Abstract
Through statistical analysis of datasets describing single cell shape following systematic gene depletion, we have found that the morphological landscapes explored by cells are composed of a small number of attractor states. We propose that the topology of these landscapes is in large part determined by cell-intrinsic factors, such as biophysical constraints on cytoskeletal organization, and reflect different stable signaling and/or transcriptional states. Cell-extrinsic factors act to determine how cells explore these landscapes, and the topology of the landscapes themselves. Informational stimuli primarily drive transitions between stable states by engaging signaling networks, while mechanical stimuli tune, or even radically alter, the topology of these landscapes. As environments fluctuate, the topology of morphological landscapes explored by cells dynamically adapts to these fluctuations. Finally we hypothesize how complex cellular and tissue morphologies can be generated from a limited number of simple cell shapes.
Keywords: cellular morphogenesis, high content screening, morphological complexity, morphological landscapes, RNAi, signaling networks
Introduction
Cellular morphogenesis, or the ability of cells to adopt particular shapes, is fundamental to organism development and homeostasis. During embryogenesis, cells must adopt particular shapes in order to migrate, form tissues, and ultimately establish the organism’s body plan. In adults, different cell shapes are necessary for cells to execute key functions that underpin homeostasis. The dysregulation of cell morphogenesis often has pathological consequences. For example, during oncogenesis, rewiring of biochemical pathways that regulate cell shape results in aberrant proliferation and metastasis, e.g. by disrupting cell-cell contact and polarity pathways that restrain the growth of normal cells [1–5]. Thus the regulation of cell shape is a process central to both physical and mental health.
The shape of many cells, particularly during development, but also in adult organisms, is highly plastic. That is, after a cell adopts one shape, it is possible for that cell to subsequently adopt another shape, and then re-adopt the original shape. Cell shape changes can occur over small or large time scales, and be of varying magnitudes. In virtually any environment, cells exhibit small fluctuations in shape due in large part to the instability of cytoskeletal structures [6]. More drastic examples of morphological plasticity include the epithelial to mesenchymal transition (EMT) and the mesenchymal to epithelial transition (MET) [7]. During EMT, non-motile epithelial cells with extensive cell-cell adhesions and apicobasal polarity become motile, protrusive, mesenchymal cells that form extensive attachments to the extracellular matrix (ECM), in MET the reverse occurs. Shape changes made by stem cells during the differentiation process are another striking example of cell shape plasticity [8–10]. But what determines the cell shape, and morphological plasticity, of a given cell is still very poorly understood.
How do cell-intrinsic and cell-extrinsic factors contribute to morphogenesis?
Cell shape is clearly a function of cell-intrinsic factors, such as gene/protein expression; and cell-extrinsic factors, such as the elasticity of the ECM [11–13], the dimensionality (1D, 2D, 3D) of the cells’ environment [14], the number of neighboring cells and cell-cell contacts, and the presence of specific stimuli. The relationship between cell-intrinsic and extrinsic factors is complex, because the effect of the environmental factors, such as matrix elasticity, on shape will reflect the signaling state of the cell at a given time. For example, cancer cells respond differently to cell tension than healthy counterparts [15]. Moreover, cell shape is not a passive victim of the environment: while cells alter their shape in response to the environment, these shape changes can lead to changes in signaling state and gene expression [9, 16–21] which in turn can alter the environment itself, e.g. by changing cell density, remodeling of the ECM, or both [22].
What is the architecture of cellular networks regulating cell shape?
That cells exhibit morphological plasticity within a defined environment, such as in a tissue, or when cultured on 2D plastic dish, suggests that cell-intrinsic factors alone determine the shape of cells. In pursuit of the goal of mapping cell-intrinsic signaling networks that regulate cellular morphogenesis, we have devised high-throughput technologies in order to systematically quantify the contribution of different genes to cell shape [23–25]. Using RNA interference (RNAi), individual genes are depleted in populations of tissue culture cells. High-resolution images of each RNAi-treated population are used as input for image processing algorithms that can identify the boundaries of single cells, and “segment” the cells in each image. Hundreds of different features are calculated that describe both the geometry of the segmented cells, nuclei, and organelles, as well as the distribution and intensity of pixels within each segment [26]. These features form the basis of “phenotypic signatures”, or “quantitative morphological signatures” (QMSs) that we have used to describe the mean shape of the population [23, 25]. When depletion of two genes by RNAi results in similar QMSs, it is highly likely these genes encode members of the same protein complex or signaling pathway, and may have an enzyme-substrate relationship [25].
How many cell shapes are there?
