Abstract
Housaku Monogatari (HM) is a plant activator prepared from a yeast cell wall extract. We examined the efficacy of HM application and observed that HM treatment increased the resistance of Arabidopsis thaliana and Brassica rapa leaves to bacterial and fungal infections. HM reduced the severity of bacterial leaf spot and anthracnose on A. thaliana and Brassica crop leaves with protective effects. In addition, gene expression analysis of A. thaliana plants after treatment with HM indicated increased expression of several plant defense-related genes. HM treatment appears to induce early activation of jasmonate/ethylene and late activation of salicylic acid (SA) pathways. Analysis using signaling mutants revealed that HM required SA accumulation and SA signaling to facilitate resistance to the bacterial pathogen Pseudomonas syringae pv. maculicola and the fungal pathogen Colletotrichum higginsianum. In addition, HM-induced resistance conferred chitin-independent disease resistance to bacterial pathogens in A. thaliana. These results suggest that HM contains multiple microbe-associated molecular patterns that activate defense responses in plants. These findings suggest that the application of HM is a useful tool that may facilitate new disease control methods.
Introduction
In nature, numerous potential pathogens such as fungi, bacteria, and viruses continually attack plants; however, disease development remains the exception. Plants are immune to most potential pathogens; this characteristic is referred to as a “nonhost resistance.” Moreover, they have the ability to reduce the disease severity of actual pathogens. Plant innate immune responses to pathogens consist of a two-layer surveillance system that comprises pattern recognition receptors (PRRs) and intracellular nucleotide binding-leucine rich repeat (NLR) proteins, which are encoded by R (resistance) genes [1]. PRRs, which are localized on the surface of plant cells, recognize microbe- or pathogen-associated molecular patterns (MAMPs or PAMPs), and intracellular NLR-type R proteins subsequently detect effectors secreted by the pathogens inside the cell. These two phases of defense induction are called MAMP- or PAMP-triggered immunity and effector-triggered immunity [2].
MAMPs are common conserved structures in microbes among pathogenic, nonpathogenic, and saprophytic microorganisms. They include chitin and ergosterol, which are the main structural components of cell walls and membranes in higher fungi, respectively; bacterial lipopolysaccarides, which are glycolipid components of the outer membranes of gram-negative bacteria; and flagellin, which is the major structural component of the bacterial motility organ [3].
MAMP-induced early defense responses include ion fluxes across the plasma membrane (Ca2+ and H+ influx) and the generation of reactive oxygen species, reactive nitrogen species, and ethylene (ET). Later MAMP responses include alterations in the plant cell wall (deposition of callose and oxidative cross-linking of polymers), biosynthesis of antimicrobial compounds (alkaloids, flavonoids/isoflavonoids, phytoalexin, terpenes, and others), and expression of pathogenesis-related (PR) genes. In addition, MAMPs trigger the activation of calcium-dependent protein kinases and mitogen-activated protein kinase cascades, which lead to transcriptional changes in numerous genes [4–6].
The budding yeast, Saccharomyces pastorianus, is the bottom fermentation yeast used in brewing. Yeast extract and a mannopeptide from yeast invertase function as MAMP and elicit plant defense responses [7, 8]. However, the yeast cell wall extract (YCWE), a by-product of beer brewing processes, has not been effectively used as an agricultural chemical. The yeast cell wall mainly consists of polysaccharides such as polymers of glucose (β-glucan) and polymers of mannose (mannoproteins) [9], which may act as MAMPs and subsequently induce plant defense responses. Using YCWE, which has been processed by treatment with cell wall-degrading enzymes, as a main ingredient, Housaku Monogatari (HM) has been developed as a compound fertilizer (http://www.asahi-fh.com/products/wholesale/good-harvest/).
Our previous study indicated that HM led to induced resistance, i.e., systemic acquired resistance (SAR) and induced systemic resistance signaling pathways [10]. However, the mechanism that enables HM to activate systemic resistance remains unknown. In the present study, we investigated the induction of the plant immune system and changes in the expression of pathogen-responsive genes following HM treatment. We observed that HM activates salicylic acid (SA) and jasmonate (JA)/ET defense signaling pathways and that HM is effective against bacterial pathogens.
Results
HM-induced gene expression
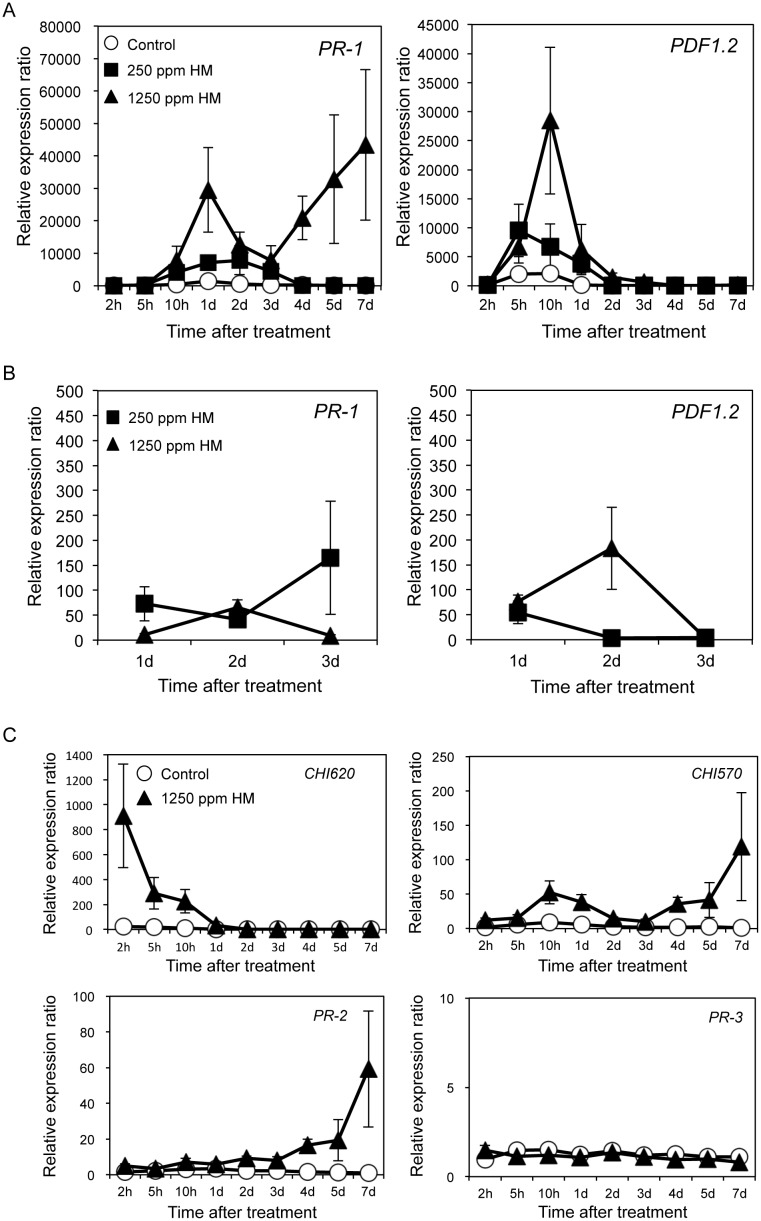
Induced resistance is associated with the expression of disease resistance marker genes. To determine whether HM acts as an inducer of induced resistance in Arabidopsis thaliana, we investigated the expression profile of defense-related genes in response to HM treatment in A. thaliana Col-0 plants by quantitative real time-polymerase chain reaction (qRT-PCR). We used the A. thaliana PR-1 gene as a marker for the SA-dependent signal transduction pathway [11] and the PDF1.2 gene, which encodes a plant defensin, as a marker for the JA/ET pathway [12].
