Abstract
Cone snails, genus Conus, are predatory marine snails that use venom to capture their prey. This venom contains a diverse array of peptide toxins, known as conotoxins, which undergo a diverse set of posttranslational modifications. Amidating enzymes modify peptides and proteins containing a C-terminal glycine residue, resulting in loss of the glycine residue and amidation of the preceding residue. A significant fraction of peptides present in the venom of cone snails contain C-terminal amidated residues, which are important for optimizing biological activity. This study describes the characterization of the amidating enzyme, peptidylglycine α-amidating monooxygenase (PAM), present in the venom duct of cone snails, Conus bullatus and Conus geographus.
PAM is known to carry out two functions, peptidyl α-hydroxylating monooxygenase (PHM) and peptidylamido-glycolate lyase (PAL). In some animals, such as Drosophila melanogaster, these two functions are present in separate polypeptides, working as individual enzymes. In other animals, such as mammals and in Aplysia californica, PAM activity resides in a single, bifunctional polypeptide. Using specific oligonucleotide primers and reverse transcription-polymerase chain reaction we have identified and cloned from the venom duct cDNA library, a cDNA with 49% homology to PAM from A. californica. We have determined that both the PHM and PAL activities are encoded in one mRNA polynucleotide in both C. bullatus and C. geographus. We have directly demonstrated enzymatic activity catalyzing the conversion of dansyl-YVG-COOH to dansyl-YV-NH2 in cloned cDNA expressed in Drosophila S2 cells.
Keywords: Posttranslational modification, Conotoxins, Peptidylglycine α-amidating, monooxygenase
1. Introduction
Cone snails (genus Conus) comprise ~700 species of venomous predatory mollusks (Olivera, 2006). They synthesize a wide array of short peptides (variously called, conotoxins or conopeptides) that target ion channels (both ligand and voltage gated), receptors and transporters to immobilize prey or for defense. An individual cone snail species synthesizes anywhere from 50 to 200 unique conotoxins (Olivera, 1997). However, using liquid chromatography and electrospray ionization spectroscopy for venom analysis, Davis et al. estimated the number of peptides to be greater than one thousand (Davis et al., 2009), a large number of which arises most likely from variable peptide processing of a primary translation product (Dutertre et al., 2013). With an estimated more than 700 different cone snail species, conotoxins provide a large repertoire of natural compounds of pharmaceutical interest. The exquisite specificity of these molecules have made them highly desirable for the functional dissection of ion channel components and composition in the nervous system in addition to providing lead compounds for drug discovery.
The venom duct is a highly specialized tissue that has evolved to carryout a large number of biosynthetic reactions dedicated to the production of a lethal ‘cocktail’ of short peptides. A feature of these peptides is that they contain posttranslational modifications that are essential for their function. Posttranslational modifications are found in the majority of conotoxins and these enhance biological activity (O. Buczek et al., 2005; AG. Craig et al.,1999). To enumerate a few, posttranslational modifications include proteolytic cleavage, disulfide bond formation, gamma-carboxylation of glutamate, hydroxylation and isomerization of proline, bromination of tryptophan, epimerization of aminoacids. The substrates for the modifications are the immature conopeptides. The primary translation product of the conotoxin mRNA include a signal sequence for peptide secretion and propeptide both of which are removed from the mature peptide. The role of the propeptide has not been elucidated except in the case of gamma-carboxylation in which it has been shown to enhance the efficiency of carboxylation (Bandyopadhyay et al.,1998). While the enzymes involved in proteolytic processing, disulfide bond formation, gamma-carboxylation of glutamate (Bandyopadhyay et al., 2002; Czerwiec et al., 2002), isomerization of proline has been characterized in conus venom duct the enzymes for the rather infrequent modifications, bromination of tryptophan and epimerization of amino acids remain to be identified. In order to understand the biosynthesis of active venom components we have undertaken to isolate the enzymes involved in the posttranslational modification of the conopeptides. Given the large cargo of structurally diverse secreted modified peptides processed in the venom duct, we anticipate the presence of multiple isoforms of enzymes involved in the posttranslational modifications that have coevolved for efficient synthesis of the mature toxins. One of the most common types of posttranslational modification many conotoxins undergo is C-terminal amidation. Table 1 shows several conotoxins that are posttranslationally amidated.
Table 1.
Examples of amidated conotoxins. Conceptual translation product of cDNA encoding conotoxins and the sequences of the corresponding mature conotoxins containing posttranslational modifications in addition to amidation. (γ = gamma-carboxy glutamate; O = 4-hydroxy proline; w = D-tryptophan).
| Conotoxin | Primary translation product | Mature conotoxin |
|---|---|---|
| GI | MFTVFLLVVLATTVVSFPSERASDGRDDTAKDEGSDMEKLV EKKECCNPACGRHYSCGR |
ECCNPACGRHYSC(NH2)a |
| AuIB | MFTVFLLVVLATTVVSFTSDRASDGRKDAASGLIALTMKGCCSYPPCFATNPDCGRRR | GCCSYPPCFATNPDC(NH2)b |
| SrIA | MGMRMMFTVFLLVVLATTVVSFTSDSAFDSRNVAANDKVSD MIALTARRTCCSRPTCRMEYPELCGGRR |
RTCCSROTCRMγYPγLCG(NH2)c |
| GIIIA | MSKLGVLLTICLLLFPLTALPMDGDEPANRPVERMQDNISS EQYPLFEKRRDCCTPPKKCKDRQCKPQRCCAGR |
RDCCTOOKKCKDRQCKOQRCCA(NH2)d |
| Conantokin G | MHLYTYLYLLVPLVTFHLILGTGTLDDGGALTERRSADATA LKAEPVLLQKSAARSTDDNGKDRLTQMKRILKQRGNKAR GEEEVQENQELIREKSNGKR |
GEγγVQγNQγLIRγKSN(NH2)e |
| BuIIIA | MMSKLGVLLTICLLLFPLFALPQDGDQPADRPAERMQDDISSEQNSLLEKR VTDRCCKGKRECGRWCRDHSRCCGRR |
VTDRCCKGKRECGRWCRDHSRCC(NH2)f |
| Contryphan M | MGKLTILVLVAAVLLSTQVMVQGDRDQPADRNAVPRDDNPGRARRKRMKVL NESECPWHPWCG |
NγSγCPwHPWC(NH2)g |
| Gm5.2 | MRCLPVFVILLLLIASAPSVDAQPKTKDDVPLAPLHDNIRS TLQTLRKKVCCRPVQDCCSGK |
VCCRPVQDCCS(NH2)h |
| TxIXA | MHLSLARSAVLMLLLLFALGNFVVVQSGQITRDVDNGQLTDNRRNLQSKWKPVSLYMSRR GCNNSCQEHSDCESHCICTFRGCGAVNG |
GCNNSCQγHSDCγSHCICTFRGCGAVN(NH2)i |
| TxX | MSGHTSVSFLLLSIVALGMVATVIC SCDSEFSSEFCERPEESCSCSTHTCCHWARRDQCMKPQRCISAQKGN GRRRLIHMQ |
