Abstract
Nearly half a century ago insect herbivores were found to induce the formation of green islands by manipulating cytokinin (CK) levels. However, the response of the CK pathway to attack by chewing insect herbivores remains unclear. Here, we characterize the CK pathway of Nicotiana attenuata (Torr. ex S. Wats.) and its response to wounding and perception of herbivore-associated molecular patterns (HAMPs). We identified 44 genes involved in CK biosynthesis, inactivation, degradation, and signaling. Leaf wounding rapidly induced transcriptional changes in multiple genes throughout the pathway, as well as in the levels of CKs, including isopentenyladenosine and cis-zeatin riboside; perception of HAMPs present in the oral secretions (OS) of the specialist herbivore Manduca sexta amplified these responses. The jasmonate pathway, which triggers many herbivore-induced processes, was not required for these HAMP-triggered changes, but rather suppressed the CK responses. Interestingly CK pathway changes were observed also in systemic leaves in response to wounding and OS application indicating a role of CKs in mediating long distance systemic processes in response to herbivory. Since wounding and grasshopper OS elicited similar accumulations of CKs in Arabidopsis thaliana L., we propose that CKs are integral components of wounding and HAMP-triggered responses in many plant species.
Keywords: Arabidopsis thaliana, cytokinin, herbivore-associated molecular patterns, herbivory, insect, jasmonic acid, Manduca sexta, Nicotiana attenuata, wounding
INTRODUCTION
Plant survival in nature strongly relies on the ability of plants to adjust their physiology to maximize their fitness in environments that frequently change. The detection of specific herbivore-derived cues, the so-called herbivory-associated molecular patterns (HAMPs), allows plants to distinguish herbivore attack from wounding and often leads to the activation of herbivore-specific responses (Erb et al. 2012). HAMP perception is known to occur in many plants, including maize (Zea mays), thale cress (Arabidopsis thaliana), soybean (Glycine max), and wild tobacco (Nicotiana attenuata) (Erb et al. 2012).
Nicotiana attenuata is an ecological model organism for analyzing plant responses to herbivory. The interaction with the lepidopteran herbivore Manduca sexta, a main defoliator in the plant's natural environment, has been intensively studied. N. attenuata specifically responds to fatty acid-amino acid conjugates (FACs), which are major HAMPs present in M. sexta oral secretions (OS; Bonaventure et al. 2011). Analysis of FAC-triggered responses has provided important insights into HAMP recognition and signaling, as well as into the diverse defense and tolerance strategies that plants use against herbivore attack (Bonaventure et al. 2011). FAC perception in N. attenuata triggers the biosynthesis of oxylipins, including jasmonic acid (JA) and the JA-isoleucine conjugate (JA-Ile; Kallenbach et al. 2010). Oxylipins play a central role in the regulation of most anti-herbivore defenses in plants (De Geyter et al. 2012). JA-Ile, the active jasmonate, is perceived by the ubiquitin-E3 ligase complex protein CORONATINE INSENSITIVE 1 (COI1), leading to the degradation of JASMONATE ZIM-DOMAIN (JAZ) proteins, which are negative transcriptional regulators of JA-responsive genes (Chini et al. 2009).
However, the oxylipin sector is not the only hormonal pathway that is involved in the regulation of herbivory-specific responses. Other phytohormones, which respond to wounding and HAMP perception, are ethylene, abscisic acid, or salicylic acid (Erb et al. 2012). In addition to these well-studied defense hormones, the roles of growth-related hormones, such as auxins, brassinosteroids, cytokinins (CKs), and gibberellins are much less understood (Erb et al. 2012). Our lack of knowledge of the role of these hormones in biotic interactions can mainly be attributed to difficulties in measuring these low abundant compounds and their common characterization as “growth-related hormones” putting them out of the scope of traditional defense pathway-oriented plant-herbivore interaction research.
It has long been suspected that CKs function in plant-insect interactions. Some insects like leaf miners have been shown to use CKs to modify the tissue surrounding their mines, resulting in the well-described phenomenon of “green islands” (Engelbrecht 1968) or certain sawflies that can induce “leaf galls” (Elzen 1983). In addition to the manipulation of CKs by insect herbivores, an increasing number of studies have provided evidence for an active role of CKs in regulating plant defense responses against herbivores (Giron et al. 2013). Transcriptional studies in N. attenuata identified the transcripts of the CK-induced gene 2 (CIG2) and CK-regulated kinase 1 (CRK1) to be induced by M. sexta feeding and FACs, respectively, indicating that the CK pathway may play a role in plant responses to herbivores (Hui et al. 2003; Gilardoni et al. 2010). Although CRK1 was previously shown to be negatively regulated by CKs, it was also reported to be responsive to auxin and abscisic acid and the function of this receptor kinase in hormone signaling was only hypothesized (Schäfer and Schmülling 2002). CIG2, in contrast, is not responsive to other tested phytohormones except CKs, which act as positive regulators (Kimura et al. 2001). It is questionable how far the information about CIG2 and CRK1, which were derived from hormone application experiments with cell cultures reflect the in vivo responses to endogenous CK dynamics. Most claims in the literature on the response of the CK pathway to HAMP perception or defoliation by insect herbivores are based on the indirect evidence of Hui et al. (2003) and Gilardoni et al. (2010), whereas less is known about actual changes in CK biosynthesis, metabolites and signaling elements.
Cytokinin metabolism and signaling is complex and a simplified overview is provided in Figure 1 (abbreviations are summarized in Table S1). In brief, CKs are synthesized by the transfer of an isopentenyl moiety to an adenosine (di/tri) phosphate or tRNA. This rate-limiting step is catalyzed by isopentenyltransferases (IPTs), whereas the CK nucleoside 5′-monophosphate phosphoribohydrolases (LOGs) are responsible for the release of the free CK bases from the CK nucleotides (Kurakawa et al. 2007; Kuroha et al. 2009). Active CKs, such as isopentenyladenine (IP), trans-zeatin (tZ), cis-zeatin (cZ), dihydrozeatin (DHZ), and their ribosides (IPR, tZR, cZR, and DHZR, respectively) can bind to specific CK receptors (Stolz et al. 2011; Lomin et al. 2012; Shi and Rashotte 2012); a class of partially redundant CHASE (cyclase/histidine kinase associated sensing extracellular) domain-containing histidine kinases (CHKs). After CK binding and auto-phosphorylation of a conserved His, the phosphoryl group is transferred to another conserved Asp and transmitted to histidine-containing phosphotransfer proteins (HPTs), which finally phosphorylate-specific response regulators (RRs) responsible for the output of the CK-pathway (Hwang et al. 2012). While the type-B RRs (RRB) function as transcription factors, the type-A RRs (RRA) are known as negative feedback regulators of the CK pathway (Hwang et al. 2012). Various glucosyltransferases (e.g., ZOG and UGT) are responsible for the formation of reversible inactivation products like O-glucoside (∼OG) and riboside O-glucoside (∼ROG) or irreversible inactivation products such as N7-glucoside (∼7G) and N9-glucoside (∼9G), whereas CK degradation is carried out by CK oxidase/dehydrogenases (CKX; Mok and Mok 2001; Schmülling et al. 2003). Many of these genes involved in CK metabolism and signaling were found to be transcriptionally regulated in response to CKs, including CKX, CK-glucosyltransferase, CK-receptor, HPT, and RR genes (Brenner et al. 2012; Bhargava et al. 2013).
Figure 1. Cytokinin metabolism and signaling.
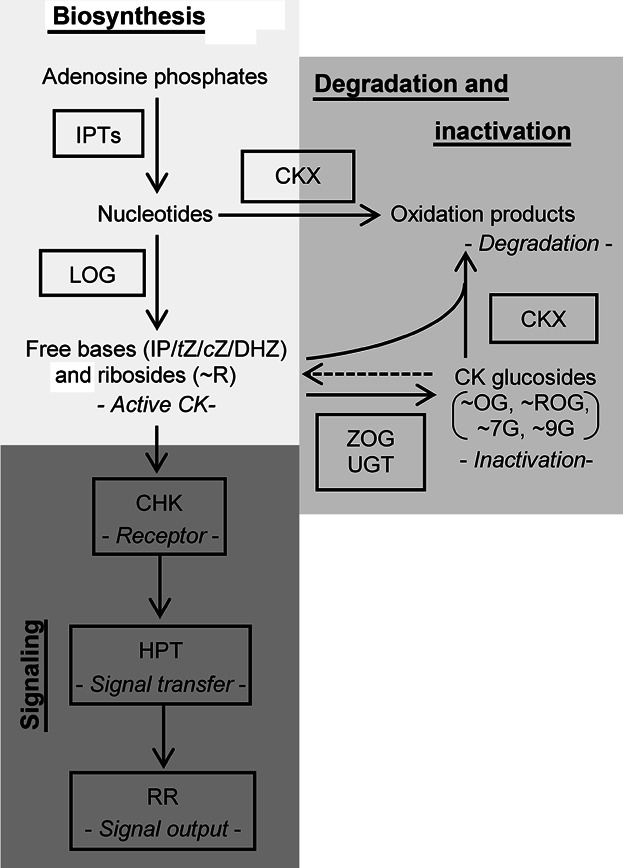
Cytokinins (CK) biosynthesis is initiated by the activity of isopentenyltransferases (IPT). Monophosphorylated CK nucleotides can be directly converted into free bases by the activity of cytokinin nucleoside 5′-monophosphate phosphoribohydrolases (LOG). Active CKs such as isopentenyladenine (IP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DHZ) and their respective ribosides (∼R) can be inactivated by CK oxidases/dehydrogenases (CKX) or by the formation of sugar conjugates like O-glucosides (∼OG), riboside O-glucosides (∼ROG), N7-glucosides (∼7G), and N9-glucosides (∼9G) through the activity of glycosyltransferases, like zeatin O-glucosyltransferase or N-glucosyltransferases (ZOG, UGT). CK are perceived by histidine kinases (CHK), which phosphorylate histidine phosphotransfer proteins (HPT). HPTs relay the signal to type-B response regulators (RR), which stimulate transcription of CK-response genes and type-A RRs that in turn mediate the feedback inhibition of the signal. The color code (light gray: CK biosynthesis, medium gray: CK degradation and inactivation, dark gray: CK signaling) is used consistently in all figures.
Here, we used the model plant N. attenuata, from which we identified homologues of known CK-related genes, from 454 transcripts to test: (i) if the CK pathway responds to wounding and HAMPs; (ii) if JAs influence CK levels and signaling during herbivory; and (iii) if the CK pathways is activated in non-treated systemic tissues. Our results demonstrate that CKs function as an integral component of the herbivory-induced signaling cascade.
