 1.4/4 peptides). These results indicate a 12 ± 1.0% variance between normal and tumor samples due to processing. This variance is well below the 1.5 times or 150% increase in peptides and spectral counts we have established as criteria to be considered as a glycoprotein with tumor-specific E-PHA reactivity.
Biological Functional Annotation
Proteins in Table 1 were uploaded to DAVID 2009 (the Database for Annotation, Visualization and Integrated Discovery) for analysis.
Table 1. Membrane Proteins Enriched by E-PHA Capture from Ovarian Carcinoma Tissuesa.
| IPIc | gene code | mol wt (kDa) | % coverage | protein name | unique peptides | subtype | localization | function | HPA/stainingb |
|---|---|---|---|---|---|---|---|---|---|
| IPI00478003 | A2M | 163 | 40 | alpha 2-macroglobulin | 39 | both | extracellular | immune | |
| IPI00291373 | ABCD1 | 81 | 4.5 | ATP-binding cassette sub family D | 2 | both | membrane | transport | |
| IPI00437751 | ACE | 149 | 3.2 | angiotensin-converting enzyme, CD143 | 4 | endo | membrane | enzyme | low (3/11) |
| IPI00013897 | ADAM10 | 84 | 4 | disintegrin and metalloproteinase domain-containing protein 10, CD156c | 2 | endo | membrane | enzyme | low/med (5/1 of 11) |
| IPI00021812 | AGR2 | 19 | 20 | anterior gradient protein 2 homologue | 3 | endo | extracellular | protein binding | high (3/10) |
| IPI00015102 | ALCAM | 65 | 5.6 | CD166 antigen | 3 | both | membrane | cell adhesion | high/med/low (2/3/6/of 12) |
| IPI00004457 | AOC3 | 84 | 17.3 | membrane primary amine oxidase | 7 | both | membrane | enzyme | low (1/11) |
| IPI00022391 | APCS | 24 | 31.8 | serum amyloid P-component | 6 | both | extracellular | protein binding | low (3/11) |
| IPI00031131 | APMAP | 46 | 18 | adipocyte plasma membrane-associated protein | 7 | both | membrane | enzyme | high/med/low (1/6/1/of 9) |
| IPI00021841 | APOA1 | 29 | 47 | apolipoprotein A-I | 13 | both | extracellular | transport | low (1/12) |
| IPI00021842 | APOE | 36 | 29 | apolipoprotein E | 7 | both | membrane | transport | high/med/low (3/3/3/of 12) |
| IPI00418446 | ASAH1 | 43 | 21 | acid ceramidase | 8 | both | lysosome | enzyme | med/low (4/5/of 12) |
| IPI00418431 | ASPN | 44 | 28 | asporin | 10 | both | extracellular | signaling | high/med/low (1/5/1/of 12) |
| IPI00006482 | ATP1A1 | 35 | 25 | sodium/potassium-transporting ATPase subunit alpha-1 | 19 | both | membrane | transport | high/med/low (5/6/1 of 12) |
| IPI00002406 | BCAM | 67 | 24 | basal cell adhesion molecule, CD239 | 10 | both | membrane | cell adhesion | high/med/low (6/4/2/of 12) |
| IPI00218200 | BCAP31 | 27 | 23 | B-cell receptor-associated protein 31 | 8 | both | membrane, ER | transport | med (12 of12) |
| IPI00010790 | BGN | 41 | 28 | biglycan | 6 | both | extracellular | signaling | low (2/11) |
| IPI00019906 | BSG | 29 | 26 | basigin (CD147) | 5 | both | membrane | signaling | high/med/low (3/5/3/of 12) |
| IPI00026241 | BST2 | 23 | 13.8 | bone marrow stromal antigen, CD317 | 2 | ser | membrane, GPI | immune | high/med/low (3/8/1/of 12) |
| IPI00418163 | C4B | 192 | 16 | complement component 4B | 24 | both | membrane | immune | med/low (3/1/of 12) |
| IPI00029260 | CD14 | 40 | 15 | monocyte differentiation antigen | 4 | both | membrane, GPI | immune | |
| IPI00104074 | CD163 | 125 | 9.5 | scavenger receptor cysteine-rich type 1 protein M130 | 8 | both | membrane | immune | |
| IPI00374740 | CD47 | 35 | 6 | leukocyte surface antigen CD47 | 2 | both | membrane | signaling | high/med/low (6/2/2/of 11) |
| IPI00011302 | CD59 | 14 | 70 | CD59 glycoprotein | 8 | both | membrane, GPI | immune | med/low (1/3/of 11) |
| IPI00022933 | CD74 | 33 | 16.9 | HLA DR antigen | 3 | both | membrane | immune | high/med (7/4/of 11) |
| IPI00215997 | CD9 | 25 | 15.3 | CD9 antigen | 2 | both | membrane | cell adhesion | high/med/low (3/8/1/of 12) |
| IPI00024046 | CDH13 | 83 | 3.4 | cadherin-13 | 2 | ser | membrane, GPI | cell adhesion | |
| IPI00000513 | CDH1 | 90 | 5 | E-cadherin | 3 | endo | membrane | cell adhesion | high (12 of 12) |
| IPI00019591 | CFB | 140 | 10 | complement factor B | 10 | both | membrane | immune | med/low (6/4/of 10) |
| IPI00141318 | CKAP4 | 66 | 47 | cytoskeleton-associated protein 4 | 27 | both | membrane | cell proliferation | high/med/low (5/2/3 of 12) |
| IPI00291262 | CLU | 52 | 22 | clusterin | 9 | both | membrane extracellular | immune | med/low (3/2/of 10) |
| IPI00297646 | COL1A1 | 139 | 10 | collagen alpha-1(I) chain | 12 | both | extracellular | signaling | low (1/of 9) |
| IPI00017292 | CTNNB1 | 85 | 15.5 | beta-catenin | 11 | both | membrane | signaling | high/med (7/4/of 11) |
