Abstract
The venom peptides (i.e., conotoxins or conopeptides) that species in the genus Conus collectively produce are remarkably diverse, estimated to be around 50,000 to 140,000, but the pace of discovery and characterization of these peptides have been rather slow. To date, only a minor fraction have been identified and studied. However, the advent of next-generation DNA sequencing technologies has opened up opportunities for expediting the exploration of this diversity.
The whole transcriptome of a venom duct from the vermivorous marine snail C. pulicarius was sequenced using the 454 sequencing platform. Analysis of the data set resulted in the identification of over eighty unique putative conopeptide sequences, the highest number discovered so far from a Conus venom duct transcriptome. More importantly, majority of the sequences were potentially novel, many with unexpected structural features, hinting at the vastness of the diversity of Conus venom peptides that remains to be explored. The sequences represented at least 14 major superfamilies/types (disulfide- and non-disulfide-rich), indicating the structural and functional diversity of conotoxins in the venom of C. pulicarius. In addition, the contry-phans were surprisingly more diverse than what is currently known. Comparative analysis of the O-superfamily sequences also revealed insights into the complexity of the processes that drive the evolution and diversification of conotoxins.
Keywords: Conotoxin, Conopeptide, Toxin, Transcriptome
1. Introduction
The venom of marine gastropods (members of the genus Conus, also known as cone snails) contains a mixture of diverse, small, highly-structured peptides commonly referred to as conotoxins or conopeptides which, when injected by the snail into its target (primarily prey, but could also be their predators and competitors), bind to specific molecular receptors in the envenomated target. This results in the disruption of specific physiological processes in the target and elicits physiological effects such as paralysis. It is estimated that each Conus species produces 100–200 different venom peptides, and that there is little or no overlap in the specific kinds of peptides that the different species produce (Olivera, 2002).
Determining the inventory of peptides in the venoms of cone snails is interesting both from a biological and biomedical/biotechnological perspective. Because each species has its own repertoire of peptides that reflect its ecological niche, identification and enumeration of the peptides in the venom may thus provide a “molecular readout” of each species’ biotic interactions (Olivera, 2002 and references cited therein). This molecular-level information provides insights on various aspects of the species’ biology, ecology, and evolution and facilitates studies on their “exogenome” (Olivera, 2006), including the evolution of the toxins and the molecular mechanisms that generate their diversity.
On the other hand, considering the enormous potential of conotoxins as lead compounds or drugs (Terlau and Olivera, 2004; Olivera and Teichert, 2007), an inventory of peptides in Conus venoms (i.e., the “venome”) would facilitate a systematic investigation of venome components and of their pharmacological properties and thus would significantly facilitate the identification of drug leads if not development of biomedical applications. Indeed, a number of these peptides are currently in advanced stages of clinical trials while others have become established experimental tools in pharmacological research (Olivera, 2006).
Because the peptides are encoded by genes and are synthesized in a specialized toxin-producing tissue, the venom duct (see Olivera, 2002), the cloning and sequencing of clones from venom duct cDNA libraries (i.e., the transcriptome) have become one of the methods of choice in the discovery of novel venom peptides. Thus, hundreds of Conus peptides have been discovered using this “transcriptomics” approach (e.g., Conticello et al., 2001; Garrett et al., 2005; Holford et al., 2009; Peng et al., 2006, 2007; Pi et al., 2006a, b; Liu et al., 2009). The advent of the next generation sequencing technologies (Margulies et al., 2005; Schuster, 2008), however, provides a means for accelerating the transcriptomics-based approach. In particular, shotgun sequencing of transcriptomes can theoretically reveal a complete or near-complete inventory of conotoxin genes expressed in the venom duct.
In this study, we utilized the 454 next generation sequencing platform (Margulies et al., 2005) to carry out whole-transcriptome sequencing of the venom duct of the tropical vermivorous gastropod Conus pulicarius. The sequences were then analyzed to identify putative conotoxins in the venom of this species.
2. Materials and methods
2.1. mRNA extraction from C. pulicarius venom duct
The venom duct from C. pulicarius was kindly provided by Dr. Jason S. Biggs. The tissue was harvested and stored in RNAlater (Ambion, Austin, Tx) as described in Biggs et al. (2008). Total RNA was isolated using TRIzol Plus RNA purification system (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendation.
2.2. cDNA synthesis and whole-transcriptome shotgun sequencing
cDNA was synthesized from total RNA using the SMART cDNA Library kit (Clontech) following the manufacturer’s recommendations, except that, instead of the primers provided in the kit, the following primers were used: (a) modified CDSIII/3′ cDNA Synthesis Primer, 5′ TAG AGA CCG AGG CGG CCG ACA TGT TTT GTT TTT TTT TCT TTT TTT TTT VN-3′, and (b) modified CDSIII/3′ PCR Primer, 5′ TAG AGG CCG AGG CGG CCG ACA TGT TTT GTC TTT TGT TCT GTT TCT TTT VN-3′. The generated cDNA was sequenced using the GS-FLX instrument (Roche, IN, USA) following the manufacturer’s instructions (supplied with the system/kits).
2.3. Sequence processing and analysis
The raw sequence reads were processed to remove primer sequences. The primer-trimmed sequences were then assembled on a small cluster of computers running Linux using the Forge-G assembler software (http://www.cebitec.uni-bielefeld.de/forge/wiki/ForgeG, version 20070801) and the LAM/MPI message passing library (http://www.lam-mpi.org/). Forge-G was chosen for its modest memory requirements and its demonstrated ability to assemble 454 sequence data.
