Abstract
The use of anion-exchange chromatography was investigated (and parameters compared) as an alternative method to concentrate and purify bacterial viruses. Chromatography was performed with Convective Interactive Media® monoliths, with three different volumes and two matrix chemistries. Eleven morphologically distinct phages were tested, infecting five different bacterial species. For each of the phages tested, a protocol was optimized, including the choice of column chemistry, loading, buffer and elution conditions. The capacity and recovery of the phages on the columns varied considerably between phages. We conclude that anion-exchange chromatography with monoliths is a valid alternative to the more traditional CsCl purification, has upscaling advantages, but it requires more extensive optimization.
Keywords: Bacteriophages, purification, anion-exchange chromatography
1. Introduction
Many applications in bacteriophage research (e.g. genomics, proteomics and crystallography) require pure and highly concentrated phage suspensions (Lavigne et al. 2009; Rossmann et al. 2005). Also for the use in phage therapy, purification steps are needed, depending on the type of application, medical (topical or systemic), agricultural or in veterinary applications (Gill and Hyman, 2010). Traditionally, this has been achieved by polyethylene glycol precipitation and subsequent CsCl gradient ultracentrifugation (Boulanger 2009; Yamamoto et al. 1970). In most cases this method gives a relatively low yield but a high quality phage preparation, yet for some phages it does not work. Phages either get damaged by the centrifugal forces, suffer from osmotic shock or interact with CsCl and lose their infectivity (Carlson, 2005). Although the latter two situations may be remedied by using another type of gradient (e.g. sucrose gradient (Serwer et al. 1978)), gradient separations in general are cumbersome and do not easily permit upscaling of the process.
The inability of some phages to be purified with CsCl gradients is common knowledge in the lab, but little has been published on specific phages exhibiting this behaviour (Table 1). In all the known CsCl-inactivated phages, no trend could be found concerning morphology, host or isolation source.
Table 1.
Bacteriophages purified with CIM® monolithic columns.
| Phage | Phage family (morphotype) | Host species | Host strain | Growth medium | Loading suspension | Optimized Column | Optimized Buffer set | Elution of pure phage fraction | Capacity (pfu/disk) | Recovery of phage in pure fraction | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dickeya phage LIMEstone1 | Myoviridae (ViI-like) | “D. solani” | GBBC 2072 | LB | LB lysate undiluted | CIM® QA disk | Tris(a) | 0.6 M NaCl | >2.5 × 1012 | 99.9% | Adriaenssens et al. 2012 |
| Dickeya phage LIMEstone2 | Myoviridae (ViI-like) | “D. solani” | GBBC 2072 | LB | LB lysate diluted in phosphate buffer (1/2) | CIM® DEAE disk | phosphate | 0.6 M NaCl | >2 × 1011 | 70% | Adriaenssens et al. 2012 |
| Staphylococcus phage ISP | Myoviridae (Twort-like) | S. aureus subsp aureus | Rosenbach ATCC 6538 | MH | MH lysate undiluted | CIM® DEAE disk | Tris(a) | 0.8 M NaCl | > 1 × 1011 | 35 – 65% | Merabishvili et al. 2009; Vandersteegen et al. 2011 |
| Pseudomonas phage ΦE2005-A | Myoviridae (PB1-like) | P. aeruginosa | EAMS2005-A | 25% TSB | TSB lysate diluted in 2× Tris(a) buffer (1/1) | CIM® QA-8f | Tris(a) | 0.25 M NaCl | 1 × 1012 | 40 – 70% | R. Donlana |
| Pseudomonas phage ΦPaer14 | Myoviridae (PB1-like) | P. aeruginosa | Paer14 | 25% TSB | TSB lysate diluted in 2× Tris(a) buffer (1/1) | CIM® QA-8f | Tris(a) | 0.25 M NaCl | 1 × 1012 | 40 – 70% | R. Donlan |
| Pseudomonas phage ΦE2005-C | Myoviridae (PB1-like) | P. aeruginosa | EAMS2005-C | 25% TSB | TSB lysate diluted in 2× Tris(a) buffer (1/1) | CIM® QA-8f | Tris(a) | 0.25 M NaCl | 1 × 1012 | 40 – 70% | R. Donlan |