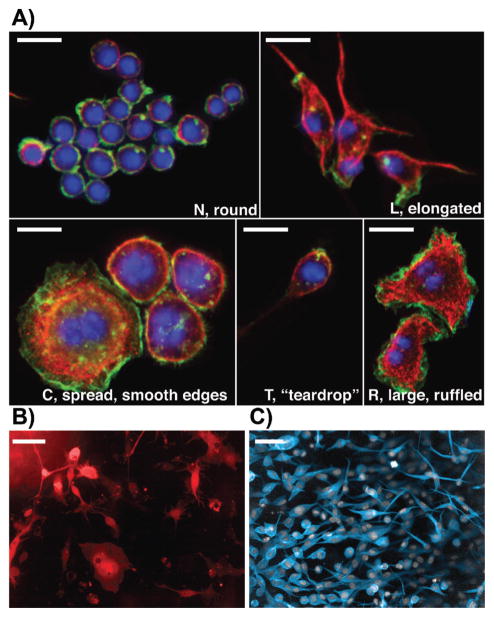
One concern with the use of population-level measures, such as the mean, to describe phenotypes such as cell shape, is that the inherent plasticity in cell shape, and the resulting morphological heterogeneity is poorly captured by such measures [23, 27, 28]. For example, if populations are composed of many distinct morphological subtypes, the mean QMS may not describe the shape of any single cell in the population. However, the single-cell nature of image-based datasets allows us to assess the shape heterogeneity of normal and wild-type populations, as well as the heterogeneity of populations following gene depletion. In fact, we have found that many wild-type cell populations exhibit considerable morphological heterogeneity in both the number and diversity of cell shapes that are present. Perhaps surprisingly, our analyses of different Drosophila and human cell lines show the number of shapes in some populations tends to range from two to seven shapes. Thus there is rarely one cell shape, or hundreds/thousands of cell shapes present in a population. For example, single cell quantification of cell shape reveals that whereas the majority of Drosophila Kc cells are predominantly highly rounded cells of approximately 10–15 μm in diameter (“N” or normal cells), the wild-type population also includes cells that are elongated or bipolar (“L” cells), teardrop shaped (“T” cells), large and smooth-edged (“C” cells), or large and ruffled (“R” cells) (Fig. 1; [23]). Importantly, using a number of different methods, including Principal Component Analysis (PCA), Gaussian Mixture Models (GMM) and Support Vector Machine derived classification schemes, we have shown that these five shapes are quantitatively discrete, and thus are five stable states in high-dimensional morphological space [23]. Quantitative methods reveal that Drosophila wild-type BG-2 cells adopt 6 shapes (Fig. 1B; [29]) and human melanoma cells cultured in 3D matrices adopt 2 shapes (Fig. 1C; [23]). In the case of BG-2 and melanoma cells, these shapes also appear to be discrete (Fig. 1). The distinctness of shapes in certain populations has led us to propose the concept of morphological complexity instead of heterogeneity. A population of cells with high morphological complexity is one that has many quantitatively distinct shapes and is also highly heterogeneous. In contrast, cells that vary continuously around a single shape may be heterogeneous, but are not morphologically complex.
Figure 1.
Morphological complexity in different cell lines. A: The five shapes adopted by wild-type Drosophila Kc Hemocytes [23]. We have termed the shapes “N”, “L”, “C”, “T”, and “R”. Cells were fixed and labeled with Hoechst (blue), phalloidin (green), and anti-tubulin antibody (red). All scale bars represent 20 μm. B: Drosophila BG-2 neuronal cells. BG-2 cells are very heterogeneous, and we have identified six different shapes [29]. BG-2 cells were transfected with EGFP (red) in order to label the entire cell body. Scale bar represents 20 μm. C: WM266.4 melanoma cells cultured on collagen and labeled with CellTracker dye and DAPI. Melanoma cells adopt two types of shape: rounded and elongated. Scale bar represents 50 μm.
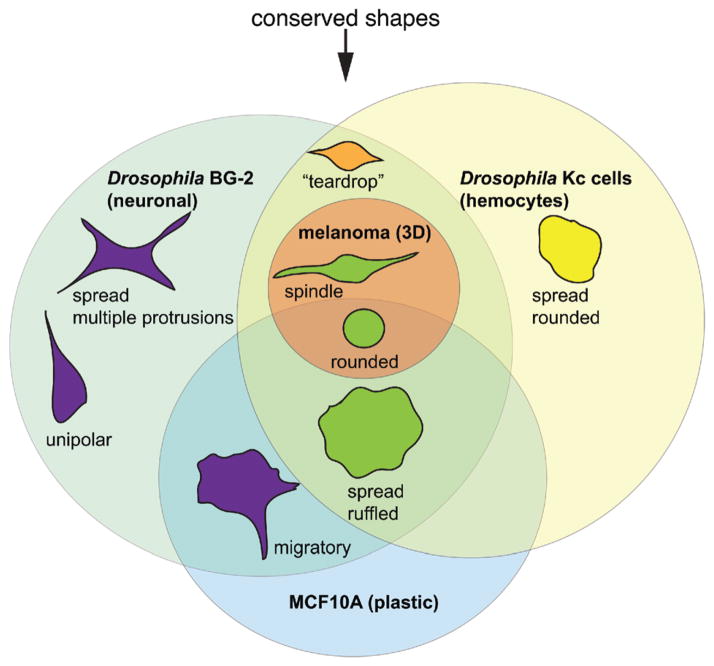
Other groups have reported that migrating fish keratocytes [30] and Dictyostelium [31] cells also exist in a low-dimensional shape space. Despite their different origins, many cell lines adopt shapes that are strikingly similar. For example, melanoma cells cultured in 3D ECM, Drosophila hemocytes, and Drosophila neuronal cells all can adopt rounded and elongated/bipolar shapes (Fig. 2). Moreover, we see many of the shapes observed in Drosophila and melanoma cells lines in MCF10A breast epithelial cells (Fig. 2, unpublished observations). Thus across many species, the number and types of shapes that are adopted by cells is relatively low, and many shapes appear conserved. However, we note that quantitative measurements of shape are still lacking for many different cell types cultured in a variety of conditions, and other cells could potentially explore shape space in different fashions.