| IPI00295741 | CTSB | 38 | 18 | cathepsin B | 4 | both | lysosome | enzyme | high/med/low (1/5/1 of 11) |
| IPI00011229 | CTSD | 44 | 24 | cathepsin D | 7 | both | lysosome | cell adhesion | high/med/low (1/3/7/of 12) |
| IPI00002714 | DKK3 | 38 | 7.5 | Dickkopf-related protein | 2 | endo | extracellular | signaling | med/low (2/5/of 12) |
| IPI00029658 | EFEMP1 | 55 | 20 | EGF-containing fibulin-like extracellular matrix protein 1 (fibulin-3) | 7 | both | extracellular | cell adhesion | |
| IPI00018274 | EGFR | 134 | 2.6 | epidermal growth factor receptor | 2 | ser | membrane | signaling | med/low (1/2/of 12) |
| IPI00013079 | EMILIN | 106 | 30 | emilin-1 | 28 | both | extracellular | cell adhesion | |
| IPI00219625 | ENG | 68 | 13 | endoglin, CD105 | 6 | ser | membrane | cell adhesion | low (1/of 12) |
| IPI00296215 | EPCAM | 21 | 27 | epithelial cell adhesion molecule, CD326 | 3 | both | membrane | cell proliferation | high/med (7/5/of 12) |
| IPI00298105 | EPHA3 | 110 | 4.2 | ephrin receptor | 3 | endo | GPI | signaling | low (11/of 12) |
| IPI00010951 | EPHX1 | 53 | 33 | eopxide hydrolase | 14 | both | membrane, ER | enzyme | med (1/of 12) |
| IPI00409635 | ESYT2 | 98 | 8 | extended synaptotagmin-2 | 6 | ser | membrane | protein binding | low (10/of 10) |
| IPI00382428 | FBLN5 | 60 | 14 | fibulin 5 | 7 | both | extracellular | cell adhesion | |
| IPI00328113 | FBN1 | 312 | 35 | fibrillin-1 | 75 | both | extracellular | signaling | med (1/of 11) |
| IPI00294578 | GGT2 | 77 | 15 | gamma-glutamyltransferase 2 | 7 | both | membrane | enzyme | low (1/of 12) |
| IPI00002243 | GGT5 | 62 | 11 | gamma-glutamyltransferase 5 | 5 | both | membrane | enzyme | med/low (1/2/of 11) |
| IPI00016801 | GLUD1 | 61 | 38 | glutamate dehydrogenase 1 | 17 | both | membrane, mito | enzyme | high/med (11/1/of 12) |
| IPI00001592 | GPNMB | 64 | 4.4 | transmembrane glycoprotein NMB | 2 | endo | membrane | cell adhesion | low (3/of 12) |
| IPI00004901 | GPRC5C | 48 | 5 | G-protein coupled receptor family C group 5 member C | 3 | endo | membrane | signaling | med/low (2/1/of 9) |
| IPI00024284 | HSPG2 | 468 | 3.1 | basement membrane heparin sulfate proteoglycan | 10 | both | membrane | protein binding | |
| IPI00289819 | IGF2R | 274 | 1.8 | insulin-like growth factor receptor 2 | 4 | both | membrance | transport | high/med/low (1/4/4 of 11) |
| IPI00217563 | ITGB1 | 88 | 22 | integrin beta-1, CD29 | 15 | both | membrane | signaling | high/med/low (3/6/3/of 12) |
| IPI00291792 | ITGB2 | 85 | 16 | integrin beta-2, CD18 | 10 | both | membrane | signaling | |
| IPI00032328 | KNG1 | 71 | 8 | kininogen-1 | 4 | both | extracellular | protein binding | med (1 of 12) |
| IPI00334532 | L1CAM | 140 | 1.9 | neural cell adhesion molecule L1, CD171 | 2 | endo | membrane | cell adhesion | med/low (3/5/of 12) |
| IPI00884105 | LAMP1 | 45 | 7 | lysosome-associated membrane glycoprotein 1, CD107a | 3 | both | membrane | transport | high/med/low (2/9/1/of 12) |
| IPI00026530 | LMAN1 | 57 | 24 | ERGIC-53 | 9 | both | membrane | transport | high/med/low (6/4/1/of 12) |
| IPI00009950 | LMAN2 | 40 | 6 | vesicular integral-membrane protein VIP36 | 2 | endo | membrane | transport | low (6/of 10) |
| IPI00020557 | LRP1 | 504 | 5.4 | Prolow-density lipoprotein receptor-related protein 1, CD91 | 20 | both | membrane | signaling | med/low (1/1/of 11) |
| IPI00024292 | LRP2 | 522 | 2.5 | low-density lipoprotein receptor-related protein 2 | 10 | both | membrane | signaling | low (3/10) |
| IPI00301202 | MAGT1 | 28 | 8 | magnesium transporter 1 | 2 | endo | membrane | transport | high/med (2/10/of 12) |
| IPI00328156 | MAOB | 46 | 20 | amine oxidase [flavin-containing] B | 9 | both | membrane | enzyme | high/med/low (3/4/5/of 12) |
| IPI00016334 | MCAM | 71 | 8 | cell surface glycoprotein MUC18 | 4 | both | membrane | cell adhesion | med/low (1/1/of 11) |
| IPI00022621 | MFAP2 | 21 | 12 | microfibril-associated glycoprotein 2 | 2 | both | extracellular | cell adhesion | low (1/of 10) |
| IPI00002236 | MFGE8 | 43 | 17 | lactadherin | 5 | both | membrane | cell adhesion | low (3/of 12) |
| IPI00029046 | MLEC | 32 | 19 | malectin | 4 | both | membrane, ER | protein binding | low (3/of 12) |
| IPI00247063 | MME | 85 | 9.4 | neprilysin, CD10 | 6 | endo | membrane | enzyme | |
| IPI00027509 | MMP9 | 78 | 6.5 | matrix metalloproteinase 9 | 3 | endo | extracellular | enzyme | |
| IPI00013955 | MUC1 | 60 | 8 | mucin 1 | 4 | endo | membrane | cell adhesion | med/low (7/3/of 10) |
| IPI00021048 | MYOF | 234 | 18 | myoferlin | 29 | both | membrane | transport | high/med/low (6/3/2/of 11) |