| Pseudomonas phage ΦM4 | Myoviridae (KPP10-like) | P. aeruginosa | M4 | 25% TSB | TSB lysate diluted in 2× Tris(a) buffer (1/1) | CIM® QA-8f | Tris(a) | 0.56 M NaCl | 1 × 1012 | 40 – 70% | Lindberg & Latta 1974 |
| Burkholderia phage Phi208 | Podoviridae | B. thailandensis | DW503 | LB | Suspension dialyzed against Tris(b) buffer | CIM® QA disk | Tris(b) | 0.3 M NaCl | 1.7 × 109 | 70% | This study |
| Pseudomonas phage Φ15 | Podoviridae (T7-like) | P. putida | PpG1 | LB | LB lysate diluted in Tris(a) buffer (1/2) | CIM® DEAE disk | Tris(a) | 0.3 M NaCl | 8 × 1010 | 87% | Cornelissen et al. 2011; Shaburova et al. 2009 |
| Pseudomonas phage ΦPaer4 | Podoviridae (LUZ24-like) | P. aeruginosa | Paer4 | 25% TSB | TSB lysate diluted in 2× Tris(a) buffer (1/1) | CIMac™ QA CIM® QA-8f | Tris(a) | 0.3 M NaCl | Ac QA: 5×108 to 1×109 QA-8f: > 1 × 1012 | 40 – 70% | R.Donlan |
| Pseudomonas phage LUZ19 | Podoviridae (φKMV-like) | P. aeruginosa | PAO1 | LB | LB lysate diluted in Tris(a) buffer (1/2) | CIM® DEAE disk | Tris(a) | 0.6 M NaCl | >1.2 × 1012 | 70% | Ceyssens et al. 2009 |
Rodney Donlan, CDC Biofilm Lab, Division of Healthcare Quality Promotion, Centers for Disease Control and Prevention, Atlanta, GA.
An alternative method for the purification of bacteriophages is chromatography. In 1953, Puck and Sagik proved that phages T1 and T2 could bind to anionic (nalcite) or cationic (dowex) resins, the latter only in the presence of salts, originally to study the attachment of phages to the host cell. Anion-exchange chromatography of phages for purification on ECTEOLA columns has been described as early as 1957 (Creaser and Taussig, 1957; Taussig and Creaser, 1957). For the lipid-containing phage PRD1, another method was designed, using commercial Memsep cartridges with quaternary methylamine and diethylaminoethyl (Walin et al. 1994). More recently, Convective Interactive Media® (CIM®) monoliths have become commercially available and have been shown to effectively purify a number of phages, including Escherichia phages T4, lambda and M13, and Staphylococcus phage VDX-10 (Kramberger et al. 2010; Smrekar et al. 2008, 2011). In these reports, two types of anion-exchange matrices have been examined, quaternary amine (QA) and diethyl amine (DEAE), the latter only for VDX-10. As Kramberger and colleagues (2010) showed, the same purification conditions apply when scaling up, making anion-exchange chromatography purification ideal for large-scale production of bacteriophage suspensions.
In this paper, we describe the purification of 11 morphologically distinct phages which infect a range of bacterial hosts using CIM® monolith anion-exchange chromatography. Columns with two monolith types, and different volumes were tested.
2. Materials & Methods
2.1. Phage amplification
Dickeya phages LIMEstone1 and LIMEstone2, Pseudomonas phages ∅15, LUZ19, ∅Paer4, ∅E2005-A, ∅Paer14, ∅E2005-C and ∅M4 and Staphylococcus phage ISP (see Table 1) were amplified in liquid culture, the first four in LB broth (10 g/l Tryptone, 5 g/l yeast extract, 10 g/l NaCl), the following five in 25% TSB broth (Becton, Dickinson and Company, Sparks, USA) and ISP in Mueller Hinton (MH) broth (Becton, Dickinson and Company, Sparks, USA) (Table 1). Phages were added to an exponential phase shaking culture of their respective bacterial host at 106 – 107 cfu/ml and incubated at 37°C (non-Dickeya phages) or 28°C (Dickeya phages) until lysis occurred (culture visibly cleared). The resulting lysate was clarified further by adding 0.5 to 2% (v/v) of chloroform, decanting, centrifugation and filtration of the supernatant (0.2 μm pore size). Burkholderia phage phi208 was amplified by confluent lysis on LB agar plates at 37°C.