SCDSγFSSγFCγRPγγSCSCSTHTCCHWARRDQCMKPQRCISAQKGN(NH2)j |
The posttranslational modification of the C-terminus into an amide is catalyzed by the amidating, copper requiring enzyme peptidylglycine α-amidating monooxygenase (PAM). It performs two distinct functions, as shown in Fig. 1. The first function, peptidyl α-hydroxylating monooxygenase (PHM) hydroxylates glycine at the C-terminal end of a given peptide (Eipper et al., 1992; Prigge et al., 2000). The second function, peptidylamido-glycolate lyase (PAL), then cleaves the hydroxylated glycine (Eipper et al., 1992; Prigge et al., 2000). This results in a C-terminally amidated peptide with glyoxylate as a byproduct. These two functions have been found in separate genes in sea anemone (Hauser et al., 1997), in arthropods (Drosophila melanogaster, Apis mellifera, (Kolhekar et al.,1997c; Zabriskie et al.,1994), whereas single genes encoding both functions have been identified in pla-cozoa (Trichoplax adhaerens Accession no. XP_002110995), a member of the phylum cnidaria, (Acropora millepora nematoda, (Attenborough et al., 2012), molluscs (Aplysia californica, Lymnaea stagnalis, (Fan et al., 2000; Spijker et al., 1999) platyhelminthes (Schistosoma mansoni (Mair et al., 2004), echinoderms (Strongylocentrotus purpuratus Accession no. XP_784943), urochordates (Ciona intestinalis Accession no. XP_002130721) and vertebrates, (Bos taurus, Rattus norvegicus, Xenopus laevis, (Eipper et al.,1987; Mizuno et al., 1987; Ohsuye et al., 1988). From a phylogenetic analyses of PAM sequences (Attenborough et al., 2012). Attenborough et al., suggest that bifunctional PAMs predate the evolution of the nervous and endocrine systems and the ancestral function may have been to amidate epitheliopeptides.
Fig. 1.
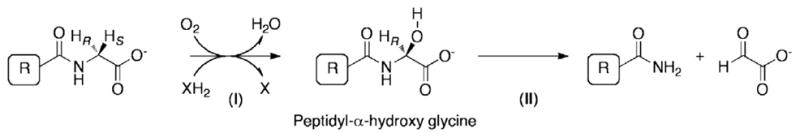
Schematic of reactions carried by PAM. Step I: PHM function hydroxylates glycine in the presence of oxygen, copper and ascorbate. Step II: PAL function cleaves the hydroxyglycine residue yielding an amidated product and glyoxylate.
C-terminal amidation has been reported to improve the stability of peptide ligands, and to increase their affinity for their receptor targets, thereby enhancing biological activity (Merkler, 1994). Eukaryotic peptide hormones and neuropeptides such as oxytocin, vasopressin, thyrotropin-releasing hormone and neuropeptide Y are amidated at the carboxy terminus. Neuropeptides (RYIRF-NH2, GYIRF-NH2, and YIRF-NH2) that are potentially excitatory in schistosome muscles identified in flat-worms require the C-terminal amidated phenylalanine for activity (Day et al., 1994, 1997). Inactivation of the PAM gene is lethal in Drosophila and mouse. PAM(±) heterozygous mice are viable and fertile, however, they exhibit enhanced anxiety like behavior and cannot maintain body temperature. Drosophila mutant nemy and knockdown of PHM activity was associated with deficits in memory retention. nemy codes for the Drosophila homolog of cytochrome B561, a transmembrane protein present in synaptic vesicles and large dense core vesicles. It uses extravesicular ascorbate to reduce intravesicular semi-dehydroascorbate to ascorbate that acts as a cofactor for DbH and PHM activity (Iliadiadia et al., 2008; Kent and Fleming, 1987). PAM also affects gene expression by mediating information transfer between secretory granules and the nucleus. A cytosolic fragment derived from a cleavage in the transmembrane region of PAM is transported into the nucleus where it has been shown to affect the level of transcripts encoding aquaporin I (Aqp I) and secretory leukocyte trypsin inhibitor (Slpi). AqpI plays an essential role in the biogenesis of secretory granules. Reduction in the levels of Slpi which inhibits elastase, cathepsin G, trypsin and chymotrypsin is likely to alter the processing of proteins and peptides in the cells (Yin et al., 2011).
PAM is localized in the trans-golgi network and secretory vesicles in which the neuropeptides are processed (Mair et al., 2004; Milgram et al., 1997; Oyarce and Eipper, 2000). Integral membrane PAM accumulates in the peri-nuclear region overlapping the Golgi complex and trans-Golgi network (TGN). The targeting is dependent on the information of the carboxyterminal cytoplasmic tail. However, when PHM and PAL are independently expressed as soluble proteins they are efficiently targeted to dense core vesicles in AtT-20 cells. Immunoelectronmicroscopy reveals that the membrane PAM is present in tubulovesicular structures that make up the TGN (Iliadiadia et al., 2008; Milgram et al., 1997).
The different isoforms of PAM present in cells arise mainly from alternative splicing and endoproteolytic cleavage of the primary translation product. In one of the transcripts encoding PAM in rats and humans the PHM and PAL functions are separated by an exon (exon A) that contain two adjacent basic amino acids. After endoproteolytic cleavage at this site by tissue specific protease a soluble form of PHM is generated. In addition, cleavage at the membrane anchor site results in soluble PAM or PAL. The tetraploid, X. laevis has two different copies of PAM gene. Unlike rats and humans the PHM and PAL functions in Xenopus, C. elegans, A. californica are not separated by an exon susceptible to endoproteolytic cleavage. In L. stagnalis PAM contain four tandem copies encoding PHM function separated from a single copy encoding PAL by exon A (Prigge et al., 2000; Spijker et al., 1999).
In this report we describe the cDNA structure of peptidylglycine α-amidating monooxygenase expressed in the venom ducts of two marine snails in the genus Conus, Conus bullatus and Conus geographus and the enzymatic activity encoded by the cDNA from C. bullatus.
2. Materials and methods
2.1. Cloning PAM from C. bullatus venom duct
Specific primers were synthesized based on the partial sequence of the amidating enzyme identified from the C. bullatus transcriptome sequences (Hu et al., 2011). cDNA for 5′ and 3′ RACE experiments were synthesized from total RNA isolated from the venom duct of C. bullatus, using SMARTer™ RACE cDNA amplification kit (Clontech) according to the manufacturer’s instructions. 5′ and 3′ gene fragments were assembled to obtain the nucleotide sequences encoding the complete PAM enzyme ascertained by comparison to the known PAM sequence of A. californica. Oligonucleotide primers corresponding to the 5′ and 3′ UTR sequences of the assembled PAM gene, were used to PCR amplify the complete PAM gene from C. bullatus cDNA.