| IPI00021983 | NCSTN | 78 | 4.5 | nicastrin | 2 | ser | membrane | signaling | high/med/low (2/6/1/of 10) |
| IPI00329352 | NOMO3 | 139 | 9 | nodal modulator 3 | 9 | both | membrane | signaling | |
| IPI00465159 | NOTUM | 49 | 11 | protein notum homology | 4 | endo | extracellular membrane, | enzyme | |
| IPI00009456 | NT5E | 63 | 16 | 5′-nucleotidase, CD73 | 8 | both | GPI | enzyme | high/med/low (3/4/5/of 12) |
| IPI00022255 | OLFM4 | 57 | 57 | olfactomedin-4 | 9 | endo | extracellular | cell adhesion | |
| IPI00024621 | OLFM3 | 43 | 12 | olfactomedin-3 | 4 | ser | extracellular | cell adhesion | |
| IPI00013569 | PAPPA2 | 198 | 3.3 | pappalysin-2 | 4 | both | extracellular | enzyme | low (5/of 12) |
| IPI00384280 | PCYOX1 | 56 | 14 | prenylcysteine oxidase 1 | 6 | both | lysosome | enzyme | med/low (2/2/of 11) |
| IPI00220739 | PGRMC1 | 22 | 34 | membrane-associated progesterone receptor component 1 | 5 | both | membrane | protein binding | med/low (6/6/of 12) |
| IPI00004573 | PIGR | 83 | 16 | polymeric immunoglobulin receptor | 8 | both | membrane extracellular | protein binding | high (4/11) |
| IPI00019580 | PLG | 90 | 24 | plasminogen | 14 | both | extracellular | enzyme | low (2/of 12) |
| IPI00299116 | PODXL | 55 | 8.6 | podacalyxin-like protein | 4 | both | membrane | cell adhesion | med (2/of 12) |
| IPI00007960 | POSTN | 93 | 20 | periostin | 12 | both | extracellular | cell adhesion | |
| IPI00020987 | PRELP | 44 | 33 | prolargin | 10 | both | extracellular | protein binding | low (2/of 12) |
| IPI00012540 | PROM1 | 97 | 2.7 | prominin-1, CD133 | 2 | endo | membrane | signaling | high/med/low (3/3/3/of 12) |
| IPI00006865 | SEC22B | 25 | 26 | vesicle-trafficking protein SEC22b | 4 | both | membrane | transport | |
| IPI00553177 | SERPINA1 | 40 | 17 | alpha-1-antitrypsin | 5 | both | extracellular | protein binding | low (1/of 10) |
| IPI00006114 | SERPINF1 | 46 | 16 | pigment-epithelium derived factor | 6 | endo | extracellular | cell proliferation | med/low (2/2/of 12) |
| IPI00019385 | SSR4 | 42 | 23 | translocon-associated protein subunit delta precursor | 3 | both | membrane | transport | high/med/low (1/10/1/of 12) |
| IPI00219682 | STOM | 31 | 40 | erythrocyte band 7 integral membrane protein | 8 | both | membrane | transport | |
| IPI00296099 | THBS1 | 129 | 26 | thrombospondin-1 | 25 | both | extracellular | cell adhesion | high/med (5/6/of 12) |
| IPI00018769 | THBS2 | 129 | 28 | thrombospondin-2 | 24 | both | extracellular | cell adhesion | med/low (4/2/of 10) |
| IPI00022892 | THY1 | 16 | 27 | Thy-1 membrane glycoprotein, CD90 | 3 | both | membrane, GPI | signaling | med (1/of 12) |
| IPI00640293 | TIMP3 | 24 | 34 | metalloproteinase inhibitor 3 | 6 | both | extracellular | protein binding | high/med/low (2/5/1/of 11) |
| IPI00028055 | TMED10 | 21 | 13 | transmembrane emp24 domain-containing protein 10 | 2 | both | membrane | transport | high/med/low (1/8/3/of 12) |
| IPI00298362 | TNFRSF11B | 46 | 9.4 | tumor necrosis superfamily receptor | 3 | both | extracellular | signaling | high/med/low 3/6/3/of 12) |
| IPI00554617 | TPP1 | 61 | 7 | tripeptidyl-peptidase 1 | 3 | both | lysosome | enzyme | med/low (1/1/of 10) |
| IPI00298971 | VTN | 54 | 23 | vitronectin | 9 | both | extracellular | cell adhesion |
Two independent peptide matches required from two separate samples. Tumor-specific E-PHA enrichment required 1.5 times increase in spectral counts and peptide abundance in tumor tissues compared with nonmalignant tissues.
Human Protein Atlas ovarian cancer tissue staining data (www.proteinatlas.org).
International Protein Index Database.
ShRNA Lentivirus Cloning, Lentivirus Production, Cell Transduction, and Cell Proliferation Assays
Two target sequences were tested for GnT-III suppression in human cells. One sequence gave >75% reduction in GnT-III expression in human ovarian adenocarcinoma cells (NM_002409, 5′ ctacgaccggttccactac). Briefly, the target sequence along with restriction site linkers and shRNA loop sequence was cloned into the pSIH-H1shRNA vector (System Biosciences). Lentivirus was produced in 293TN cells, and OVCAR3 cells were transduced as previously described.6 OVCAR3 cells stably expressing GnT-III siRNA target sequence were transfected transiently with HA-tagged mouse GnT-III cDNA expression plasmid (gift from Pamela Stanley) or empty cDNA vector. Subconfluent cells were serum-starved for 24 h prior to the addition of 25 ng/mL EGF for 24 h. Cell proliferation was measured using the CellTiter Aqueous One solution (Promega) according to the manufacturer’s recommendations and absorbance was read at 490 nm. The ± SEM represents results from three separate experiments.
