Abstract
Single walled carbon nanotubes (SWCNTs) are utilized in many areas, accompanied with the ever rising safety concerns. Coating the SWCNTs by serum albumin has shown promises in reduction of their cytotoxicity. The cause of toxicity reduction could be due to the blockage of cellular protein adsorption by bovine serum albumin (BSA). Here, our study explored the mechanism of toxicity reduction from the point of view of protein adsorption. Different loadings of BSA led to varied surface coverage of the SWCNTs, which was positively related to the level of cytotoxicity. In addition, the BSA-coated SWCNTs were tested for their surface morphology change, cellular uptake, and adsorption of cellular proteins. BSA could be competed off the SWCNT surface by the cytosol proteins, and thus a higher BSA loading was needed to provide better protection to the cells. Cellular uptake was also reduced with a higher BSA loading. Moreover, the BSA coating changed the surface property of SWCNTs, and as a consequence, altered the types of proteins adsorbed by the SWCNTs. Our results support that adsorption of BSA reduces cellular uptake of SWCNTs as well as adsorption of cellular proteins on SWCNTs, both contributing to the much lower cytotoxicity observed for the BSA-coated SWCNTs.
Keywords: SWCNTs, surface coating, protein adsorption, cellular uptake, cytotoxicity
1. Introduction
Single-walled carbon nanotubes (SWCNTs) are expected to be one of the most widely used engineering nanomaterials.[1] The unique physicochemical properties have made them popular materials in electronics,[2] biosensors and transistors [3]. They have also been adopted as drug carriers for targeted delivery[4] and nano-probes for biomedical imaging[5]. These increasing applications of SWCNTs are greatly increasing the risk of SWCNT exposure to human and environments.[6] The safety of such materials has become more and more of a concern[7]. Toxicity investigation has been reported both in vivo and in vitro, and the different toxic effects observed have been attributed to factors including, impurities, surface modification, shape, length, agglomeration, etc.[8]
Proteins are the main functional molecules inside biological species, and the adsorption of proteins to SWCNTs could induce conformational changes in proteins, altering their function. For example, SWCNTs have been reported to be able to intrude into the hydrophobic core of proteins, which could either enhance the protein’s activity[9] or deactivate the protein.[10] Another report also showed CNT could also serve as a K+ channel blocker in a dose-dependent manner.[11] Thus, to protect cells, one possible way is to reduce the contact between SWCNTs and proteins, which can be done through surface modification. Strategies for modifying the SWCNT surface, has been a recent focus of SWCNT research.[12] The modification is usually to change the surface charge and hydrophobicity.[13] Non-covalent modification has been used very often to obtain better dispersions of SWCNTs.[14]
Binding of SWCNTs to proteins is mostly governed by π-π stacking,[15] electrostatic attraction [16] and hydrophobic interactions.[17] Therefore, one approach to modify the SWCNTs surface is by wrapping the SWCNTs with a layer of inert polymers which could mask the SWCNTs from being seen by the inner cellular biomolecules including proteins, nucleic acids, and metabolites. Reports have shown that SWCNTs pre-incubated with blood proteins induced less cytotoxicity on two different cell lines.[17] SWCNTs coated by bovine serum albumin (BSA) imposed no obvious toxicity to cells.[18] Albumin is chosen for protection of SWCNTs because it is the most abundant protein in the circulation system for the sake of in vivo applications of SWCNTs, as well as in the cell culture media used during in vitro study.[19] It is also a good dispersant for SWCNTs, which could minimize possible aggregation and agglomeration of SWCNTs.[20] We hypothesized that, the reduced toxicity effect from the BSA-coated SWCNTs could be caused by prevention of adsorption of certain cellular proteins to the SWCNTs.
Herein, in the present work, toxicity of SWCNTs coated with different amounts of BSA to the fibroblast cells was studied, and cytosol proteins that interacted with the non-coated and BSA-coated SWCNTs were identified. Competition of BSA and the cytosol protein binding on the SWCNTs surface was investigated. The results were correlated with the toxic effects observed for the SWCNTs coated with different amounts of BSA for better understanding the fundamental protection mechanism of BSA. We expected the study could reveal important protein contributors to the toxic effect caused by SWCNTs.
2. Results and Discussion
SWCNTs have been reported to cause damage to cells through different ways, including oxidative stress, inflammatory responses, and DNA damage.[21] Reports have shown that BSA is a good dispersant for SWCNTs which could disperse up to 300 µg/mL and showed negligible toxic effects to human mesenchymal stem cells (hMSCs) and HeLa cells.[18] We tested two different SWCNTs : BSA mass ratios, 1: 1 and 1: 4, to reveal the changes in cellular uptake and protein adsorption pattern caused by BSA adsorption. By gradually increasing the surface coverage of SWCNTs via the BSA coating, we hoped to identify the particular cellular protein-SWCNT interaction that contributes significantly to the toxic effect of the SWCNTs.