Figure 2.
Different cell types can adopt similar shapes. Although the shape space explored by different cell types is diverse, some shapes, such as the rounded or large/flattened shape, are routinely observed. We propose that these shapes are “conserved”.
The low intra- and inter-cell line complexity is perhaps counterintuitive given the diversity of cell shapes observed across nature, but it is consistent with the notion that there exists biophysical constraints on the number of possible configurations of conserved polymers made of actin or tubulin across a wide variety of environmental conditions (e.g. different substrates, osmotic pressures, pH, etc.). This suggests that through the evolution of a small number of genes (actin, tubulin), cells evolved a limited number of shapes such as the spread, elongated, or round shapes that can be used in a variety of different contexts and take advantage of physical laws such as the tight packing of hexagons [32]. We propose that these limited numbers of shapes represent conserved shape templates that can be adopted by many different cell types, which can then be tailored by additional factors in order for cells to adopt more specialized forms.
How does gene inhibition affect morphological complexity?
Analysis of morphological complexity following genome-scale RNAi has revealed another surprising aspect of cell shape, in that gene depletion rarely, if ever, alters the shape space that cells explore, but instead often enriches for particular shapes that are present at low frequencies in wild-type populations [23, 29]. For example, in Kc cells while the bulk of cells are rounded (“N”), gene depletion by RNAi leads to enrichment of “L”, “C”, “T”, and “R” shapes. [23]. The analysis of human RNAi screens also hints that enrichment of pre-existing mutant shape phenotypes in genetic screens is a common phenomenon [33]. Moreover, we have found that depletion of genes required for the morphogenesis of metastatic melanoma shape often leads to the enrichment of one of two pre-existing shapes (elongated or rounded), rather than to new mutant shapes [23].
In order to explain the enrichment of shapes following RNAi, we propose that wild-type cells normally transition between a limited number of cell shapes, and that cells become “trapped” in a particular shape following RNAi, leading to enrichment of that shape [23]. Through live-cell imaging of Drosophila BG-2 neuronal and metastatic melanoma cells we observe that cells do indeed make dynamic and reversible transitions between different shapes [29]. However, we still cannot formally exclude the possibility that in some cell lines particular genes are needed for the morphogenesis or survival of a sub-population that co-exist with, but are not derived from, each other.
We hypothesize that cells could either make switch-like or continuous transitions between shapes. If cells make switch-like transitions, intermediate shapes that exist between stable shapes are highly unstable, and thus not frequently observed. In contrast, many intermediate shapes would be observed in populations making continuous transitions. Qualitatively, Drosophila BG-2 and melanoma cells, make switch-like transitions on the order of minutes to hours [23, 29]. To quantitatively assess the nature of transitions in large populations based on static imaging data, we have developed a statistic termed the RIFT score (rate of intermediate forms or transitions) [23]. When many intermediates are found, transitions can be considered continuous; when few exist transitions are switch-like. Upon calculation of a RIFT score for hundreds of different populations following RNAi in Drosophila hemocytes, we found that the majority of populations, including wild-type cells, can indeed be considered to be undergoing switch-like transitions [23]. Taken together, these data suggest that there may be only a limited number of discrete cell shapes present in a population in part because many intermediate shapes are unstable.
What does morphological space look like?
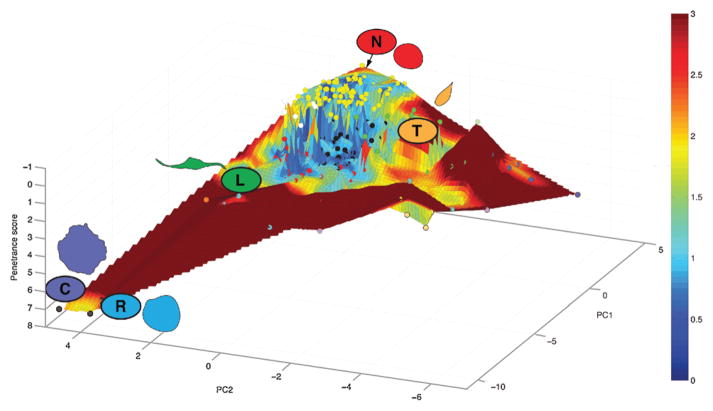
If in a given state space, some shapes are more stable than others, can this space be visualized? We introduce the concept of regression models to plot the shape space, or morphological “landscapes” that cells can explore (Fig. 3). In order to generate morphological landscapes, we first reduce high-dimensional cell shape data from genome-scale RNAi datasets using Principal Component Analysis (PCA). The average QMS of each RNAi-treated population is then projected into two dimensional PC space as points in the x,y plane (circles in Fig. 3). In our morphological landscapes, the third axis is defined by the penetrance of each RNAi, or the percentage of “non-normal” (in this case non-rounded) cells present in a population following gene depletion. Triangle based linear interpolation is performed to generate a surface covering the space between the actual nodes in the x, y, and z dimensions. The x and y dimensions correspond to the shape space explored by cells following systematic gene depletion. The direction of the z-axis is selected so that the peak of the landscape (smallest penetrance) represents the rounded shape that predominates in wild-type populations. Valleys in the landscape correspond to highly penetrant other shapes. Thus the distance between phenotypes on the z-axis corresponds to a phenotypic “potential”, or the amount of energy that must be input into the system in order to switch between two phenotypes. The higher the potential, the more unlikely it is that a cell is able to climb up from a valley to another point in the landscape following RNAi, which thus increases the overall penetrance of the population. We further color the landscape by calculating the average distance to three nearest neighbors nodes of each point in the landscape. A blue color indicates that the point is very close to a group of clustered nodes, and thus these are regions in the shape space where cells are favored to exist. A red color indicates that these are regions of shape space that are very rarely observed in the dataset, and are thus improbable shapes.