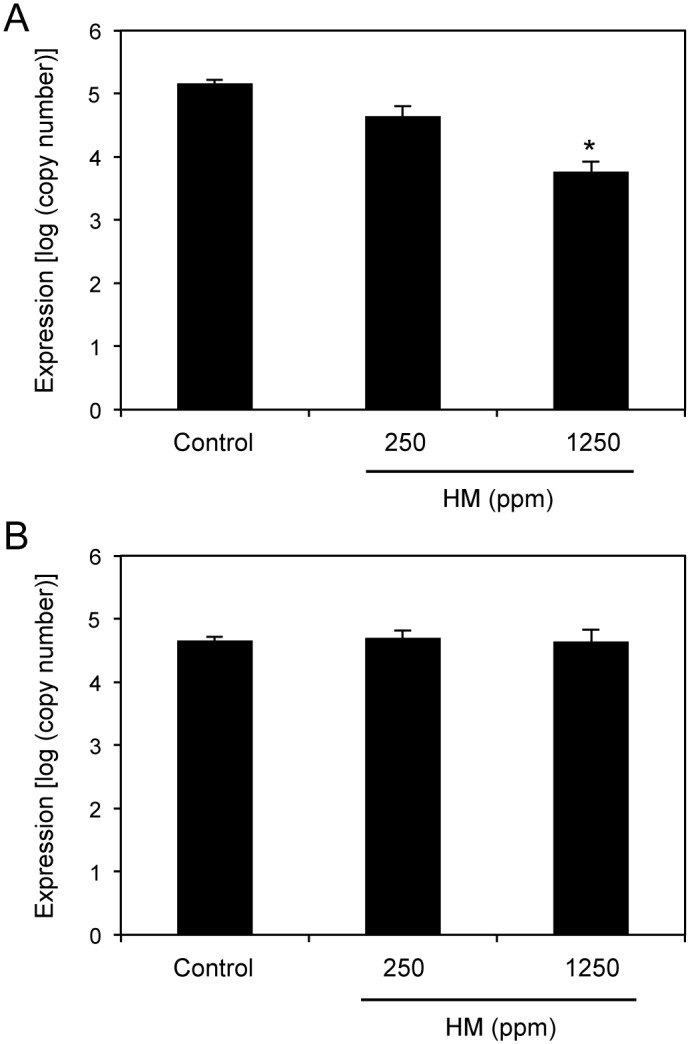
Expression of the PR-1 gene increased over time in Col-0 plants sprayed with 1250 ppm HM, with a peak at day 1, whereas the expression level continued to rise for 2 days in plants sprayed with 250 ppm HM (Fig. 1A). The expression of PDF1.2 initially increased with a peak at 10 h and 5 h in Col-0 plants sprayed with 1250 ppm and 250 ppm HM, respectively, which was earlier than that of the PR-1 gene. The timing of induction differed between the two representative marker genes.
Figure 1. Expression of defense-related genes after treatment with HM.
The 28- to 30-day-old Arabidopsis thaliana Col-0 plants were sprayed (A, C) or soil drenched (B) with 250 and/or 1250 ppm. Aboveground tissues were collected 0–7 (A, C) or 0–3 (B) days after treatment. Total RNA was extracted, and first-strand cDNA was synthesized for expression analysis. Expression levels of the PR-1, PDF1.2, PR-2 (glucanase; BGL2), basic chitinase PR-3, and chitinase (At2g43620; CHI620, At2g43570; CHI570) genes were monitored by qRT-PCR. The expression level of each gene was normalized against the expression level of CBP20, which is constitutively expressed. Relative expression ratios are shown as fold induction relative to the expression level at 0 h. Each experiment was repeated at least three times. Bars indicate the standard error (SE). The nucleotide sequences of the gene-specific primers for each gene are listed in Table 1.
The expression of the PR-1 gene increased with time in Col-0 plants where the soil was drenched with 250 ppm HM (Fig. 1B). In addition, the expression of PR-1 and PDF1.2 increased with a peak at 2 days in Col-0 plants where the soil was drenched with 1250 ppm HM. The transcription levels of PR-1 and PDF1.2 were much higher in the Col-0 plants sprayed with HM than in plants where the soil was drenched.