Microscopy
E-PHA Lectin Staining
Cells were grown to subconfluence in chamber slides before fixing in 3% formaldehyde/1× phosphate buffered saline (PBS) for 10 min. Cells were blocked using 1% bovine serum albumin (BSA)/1XPBS-0.2% tween-20 (PBST) prior to the addition of biotin-labeled E-PHA (Vector Laboratories) diluted 1:300 in 1%BSA/1XPBST. Bound lectin was detected using fluorescein-tagged streptavidin (Invitrogen) diluted 1:500 in 1% BSA/1XPBST.
EGF Internalization
Cells were grown to subconfluence and serum starved prior to analysis. Cells were placed on ice for 1 h in the presence of fluorescein tagged EGF (50 ng/mL) (Invitrogen). Cells were placed back at 37 °C for 30 min before the removal of unbound EGF with ice-cold 20 mM acetic acid/0.5 M NaCl. Cells were fixed prior to detection of early endosomes using anti-EEA1 (1:400) (Invitrogen), followed by rhodamine antimouse IgG (1:250). Nuclei were visualized using Dapi counterstain.
Results
Glycoproteins at the Cell–Cell Boundary of Ovarian Cancer Cells Are Bound by E-PHA in a GnT-III-Dependent Manner
In previous studies we have used the plant lectin E-PHA (Phaseolus vulgaris Erythroagglutinin) to capture soluble or extracellular glycoproteins with bisecting glycosylation from human ovarian cancer tissues due to the elevation of this form of glycosylation in human epithelial EOC.7,8 In this study, we are comparing the analysis of both serous and endometrioid ovarian cancer, the two most abundant histological subtypes of EOC, focusing on the identification of glycoproteins located in the membrane fraction that have tumor-specific bisecting glycosylation. Bisecting glycosylation is amplified in ovarian cancer due to increased expression of the glycosyltransferase GnT-III.7,9 Therefore, to explore the localization of glycoproteins with bisecting glycosylation and validate the specificity of E-PHA for this glycan structure, we have created cell lines using the human ovarian adenocarcinoma cell line known as OVCAR3 that express either a control shRNA sequence that targets no human gene (control siRNA) or a GnT-III shRNA target sequence (GnT-III siRNA). Constitutive expression of the GnT-III siRNA reduced the expression of GnT-III by >75% compared with control siRNA (Figure 1A). E-PHA binding in OVCAR3 cells stably expressing the control siRNA is strong, localized to the cell–cell junctions, indicating a concentration of glycoproteins at the cell periphery with bisecting glycosylation. The cell boundaries of OVCAR3 cells stably expressing GnT-III siRNA are not visible, indicating a loss of E-PHA accumulation and a reduction of bisecting glycosylation (Figure 1B). Therefore, E-PHA binding to glycoproteins in OVCAR3 cells requires the expression of GnT-III, and glycoproteins with bisecting glycosylation are concentrated at the cell periphery.
Figure 1.
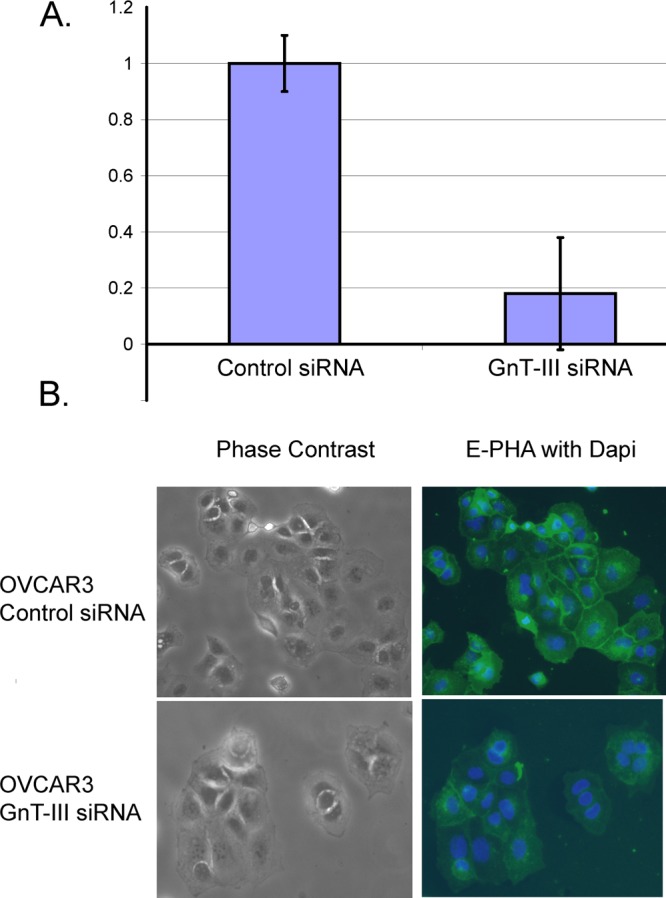
E-PHA binds bisecting glycans in a GnT-III dependent manner. (A) Real-time PCR measurement of GnT-III mRNA levels in OVCAR3 human ovarian cancer cells expressing control siRNA or GnT-III siRNA. GnT-III levels were normalized to RPL4 and the control siRNA level was set to 1.0 for comparison; error bars represent the mean ± SE for triplicate measurements. (B) E-PHA staining of glycoproteins with bisecting glycosylation in OVCAR3 cells; nuclei are stained with DAPI, magnification 20×.