2.1 Cell toxicity and ATP Production
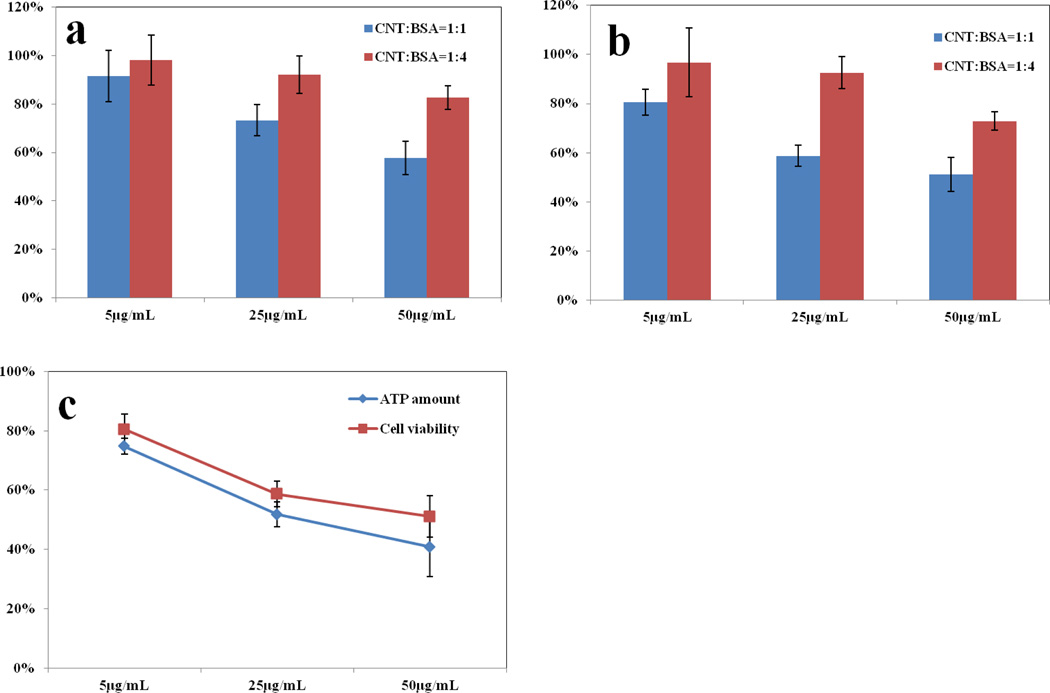
After being coated with different amounts of BSA, the SWCNTs dispersed very well in the FBS free culture media. Cell viability was tested after 6 and 12 hrs incubation and the results were shown in Figure 1. Regardless of whether a 1 : 1 or 1 : 4 SWCNT : BSA ratio was used, when a higher amount of SWCNTs or a longer time was used during incubation, the cell viability was lower. This tells us that the SWCNTs indeed caused harmful effects to the cells under our experimental conditions. But, the SWCNTs coated with a higher BSA amount (1:4 ratio), had less cytotoxicity than the ones coated at the ratio of 1:1. No viability reduction was observed with 25 and 5 µg/mL of SWCNTs coated by 4-fold more BSA when the incubation time extended to 12 hrs, but an additional 10% loss in viability was seen using the SWCNTs coated at a 1:1 SWCNT:BSA ratio. Thus, a higher BSA amount did reduce SWCNT cytotoxicity.
Figure 1.
Cell viability and ATP production after treatment with BSA-coated SWCNTs. SWCNT concentration is set to 50µg/mL, 25µg/mL and 5µg/mL and premixed with two BSA mass ratios of 1:1 and 1:4, and the mixture was then added to cell culture media and incubated for 6 hrs (a) and 12 hrs (b). Cell viability is measured by comparing the absorbance at 490 nm with negative control. c) ATP production is measured at 12 hrs with the SWCNTs coated with BSA at a mass ratio of 1:1, and compared with the cell viability test result. In (a) and (b), the cell viability difference between the two coating ratios was significant at the significance level of 0.05 as indicated by the Student’s t-test, except for the incubation with the lowest SWCNT concentration of 5 µg/mL.
Several studies pointed out the toxicity of CNTs came from oxidative stress, which could be caused by damages to the mitochondria.[22] In addition, two cell death pathways, apoptosis and necrosis, both involve the malfunction of mitochondria. Since mitochondria are the major energy production organelles in the cell, thus, its proper functioning can be viewed from the level of ATP production. Thus, we measured the ATP production levels to see whether ATP reduction incurred prior to the on-set of cell death, which could link the cell death directly to the malfunction of mitochondria. However, both ATP production and cell viability occurred simultaneously, which implies that the decrease in ATP level was caused by cell death rather than direct mitochondrial damage. No acute damage to the mitochondria was induced by SWCNTs before they killed the cells by other mechanisms.
2.2 Morphology of BSA-coated SWCNTs and the Amounts of BSA Loaded
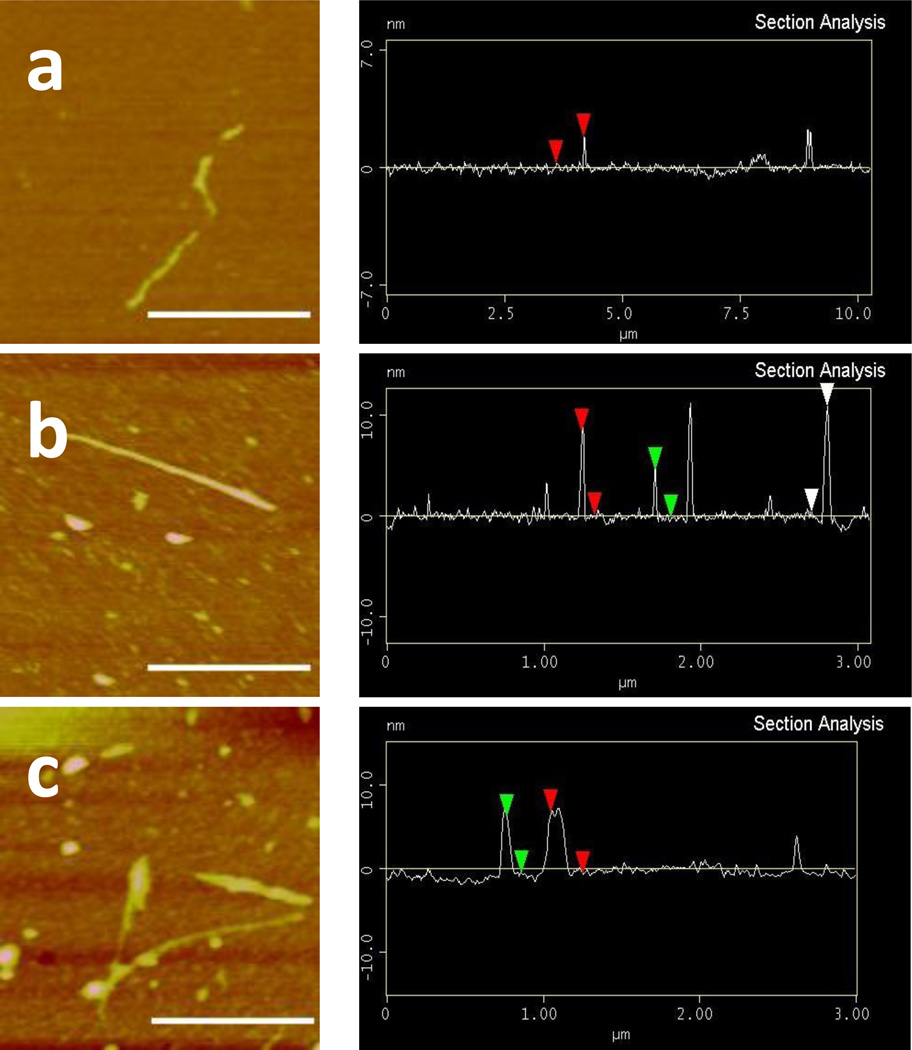
As discussed above, one possible outcome of coating the SWCNTs with BSA is to block their interaction with proteins, which requires a complete coverage of the nanotube surface by BSA. The globular structure of BSA in pH 7.4 has the dimensions of 14×4×4 nm.[23] The diameter of the SWCNTs is between 1–5 nm according to the product information sheet provided by the manufacturer. By being wrapped with a bulky protein like BSA, the diameter of the SWCNTs should increase. AFM is an appropriate technique to visualize such increase and from the increase in CNT diameter, we could evaluate the degree of surface coverage.