To investigate the signal transduction pathways activated by HM, we examined other defense-related genes induced in response to treatment with HM using A. thaliana as a model system. Thus, we used qRT-PCR to analyze the effects of HM on the expression of the following A. thaliana putative marker genes: PR-2 (glucanase; BGL2), basic chitinase PR-3 (At3g12500), and chitinase (At2g43620; CHI620, At2g43570; CHI570). The expression levels of these genes were induced by 1250 ppm HM treatment (Fig. 1C). The expression levels of chitinase (CHI620, CHI570) and glucanase (PR-2), which are antipathogenic enzymes, also increased after HM treatment. The expression level of CHI620 increased early in Col-0 plants sprayed with HM. Thus, the plants recognized the HM components as MAMPs and increased the expression of defense-related genes.
HM protects Arabidopsis against Pseudomonas syringae pv. maculicola
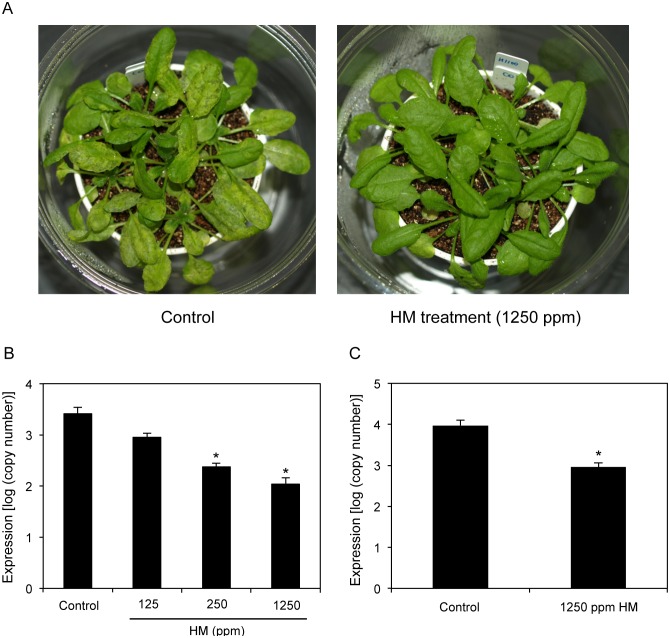
Induced resistance is characterized by the induction of disease resistance to various pathogens. Previously, induced resistance in A. thaliana has been shown to be effective against disease caused by P. syringae pv. tomato [13]. To determine whether HM acts as an inducer of induced resistance in A. thaliana Col-0, the plants were sprayed with water or HM 2 days prior to P. syringae pv. maculicola (Psm) inoculation. As shown in Fig. 2A, treatment with 1250 ppm HM decreased the disease symptoms caused by the pathogen. In contrast, water-treated plants inoculated with Psm exhibited chlorotic leaf spotting. In addition, bacterial growth was reduced in Col-0 plants sprayed with 1250 ppm HM. At 3 days postinoculation (dpi), the HM-treated plants contained 10-fold lower bacterial titers than the water-presprayed control leaves (Fig. 2B). Thus, reduced bacterial growth was responsible for the decrease in the disease symptoms. Furthermore, this concentration of HM did not cause any phytotoxicity. Moreover, bacterial growth decreased in Col-0 plants where the soil was drenched with 1250 ppm HM (Fig. 2C).
Figure 2. Effects of HM application on P. syringae pv. maculicola in Arabidopsis plants.
The 35-day-old A. thaliana Col-0 plants were sprayed (A, B) or soil drenched (C) with water (control) or the indicated concentrations of HM at 2 days prior to spray inoculation with a bacterial suspension (108 cfu mL−1) of P. syringae pv. maculicola. Symptoms were observed 3 days after inoculation with the pathogen (A). Pathogen growth was determined 3 days after inoculation by assessing P. syringae pv. maculicola-rpoD mRNA by qRT-PCR (B, C). Bars indicate the standard error (SE). The asterisk indicates a significant difference compared with the control (Dunnett’s method [35], P < 0.05). The experiment was repeated at least twice with similar results.
To determine whether HM treatment reduced bacterial growth in a dose-dependent manner, HM was applied at increasing concentrations and bacterial growth was measured (Fig. 2B). Compared with the control, HM concentrations as low as 250 ppm reduced bacterial growth in a dose-dependent manner.
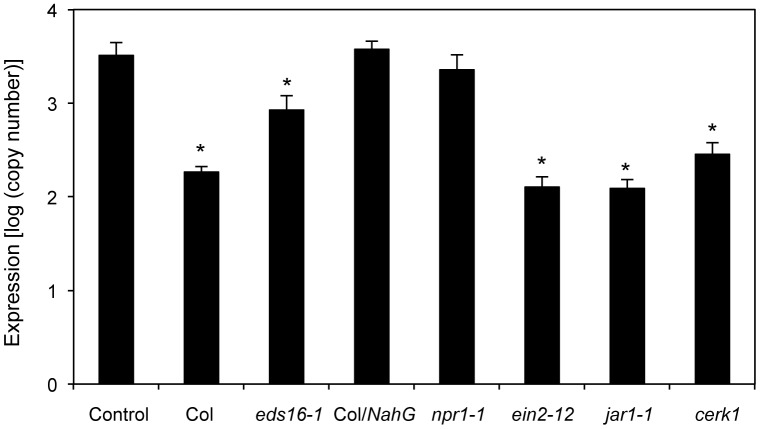
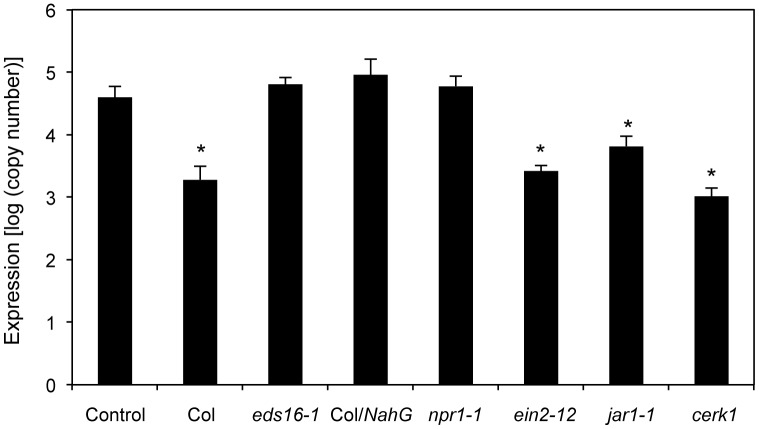
We investigated whether the defense-related hormone SA plays a role in HM-induced activation of SAR using NahG transgenic Arabidopsis, which fails to accumulate SA [14]; eds16–1, which does not produce SA [15]; and npr1–1, which fails to activate PR gene expression [16]. Both JA and ET have been suggested to play important roles in plant defense against pathogen infection. To determine the roles of JA and ET in HM-induced resistance, we tested A. thaliana mutants that varied in their ability to respond to methyl jasmonate (jar1–1 is insensitive to JA [17]) or ET (ein2–12 is insensitive to ET [18]). HM was applied to these plants, and their resistance levels to Psm were determined. These mutants were treated with 1250 ppm HM at 2 days prior to bacterial inoculation. At 3 dpi, mutants deficient in the SA signaling pathways exhibited decreased induced resistance to Psm infection, whereas mutants deficient in the JA or ET signaling pathways were protected in a manner similar to the wild-type Col-0 (Fig. 3). The water-pretreated controls exhibited increased bacterial growth. Thus, HM appears to activate disease resistance to Psm via a pathway that is dependent on SA. In addition, HM has no direct toxic effect on bacteria.