The resulting sequences/contigs were then analyzed primarily through comparison with similar sequences in the Swissprot database. Searches for similar sequences were carried out using the BLAST (Basic Local Alignment Search Tool) software (Altschul et al., 1990). Standalone BLAST executables were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/) and installed on local desktop computers. A reference database was constructed by adding selected non-redundant conotoxin sequences (downloaded from the Conoserver database http://research1t.imb.uq.edu.au/conoserver/) to a local copy of the UniProtKB/Swiss-Prot Database (release 15.4, downloaded from the UniProt web site, http://www.uniprot.org/downloads). This reference database was formatted using the formatdb software from the downloaded copy of the BLAST executables. Searches for similar sequences in the reference database were made using the blastx option of blastall which was run locally; the output files (in XML format) were processed using custom Python scripts to identify the contigs with hits to conotoxin sequences and to generate files that display the alignment of the sequences of the conotoxin hits with the full sequences of the contigs (translated from three reading frames). These contigs (including those with low scores) were then assigned into categories using the classification of the highest-scoring conotoxins that matched the contigs as a guide to facilitate sequence alignment and comparison. Where necessary, the full precursor sequences of the best-matching conotoxins (the reference sequences) were manually added to the alignment. The alignments were individually inspected to evaluate their quality; the sequence of the reference conotoxins was used as guide to detect frameshifts and to infer the correct translation of the sequences. Sequences that appear to be good conotoxin candidates on the basis of sequence similarity or structural characteristics (i.e., presence and arrangement of multiple Cys residues) were then subjected to multiple sequence alignment (in separate groups according to presumed conotoxin type) and based on this alignment unique peptide sequences which were either full-length or nearly full-length were identified and compiled into a non-redundant list.
To analyze the diversification of the O-superfamily sequences in Conus, all O-superfamily sequences (mature peptide region) in the Conoserver database were downloaded and, together with the C. pulicarius O-superfamily mature-region sequences generated in the study, were aligned using the software MUSCLE (Edgar, 2004). The resulting alignment was separated into clusters based on overall sequence similarity and length, and the sequence alignment in each cluster was then refined by eye. To generate a cladogram for the species represented in the O-superfamily dataset, 16S rRNA gene sequences for these species were downloaded from GenBank and aligned using the software ClustalW (Larkin et al., 2007). The cladogram was then constructed through Maximum Likelihood analysis as implemented in the software PhyML (Guindon and Gascuel, 2003). The following options were used: Subtree Pruning and Regrafting for the tree topology search algorithm and GTR+Γ+I (discrete gamma model with 4 categories) as the model of nucleotide substitution. Where the option is allowed, the other parameters were set to be optimized by the software.
3. Results and discussion
3.1. Identification of conotoxin sequences from the sequencing reads
Using the 454 Next-Generation DNA sequencing technology, sequencing of the C. pulicarius venom duct transcriptome library yielded 359,213 DNA reads and associated quality scores (minimum length: 36, median length: 228, maximum length: 393, total yield: 73,502,057 nucleotides). Primer trimming reduced this data set to 333,478 reads (minimum length: 30, median length: 186, maximum length: 393, total yield: 52,886,072 nucleotides). A total of 81,668 contigs were assembled from these sequence reads. The frequency distributions of the lengths and average read coverage (per bp) of the contigs are shown in Tables 1 and 2. Majority of the contigs (~98%) were less than 300 bp in length, and those that were longer than 500 bp comprised less than 1% of the total. Majority of the contigs (>99%) had relatively low average read coverage (<20), with those having an average read coverage of only 1 accounting for a major proportion (86%) of the total. Contigs with a relatively high average read coverage (>200) accounted for only a minuscule proportion (0.13%) of the total.
Table 1.
Frequency distribution of the lengths of the contigs.
| Length (bp) | Number of contigs | % of total | % of total, cumulative |
|---|---|---|---|
| ≤100 | 50,379 | 61.688 | 61.688 |
| >100, ≤200 | 18,535 | 22.696 | 84.383 |
| >200, ≤300 | 11,294 | 13.829 | 98.212 |
| >300, ≤500 | 1020 | 1.249 | 99.461 |
| >500, ≤1000 | 324 | 0.397 | 99.858 |
| >1000 | 116 | 0.142 | 100.000 |
Table 2.
Frequency distribution of the average read coverage of the contigs.
| Ave. read coverage (per bp) | Number of contigs | % of total | % of total, cumulative |
|---|---|---|---|
| 1 | 70,430 | 86.239 | 86.239 |
| >1, ≤10 | 10,239 | 12.537 | 98.777 |
| >10, ≤20 | 467 | 0.572 | 99.349 |
| >20, ≤100 | 423 | 0.518 | 99.867 |
| >100, ≤200 | 77 | 0.094 | 99.961 |
| >200, ≤400 | 29 | 0.036 | 99.996 |
| >400 | 3 | 0.004 | 100.000 |
Of the 81,668 contigs, 1567 showed high similarity at the amino acid sequence level with conotoxin sequences in our reference database (construction of this reference database is described in the Materials and methods section). After evaluation of the scores and the quality of the match and comparison of the deduced peptide sequences, 82 unique putative conotoxin sequences were identified. Majority of these sequences were full-length but some were truncated at the N-terminus and a few at the C-terminus. A few were also identical with respect to the mature region but were considered unique owing to some divergence at the pre-pro region. A number of other sequences showed some sequence similarity with conotoxins but were either too short (hence could not be reliably identified as conotoxin sequences) or were duplicates of the selected representatives.
Inference of the peptide sequence from the nucleotide sequence of a large number of contigs (38 of the 82, or 46.3%) required reading from more than one translation frame, which was apparent upon alignment and comparison of the translation of the sequences from three reading frames with the sequence of the reference conotoxin.
Of the 82 unique sequences, only three (peptides #82290, #9860 and #70172; see Supplementary data) were found to be identical to previously known conotoxin sequences (Pu5.5 precursor, Pu6.1 precursor, and PuIIA precursor, respectively), all of which were conotoxins from the same species used in this study, C. pulicarius. The rest of sequences (i.e., 78) differed in at least one position from the highest-scoring conotoxin sequence and hence were considered novel, although the actual number of truly novel sequences could be lower considering the possibility that the observed single-residue mutations could be sequencing artifacts. One of the sequences, peptide #73307, was found to have a putative mature region that is identical in sequence to that of the Conus arenatus conotoxin ArMMSK-01, but two substitutions were observed in the preproregion.
3.2. Diversity of conotoxins: comparison of transcriptomes
The number of unique conotoxin sequences identified in this study, i.e., 82, was the highest so far reported for Conus transcriptome sequencing (Table 3, last column). The highest number reported by other studies was 42 (Pi et al., 2006b). Except for the study by Hu et al. (2011), these studies were all based on the cDNA-cloning and sequencing approach, which could account for the relatively lower number of conotoxins observed. Hu et al. (2011) employed the next generation sequencing approach (but different sequencing platform) but identified only 30 unique sequences owing to problems related to the quality of the data (other candidate conopeptide sequences were observed but were too short to be reliably identified).