Figure 2 shows the representative AFM images of the bare SWCNTs and the SWCNTs coated with 1- or 4-a mass ratios of 1:1 and 1:4 (SWCNT : BSA). Without BSA coating, the SWCNTs captured on that image had a height of 1.5 nm (this and the following dimensions were the average values after counting tubes in multiple images, n > 5) (Figure 2a). After coated with BSA at a mass ratio of 1:1, the height increased to 4.6 nm (Figure 2b). This height increase is similar to two of the dimensions of BSA, suggesting the formation of a monolayer of the BSA coating. The coating formed at this mass ratio was quite uniform, giving out a smooth surface. Increasing the amount of BSA by 4 folds, the height of the SWCNTs increased to an average of 7.6 nm (Figure 2c). Two layers of BSA may have been formed on the SWCNTs but the coating was less uniform, giving out more roughness on the surface. The thicker BSA layer could then shield the original surface of SWCNTs better.
Figure 2.
AFM images and height analysis of SWCNTs without BSA (a), with 1:1 ratio of BSA (b) and with 1:4 ratio of BSA (c). The scale bar was 500 nm.
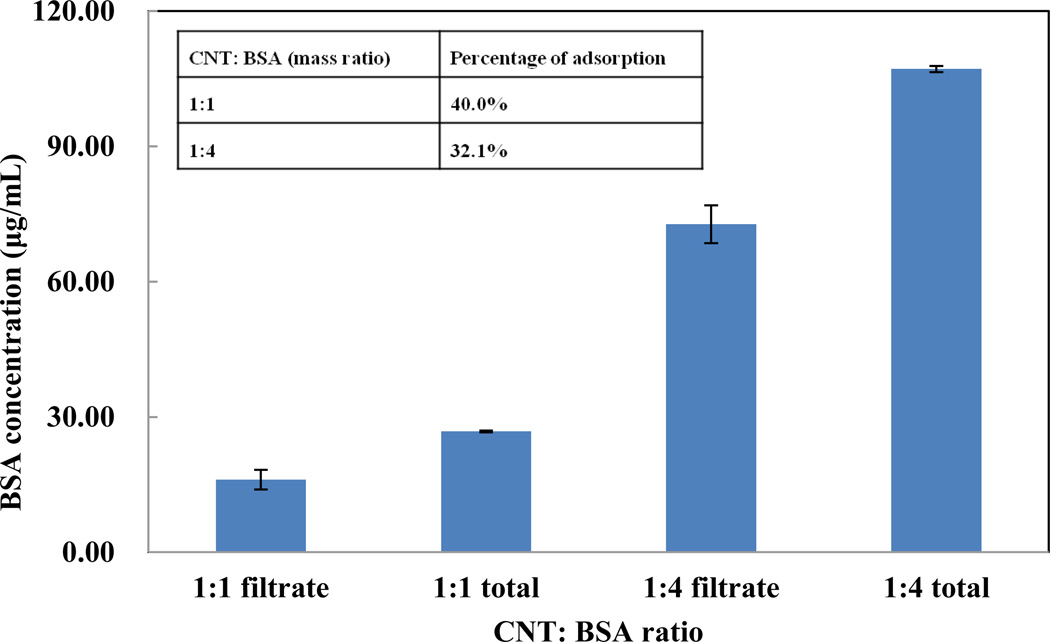
Although AFM images can tell us the morphology of the BSA-coated SWCNTs, they cannot reveal the coating coverage because only a few tubes could be observed in each image. To get a better idea of the surface coverage, we designed a filter-based assay to quantify the amount of BSA adsorbed on the SWCNTs. After 30 mins incubation, the mixture of SWCNTs and BSA was loaded on the ultracentrifuge filer with a MWCO of 300 kDa. Such filters have a large enough pore size to allow the passage of the free BSA molecules, but retain the long SWCNTs with the adsorbed BSA. The amounts of both the total BSA used for coating and the BSA recovered in the filtrate were determined by the BCA assay; and the results were displayed in Figure 3 with an inserted table.
Figure 3.
Amount of BSA adsorbed on 25 µg/mL SWCNTs in PBS (pH=7.4). Filtrate stands for filtrate collected after filtration and total means the initial BSA added. 1:1 means SWCNTs: BSA=1:1(mass ratio) and 1:4 means SWCNTs: BSA=1:4 (mass ratio).