Figure 3.
A morphological landscape for Drosophila Kc hemocytes. We have previously described the average quantitative morphological signature (QMS) of cellular populations following depletion of 282 different genes and two control populations [23]. Here, the average QMS of each cell population is represented by a node in a 3D space, where the x and y coordinates represent the first two principal components (PCs) of the QMS vector, and the z-axis is the z-score for penetrance. The z-axis has a reversed direction as smaller numbers are on top, thus a population of completely rounded (or “normal”) cells would be at the peak of the landscape; and populations enriched with mutant phenotypes are located in broad “high valleys”. The color of each node corresponding to the cluster assignment defined in Yin et al. [23], and the surface connecting all the nodes are fitted using linear interpolation, and for each point within the surface, the sum of the distance to three nearest nodes is quantified as the color of the point. Blue coloring indicates that the nodes are in close proximity to each other, whereas increasing levels of red shading indicates increasing distance to any other node in PC space. A cutoff was applied to the distance/color measurements to preserve the detailed geographical information close to the peak of the landscape.
Our landscapes are somewhat analogous to landscapes used to describe how genes search fitness space, through evolution, to find peaks where there is optimal performance of a specific function. (We here refer to fitness as referred to by Kaufman [34] which could be, for example, the velocity of a reaction catalyzed by a specific protein in a specified set of conditions, and where the distribution of velocities catalyzed across protein sequence space constitutes a fitness landscape). In our landscapes cells can be thought of as searching shape space by signaling to find the optimal “rounded” shape. As wild-type cells traverse these landscapes in search of the rounded shape, they often become stabilized in defined attractor regions or “high valleys” of shape (Fig. 4). The inability to activate specific signaling pathways, as in RNAi experiments, results in cells both becoming trapped in certain attractor regions, and thus being unable to “climb” to the optimal rounded shape (Fig. 4). Within each stable state, cells can sample local regions of morphological space, leading to small variations in shape (Fig. 4).
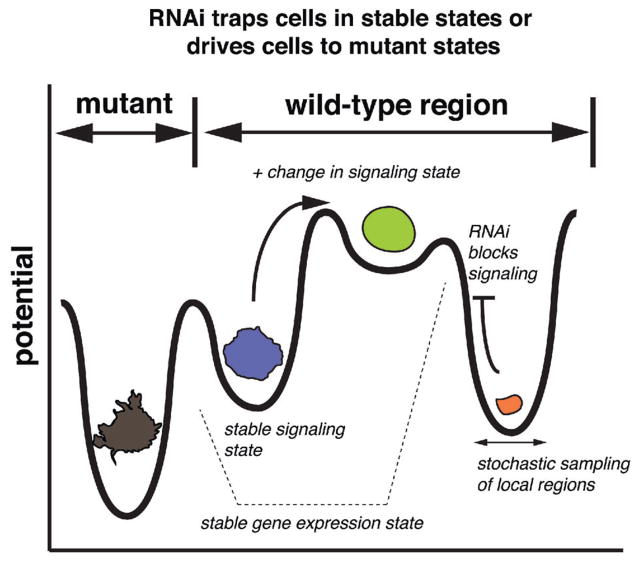
Figure 4.
A revised model of cell morphogenesis. Wild-type cells exist as discrete, stable shapes, which can be considered “high valleys” in a shape space. In order to transition between shapes, there must be an input of signaling activity. RNAi blocks signaling activity thus trapping cells in valleys. Inhibition of some genes pushes cells into stable valleys that are not explored by wild-type populations.
A morphological landscape where cells search for a peak, but can fall into different attractor regions, also has similarities with landscapes used famously by Conrad Waddington to describe the canalization of cell/organism fates [35]. In Waddington’s models, the x-axis represents the phenotype, the y-axis represents time, and the z-axis describes the system’s potential. In Waddington spaces, fates become restricted to one of a limited number of discrete canals, or attractor regions as time passes (e.g. during development). Cells/organisms cannot easily jump from one canal to another because of energetic barriers between canals, which are also important in buffering the determination of cell/organism fates to environmental and genetic flux. This heuristic is largely used to explain stereotypic outcomes of development and differentiation despite the usually large differences in genetic and environmental backgrounds. The morphological landscapes that we have generated also have differences from Waddington landscapes. In morphological landscapes cells can climb up the landscape towards peaks through the activation of signaling pathways, whereas in Waddington landscapes cells/organisms only fall downhill. Furthermore, during exploration of morphological space, cells “fall” into attractor regions of different discrete shapes due to changes in signaling dynamics (e.g. following RNAi of a kinase gene), rather than due to the passage of time as in Waddington landscape. We propose that signaling activity is the equivalent to the input of energy, which allows cells to go up gradients of potential.