2.2. Cloning PAM from C. geographus venom duct
Partial sequence of PAM was identified in the transcriptome sequence (Hu et al., 2012) of the venom duct of C. geographus. Using specific primers and 5′ and 3′ RACE experiments we assembled the complete sequence of PAM as described above for C. bullatus.
2.3. PAM expression in S2 cells
C. bullatus PAM gene sequences were inserted into the vector pRmHa-3 (Bunch et al., 1988) for expression in Drosophila S2 cells (Walker et al., 2001). In this construct expression of PAM is under the control of the inducible metallothionein promoter. Plasmid DNA encoding PAM along with pCoHygro (20:1) was used to cotransfect Drosophila S2 cells using Cellfectin® reagent (Invitrogen life technologies) according to the vendor’s instructions. Cells were selected for hygromycin resistance by growing the transfected cells in Schneider’s media with 10% FBS in the presence 700 μg/mL of hygromycin. A control cell line resistant to hygromycin was also established from Drosophila S2 cells transfected with only pCoHygro. The expression of PAM was induced by growing cells in the presence of 0.7 mM CuSO4. Cells were harvested three days after induction.
Cells were suspended in lysis buffer (1 mM pepstatin, 1 mM lima bean trypsin inhibitor, 1 mM benzamide HCl, 1 mM PMSF freshly dissolved in isopropanol, 20 mM NaTES [N-tris (hydroxymethyl) methyl-2-aminoethanesulfonic acid, sodium salt] pH 7.0, 10 mM D-Mannitol, 1% Triton X100) (Kolhekar et al., 1997b). Suspended cells were lysed by freezing and thawing 3× in a dry ice and ethanol bath, followed by sonication. The lysates were then centrifuged at 2000 rpm for 15 min at 4 °C. The supernatant was subsequently centrifuged at 100,000 g for one hour at 4 °C. PAM activity was recovered from the pellet. The pellet was suspended in lysis buffer and mildly sonicated. This was used as source of enzyme for subsequent studies.
2.4. PAM activity
Lysates were used in a PAM enzyme assay (600 μL of reaction buffer contains 50 μL cell lysate – roughly 3.13 × 106 cells, 50 mM NaTES pH 6.5, 0.01% Tween 20, 2.5 mM freshly made L-ascorbic Acid, 2 μM CuSO4, and 150 μM dansyl-YVG substrate (GenScript, NJ, USA) and 100 μg/mL catalase) to determine enzymatic activity according to methods of Suzuki et al. (1990) and Jones et al. (1988). The reaction mixture was incubated at 37 °C. Aliquots were removed at times 0, 0.17, 0.67,1, 3, 6, 9, 12,15, 18, 21, and 24 h and the reaction was quenched by adding EDTA to a final concentration of 50 mM. Each aliquot was spun through a Amicon® Ultra (Ultracel®-3K membrane) (Millipore) filter for 30 min at 13,000 rpm at room temperature before being stored at −80 °C. This removes proteins and other particulate matter from the reaction before the analysis.
The conversion of the substrate (dansyl-YVG) to the product (danyl-YV-NH2) was monitored by Reversed-Phase High Performance Liquid Chromatography (HPLC) using a Vydac C18 (5 μm, 250 mm × 4.6 mm) (Grace Davison Discovery Sciences™) analytical column. The progress of the reaction was monitored over a linear gradient ranging from 22% to 25% of solvent B90 in 20 min. The HPLC solvents were: 0.08% Trifluoroacetic Acid (TFA) in water (solvent A) and 0.08% TFA (v/v) in 90% aqueous acetonitrile (ACN) (solvent B). The C18 column was used at 22 °C with a flow rate of 1 mL/min. The area under the peaks were used to quantify amounts of substrate and product over time. Chromatographs were recorded at 220 nm. Peaks were collected and analyzed by Electrospray Ionization Mass Spectrometry (ESIMS) to identify components of the reaction mixture.
The percentage of substrate converted to amidated product was normalized to the protein concentrations of lysates. Bovine Serum Albumin (BSA) was used as a standard. Protein concentration was determined using Bio-Rad DC Protein Assay kit and protocol (BIO-RAD Laboratories, CA, USA) using bovine serum albumin as a standard.
Product formation was also monitored at five different concentrations (16.7 μM, 50 μM, 150 μM, 450 μM, and 1.35 mM) of the substrate, dansyl-YVG, using cell lysates from PAM isoforms 1 and 2 as the source of enzyme. Product formation was determined after 21 h of incubation.
3. Results
3.1. PAM from C. bullatus
We have assembled and cloned two isoforms of PAM from C. bullatus (designated here as Bullatus 1 and Bullatus 2). Fig. 2 shows the alignment of amino acids and nucleotides for the two clones. While Bullatus 1 is predicted to be 721 aa long, Bullatus 2 is 747 aa. Fig. 2 shows that the amino acid sequences are almost completely conserved with differences mainly at the carboxy terminus (19 aa differences out of 721 aa; 2.6% divergence). The major differences in the nucleotide sequences are also at the 3′ end of the cDNA (Supplementary data-Fig S1). Beginning with nt 2298 in Bullatus 2 (Fig S1) the dinucleotide CA is repeated 22 times, however, there is a deletion of 15nt in Bullatus 1 in this region. In addition, in Bullatus 1 there is a duplication of a tetranucleotide TGTT at nt 2372. One copy of the tetranucleotide is also deleted from Bullatus 2 resulting in a change in the reading frame leading to an open reading frame of 747 aa in contrast to 721 aa observed for Bullatus 1.
Fig. 2.
Comparison of the amino acid sequences of Conus bullatus 1 and 2, and Conus geographus 1.
Analysis of the hydrophobicity profile of both Bullatus 1 and Bullatus 2 suggests the presence of a hydrophobic patch of amino acids at the C-terminus that may serve to anchor the proteins to membranes.
Pairs of dibasic amino acid residues, RK at positions 434 and 435 and KK, at positions 623 and 624 are potential sites for endoproteolytic cleavages for producing soluble mono-and bi-functional enzymes respectively.
3.2. PAM from C. geographus
We have assembled three different species of PAM cDNA from C. geographus. However, comparison of the amino acid sequence with that obtained from C. bullatus suggested that only one of them, Geographus 1, contain the complete sequence of the amidating enzyme. The other two sequences (Geographus 2, and 3) are truncated and preclude complete enzymatic activity. Geographus 2 is 610 amino acids, and Geographus 3, 602 amino acids compared to Geographus 1 and Bullatus 1 that are 769 and 721 amino acids respectively. Comparison of the nucleotide and amino acid sequences of Bullatus 1 and 2 and Geographus 1 are shown in Fig. 2 and S1 respectively. The protein sequence was analyzed for the presence of signal sequence using SignalP 4.0 (Petersen et al., 2011). Signal sequence cleavage site is predicted between amino acids 20 and 21 (SAS-DA).