Up to 10 µg and 32.1 µg of BSA were absorbed by 25 µg SWCNT with a coating ratio of 1:1 and 1:4, respectively. The surface area of SWCNTs could commonly reach 600 m2g−1.[24] Assuming that each BSA molecule is adsorbed on the SWCNTs with an area of 14 nm × 4 nm, we estimated that, only 34.8% of the surface of 25 µg SWCNTs was covered by BSA, but the coverage went up to 119.9% for the 1:4 coating ratio. The more than 100% coverage supports our observation made by AFM that multiple BSA layers could be formed on the SWCNTs coated by BSA at the 1:4 mass ratio. Our result revealed that, only less than half of the surface area of the SWCNTs could be covered when using the 1:1 coating ratio. The exposed surface may induce toxic effects to the cells, leading to the higher rate of cell death.
2.3 Cystosol Protein Competition with BSA to the SWCNTs Surface
BSA adsorption on CNT has been reported to be mainly due to hydrophobic reaction and π-π stacking;[25] and the adsorption could be reversible.[17] The adsorbed BSA could be competed off the surface of SWCNTs by cellular proteins during and/or after cell entry. The functions of the adsorbed cellular proteins could then be altered and induce adverse effects. Thus, it is necessary to study the stability of BSA adsorption at the presence of cellular proteins.
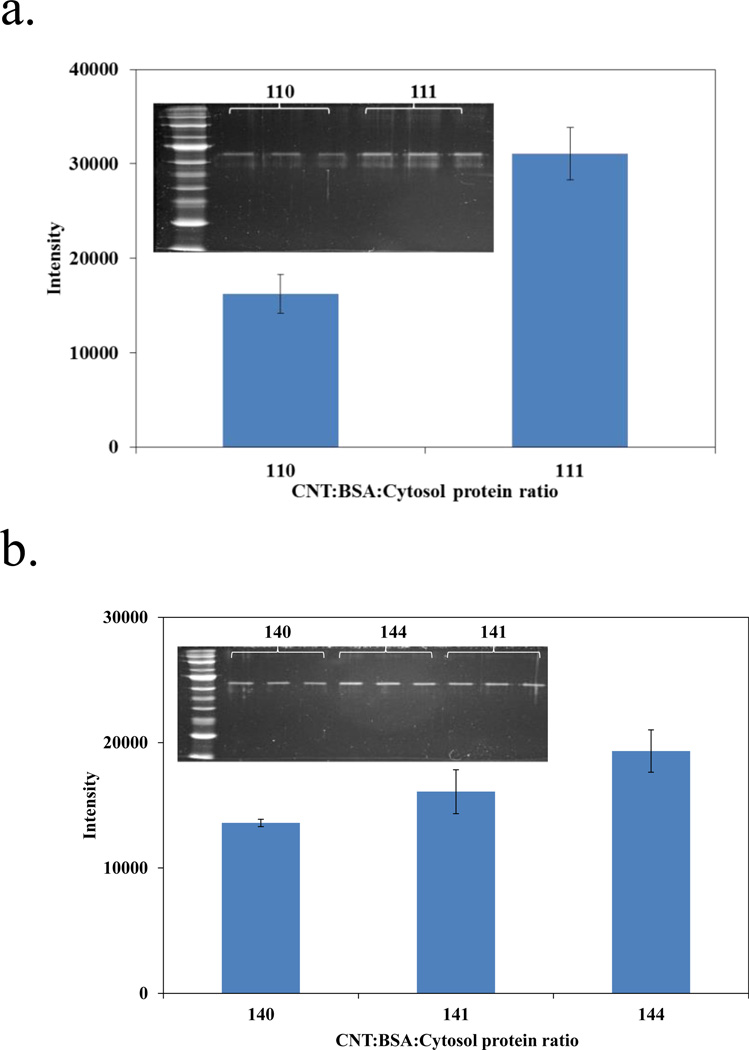
Most studies have shown that the major location of SWCNTs in the cell could be in the cytoplasm.[18] We chose the cytosol proteins extracted from the fibroblast cells because they represent the first defense line against the exposure to SWCNTs which enter the human body via inhalation or skin adsorption. After incubating the SWCNTs with BSA for 30 minutes, we added the cytosol proteins to the mixture to a final concentration of 0, 25 and 100 µg/mL. Two hours later, the mixture was loaded on the ultracentrifuge filter with the MWCO value of 300 kDa. The desorbed BSA in the filtrate was analyzed by SDS-PAGE and visualized by SYPRO Ruby staining. ImageJ was used for semi-quantification of the BSA amount. Each sample was labeled with 3 numbers (for example, 101, 111 etc), representing the mass ratio of the three components, SWCNTs, BSA, and the cytosol protein extract, used in the incubation. For example, sample 101 contained equal amounts of the SWCNT (1) and cytosol protein extracts (1), but had no BSA (0). From Figure 4, we can see that, for both the BSA-coated SWCNTs, more BSA molecules were competed off the SWCNTs surface with the increase of cytosol protein amounts used in incubation. From the band intensity change, obviously more BSA molecules came off from the SWCNTs with a 1:1 coating ratio. Even when an equal amount of cytosol proteins was used to compete with the BSA coating, i.e. in the sample labeled as 144, both the BSA band intensity and the percent change in the band intensity were lower, compared to the incubation without the cytosol proteins were observed, compared to those seen in the sample of 111. Our result indicated that the higher BSA loading was more resistant to being replaced by proteins in the environment than the lower BSA loading used in coating. Nevertheless, the cytosol proteins that replaced the BSA molecules for binding to the SWCNT surface upon cell entry may be the key inducers for the toxicity of SWCNTs.
Figure 4.
Quantification of BSA in filtrate upon competition with different concentrations of cytosol protein. a) Incubate 1:1 ratio BSA-SWCNT mixture with no cytosol protein(110) and one time cystol protein(111). b) Incubate 1:4 ratio BSA-SWCNT mixture with no cytosol protein(140) and one time cystol protein(141) and four times cytosol protein (144). Inset graphs show the SDS-PAGE picture of BSA.