A notable aspect to the morphological landscapes we have derived using Drosophila hemocyte RNAi data are the large regions of shape space from which cells appear to be largely excluded (shaded red in Fig. 3). We propose that these regions of shape space include shapes whose adoption would require the polymerization of cytoskeletal filaments or membranes to be curved in ways that are biophysically unstable. The restriction of cells to specific regions of potential shape space may underpin the robustness of shape to environmental and genetic fluctuation.
How do cells explore morphological landscapes?
Importantly, because the same five morphologies are consistently seen in the majority of populations treated following RNAi, this suggests that many single kinase/phosphatase genes, or signaling pathways, do not act to largely define shape of the landscape itself, but rather the signaling state of a cell in a given environment determines the morphological attractor(s) that the cell will be drawn to. This means that each morphological attractor may also be an attractor in signaling network state space – where “signaling network state” describes the dynamics of all signaling proteins at any given time (Fig. 4). Notably, different signaling states need not be defined by discrete differences in gene expression states, or changes in the environment, and can be due solely to the dynamical nature of signaling networks [34]. Although it is still difficult to comprehensively define a signaling network state, and whether such states would exist as discrete attractors in state space is still unclear, both experimental [36, 37] and theoretical work [34] largely agrees with the idea that there exists a limited number of signaling networks states that could underpin discrete morphologies. The idea that each stable shape is driven by a corresponding stable signaling network state is in fact consistent with a very simple Boolean model that predicts cell shape based on signaling states [41]. Transitions between stable morphology/signaling network states could be made in stochastic or deterministic fashions (e.g. following exposure of cells to an informational signal) that alters signaling dynamics in a manner similar to conversions between other signaling states – such as ultrasensitive engagement of signaling activity that leads to a transition between bistable states [38].
Do different shapes express different genes?
While we propose that different signaling states drive different shapes, it is unclear whether each morphological and signaling state is driven by large-scale changes in gene expression. On the one hand, the discretized, low-dimensional exploration of morphological space is highly reminiscent of the limited number of transcriptional states that are explored by cells as described by gene expression profiling [39], and the limited number of gene expression states. Certainly, large-scale changes in gene expression are consistent with, and often causal to, longer-term changes in cell shapes that occur during morphogenetic processes such as during EMT [40, 41]. In some cases changes in transcriptional programs may be dependent on, and not necessarily be causal to, changes in morphology [42–46], and in such cases different gene expression states would still be associated with different cell shapes. However, some cells can transition between shapes in very transient, rapid, and reversible fashions over minute time scales. Such rapid changes in shape are unlikely to be correlated with changes in gene expression states. For example, WM266.4 cells switch between rounded and spindle morphologies in 3D culture at 5–10 min intervals [23]. Thus while it needs to be formally tested, it is unlikely that differences in gene expression and gene expression states can completely explain all discrete differences in shapes.
A hypothesis that unifies the different nature of gene expression states, signaling, and shape states is that for a cell in a given environment there exist a limited number of stable gene expression states, each of which may correspond to a limited number of stable signaling states; and some of these different signaling states may correspond to different stable morphological states (Fig. 4). Thus, this model would suggest that different morphologies may or may not differ in their gene expression states, but that they definitely are distinct in terms of their signaling state.
How does the environment act on morphological landscapes?
While the interplay between cell-intrinsic and extrinsic factors on cell morphogenesis is well appreciated, our observation casts this relationship in a new light. Based on data collected in a stable, well-defined environments, we propose that cell-intrinsic factors, such as the dynamics of signaling networks, gene expression, and biophysical constraints, restrict cell morphogenesis to a low-dimensional landscape consisting of a limited number of attractors. We hypothesize that cell-extrinsic factors such as informational stimuli and mechanical forces act upon these landscapes in different ways. Because RNAi-mediated depletion of many signaling genes rarely alters the shape of these landscapes, we suggest that informational stimuli, such as cytokines and growth factors, act to promote transitions between stable states in a landscape. In contrast, mechanical forces (e.g. imposed by matrix elasticity, cell-cell contacts, fluid stress) can act upon these landscapes to mold their contours. In some cases the environmental effects could be quite drastic. For example, we have shown the effect of matrix elasticity on the morphological landscape of melanoma cells. On very stiff plastic environments, WM266.4 cells adopt essentially one shape, and make continuous variations around that shapes, but on soft surfaces they exist in two stable states [23]. In fact, we would argue that the use of plastic in tissue culture experiments has largely obscured the fact that cells can exist in discrete states.
How does diversity arises from simplicity?
We suggest that in fact the environment also has a key role to play in generating the morphological diversity of cells, tissues, and organisms from cell shapes that are intrinsically simple and low-dimensional. Our observations suggest that cells did not evolve hundreds to thousands of genes, such as those encoding signaling proteins, in order to generate different shapes. Rather cell-intrinsic factors are capable of generating a limited number of basic shapes or “shape building blocks” that are very conserved. The generation of these shapes could result from the actions of a very small number of genes, especially genes that encode conserved cytoskeleton proteins such as actin and tubulin. Although actin and tubulin proteins can polymerize in a variety of structures, these structures are likely to be limited, leading to only a small number of shapes. We propose that in order to generate further diversity from these basic shapes, metazoan cells have not necessarily evolved many new genes to encode these shapes, but rather have evolved cell-extrinsic factors that act to control the exploration of landscape, and alter the topology of these landscapes.