RESULTS
CK metabolism and signaling genes in N. attenuata
Genes involved in CK biosynthesis and signaling have previously been described in several plant species (Pils and Heyl 2009; Frébort et al. 2011), with in-depth studies in A. thaliana (e.g., Frébort et al. 2011), rice (Oryza sativa; e.g., Tsai et al. 2012), and a moss (Physcomitrella patens; Ishida et al. 2010). Here, we present a comprehensive description and phylogenetic analysis of these genes in a Solanaceous plant. We used 454 transcriptome sequencing data obtained from RNA extractions of various plant tissues to identify the gene homologs in N. attenuata. We used cloned cDNA sequences, as well as homology search to protein sequences in public databases and identified 44 genes involved in CK metabolism and signaling. Given that many annotated genomes are available now in easy to access databases, such as NCBI (http://www.ncbi.nlm.nih.gov/) or Phytozome (http://www.phytozome.org/), we mined 15 proteomes, including green algae, lower land plants, monocots, and dicots and compared the results about gene number expansion and reduction to previous studies (e.g., Pils and Heyl 2009; Tsai et al. 2012). Our analysis also includes the CK biosynthesis genes, which were often excluded from previous phylogenetic studies of the CK pathway. An overview of the investigated gene families is shown in Figure 1; their phylogenetic relationships are shown in Figures S1B–S8B.
Comparisons of gene numbers among different plants revealed that genes involved in CK biosynthesis (IPT, LOG), inactivation (ZOG), and signaling output (RR) were more susceptible to rapid intra-species gene gains than were the CK perception (CHK) and signal transfer proteins (HPT), which represent the core elements of the CK phosphorelay. The genes involved in CK degradation (CKX) also belong to genes with more restrained intra-species duplication (Figures S1B–S8B). Our data-mining effort in green plants also shows that the genes encoding for the CK inactivation and degradation enzymes ZOG and CKX appear for the first time in land plants.
Regulation of CK-related genes by wounding and simulated herbivory
To investigate if these genes are transcriptionally regulated by wounding and perception of M. sexta-derived cues, we mined our recently established herbivory-regulated microarray dataset (Onkokesung et al. 2012). In this microarray experiment, time kinetics of labeled copy RNA probes from leaves 1, 5, and 17 h after treatment with wounding and water (W + W) or 0.5, 1, 5, 9, 13, 17, and 21 h after treatment with wounding and the immediate application of M. sexta OS to the puncture wounds (W + OS), as well as from untreated control leaves were hybridized to a N. attenuata-specific Agilent microarray platform (GEO microarray repository, GPL13527). Since W + OS treatments mimic plant responses to actual M. sexta attack (Halitschke et al. 2001), these treatments allow for the rigorous discrimination of wound-induced responses (W + W) from those elicited by herbivore perception (W + OS). Figure 2 provides an overview of highly regulated transcripts, whereas the detailed results from all analyzed genes are shown in Figures S1–S8. We independently determined transcript expression of selected genes by quantitative PCR (qPCR) to confirm the microarray results (Figures S9, S10).
Figure 2. Wounding and herbivory regulate transcript accumulations of cytokinin-related genes.
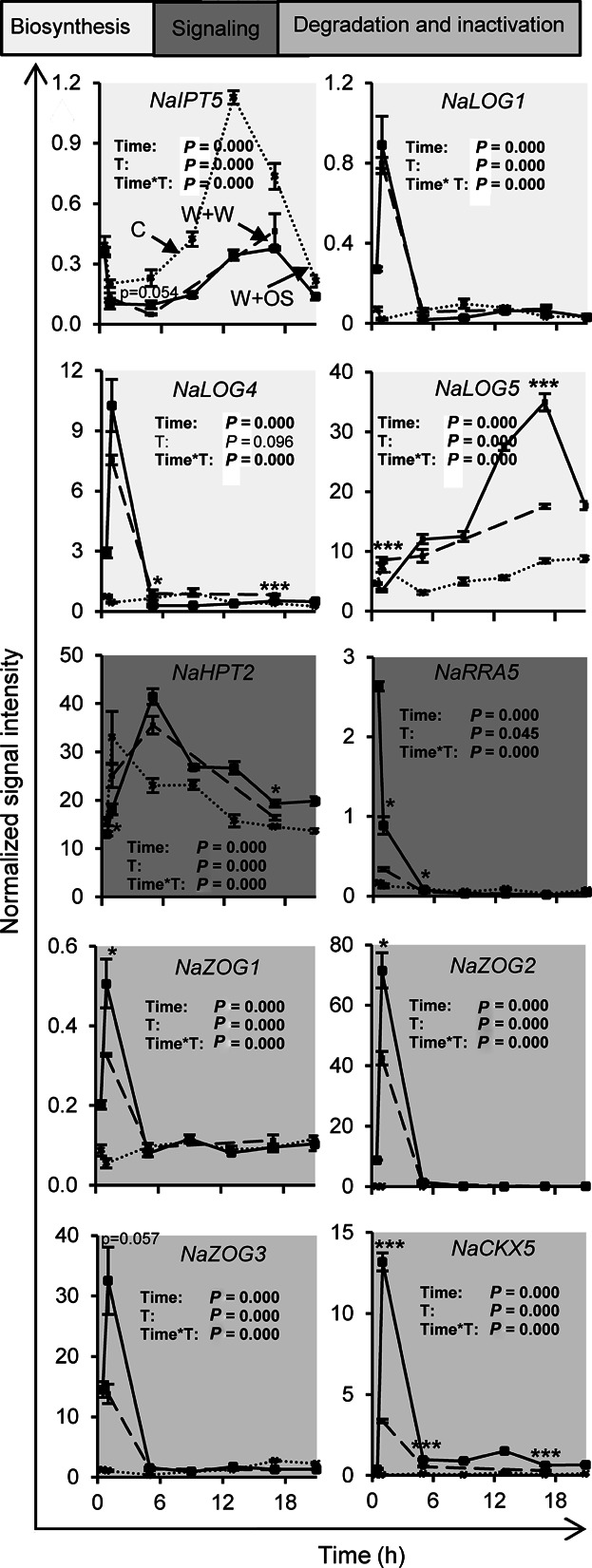
Transcript accumulations were measured in leaves of Nicotiana attenuata at different time points after wounding and application of water (W + W; dashed line; 1, 5, and 17 h) or Manduca sexta oral secretions (W + OS; solid line; 0.5, 1, 5, 9, 13, 17, and 21 h) to the puncture wounds, as well as in untreated control leaves (C; dotted line; 0.5, 1, 5, 9, 13, 17, and 21 h). Data are obtained from kinetic analysis conducted with microarrays. Time and treatment (C and W + OS; T) effects and their interaction (Time*T) were analyzed by univariate ANOVA, except for NaLOG4, NaHPT2, NaRRA5, NaCKX5, and NaZOG2 data which were analyzed by generalized least squares model instead. Asterisks indicate significant differences between W + W and W + OS-treated samples at the same time point (independent samples t-test: *P ≤ 0.05, ***P ≤ 0.001). Error bars are standard errors (n = 3). For overview of transcript accumulation of additional cytokinin-related N. attenuata genes and their phylogeny see Figures S1–S8.
Figure 2 shows the transcript levels of genes with high homologies to CK biosynthesis enzymes, such as NaIPT5 and several LOGs (NaLOG1, NaLOG4, and NaLOG5), and also the transcripts of signaling components including the phosphotransfer protein, NaHPT2, and the RR, NaRRA5, to be differentially regulated by W + W and W + OS, when compared to untreated leaf tissues. The putative inactivation and degradation enzymes, NaCKX5, NaZOG1, NaZOG2, and NaZOG3, were particularly induced by W + W and W + OS treatments. While the transcript levels of some genes were quickly regulated within the first hours after single treatments (e.g., NaLOG1, NaLOG4, NaRRA5, NaCKX5, NaZOG1, NaZOG2, and NaZOG3), long-term effects, lasting frequently for more than 20 h, were also observed (e.g., NaIPT5, NaLOG5, and NaHPT2). Treatment-dependent up- as well as downregulations of transcripts were observed. In addition to genes such as NaIPT5, whose transcripts responded to wounding irrespective of the presence of OS, other transcripts (e.g., NaRRA5) were highly responsive to OS-derived cues, which highly amplified wound-induced accumulations. Interestingly downregulation of NaIPT5 transcripts and upregulation of NaRRA5 and NaCKX5 transcripts could also be achieved by tZ and cZR application to N. attenuata leaves (Figure S11). Given the rapid and strong regulation of the CK biosynthesis and signaling pathway at the transcriptional level, these data suggest that wounding and OS perception strongly affect the accumulation of CK metabolites.
CK levels are regulated by wounding and simulated herbivory
Another kinetic experiment was performed (lasting 4 h after treatments) to analyze rapid changes in CK metabolites to the W + W and W + OS treatments (Figures 3, S12). The levels of the active CKs, namely IP, IPR, and cZR, as well as the CK inactivation forms, tZROG, cZROG, and tZ7G responded particularly strong to wounding and OS application (Figure 3). While IP and IPR levels peaked already at 30 min after wounding and OS application and declined afterwards, cZR, tZROG, cZROG, and tZ7G levels had accumulated at 1 h and remained at elevated levels at least for the 4 h duration of the analysis. IP, tZROG, cZROG, and tZ7G levels attained maximum increases of 25%–50%, IPR levels doubled and cZR levels increased fourfold. Similar to the observed transcript changes, OS application to the puncture wounds further elevated some of the wound-induced CK levels compared to wounding alone. OS application increased the wound-induced IPR, tZROG, and cZROG levels by approximately 25% and cZR levels were elevated by 75%, when compared to wounding alone. These results were consistent with the analysis of the transcripts demonstrating wound- and HAMP perception-specific regulation of CK levels.
Figure 3. Wounding- and herbivory-induced changes in cytokinin levels.