Table 3.
Comparison of the number of unique conotoxin (peptide) sequences in the venom duct transcriptomes of different Conus species identified via whole transcriptome shotgun sequencing (this study) or EST sequencing (other studies). NGS, Next-Generation Sequencing Technology.
| Species | Number | Sequencing approach | Reference |
|---|---|---|---|
| C. arenatus | 29 | Sequencing of individual cDNA clones | Conticello et al. (2001) |
| C. pennaceus | 21 | Sequencing of individual cDNA clones | Conticello et al. (2001) |
| C. textile | 30 | Sequencing of individual cDNA clones | Conticello et al. (2001) |
| C. striatus | 19 | Sequencing of individual cDNA clones | Pi et al. (2006a) |
| C. litteratus | 42 | Sequencing of individual cDNA clones | Pi et al. (2006b) |
| C. leopardus | 7 | Sequencing of individual cDNA clones | Remigio and Duda (2008) |
| C. bullatus | 30a | NGS | Hu et al. (2011) |
| C. pulicarius | 82 | NGS | This study |
Other peptide sequences were observed but were too short to be reliably identified.
All identified putative conotoxin sequences were classifiable into various currently recognized conotoxin Cys frameworks/superfamilies, primarily using the Conoserver classification scheme as reference, based on their similarity to previously described conotoxins. This was expected as the screening procedure was designed to identify candidate conotoxins based on general sequence similarity to the known ones. Thus, these sequences showed a high degree of conservation with respect to the prepro region of the reference sequences, and Cys residues were observed to form a pattern that correspond to known conotoxin Cys frameworks. In many sequences, residues in the inter-Cys loops also showed similar, if not conserved, characteristics with respect to corresponding residues in the known conotoxins. However, for most sequences, the putative mature regions exhibited considerable diversity and were moderately to highly divergent not only in the sequence but also length of the inter-Cys regions. In fact, a number of the putative novel conopeptides appeared to be sufficiently distinct that they could potentially represent new conotoxin groups.
The sequences were assigned to the different conotoxin superfamilies, as shown in Table 4 (last column). We found representatives from 14 conotoxin superfamilies, indicating the high diversity of putative conotoxins in the C. pulicarius venom duct transcriptome. The most well-represented group was the O-Superfamily, with 41 sequences accounting for 46% of the total unique sequences observed; these sequences were further classified into the O1-Superfamily (36 sequences) or O3-Superfamily (5 sequences). Sequences representing the M-, P-, T-, I-, and V-Superfamilies as well as the non-disulfide-rich groups Conantokin, Contulakin, Contryphan and Conkunitzin, were also observed.
Table 4.
Comparison of the relative frequency of various conotoxin types (according to Cys framework) found in various Conus venom duct transcriptomes.
| Framework | Cys pattern | Super-family | C.str1 | C.lit2 | C.tex3 | C.pen3 | C.are3 | C.bul4 | C.pul5 |
|---|---|---|---|---|---|---|---|---|---|
| I | CC-C-C | A | 5 | 3 | 10 | ||||
| III | CC-C-C-CC | M | 1 | 8a | 1 | 3 | 4 | ||
| V | CC-CC | T | 1 | 13 | 5 | ||||
| VI/VII | C-C-CC-C-C | O | 10 | 8b | 24 | 9 | 26 | 14 | 41d |
| VIII | C-C-C-C-C-C-C-C-C-C | S | 1 | ||||||
| IX | C-C-C-C-C-C | P | 3 | 5 | 12 | 3 | 3 | ||
| XI | C-C-CC-CC-C-C | I | 10e | ||||||
| XIV | C-C-C-C | J, L | 2 | 1 | 5 | ||||
| XV | C-C-CC-C-C-C-C | V | 1c | 1 | |||||
| XVI | C-C-CC | 1c | |||||||
| XIX | C-C-C-CCC-C-C-C-C | 1 | |||||||
| Conantokin | Non-disul3de-rich | 3 | |||||||
| Contryphan | Non-disul3de-rich | 1 | 1 | 1 | 6 | ||||
| Contulakin | Non-disul3de-rich | 2 | 1 | ||||||
| Conkunitzin | Non-disul3de-rich | 1 | 2 |
Pi et al. (2006a) (C.str, C. striatus);
Pi et al. (2006b) (C.lit, C. litteratus);
Conticello et al. (2001) (C. tex, C. textile; C.pen, C. pennaceus; C.are, C. arenatus);
Hu et al. (2011)(C. bul, C. bullatus);
this study (C.pul, C. pulicarius).
Includes sequences with 3 Cys residues;
includes sequences with 2 Cys residues.
The pre-region (i.e., signal peptide) sequence indicates that this peptide belongs to a different superfamily.
Includes O1 and O3 sequences.
Includes O1 and O3 sequences.
Table 4 also shows the comparison of the data from C. pulicarius with those from other Conus transcriptomes. Our data set was the most diverse in terms of number and conotoxin type found for a Conus venom duct transcriptome. More interestingly, the relative abundance of the various types differed among the species. O-Superfamily members appeared to be the most abundant form in C. pulicarius, Conus arenatus (a species that is phylogenetically closely related to Conus pulicarius) and in the species C. textile and C. striatus (which are relatively more phylogenetically distant). In contrast, the relatively more abundant types in Conus litteratus and Conus pennaceus were the T-Superfamily and the P-Superfamily peptides, respectively.