How does tuning of landscapes facilitate transitions between shapes?
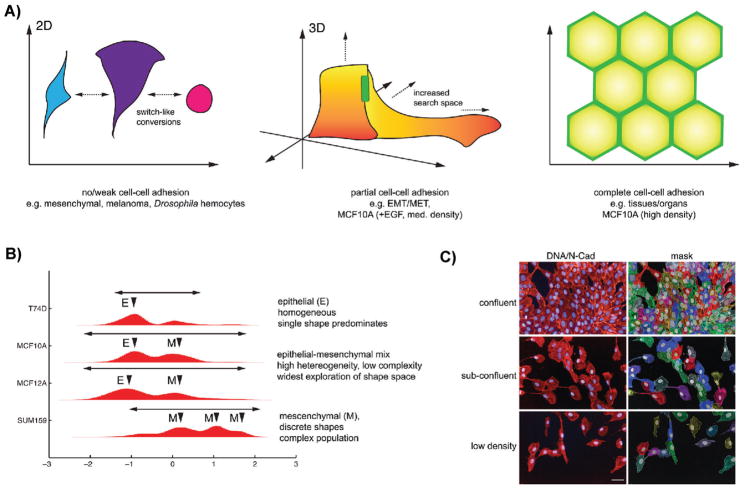
Amongst the mechanisms that we propose that metazoan cells use to generate diversity are cell-cell interactions [47], such as adherens junctions (AJs), and cell-ECM adhesions that allow cells to modify intrinsically determined shape space. While cells produce cell-cell junction and ECM proteins, we consider these factors to be “cell-extrinsic” as they modify the environment of a single cell. For example, in the absence of cell-cell interactions cells are typically flattened, and variations in morphology are primarily two-dimensional [48] (Fig. 5A), and we propose the shape space explored by cells is dictated largely by autonomous biophysical constraints, which limit cells to a conserved set of shapes. In fact, Drosophila or human cells cultured on plastic, or human melanoma cells on soft substrates -- none of which make extensive AJs, exist in a limited number of discrete morphologies -- We hypothesize that the ability to generate cell-cell adhesions changes the cell’s environment, facilitates lateral growth, the establishment of apicobasal polarity, and the exploration of morphological space in three dimensions (Fig. 5A). Moreover, when cell-cell contacts do not occupy the entire lateral region of the cell, cell-cell adhesion can serve as anchors from which cells can generate protrusive forces and thus adopt shapes that might not otherwise be possible. Thus cell-cell contacts can both increase the size of the morphological space available to cells, and modify the shape of these landscapes. We propose that modification of the cellular environment by the generation of cell-cell contacts, and thus alteration of the morphological landscape available to a cell, could increase the heterogeneity, if not necessarily the complexity, of a population; that is the generation of cell-cell contacts would result in populations that are still variations of mesenchymal or epithelial shapes, because attractor regions in morphological landscapes are “widened” by factors such as cell-cell adhesion.
Figure 5.
Cells can explore greater regions of shape space by making cell-cell adhesions. A: In the absence of cell-cell adhesions, cell autonomous factors such as cytoskeletal dynamics largely dictate cell shape resulting in switch-like conversions between discrete shapes. By making cell-cell adhesions, cells can explore greater regions of shape space. Attractor regions can become wider and space between the regions can decrease. When cells are completely surrounded by other cells and cell-cell adhesion is “saturating”, such as in epithelial tissues, cells are highly restricted in the shape space and multiple attractors merge creating one single attractor in shape space. B: Shape quantification of different breast cell lines. T247D grow in colonies and primarily adopt an epithelial (E) shape. Subconfluent MCF10A and MCF12A cells, which exhibit partial cell-cell adhesions are a mix of Epithelial (E) and Mesenchymal (M) morphologies, and in addition explore large regions of shape space. SUM159 make poor cell-cell adhesions, and adopt primarily Mesenchymal (M) shapes. These populations appear qualitatively more discrete capered to MCF10A and MCF1012A populations. C: MCF10A cells cultured at different densities, and thus making different degrees of cell-cell adhesions explore shape space in different ways. In the left hand panels DNA is labeled in blue and N-cadherin is labeled in red. In the right hand panel cells have been segmented. Scale bar is 50 μm.
In some cases the widening of the attractor regions may have the effect of decreasing the barrier of potential cells must overcome in order to cross from one attractor to the next, which thus facilitates transitioning between two attractor states. For example, during EMT, the shape of partially adhered epithelial cells at the edge of tissue may be much more variable than that of epithelial counterparts which are completely surrounded by other cells. The partially adhered nature of the edge cells widens the attractor region in which they exist to the point that the epithelial attractor is brought in closer proximity in morphological space to a mesenchymal attractor, and thus increasing the probability of transition to a mesenchymal form. The increased degrees of morphological freedom that are available to partially adhered cells may also underpin the metastatic process, as we would predict that partially adhered cells at the invasive front of tumor would also be able to search larger regions of shape space than those embedded in the tumor body. Consistent with this model, we have observed that MCF10A or MCF12A epithelial cells cultured at sub-confluent conditions, transition between epithelial and mesenchymal shapes and explore larger regions of shape space than mesenchymal cells alone – and appear much more heterogeneous (Fig. 5B,C).