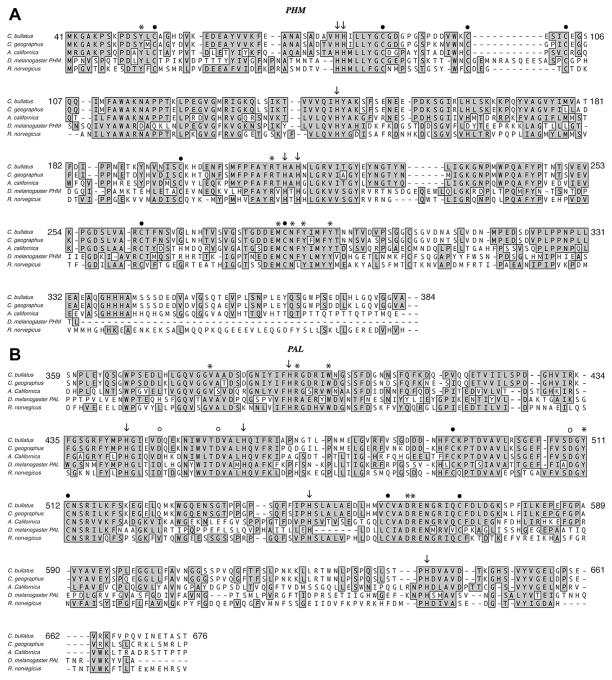
The catalytic domains of PHM and PAL for the amidating enzyme encoded by C. bullatus and C. geographus are highly homologous to the corresponding domains identified in enzymes from other sources. Fig. 3A and B shows the alignment of the amino acids of PHMcc and PALcc catalytic domains in PAM sequences from A. californica, C. bullatus (Bullatus 1), C. geographus (Geographus 1) and R. norvegicus and monofunctional PHM and PAL enzymes from D. melanogaster (Kolhekar et al., 1997c) (Fan et al., 2000; Han et al., 2004). Table 2 shows the percent identity and similarity among the proteins from the different sources. The PHM encoding segment of the enzyme consists of two domains, each containing a copper residue. From the multiple sequence alignment it is apparent that the conserved residues H79, H80 and H144 correspond to the copper coordination residues determined by X-ray crystallography for domain I, CuA, so also are the residues, H213, H215 and M284 in domain II which interact with CuB. By site directed mutagenesis Yonekura et al. identified these histidine residues as being essential for enzymatic activity of rat PHM (H. Yonekura, 1996). These residues are conserved in PHM identified from different sources. Pairs of conserved cysteine residues (C54, C99; C86, C103; C197, C304; and C263, C285) correspond to the cysteine residues involved in the four disulfide linkages identified in the catalytic domain of rat PHM (Eipper et al., 1995; Kolhekar et al., 1997a; Prigge et al., 1997). By homology to the rat enzyme the conserved residues Y52, R211, N286, Y288, and Y292, are expected to be involved in some aspect of substrate binding.
Fig. 3.
A. Multiple sequence alignment of PHM sequences from C. bullatus (Bullatus 1) (AE 184584.1), C. geographus (Geographus 1), A. californica (AF14027.1), D. melanogaster (NP_477225), R. norvegicus (NP_037132). (●) indicates cysteine residues involved in disulfide linkages, (↓) indicates histidine residues important for coordination of copper, (*) indicates residues important for substrate binding. B. Multiple sequence alignment of PAL sequences from C. bullatus (Bullatus 1) (AE 184584.1), C. geographus (Geographus 1), A. californica (AF14027.1), D. melanogaster (AAF47043.2), R. norvegicus (NP_037132). (●) indicates cysteine residues involved in disulfide linkages, (↓) shows H444, H547, H641 important for coordination of zinc, H393 and H462 and (°) D449, D458, and D509 serve structural roles, and (*) indicates residues Y511, R563, D562, and R394 important for catalysis.
Table 2.
Comparison of amino acid identity and similarity of PHM and PAL domains of C. bullatus (Bullatus 1), C. geographus (Geographus 1), A. californica, D. melanogaster and R. norvegicus.
|
C. bullatus
|
A. californica
|
D. melanogaster
|
R. norvegicus
|
|||||
|---|---|---|---|---|---|---|---|---|
| Percent identity | Percent similarity | Percent identity | Percent similarity | Percent identity | Percent similarity | Percent identity | Percent similarity | |
| PHMCC | ||||||||
| Bullatus 1 | 100 | – | 56.2 | 75.4 | 29.2 | 46 | 36 | 55.4 |
| Geographus 1 | 89.4 | 94.1 | 55.1 | 74.6 | 29.9 | 47.8 | 33.7 | 59.3 |
| PALCC | ||||||||
| Bullatus 1 | 100 | – | 54.4 | 65.1 | 28.3 | 36.2 | 35.2 | 44.3 |
| Geographus 1 | 88.7 | 89.3 | 42.8 | 53.5 | 35.6 | 43.4 | 36.2 | 45.6 |
Multiple sequence alignment of the PAL domain is shown in Fig. 3B. The rat PAL catalytic core (PALcc) has been mapped and extends from aa 498 to aa 820 (Kolhekar et al., 2002). The PAL domain for Conus has been inferred from sequences homologous to the rat PALcc. While both Bullatus 1 and 2 and Geographus 1 include sequences that are homologous to drosophila PAL and expected to encode PAL activity, Geographus 2 and 3 are truncated. In the absence of experimental determinations we cannot infer if Geographus 2 and 3 also exhibit PAL activity. Multiple sequence alignment and comparison to drosophila PAL (Fig. 3B) suggests that PAM activity in Conus is contained within the amino terminal 664 amino acids. In this region the amino acid sequences of PAM from C. bullatus (Bullatus 1) and C. geographus (Geographus 1) vary by 8% (53 out of 664 amino acids). Structural and functional roles of different amino acids in rat PALcc have been determined. These residues are conserved in the enzymes from C. bullatus and C. geographus. These include the four cysteine residues involved in disulfide formation, (Cys 492, 512, 559 and 570), histidine residues that coordinate a zinc ion at the active site (H444, H547, H641), or have structural roles (H393, H462) and other residues having catalytic roles (Y511, R563, D562, R394), structural roles (D449, D458, and D509) (Attenborough et al., 2012; Chufan et al., 2009; De et al., 2006; Kolhekar et al., 2002).
