Abstract
Intrinsically disordered proteins (IDPs) are involved in diverse cellular functions. Many IDPs can interact with multiple binding partners, resulting in their folding into alternative ligand-specific functional structures. For such multi-structural IDPs, a key question is whether these multiple structures are fully encoded in the protein sequence, as is the case in many globular proteins. To answer this question, here we employed a combination of single-molecule and ensemble techniques to compare ligand-induced and osmolyte-forced folding of α-synuclein. Our results reveal context-dependent modulation of the protein’s folding landscape, suggesting that the codes for the protein’s native folds are partially encoded in its primary sequence, and are completed only upon interaction with binding partners. Our findings suggest a critical role for cellular interactions in expanding the repertoire of folds and functions available to disordered proteins.
Keywords: Biophysics, intrinsically disordered protein, protein folding, protein-ligand interactions, single-molecule FRET
Intrinsically disordered proteins (IDPs) are structurally dynamic entities accessing a broad conformational space. They are abundantly represented in all kingdoms of life and account for approximately one-third of the eukaryote proteome.[1] Many IDPs undergo disorder-to-order transitions upon binding to their cognate ligands, allowing them to carry out multifarious biological functions.[2] They are functionally prevalent in cellular recognition, signaling, and regulation,[2–3] whereas their dysfunction has been linked to several human diseases.[4]
Globular proteins fold via a thermodynamic process that involves formation of primary sequence-encoded intramolecular interactions in the context of the solution environment.[5] In contrast, IDPs lack sufficient structure-stabilizing interactions, and thus remain in their disordered unfolded states. From a mechanistic point of view, at least for a subset of disordered proteins, the inability to adopt compact folded structures stems from their unfavorable folding energetics and conformational instability at physiological conditions. Consequently, these proteins exhibit destabilized folded states, with the denatured states being the major populated ensembles, and interaction with partners being able to stabilize the folded state. Because folding for this class of IDPs is mostly encoded in their primary sequence, the folded-denatured equilibrium can also be modulated to favor population of the native folded structure (forced folding) using general stabilizing agents such as osmolytes, one of the major types of chemical chaperones.[6]
Previous biophysical forced-folding studies have reported on IDPs with putative single native folded states.[6] In contrast, several IDPs can adopt multiple alternative structures upon binding to different partners, as in the case of the primarily neuronal protein α-synuclein,[7] and the hub proteins p53 and E1A.[8] Indeed, the plasticity of binding-induced folding is believed to be important for IDPs that carry out multiple functions.[9] For such IDPs, a key question that arises is whether the ligand-induced folds are still mostly encoded in the protein’s primary sequence or not. In one scenario, the intrinsic folding landscape of the protein could exhibit multiple native folded minima,[10] and binding to different ligands would stabilize individual native folds. Alternatively, the fold for some or all of the ligand-bound structures could be incompletely encoded in the IDP’s primary sequence, and partner interactions would complete the codes that allow formation of different structures. Here, we distinguish between these possibilities in the context of α-synuclein, a Parkinson’s disease-linked IDP that can fold into two alternative helical structures upon its interaction with lipid membranes and mimics.[7, 11] We test if one or both of these alternative folds are primarily encoded in the protein’s sequence by carrying out forced folding experiments using the protecting osmolyte trimethylamine N-oxide (TMAO), and comparing results with those from ligand-induced folding experiments.
α-Synuclein is a small acidic protein that is predominantly expressed in neuronal cells. Whereas the exact function of this protein remains elusive, it is known to be localized in presynaptic nerve termini and thought to be involved in vesicular transport and neurotransmitter release.[12] A number of biophysical studies suggested that upon interaction with different lipid membranes and mimics, the protein undergoes coupled binding and folding, and forms partially folded structures with high α-helical content.[13] Further thermodynamics, kinetics and structural studies revealed that the protein adopts multiple folded states and can switch between conformations depending on the protein’s physicochemical environment.[7, 11, 14] To assess whether similar multistate behavior involving the same folded states exist in the TMAO-induced folding of α-synuclein, we employed a combination of ensemble and single-molecule spectroscopic methods to probe the protein’s conformations.