2.4 Quantification of Cellular Uptake of BSA Coated SWCNTs
Cell entry should be tested because it should precede any replacement of BSA and adsorption of the cytosol proteins on SWCNTs. It has been reported that SWCNTs enter cells mainly through an energy consuming process called endocytosis.[24] Previous works have shown that protein corona could greatly influence the cellular uptake of nanoparticles.[27] The impact could be related to the degree of perturbation to the cellular membranes. It has been reported that with the presence of high amounts of human serum albumin, the lipid bilayer vesicles experienced less perturbation from the nanoparticles.[28] Endocytosis of SWCNTs will likely start with an initial nonspecific adsorption of the hydrophobic part of SWCNTs on cell membrane.[4b] The bare nanoparticle surface may have higher affinity towards cell membrane comparing with the nanoparticle surrounded by protein ‘corona’.[29] Serum albumin is a common protein serving as a nonspecific transporter and osmotic pressure maintainer, and as a passive blocking protein to reduce nonspecific adsorption. Therefore, the coating of BSA on SWCNTs may possibly reduce cell entry through endocytosis, because the less hydrophobicity of the SWCNT surface after BSA coating could cause lower nonspecific interaction with the cell membrane.
To evaluate the cellular uptake of the BSA-coated SWCNTs, 12 mL of 10 µg/mL SWCNTs with the BSA coating ratio of 1:1 and 1: 4 were used to incubate with the fibroblast cells. Since incubation longer than 6 hours could induce significant cell death, a shorter incubation time of 3 hrs was employed in this test to ensure good cell viability during the uptake experiment. Following the incubation, the cells were washed to remove the free SWCNTs and treated with the mixture of 1 M NaOH and DMSO. Such a strong solution completely broke down the cells and also provided good suspension of the SWCNTs. The freely suspended SWCNTs were then quantified by NIR spectroscopy at the wavelength of 1050 nm, which has been proved in our previous work to be the center of SWCNT’s distinct absorption region S22 (900–1100 nm).[30] At this wavelength, the absorbance showed good linearity with the SWCNTs concentration (Figure S1 (showing both spectra and the calibration plot)), even when measured by spiking the known amounts of SWCNTs to the cell lysate. We then calculated the amount of SWCNT interacting with the cells either through cellular uptake or membrane adsorption. When coating the SWCNTs with an equal mass of BSA, 25.4±0.6% of all the SWCNTs added was taken by the cells. This percentage dropped to 18.5±0.8% when the SWCNTs were coated by 4-fold more BSA. Therefore, a thicker BSA coating may prevent the SWCNTs from attaching to the cell membrane or entering the cells, which may in turn reduce the cytotoxicity of SWCNTs.
2.5 Identification of Cytosol Proteins with High Affinity to the SWCNTs
Our results by far have pointed out that the SWCNTs could enter the cells or attach to the cell membrane; and some of the BSA coating could be replaced by cytosol proteins when the BSA-coated SWCNTs were incubated in cell lysate. Since the SWCNT-bound cytosol proteins could contribute to the cytotoxicity of SWCNTs, we isolated them for identification after incubating the cell lysate with the SWCNTs coated with BSA at either the 1:0, 1:1 or 1:4 ratio, again using the 300 MWCO filter. The unbound proteins were removed by through washing with three times of 1×PBS buffer, and the adsorbed cytosol proteins remained on top of the filter were digested by trypsin. The peptides were collected as the filtrate and analyzed by LC-MS/MS.
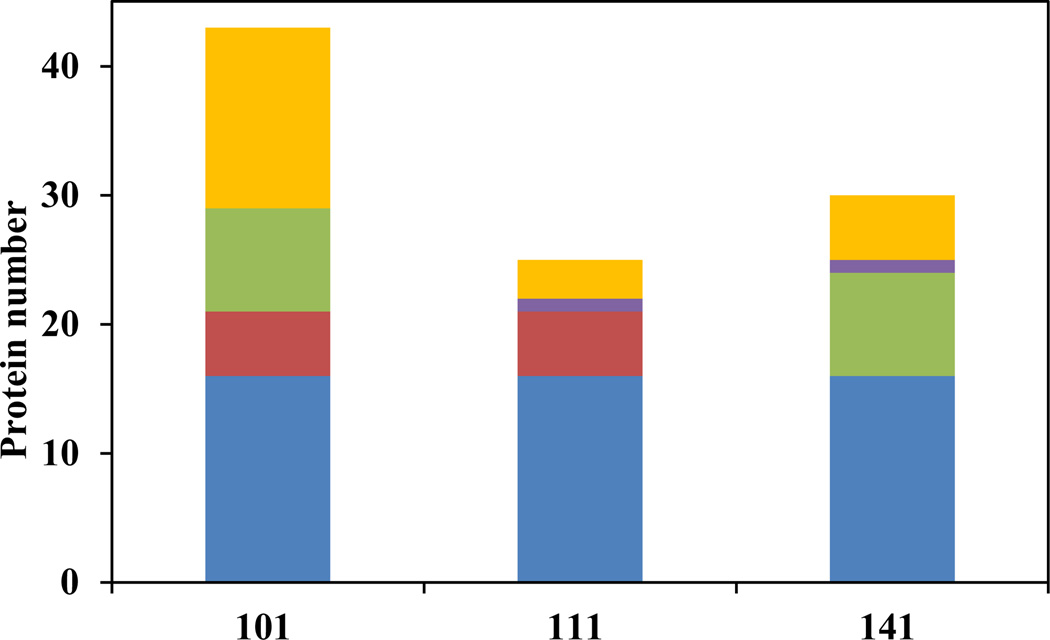
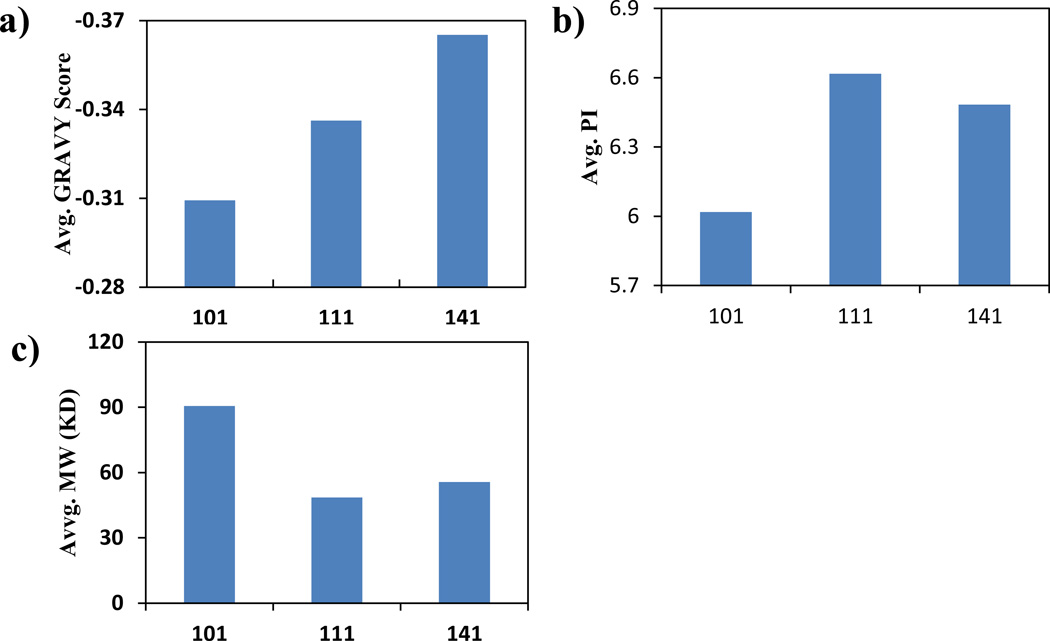