3.3. Enzymatic activity of PAM encoded by C. bullatus
We examined the ability of cell lysates from Drosophila S2 cells carrying the C. bullatus PAM genes to catalyze the conversion of dansyl-YVG(COOH) to dansyl-YV-NH2. Fig. 4 shows the HPLC chromatogram for the analysis of the reaction products. Peaks 1, 2, and 3 were analyzed by ESIMS. Cells carrying Bullatus 2 have poor activity, however, both the product and intermediate can be observed in contrast to Bullatus 1 in which the amidated product is the only species that can be identified. Based on the predicted masses peak1 corresponds to the substrate (dansyl-YVG), peak 2 the intermediate (dansyl-YVG(OH)) and peak 3 (dansyl-YV-NH2). The material from peak 3 also coelutes with synthetically prepared dansyl-YV-NH2. In the control cell lysates peaks 2 and 3 are absent. The enzymatic activity is localized in the 100,000 g pellet of the cell lysate.
Fig. 4.
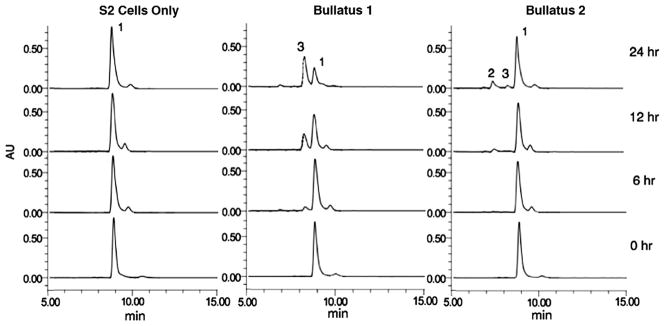
HPLC chromatogram of amidation activity (monitored at 220 nm with dansyl-YVG as substrate) carried out by lysates of control cell lines (S2 cells with empty vector) compared to activity of S2 cells expressing Bullatus 1 and Bullatus 2 isoforms at time intervals 0, 6, 12, and 24 h. Peaks 1, 2 and 3 represent the substrate, hydroxyglycine intermediate and the amidated product respectively.
Fig. 5 shows the time course of the reaction carried out by cell lysates expressing Bullatus 1 and Bullatus 2. In the case of Bullatus 1 the majority of the substrate is converted to the final amidated product. Fig. 6 shows the accumulation of product as a function of substrate concentration.
Fig. 5.
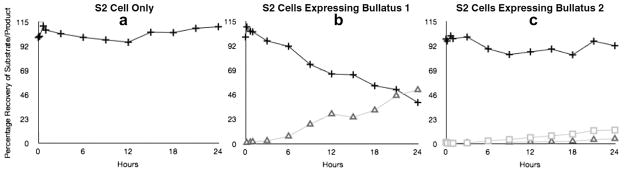
Time course of amidation reaction by Bullatus 1 and Bullatus 2. (a) Control S2 cell lysate; (b) S2 cells expressing Bullatus 1; (c) S2 cells expressing Bullatus 2. (+ substrate; □ intermediate; △ Product).
Fig. 6.
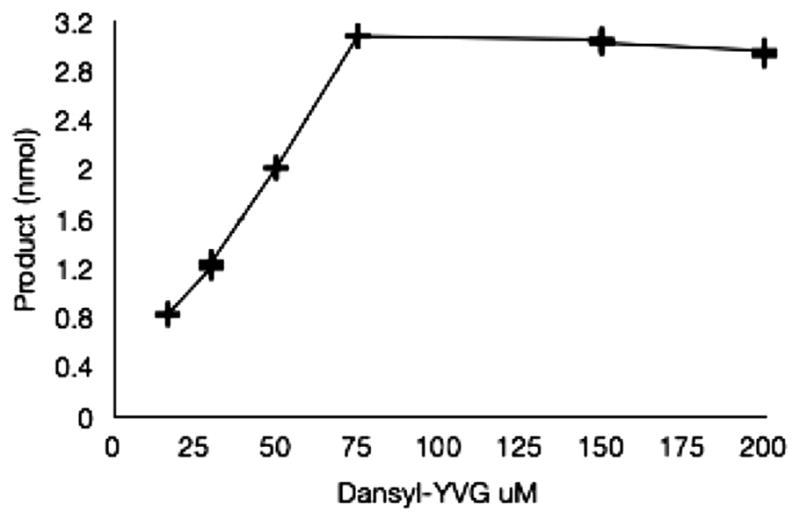
Synthesis of amidated product (dansyl-YV-NH2) by Bullatus 1 as a function of substrate (dansyl-YVG) concentration. Reactions were carried out in duplicate for 21 h. The variation in yield at each concentration of substrate was within ±15%.
4. Discussion
The venom duct of cone snails is a highly peptidergic organ that express 100–200 different peptide toxins used by the snail for envenomation of its prey, predator or competitor. A large fraction of venom peptides are posttranslationally modified. The most frequently encountered modification found is C-terminal amidation catalyzed by the enzyme peptidylglycine α-amidating monooxygenase (PAM). We have identified the PAM encoding cDNA from the venom ducts of C. bullatus and C. geographus. Two isoforms of the enzyme with differences at the C-terminus were identified in C. bullatus and three in C. geographus. In C. bullatus the sequences at the C-terminus suggest that the enzymes may be associated with membranes. A bi-functional single mRNA encoded both the PHM and PAL functions. We have not detected any RNA encoding only PHM or PAL functions. Since we have not probed the expression of proteins in the cells of the venom duct, our experiments were unable to identify any mono-functional enzyme, soluble or membrane bound, generated by endoproteolytic cleavage of the complete PAM protein.
Prohormone convertases present in the venom duct (Hu et al., 2011) could play a role in producing soluble mono- or bi-functional enzyme. Solubilization may increase the turnover number and affinity for substrates as has been observed in the case of rat PHM (Husten and Eipper, 1994; Husten et al., 1993). We speculate that the truncated cDNAs identified in C. geographus are probably involved in the synthesis of mono-functional PHM enzyme rather than being cloning artifacts arising from aberrant initiation of cDNA synthesis. Furthermore, the mono-functional enzymes may be involved in the amidation of specific conotoxin precursors.
Analysis of the reaction products of dansyl-YVG and Bullatus 1 shows the presence of the final product dansyl-YV-amide and no dansyl-YVG(OH) intermediate (Fig. 4). It has been reported that the intermediate is spontaneously converted to the final product at basic pH (Kolhekar et al., 1997b). In addition Kolhekar et al. (1997b) has also described the isolation of the hydroxyglycine containing intermediate in the presence of 0.1% TFA. We surmise that the absence of the intermediate in our experiments is not due to the non-enzymatic spontaneous conversion of the intermediate to the amidated final product during the analysis, rather to the fast conversion by PAL.