The relative abundance of α-conotoxins (or A-Superfamily conotoxins) also differed among these species (Table 4). Although the total number of unique peptides found in C. striatus, C. litteratus, and Conus bullatus was roughly only half (or even less) of what we found in C. pulicarius, α-conotoxins were found in these species, and in fact comprised a significant fraction in C. bullatus, whereas none was found in C. pulicarius which was sampled more extensively. This could be an artifact of sampling (i.e., this reflected the level of expression of α-conotoxin genes in the venom duct at the time it was sampled) but other explanations were also possible, such as the possibility that C. pulicarius might in general express α-conotoxin genes in relatively lower amounts in the venom duct, or that α-conotoxins are encoded by relatively fewer genes in its genome. Previous studies suggested that α-conotoxins were expressed in C. pulicarius venom ducts (Biggs et al., 2008; Yuan et al., 2007), but both studies detected the transcripts via PCR amplification (using α-conotoxin signal sequence-specific primers). Interestingly, Biggs et al. (2008) observed that α-conotoxins were expressed in the salivary gland of C. pulicarius and that those expressed in the venom duct had rather divergent signal sequences indicating relaxed evolutionary constraints. Whether this implies that the α-conotoxins are therefore poorly represented in the C. pulicarius venom duct remains to be investigated.
We also noted that the C. pulicarius venom duct transcriptome was relatively enriched for non-disulfide-rich conopeptides, i.e., Conantokin, Contulakin, Contryphan and Conkunitzin. In contrast, these peptides were either not observed or poorly represented in the other reported transcriptomes (Table 4).
The variation in the relative abundance of different conotoxins across species has important implications for studies on Conus venom peptides. It highlights the need to understand not only the functional characteristics of the individual peptides but also how the overall composition of the venom itself evolved to optimize the combined effects of the peptides’ different pharmacological activities. The importance of understanding these synergistic effects is exemplified by the concept of “toxin cabals”, i.e., groups of toxins that synergistically act together (Olivera and Cruz, 2001). As suggested by the comparison in Table 4, the sequencing and comparison of venom duct transcriptomes can yield insights into the variation of venom composition across species and could facilitate the investigation of potential synergism among the venom components.
3.3. Structural and potential functional diversity
Representative sequences (out of the 82 sequences) that we deduced via conceptual translation from the transcriptome data are shown in Fig. 1. These sequences illustrate the observed diversity of the novel peptides at the sequence level.
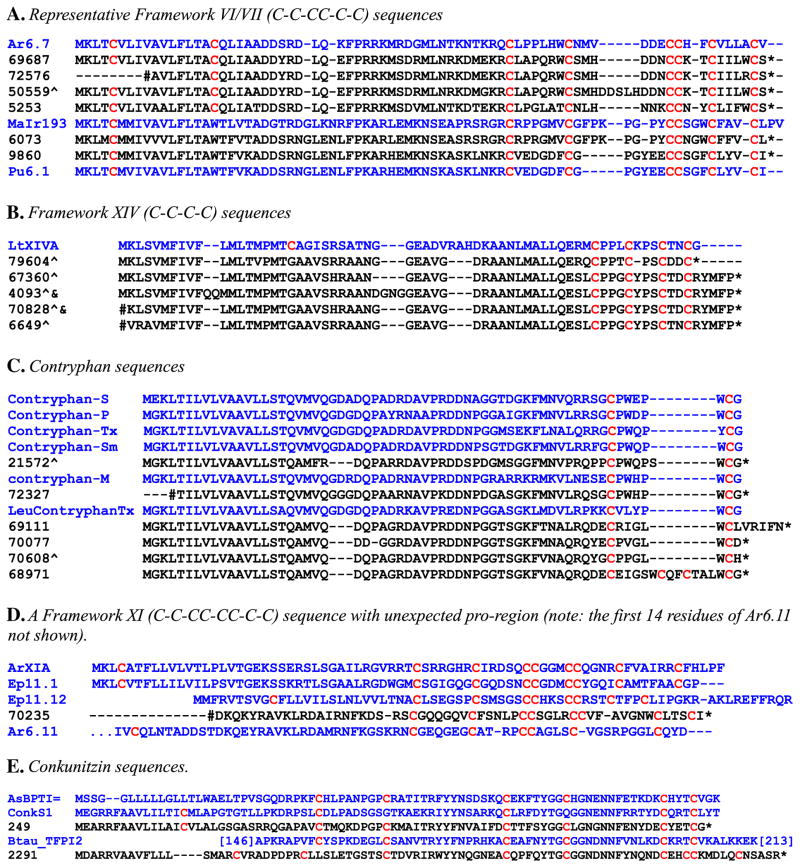
Fig. 1.
Representative conotoxin sequences from the C. pulicarius venom duct transcriptome. The putative conotoxins (in black) are aligned with reference conotoxins, i.e., precursor conopeptide sequences obtained from the reference database (see Materials and methods) to which they are most similar (in blue). The sequences are labeled using the code of the contig from which they were conceptually translated. The names of the reference sequences are from the Conoserver database except that the postfix, i.e., “precursor”, is omitted. Symbols used: ^ = conceptual translation of this contig requires frameshifting (see Materials and methods) to optimize alignment with the reference sequence; # = marks 5′-end of contig (i.e., sequence truncated at the N-terminus); & = the putative mature region of the peptide is identical to that of the peptide above it. AsBPTI*, AsBPTI-like sequence; ConkS1, Conkunitzin S1; Btau_TFPI2, Bos taurus tissue factor pathway inhibitor 2 (only residues 146–213 are shown.). Swiss-Prot Accession numbers: TFPI2, Q7YRQ8; AsBPTI*, B5L5M7.
Sequences that showed the Cys Framework VI/VII pattern are shown in Fig. 1A, aligned to reference conotoxins. The high sequence similarity between the C. pulicarius sequences and known (reference) conotoxins suggests that the peptides also share similar functional characteristics. However, notwithstanding the high similarity observed, the target receptor of the putative conopeptides could not be predicted as the target receptors of many reference conotoxins are not yet known.
A number of reference conotoxins, however, have known targets (i.e., have previously been reported to specifically bind certain receptors). Since some peptides we discovered were highly similar in sequence to these conotoxins, similar functional characteristics could be postulated. A set of sequences having the Cys Framework XIV (C-C-C-C), peptides #79604, #67360, #4093, #70828, and #6694 (alignment shown in Fig. 1B), were highly similar to conotoxin LtXIVA from C. litteratus. The synthetic form of LtXIVA had been shown to have analgesic activity in mice and to inhibit neuronal-type nicotinic acetylcholine receptors (nAchRs), which was used as basis by Peng et al. (2006) to classify the peptide as an αL-conotoxin. This raises the possibility that the new peptides could also be nAchR ligands. In fact, peptides #4093 and #70828 were identical to peptide #67360 in the sequence of their putative mature regions, and only slightly differed from the latter in the prepro-region (owing to the presence of indels).