The protein adsorption experiments were repeated four times and only the proteins identified in at least 3 out of the 4 repeats were reported and analyzed here (Supporting Information Table S1). A total of 43 proteins were identified to be absorbed by the bare SWCNTs. This number dropped to 25 for the BSA-coated SWCNTs at the 1:1 ratio; and to 30 for the sample coated at the 1:4 ratio (Figure 5). Comparing with the uncoated SWCNTs, the ones coated by BSA adsorbed much fewer proteins, which supports that the BSA coating could serve as a passive layer to reduce the adsorption of cytosol proteins. We first analyzed the general physicochemical properties of the bound proteins: hydrophobicity, pI, and Mw (Figure 6). The hydrophobicity is represented by the GRAVY score averaged from all proteins found in different samples (Figure 6a). With increasing amounts of BSA used in the coating, a more negative GRAVY score was observed. Although the absolute changes in the GRAVY score were small along with quite large variances, the data suggested an overall decrease of the hydrophobicity of the bound proteins, agreeing with what was reported previously.[31] This result supports the speculation that BSA covers up the hydrophobic surface of SWCNTs, making the tubes more hydrophilic and thus more stably suspended in aqueous solutions.[32] The average pI value for the uncoated SWCNTs was slightly lower than those found in the BSA-coated tubes (Figure 6b), but mainly the protein coronas formed on all SWCNTs carried no significant charges with the average pI value close to 7. The more distinct change was in the average Mw of the adsorbed proteins (Figure 6c). A much higher average Mw around 90 kDa was found for the proteins adsorbed by the uncoated SWCNTs, but those for the BSA-coated SWCNTs were around 45–55 kDa. The more than 35% or even 100% surface coverage by BSA did not leave large enough bare tube surfaces for the bulky proteins to bind. To displace the BSA molecules off the tubes, space hindrances would need to be overcome, which is not favored for larger proteins.
Figure 5.
Adsorbed cytosol protein distribution in different SWCNT-BSA coating ratios (101: SWCNTs: BSA: Cytosol Protein =1:0:1, 111: SWCNTs: BSA: Cytosol Protein =1:1:1, 141: SWCNT: BSA: Cytosol Protein =1:4:1). The proteins shared by all three coating ratios were in blue, shared by two were in red, green and purple. The unique proteins with each coating ratios were in yellow.
Figure 6.
The calculated average score of proteins adsorbed with different coating ratios. a) average GRAVY score; b) average PI, c) average molecular weight.
We also looked at the individual proteins found in each incubation situation, hoping to reveal proteins potentially linked to the cytotoxicity of SWCNTs. Among all three samples, there are consistently 16 proteins being identified. These proteins could be sensitive to the shape of the SWCNTs but not the surface property. For example, actin, the actin binding transgelin, and tubulin were included in this group. Actin and tubulin can form structural filaments or tubular structures,[33] which may align better with the long and thin SWCNTs. Binding to actin, have been frequently reported to be the target molecules or structures for CNTs entering cells,[34] and the binding could be mediated via hydrophobic interactions. [31a] Three nucleus proteins were found to bind to all three SWCNTs: Nucleolin, Nucleophosmin, and Nucleosome assembly protein 1-like 1. They are all involved in chromosome formation. Binding to the cytoskeleton proteins like actin and tubulin, and to proteins involved chromosome formation could both affect cell proliferation and reduce cell viability.
There are 11 proteins uniquely found only in the incubation with the uncoated SWCNTs. Such proteins could be potentially of great importance in causing the cytotoxicity of bare carbon nanotubes without protein corona. Among them, clathrin controls the clathrin-medicated endocytosis, which is reported to be one of the cell uptake mechanism for SWCNTs.[35] Another protein, Eukaryotic translation initiation factor 5A-1 was considered to be important in apoptosis process.[36] Prevention of adsorption of these proteins by the BSA coating could definitely contribute to the reduction of the observed cytotoxicity of the BSA-coated SWCNTs.