It is surprising that C. bullatus PAM, Bullatus 1 and Bullatus 2, differ drastically in their activities. In the PHM plus PAL domains the enzymes differ at eight residues (Fig. 2). While none of these residues have been demonstrated to be essential for enzymatic activity their role cannot be dismissed a priori. One of the changes, the mutation of a cysteine to tyrosine (C559Y) may be important. The cysteine residue is conserved in PAM identified from other sources (Fig. 3B). It is involved in forming a disulfide linkage in the tertiary structure of the enzyme. In the absence of the disulfide linkage the protein may be unstable and have poor activity. Physiologically, however, the mutation may be useful. The mutation C559Y is in the PAL domain. Proteins carrying this mutation may undergo proteolytic cleavage (eg at RK, aa 434–435) to give a mono-functional PHM enzyme specific for conotoxin amidation. The enzyme may be specific for a conopeptide substrate and hence the poor activity on dansyl-YVG. Other cellular proteins may bind to the additional amino acid sequences at the carboxy-terminus of Bullatus 2 and inhibit its activity or unintended mutations were generated during the construction of the cell line resulting in poor activity. The poor activity of the enzyme results in slow utilization of the intermediate and hence our ability to detect it.
As shown in Table 1 amidated residues are in close proximity to other posttranslational modifications. The amino acid residues targeted for amidation is unlikely to be in the optimum configuration necessary for modification by the same enzyme. Multiple mono- or bi-functional enzymes that can “read” the contexts of the different glycine residues may accomplish the modification. We anticipate additional molecular forms of the enzyme in the cell, arising as products of additional genes, alternate splicing and proteolytic processing.
PAM has been identified in two other molluscan species, L. stagnalis (Spijker et al., 1999) and A. californica (Fan et al., 2000). The primary transcript of Lymnaea PAM consists of four tandem divergent PHM domains adjacent to a PAL domain (LPAM-2) or separated from a single copy of PAL by exon A (LPAM-1). The LPAM arouse by intragenic duplication and subsequent mutations leading to different kinetic features of the isozymes. Endoproteolytic cleavage of the LPAM polypeptides generates a mixture of monofunctional isozymes. The enzymes exhibit distinct specificities determined by the amino acid preceding the glycine residue. PAM has been localized to neurons expressing amidated peptides. The colocalization of the appropriate PAMs and cognate substrates assures the efficient synthesis of active neuropeptides.
PAM in Aplysia is encoded by a bifunctional polypeptide in which the PHM and PAL functions are contiguous. PAM is expressed in tissues that are rich in amidated peptides, for example in the head and abdominal ganglia and other endocrine and exocrine organs. The highest specific α-amidating activity is observed in the abdominal ganglion that contains peptidergic bag neurons.
In this communication we have described the identification of an amidating enzyme from the venom duct of cone snails and its activity on a model peptide. We are now examining the ability of the enzyme to amidate different conopeptides. In the context of conotoxin biosynthesis it is important to determine the relative localization of the conopeptides and the amidating enzyme and the molecular form of the enzyme. The amino acid sequences of C. bullatus and C. geographus PAM reported will be used to synthesize peptides to produce antibodies for use in the subcellular localization of the enzyme and to identify different molecular forms of the protein present in the venom duct and other organs of the snail.
Supplementary Material
Acknowledgments
We thank Julita S. Imperial, Samuel S. Espino and Vernon D. Tweed for discussions regarding the analysis of the products of the amidation reaction, University of Utah core facilities for DNA sequencing and oligonucleotide sequencing and William Low at the Salk Institute for Biological Studies for mass spectrometric analysis. S.Ul-H and DMB and PKB carried out the molecular cloning and nucleotide sequence analysis of PAM, S.Ul-H and JG carried out the experiments on enzyme activity, LQ, HH, and MY carried out the analysis of venom duct transcriptome from which the initial sequences of the oligonucleotides for PCR were designed. S.Ul-H, DMB, JG, PKB and BMO wrote the manuscript. We had help from Terry Merritt and My Hyunh in preparing the manuscript. PKB and BMO planned and supervised the experiments. The project was supported by grants, NIH program project GM48677 (S.Ul-H, JG, BMO, PKB) and 5R01GM099939 (LQ, HH, PKB, MY)
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.toxicon.2013.08.054.
Footnotes
Ethical statement
On behalf of the authors, Pradip K Bandyopadhyay declares that: (a) the material has not been published in elsewhere; (b) the paper is not currently being considered for publication elsewhere; (c) all authors have participated in this work and responsible for its content; (d) no animal experimentation was carried out as a part of the results reported here.
Conflict of interest
The authors declare no competing interests.
References
- Attenborough RMF, Hayward DC, Kitahara MV, Miller DJ, Ball EE. A “neural” enzyme in nonbilaterian animals and algae: pre-neural origins for peptidylglycine α-amidating monooxygenase. Mol Biol Evol. 2012;29 (10):3095–3109. doi: 10.1093/molbev/mss114. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay PK, Colledge CJ, Walker CS, Zhou LM, Hillyard DR, Olivera BM. Conantokin-G precursor and its role in gamma-carboxylation by a vitamin K-dependent carboxylase from a Conus snail. J Biol Chem. 1998;273:5447–5450. doi: 10.1074/jbc.273.10.5447. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay PK, Garret JE, Shetty RP, Keate T, Walker CS, Olivera BM. gamma-Glutamyl carboxylation: an extracellular posttranslational modification that antedates the divergence of molluscs, arthropods and chordates. PNAS. 2002;99:1264–1269. doi: 10.1073/pnas.022637099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. Conotoxins and the post-translational modification of secreted gene products. Cell Mol Life Sci. 2005;62 (24):3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch T, Grinblat Y, Goldstein LSB. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Begley GS, Czerwiec E, Stenberg LM, Jacobs M, Kalume DE, Roepstorff P, Stenflo J, Furie BC, Furie B. Precursors of novel Gla-containing conotoxins contain a carboxy-terminal recognition site that directs gamma-carboxylation. Biochemistry. 2005;44:9150–9159. doi: 10.1021/bi0503293. [DOI] [PubMed] [Google Scholar]