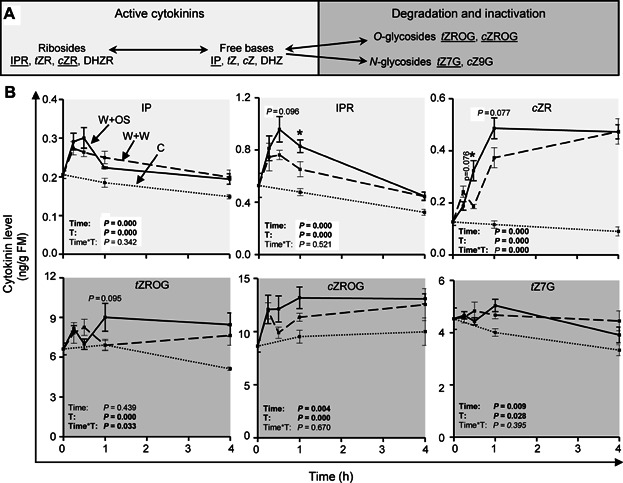
(A) Cytokinin metabolism overview. (B) Isopentenyladenine (IP), isopentenyladenosine (IPR), cis-zeatin riboside (cZR), trans-zeatin riboside O-glucoside (tZROG), cis-zeatin riboside O-glucoside (cZROG), and trans-zeatin N7-glucoside (tZ7G) levels in leaves of Nicotiana attenuata at different time points after wounding and application of water (W + W, dashed line) or Manduca sexta oral secretions (W + OS, solid line) to the puncture wounds, as well as in untreated control leaves (C, dotted line). Time and treatment (C, W + W and W + OS; T) effects and their interaction (Time*T) were analyzed by univariate ANOVA, except for cZROG data which were analyzed by a generalized least squares model. Asterisks indicate significant differences between W + W and W + OS-treated samples at the same time point (independent samples t-test: *P ≤ 0.05). Error bars are standard errors (n = 5). For additional cytokinins see Figure S12. FM, fresh mass.
Fatty acid-amino acid conjugates are the main HAMPs in the OS of M. sexta (Halitschke et al. 2001). We used synthetic FACs to evaluate if they could account for the inductions of the CK pathway. Figure 4 shows that FACs are sufficient to elevate wound-induced CK levels, as well as NaRRA5 transcript accumulation. Transcript levels of NaRRA5 homologues have been used as marker genes for analyzing CK-dependent responses in several plant species (e.g., Kurakawa et al. 2007; Stolz et al. 2011). Since NaRRA5 was also highly responsive to CK application in N. attenuata (Figure S11), it was used as CK-responsive marker gene in this investigation.
Figure 4. Fatty acid-amino acid conjugates increase active cytokinin levels and signaling.
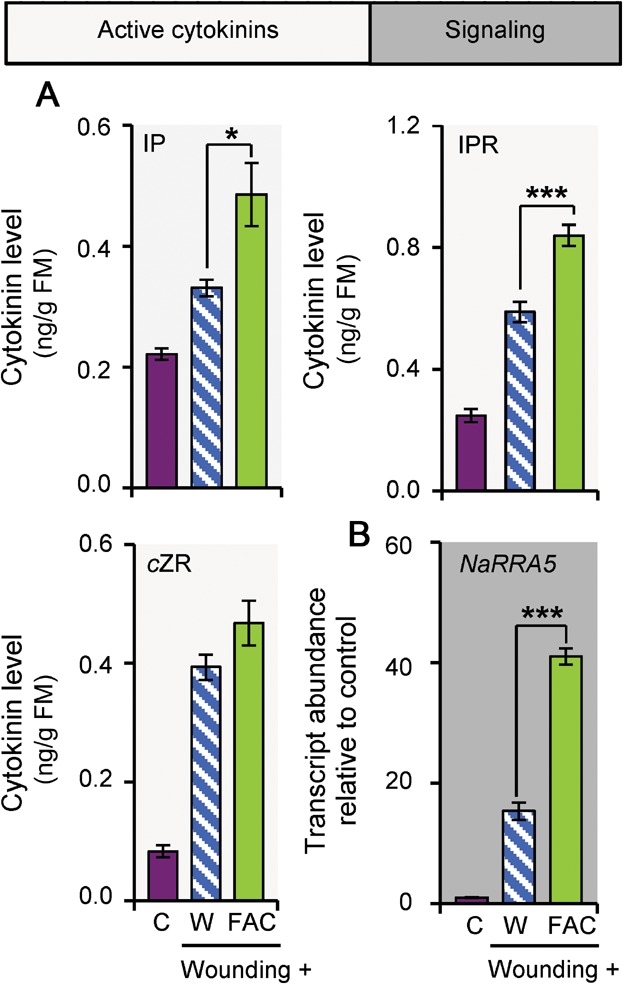
(A) Isopentenyladenine (IP), isopentenyladenosine (IPR), and cis-zeatin riboside (cZR) levels in Nicotiana attenuata 30 min after wounding and application of water (W) or the fatty acid-amino acid conjugate (FAC) N-linolenoyl-glutamate to the puncture wounds, as well as in untreated control leaves (C). (B) Response regulator 5 (NaRRA5) transcript accumulation under the conditions described in (A). Asterisks indicate significant differences between FAC application compared to water applications to wounded leaves (independent samples t-test: *P ≤ 0.05, ***P ≤ 0.001). Error bars are standard errors (n = 5). FM, fresh mass.
JA affects CK levels and signaling
Many herbivory-induced responses in N. attenuata (and most other plant species) are JA dependent (Halitschke and Baldwin 2003; Paschold et al. 2007; Stitz et al. 2011). We tested if JA also regulates the CK pathway. We analyzed different transgenic lines impaired in JA and JA-Ile biosynthesis and perception and performed JA supplementation experiments (Figure 5). Our results revealed that JAs partially regulate CK levels and signaling in W + OS-treated leaves. Transgenic lines with an impaired JA pathway showed more pronounced NaRRA5 transcript accumulations, while treatments with methyl-JA (MeJA) attenuated NaRRA5 transcript levels (Figure 5C, D). The iraoc line, which is silenced in the allene oxide cyclase (AOC) an early step of the JA biosynthesis and the ircoi1 line, which is deficient in JA signaling, showed mild reductions in IPR and stronger reductions in cZR and tZROG levels (Figure 5A). MeJA treatment strongly reduced the herbivory-elicited accumulation of IPR, but significantly increased cZR and tended to increase tZROG levels (Figure 5B). The control and wound-induced CK levels and NaRRA5 transcripts were regulated in similar ways by the treatments (Figure 5).
Figure 5. Herbivory-induced cytokinin levels and signaling are regulated by jasmonates.
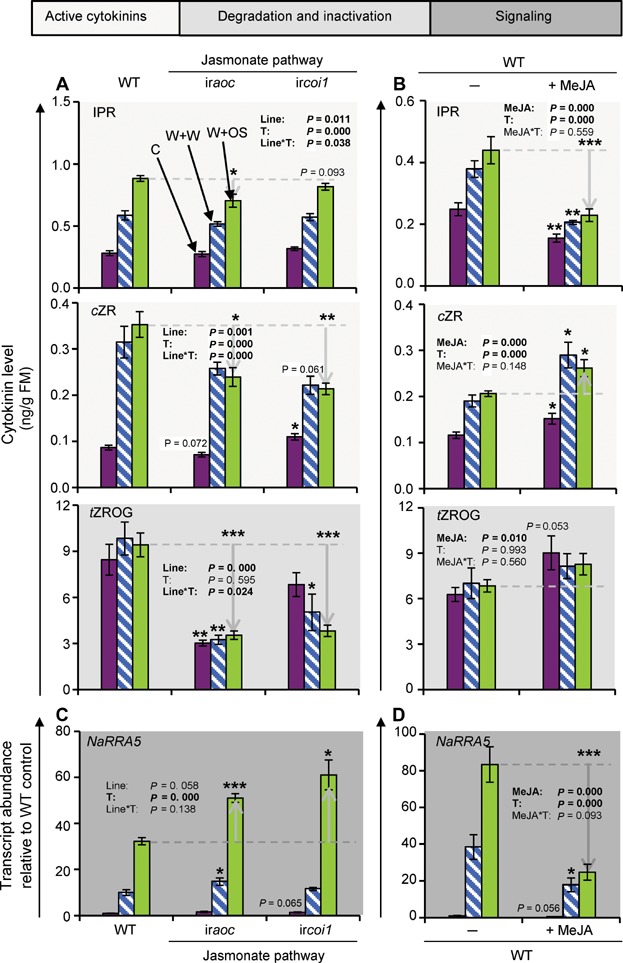
(A) Isopentenyladenosine (IPR), cis-zeatin riboside (cZR) and trans-zeatin riboside O-glucoside (tZROG) levels in leaves of Nicotiana attenuata 30 min after wounding and application of water (W + W, white bars with blue lines) or Manduca sexta oral secretions (W + OS, lime-green bars) to the puncture wounds and in untreated control leaves (C, purple bars). Measurements were performed in leaves of wild-type (WT) plants and RNAi lines silenced in AOC or COI1 expression. (B) IPR, cZR, and tZROG levels in leaves of N. attenuata 30 min after W + W (white bars with blue lines) or W + OS treatment (lime-green bars) and in untreated control leaves (C; purple bars). Measurements were performed in leaves of WT plants with and without a 24 h methyl-jasmonate (MeJA; 150 µg per leaf) pretreatment. (C) Response regulator 5 (NaRRA5) transcript abundance under the conditions mentioned in (A). (D) NaRRA5 transcript abundance under the conditions mentioned in (B). Line/MeJA and treatment (C, W + W and W + OS; T) effects and their interaction (Line*T and MeJA*T, respectively) were analyzed by univariate ANOVA, except for NaRRA5 (C) data which were analyzed by a generalized least squares model. Asterisks indicate significant differences between same treatments from RNAi lines and WT plants, and plants with and without a MeJA pretreatment (independent samples t-test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). Error bars are standard errors (A, C n ≥ 4; B, D n ≥ 5). FM, fresh mass.
Wounding and simulated herbivory induce systemic CK pathway changes
In addition to local CK pathway changes, we also found wound and herbivory-induced alterations in the abundance of CK-related transcript (Figures 6, S13, S14) and CK levels (Figure 7) in systemic leaves and root tissues. Figure 6 shows some representative changes and additional information can be found in Figures S13 and S14. In addition to the changes in the transcripts of the CK biosynthesis genes like NaLOG4, we also observed changes in the transcripts of the CK signaling elements and CK-inactivation/degradation enzymes, including NaRRA5 and NaCKX5, respectively (Figures 6, S13, S14). The levels of cZR slightly increased systemically after wounding, whereas tZR levels decreased (Figure 7).
Figure 6. Wounding and herbivory regulate systemic transcript accumulation of cytokinin-related genes.