Moreover, one protein was found in the 101 and 111 incubations but not in the 141 one. It is Peroxiredoxin-1, which is critical in eliminating peroxides generated during metabolism and interacting with signaling molecules.[37] Oxidative stress is often observed in the SWCNTs infected cells. Then recruiting of this particular protein may induce two possible impacts, depending on whether the protein could be deactivated upon adsorption. If the protein can be adsorbed in its active form, then accumulation of peroxiredoxin-1 on the surface of SWCNTs may eliminate the peroxides generated in situ. However, if it gets deactivated upon adsorption, it may increase local oxidative stress since its anti-oxidant role is interrupted.
3. Conclusion
Our results support that, the cause of the reduced cytotoxicity of SWCNTs coated with BSA could be via reduction of interaction between the SWCNTs and the cellular proteins. BSA would wrap around the tubes and prevent strong adsorption of the cytosol proteins on the SWCNTs. Since some of the potential adsorbents carry out important cellular functions such as maintaining cell structure and reducing levels of peroxide, prevention of the adsorption of these proteins could lead to enhanced cell viability upon the attack by SWCNTs. Our study is meaningful for the future engineering safe nanomaterials for applications in biotechnologies and drug deliveries; also implies the potential behaviors of biopolymer-coated nanomaterials in the environment.
4. Experimental Section
Chemicals
All chemicals were purchased from commercial suppliers without further treatment, unless otherwise noted. Carboxylic acid-functionalized SWCNTs (SWCNT-COOH, D = 4–5 nm× L = 0.5–1.5 µm), bovine serum albumin (BSA) and Somatic cell ATP releasing reagent were purchased from Sigma (St. Louis, MO). CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS) kit and ENLITEN®ATP Assay System were purchased from Promega (Madison, WI). Pierce™ Bicinchoninic Acid (BCA) Protein Assay Kit was purchased from Thermo Scientific (Rockford, IL). Dulbecco's Modified Eagle's Medium (DMEM), penicillin-streptomycin solution and fetal bovine serum (FBS) for cell culture were obtained from American Type Culture Collection (ATCC, Manassas, VA). The water and organic solvent used in this research were all HPLC grade from Fisher Scientific (Fair Lawn, NJ). All the salts used to prepare the buffer were also purchased from Fisher Scientific (Fair Lawn, NJ). The human fibroblast cell line GM00637 was obtained from the Coriell Institute for Medical Research (Camden, NJ). Vivaspin 500 (300,000 Mw cut-off value) centrifugal spin columns were purchased from Sartorius Stedim Biotech (Göttingen, Germany). SYPRO® Ruby Protein Gel Stain was purchased from Life Technologies (Carlsbad, CA).
Preparation of BSA-coated SWCNTs
The SWCNT powder was first dispersed in DI water with mild sonication to make a solution of 100 µg/mL. The stock was kept in 4°C until usage. A 1 mg/mL BSA solution was prepared in the phosphate buffer (10mM, pH at 7.4). Then the SWCNTs and BSA were mixed in a 2-mL centrifuge tube and diluted with the phosphate buffer to a final volume of 1 mL, and sonicated mildly in ice water bath for 30 minutes. For both the 1:1 and 1:4 (SWCNT : BSA) ratios, the final concentration of SWCNTs was 25 µg/mL. For the cytotoxicity and cellular uptake test, the preparation is similar, but SWCNTs and BSA were directly dispersed in DMEM without FBS instead of in the phosphate buffer.
Cytotoxicity, ATP Assay and Cellular Uptake of SWCNTs
The cells were cultured in DMEM supplemented with 10% non-inactivated FBS, 100 µg/mL penicillin and 100 µg/mL streptomycin, and maintained in a 5% CO2 humidified atmosphere at 37 °C. Around 5×103 cells were added to the wells of a 96-well plate and incubated overnight to reach around 70% confluence, followed with the addition of 200 µL BSA-coated SWCNTs at the concentration of 50 µg/mL, 25µg/mL and 5µg/mL in DMEM without FBS. After 6 or 12 hrs incubation, the SWCNTs solutions were removed, and the cells were washed with PBS (pH 7.4) buffer for three times.
For the cell proliferation assay, MTS solution was added to each well of plate following the manual protocol, followed by additional 1-hrs incubation at 37 °C. The absorbance was recorded at 490 nm, and the mean absorbance of control cells (no BSA or SWCNTs added) served as the reference value of 100 % for calculating cellular viability.
To test ATP production, after 12 hrs incubation, the cells were washed three times. The Somatic cell ATP releasing reagent was first introduced to release ATP from cells. Then, ATP concentration was detected using the ENLITEN®ATP Assay System, following the standard protocol. The mean luminescence of control cells were used as 100% percent to calculate ATP percentage.
Cellular uptake of SWCNTs was measured by Near-Infrared (NIR) absorption spectroscopy using the previously reported method.[7] Twelve mL of the 10 µg/mL BSA-coated SWCNT prepared in DMEM was added to the cell culture flask containing about 5 million cells and incubated for 3 hours. Then, cells were washed three times with ice-cold PBS to remove the free SWCNTs in media. Three mL of 1M NaOH and DMSO (2:1) were used to dissolve the cells and disperse SWCNTs. The concentration of SWCNTs was determined by the NIR spectrometer (Varian Cary 500, Palo Alto, California) at the wavelength of 1,050 nm.
Atomic Force Microscopy of SWCNTs
The samples of the uncoated SWCNTs and SWCNTs coated by BSA at different mass ratio were deposited onto silica wafer, which was cleaned with acetone and isopropanol and then dried by nitrogen. Morphology of BSA on SWCNT was analyzed by a Veeco D3100 atomic force microscope (Bruker, Camarillo, CA). Images were acquired in tapping model, using the RTESP AFM probe obtained from Bruker. The scanning rate was 0.5 Hz, with integral gain of 0.1 and proportional gain of 0.2. The amplitude set-point was set at 1.327 V, and the tip velocity was 0.975 µm/s. Image analysis was performed with NanoScope V530 (Bruker).