Figure 3. Induction of resistance to P. syringae pv. maculicola by HM application in Arabidopsis defense signaling-defective mutants.
The 35-day-old A. thaliana mutants were sprayed with water (control) or 1250 ppm HM at 2 days prior to spray inoculation with a bacterial suspension (108 cfu mL−1) of P. syringae pv. maculicola. Pathogen growth was determined 3 days after inoculation by assessing P. syringae pv. maculicola-rpoD mRNA by qRT-PCR. Bars indicate the standard error (SE). The asterisk indicates a significant difference compared with the control (Dunnett’s method [35], P < 0.05). The experiment was repeated at least three times with similar results.
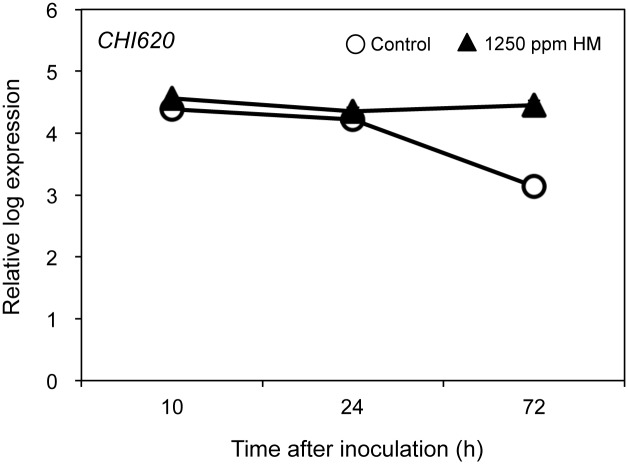
We investigated whether HM potentiates defense responses during infection with pathogens. Treatment with HM potentiated Arabidopsis plants inoculated with Psm to induce expression of chitinase (CHI620) gene more intensively (Fig. 4). Thus, defense responses are boosted in HM-treated plants during the infection with Psm.
Figure 4. Expression of chitinase genes in HM-treated Arabidopsis plants during infection with P. syringae pv. Maculicola.
The 35-day-old A. thaliana Col-0 plants were sprayed with water (control) or 1250 ppm HM at 2 days prior to spray inoculation with a bacterial suspension (108 cfu mL−1) of P. syringae pv. maculicola. Aboveground tissues were collected 10, 24, 72 h after inoculation. Total RNA was extracted, and first-strand cDNA was synthesized for expression analysis. Expression levels of the chitinase gene (At2g43620; CHI620) were monitored by qRT-PCR. The expression level of each gene was normalized against the expression level of CBP20, which is constitutively expressed. Each experiment was repeated at least twice with similar results. Bars indicate the standard error (SE). The nucleotide sequences of the gene-specific primers for each gene are listed in Table 1.
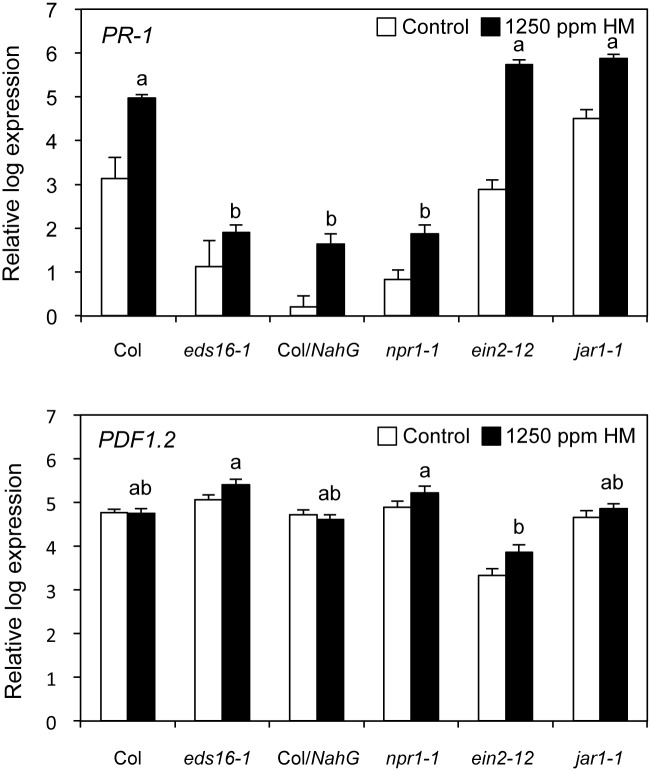
Furthermore, the expression of PR-1 and PDF1.2 genes in SA/ET/JA signaling-defective mutants were investigated for analyzing HM-primed defense responses after infection with Psm.