Previous studies have shown that α-synuclein forms aggregates and/or oligomers in the presence of high concentrations of TMAO at physiological pH conditions.[15] To avoid potential experimental complications, all measurements here were carried out at pH 10.5. The theoretical isoelectric point of the protein is 4.67, assuming model compound pKas.[16] Thus, at alkaline pH, α-synuclein exhibits higher net charge relative to more neutral pH conditions, resulting in increased electrostatic repulsion between individual proteins and reduced propensity for oligomerization and/or aggregation.[17]
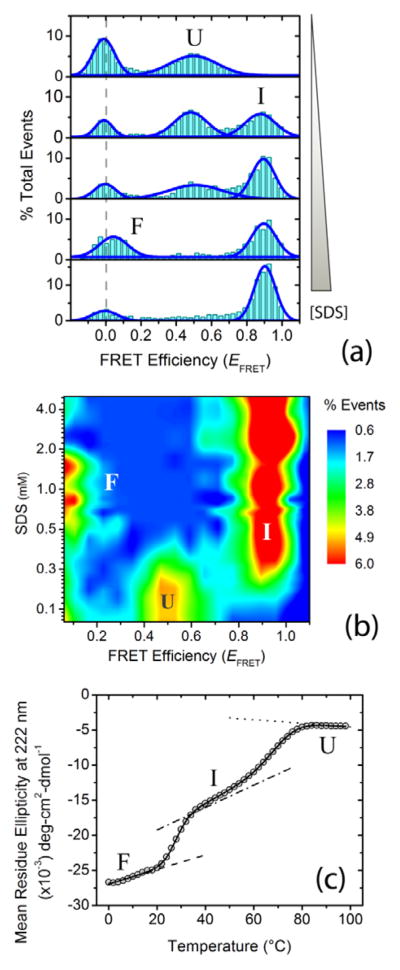
Upon interaction with the lipid mimic SDS at physiological pH conditions, we previously showed that α-synuclein undergoes a multistate disorder-to-order transition, forming partially folded structures with high α-helical content.[11] To assess whether this multistate behavior is retained at alkaline pH (i.e., pH 10.5 ± 0.1), we probed the binding-induced folding landscape of α-synuclein by monitoring changes in protein dimensions utilizing the distance-dependence of single-molecule Förster resonance energy transfer (smFRET). The isothermal smFRET SDS titration was performed with approximately 100 pM α-synuclein dual-labeled at residue positions 7 and 84 (Alexa 488/594 dye pairs), employing the previously described home-built confocal single-molecule diffusion setup.[7] Bursts of donor and acceptor fluorescence were recorded from individual molecules as freely diffusing dual-labeled proteins passed through the sub-fL detection volume. The data were then analyzed to generate smFRET histograms, providing a scheme for direct visualization of individual conformational states (Figure 1a). Similar to what we observed previously at physiological pH (i.e., pH 7.5 ± 0.05),[7] the single-molecule isothermal SDS titration data at alkaline pH revealed switching of the protein between three different conformations depending on SDS concentration (i.e., unfolded (U), broken helix (I) and elongated helix (F) conformations; Figure 1a,b). To further characterize the multistate transitions, we used UV-CD spectroscopy, and carried out thermal unfolding of SDS-folded α-synuclein using 20 μM α-synuclein and 1mM SDS at pH 10.5 ± 0.1, and monitored changes in ellipticity at 222 nm, a reporter for α-helicity in proteins.[18] Similar to the single-molecule titration data, a multistate folding-unfolding transition of the protein was observed at the employed solution condition (Figure 1c). The presence of similar conformations and comparable multistate behavior at physiological and alkaline pH conditions in α-synuclein SDS-induced folding manifests the robustness of the nature and mechanism of ligand-induced folding for the protein.
Figure 1.