E-PHA Capture of Tumor-Specific Glycoproteins from Serous and Endometrioid Ovarian Cancer Tissues Reveals Common Glycoprotein Targets for the Two Most Common Types of EOC
Fresh frozen tissue samples (Supplementary Table 1 in the Supporting Information) from EOC (n = 3), serous ovarian cancer (n = 3), nonmalignant ovary (n = 2), or isolated nonmalignant ovarian surface epithelial cells (n = 1) were each subjected to membrane protein isolation and E-PHA lectin capture. A diagram of the extraction and analysis scheme for glycoproteomics is shown in Figure 2A. Comparison analysis of each membrane extracted sample before and after E-PHA capture enabled the identification of glycoproteins that are potential acceptors for bisecting glycosylation. Glycoproteins that were enriched following E-PHA capture by at least 1.5 times (150% by spectral counts) in tumor tissues relative to nonmalignant controls are listed in Table 1. In Table 1, we provide Human Protein Atlas (HPA)10 antibody immunohistochemistry staining data (for ovarian cancer tissues) for each E-PHA enriched glycoprotein. Over 80% of the glycoproteins in Table 1 have positive antibody immunohistochemistry staining in ovarian carcinoma tissues (data from the HPA annotated in Table 1).10 The majority (81%) of glycoproteins enriched by E-PHA are present in both histological subtypes of EOC, suggesting that the glycosylation changes on these proteins are important for the survival and growth of ovarian cancer cells (Figure 2B). The overall functional annotation (Figure 2C) of glycoproteins enriched by E-PHA capture reveals that the largest functional categories are cell adhesion, regulation of signaling, enzymatic functions, and control of protein transport (Figure 2C). Several glycoproteins have reported activities in the Notch11−15 (LRP2, POSTN, THBS1, THBS2, MFAP4, OLFM4, NCSTN), TGFβ,16,17(ASPN, FBN1, ENG), and Wnt18 (COL1A1, CTNNB1) signaling pathways. These pathways are all implicated in control of cancer stem cell populations.
Figure 2.
(A) Schematic flow of glycoproteomic experiments. Membrane proteins were analyzed by nanoflow-ESI–RPLC–MS/MS before E-PHA capture to verify equivalent protein quality and quantity. Biotinylated E-PHA was added to membrane extracted proteins, and glycoproteins with bisecting glycans were captured using streptavidin magnetic resin prior to nanoflow-ESI–RPLC–MS/MS. Membrane glycoproteins enriched in tumor tissues relative to nonmalignant tissues by E-PHA capture by at least 1.5 times or 150% by spectral counts were identified and are listed in Table 1. (B) Venn diagram showing the percentage of glycoproteins in Table 1 from endometrioid only (13% purple), serous only (6% blue), or shared (81% blue/purple). (C) Functional annotation of E-PHA enriched membrane glycoproteins using DAVID 2009.
E-PHA Captured Glycoproteins from Serous and Endometrioid Ovarian Cancer Share a Unique Truncated N-Linked Glycan Structure Not Found in Control Nonmalignant Samples
The membrane-extracted proteins from endometrioid and serous ovarian tissues were subjected to PNGaseF treatment following E-PHA capture. Membrane proteins extracted from nonmalignant ovarian surface epithelial cells have low levels of binding to E-PHA; therefore, equivalent levels of total membrane protein were used for the release of N-linked glycans. The N-linked glycans were released from trypsin and chymotrypsin digests using PNGaseF before permethylation and analysis using mass spectrometry methods. We have identified nine very unique and unusual bisecting N-linked glycan structures that are present in both endometrioid and serous EOC tissues (Figures 3 and 4) that were not observed in control nonmalignant ovarian epithelial cells. The full MS reveals the presence of abundant high mannose glycan structures (Supplemental Figure 1 in the Supporting Information) that are marked with asterisks in Figure 3. MS6 analysis was necessary to confirm the presence of the bisecting glycan structures (Supplemental Figures 2–4 in the Supporting Information). The fragment ion observed at m/z 444 for MS6 clearly demonstrates the presence of the bisected mannose, of which three of the hydroxyl groups were substituted with three HexNAc (Supplemental Figures 2–4 in the Supporting Information). Although, some of the bisecting N-linked glycan structures contain isobaric forms of N-glycans capped with terminal HexNAc, which demonstrates a specific fragment ion at m/z 458 for MS6 (Supplemental Figure 2 in the Supporting Information). Thus, the sequential MSn approach using permethylated N-glycans effectively discriminates bisecting N-glycan structures from other N-glycans with terminal HexNAc. The most prevalent bisecting N-linked structures are shown on the TIM chromatogram (Figure 3). These structures are dramatically truncated with minimal branching with reductions in distal galactose and sialic acid modifications. Most of the bisecting N-linked glycans also have core fucosylation, another neutral N-linked glycan structure known to be amplified in EOC.7,19 The unusual lack of branched glycans with the presence of core fucosylation may represent a biomarker useful for the development of future therapeutic and diagnostic applications.
Figure 3.
Prevalence of unique bisecting N-linked glycan structures in ovarian carcinoma tissues. PNGaseF released N-linked glycan structures from glycoproteins enriched by E-PHA capture. Glycans were released from pooled E-PHA captured glycoproteins from endometrioid (n = 3), serous (n = 3), or total membrane proteins from nonmalignant control surface epithelial cells (n = 1). TIM profile was filtered with neutral loss of HexNAc and bisecting N-linked glycan structures were identified by MSn analysis manually. Peaks labeled M7, M8, and M9 indicate high mannose type N-linked glycans.
Figure 4.

Bisecting N-linked glycan structures identified from human endometrioid and serous EOC. Bisecting structures identified from endometrioid and serous ovarian cancer tissues by MSn analysis are summarized. Represented glycan structures are in accordance with the guidelines from the Consortium for Functional Glycomics (CFG): blue square, N-acetylglucosamine (GlcNAc); green circle, mannose (Man); yellow circle, galactose (Gal); red triangle, fucose (Fuc).