Accumulation of PR-1 transcripts in NahG transgenic plants, eds16–1 and npr1–1 mutants were lower than those in Col-WT, ein2–12 and jar1–1 mutants during the early phase of infection (10 h post inoculation) (Fig. 5). In addition, PDF1.2 transcripts accumulated in NahG, eds16–1 and npr1–1 plants, but accumulation of PDF1.2 transcripts was low level in ein2–12 mutant (Fig. 5). Therefore, accumulation of PR-1 mRNAs depended on activation of SA signaling defense pathways during infection with Psm.
Figure 5. HM-primed defense responses after the infection with P. syringae pv. maculicola in Arabidopsis defense signaling-defective mutants.
The 35-day-old A. thaliana Col-0 plants and defense signaling-defective mutants were sprayed with water (control) or 1250 ppm HM at 2 days prior to spray inoculation with a bacterial suspension (108 cfu mL−1) of P. syringae pv. maculicola. Aboveground tissues were collected 10 h after inoculation. Total RNA was extracted, and first-strand cDNA was synthesized for expression analysis. Expression levels of the PR-1 and PDF1.2 genes were monitored by qRT-PCR. The expression level of each gene was normalized against the expression level of CBP20, which is constitutively expressed and is shown as the reference value. Each experiment was repeated at least twice with similar results. Bars indicate the standard error (SE). Means labeled with the same letters are not statistically different at the 5% confidence level based on Tukey’s test. The nucleotide sequences of the gene-specific primers for each gene are listed in Table 1.
Does HM activate chitin signaling?
Chitin is a representative MAMP molecule for various fungi, and its perception by PRRs triggers various plant defense responses [19, 20]. Chitin elicitor receptor kinase 1 (CERK1) is an essential molecule for chitin perception and chitin elicitor signaling in A. thaliana [21]. In the present study, compared with control plants, an HM-treated cerk1 mutant exhibited increased resistance to Psm (Fig. 3).
HM-induced disease resistance to Colletotrichum higginsianum
To test whether HM acts as an inducer of resistance to fungal pathogens, A. thaliana plants were sprayed with water or HM at 2 days prior to C. higginsianum inoculation. The results showed that the infection process was not completely stopped by HM treatment; however, the disease incidence was moderately reduced (Fig. 6A). In contrast, no enhanced resistance to C. higginsianum was detected in the Col-0 plants grown in soil drenched with 1250 ppm HM (Fig. 6B).
Figure 6. Effect of HM application on Colletotrichum higginsianum in Arabidopsis plants.
The 28- to 30-day-old A. thaliana Col-0 plants were sprayed (A) or soil drenched (B) with water (control) or the indicated concentrations of HM at 2 days prior to spray inoculation with a spore suspension (5 × 105 spores mL−1) of C. higginsianum. Pathogen growth was determined 5 days after inoculation by assessing C. higginsianum actin mRNA by qRT-PCR. Bars indicate the standard error (SE). The asterisk indicates a significant difference compared with the control (Dunnett’s method [35], P < 0.05). The experiment was repeated at least twice with similar results.
HM was applied to transgenic plants and mutants, and their resistance to C. higginsianum was evaluated (Fig. 7). Enhanced resistance to C. higginsianum was not detected in NahG transgenic plants and eds16–1 and npr1–1 mutants that had been pretreated with 1250 ppm HM at 2 days prior to fungal inoculation. In contrast, HM-treated ein2–12 and jar1–1 mutants did not support the fungal growth, whereas water-pretreated controls exhibited increased fungal growth. Thus, HM appears to activate disease resistance to C. higginsianum via an SA-dependent pathway. Compared with control plants, the HM-treated cerk1 mutant also exhibited increased resistance to C. higginsianum (Fig. 7). In addition, HM exhibited no direct toxic effect on fungi.
Figure 7. Induction of resistance to C. higginsianum by HM application in Arabidopsis defense signaling-defective mutants.
The 28- to 30-day-old A. thaliana mutants were sprayed with water (control) or 1250 ppm HM at 2 days prior to spray inoculation with a spore suspension (5 × 105 spores mL−1) of C. higginsianum. Pathogen growth was determined 5 days after inoculation by assessing C. higginsianum actin mRNA by qRT-PCR. Bars indicate the standard error (SE). The asterisk indicates a significant difference compared with the control (Dunnett’s method [35], P < 0.05). The experiment was repeated at least twice with similar results.
HM protects Brassica crops against P. cannabina pv. alisalensis
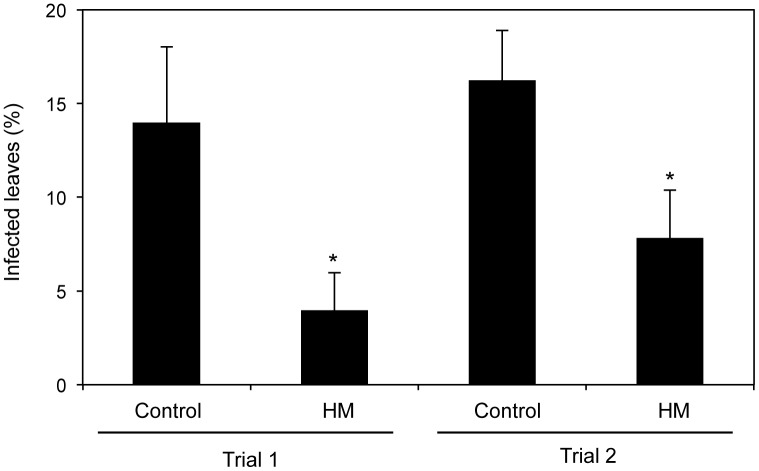
P. syringae pv. alisalensis was recently renamed as P. cannabina pv. alisalensis (Pca) [22]. To test whether HM induces resistance in Brassica rapa var. chinensis, plants treated with HM were inoculated by spraying with Pca. The results revealed that HM protects Brassica crops against Pca (Fig. 8). Thus, HM appears to activate disease resistance in Brassica crops.