2.2. Concentration using CIM® monoliths
Three different buffer systems were used for loading the phages on the columns: Tris(a) buffer (20 mM Tris-HCl, pH 7.5), Tris(b) buffer (50 mM Tris-HCl, pH 7.5, 8 mM MgSO4) or phosphate buffer (125 mM Na2HPO4, pH 7.2). For elution, 1 to 2 M NaCl was added to the loading buffer, depending on the phage. The anion-exchange chromatography columns used were the CIM® QA and DEAE disks, the CIMacTM QA column and the CIM® QA-8f mL Tube Column (BIA Separations, Ljubljana, Slovenia). The columns were attached to an ÄKTA™ FPLC™ system (GE Healthcare, Little Chalfont, UK) with a P900 pump system and analyzed with UNICORN™ 5.01 software.
2.3. Phage enumeration
Phages were enumerated with plaque assays using the traditional agar-overlay method (Adams, 1959).
3. Results
3.1. Purification of bacteriophages using CIM® monolithic columns
A set of phages (Table 1), with different morphologies and which infect different hosts, was concentrated and purified on CIM® monolithic columns. Dickeya phages LIMEstone1 and LIMEstone2, Pseudomonas phages ∅15 and LUZ19, Staphylococcus phage ISP and Burkholderia phage phi208 were tested on the laboratory scale anion-exchange columns, the CIM® QA Disk Monolithic Column (QA: strong anion exchanger) and/or the CIM® DEAE Disk Monolithic Column (DEAE: weak anion exchanger). Pseudomonas phage ∅Paer4 was tested on the CIMac™ QA-0.1 mL Analytical Column and the industrial scale CIM® QA-8f mL Tube Monolithic Column. The other Pseudomonas phages ∅E2005-A, ∅Paer14, ∅E2005-C and ∅M4 were tested on the CIM® QA-8f mL Tube Monolithic Column.
Optimizing the purification of a phage with anion-exchange chromatography is a stepwise process, in which different parameters need to be taken into consideration, e.g. binding, elution, capacity of the column and phage recovery.
3.1.1. Binding conditions
In the first step, the specific binding conditions for each phage were determined, using a one-step gradient loading and elution approach. A small volume of phage suspension was loaded on a column (usually 2 ml) and the flow-through (FT) fraction was collected as a whole. The particles were eluted in one step with 100% elution buffer and this fraction (E) was also collected. The aim was to have no phage in the FT fraction. Various methods to achieve binding can be used. (1) For phages ISP and LIMEstone1, this was accomplished by simply loading filtered (0.22 μm) lysate onto the QA and DEAE disks. (2) The other phages had to be diluted in their respective loading buffers (Table 1) to reduce ionic strength and promote binding of the phage particles on the column matrix or dilute lysate proteins which might bind to the matrix. (3) In the case of phage phi208, the phage suspension was dialyzed against the loading buffer.
3.1.2. Elution
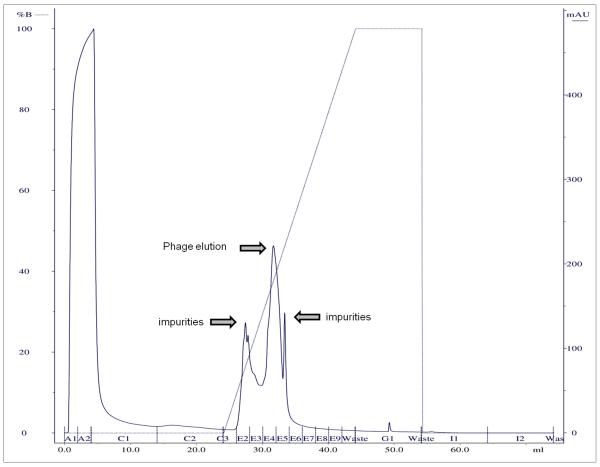
In a next step, a linear elution gradient was used to calculate the most optimal concentration of elution buffer, i.e. the NaCl concentration of the buffer (Figure 1, Table 1). Again, a small volume of phage suspension was loaded on a column under conditions as optimized in the first step. The E fractions were divided among the elution gradient and the corresponding phage titers were determined. Using a loading system with UV or conductivity detectors, peaks were visible when phages and/or impurities were eluted. Combining this information with the titers of the E fractions, the concentration of elution buffer could be calculated for washing away impurities and for the actual elution of phage. For the example of phage ISP in Figure 1, phage elution started at approximately 35% buffer B and therefore 40% buffer B was chosen for phage elution. A higher elution concentration might have had a higher yield of phage, but purity decreases as more unwanted particles are co-eluted. NaCl elution concentrations ranged between 0.25 M for phages ∅E2005-A and ∅Paer14, and 0.8 M for phage ISP; elution for the other phages was intermediate (Table 1). For complete elution of all particles after purification, 1 M of NaCl was sufficient for phages LIMEstone1, LIMEstone2, ∅15 and phi208; for ∅Paer4, ∅E2005-A, ∅E2005-C and ∅Paer14 1.5 M NaCl was used; phages LUZ19 and ISP required 2 M NaCl.