Ligand-induced multistate folding of the intrinsically disordered protein α-synuclein. Single-molecule and ensemble experiments were performed in αβγ buffer (i.e., 0.2 M sodium chloride, 10 mM sodium acetate, 10 mM monosodium phosphate, 10 mM glycine) at pH 10.5 ± 0.1. (a) Representative single-molecule Förster resonance energy transfer (smFRET) histograms as a function of increasing concentration (0, 0.2, 0.3, 0.6 and 5.0 mM) of the ligand and lipid mimic sodium dodecyl sulfate (SDS). The dashed gray line shows the expected zero peak center position. Peaks for U, I and F states are indicated. (b) [Ligand]-FRET efficiency (EFRET) contour map, color-coded on the basis of fractional population. (c) Thermal unfolding of α-synuclein folded using 1mM SDS, monitored using far-UV CD spectroscopy. The solid curve represents the nonlinear least-squares (NLS) fit of the data to a three-state model;[11] the dotted, dash-dotted and dashed lines are the baselines for U, I and F states.
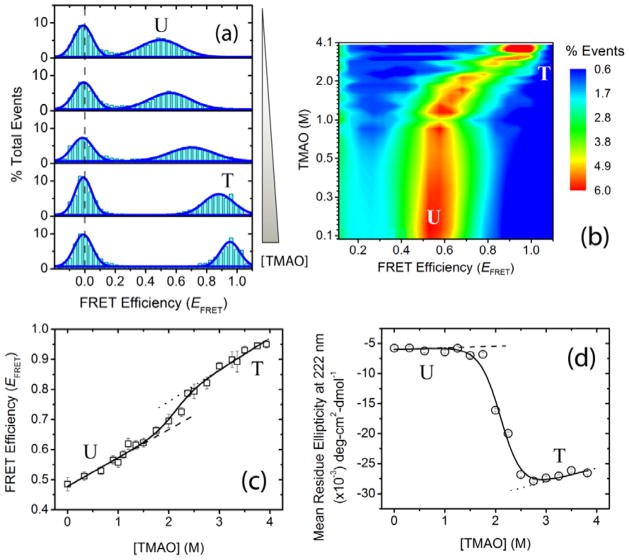
Having ascertained the multistate behavior of ligand-induced folding of α-synuclein at alkaline pH, we next studied osmolyte forced-folding of the protein at similar experimental conditions. Osmolytes are small-molecule organic compounds used by biological systems to perturb the physicochemical environment of biomacromolecules and counteract the deleterious effects of extreme conditions such as high osmotic and hydrostatic pressures, dehydration, and high or low temperatures.[19] These chemical chaperones modulate protein structural landscapes in a non-specific manner and exhibit global effects on cellular proteostasis.[20] Among the well-studied osmolytes, TMAO was found to be one of the most efficient as a protecting agent, and has been shown to reverse the effect of denaturants on proteins,[21] and fold mutation-destabilized as well as partially folded or unfolded proteins in vitro.[22] Furthermore, a recent in vivo study showed that TMAO can assist intracellular folding of mutant forms of globular proteins in Escherichia coli and Saccharomyces cerevisiae.[23] To characterize the effects of TMAO on the α-synuclein folding landscape, we carried out isothermal smFRET osmolyte titration of the protein (described in the Supporting Information). In contrast to the multistate SDS-induced α-synuclein folding, the single-molecule isothermal TMAO titration data showed a single non-zero smFRET peak that shifted continuously as a function of TMAO concentration (Figure 2a,b). Such a behavior could indicate a two-state transition with rapidly interconverting conformations, or a more complex mechanism. To distinguish between the possibilities, we plotted the mean non-zero smFRET peak position against osmolyte concentration (Figure 2c). A sigmoidal transition profile with positively sloping pre- and post-transition baselines suggested a two-state folding mechanism. Here, by leveraging the high pH condition that disfavors protein aggregation, we next monitored α-helix formation by far UV-CD spectroscopy (Figure 2d). Similar to the smFRET data, a clear cooperative two-state folding transition to a helical state was observed in the ensemble experiment.
Figure 2.