Figure 8. HM application induced resistance against P. cannabina pv. alisalensis in Brassica rapa var. chinensis.
Brassica rapa var. chinensis seeds were sown in soil mixed with 5 g L−1 of raw HM material powder. Two-week-old seedlings were inoculated at approximately the one true leaf stage by spraying with a bacterial suspension (106 cfu mL−1) of P. cannabina pv. alisalensis. Observations of bacterial leaf spot symptoms were made at 7 dpi. The percentage of infected leaves (IL) was calculated as follows: IL = (number of leaves with any chlorotic leaf spots)/(total number of leaves), Data represent mean ± SE (Trial 1: n = 12, Trial 2: n = 23). Asterisks indicate significant differences compared with the controls (Welch’s t-test [36]).
Discussion
YCWE has not been tested in practice as an agricultural chemical. However, it may act as MAMP and lead to induced resistance in plants. The mode of action of YCWE is not well known in plants; therefore, we analyzed the protective effect of this product against bacterial and fungal pathogens in A. thaliana and Brassica plants. In A. thaliana and Brassica plants, we observed that HM prepared from YCWE induced the expression of plant defense-related genes and resistance to Psm and Pca, which are similar pathogens that are combated via SA-induced resistance. The infection process of the hemibiotrophic fungal pathogen C. higginsianum was not completely stopped by HM treatment; however, plants treated with HM were moderately protected against the pathogen. Thus, HM protects different Brassicaceae species against bacterial and fungal pathogens, demonstrating the broad range of activity of this product. Importantly, HM has no direct toxic effect on bacteria or fungi. HM-mediated resistance is probably based on the activation of host resistance mechanisms.
Yeast cell walls comprise polysaccharides, i.e., approximately 40% of mannoproteins, approximately 60% of β-glucan, and approximately 2% of chitin [9, 23], whereas HM contains approximately 1.3% of chitin. Chitin-induced defense responses were completely suppressed in the cerk1 mutant [21]; thus, the present study indicated that HM-induced resistance involved chitin-independent disease resistance to bacterial and fungal pathogens in A. thaliana.
The NahG transgenic plants and eds16–1 and npr1–1 mutants exhibited reduced induced resistance to Psm and C. higginsianum after HM treatment, indicating that SA synthesis and NPR1 play a role in HM-elicited defense responses of A. thaliana against Psm and C. higginsianum. In addition, the results demonstrated that HM does not exhibit a direct antibiotic effect on Psm and C. higginsianum. The impaired synthesis of SA, EDS16, and NPR1 in defense mechanisms may be disadvantageous for plants. In contrast, the defective mutants of JA and ET signaling pathways, jar1–1 and ein2–12, respectively, were protected against Psm and C. higginsianum with an effect resembling the wild-type Col-0 control. The plant hormones JA and ET have been implicated in an alternative defense transduction pathway that is separate from SA [24]. HM-induced resistance to these pathogens was not dependent on the sensitivity to JA and ET, which excludes the involvement of JA- and ET-mediated defense signaling mechanisms. A previous study revealed that β-aminobutyric acid (BABA), which is known to be a disease resistance inducer, did not protect transgenic Arabidopsis NahG plants and npr1–1 mutants against P. syringae pv. tomato DC3000 (Pst DC3000) [25]. Lawton et al. revealed that benzothiadiazole (BTH)-treated A. thaliana plants exhibited resistance to Pst DC3000 [26]. In addition, BTH treatment induced disease resistance in NahG transgenic A. thaliana plants, suggesting that the action of BTH does not require SA accumulation [26]. However, in contrast to BTH, HM and BABA require SA accumulation and enhance resistance via plant defense responses.
The protective action of HM against Psm and C. higginsianum seems to act as a disease resistance inducer or a plant defense activator but not as a bactericide and fungicide. Therefore, we analyzed the expression of plant defense-related genes induced by HM treatment. The results indicated that HM treatment elicited the induction of several defense-signaling pathways, including SA, JA, and ET signaling. There is antagonistic signaling cross-talk between the defense signal transduction pathways in A. thaliana and tobacco [27–30]. The maximum induction timing for two representative marker genes of plant defense responses, PR-1 and PDF1.2, differed in A. thaliana treated with HM. Thus, HM treatment appeared to induce early activation of the JA/ET signaling pathway and late activation of the SA signaling pathway. The timing of activation of these signaling pathways may be essential for plant resistance to pathogenic infections. Therefore, the activation of SA and JA/ET defense signaling pathways at different times and our analyses using signaling mutants of these pathways suggest that HM could control various diseases.
HM-primed and -induced defense responses after the infection with Psm were investigated by analyzing the expression of defense-related genes in defense signaling-defective mutants and wild-type plants. The expression of PR-1 gene in response to Psm was reduced in HM-treated SA signaling-defective mutants indicating susceptible to Psm. On the contrary, PR-1 transcripts were potentiated in ET/JA signaling-defective mutants indicating resistance to Psm. As a consequence, SA-mediated defense responses are involved in resistance to Psm.
In addition, expression of CHI620 gene was strongly induced in resistance interaction of HM-treated plants with Psm, but weekly induced in susceptible interaction. Therefore, HM treatment prime and boost defense responses leading to restriction of Psm growth.
Our results indicate that HM reduced the severity of bacterial leaf spot and anthracnose on A. thaliana and Brassica crop leaves with protective effects. In addition, HM facilitated induced resistance to pathogens on plant leaves, which enhanced the expression of plant defense-related genes, i.e., PR gene expression. Our experiments suggest that HM could be a useful tool in the effective control of plant disease.
Materials and Methods
Preparation of Housaku Monogatari (HM)
HM was produced using a by-product, the yeast cell wall extract (YCWE), prepared from the budding yeast Saccharomyces pastorianus during the beer brewing process. The yeast slurry collected after beer brewing was treated with protease YL-15 (Amano Enzyme Inc., Nagoya, Japan). YCWE was obtained by removing the supernatant following centrifugation. To digest YCWE to a moderate molecular size, YCWE was treated again with 0.5% YL-15 at 55°C for 18 h and powdered HM was produced by drum drying. Similar to general yeast cell wall composition [9], HM contains polysaccharides (15%–25% of β-glucan, 5%–15% of α-glucan, 10%–20% of mannan, and 0.5%–2% of chitin).