Novel Role for Bisecting Glycosylation in Protein Transport
Our glycoproteomic analysis of E-PHA captured membrane glycoproteins from EOC has led to the identification of potential functions that bisecting glycosylation changes may impact. Several proteins enriched by E-PHA have potential functions in the control of protein transport (BCAP31, LMAN1, LMAN2, LRP2, MYOF, SEC22B, SSR4, and TMED10). To explore this area of functional significance further, we exposed the OVCAR3 cell lines, control siRNA, or GnT-III siRNA to fluorescently tagged EGF to track the transport of receptors that receive bisecting glycosylation and bind EGF, such as EGFR. EGF was internalized in both cells; however, in control cells, the EGF is distributed to several intracellular vesicles, and in GnT-III siRNA cells, the EGF is primarily localized in a perinuclear compartment (Figure 5). EGF is active in the cells demonstrated by the ligand-induced phosphorylation observed (Supplementary Data in the Supporting Information, Figure 5, last panel). The early endosomal marker EEA1 also displays a change in intracellular localization in GnTIII siRNA cells compared with control cells. These results suggest that bisecting glycosylation is promoting transport of certain proteins in ovarian cancer cells. To confirm that this protein transport change in GnT-III siRNA cells is due to bisecting glycosylation, we show that we can rescue this defect with the expression of mouse GnT-III in GnT-III siRNA stable OVCAR3 cells. Mouse GnT-III is not targeted by our constitutively expressed human GnT-III shRNA target sequence. Stable GnT-III siRNA OVCAR3 cells were transiently transfected with full-length mouse GnT-III fused to an HA-tag. Immunofluorescence microscopy demonstrates that cells transiently transfected show positive E-PHA staining (green, FITC, Figure 6A)) and positive HA-tag staining (red, rhodamine, Figure 6A), indicating that the mouse GnT-III is active in human cells. Introduction of the mouse GnT-III gene into OVCAR3 cells stably expressing GnT-III siRNA can rescue the transport of EGF-bound glycoproteins (Figure 6B). Fluorescent EGF and rhodamine-stained EEA1 are transported into different intracellular vesicles in rescued cells (Figure 6B top panel), while EGF and EEA1 show stalled movement in a perinuclear compartment without rescue in the GnT-III siRNA cells without mouse GnT-III (Figure 6B, bottom panel). These experiments confirm a novel role for GnT-III expression and therefore bisecting glycans in promoting vesicular transport in human ovarian adenocarcinoma cells. Downstream signaling induced by EGF is suppressed in GnT-III siRNA cells (Supplementary Data in the Supporting Information, Figure 6). OVCAR3 cells stably expressing GNT-III siRNA have reduced basal proliferation and show no change in cell proliferation following EGF addition (Figure 6C). Transfection of mouse GNT-III increases basal cell proliferation and restores the proliferation response to EGF (Figure 6C). These data indicate that GNT-III expression and bisecting glycosylation promote the vesicular transport and activity of EGF-bound receptors such as EGFR.
Figure 5.

Protein transport is impaired in GnT-III siRNA cells. (A) OVCAR3 control siRNA cells and GnT-III siRNA cells were serum-starved prior to incubation with fluorescein-EGF (50 ng/mL) for 1 h on ice. Cells were placed in a 37 °C incubator for 30 min prior to removal of EGF that was not internalized by an acid wash. Cells were fixed and stained using an EEA1 monoclonal antibody. Nuclei are stained with DAPI, magnification 40×.
Figure 6.
Protein transport and response to EGF is restored with mouse GnT-III expression. (A) OVCAR3 cells expressing GnT-III siRNA constitutively were transiently transfected with mouse HA-GnT-III (gift from Pamela Stanley). Cells expressing mouse HA-GnT-III show positive E-PHA staining and HA antibody staining indicating expression and activity in human cells. Nuclei are stained with DAPI, magnification 20×. (B) Transport of EGF fluorescein-bound receptors is rescued by mouse HA-GnT-III expression. OVCAR3 GnT-III siRNA cells were transiently transfected with mouse HA-GnT-III (top panel) or empty vector (bottom panel). Cells were serum-starved for 2 h 1 day after transfection prior to incubation with fluorescein-EGF, as described for Figure 4. (C) OVCAR3 GnT-III siRNA cells were transiently transfected with vector-only or mouse HA-GnT-III prior to plating into 96-well plates. Cells were serum-starved and incubated with vehicle only or EGF 35 ng/mL for 24 h. Cell proliferation was measured using CellTiter Aqueous One (Promega) reagent. Error bars represent the ± SEM from three separate experiments.
Discussion
The mechanisms that lead to overexpression of GnT-III in ovarian cancer may involve epigenetic mechanisms.9 A recent study of human ovarian cancer and nonmalignant ovarian epithelial cell lines demonstrated that the Mgat3 gene could be induced following treatment with an inhibitor of DNA methyltransferase. These results provide strong evidence that the increased expression of Mgat3 transcripts in ovarian cancer may be due to epigenetic dysregulation of hypermethylation.
The normal functional roles of bisecting glycans on glycoproteins in the kidney and brain are not well understood. The overexpression of GnT-III in ovarian cancer offers an opportunity to learn more about this form of glycosylation and discover new mechanisms that contribute to ovarian tumor growth.
We have discovered a new function for GnT-III expression and bisecting N-linked glycosylation in promoting protein transport. Suppression of GnT-III expression in the human ovarian cancer cell line OVCAR3 results in a dramatic change in the intracellular localization of EGF bound receptors. We have confirmed that EGFR is glycosylated by GnT-III in OVCAR3 cells (Supplementary Data in the Supporting Information, Figure 5, top panel). EGF was capable of activating the EGFR receptor, and there are no differences in EGFR autophosphorylation levels between control siRNA and GnT-III siRNA OVCAR3 cells (Supplementary Figure 5 in the Supporting Information), suggesting that receptor dimerization and endocytosis are not influenced by bisecting glycosylation. However, the transport of vesicles containing EGF bound receptor is slowed in cells with reduced GnT-III expression, allowing the EGF to accumulate in a perinuclear compartment. The accumulation of EGF-bound receptor in this perinuclear compartment reduces downstream EGF-induced signaling (Supplementary Data in the Supporting Information, Figure 6) and inhibits proliferative responses to EGF (Figure 6C). Our ability to rescue the transport and activity of EGF-bound receptors with the expression of mouse HA-GnT-III (Figure 6A–C) supports a role for GnT-III expression and bisecting glycosylation as a factor promoting growth factor receptor transport and activity. Our glycoproteomic analysis from ovarian cancer tissues also supports a role for bisecting glycans in protein transport as several proteins with known transport functions are enriched by E-PHA. Therapeutic strategies to block GnT-III expression, enzymatic activity, or growth factor receptor interactions with transport proteins may be new avenues to reduce ovarian cancer cell responses to growth factor ligands by slowing the movement of receptors within the cell.