It must be noted that these peptides, together with LtXIVA, differed from other Framework 14 conotoxins which have shorter inter-Cys loops (Zugasti-Cruz et al., 2008) and from the other J-Superfamily conotoxins which, although showing the same C-C-C-C framework, have a different sequence in the prepro-region (Imperial et al., 2006); hence, the new peptides likely belong to a new superfamily. We refer to this new conotoxin group as the J2-Superfamily rather than the L-Superfamily as proposed by Peng et al. (2006) to emphasize the structural similarity (i.e., Framework XIV Scaffold) of the two superfamilies.
Among the putative contryphans, which were identified based on high sequence similarity to known contryphans (Fig. 1C), an unexpectedly high level of diversity was observed. Six unique sequences showed greater predicted sequence diversity in this species than has been reported so far for all other Conus species combined. Most previously described contryphans fall into two well-defined classes, the standard contryphans and the Leu contryphans; peptides in both classes are highly post-translationally modified, a notable feature being the presence of either D-Trp (in standard contryphans) or D-Leu (in Leu contryphans). The putative C. pulicarius contryphan complement includes one standard contryphan (peptide #72327) and a second standard contryphan (peptide #21572) that is unusual in having an extra amino acid (Ser) in the otherwise conserved inter-Cys loop of five amino acids that in all standard contryphans has the sequence CPWXOW*C (where W is D-Trp, O is 3-OH Pro, and W* is either Trp or Br-Trp) and in having a pre-pro region that is shorter by three residues.
The other four members of the contryphan family were observed to be much more strikingly divergent from previously identified contryphans, and from each other, suggesting that these C. pulicarius contryphans may have a novel function. It is notable that except for the standard contryphan (peptide #72327), all other contryphan precursor sequences are shorter than standard contryphans by 3 amino acids in their propeptide region (an otherwise conserved GG/DG sequence is deleted). In the mature region of three of these sequences (peptides #69111, #70077, and #70608), the residue that is aligned with D-Trp or D-Leu is Iso, Val, or Pro, respectively; whether these are also modified into the D-isomer remains to be seen. These observations highlight the need to determine the patterns of modification in the mature contryphans in C. pulicarius venom as they may provide general insights into the mechanisms of post-translational modification of residues in the mature region (such as the isomerization of specific amino acids into their D-forms) as well as clues as to which sequence features might be the determinants, hence of use for predicting which residues are targets, of such modifications (Buczek et al., 2008).
One sequence (peptide #68971) that could be classified as a contryphan (it has a prepro sequence that is characteristic of contryphans) differed from the other putative contryphan sequences in having not two but four Cys residues in the predicted mature region (thus forming the Framework XIV pattern C-C-C-C characteristic of the J-Superfamily). The two extra Cys residues were in a seven-residue segment WCQFCTA found between the two other (conserved) Cys residues and which is absent in the other sequences. The functional consequence of this structural modification cannot be predicted based on the sequence alone hence it would be of interest to investigate its potential pharmacological activity experimentally.
A similar incongruence between the mature and the prepro region sequences was also observed in a number of other sequences. A peptide, #70235 (aligned with reference conotoxins in Fig. 1D), was observed to have the characteristic I-Superfamily Cys pattern (C-C-CC-CC-C-C) in the mature region, but the pro-region (the sequence was truncated at the N-terminus) was highly similar in sequence to that of O1-Superfamily peptide Ar6.11 (from C. arenatus). Interestingly, a similar observation, although of an opposite pattern, had previously been reported. Yuan et al. (2009) used PCR primers that were designed based on the conserved I3-Superfamily signal sequence to clone two peptide which they referred to as Pu6.1 (from C. pulicarius) and Lt6.4 (from C. litteratus). The peptides thus carried the expected signal sequences characteristic of the I3-Superfamily; however, the mature regions had only 6 Cys that form the Framework VI/VII pattern, C-C-CC-C-C, which is characteristic of the O-Superfamily.
Two sequences, peptides #249 and #2291, which showed high sequence similarity to conkunitzin-S1 (Fig. 1E), were noteworthy in that unlike Conkunitzin-S1 which has only four Cys residues in its mature region, these two sequences contain more than four Cys residues in the corresponding region. Peptide #249 contains six Cys residues in the putative mature region, the typical number in Kunitz proteins. Comparison of this peptide sequence with two other Kunitz proteins (Kunitz/BPTI-like toxin from the Australian copperhead, Austrelaps superbus, and the Tissue factor pathway inhibitor 2 [TFPI-2] from Bos taurus) also revealed high similarity in terms of sequence and length of inter-Cys loops (Fig. 1E). The second peptide, #2291, though exhibiting some sequence similarity in certain regions, appeared distinct in terms of number of Cys residues and length of inter-Cys loops.
3.4. Transcriptome sequences as basis for evolutionary insights: comparative analysis of the O-superfamily sequences
The large number of venom peptide sequences from the C. pulicarius venom duct transcriptome provided useful data for obtaining insights into the diversification of rapidly evolving genes in a biodiverse lineage. To reveal patterns of diversification, we analyzed the sequences using a comparative approach in a phylogenetic context. Because only a few sequences were obtained for most conotoxin superfamilies, our analysis focused only on the group with the most number of sequences, the O-superfamily.
We first aligned the mature-region of the C. pulicarius sequences to those of the O-Superfamily sequences from the Conoserver database (we recognized the potential biases of individual researchers/laboratories with regard to the cloning of genes or isolation of peptides but since the data in the database originated from multiple laboratories, we made the assumption that the collective effort of the different laboratories would roughly approximate a random sampling of conotoxins); the total number of sequences was 390, representing 49 Conus species. Based on overall sequence similarity, the multiple sequence alignment was then separated into sequence clusters and the alignment of each cluster was refined and columns containing only gaps were removed; 33 clusters were identified. Because of the very high degree of polymorphism of the sequences in both sequence and length, alignment of homologous residues for the whole set of sequences could not be made with confidence hence we chose to group the sequences into clusters rather than construct a phylogeny for these sequences. We then counted the number of sequences observed in each cluster for each species. To discover potential correlation between the distribution of the sequences and the phylogenetic relationships among the species, a phylogenic tree for these species was constructed based on available 16S rDNA sequences in GenBank. The results are shown in Fig. 2. Because only 47 of the 49 species in the alignment have 16S rRNA sequence in GenBank, only 47 species (and 380 sequences) were included in the analysis. The number of sequences observed in each cluster ranged from 1 to 63.