Plant materials
Arabidopsis thaliana accession Columbia-0 (Col-0) was obtained from RIKEN BRC (Tsukuba, Japan). Before inoculation, the plants were grown in Soil-mix (Sakata Seed Corp., Yokohama, Japan) and expanded vermiculite (1.5–2 mm granules) at a ratio of 1:2 for 28–35 days in a growth chamber at 22°C with 12-h light.
Brassica rapa var. chinensis (cv. Seitei, Sakata Seed Co., Yokohama, Japan) plants were grown in Yosaku N-15 (JCAM AGRI.CO., LTD, Tokyo, Japan) in a growth chamber with 75% relative humidity at 26°C during the daylight hours.
HM treatment
A. thaliana plants were treated with water (control) or the indicated concentration of HM, containing a spreader (0.01%) for spraying but not for soil drenching.
Expression analysis of defense-related genes in Arabidopsis plants
The expression levels of the PR-1 and PDF1.2 genes were monitored by quantitative real time-polymerase chain reaction (qRT-PCR), as described previously [31]. PR-2 (glucanase; BGL2), basic chitinase PR-3, and chitinase (At2g43620; CHI620, At2g43570; CHI570) gene expression levels were also monitored by qRT-PCR. The primers used for qRT-PCR are listed in Table 1.
Table 1. PCR primers used in the present study.
| Forward primer | Reverse primer | |
|---|---|---|
| PR-1 | CCC ACA AGA TTA TCT AAG GGT TCA C | CCC TCT CGT CCC ACT GCA T |
| PDF1.2 | CCA TCA TCA CCC TTA TCT TCG C | TGT CCC ACT TGG CTT CTC G |
| PR-2 | CTT CAA CCA CAC AGC TGG A | GCA TTC GCT GGA TGT TTT GT |
| PR-3 | GGC AAA CGC TAC TAC GGA AG | AAG CGA TCA CTG CGT CGT T |
| CHI620 | GCT AGA GGG AAA TAC TGC TCA C | GAG TCC GAG GAA CTT TCC AG |
| CHI570 | CCA AGA AAC AGG GTT CAT GTG T | TAG TAG CCC TTT CCT TGT GC |
Pseudomonas syringae pv. maculicola infection and quantification of rpoD mRNA
Spontaneous rifampicin-resistant colonies of P. syringae pv. maculicola (MAFF302783Rif4) (Psm) were obtained by culturing the strain MAFF302783 in King’s B medium containing 100 μg mL−1 of rifampicin. Psm was grown in liquid King’s B medium containing rifampicin (25 μg mL−1). Bacteria were harvested by centrifugation, and the cell pellets were washed with 10 mM MgSO4 before resuspension in 10 mM MgSO4 at a concentration of 1 × 108 cfu mL−1 for in planta growth assays. Five-week-old Arabidopsis Col-0 plants (susceptible to Psm) and the mutants were used in virulence assays. The plants were inoculated by spraying the leaves with the bacterial suspension. The inoculated plants were then placed in a growth chamber with 100% relative humidity at 22°C (12-h light cycle). Bacterial growth was determined 3 days after inoculation, and pathogen growth was determined by measuring the rpoD mRNA level by qRT-PCR, as described previously [32].
Colletotrichum higginsianum infection and quantification of actin mRNA
Colletotrichum higginsianum Saccardo isolates (MAFF305635) were obtained from the MAFF Genebank project, Japan. The 28- to 30-day-old Arabidopsis Col-0 plants were inoculated as described previously [33].
Plants inoculated with C. higginsianum were harvested at 5 days postinoculation (dpi) for qRT-PCR, and C. higginsianum was quantified as described previously [34].
P. cannabina pv. alisalensis infection
B. rapa var. chinensis seeds were sown in soil mixed with 5 g L−1 of HM raw material powder. Two-week-old seedlings at approximately the one true leaf stage were inoculated by spraying with a bacterial suspension (106 cfu mL−1) of P. cannabina pv. alisalensis. Observations of bacterial leaf spot symptoms were made at 7 dpi. The percentage of infected leaves (IL) was calculated as follows:
IL = (number of leaves with any chlorotic leaf spots)/(number of leaves)
Acknowledgments
We thank Mses Yasuyo Katayama, Shoko Miyashita, Yukiko Kurosaki, Chiaki Watanabe, and Masami Miyamoto (RIBS) for their excellent technical assistance. We also thank Nagano Vegetable and Ornamental Crops Experiment Station for providing P. cannabina pv. alisalensis. We are grateful to Dr. Naoto Shibuya (Meiji Univ.) for providing cerk1 mutant.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Industrial Technology Research Grant Program in 2004 and 2009 from the New Energy and Industrial Technology Development Organization (NEDO) of Japan to YN, and by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 24580071 to YN and 25450523 to MN. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. TM, CI, TH, ST and KK are employed by Asahi Group Holdings, Ltd. Asahi Group Holdings, Ltd. provided support in the form of salaries for authors TM, CI, TH, ST and KK, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- 2. Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814. 10.1016/j.cell.2006.02.008 [DOI] [PubMed] [Google Scholar]
- 3. Zipfel C, Felix G (2005) Plants and animals: a different taste for microbes? Curr Opin Plant Biol 8: 353–360. 10.1016/j.pbi.2005.05.004 [DOI] [PubMed] [Google Scholar]
- 4. de Torres M, Sanchez P, Fernandez-Delmond I, Grant M (2003) Expression profiling of the host response to bacterial infection: the transition from basal to induced defense responses in RPM1-mediatedresistance. Plant J 33: 665–676. 10.1046/j.1365-313X.2003.01653.x [DOI] [PubMed] [Google Scholar]
- 5. Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, et al. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- 6. He P, Shan L, Sheen J (2007) Elicitation and suppression of microbe-associated molecular pattern-triggered immunity in plant–microbe interactions. Cell Microbiol 9: 1385–1396. 10.1111/j.1462-5822.2007.00944.x [DOI] [PubMed] [Google Scholar]
- 7. Basse CW, Fath A, Boller T (1993) High affinity binding of a glycopeptides elicitor to tomato cells and microsomal membranes and displacement by specific glycan suppressors. J Biol Chem 268: 14724–14731. [PubMed] [Google Scholar]