- Chufán EE, De M, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of the lyase domain of the amidating enzyme. Structure. 2009;17:965–973. doi: 10.1016/j.str.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AG, Bandyopadhyay P, Olivera BM. Post-translationally modified peptides from Conus venoms. Eur J Biochem. 1999;264:271–275. doi: 10.1046/j.1432-1327.1999.00624.x. [DOI] [PubMed] [Google Scholar]
- Cruz LJ, Gray WR, Olivera BM, Zeikus RD, Kerr L, Yoshikami D, Moczydlowski E. Conus geographus toxins that discriminate between neuronal and muscle sodium channels. J Biol Chem. 1985;260:9280–9288. [PubMed] [Google Scholar]
- Czerwiec E, Begley GS, Bronstein M, Stenflo J, Taylor K, Furie BC, Furie B. Expression and characterization of recombinant vitamin K-dependent gamma-glutamyl carboxylase from an invertebrate Conus textile. Eur J Biochem. 2002;269:6162–6172. doi: 10.1046/j.1432-1033.2002.03335.x. [DOI] [PubMed] [Google Scholar]
- Davis J, Alun Jones A, Lewis RJ. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides. 2009;30:1222–1227. doi: 10.1016/j.peptides.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Day TA, Maule AG, Shaw C, Halton DW, Moore S, Bennett JL, Pax RA. Platyhelminth FMRFamide related peptides (FaRPs) contract Schistosoma mansoni (Trematoda: Digenea) muscle fibres in vitro. Parasitology. 1994;109:455–459. doi: 10.1017/s0031182000080707. [DOI] [PubMed] [Google Scholar]
- Day TA, Maule AG, Shaw C, Pax RA. Structure–activity relationships of FMRFamide-related peptides contracting Schistosoma mansoni muscle. Peptides. 1997;18:917–921. doi: 10.1016/s0196-9781(97)00073-9. [DOI] [PubMed] [Google Scholar]
- De M, Bell J, Blackburn NJ, Mains RE, Eipper BA. Role of an essential tyrosine in peptide amidation. J Biol Chem. 2006;281:20873–20882. doi: 10.1074/jbc.M513886200. [DOI] [PubMed] [Google Scholar]
- Dutertre S, Jin AH, Kaas Q, Jones A, Alewood PF, Lewis RJ. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol Cell Proteomics. 2013;12 (2):312–329. doi: 10.1074/mcp.M112.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper BA, Park LP, Dickerson IM, Keutmann HT, Thiele EA, Rodriguez H, Schofield PR, Mains RE. Structure of the precursor to an enzyme mediating COOH-terminal amidation in peptide biosynthesis. Mol Endocrinol. 1987;1:777–790. doi: 10.1210/mend-1-11-777. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: peptide α-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- Eipper BA, Quon AS, Mains RE, Boswell JS, Blackburn NJ. The catalytic core of peptidylglycine alpha-hydroxylating mono-oxygenase: investigation by site-directed mutagenesis, Cu X-ray absorption spectroscopy, and electron paramagnetic resonance. Biochemistry. 1995;34:2857–2865. doi: 10.1021/bi00009a016. [DOI] [PubMed] [Google Scholar]
- Fan X, Spijker S, Aklal DB, Nagle GT. Neuropeptide amidation: cloning of bifunctional α-amidating enzyme from Aplysia. Mol Brain Res. 2000;82:25–34. doi: 10.1016/s0169-328x(00)00173-x. [DOI] [PubMed] [Google Scholar]
- Gray WR, Luque A, Olivera BM, Barrett J, Cruz LJ. Peptide toxins from Conus geographus venom. J Biol Chem. 1981;256:4734–4740. [PubMed] [Google Scholar]
- Han M, Park D, Vanderzalm PJ, Mains RE, Eipper BA, Taghert PH. Drosophila uses two distinct neuropeptide amidating enzymes, dPAL1 and dPAL2. J Neurochem. 2004;90 (1):129–141. doi: 10.1111/j.1471-4159.2004.02464.x. [DOI] [PubMed] [Google Scholar]
- Hansson K, Ma X, Eliasson L, Czerwiec E, Furie B, Furie BC, Rorsman P, Stenflo J. The first gamma-carboxyglutamic acid-containing contryphan. A selective L-type calcium ion channel blocker isolated from the venom of Conus marmoreus. J Biol Chem. 2004;279:32453–32463. doi: 10.1074/jbc.M313825200. [DOI] [PubMed] [Google Scholar]
- Hauser F, Williamson M, Grimmelikhuijzen CJP. Molecular cloning of a peptidylglycine α-hydroxylating monooxygenase from sea anemones. Biochem Biophys Res Commun. 1997;241:509–512. doi: 10.1006/bbrc.1997.7854. [DOI] [PubMed] [Google Scholar]
- Holford M, Zhang MM, Gowd KH, Azam L, Green BR, Watkins M, Ownby JP, Yoshikami D, Bulaj G, Olivera BM. Pruning nature: biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus. Toxicon. 2009;53:90–98. doi: 10.1016/j.toxicon.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. Characterization of the Conus Bullatus genome and its venom duct transcriptome. BMC Genomic. 2011:12. doi: 10.1186/1471-2164-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. Elucidation of the molecular envenomation strategy of the cone snail Conus geographus through transcriptome sequencing of its venom duct. BMC Genomics. 2012:13. doi: 10.1186/1471-2164-13-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husten EJ, Eipper BA. Purification and characterization of PAM I, an integral membrane protein involved in peptide processing. Arch Biochem Biophys. 1994;312:487–492. doi: 10.1006/abbi.1994.1336. [DOI] [PubMed] [Google Scholar]
- Husten EJ, Tausk FA, Keutmann HT, Eipper BA. Use of endo-protease to identify catalytic domains, linker regions, and functional interactions in soluble peptidyl glycine α-amidating monooxygenase. J Biol Chem. 1993;268:9709–9717. [PubMed] [Google Scholar]
- Iliadiadia KG, Avivib A, Iliadia NN, Knighta D, Korolb AB, Nevob E, Taylorc P, Moranc MF, Kamyshevf NG, Bouliannea GL. nemy encodes a cytochrome b561 that is required for Drosophila learning and memory. PNAS. 2008;105:19986–19991. doi: 10.1073/pnas.0810698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BN, Tamburini PP, Consalvo AP, Young SD, Lovato SJ, Gilligan JP, Jeng AY, Wennogle LP. A fluorometric assay of peptidyl α-amidation activity using high-performance liquid chromatography. Anal Biochem. 1988;168:272–279. doi: 10.1016/0003-2697(88)90318-1. [DOI] [PubMed] [Google Scholar]
- Kent UM, Fleming PJ. Purified cytochrome b561 catalyzes transmembrane electrontransfer for dopamine beta-hydroxylase and peptidyl glycine alpha-amidating monooxygenase activities in reconstituted systems. J Biol Chem. 1987;262:8174–8178. [PubMed] [Google Scholar]