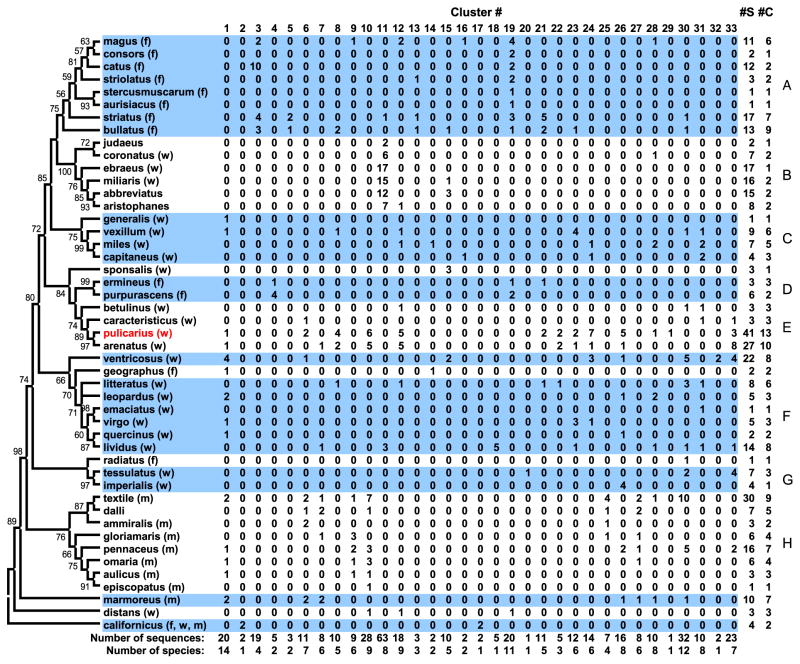
Fig. 2.
Distribution of the O-superfamily sequences from C. pulicarius and other Conus species across 33 O-superfamily sequence clusters (columns numbered 1 to 33). The procedure used to group the sequences into clusters is described in the Materials and methods. The data were obtained from Conoserver, except for the C. bullatus data which were obtained from Hu et al. (2011). Each row lists the count of O-Superfamily sequences for a particular Conus species (indicated by the row label) that fall under each O-superfamily cluster (indicated by the column number). The species are arranged according to their phylogenetic relationships as indicated by the cladogram; the type of prey is indicated inside the parentheses (f = fish, w = worm, m = mollusk). Groups/clades with 2 or more species are labeled (A through H). The numbers close to the nodes are clade support values as generated by PhyML. #C, column indicating the number of O-superfamily clusters in which a given species has 1 or more sequences; #S, column indicating the total number of sequences (summed over all clusters) for each species. GenBank Accession Numbers for 16S rRNA sequences included in the figure: C. abbreviatus, AF174140; C. ammiralis, EU682299; C. arenatus, AF103817; C. aristophanes, AY381997; C. aulicus, EU794324; C. aurisiacus, EU078943; C. betulinus, AF143999; C. bullatus, AF126016; C. californicus, AF036534; C. capitaneus, AF126014; C. caracteristicus, AF126017; C. catus, AF174154; C. consors, AF160721; C. coronatus, AF126019; C. dalli, EU078935; C. distans, AF036532; C. ebraeus, AF086613; C. emaciatus, AF126018; C. episcopatus, AF126166; C. ermineus, AF036530; C. generalis, AF160722; C. geographus, AF126165; C. gloriamaris, AF126168; C. imperialis, AF108828; C. judaeus, EU492441; C. leopardus, AF174175; C. litteratus, AF126170; C. lividus, AF086611; C. magus, EU078939; C. marmoreus, EU794330; C. miles, AF108821; C. miliaris, AF143998; C. omaria, AF108823; C. pennaceus, AF174190; C. pulicarius, AF143992; C. purpurascens, AF480308; C. quercinus, AJ717603; C. radiatus, AF160724; C. sponsalis, AF143993; C. stercusmuscarum, AF103813; C. striatus, EU078945; C. striolatus, AF174201; C. tessulatus, AF160715; C. textile, EU078936; C. ventricosus, AY726487; C. vexillum, AF108822; C. virgo, EU794334.
Two major patterns were apparent from the results. When comparison was made across the sequence clusters (species with <14 sequences were not considered as the small number of sequences might not be enough to reveal a pattern), it appeared that for some species their O-superfamily conotoxins were highly divergent and hence were spread out over at least 7 clusters; examples include C. pulicarius with 41 sequences in 13 clusters; C. arenatus, with 27 sequences in 10 clusters; Conus lividus and C. textile with 14 and 30 sequences, respectively, in 8 and 9 clusters, respectively. For other species, their O-superfamily conotoxins were highly similar, hence found mostly in only 1 cluster. Conus ebraeus, Conus miliaris, and Conus abbreviatus, which are all Clade B species, had 17, 16, and 15 O-superfamily sequences, respectively, but which were mainly found in only 1 or 2 clusters, most if not all of which were observed in only one cluster (Cluster 11).