Figure 1.
Linear gradient output diagram of the purification of phage ISP.
3.1.3. Step gradient, capacity and recovery
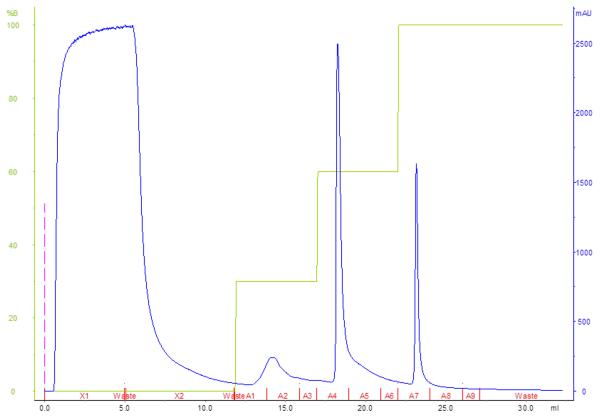
Based on the data previously collected, a step-wise gradient was designed for each phage, using different concentrations of the elution buffer to wash away impurities, elution of phage and removal of the remaining phage and impurities of the column (example in Figure 2). Depending on the delay of the FPLC system (between the UV detector and the fractionator), the concentration of elution buffer used, was somewhat lower than initially expected from the linear gradient. This could be minimalized by lengthening the linear gradient to better calculate the concentration of elution buffer for each step. From this step-wise gradient, the capacity of the column could be determined by loading an excess of phage and collecting the FT in different fractions. Different phages gave markedly different capacities on the same columns. For example, the laboratory scale DEAE disk could bind over 2 × 1011 pfu of LIMEstone2 (2.1 × 1011 pfu added to disk, capacity not reached), but only 8 × 1010 pfu of ∅15 (9.3 × 1010 pfu added, capacity reached). Often, on a laboratory scale, the maximum capacity of the columns was not reached as the limiting factor was the phage amplification step combined with the amount of suspension that could be loaded on the system. In the case of Pseudomonas phage ∅Paer4, the analytical QA column had a maximum capacity of 1 × 109 pfu, which was deemed too low and the process was scaled up to the industrial scale QA-8f tubes. Here, a yield of 1 × 1012 pfu was achieved, similar to that of ∅E2005-A, ∅Paer14, ∅E2005-C and ∅M4. This 1000-fold higher yield between the columns is only partly explained by their difference in volume and we suspect that the matrix and column build are also responsible (disk versus tube, respectively).
Figure 2.
Step gradient output diagram of the purification of phage LIMEstone2 on a DEAE disk. Phage were eluted in fractions A4 and A5.
The recovery of phages in the pure elution fraction was also calculated as the ratio of total phage found in the pure fractions to the total number of phage loaded on the column (Table 1). Generally, a considerable loss of phage was witnessed, from 30 to 65% of phages could be washed away in the FT fractions, in other E fractions or were too strongly bound to the column matrix. Only phage LIMEstone1 showed a recovery of 99.9%.
4. Discussion
A number of observations can be made from comparing the different bacteriophages and the different columns.