Osmolyte-induced two-state folding of α-synuclein. Single-molecule and ensemble experiments were performed in αβγ buffer at pH 10.5 ± 0.1. (a) Representative smFRET histograms as a function of increasing trimethylamine N-oxide (TMAO) concentration (0, 1, 2, 3, and 3.9 M). The dashed gray line shows the expected zero peak position; peaks for U and T protein states are indicated. (b) [TMAO]-EFRET contour map, color-coded on the basis of fractional population. (c) α-Synuclein isothermal smFRET TMAO titration data. (d) Osmolyte-forced folding of α-synuclein using TMAO, monitored via far-UV CD spectroscopy. The solid curves in (c) and (d) represent the global NLS fits of the data to a two-state linear extrapolation model (LEM);[11] the dashed and dotted lines are the baselines for U and T, respectively.
To obtain thermodynamic parameters for the TMAO-induced folding transition, data from both smFRET and far-UV CD experiments were fitted globally to a two-state linear extrapolation model (LEM) using non-linear least squares fitting in OriginPro 8.0 (OriginLab, Northampton, MA).[24] Comparable thermodynamic parameters were obtained when the folding was probed by fluorescence anisotropy, and the data were fitted independently to the same two-state model (Supplementary Figure 1). The similarity among the thermodynamic parameters obtained from experiments carried out at different protein concentration regimes (from ~100 pM for the single-molecule experiments to 20 μM for the far-UV CD experiments) is consistent with the observed two-state transitions being due to folding of monomeric α-synuclein.
Osmolytes perturb protein conformational stability mainly through their preferential exclusion from the amide backbone and favor the formation of compact conformations.[25] Our results on the TMAO-induced folding of α-synuclein clearly show that forced compaction of the protein leads to cooperative structure formation (Figure 2). This TMAO-forced folded structure, designated here as T, has high α-helicity similar to that of the extended helix conformation (F), while also exhibiting high EFRET similar to that of the broken helix conformation (I). Furthermore, unlike the SDS titration results, the absence of multiple non-zero smFRET peaks in the single-molecule osmolyte titration at the transition region suggests rapid inter-conversion (≪ 0.5 ms) of the protein between unfolded and force-folded states (Figure 2a,b). Thus, the T state is also conformationally more dynamic than the I and F states. In addition, the positive sloping pre- and post-transition baselines of the smFRET data suggests compaction of both unfolded and forced-folded ensembles as a function of increasing TMAO concentrations (Figure 2c). The compaction observed here follows the same trend as in our previous results under physiological pH conditions (i.e., pH 7.5 ± 0.05).[26]
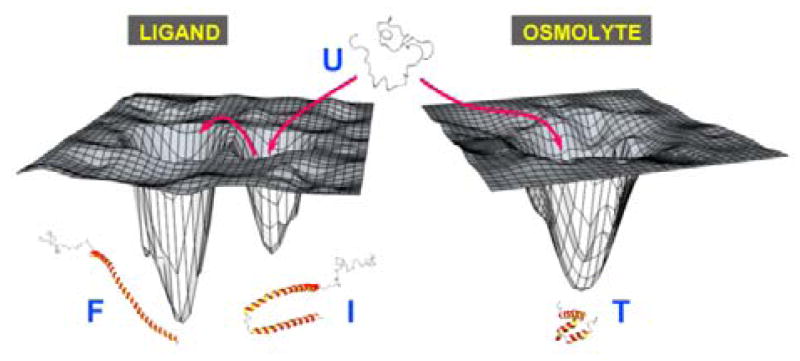
While ligand- and osmolyte-induced folding have been studied for a simpler IDP system where different folding agents induce similar folds,[6b,d] an analogous comparison has not been reported for IDPs that display complex multistate ligand-induced folding. Our results now clearly demonstrate that forced folding of α-synuclein does not result in transition of the protein to either of its ligand-induced folds, but rather to a different one that is compact, yet helical and dynamic (Figure 3). Therefore, the amino-acid sequence for α-synuclein does not fully encode the ligand-induced structures, and ligand interactions are needed to complete the folding code. This is especially interesting from a broader IDP context, because many of these proteins adopt folded conformations to carry out their biological functions and similar mechanisms are likely to be operative for many of them. Because of the extremely low probability for adopting the energetically unfavorable high-affinity binding-competent species, binding-induced folding of this class of IDPs is likely to follow an induced folding mechanism that involves formation of encounter complexes as observed for ligand-induced folding of α-synuclein.[14]
Figure 3.