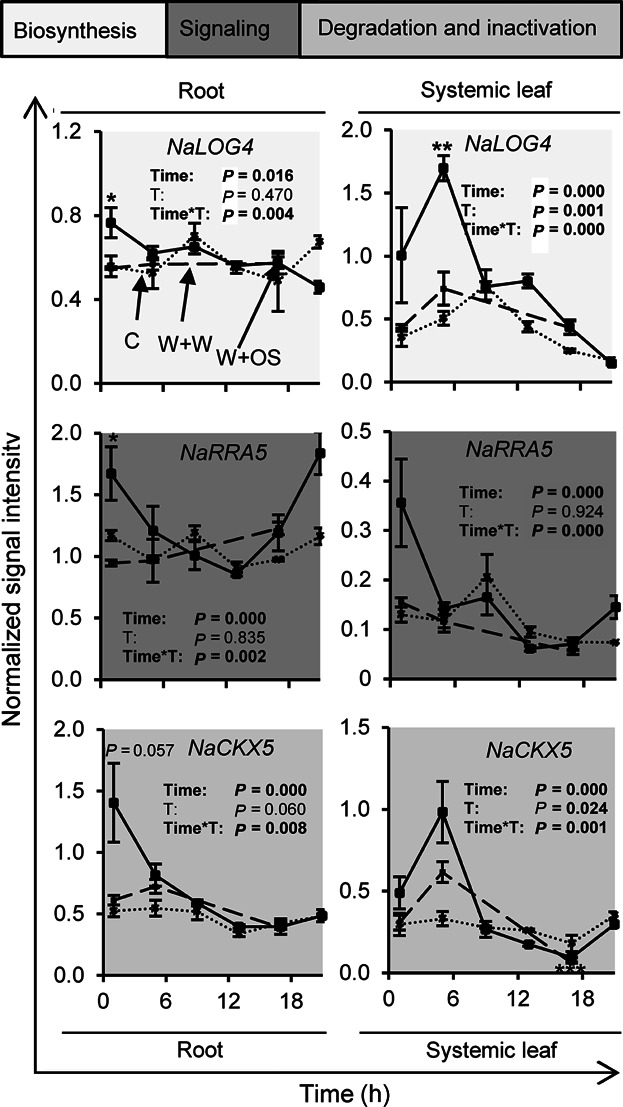
Transcript accumulation was measured in roots and systemic leaves of N. attenuata at different time points after wounding and application of water (W + W; dashed line; 1, 5 and 17 h) or M. sexta oral secretions (W+OS; solid line; 1, 5, 9, 13, 17 and 21 h) to the puncture wounds, as well as in untreated control leaves (C; dotted line; 1, 5, 9, 13, 17 and 21 h). Data are obtained from kinetic analysis conducted with microarrays. Time and treatment (C and W + OS; T) effects and there interaction (Time*T) were analyzed by a generalized least squares model. Asterisks indicate significant differences between W + W and W + OS-treated samples at the same time point (independent samples t test: *P ≤0.05, **P ≤0.01, ***P ≤0.001). Error bars are standard errors (N=3). For additional transcript information see Figure S13 and S14.
Figure 7. Wounding and herbivory regulate systemic changes in cytokinin levels.
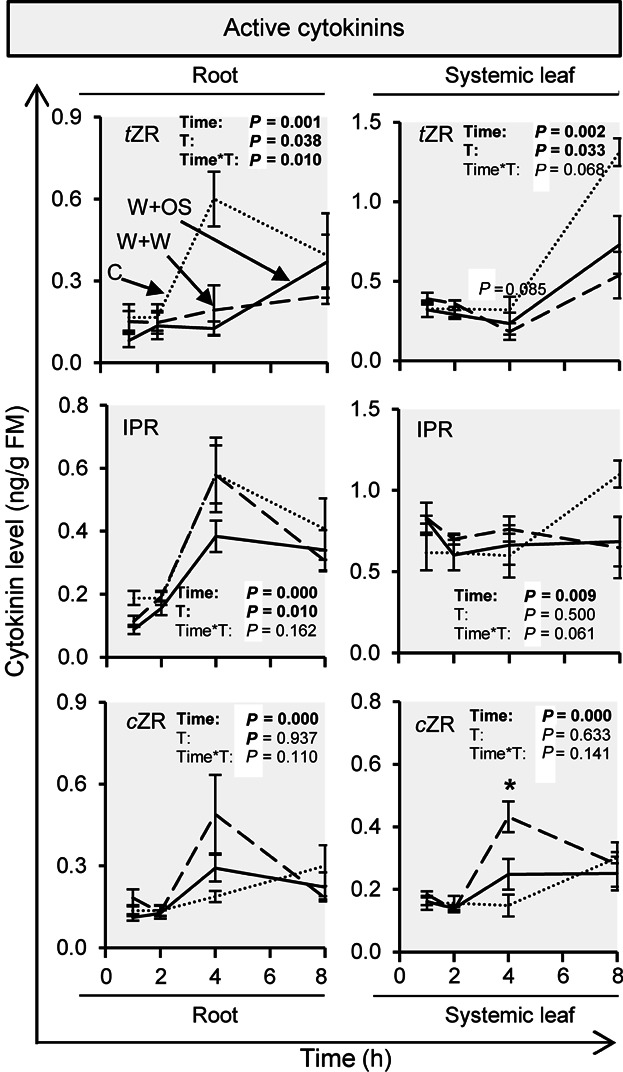
Cis-zeatin riboside (cZR) and trans-zeatin riboside (tZR) levels were measured in roots and systemic leaves of Nicotiana attenuata at different time points after wounding and application of water (W + W; dashed line) or Manduca sexta oral secretions (W + OS; solid line) to the puncture wounds, as well as in untreated control leaves (C; dotted line). Time and treatment (C and W + OS; T) effects and there interaction (Time*T) were analyzed by a generalized least squares model. Asterisks indicate significant differences between W + W and W + OS-treated samples at the same time point (independent samples t-test: *P ≤ 0.05). Error bars are standard errors (n ≥ 4).
Wounding and HAMP-mediated CK level changes in Arabidopsis
To investigate if HAMP-induced CK levels are also induced in other plants, we wounded leaves of A. thaliana and treated them with water or grasshopper OS (OSGH). While A. thaliana leaves do not respond to FACs, they do perceive OSGH, which results in an amplification of wound-induced defense responses (Schäfer et al. 2011). Wounding alone increased the IPR levels in leaves by 35%, cZ level by 115%, cZR level by 86%, and cZROG level by 73%, when compared to untreated leaves. OSGH application to puncture wounds significantly elevated the W + W-induced changes in IPR, cZ, and cZR levels and marginally increased cZROG levels. W + OSGH treatment resulted in 2.3, 18.1, 7.3, and 2.1 times as much IPR, cZ, cZR, and cZROG accumulation, respectively, when compared to control levels (Figure 8).
Figure 8. Wounding and herbivory induce changes in cytokinin levels in leaves of Arabidopsis thaliana.
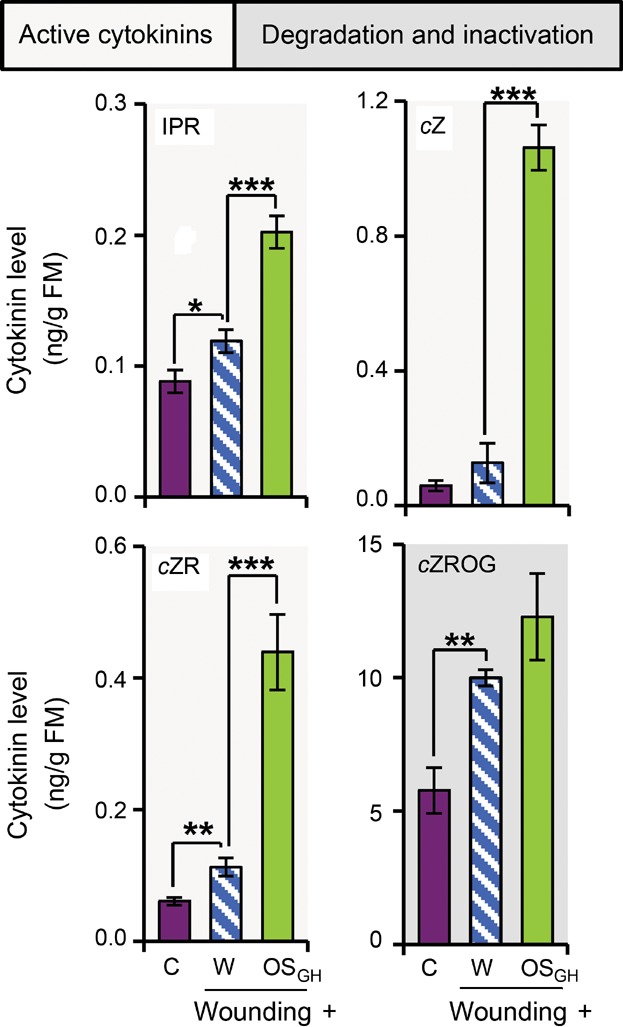
Isopentenyladenosine (IPR), cis-zeatin (cZ), cis-zeatin riboside (cZR), and cis-zeatin riboside O-glucoside (cZROG) levels in leaves 30 min after wounding and application of water (W) or grasshopper oral secretions (OSGH) to the puncture wounds, as well as in untreated control leaves (C). Asterisks indicate significant differences between W and OSGH application to wounding sites or between wounding and W application compared to C, as indicated (independent samples t-test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). Error bars are standard errors (n ≥ 4). FM, fresh mass. [Correction made on 23 January 2015, after first online publication: An asterisk, which has been erroneously added to Figure 8, has been removed from the figure.]
DISCUSSION
Here, we addressed previous speculations about the responsiveness of CKs to herbivore attack (Giron et al. 2013), by conducting an analysis of the CK pathway in the Solanaceous plant, N. attenuata. We show that the CK pathway responds to wounding and perception of HAMPs in local and systemic tissues and demonstrate its intimate interaction with the JA pathway. These results establish CKs as new components of the herbivore response in plants.
The CK pathway responds to wounding and simulated herbivory
More than 90 years ago, Haberlandt (1921) reported that wound-induced hormones trigger cell division. Cell division is well known to be regulated by CKs (Miller et al. 1955) and wound-induced increases in CK activities, which likely promote tissue healing, were reported, for example, for potato and cucumber (Conrad and Kohn 1975; Mitchell and van Staden 1983; Crane and Ross 1986); however, due to limitations in analytical chemistry at the time of these studies, a comprehensive analysis of changes in CK metabolites was not possible. Here, we confirm that wounding rapidly changes the levels of CKs, including active CKs (IP, IPR, and cZR) and inactivation products (tZROG, cZROG, and tZ7G) (Figure 3). Interestingly, OS elicitation specifically amplified many of the wound-induced increases. Changes in CK levels were accompanied by a dramatic regulation of many genes involved in CK metabolism and signaling. We found that wounding and FAC application was sufficient to mimic W + OS-induced changes in CK levels and expression of the CK-marker gene NaRRA5 (Figure 4), demonstrating that regulation of the CK pathway is one of the earliest HAMP-induced hormone responses in plants.