While it may be advantageous for cells tune landscapes to facilitate transitions between shapes, there likely exists a threshold where the barriers between attractor regions are reduced to the point where they merge into a single attractor. For example, as cells form cell-cell adhesion this initially may allow great exploration of morphological space and facilitates transitions between epithelial and mesenchymal shapes. However, when the number of cell-cell adhesion increases to the point where cells are completely flanked by neighbors, the space between attractors is reduced to the point where they merge in into a single attractor, and the number of possible shapes that can be adopted is markedly reduced, and populations are composed of largely homogeneous shapes (Fig. 5A) [32]
Cell-cell adhesions represent only one of numerous mechanisms that cells have evolved in order to sculpt cell-intrinsic morphological landscapes. The evolution of numerous ECM components certainly represents another means by which can cells sculpt landscapes. Moreover, in some cases cells may be able to explore morphological spaces in both local and “global”, or cell-wide, fashions. For example, growing axons may explore shape space in a different manner than the body of neurons.
Dynamic adaption of morphological landscapes to environmental fluctuations
We propose that in a fixed environment, such as in tissue culture, or in regions of tissue that experience largely similar environments (e.g. similar matrix elasticity), it is cell-intrinsic factors, such as signaling dynamics, that are the principal drivers of transitions between stable states. When microenvironmental conditions are largely held constant, cell-extrinsic factors can tune the topology of an intrinsically determined landscape to facilitate transitions. In vivo, however, many changes in cell shape could in fact lead to radical changes in the environment and large-scale alterations in the topology of morphological landscape that is searchable by cells. These large-scale changes represent a means by which cell-extrinsic factors can impact cellular plasticity. Such a model is consistent with recent ideas suggesting that the ECM regulates local versus global changes in morphogenesis [49]. As a simple example, if a cell adopts a migratory form, that alters its position in a tissue, the difference in cell-extrinsic factors between the new and previous microenvironment -- such as substrate stiffness or concentration of a particular stimulus -- could alter the cell’s potential shape space such that a new set of attractor regions emerge. Thus the cell, and the morphological landscape at a given time, dynamically adapts to environmental fluctuations.
In fact, behaviors involving changes in morphology over relatively large time and space scales that are seemingly complex, such as chemotaxis, durotaxis, vascularization, and metastasis, could in part be explained by cells instead proceeding through a series of landscapes. As cell migrate, and their environment changes, the potential shapes that are available to a cell is restricted to few discrete shapes.
Conclusions and perspectives
Despite the long and illustrious history of cell shape research, we still have very little understanding of how genetic and environmental forces shape cells in stereotypic, consistent, and robust fashions during development and homeostasis. However, we anticipate that continual use of rigorous statistical methods to analyze datasets derived by imaging cells in a variety of conditions will provide new, and sometimes surprising, insights into how shape is determined – particularly as imaging in vivo becomes easier and faster.
Through our studies, as well as those of others [23, 29–31, 50], a picture is emerging that very simple mechanisms may underpin morphogenesis. Specifically, that there is only a limited number of cell shapes, and the morphological landscapes cells explore in a given environment are thus composed of only a few attractor regions. Cell-extrinsic factors can tune, or even alter the topology of these landscapes to generate complexity. These ideas advance our understanding of development and homeostasis, and open new therapeutic avenues. For example in the case of metastasis, strategies to manipulate the shape space available to tumor cells -- or drive metastatic cells towards certain attractor shapes such that highly motile and invasive shapes become less probable -- may provide clinical benefit.
Acknowledgments
We thank Alexis Barr and Louise Evans for comments on the manuscript. Research in the Bakal laboratory is supported by grants from the Biotechnology and Biological Sciences Research Council (BB/J017183/1, and BB/I002510/1) and Cancer Research UK (C37275/A13478). Research in the Wong laboratory is supported by grants from the National Institutes of Health (NIH U54 CA149196, and NIH R01 CA121225), a John S. Dunn Research Foundation grant and a TT & WF Chao Foundation grant. Chris Bakal is a Research Career Development Fellow of the Wellcome Trust.
Abbreviations
- ECM
extracellular matrix
- EMT
epithelial to mesenchymal transition
- MET
mesenchymal to epithelial transition
- QMS
quantitative morphological signatures
Footnotes
None of the authors have any conflict of interest to disclose.