- 8. Obara N, Mitsuhara I, Seo S, Ohashi Y, Hasegawa M, et al. (2007) Mechanism of PR gene expression by treatment of tobacco leaves with yeast extract (AGREVO EX). Jpn J Phytopathol 73: 94–101. 10.3186/jjphytopath.73.94 [DOI] [Google Scholar]
- 9. Klis FM, Boorsma A, De Groot PWJ (2006) Cell wall construction in Saccharomyces cerevisiae . Yeast 23: 185–202. 10.1002/yea.1349 [DOI] [PubMed] [Google Scholar]
- 10. Minami T, Yanaka T, Takasaki S, Kawamura K, Hiratsuka K (2011) In vivo bioluminescence monitoring of defense gene expression in response to treatment with yeast cell wall extract. Plant Biotechnol 28: 481–484. 10.5511/plantbiotechnology.11.1020a [DOI] [Google Scholar]
- 11. Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, et al. (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819. 10.1105/tpc.8.10.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, et al. (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323. 10.1105/tpc.8.12.2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, et al. (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656. 10.1105/tpc.4.6.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. 10.1126/science.266.5188.1247 [DOI] [PubMed] [Google Scholar]
- 15. Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, et al. (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24: 205–218. 10.1046/j.1365-313x.2000.00870.x [DOI] [PubMed] [Google Scholar]
- 16. Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592. 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89: 6837–6840. 10.1073/pnas.89.15.6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guzman P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. 10.1105/tpc.2.6.513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shibuya N, Minami E (2001) Oligosaccharide signalling for defense responses in plant. Physiol Mol Plant Pathol 59: 223–233. 10.1006/pmpp.2001.0364 [DOI] [Google Scholar]
- 20. Silipo A, Erbs G, Shinya T, Dow JM, Parrilli M, et al. (2010) Glyco-conjugates as elicitors or suppressors of plant innate immunity. Glycobiology 20: 406–419. 10.1093/glycob/cwp201 [DOI] [PubMed] [Google Scholar]
- 21. Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis . Proc Natl Acad Sci USA; 104:19613–19618. 10.1073/pnas.0705147104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bull CT, Manceau C, Lydon J, Kong H, Vinatzer BA, et al. (2010) Pseudomonas cannabina pv. cannabina pv. nov., and Pseudomonas cannabina pv. alisalensis (Cintas Koike and Bull, 2000) comb. nov., are members of the emended species Pseudomonas cannabina (ex Sutic & Dowson 1959) Gardan, Shafik, Belouin, Brosch, Grimont & Grimont 1999. Syst Appl Microbiol 33: 105–115. 10.1016/j.syapm.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 23. Aguilar-Uscanga B, François JM (2003) A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett Appl Microbiol 37: 268–274. 10.1046/j.1472-765X.2003.01394.x [DOI] [PubMed] [Google Scholar]
- 24. Thomma BP, Eggermont K, Penninckx IA, Mauch-Mani B, Vogelsang R, et al. (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111. 10.1073/pnas.95.25.15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zimmerli L, Jakab G, Metraux JP, Mauch-Mani B. (2000) Potentiation of pathogen-specific defense mechanisms in Arabidopsis by β-aminobutyric acid. Proc Natl Acad Sci USA 97: 12920–12925. 10.1073/pnas.230416897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, et al. (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82. 10.1046/j.1365-313X.1996.10010071.x [DOI] [PubMed] [Google Scholar]
- 27. Sano H, Seo S, Koizumi N, Niki T, Iwamura H, et al. (1996) Regulation by cytokinins of endogenous levels of jasmonic and salicylic acids in mechanically wounded tobacco plants. Plant Cell Physiol 37: 762–769. 10.1093/oxfordjournals.pcp.a029011 [DOI] [Google Scholar]
- 28. Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, et al. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120. 10.1016/S0092-8674(00)00213-0 [DOI] [PubMed] [Google Scholar]
- 29. Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, et al. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15: 760–770. 10.1105/tpc.009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yasuda M, Ishikawa A, Jikumaru Y, Seki M, Umezawa T, et al. (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis . Plant Cell 20:1678–1692. 10.1105/tpc.107.054296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Narusaka Y, Narusaka M, Abe H, Hosaka N, Kobayashi M, et al. (2009) High-throughput screening for plant defense activators using a β-glucuronidase-reporter gene assay in Arabidopsis thaliana . Plant Biotechnol 26: 345–349. 10.5511/plantbiotechnology.26.345 [DOI] [Google Scholar]
- 32. Narusaka M, Shiraishi T, Iwabuchi M, Narusaka Y (2011) rpoD gene expression as an indicator of bacterial pathogens in host plants. J Gen Plant Pathol 77: 75–80. 10.1007/s10327-011-0298-x [DOI] [Google Scholar]
- 33. Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, et al. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226. 10.1111/j.1365-313X.2009.03949.x [DOI] [PubMed] [Google Scholar]
- 34. Narusaka M, Shiraishi T, Iwabuchi M, Narusaka Y (2010) Monitoring fungal viability and development in plants infected with Colletotrichum higginsianum by quantitative reverse transcription-polymerase chain reaction. J Gen Plant Pathol 76: 1–6. 10.1007/s10327-009-0211-z [DOI] [Google Scholar]
- 35. Dunnett CW (1955) A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc 50: 1096–1211. 10.1080/01621459.1955.10501294 [DOI] [Google Scholar]
- 36. Welch BL (1947) The generalization of “Student’s” problem when several different population variances are involved. Biometrika 34: 28–35. 10.1093/biomet/34.1-2.28 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.