Connecting specific changes in transcript levels of genes involved in CK metabolism with concomitant changes in CK levels is intricate, since the transcript levels are expected to be influenced by multiple factors, including tissue disruption and FAC perception, CK-mediated feedback regulation (Brenner et al. 2012; Bhargava et al. 2013) and by the interaction with other herbivory induced phytohormones (Erb et al. 2012; El-Showk et al. 2013). In addition, changes in CK levels could also be a result of changes in their transport rates away from or to the specific tissue. Posttranslational regulation might also play an important role, as reported for the CK-dependent increase in CKX activity (Motyka et al. 1996). Increased levels of NaRRA5 (Figure 2) are likely related with the rapid changes in the active CKs IP, IPR, and cZR (Figures 3, S11). Additionally, we assume that the rapid increases in some LOG transcripts (Figures 2, S1) might play a role in elevating CK levels and sustaining them by counteracting CK-inactivation processes, which are indicated by the elevation in CK-glucoside levels (Figure 3). Based on the literature (Brenner et al. 2012; Bhargava et al. 2013) and the timing of increases in ZOG and CKX transcript accumulation (Figures 2, S7, S8), these genes could be also induced by the observed increase in IP, IPR, and cZR (Figure 3) levels. That CKs themselves are mediating some of the observed transcript changes is supported by Figure S11, showing similar changes in NaIPT5, NaRRA5, and NaCKX5 transcript accumulation after external CK application. Changes in ZOG expression might account for the increase in CK inactivation products (Figure 3). Additionally changes in cytochrome P450 monooxygenases involved in the conversion of IP-type to tZ-type CKs (Takei et al. 2004), the postulated zeatin cis-trans isomerase (Bassil et al. 1993) or ß-glucosidases responsible for the release of CKs from CK O-glucosides (Brzobohaty et al. 1993) could play a role, but were not further addressed in this study.
Since CKs have been shown to increase a plant's resistance to pathogens (Choi et al. 2010; Großkinsky et al. 2011; Argueso et al. 2012), future experiments should investigate a putative role of CKs in preventing pathogen infections after wounding and especially after attack by herbivores, which frequently are the vectors of pathogens (Sobek and Munkvold 1999; Frisinghelli et al. 2000). Regulation of CK metabolism and signaling might also play an important role for herbivory-induced changes in primary metabolism and source-sink regulation (Quilliam et al. 2006; Ferrieri et al. 2013; Machado et al. 2013). Additionally, CKs might be directly involved in assisting herbivore-induced defense responses as proposed, for example, by Dervinis et al. (2010) and Smigocki et al. (2000). Using transgenic plants with altered CK levels or signaling will allow these questions to be addressed.
Regulation of the CK pathway after herbivory
CKs cross-talk with other hormones, including auxins, gibberellins, abscisic acid, and ethylene (Naseem et al. 2012; El-Showk et al. 2013), but aside from a few reports about CK-mediated effects on JAs (Sano et al. 1996; Dervinis et al. 2010), very little is known about the JA-CK interaction. JAs are widely accepted as one of the main regulators of wound- and HAMP-induced responses (Erb et al. 2012). We found that JA pathway manipulations had an influence on CK levels and signaling; however, a functional JA pathway was not required for the induction of W + W and W + OS-induced CK responses. Instead of increasing CK responses JA had a suppressive effect on CK signaling after simulated herbivory (NaRRA5; Figure 5C, D). The elevated CK signaling response (NaRRA5; Figures 5C) in JA pathway-impaired plants is not explained by the levels of the measured active CKs (Figures 5A, S15); therefore in addition to changes in CK metabolism, CK signaling might also be affected, as reported for the JA-interaction with other phytohormones such as auxin or abscisic acid (Pauwels et al. 2010). JA supplementation resulted in downregulation of the shoot derived active CK IPR, while it promoted the accumulation of cZR (Figure 5A, B), a CK associated with stress responses (Gajdošová et al. 2011). JAs also trigger CK inactivation processes, as indicated by O-glucosylation of tZ. While the JA supplementation experiments (Figure 5B) indicate a negative effect of JA on the IPR accumulation, IPR levels were not increased, but slightly reduced in iraoc and ircoi1 plants, which are impaired in the JA pathway (Figure 5A). This might be explained by the very low levels of active JA-Ile in the absence of wound or HAMP induction and the chronological order of events after W + OS treatment: that IPR accumulates before JA-Ile in N. attenuata (maximum accumulation: IPR, 30 min, see Figure 3; JA-Ile, 1 h, compare Meldau et al. (2011). We measured CKs only at a single time-point in JA-deficient lines and after MeJA treatments. Kinetic measurements are required to identify the critical steps in CK metabolism that are regulated by JAs. Additionally, the effect of multiple elicitations should be investigated since JA accumulation from previous elicitations might influence CK response, thereby representing a potential mechanism to distinguish between early and late herbivory-induced responses or attack frequency-dependent responses. Interestingly JA-mediated effects on the CK pathway were also observed in untreated control leaves (Figure 5B, D) indicating that herbivory is not required for the observed JA-CK interaction.
Cytokinins are long known for their role in inhibiting leaf senescence (Richmond and Lang 1957), for growth promotion of leaf tissue (Miller 1961) and to induce cell division (Miller et al. 1955). JAs were shown to counteract the senescence-inhibiting activities of the CK kinetin, to induce premature senescence (Ueda and Kato 1980; He et al. 2002), suppress leaf growth (Attaran et al. 2014), as well as to repress cell cycle propagation (Noir et al. 2013), which indicates a more general negative effect of JA on the CK pathway activity than our data on herbivory-induced signaling suggests.
The CK pathway in long distance systemic responses
Cytokinins not only play a role in regulating local processes, but were also found as important systemic regulators, for example, for the adjustment of the nitrogen-foraging strategy (Ruffel et al. 2011) and nitrogen-dependent root-shoot signaling (Takei et al. 2001). We observed that W + W and W + OS treatment of leaves resulted in CK pathway changes in other untreated leaves and the roots of the same plants (Figures 6, 7). Therefore, our data indicate CKs as an integral component of the systemic response after herbivore attack. The wound and herbivory-induced transcript changes shown in Figures 6, S13, and S14 indicate that the CK-biosynthesis and the CK-inactivation/degradation could be affected systemically after W + W and W + OS treatment, as well as the CK signaling output (NaRRA5). Effects on CK sensitivity or signal transduction might explain why the OS-mediated effects differ between CK-related transcripts and CK metabolites, but it is also possible that the systemic CK response is mainly wound-dependent. Since CKs are known to be specifically transported either by the phloem (IP-type CKs) or the xylem (tZ-type CKs; Kudo et al. 2010) between different plant parts, changed CK transport might also be involved in shaping systemic CK patterns.
With regard to the work of Dervinis et al. (2010), systemic CK level changes might play a role in systemic priming of plant-defense responses after herbivore attack. Additionally, CKs are known to be involved in many abiotic stress responses (Jeon et al. 2010; Nishiyama et al. 2011) and therefore could play a role in integrating these different signals on a whole plant scale.
The CK pathway proteins show diverse evolutionary patterns
The response of the CK pathway to HAMPs, which are only perceived by particular plant species (Erb et al. 2012) raised the question if the genes in the CK pathway of N. attenuata went through a similar evolutionary history as observed in other plant species. To answer this we constructed a phylogeny of the CK pathway and analyzed it in respect to established concepts of the CK pathway evolution.
Evolutionary analyses have previously been used to support CK signaling models and gene function analysis (Gruhn and Heyl 2013). We used our available 454 sequences for data mining and confirmed most of the evolutionary patterns found in previous studies (Pils and Heyl 2009; Tsai et al. 2012). We found that HPTs have notably expanded in Poplar (10 copies) in comparison to the remaining dicot sequences available, where a stable basal number of five copies is found, as described by Pils and Heyl (2009). We corroborated that HPTs, together with CKXs and CHKs have stable numbers across all green plant species analyzed and further observed that each gene has a homolog in monocots and dicots, indicating conserved evolution after duplication of these genes in the flowering plants. Our findings for HPTs contrast with the results from Tsai et al. (2012), which described gene expansion for this gene family. The discrepancy may result from the fact that we analyzed more species than they did. A higher similarity of CK receptors to the homologs in monocot/dicot species had also been shown by Tsai et al. (2012) and Pils and Heyl (2009). Both studies also showed that RRs had expanded after the separation of lower land plants and flowering plants. In addition to the confirmation of results from previous publications, we observed a similar gene expansion pattern in the CK biosynthesis genes (IPTs and LOGs) and in the first step of CK inactivation (ZOGs).
Rapid evolution due to duplication can result in functional diversification. We expect this to occur in RRs, which are reported to play roles as both negative and positive stress regulators (Wohlbach et al. 2008). Interestingly, gene number expansion also appears to play a role in CK deactivation enzymes, such as the glucosyltransferases that do not have homologs in green algae and lower land plants, which indicates that these proteins duplicated after their emergence in flowering plants, allowing them to develop specialized functions. A higher specificity in N-glucosylation enzymes had already been described by Sakakibara (2006). In the case of IPTs, evidence for functional diversification also exists (Miyawaki et al. 2006), which might be partly explained by maintaining different gene copies of these genes in the genome. Relaxed selection might also result in amplification of gene numbers of one family due to a process called genomic drift (Nei 2007), which occurs if gene dosage effects can be neglected. We cannot rule out relaxed selection in case of LOGs, although this family most probably is undergoing changes in gene birth-and-death dynamics, as has been shown for similar enzymes in A. thaliana (Kuroha et al. 2009). From this analysis, we confirm that N. attenuata's CK pathway genes have evolved in similar ways to homologous genes in other species.
CKs are also regulated by herbivore perception in Arabidopsis
From the gene phylogenetic analysis we observe that among other important model plants for the analysis of plant defense against herbivores, A. thaliana, has similar evolutionary patterns for CK biosynthesis and regulation genes. Therefore, an analysis of the CK response in this plant is interesting, especially as it was previously shown to be unresponsive to FACs, but to respond instead to lipases and other unknown elicitors present in OSGH (Schmelz et al. 2009; Schäfer et al. 2011). We found that A. thaliana showed similar qualitative changes in CK levels when treated with OSGH as observed for N. attenuata treated with OS from M. sexta (Figure 8). The elevation of IPR and cZR in W + OSGH-treated leaves, as compared to those elicited by wounding alone, was particularly noteworthy. This indicates that the patterns of HAMP-induced CK changes are likely conserved among different plant families. A broader analysis of herbivore-induced CK responses in plants from additional plant families and ecological backgrounds could be useful to understand the conservation patterns of the CK-pathway genes and their potential functional implication.