Considering the results for the different phages, it is clear that the protocol needs to be optimized for each phage individually. The protocol may be almost identical for similar phages, for example Pseudomonas phages ∅E2005-A, ∅Paer14 and ∅M4, or the optimal conditions might have other column chemistries and buffer conditions, as for Dickeya phages LIMEstone1 and LIMEstone2.
For each phage, the appropriate column type and volume needs to be chosen, depending on the required titer of the end product. The laboratory scale colums of 0.34 ml used in this study gave yields of 3 to 5 ml of 1011 to 1012 pfu/ml (LIMEstone1, LIMEstone2, LUZ19) which is sufficient for most small scale experiments. The analytical scale column CIMac™ QA produced a lower titer than desirable for phage ∅Paer4 (maximum capacity of 1 × 109 pfu/column, while at least 1011 pfu/column is necessary) and the process was successfully scaled up to the industrial scale 8 ml CIM® QA-8f column with the same optimized conditions. Consequently, it is always possible to first optimize the protocol on a laboratory scale or analytical scale column, and then a larger volume industrial scale column can be used for large-scale applications. The anion-exchange columns can be used more than once, although, after multiple usages it was noted that the capacity may sometimes be reduced. A new CIM® DEAE laboratory scale column had a capacity of more than 1.2 × 1012 LUZ19 phage particles, whereas an older column, which was used several times for different phages, could not retain more than 9.7 × 1010 pfu. Perhaps some particles are bound too strongly to the column matrix and cannot be eluted, even when using a high molarity of NaCl solution. To keep the columns in optimal condition, it is recommended to regenerate the counter-ions before and after every use, according to manufacturer’s instructions.
Loss of phage titer was almost always observed during the purification process. This was also observed by Smrekar et al. (2008) and Kramberger et al. (2010) with recoveries which ranged from 60 to 70%. In our study, recovery seems lower for some phages, but we looked at the purest elution fraction alone, and disregarded phage loss in the other fractions. It has proven possible to increase capacity to 100% by drastically reducing the NaCl concentration at loading (Smrekar et al. 2010). However, this also increases the volume of phage suspension when diluting or adds an extra step when dialyzing. Loss of phage can also consistently occur for CsCl-gradient centrifugation purification. This can happen at many stages in the purification process, before the centrifugation step during PEG precipitation and/or resuspension, during centrifugation because of interaction with CsCl or in the dialysis step after centrifugation. Moreover, for phages ∅15, LIMEstone1 and LIMEstone2 the latter resulted in dramatic phage losses of up to 5 orders of magnitude. For these phages, the use of the monoliths is an excellent alternative purification method on a laboratory scale.
In principle, it should be possible to separate a mixture of two phages with different elution conditions on the anion-exchange columns. However, for a number of phages a small titer of residual phage particles (103 to 105 pfu/ml) was found in most of the fractions of the elution process (LIMEstone1, LIMEstone2, ISP, LUZ19 and ∅Paer4). Use of the columns for this purpose holds therefore the risk of contamination. When reusing columns with different phages, washing with 1 M NaOH proved to remove all viable phage particles from the matrix and from the FPLC system.
A comparison between the anion-exchange chromatography method using CIM® monoliths and traditional CsCl gradient ultracentrifugation, factors in many parameters. When looking at yield only, CsCl purification can generally reach higher yields per sample than the 0.34 ml and 8 ml columns used in this study. However, because of the centrifugation step in the CsCl method, the volume of phage suspension used in each sample is constricted, while for the chromatography method an unlimited volume of phages can be loaded on each column (with the appropriate FPLC or HPLC pump). This offers an extra advantage for phages which do not amplify well in the previous step of liquid or plate amplification. Also, the CIM® monoliths’ scalability under previously optimized conditions would permit higher titers to be reached when using the larger industrial-scale columns which were not investigated in this study.
After the optimization process, the chromatographic method is faster than CsCl purification. Layering of CsCl gradients is a time-consuming process, followed by a centrifugation step that lasts 1 to 3 hours, finishing with dialysis of the phage suspension which in turn takes several hours. Starting from loading the phage suspension on a column, the whole chromatography process usually does not take longer than an hour, depending on the volume loaded and flow rate used, and the resulting phage elution suspension can be stored directly.