- Kolhekar AS, Keutmann HT, Mains RE, Quon ASW, Eipper BA. Peptidylglycine alpha-hydroxylating monooxygenase: active site residues, disulfide linkages, and a two-domain model of the catalytic core. Biochemistry. 1997a;36:10901–10909. doi: 10.1021/bi9708747. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Mains RE, Eipper BA. Peptidylglycine α-amidating monooxygenase: an ascorbate-requiring enzyme. Meth Enzymol. 1997b;279:35–43. doi: 10.1016/s0076-6879(97)79007-4. [DOI] [PubMed] [Google Scholar]
- Kolhekar AS, Roberts MS, Jiang N, Johnson RC, Mains RE, Eipper BA, Taghert PH. Neuropeptide amidation in Drosophila: separate genes encode the two enzymes catalyzing amidation. J Neurosci. 1997c;17:1363–2476. doi: 10.1523/JNEUROSCI.17-04-01363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolhekar AS, Bell J, Shiozaki EN, Jin L, Keutmann HT, Hand TA, Mains RE, Eipper BA. Essential features of the catalytic core of peptidyl-α-hydroxyglycine α-amidating lyase. Biochemistry. 2002;41:12384–12394. doi: 10.1021/bi0260280. [DOI] [PubMed] [Google Scholar]
- Lirazan MB, Hooper D, Corpuz GP, Ramilo CA, Bandyopadhyay P, Cruz LJ, Olivera BM. The spasmodic peptide defines a new conotoxin superfamily. Biochemistry. 2000;39:1583–1588. doi: 10.1021/bi9923712. [DOI] [PubMed] [Google Scholar]
- López-Vera E, Aguilar MB, Schiavon E, Marinzi C, Ortiz E, Restano Cassulini R, Batista CV, Possani LD, Heimer de la Cotera EP, Peri F, Becerril B, Wanke E. Novel alpha-conotoxins from Conus spurius and the alpha-conotoxin EI share high-affinity potentiation and low-affinity inhibition of nicotinic acetylcholine receptors. FEBS J. 2007;274:3972–3985. doi: 10.1111/j.1742-4658.2007.05931.x. [DOI] [PubMed] [Google Scholar]
- McIntosh JM, Olivera BM, Cruz LJ, Gray WR. Gamma-carboxyglutamate in a neuroactive toxin. J Biol Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- Mair GR, Niciu MJ, Stewart MT, Brennan G, Omar H, Halton DW, Mains R, Eipper BA, Maule AG, Day TA. A functionally atypical amidating enzyme from the human parasite Schistosoma mansoni. FASEB J. 2004;18:114–121. doi: 10.1096/fj.03-0429com. [DOI] [PubMed] [Google Scholar]
- Merkler DJ. C-terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extended peptides and the importance of the amide to bioactivity. Enzyme Microb Technol. 1994;16:450–456. doi: 10.1016/0141-0229(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Milgram SL, Kho ST, Martin GV, Mains RE, Eipper BA. Localization of integral membrane peptidylglycine a-amidating monooxygenase in neuroendocrine cells. J Cell Sci. 1997;110:695–706. doi: 10.1242/jcs.110.6.695. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Ohsuye K, Wada Y, Fuchimura K, Tanaka S, Matsuo H. Cloning and sequence of cDNA encoding a peptide C-terminal α-amidating enzyme from Xenopus laevis. Biochem Biophys Res Commun. 1987;148:546–552. doi: 10.1016/0006-291x(87)90911-9. [DOI] [PubMed] [Google Scholar]
- Ohsuye K, Kitano K, Wada Y, Fuchimura K, Tanaka S, Mizuno K, Matsuo H. Cloning of cDNA encoding a new peptide C-terminal α-amidating enzyme having a putative membrane -spanning domain from Xenopus laevis skin. Biochem Biophys Res Commun. 1988;150:1275–1281. doi: 10.1016/0006-291x(88)90767-x. [DOI] [PubMed] [Google Scholar]
- Olivera BM. Conus venom peptides, receptor and ion channel targets and drug design: 50 million years of neuropharmacology (EE Just lecture 1996) Mol Biol Cell. 1997;8:2101–2109. doi: 10.1091/mbc.8.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281 (42):31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Oyarce AM, Eipper BA. Cell type-specific storage of dopamine beta-monooxygenase. J Biol Chem. 2000;275:3270–3278. doi: 10.1074/jbc.275.5.3270. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Kolhekar AS, Eipper BA, Mains RE, Amzel LM. Amidation of bioactive peptides: the structure of peptidylglycine alpha-hydroxylating monooxygenase. Science. 1997;278:1300–1305. doi: 10.1126/science.278.5341.1300. [DOI] [PubMed] [Google Scholar]
- Prigge ST, Mains RE, Eipper BA, Amzela LM. New insights into copper monooxygenases and peptide amidation: structure, mechanism, and function. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spijker S, Smit AB, Eipper BA, Malik A, Mains RE, Geraerts PM. A molluscan peptide a-amidating enzyme precursor that generates five distinct enzymes. FASEB J. 1999;13:735–748. doi: 10.1096/fasebj.13.6.735. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Shimoi H, Iwasaki Y, Kawahara T, Matsuura Y, Nishikawa Y. Elucidation of amidating reaction mechanism by frog amidating enzyme, peptidylglycine α-hydroxylating mono-oxygenase, expressed in insect cell culture. EMBO. 1990;9:4259–4265. doi: 10.1002/j.1460-2075.1990.tb07874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CS, Shetty RP, Clark K, Kazuko SG, Letsou A, Olivera BM, Bandyopadhyay PK. On a potential global role of vitamin K-dependent α-carboxylation in animal systems. J Biol Chem. 2001;276:7769–7774. doi: 10.1074/jbc.M009576200. [DOI] [PubMed] [Google Scholar]
- Walker CS, Steel D, Jacobsen RB, Lirazan MB, Cruz LJ, Hooper D, Shetty R, DelaCruz RC, Nielsen JS, Zhou LM, Bandyopadhyay P, Craig AG, Olivera BM. The T-superfamily of conotoxins. J Biol Chem. 1999;274:30664–30671. doi: 10.1074/jbc.274.43.30664. [DOI] [PubMed] [Google Scholar]
- Watkins M, Olivera BM, Hillyard DR, McIntosh JM, Jones RM. Patent number US6797808. Alpha-Conotoxin Peptides. 2004 Sep 28;
- Yin P, Bousquet-Moore D, Annangudi SP, Southey BR, Mains RE, Eipper BA, Sweedler JV. Probing the production of amidated peptides following genetic and dietary copper manipulations. PLoS ONE. 2011;6(12) doi: 10.1371/journal.pone.0028679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura H, Anzai T, Kato I, Furuya Y, Shizuta S, Takasawa S, et al. Identification of the five essential histidine residues for peptidylglycine monooxygenase. Biochem Biophys Res Commun. 1996;218:495–499. doi: 10.1006/bbrc.1996.0088. [DOI] [PubMed] [Google Scholar]
- Zabriskie TM, Klinge M, Szymanski CM, Cheng H, Vederas JC. Peptide amidation in an invertebrate: purification, characterization, and inhibition of peptidylglycine α-hydroxylating monooxygenase from the heads of honeybees (Apis mellifera) Arch Insect Biochem Physiol. 1994;26:27–48. doi: 10.1002/arch.940260104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.