MATERIALS AND METHODS
Plant material and growth
The generation of the stable transformed iraoc (line number A-07-457) and ircoi1 (line number A-04-249) plants was described by Kallenbach et al. (2012) and Paschold et al. (2007), respectively. As WT we used plants from an inbred “Utah” line of Nicotiana attenuata (Torr. ex S. Wats.). N. attenuata seed germination and growth under glasshouse conditions was performed as described elsewhere (Krügel et al. 2002). In brief, after 10 d on Gamborg's B5 medium (Sigma-Aldrich, Taufkirchen, Germany, http://www.sigmaaldrich.com) with phytagel (Sigma) and 12 d in TEKU JP 3050 104 pots plants were transferred to 1 L pots filled with sand. Plants were kept under glasshouse conditions with 26–28 °C under 16 h supplemental light from Master Sun-T PIA Agro 400 or Master Sun-T PIA Plus 600 W Na lights (Philips, Turnhout, Belgium). Fertilization was done by flood irrigation with additions of 240 g Ca(NO3)2 • 4H2O (Merck, Schwalbach, Germany, http://www.merck-chemicals.com/) and 120 g Ferty B1 (Planta Düngemittel, Regenstauf, Germany, http://www.plantafert.com/) in a 400 L watering tank.
Arabidopsis thaliana plants (ecotype Col-0) were grown on a substrate consisting of 80% Fruhstorfer Nullerde, 10% vermiculite, and 10% sand, fertilized with Triabon (1 g/L) and Osmocote Exact Mini (1 g/L) in a growth chamber at 21 °C, 60% humidity with 10 h light/day with an intensity of 190–220 µmol/m2 per s.
Leaf treatments
Standardized wound treatments in N. attenuata were performed by rolling a fabric pattern-wheel three times on each side of the leaf and subsequently applying 20 µL water to the punctured holes (W + W). Herbivory was simulated by applying herbivore OS instead of water (W + OS) and for FAC treatments, N-linolenoyl-l-glutamate at a concentration similar to 1:5 diluted M. sexta OS was used (27.6 ng/µL; Hettenhausen et al. 2013). These treatments were performed in the morning (09.00–10.00 hours). In contrast to N. attenuata, A. thaliana leaves were treated with only 5 µL water or OSGH. After incubation for the indicated time, the leaf tissue was harvested and immediately frozen in liquid nitrogen and stored at −80 °C.
Manduca sexta and Schistocerca gregaria
Manduca sexta larvae were obtained from in-house colonies and S. gregaria from Bugs International, Irsingen/Unterfeld, Germany (http://www.bugs-international.com/). M. sexta larvae were fed on N. attenuata and S. gregaria on A. thaliana before OS/OSGH collection, which was performed according to Turlings et al. (1993) with the modifications of Alborn et al. (2003). OS and OSGH were diluted 1:5 in water before application.
CK spray application
For spray application, tZ and cZR were dissolved in 80% (v/v) EtOH (tZ, 1 mg/mL; cZR, 5 mg/mL) and diluted in an aqueous solution of 0.02% (v/v) Tween-20 to 1 µmol/L and 5 µmol/L, respectively. Spray application of tZ, cZR and the corresponding buffer control was done three times per day over 3 d.
MeJA pretreatment
MeJA treatment was done according to Baldwin (1996), by applying 20 µL of a 7.5 µg/µL MeJA containing lanolin paste to the adaxial side of the base of a leaf. Lanolin without MeJA was applied as control. MeJA applications were performed 1 d before the start of the experiments.
Phylogenetic analysis
Cloned CK biosynthesis and receptor gene sequences from cDNA were confirmed using sequences of N. attenuata 454 sequenced transcripts obtained as described in Gase and Baldwin (2013). Confirmed sequences were then used to perform a blastx search to the proteomes of all green plants (Viridiplantae) at NCBI. For illustration purposes, we show proteins present in at least one (ideally all) of the representative species for each phyletic group: green algae (Volvox carteri, Micromonas pusilla, Ostreococcus lucimarinus, Ostreococcus tauri, and Chlamydomonas reinhardtii), lower-land plants (P. patens, Selaginella mollendorfii), monocots (O. sativa cv. Japonica, Sorghum bicolor, and Z. mays), and dicots (Vitis vinifera, Solanum lycopersicum, A. thaliana, G. max, Ricinus communis, and Populus trichocarpa).
For each protein family, multiple sequence alignments were generated using MUSCLE (Edgar 2004). The phylogenetic relationships were analyzed using the Neighbor-Joining method (Saitou and Nei 1987) as implemented in MEGA5 (Tamura et al. 2011). We used 1000 bootstrap replicates for tree support. Evolutionary distances were computed using the JTT matrix-based method (Jones et al. 1992) with a gamma distribution (shape parameter = 1) for rate site variation. For easier understanding, we display only the topology of the trees. Genes were named according to Heyl et al. (2013).
Microarray analysis
Microarray analysis was done as described by Onkokesung et al. (2012). Analysis was done with leaves treated for 1, 5, and 17 h with W + W or 0.5 (only local leaves), 1, 5, 9, 13, 17, and 21 h with W + OS, as well as in untreated control leaves. For hybridization we used a N. attenuata-specific Agilent microarray platform (GEO microarray repository, GPL13527). Microarray data (Figures 2, S1–S8) were confirmed by qPCR analysis of representative genes in samples from an independent experiment (Figures S9, S10).
qPCR analysis
RNA extraction was performed with TRIzol (Invitrogen, Darmstadt, Germany), according to the manufacturer's instructions. cDNA was synthesized by reverse transcription using oligo(dT) primer and RevertAid reverse transcriptase (Invitrogen). qPCR was performed using actin as standard on a Stratagene Mx3005P qPCR machine using a SYBR Green containing reaction mix (Eurogentec, Cologne, D, http://www.eurogentec.com/; qPCR Core kit for SYBR Green I No ROX). The primer sequences are provided in Table S2.
CK analysis
Cytokinins were extracted from plant tissue with acidified aqueous methanol followed by two solid-phase extraction (SPE) steps. Measurement was done by LC-MS/MS. The method was adapted according to Dobrev and Kamı'nek (2002) with the modifications by Kojima et al. (2009) and Schäfer et al. (2013).
In brief, 100 mg frozen plant material was extracted twice with 800 µL MeOH:H2O:HCOOH (15:4:1) at −20 °C. Labeled internal standards were supplemented in the first extraction step. The first SPE step was performed on a Multi 96 HR-X column (96 × 25 mg; Macherey-Nagel, Düren, Germany, http://www.mn-net.com/) conditioned with extraction buffer. The methanol in the column eluent was evaporated and after replenishment with 850 µL 1 N HCOOH a second SPE step was performed using a Multi 96 HR-XC column (96 × 25 mg; Macherey-Nagel) conditioned with 1 N HCOOH. After several washing steps (consecutively 1 mL 1 N HCOOH, 1 mL MeOH, and 1 mL 0.35 N NH4OH) the CK-ribosides, free bases, and glucosides were eluted with 1 mL 0.35 N NH4OH in 60% (v/v) MeOH. After evaporation, samples were reconstituted in 50 µL 0.1% (v/v) acetic acid.
Extraction was performed in 96-well BioTubes (1.1 mL individual tubes, Arctic White LLC, Bethlehem, PA, USA, http://www.arcticwhiteusa.com/) and Nunc 96-Well Deep Well Plates (Thermo Scientific, Waltham, MA USA, https://www.thermoscientific.com/), evaporation under constant nitrogen flow in an Evaporator system (Glas-Col, Terre Haute, IN, USA, http://www.glascol.com/) and SPE using a Chromabond Multi 96 vacuum chamber (Macherey).
Chromatography was performed on an Agilent 1200 HPLC system (Agilent Technologies, Santa Clara, CA, USA, http://www.home.agilent.com). For separation a Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 µm, Agilent Technologies) was used. The mobile phase comprised solvent A (water, 0.05% (v/v) HCOOH) and solvent B (acetonitrile) with the elution profile: 0–0.5 min, 95% A; 0.5–5 min, 5%–31.5% B in A; 5.01–6.5 min 100% B and 6.51–9 min 95% A, with a flow rate of 1.1 mL/min. The column temperature was maintained at 25 °C. The liquid chromatography was coupled to an API 5000 tandem mass spectrometer (Applied Biosystems, Darmstadt, Germany, http://www.invitrogen.com/site/us/en/home/brands/Applied-Biosystems.html?CID=fl-AppliedBiosystems) equipped with a Turbospray ion source. For detection, the mass spectrometer was operated in positive ionization mode multi-reaction-monitoring modus to monitor analyte parent ion → product ion (Table S3). Settings were as follows: ion spray voltage, 5,500 eV; turbo gas temperature, 700 °C; nebulizing gas, 70 psi; curtain gas, 25 psi; heating gas, 60 psi; and collision gas, 6 psi. Both Q1 and Q3 quadrupoles were maintained at unit resolution. Analyst 1.5 software (Applied Biosystems) was used for data acquisition and processing. tZ, tZR, tZROG, tZ7G, cZ, cZR, cZROG, cZ9G, DHZ, DHZR, IP, and IPR were quantified by using deuterated internal standards (Table S3; Olchemim, Olomouc, CZ, http://www.olchemim.cz/).
Chemicals
EtOH and MeOH were purchased by Merck, acetonitrile by VWR (Darmstadt, D, http://www.vwr.com/), lanolin, Tween-20, as well as MeJA by Sigma–Aldrich and HCOOH for chromatography by Fisher Scientific (Schwerte, D, http://www.de.fishersci.com/de/), otherwise by Riedel-de Haën (Seelze, D, http://www.riedeldehaen.de/), acetic acid by Carl Roth (Karlsruhe, D, http://www.carlroth.com/) and CK standards by Olchemim.
Statistical analysis
Data were analyzed by SPSS Statistics 17.0 (IBM, Ehningen, D, http://www.01.ibm.com/software/de/analytics/spss/) with independent (unpaired) samples t-test and univariate ANOVA. If homoscedasticity could not be achieved by transformation, datasets were analyzed by a generalized least squares (GLS) model using R 3.0.1 (http://www.r-project.org). The used statistical analysis methods are indicated in the figure legends. In Figures 3 and S12, the 0 h control was also used for the 15 and 30 min time point comparisons. In Figures 2, S1–S8, S10, S13, and S14, the W + W treatment was excluded from the analysis by univariate ANOVA and GLS model. The number of biological replicates per experiment is indicated in the figure legends. The presented data are supported by at least two independent experiments with similar results.