References
- 1.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–6. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 2.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeichi M. Morphogenetic roles of classic cadherins. Curr Opin Cell Biol. 1995;7:619–27. doi: 10.1016/0955-0674(95)80102-2. [DOI] [PubMed] [Google Scholar]
- 4.Wildenberg GA, Dohn MR, Carnahan RH, Davis MA, et al. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–39. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Bustelo XR. Intratumoral stages of metastatic cells: a synthesis of ontogeny, Rho/Rac GTPases, epithelial-mesenchymal transitions, and more. BioEssays. 2012;34:748–59. doi: 10.1002/bies.201200041. [DOI] [PubMed] [Google Scholar]
- 6.Kueh HY, Mitchison TJ. Structural plasticity in actin and tubulin polymer dynamics. Science. 2009;325:960–3. doi: 10.1126/science.1168823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 8.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 9.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 10.Deng J, Petersen BE, Steindler DA, Jorgensen ML, et al. Mesenchymal stem cells spontaneously express neural proteins in culture and are neurogenic after transplantation. Stem Cells. 2006;24:1054–64. doi: 10.1634/stemcells.2005-0370. [DOI] [PubMed] [Google Scholar]
- 11.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–5. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–19. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 14.Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–9. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paszek MJ, Zahir N, Johnson KR, Lakins JN, et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–54. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–9. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 17.Chen CS, Mrksich M, Huang S, Whitesides GM, et al. Geometric control of cell life and death. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 18.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, et al. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–95. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 19.Mammoto A, Connor KM, Mammoto T, Yung CW, et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–8. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.North TE, Goessling W, Peeters M, Li P, et al. Hematopoietic stem cell development is dependent on blood flow. Cell. 2009;137:736–48. doi: 10.1016/j.cell.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farge E. Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr Biol. 2003;13:1365–77. doi: 10.1016/s0960-9822(03)00576-1. [DOI] [PubMed] [Google Scholar]
- 22.Wolf K, Friedl P. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 2011;21:736–44. doi: 10.1016/j.tcb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Yin Z, Sadok A, Sailem H, McCarthy A, et al. A screen for morphological complexity identifies regulators of switch-like transitions between discrete cell shapes. Nat Cell Biol. 2013;15:860–71. doi: 10.1038/ncb2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin Z, Zhou X, Bakal C, Li F, et al. Using iterative cluster merging with improved gap statistics to perform online phenotype discovery in the context of high-throughput RNAi screens. BMC Bioinformatics. 2008;9:264. doi: 10.1186/1471-2105-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bakal C, Aach J, Church G, Perrimon N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science. 2007;316:1753–6. doi: 10.1126/science.1140324. [DOI] [PubMed] [Google Scholar]
- 26.Evans L, Sailem H, Vargas PP, Bakal C. Inferring signalling networks from images. J Microscopy. 2013;252:1–7. doi: 10.1111/jmi.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference? Cell. 2010;141:559–63. doi: 10.1016/j.cell.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pelkmans L. Cell Biology. Using cell-to-cell variability--a new era in molecular biology. Science. 2012;336:425–6. doi: 10.1126/science.1222161. [DOI] [PubMed] [Google Scholar]
- 29.Sailem H, Bousgouni V, Cooper S, Bakal C. Cross-talk between Rho and Rac GTPases drives deterministic exploration of cellular shape space and morphological heterogeneity. Open Biol. 2014;4:130132. doi: 10.1098/rsob.130132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keren K, Pincus Z, Allen GM, Barnhart EL, et al. Mechanism of shape determination in motile cells. Nature. 2008;453:475–80. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tweedy L, Meier B, Stephan J, Heinrich D, et al. Distinct cell shapes determine accurate chemotaxis. Sci Rep. 2013;3:2606. doi: 10.1038/srep02606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson MC, Patel AB, Nagpal R, Perrimon N. The emergence of geometric order in proliferating metazoan epithelia. Nature. 2006;442:1038–41. doi: 10.1038/nature05014. [DOI] [PubMed] [Google Scholar]
- 33.Jones TR, Carpenter AE, Lamprecht MR, Moffat J, et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci USA. 2009;106:1826–31. doi: 10.1073/pnas.0808843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kauffman SA. Self-Organization and Selection in Evolution. New York, Oxford: Oxford University Press; 1993. The Origins of Order. [Google Scholar]
- 35.Waddington CH. The Strategy of Genes. Allen & Unwin; 1957. [Google Scholar]
- 36.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–8. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 37.Janes KA, Albeck JG, Gaudet S, Sorger PK, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–53. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 38.Pomerening JR. Uncovering mechanisms of bistability in biological systems. Curr Opin Biotechnol. 2008;19:381–8. doi: 10.1016/j.copbio.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Huang S. Reprogramming cell fates: reconciling rarity with robustness. BioEssays. 2009;31:546–60. doi: 10.1002/bies.200800189. [DOI] [PubMed] [Google Scholar]
- 40.Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139:3471–86. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 41.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 42.Desprat N, Supatto W, Pouille PA, Beaurepaire E, et al. Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Dev Cell. 2008;15:470–7. doi: 10.1016/j.devcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–72. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 44.Dupont S, Morsut L, Aragona M, Enzo E, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 45.Mammoto A, Mammoto T, Ingber DE. Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci. 2012;125:3061–73. doi: 10.1242/jcs.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajakyla EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases. 2014;5:e27539. doi: 10.4161/sgtp.27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jamora C, Fuchs E. Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol. 2002;4:E101–8. doi: 10.1038/ncb0402-e101. [DOI] [PubMed] [Google Scholar]
- 48.Etienne-Manneville S. Control of polarized cell morphology and motility by adherens junctions. Semin Cell Dev Biol. 2011;22:850–7. doi: 10.1016/j.semcdb.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 49.Daley WP, Yamada KM. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr Opin Genet Dev. 2013;23:408–14. doi: 10.1016/j.gde.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–88. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]