When looking at the price tag, both methods require an expensive piece of equipment, an HPLC or FPLC for chromatography and an ultracentrifuge for CsCl purification. Apart from that, the amount of CsCl needed to process one phage sample is cheaper than one column, but the latter can be reused a number of times, making it cheaper after several reuses.
In conclusion, the technique of anion-exchange chromatography with CIM® monoliths offers a valid alternative for traditional benchtop purification methods, especially for phages which prove to be unstable in these traditional methods. Additionally, the columns are easily scalable without the need for further optimization. Drawbacks are a noticeable loss of phage during the purification process and a potentially long optimization process. Therefore, the decision to use this method needs to be made for each phage separately.
Table 2.
Bacteriophages purified with CIM® monolithic columns.
| Phage name | Morphology | Host | Isolated from | Troubles with CsCI gradient purification | Reference |
|---|---|---|---|---|---|
| LIMEstonel | Myoviridae (Vil-like) | “Dickeya solani” | soil | Drop in titer (up to 105pfu) after CsCl purification | This study |
| LIMEstone2 | Myoviridae (Vil-like) | ||||
| ∅15 | Podoviridae (T7-like) | Pseudomonas aeruginosa | Drop in titer (up to 103 pfu) after CsCl purification | This study | |
| Ac6, Ac7 | Acinetobacter baumannii | ? | No band is formed in CsCl gradient | M Merabishvili, personal communication | |
| LDR2 | P. aeruginosa | Water? | Drop in titer after CsCl purification | PJ Ceyssens, personal communication | |
| 0305∅8-36 | Myoviridae (giant) | Bacillus thuringiensis | Drop in titer after CsCl purification due to tail sheath contraction | Pathria et al, 2012 | |
| G | Myovirodae (giant) | B. megatherium | Soil | Serwer |
Acknowledgements
EMA and AC would like to thank Frenk Smrekar for his advice with the optimization of the purification protocols for LIMEstone1 and ∅15. DV and AC were funded by a PhD grant of the Agency for Innovation by Science and Technology (IWT) Flanders. SML and AJG thank Dr. Thomas Barker (Georgia Institute of Technology) for use of his lab’s FPLC equipment and funding from the Wallace H. Coulter Translational/Clinical Research Grant Program.
Abbreviations
- CIM
Convective Interactive Media
- QA
quaternary amine
- DEAE
diethyl amine
- FT fraction
flow-through fraction
- E fraction
elution fraction
Reference List
- Adams MH. Bacteriophages. Interscience Publishers Inc.; New York: 1959. pp. 14–15. [Google Scholar]
- Adriaenssens EM, Van Vaerenbergh J, Vandenheuvel D, Dunon V, Ceyssens PJ, De Proft M, Kropinksi AM, Noben JP, Maes M, Lavigne R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by ‘Dickeya solani’. PLoS.One. 2012;7:e33227. doi: 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger P. Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from ghost particles. In: Clokie MR, Kropinksi AM, editors. Bacteriophages: Methods and Protocols. Humana Press; New York: 2009. pp. 227–238. 2009. [DOI] [PubMed] [Google Scholar]
- Carlson K. Appendix: Working with bacteriophages: common techniques and methodological approaches. In: Kutter EM, Sulakvelidze A, editors. Bacteriophages: biology and application. CRC Press; Boca Raton, FL: 2005. 2005. [Google Scholar]
- Ceyssens PJ, Noben JP, Ackermann HW, Verhaegen J, De Vos D, Pirnay JP, Merabishvili M, Vaneechoutte M, Chibeu A, Volckaert G, Lavigne R. Survey of Pseudomonas aeruginosa and its phages: de novo peptide sequencing as a novel tool to assess the diversity of worldwide collected viruses. Environ.Microbiol. 2009;11:1303–1313. doi: 10.1111/j.1462-2920.2008.01862.x. [DOI] [PubMed] [Google Scholar]