Accession numbers
The data from the Transcriptome Shotgun Assembly project have been deposited at DDBJ/EMBL/GenBank under the accession GBGF00000000. The version described in this paper is the first version, GBGF01000000.
Acknowledgments
We thank Michael Reichelt, Mario Kallenbach, Klaus Gase, Matthias Schöttner, Thomas Hahn, Antje Wissgott, Susanne Kutschbach, Wibke Kröber, and Eva Rothe for technical assistance and Tamara Krügel, Andreas Weber, and Andreas Schünzel from the glasshouse team for plant cultivation. Schäfer, Navarro-Quezada, and Baldwin are funded by the Max-Planck-Society, Meza-Canales by the DAAD, and Vanková by the Czech Science Foundation, project no. 206/09/2062. Meldau and Brütting are funded by Advanced Grant no. 293926 of the European Research Council to Baldwin. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
SUPPORTING INFORMATION
Additional supporting information can be found in the online version of this article:
Figure S1. Wounding and herbivory regulate the expression of LOG-family genes
Figure S2. Wounding and herbivory regulate the expression of IPT-family genes
Figure S3. Wounding and herbivory regulate the expression of cytokinin receptor genes
Figure S4. Wounding and herbivory regulate the expression of HPT-family genes
Figure S5. Wounding and herbivory regulate the expression of type-A RR-family genes
Figure S6. Wounding and herbivory regulate the expression of type-B RR-family genes
Figure S7. Wounding and herbivory regulate the expression of CKX-family genes
Figure S8. Wounding and herbivory regulate the expression of ZOG-family genes
Figure S9. Confirmation of microarray expression data
Figure S10. Confirmation of NaIPT5 microarray data
Figure S11. Cytokinin-spraying changes NaIPT5, NaRRA5, and NaCKX5 transcript accumulation
Figure S12. Wounding and herbivory-induced changes in cytokinin levels
Figure S13. Wounding and herbivory regulate transcript accumulation of cytokinin-related genes in the root
Figure S14. Wounding and herbivory regulate transcript accumulation of cytokinin-related genes in systemic leaves
Figure S15. Trans-zeatin riboside levels are reduced in jasmonic acid pathway impaired transgenic plants
Table S1. Abbreviations
Table S2. Sequences of primers used for qPCR
Table S3. Multi-reaction-monitoring settings for cytokinin quantification in positive ionization mode
REFERENCES
- Alborn HT, Brennan MM, Tumlinson JH. Differential activity and degradation of plant volatile elicitors in regurgitant of tobacco hornworm (Manduca sexta) larvae. J Chem Ecol. 2003;29:1357–1372. doi: 10.1023/a:1024209302628. [DOI] [PubMed] [Google Scholar]
- Argueso CT, Ferreira FJ, Epple P, To JPC, Hutchison CE, Schaller GE, Dangl JL, Kieber JJ. Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 2012;8:e1002448. doi: 10.1371/journal.pgen.1002448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran E, Major I, Cruz J, Rosa B, Koo A, Chen J, Kramer D, He SY, Howe G. Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 2014;165:1302–1314. doi: 10.1104/pp.114.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin I. Methyl jasmonate-induced nicotine production in Nicotiana attenuata: Inducing defenses in the field without wounding. In: Städler E, Rowell-Rahier M, Bauer R, editors. Proceedings of the 9th International Symposium on Insect-Plant Relationships, Vol 53. the Netherlands: Springer; 1996. pp. 213–220. [Google Scholar]
- Bassil NV, Mok D, Mok MC. Partial purification of a cistrans-isomerase of zeatin from immature seed of Phaseolus vulgaris L. Plant Physiol. 1993;102:867–872. doi: 10.1104/pp.102.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang YH, Schaller GE, Loraine A, Kieber JJ. Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-seq in Arabidopsis. Plant Physiol. 2013;162:272–294. doi: 10.1104/pp.113.217026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaventure G, Van Doorn A, Baldwin IT. Herbivore-associated elicitors: FAC signaling and metabolism. Trends Plant Sci. 2011;16:294–299. doi: 10.1016/j.tplants.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Brenner WG, Ramireddy E, Heyl A, Schmülling T. Gene regulation by cytokinin. Front Plant Sci. 2012;3:8. doi: 10.3389/fpls.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohaty B, Moore I, Kristoffersen P, Bako L, Campos N, Schell J, Palme K. Release of active cytokinin by a β-glucosidase localized to the maize root meristem. Science. 1993;262:1051–1054. doi: 10.1126/science.8235622. [DOI] [PubMed] [Google Scholar]
- Chini A, Boter M, Solano R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009;276:4682–4692. doi: 10.1111/j.1742-4658.2009.07194.x. [DOI] [PubMed] [Google Scholar]
- Choi J, Huh SU, Kojima M, Sakakibara H, Paek KH, Hwang I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev Cell. 2010;19:284–295. doi: 10.1016/j.devcel.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Conrad K, Kohn B. Zunahme von cytokinin und auxin in verwundetem Speichergewebe von Solanum tuberosum. Phytochemistry. 1975;14:325–328. [Google Scholar]
- Crane KE, Ross CW. Effects of wounding on cytokinin activity in cucumber cotyledons. Plant Physiol. 1986;82:1151–1152. doi: 10.1104/pp.82.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter N, Gholami A, Goormachtig S, Goossens A. Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 2012;17:349–359. doi: 10.1016/j.tplants.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Dervinis C, Frost CJ, Lawrence SD, Novak NG, Davis JM. Cytokinin primes plant responses to wounding and reduces insect performance. J Plant Growth Regul. 2010;29:289–296. [Google Scholar]
- Dobrev PI, Kamı'nek M. Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J Chromatogr A. 2002;950:21–29. doi: 10.1016/s0021-9673(02)00024-9. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S, Ruonala R, Helariutta Y. Crossing paths: Cytokinin signalling and crosstalk. Development. 2013;140:1373–1383. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- Elzen GW. Cytokinins and insect galls. Comp Biochem Physiol Part A Physiol. 1983;76:17–19. [Google Scholar]
- Engelbrecht L. Cytokinin in den grünen Inseln des Herbstlaubes. Flora. 1968;159:369–374. [Google Scholar]
- Erb M, Meldau S, Howe GA. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012;17:250–259. doi: 10.1016/j.tplants.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrieri AP, Agtuca B, Appel HM, Ferrieri RA, Schultz JC. Temporal changes in allocation and partitioning of new carbon as 11C elicited by simulated herbivory suggest that roots shape aboveground responses in Arabidopsis. Plant Physiol. 2013;161:692–704. doi: 10.1104/pp.112.208868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frébort I, Kowalska M, Hluska T, Frébortová J, Galuszka P. Evolution of cytokinin biosynthesis and degradation. J Exp Bot. 2011;62:2431–2452. doi: 10.1093/jxb/err004. [DOI] [PubMed] [Google Scholar]
- Frisinghelli C, Delaiti L, Grando MS, Forti D, Vindimian ME. Cacopsyllacostalis (Flor 1861), as a vector of apple proliferation in Trentino. J Phytopathol. 2000;148:425–431. [Google Scholar]
- Gajdošová S, Spíchal L, Kamínek M, Hoyerová K, Novák O, Dobrev PI, Galuszka P, Klíma P, Gaudinová A, Žižková E, Hanuš J, Dančák M, Trávníček B, Pešek B, Krupička M, Vaňková R, Strnad M, Motyka V. Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J Exp Bot. 2011;62:2827–2840. doi: 10.1093/jxb/erq457. [DOI] [PubMed] [Google Scholar]
- Gase K, Baldwin IT. Transformational tools for next-generation plant ecology: Manipulation of gene expression for the functional analysis of genes. Plant Ecol Divers. 2013;5:485–490. [Google Scholar]
- Gilardoni P, Schuck S, Jungling R, Rotter B, Baldwin I, Bonaventure G. SuperSAGE analysis of the Nicotiana attenuata transcriptome after fatty acid–amino acid elicitation (FAC): Identification of early mediators of insect responses. BMC Plant Biol. 2010;10:66. doi: 10.1186/1471-2229-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giron D, Frago E, Glevarec G, Pieterse CMJ, Dicke M. Cytokinins as key regulators in plant–microbe–insect interactions: Connecting plant growth and defence. Funct Ecol. 2013;27:599–609. [Google Scholar]
- Großkinsky DK, Naseem M, Abdelmohsen UR, Plickert N, Engelke T, Griebel T, Zeier J, Novák O, Strnad M, Pfeifhofer H, van der Graaff E, Simon U, Roitsch T. Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol. 2011;157:815–830. doi: 10.1104/pp.111.182931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn N, Heyl A. Updates on the model and the evolution of cytokinin signaling. Curr Opin Plant Biol. 2013;16:569–574. doi: 10.1016/j.pbi.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Haberlandt G. 1921. Wundhormone als Erreger von Zellteilungen. Beiträge zur allgemeinen Botanik, Band 2, Ausgabe 1.