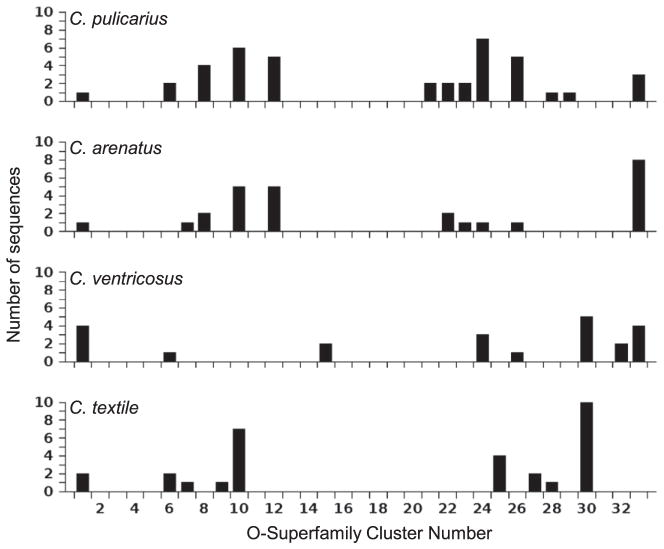
This pattern of distribution appeared to be correlated with phylogeny. We compared the data for four species of varying degree of relatedness, C. pulicarius, C. arenatus, Conus ventricosus, and C. textile (Fig. 3). These were the only species chosen because only these species have a relatively large number of sequences (41, 27, 22, and 30, respectively); the other species have fewer sequences (≤17). As Fig. 3 shows, both the distribution and relative frequency of C. pulicarius and C. arenatus sequences were very similar. In fact, 90% (9 out of 10) of the clusters where C. arenatus sequences were found also contained C. pulicarius sequences. In contrast, the distribution and relative frequency of the sequences from the other two species (C. ventricosus and C. textile) apparently differed significantly from those of either C. pulicarius or C. arenatus. Only 62.5% and 44% of the clusters where sequences from C. ventricosus and C. textile, respectively, were found were also observed to contain C. pulicarius sequences; a similar pattern was observed between these two species and C. arenatus. Moreover, the dissimilarity between C. ventricosus and C. textile is roughly equivalent to that between C. pulicarius/C. arenatus and C. ventricosus/C. textile.
Fig. 3.
Comparison of C. pulicarius with C. arenatus, C. ventricosus, and C. textile with respect to the distribution and frequency of their sequences in the different O-superfamily sequence clusters (clusters 1 to 33).
The second major pattern emerged when comparison was made across clades. From Fig. 2 it was apparent that there were O-superfamily sequence clusters in which the sequences were observed only from species of a specific clade. For example, all 16 sequences in Cluster 3 were from species that belong to Clade A, a clade of fish hunters; of the 63 sequences in Cluster 11, 94% (i.e., 59) were from species that belong to Clade B (worm-hunters). Like Cluster 11, Clusters 9, 10 and 19 contained sequences that were mainly from one clade and few sequences from species in more distant clades. In contrast, a number of clusters (e.g., Clusters 1 and 30) contained sequences from divergent clades.
These patterns could provide insights into the evolution of conotoxins. The patterns suggest that the Conus species differ in their ability to explore the sequence space to evolve novel conotoxins, i.e., some species appear to have sampled the conotoxin sequence space more extensively than the others. Another insight pertains to the ongoing diversification of conotoxins. That there are O-superfamily sequence clusters containing only sequences from closely-related species (i.e., belonging only to a single clade) while other clusters contain sequences from divergent species (i.e., belonging to divergent clades) could indicate how recently have specific groups of conotoxins evolved. Clusters in the latter category may likely represent sequences that arose in the more distant past and hence have become widely distributed in the various clades, e.g., Cluster 30 (Fig. 2) which includes 31 sequences representing majority of the clades. In contrast, there are O-superfamily sequence clusters in which the species represented in the cluster belong to only a few (i.e., ≤3) but distantly related clades, suggesting that the sequences may have evolved as a result of convergent or parallel evolution (although the possibility that this reflects loss of a specific lineage of conotoxins in the other clades cannot be discounted). C. pulicarius and C. arenatus apparently have conotoxins in this category (Cluster 10, Fig. 2). Phylogenetic analysis of the peptide sequences in Cluster 10 revealed that the C. pulicarius and C. arenatus sequences are monophyletic, forming a group distinct from that of clade H species (data not shown); this pattern is consistent with the hypothesis that the two lineages of conotoxins probably arose independently and relatively more recently, in contrast to the other conotoxins that evolved and diversified early in the evolution of the genus.
A distribution of sequences that appeared to be correlated with feeding mode is also apparent in an O-superfamily cluster, Cluster 19 (Fig. 2), which contains sequences from fish-hunting species belonging to divergent clades (Clades A and E, although a single sequence from the distantly-related species Conus distans was observed). Whether the conotoxins in this cluster have fish-specific bioactivities remains to be seen.
3.5. Other observations
The fact that we identified 82 unique (putative) conotoxin sequences from the transcriptome of C. pulicarius venom duct raises the question of whether this number, which is the highest so far reported for a Conus transcriptome, represents the entire inventory of conotoxins in the venom of this species. This question is important as it has been generally accepted that each species of Conus likely produces between 100 and 200 peptides in their venom (Terlau and Olivera, 2004). Although the number we actually observed was slightly lower, the actual number of peptides expressed could be higher. For example, as mentioned earlier, a few conopeptide sequences (likely expressed in low amounts) that are expected to be present in the C. pulicarius venom were not observed, such as the α-conotoxins (Biggs et al., 2008) and Pu1.1 (Yuan et al., 2007). In addition, three unpublished C. pulicarius sequences (referred to as L-Superfamily sequences, isolated via cDNA cloning, that are in the Conoserver database) were not detected in this study. In addition, it is also likely that a significant number of the conopeptides we identified likely undergo posttranslational modification, a common post-translational processing of conotoxins (Buczek et al., 2005a), which can give rise to two or more forms of the same peptide (and thus could be recognized as separate peaks in HPLC chromatograms). The number of conotoxin sequences we found in the transcriptome would therefore be consistent with the generally accepted estimate.
Recently, Davis et al. (2009) raised the estimate of the number of peptides in Conus venom per species 10-fold to between 1000 and 1900 based on liquid chromatography–mass spectrometry studies on the venom of three species, C. textile, Conus imperialis, and Conus marmoreus. Whether this much higher estimate is correlated with a higher number of unique conopeptide sequences in the venom duct transcriptome of these species or indicative of the variety and complexity of the post-translational peptide modification mechanisms remains to be investigated.
Our analysis also revealed a number of sequences in the transcriptome that represent other venom components. Sequences that are highly similar to the alpha and beta chains of Conodipine-M from Conus magus and proteins that are potentially involved in conotoxin biosynthesis were observed, including protein disulfide isomerase (additionally, two sequences with high similarity to bacterial disulfide bond formation proteins were also found), peptidyl-prolyl cis-trans isomerase, alpha subunits of prolyl 4-hydroxylase, Vitamin K-dependent γ-carboxylase, calreticulin and other proteins that function as chaperones, and proteins potentially involved in signal peptide cleavage (data not shown).