Schematic representation of the context-dependent folding landscapes of α-synuclein.
Akin to the widespread use of denaturants to study the unfolding properties of globular proteins, protecting osmolytes have been used for the study of IDP folding.[6] For thermodynamically unfolded disordered proteins with folding codes already present in the primary sequence, compaction leads to the formation of native folds. However, for IDPs that lack complete folding codes, forced compaction would either fail to fold the protein or induce folds that are non-native, as observed in this work for α-synuclein (Figure 3). Thus, the application of osmolyte-induced folding of IDPs warrants caution, taking into account the completeness of the folding codes of the disordered protein under consideration. One could employ a hybrid approach to study folding of the latter IDP class, wherein osmolye-induced folding is carried out in the presence of the binding partners, thus providing the missing codes along with an additional energetic push for physiologically relevant disorder-to-order transitions.
IDPs are an interesting family of proteins that came to prominence in the last decade due to their novel structural complexity and enormous diversity in function and interactions. Results from our experiments show an additional layer of complexity where at least for one important class of IDPs, parts of the codes for the native folds are encoded in ligand interactions. This could be a general mechanism employed by many multifunctional IDPs, expanding their coding repertoire to facilitate structural plasticity and interaction promiscuity. Our experiments on the ligand-induced and osmolyte-forced folding of α-synuclein, a misfolding disease-associated disordered protein, revealed folding landscapes that are modulated in a context-dependent fashion. Similar modulation mechanisms are likely to play vital roles in the biology of IDPs where different components of the protein physicochemical environment act synergistically, resulting in a complex and regulatable diversity of cellular functions.
Experimental Section
Proteins were prepared and labeled as described in the Supporting Information. All of the experiments were carried out in αβγ buffer (i.e., 0.2 M sodium chloride, 10 mM sodium acetate, 10 mM monosodium phosphate, 10 mM glycine; pH 10.5 ± 0.1). Isothermal smFRET experiments were carried out using a custom-built confocal microscopic setup described previously.[7] Details of the ensemble spectroscopic methods are described in the Supporting Information. Folding transitions were analyzed by NLS fitting of data to the LEM.[24]
Supplementary Material
Footnotes
This work was supported by Grant GM066833 (to A.A.D.) from the National Institute of General Medical Sciences, National Institutes of Health. We thank Robert L. Nussbaum and Nelson B. Cole for providing the plasmid construct for wildtype α-synuclein, Peter E. Wright for use of a CD spectrometer, and Josephine C. Ferreon for insightful discussions and critical review of the manuscript.
References
- 1.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Uversky VN. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Uversky VN, Oldfield CJ, Dunker AK. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]; b) Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Nat Chem Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uversky VN, Oldfield CJ, Dunker AK. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 5.Anfinsen CB. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 6.a) Baskakov I, Bolen DW. J Biol Chem. 1998;273:4831–4834. doi: 10.1074/jbc.273.9.4831. [DOI] [PubMed] [Google Scholar]; b) Henkels CH, Kurz JC, Fierke CA, Oas TG. Biochemistry. 2001;40:2777–2789. doi: 10.1021/bi002078y. [DOI] [PubMed] [Google Scholar]; c) Rajagopalan L, Rösgen J, Bolen DW, Rajarathnam K. Biochemistry. 2005;44:12932–12939. doi: 10.1021/bi051219z. [DOI] [PubMed] [Google Scholar]; d) Chang YC, Oas TG. Biochemistry. 2010;49:5086–5096. doi: 10.1021/bi100222h. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Li J, Motlagh HN, Chakuroff C, Thompson EB, Hilser VJ. J Biol Chem. 2012;287:26777–26787. doi: 10.1074/jbc.M112.355651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreon ACM, Gambin Y, Lemke EA, Deniz AA. Proc Natl Acad Sci U S A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ferreon ACM, Ferreon JC, Wright PE, Deniz AA. Nature. 2013;498:390–394. doi: 10.1038/nature12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson HJ, Wright PE. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 10.Gambin Y, Schug A, Lemke EA, Lavinder JJ, Ferreon ACM, Magliery TJ, Onuchic JN, Deniz AA. Proc Natl Acad Sci U S A. 2009;106:10153–10158. doi: 10.1073/pnas.0904461106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreon ACM, Deniz AA. Biochemistry. 2007;46:4499–4509. doi: 10.1021/bi602461y. [DOI] [PubMed] [Google Scholar]
- 12.a) Kahle PJ, Neumann M, Ozmen L, Muller V, Jacobsen H, Schindzielorz A, Okochi M, Leimer U, van Der Putten H, Probst A, Kremmer E, Kretzschmar HA, Haass C. J Neurosci. 2000;20:6365–6373. doi: 10.1523/JNEUROSCI.20-17-06365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, Shinsky N, Verdugo JM, Armanini M, Ryan A, Hynes M, Phillips H, Sulzer D, Rosenthal A. Neuron. 2000;25:239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 13.Pfefferkorn CM, Jiang Z, Lee JC. Biochim Biophys Acta. 2012;1818:162–171. doi: 10.1016/j.bbamem.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambin Y, VanDelinder V, Ferreon ACM, Lemke EA, Groisman A, Deniz AA. Nat Methods. 2011;8:239–241. doi: 10.1038/nmeth.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uversky VN, Li J, Fink AL. FEBS Lett. 2001;509:31–35. doi: 10.1016/s0014-5793(01)03121-0. [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger E, Hoogland C, Gattiker A, Duvaud Se, Wilkins MR, Appel RD, Bairoch A. In: The Proteomics Protocols Handbook. Walker JM, editor. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 17.a) Uversky VN, Li J, Fink AL. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]; b) Wang W, Nema S, Teagarden D. Int J Pharm. 2010;390:89–99. doi: 10.1016/j.ijpharm.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Greenfield NJ. Nat Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yancey PH. Am Zool. 2001;41:699–709. [Google Scholar]
- 20.Leandro P, Gomes CM. Mini Rev Med Chem. 2008;8:901–911. doi: 10.2174/138955708785132783. [DOI] [PubMed] [Google Scholar]
- 21.a) Baskakov I, Wang A, Bolen DW. Biophys J. 1998;74:2666–2673. doi: 10.1016/S0006-3495(98)77972-X. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Doan-Nguyen V, Loria JP. Protein Sci. 2007;16:20–29. doi: 10.1110/ps.062393707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Mello CC, Barrick D. Protein Sci. 2003;12:1522–1529. doi: 10.1110/ps.0372903. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 23.Bandyopadhyay A, Saxena K, Kasturia N, Dalal V, Bhatt N, Rajkumar A, Maity S, Sengupta S, Chakraborty K. Nat Chem Biol. 2012;8:238–245. doi: 10.1038/nchembio.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.a) Santoro MM, Bolen DW. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]; b) Ferreon ACM, Deniz AA. Methods Mol Biol. 2012;896:257–266. doi: 10.1007/978-1-4614-3704-8_17. [DOI] [PubMed] [Google Scholar]
- 25.a) Bolen DW. Methods Mol Biol. 2001;168:17–36. doi: 10.1385/1-59259-193-0:017. [DOI] [PubMed] [Google Scholar]; b) Zou Q, Bennion BJ, Dagget V, Murphy KP. J Am Chem Soc. 2002;124:1192–1202. doi: 10.1021/ja004206b. [DOI] [PubMed] [Google Scholar]; c) Cho SS, Reddy G, Straub JE, Thirumalai D. J Phys Chem B. 2011;115:13401–13407. doi: 10.1021/jp207289b. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Canchi DR, García AE. Annu Rev Phys Chem. 2013;64:273–293. doi: 10.1146/annurev-physchem-040412-110156. [DOI] [PubMed] [Google Scholar]
- 26.Ferreon ACM, Moosa MM, Gambin Y, Deniz AA. Proc Natl Acad Sci U S A. 2012;109:17826–17831. doi: 10.1073/pnas.1201802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.