- Halitschke R, Baldwin IT. Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 2003;36:794–807. doi: 10.1046/j.1365-313x.2003.01921.x. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid–amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YH, Fukushige H, Hildebrand DF, Gan SS. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002;128:876–884. doi: 10.1104/pp.010843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen C, Baldwin IT, Wu J. Nicotiana attenuata MPK4 suppresses a novel jasmonic acid (JA) signaling-independent defense pathway against the specialist insect Manduca sexta, but is not required for the resistance to the generalist Spodoptera littoralis. New Phytol. 2013;199:787–799. doi: 10.1111/nph.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyl A, Brault M, Frugier F, Kuderova A, Lindner AC, Motyka V, Rashotte AM, Schwartzenberg KV, Vankova R, Schaller GE. Nomenclature for members of the two-component signaling pathway of plants. Plant Physiol. 2013;161:1063–1065. doi: 10.1104/pp.112.213207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D, Iqbal J, Lehmann K, Gase K, Saluz HP, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (lepidoptera, sphingidae) and its natural host Nicotiana attenuata: V. microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol. 2003;131:1877–1893. doi: 10.1104/pp.102.018176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Ann Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Nakanishi H, Mizuno T. Classification of the genes involved in the two-component system of the moss Physcomitrella patens. Biosci Biotechnol Biochem. 2010;74:2542–2545. doi: 10.1271/bbb.100623. [DOI] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, Kang NY, Novak O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, Kim J. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem. 2010;285:23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kallenbach M, Alagna F, Baldwin IT, Bonaventure G. Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol. 2010;152:96–106. doi: 10.1104/pp.109.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallenbach M, Bonaventure G, Gilardoni PA, Wissgott A, Baldwin IT. Empoasca leafhoppers attack wild tobacco plants in a jasmonate-dependent manner and identify jasmonate mutants in natural populations. Proc Natl Acad Sci USA. 2012;109:E1548–E1557. doi: 10.1073/pnas.1200363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Nakano T, Taki N, Ishikawa M, Asami T, Yoshida S. Cytokinin-induced gene expression in cultured green cells of Nicotiana tabacum identified by fluorescent differential display. Biosci Biotechnol Biochem. 2001;65:1275–1283. doi: 10.1271/bbb.65.1275. [DOI] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, Ashikari M, Ueguchi-Tanaka M, Matsuoka M, Suzuki K, Sakakibara H. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krügel T, Lim M, Gase K, Halitschke R, Baldwin IT. Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology. 2002;12:177–183. [Google Scholar]
- Kudo T, Kiba T, Sakakibara H. Metabolism and long-distance translocation of cytokinins. J Integr Plant Biol. 2010;52:53–60. doi: 10.1111/j.1744-7909.2010.00898.x. [DOI] [PubMed] [Google Scholar]
- Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, Sakakibara H, Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomin SN, Krivosheev DM, Steklov MY, Osolodkin DI, Romanov GA. Receptor properties and features of cytokinin signaling. Acta Nat. 2012;4:31–45. [PMC free article] [PubMed] [Google Scholar]
- Machado RA, Ferrieri AP, Robert CA, Glauser G, Kallenbach M, Baldwin IT, Erb M. Leaf-herbivore attack reduces carbon reserves and regrowth from the roots via jasmonate and auxin signaling. New Phytol. 2013;200:1234–1246. doi: 10.1111/nph.12438. [DOI] [PubMed] [Google Scholar]
- Meldau S, Baldwin IT, Wu J. SGT1 regulates wounding-and herbivory-induced jasmonic acid accumulation and Nicotiana attenuata's resistance to the specialist lepidopteran herbivore Manduca sexta. New Phytol. 2011;189:1143–1156. doi: 10.1111/j.1469-8137.2010.03558.x. [DOI] [PubMed] [Google Scholar]
- Miller CO. Kinetin and related compounds in plant growth. Annu Rev Plant Physiol. 1961;12:395–408. [Google Scholar]
- Miller CO, Skoog F, Von Saltza MH, Strong FM. Kinetin, a cell division factor from deoxyribonucleic acid. J Am Chem Soc. 1955;77:1392–1392. [Google Scholar]
- Mitchell JJ, van Staden J. Cytokinins and the wounding response in potato tissue. Z Pflanzenphysiol. 1983;109:1–5. [Google Scholar]
- Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabata S, Sandberg G, Kakimoto T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc Natl Acad Sci USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DW, Mok MC. Cytokinin metabolism and action. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Motyka V, Faiss M, Strand M, Kaminek M, Schmülling T. Changes in cytokinin content and cytokinin oxidase activity in response to derepression of ipt gene transcription in transgenic tobacco calli and plants. Plant Physiol. 1996;112:1035–1043. doi: 10.1104/pp.112.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseem M, Philippi N, Hussain A, Wangorsch G, Ahmed N, Dandekar T. Integrated systems view on networking by hormones in Arabidopsis immunity reveals multiple crosstalk for cytokinin. Plant Cell. 2012;24:1793–1814. doi: 10.1105/tpc.112.098335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. The new mutation theory of phenotypic evolution. Proc Natl Acad Sci USA. 2007;104:12235–12242. doi: 10.1073/pnas.0703349104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, Sakakibara H, Schmülling T, Tran LSP. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23:2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Bömer M, Takahashi N, Ishida T, Tsui TL, Balbi V, Shanahan H, Sugimoto K, Devoto A. Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 2013;161:1930–1951. doi: 10.1104/pp.113.214908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onkokesung N, Gaquerel E, Kotkar H, Kaur H, Baldwin IT, Galis I. MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme A: Polyamine transferases in Nicotiana attenuata. Plant Physiol. 2012;158:389–407. doi: 10.1104/pp.111.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. Co(i)-ordinating defenses: NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant J. 2007;51:79–91. doi: 10.1111/j.1365-313X.2007.03119.x. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Perez AC, Chico JM, Bossche RV, Sewell J, Gil E, Garcia-Casado G, Witters E, Inze D, Long JA, De Jaeger G, Solano R, Goossens A. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils B, Heyl A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009;151:782–791. doi: 10.1104/pp.109.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam RS, Swarbrick PJ, Scholes JD, Rolfe SA. Imaging photosynthesis in wounded leaves of Arabidopsis thaliana. J Exp Bot. 2006;57:55–69. doi: 10.1093/jxb/erj039. [DOI] [PubMed] [Google Scholar]
- Richmond AE, Lang A. Effect of kinetin on protein content and survival of detached Xanthium leaves. Science. 1957;125:650–651. [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. Nitrogen economics of root foraging: Transitive closure of the nitrate–cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA. 2011;108:18524–18529. doi: 10.1073/pnas.1108684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinins: Activity, biosynthesis, and translocation. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Sano H, Seo S, Koizumi N, Niki T, Iwamura H, Ohashi Y. Regulation by cytokinins of endogenous levels of jasmonic and salicylic acids in mechanically wounded tobacco plants. Plant Cell Physiol. 1996;37:762–769. [Google Scholar]
- Schäfer M, Brütting C, Gase K, Reichelt M, Baldwin I, Meldau S. #x201C;Real time” genetic manipulation: A new tool for ecological field studies. Plant J. 2013;76:506–518. doi: 10.1111/tpj.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Fischer C, Meldau S, Seebald E, Oelmüller R, Baldwin IT. Lipase activity in insect oral secretions mediates defense responses in Arabidopsis. Plant Physiol. 2011;156:1520–1534. doi: 10.1104/pp.111.173567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer S, Schmülling T. The CRK1 receptor-like kinase gene of tobacco is negatively regulated by cytokinin. Plant Mol Biol. 2002;50:155–166. doi: 10.1023/a:1016087908746. [DOI] [PubMed] [Google Scholar]
- Schmelz EA, Engelberth J, Alborn HT, Tumlinson JH, Teal PEA. Phytohormone-based activity mapping of insect herbivore-produced elicitors. Proc Natl Acad Sci USA. 2009;106:653–657. doi: 10.1073/pnas.0811861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T, Werner T, Riefler M, Krupkova E, Bartrina y Manns I. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res. 2003;116:241–252. doi: 10.1007/s10265-003-0096-4. [DOI] [PubMed] [Google Scholar]
- Shi XL, Rashotte AM. Advances in upstream players of cytokinin phosphorelay: Receptors and histidine phosphotransfer proteins. Plant Cell Rep. 2012;31:789–799. doi: 10.1007/s00299-012-1229-9. [DOI] [PubMed] [Google Scholar]
- Smigocki A, Heu S, Buta G. Analysis of insecticidal activity in transgenic plants carrying the ipt plant growth hormone gene. Acta Physiol Plant. 2000;22:295–299. [Google Scholar]
- Sobek EA, Munkvold GP. European corn borer (Lepidoptera: Pyralidae) larvae as vectors of Fusarium moniliforme, causing kernel rot and symptomless infection of maize kernels. J Econ Entomol. 1999;92:503–509. [Google Scholar]
- Stitz M, Baldwin IT, Gaquerel E. Diverting the flux of the JA pathway in Nicotiana attenuata compromises the plant's defense metabolism and fitness in nature and glasshouse. PLoS ONE. 2011;6:e25925. doi: 10.1371/journal.pone.0025925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Riefler M, Lomin SN, Achazi K, Romanov GA, Schmülling T. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011;67:157–168. doi: 10.1111/j.1365-313X.2011.04584.x. [DOI] [PubMed] [Google Scholar]
- Takei K, Sakakibara H, Taniguchi M, Sugiyama T. Nitrogen-dependent accumulation of cytokinins in root and the translocation to leaf: Implication of cytokinin species that induces geneexpression of maize response regulator. Plant Cell Physiol. 2001;42:85–93. doi: 10.1093/pcp/pce009. [DOI] [PubMed] [Google Scholar]
- Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-Zeatin. J Biol Chem. 2004;279:41866–41872. doi: 10.1074/jbc.M406337200. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, Schaller GE, Kieber JJ. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012;158:1666–1684. doi: 10.1104/pp.111.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlings TCJ, McCall PJ, Alborn HT, Tumlinson JH. An elicitor in caterpillar oral secretions that induces corn seedlings to emit chemical signals attractive to parasitic wasps. J Chem Ecol. 1993;19:411–425. doi: 10.1007/BF00994314. [DOI] [PubMed] [Google Scholar]
- Ueda J, Kato J. Isolation and identification of a senescence-promoting substance from wormwood (Artemisia absinthium L.) Plant Physiol. 1980;66:246–249. doi: 10.1104/pp.66.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell. 2008;20:1101–1117. doi: 10.1105/tpc.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Wounding and herbivory regulate the expression of LOG-family genes
Figure S2. Wounding and herbivory regulate the expression of IPT-family genes
Figure S3. Wounding and herbivory regulate the expression of cytokinin receptor genes
Figure S4. Wounding and herbivory regulate the expression of HPT-family genes
Figure S5. Wounding and herbivory regulate the expression of type-A RR-family genes
Figure S6. Wounding and herbivory regulate the expression of type-B RR-family genes
Figure S7. Wounding and herbivory regulate the expression of CKX-family genes
Figure S8. Wounding and herbivory regulate the expression of ZOG-family genes
Figure S9. Confirmation of microarray expression data
Figure S10. Confirmation of NaIPT5 microarray data
Figure S11. Cytokinin-spraying changes NaIPT5, NaRRA5, and NaCKX5 transcript accumulation
Figure S12. Wounding and herbivory-induced changes in cytokinin levels
Figure S13. Wounding and herbivory regulate transcript accumulation of cytokinin-related genes in the root
Figure S14. Wounding and herbivory regulate transcript accumulation of cytokinin-related genes in systemic leaves
Figure S15. Trans-zeatin riboside levels are reduced in jasmonic acid pathway impaired transgenic plants
Table S1. Abbreviations
Table S2. Sequences of primers used for qPCR
Table S3. Multi-reaction-monitoring settings for cytokinin quantification in positive ionization mode