Supplementary Material
Acknowledgments
We thank Dr. Timothy Harkins, Genome Sequencing, Roche Applied Science, Roche, Inc. and the 454 Sequencing Center for generating the sequencing data. We would like to thank Dr. Jason S. Biggs for providing us with the venom duct. This work was supported by grant GM48677 (BMO, PB).
Appendix A. Supplementary data
Supplementary data to this article can be found online at doi:10. 1016/j.margen.2011.09.002.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Biggs JS, Olivera BM, Kantor YI. Alpha-conopeptides specifically expressed in the salivary gland of Conus pulicarius. Toxicon. 2008;52:101–105. doi: 10.1016/j.toxicon.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek O, Bulaj G, Olivera BM. Conotoxins and the post-translational modification of secreted gene products. Cell Mol Life Sci. 2005a;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczek O, Jimenez EC, Yoshikami D, Imperial JS, Watkins M, Morrison A, Olivera BM. I1-superfamily conotoxins and prediction of single D-amino acid occurrence. Toxicon. 2008;51:218–229. doi: 10.1016/j.toxicon.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Gilad Y, Avidan M, Ben-Asher E, Levy Z, Fainzilber M. Mechanisms for evolving hypervariability: the case of conopeptides. Mol Biol Evol. 2001;18:120–131. doi: 10.1093/oxfordjournals.molbev.a003786. [DOI] [PubMed] [Google Scholar]
- Davis J, Jones A, Lewis RJ. Remarkable inter- and intra-species complexity of conotoxins revealed by LC/MS. Peptides. 2009;30:1222–1227. doi: 10.1016/j.peptides.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett JE, Buczek O, Watkins M, Olivera BM, Bulaj G. Biochemical and gene expression analyses of conotoxins in Conus textile venom ducts. Biochem Biophys Res Commun. 2005;328:362–367. doi: 10.1016/j.bbrc.2004.12.178. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Holford M, Zhang MM, Gowd KH, Azam L, Green BR, Watkins M, Ownby JP, Yoshikami D, Bulaj G, Olivera BM. Pruning nature: biodiversity-derived discovery of novel sodium channel blocking conotoxins from Conus bullatus. Toxicon. 2009;53:90–98. doi: 10.1016/j.toxicon.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Bandyopadhyay PK, Olivera BM, Yandell M. Characterization of the Conus bullatus genome and its venom-duct transcriptome. BMC Genomics. 2011;12:60. doi: 10.1186/1471-2164-12-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperial JS, Bansal PS, Alewood PF, Daly NL, Craik DJ, Sporning A, Terlau H, López-Vera E, Bandyopadhyay PK, Olivera BM. A novel conotoxin inhibitor of Kv1.6 channel and nAChR subtypes defines a new superfamily of conotoxins. Biochemistry. 2006;45:8331–8340. doi: 10.1021/bi060263r. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Liu Z, Xu N, Hu J, Zhao C, Yu Z, Dai Q. Identification of novel I-superfamily conopeptides from several clades of Conus species found in the South China Sea. Peptides. 2009;30:1782–1787. doi: 10.1016/j.peptides.2009.06.036. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivera BM. Conus venom peptides: reflections from the biology of clades and species. Annu Rev Ecol Syst. 2002;33:25–47. [Google Scholar]
- Olivera BM. Conus peptides: biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Cruz LJ. Conotoxins, in retrospect. Toxicon. 2001;39:7–14. doi: 10.1016/s0041-0101(00)00157-4. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: a model for concerted pharmacological discovery. Mol Interv. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- Peng C, Tang S, Pi C, Liu J, Wang F, Wang L, Zhou W, Xu A. Discovery of a novel class of conotoxin from Conus litteratus, lt14a, with a unique cysteine pattern. Peptides. 2006;27:2174–2181. doi: 10.1016/j.peptides.2006.04.016. [DOI] [PubMed] [Google Scholar]
- Peng C, Wu X, Han Y, Yuan D, Chi C, Wang C. Identification of six novel T-1 conotoxins from Conus pulicarius by molecular cloning. Peptides. 2007;28:2116–2124. doi: 10.1016/j.peptides.2007.08.026. [DOI] [PubMed] [Google Scholar]
- Pi C, Liu Y, Peng C, Jiang X, Liu J, Xu B, Yu X, Yu Y, Jiang X, Wang L, Dong M, Chen S, Xu AL. Analysis of expressed sequence tags from the venom ducts of Conus striatus focusing on the expression profile of conotoxins. Biochimie. 2006a;88:131–140. doi: 10.1016/j.biochi.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Pi C, Liu J, Peng C, Liu Y, Jiang X, Zhao Y, Tang S, Wang L, Dong M, Chen S, Xu A. Diversity and evolution of conotoxins based on gene expression profiling of Conus litteratus. Genomics. 2006b;88:809–819. doi: 10.1016/j.ygeno.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Remigio EA, Duda TF., Jr Evolution of ecological specialization and venom of a predatory marine gastropod. Mol Ecol. 2008;17:1156–1162. doi: 10.1111/j.1365-294X.2007.03627.x. [DOI] [PubMed] [Google Scholar]
- Schuster SC. Next-generation sequencing transforms today’s biology. Nat Methods. 2008;5:16–18. doi: 10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- Yuan DD, Han YH, Wang CG, Chi CW. From the identification of gene organization of alpha conotoxins to the cloning of novel toxins. Toxicon. 2007;49:1135–1149. doi: 10.1016/j.toxicon.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Yuan DD, Liu L, Shao XX, Peng C, Chi CW, Guo ZY. New conotoxins define the novel I3-superfamily. Peptides. 2009;30:861–865. doi: 10.1016/j.peptides.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Zugasti-Cruz A, Aguilar MB, Falcón A, Olivera BM, Heimer de la Cotera EP. Two new 4-Cys conotoxins (framework 14) of the vermivorous snail Conus austini from the Gulf of Mexico with activity in the central nervous system of mice. Peptides. 2008;29:179–185. doi: 10.1016/j.peptides.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.