- Cornelissen A, Ceyssens PJ, T’Syen J, Van Praet H, Noben JP, Shaburova OV, Krylov VN, Volckaert G, Lavigne R. The T7-related Pseudomonasputida Phage phi15 displays virion-associated biofilm degradation properties. PLoS ONE. 2011;6:e18597. doi: 10.1371/journal.pone.0018597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creaser EH, Taussig A. The purification and chromatography of bacteriophages on anion-exchange cellulose. Virology. 1957;4:200–208. doi: 10.1016/0042-6822(57)90057-0. [DOI] [PubMed] [Google Scholar]
- Gill JJ, Hyman P. Phage choice, isolation and preparation. Curr. Pharm. Biotechnol. 2010;11:2–14. doi: 10.2174/138920110790725311. [DOI] [PubMed] [Google Scholar]
- Kramberger P, Honour RC, Herman RE, Smrekar F, Peterka M. Purification of the Staphylococcus aureus bacteriophages VDX-10 on methacrylate monoliths. J.Virol.Methods. 2010;166:60–64. doi: 10.1016/j.jviromet.2010.02.020. [DOI] [PubMed] [Google Scholar]
- Lavigne R, Ceyssens PJ, Robben J. Phage proteomics: applications of mass spectrometry. Methods Mol.Biol. 2009;502:239–251. doi: 10.1007/978-1-60327-565-1_14. [DOI] [PubMed] [Google Scholar]
- Lindberg RB, Latta RL. Phage typing of Pseudomonas aeruginosa: clinical and epidemiologic considerations. J.Inf.Dis. 1974;130:S33–S42. doi: 10.1093/infdis/130.supplement.s33. [DOI] [PubMed] [Google Scholar]
- Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov VN, Mast J, Van Parys L, Lavigne R, Volckaert G, Mattheus W, Verween G, De Corte P, Rose T, Jennes S, Zizi M, De Vos D, Vaneechoutte M. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS.One. 2009;4:e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puck T, Sagik B. Virus and cell interaction with ion exchangers. J. Exp. Med. 1953;97:807–820. doi: 10.1084/jem.97.6.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining X-Ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwer P, Graef PR, Garrison PN. Use of ethidium bromide fluorescence enhancement to detect duplex DNA and DNA bacteriophages during zone sedimentation in sucrose gradients: molecular weight of DNA as a function of sedimentation rate. Biochemistry. 1978;17:1166–1170. doi: 10.1021/bi00600a005. [DOI] [PubMed] [Google Scholar]
- Shaburova OV, Krylov SV, Veiko VP, Pleteneva EA, Burkal’tseva MV, Miroshnikov KA, Cornelissen A, Lavigne R, Sykilinda NN, Kadykov VA, Mesianzhinov VV, Volckaert G, Krylov VN. Search for destruction factors of bacterial biofilms: comparison of phage properties in a group of Pseudomonas putida bacteriophages and specificity of their halo-formation products. Genetika. 2009;45:185–195. [PubMed] [Google Scholar]
- Smrekar F, Ciringer M, Peterka M, Podgornik A, Strancar A. Purification and concentration of bacteriophage T4 using monolithic chromatographic supports. J.Chromatogr.B. 2008;861:177–180. doi: 10.1016/j.jchromb.2007.05.048. [DOI] [PubMed] [Google Scholar]
- Smrekar F, Ciringer M, Strancar A, Podgornik A. Characterisation of methacrylate monoliths for bacteriophage purification. J.Chromatogr.A. 2011;1218:2438–2444. doi: 10.1016/j.chroma.2010.12.083. [DOI] [PubMed] [Google Scholar]
- Taussig A, Creaser EH. Chromatographic purification of T2r bacteriophage. Biochim. et Biophys. Acta. 1957;24:448–449. doi: 10.1016/0006-3002(57)90230-5. [DOI] [PubMed] [Google Scholar]
- Vandersteegen K, Mattheus W, Ceyssens PJ, Bilocq F, De Vos D, Pirnay JP, Noben JP, Merabishvili M, Lipinska U, Hermans K, Lavigne R. Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS ONE. 2011;6:e24418. doi: 10.1371/journal.pone.0024418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walin L, Tuma R, Thomas GJ, Jr., Bamford DH. Purification of viruses and macromolecular assemblies for structural investigations using a novel ion exchange method. Virology. 1994;201:1–7. doi: 10.1006/viro.1994.1259. [DOI] [PubMed] [Google Scholar]
- Yamamoto KR, Alberts BM, Benzinger R, Lawhorne L, Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970;40:734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]