The unusual bisecting N-linked glycan structures identified for both endometrioid and serous EOC subtypes are an important discovery that may lead to the development of antibodies or strategies to target ovarian cancer cells. The prevalence and similarity of these glycan structures for both histological subtypes of ovarian cancer indicate an importance for this glycan structure in the development and survival of ovarian carcinoma cells. Also, the lack of these structures in nonmalignant tissues and cells supports the use of these structures as potential biomarkers. A recent glycomic analysis of all N-linked glycan structures for membrane glycoproteins isolated from ovarian cancer and nonmalignant ovarian cell lines also found the presence of bisecting structures specific to malignant cells.9 GnT-III expression and bisecting glycosylation have been reported to compete with branching enzymes such as GnT-V, reducing tumor growth for cancers with elevated GnT-V expression such as breast cancer.20 However, in ovarian cancer cells there is no competition with GnT-V, and the bisecting glycan structure has a growth promoting role. Furthermore, the truncated glycan structures found on bisecting N-linked glycans released from ovarian cancer tissues have not been observed in analysis of bisecting N-linked glycan structures isolated from nonmalignant cell types such as HEK293 and CHO.21,22 These results indicate that there may be additional changes in Golgi transport or other as yet unknown factors in ovarian cancer that may contribute to the production of these unusual glycan structures.
Several of the extracellular glycoproteins identified following the stringent membrane isolation procedures (THBS1, THBS2, LRP2, POSTN) are adhesive glycoproteins with reported functions as noncanonical regulators of the Notch signaling pathway.11 The extraction of these proteins with membrane protein fractions suggests high affinity interactions with membrane proteins. We have identified nicastrin (NCSTN) as a glycoprotein enriched by E-PHA specific to serous ovarian cancer. NCSTN is an essential regulator for gamma secretase activity in the Notch pathway.15 The Notch signaling pathway has been identified by The Cancer Genome Atlas (TCGA) as a pathway significant for the pathophysiology of ovarian cancer.23 The abundance of glycoproteins known to participate in the Notch signaling pathway that are enriched by E-PHA capture in tumor tissues suggest that this unusual bisecting glycan structure may be important for Notch functions in ovarian carcinoma.
High-grade serous ovarian cancer is the largest fraction of ovarian cancer patients (∼80%). The small percentage of membrane glycoproteins that we identified that are unique to this group of cancers following E-PHA capture may offer some insights into why these tumors are so aggressive.
BST2
The BST2 (CD317) is a tetherin glycoprotein that is linked to the membrane via both transmembrane and glycosylphosphatidylinositol (GPI) linkage. This protein is normally expressed in very specialized cells such as paneth cells and in the bone marrow with functions as an intrinsic immunity factor.24 The functions of BST2 in cancer are largely unknown. BST2 expression in breast cancer is associated with metastasis.25 The use of this protein as a therapeutic biomarker for multiple myeloma has been complicated due to expression in nonmalignant bone marrow cells.26 Our discovery of a unique form of glycosylation on this protein in serous ovarian cancer is novel and may allow use of BST2 as a diagnostic or therapeutic target for this solid tumor.
ENG
Endoglin or CD105 is another glycoprotein unique to the serous ovarian carcinoma tumor tissues. Endoglin is a potential marker for mesenchymal stem cell populations.27 A recent study identified endoglin as a potential GPI anchored protein biomarker for breast cancer.6 The GPI linkage may explain the increased levels of soluble endoglin in the ascites of ovarian cancer patients.28 Endoglin expression in ovarian cancer is associated with shorter survival.29 A recent study found that endoglin contributes to chemoresistance in ovarian cancer cells.30 Endoglin is typically expressed in endothelial cells at low levels. Therefore, the increased expression levels of endoglin in ovarian cancer coupled to the presence of unique bisecting glycosylation make endoglin a potential therapeutic target for serous ovarian carcinoma.
CDH13
Cadherin-13 or T (truncated) cadherin is an atypical GPI-linked cadherin. CDH13 has been referred to as a tumor suppressor that is dysregulated by epigenetic changes in methylation for many cancers including ovarian cancer.31 However, CDH13 has also been demonstrated as a stromal factor that is expressed in the tumor microvascular that can favor tumor growth.32 Also, CDH13 has been shown to associate with cell surface integrins and GRP78 to promote cell survival signaling pathways.33 Our identification of CDH13 with serous ovarian carcinoma membranes indicates that this protein is expressed in serous ovarian cancer. Future studies will be needed to study the functional impact of CDH13 in ovarian cancer cell survival.
NCSTN
Nicastrin is a component of the γ-secretase (GS) multiprotein enzyme complex responsible for activation of various cell surface membrane proteins including Notch. Nicastrin has been shown to participate in the regulation of breast cancer stem cell expansion and tumor growth.34 Our identification of nicastrin as a glycoprotein with bisecting glycosylation in serous ovarian carcinoma is novel, and future studies will be needed to determine the role of nicastrin and bisecting glycosylation on ovarian cancer growth and survival.
In conclusion, we present evidence that GnT-III expression is required for the expression of bisecting N-linked glycosylation detected using the lectin E-PHA. Because of the elevated expression levels of GnT-III in ovarian cancer, we have utilized E-PHA for the glycoproteomic analysis of membrane proteins extracted from primary ovarian carcinoma tissues (serous and endometrioid). We identified 100 proteins that were enriched in abundance in tumors demonstrated by an increase in spectral counts by at least 1.5 times following E-PHA capture. Our results suggest new functional roles for bisecting glycosylation in controlling the growth and survival of ovarian cancer. These results may lead to future studies that contribute to the development of new therapeutic interventions for the treatment of ovarian cancer.
Acknowledgments
We thank Rob Bridger and Lance Wells for preliminary technical support. This work was supported by a UO1CA168870 grant from NIH/NCI and a Marsha Rivkin Ovarian Cancer Center Pilot Award.
Supporting Information Available
Supplementary Table 1. Patient sample information. Supplementary Table 2. Identified peptides for each protein in Table 1 of the manuscript. Supplementary Figure 1. Full MS spectrum. Supplementary Figures 2–4. MSn data analysis. Supplementary Figure 5. Western blot showing EGFR glycosylation and activation in OVCAR3 cells. Supplementary Figure 6. Changes in downstream ERK signaling. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Bhaumik M.; Seldin M. F.; Stanley P. Cloning and chromosomal mapping of the mouse Mgat3 gene encoding N-acetylglucosaminyltransferase III. Gene 1995, 1642295–300. [DOI] [PubMed] [Google Scholar]
- Koles K.; et al. Identification of N-glycosylated proteins from the central nervous system of Drosophila melanogaster. Glycobiology 2007, 17121388–1403. [DOI] [PubMed] [Google Scholar]
- Abbott K. L.; et al. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J. Proteome Res. 2008, 741470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb D. L.; Fernando C. G.; Chambers M. C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007, 62654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z. Q.; et al. IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res. 2009, 883872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.; et al. Proteomic identification of glycosylphosphatidylinositol anchor-dependent membrane proteins elevated in breast carcinoma. J. Biol. Chem. 2012, 2873025230–25240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott K. L.; et al. Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics 2008, 8163210–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott K. L.; et al. Identification of candidate biomarkers with cancer-specific glycosylation in the tissue and serum of endometrioid ovarian cancer patients by glycoproteomic analysis. Proteomics 2010, 103470–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anugraham M. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: glycan structures reflect gene expression and DNA methylation status. Mol. Cell. Proteomics 2014, 13, 2213–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28121248–1250. [DOI] [PubMed] [Google Scholar]
- Wang M. M. Notch signaling and Notch signaling modifiers. Int. J. Biochem. Cell Biol. 2011, 43111550–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley C. S. Notch and wingless regulate expression of cuticle patterning genes. Mol. Cell. Biol. 1999, 1985743–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehring L. C.; et al. The extracellular matrix protein MAGP-2 interacts with Jagged1 and induces its shedding from the cell surface. J. Biol. Chem. 2005, 2802120349–20355. [DOI] [PubMed] [Google Scholar]
- VanDussen K. L.; et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 2012, 1393488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; et al. Nicastrin is required for assembly of presenilin/gamma-secretase complexes to mediate Notch signaling and for processing and trafficking of beta-amyloid precursor protein in mammals. J. Neurosci. 2003, 2383272–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi M.; Ota M.; Rifkin D. B. Matrix control of transforming growth factor-beta function. J. Biochem. 2012, 1524321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer S. K.; et al. Overexpression of endoglin modulates TGF-beta1-signalling pathways in a novel immortalized mouse hepatic stellate cell line. PLoS One 2013, 82e56116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbolina M. V.; et al. Matrix rigidity activates Wnt signaling through down-regulation of Dickkopf-1 protein. J. Biol. Chem. 2013, 2881141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T.; et al. alpha1,6fucosyltransferase is highly and specifically expressed in human ovarian serous adenocarcinomas. Int. J. Cancer 2000, 886914–919. [DOI] [PubMed] [Google Scholar]
- Song Y.; et al. The bisecting GlcNAc on N-glycans inhibits growth factor signaling and retards mammary tumor progression. Cancer Res. 2010, 7083361–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North S. J.; et al. Glycomics profiling of Chinese hamster ovary cell glycosylation mutants reveals N-glycans of a novel size and complexity. J. Biol. Chem. 2010, 28585759–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; et al. Structures and biosynthesis of the N- and O-glycans of recombinant human oviduct-specific glycoprotein expressed in human embryonic kidney cells. Carbohydr. Res. 2012, 358, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Integrated genomic analyses of ovarian carcinoma. Nature 2011, 4747353609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A. L.; et al. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol 2006, 17753260–3265. [DOI] [PubMed] [Google Scholar]
- Cai D.; et al. Up-regulation of bone marrow stromal protein 2 (BST2) in breast cancer with bone metastasis. BMC Cancer 2009, 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai Y. T.; et al. Potent in vitro and in vivo activity of an Fc-engineered humanized anti-HM1.24 antibody against multiple myeloma via augmented effector function. Blood 2012, 11992074–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J. A.; et al. Membrane proteomic analysis of human mesenchymal stromal cells during adipogenesis. Proteomics 2007, 7224181–4191. [DOI] [PubMed] [Google Scholar]
- Bock A. J.; et al. Endoglin (CD105) expression in ovarian serous carcinoma effusions is related to chemotherapy status. Tumour Biol. 2011, 323589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen R.; et al. Expression and prognostic significance of TGF-beta isotypes, latent TGF-beta 1 binding protein, TGF-beta type I and type II receptors, and endoglin in normal ovary and ovarian neoplasms. Lab. Invest. 1995, 732213–220. [PubMed] [Google Scholar]
- Ziebarth A. J.; et al. Endoglin (CD105) contributes to platinum resistance and is a target for tumor-specific therapy in epithelial ovarian cancer. Clin. Cancer Res. 2013, 191170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmelarova M.; et al. Methylation analysis of tumour suppressor genes in ovarian cancer using MS-MLPA. Folia Biol. (Prague, Czech Repub.) 2012, 586246–250. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; et al. Use of multicellular tumor spheroids to dissect endothelial cell-tumor cell interactions: a role for T-cadherin in tumor angiogenesis. FEBS Lett. 2007, 581234523–4528. [DOI] [PubMed] [Google Scholar]
- Philippova M.; et al. Identification of proteins associating with glycosylphosphatidylinositol- anchored T-cadherin on the surface of vascular endothelial cells: role for Grp78/BiP in T-cadherin-dependent cell survival. Mol. Cell. Biol. 2008, 28124004–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo Y.; et al. Nicastrin regulates breast cancer stem cell properties and tumor growth in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 2012, 1094116558–16563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