Quantification of BSA adsorption on SWCNT
After incubation of BSA with SWCNT, the mixture was separated by 300 kDa centrifugal filter. The BSA remaining in the filtrate was quantified using a BCA assay. The filtrate was incubated with BCA reagent for 30 mins at 60 °C. After cooling, the absorbance at 562 nm was measured using a Cary 50 spectrophotometer. The amount of BSA adsorbed on the SWCNTs was then determined by difference.
Cytosol protein extraction and BSA displacement test
Human fibroblast cells were first washed with PBS three times, and trypsin-EDTA was used to detach the cells from the flask. Following this, cells were washed three times with ice-cold PBS. Cell lysis buffer and protease inhibitor was added into the cells and incubated on ice for 30 min with vortexing every 10 min. After this, the lysate was centrifuged at 14,000g for 15 min, and the supernatant was collected as the cytosol protein. This supernatant was stored at −80 °C until use. The concentration was determined by BCA assay.
BSA displacement tests were carried out by mixing certain amount of cytosol protein with the mixture of BSA and SWCNTs. After incubation for 2 hrs at 4 °C, the mixture was separated by 300,000 MW centrifugal filters. SDS PAGE was carried out to determine the abundance of BSA in the filtrate. SYPRO Ruby staining was used to visualize the gel and Image J was used to quantify the intensity of the bands.
Identification of the SWCNT-protein coating
After separation of SWCNTs by a 300,000 MW filter, the proteins absorbed on the SWCNTs were digested by trypsin directly on the filter following standard protocol. In short, proteins were first denatured in boiling water for 10 min. After cooling of the sample to room temperature, DTT and IAA were added sequentially. Following this, trypsin was added to the protein and incubated at 37 °C overnight. The as prepared peptides were collected and desalted using Ziptips. The peptides were were analyzed by LC-MS/MS. The analysis of peptide mixtures was carried out on an LTQ linear ion-trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a nano electrospray ionization (NSI) source. Full MS spectra were recorded over a 300–2000 m/z range followed by four sequential data-dependent MS/MS scans. Dynamic exclusion was implemented. The 200 nL/min nano flow was achieved by split flow from a 200 µL/min delivered by a Waters 2695 HPLC pump. The peptide separation was performed on a self-packed (packed 10 cm with 3 µm C18 beads, Dr. Maisch HPLC GmbH, Germany) PicoFrit column (75 µm in tubing ID and 15 µm in tip ID, New Objective, Inc., Woburn, MA). Mobile phase A was 0.01% formic acid in water and mobile phase B was acetonitrile. The gradient started at 0% B for 10 min of enrichment and then linearly increased to 100% B within 80 min. The mobile phase was kept at isocratic conditions (100% B) for 30 min and then returned to 0% B. The acquired data was analyzed by database searching with X!Tandem. The searching criteria were set as default of the website. Four repeats were carried out for all three SWCNT : BSA : cell lysate ratios 1:0:1 (101), 1:1:1(111); and 1:4:1(141) and only the proteins that appeared at least three times in the four repeats with more than one peptide being identified each time were considered to be the reliable binders.
Supplementary Material
Acknowledgements
The project was originally supported by National Institute of Environmental Health Sciences Grant No. 1R21ES017870-01A1; and then continued under the support of City of Hope – UC Riverside Biomedical Research Initiative (CUBRI). Y. Liu is thankful for the support from the Environmental Toxicology Program, UC Riverside.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Yang Liu, Environmental Toxicology Graduate Program, University of California, Riverside, CA 92521, USA.
Lei Ren, Environmental Toxicology Graduate Program, University of California, Riverside, CA 92521, USA.
Dong Yan, Center for Nanoscale Science and Engineering, University of California, Riverside, CA 92521, USA.
Wenwan Zhong, Email: wenwan.zhong@ucr.edu, Environmental Toxicology Graduate Program and Department of Chemistry, University of California, Riverside, CA 92521, USA.
References
- 1.Ball P. Nature. 2001;414(6860):142–144. doi: 10.1038/35102721. [DOI] [PubMed] [Google Scholar]
- 2.Baughman RH, Cui C, Zakhidov AA, Iqbal Z, Barisci JN, Spinks GM, Wallace GG, Mazzoldi A, De Rossi D, Rinzler AG, Jaschinski O, Roth S, Kertesz M. Science. 1999;284(5418):1340–1344. doi: 10.1126/science.284.5418.1340. [DOI] [PubMed] [Google Scholar]
- 3.(a) Lin Y, Lu F, Tu Y, Ren Z. Nano. Lett. 2003;4(2):191–195. [Google Scholar]; (b) So H-M, Won K, Kim YH, Kim B-K, Ryu BH, Na PS, Kim H, Lee J-O. J. Am. Chem. Soc. 2005;127(34):11906–11907. doi: 10.1021/ja053094r. [DOI] [PubMed] [Google Scholar]
- 4.(a) Mundra RV, Wu X, Sauer J, Dordick JS, Kane RS. Curr. Opin. Biotechnol. 2014;28(0):25–32. doi: 10.1016/j.copbio.2013.10.012. [DOI] [PubMed] [Google Scholar]; (b) Shi Kam NW, Jessop TC, Wender PA, Dai H. J. Am. Chem. Soc. 2004;126(22):6850–6851. doi: 10.1021/ja0486059. [DOI] [PubMed] [Google Scholar]; (c) Bianco A, Kostarelos K, Prato M. Curr. Opin. Biotechnol. 2005;9(6):674–679. doi: 10.1016/j.cbpa.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Gong H, Peng R, Liu Z. Adv. Drug Delivery Rev. 2013;65:1951–1963. doi: 10.1016/j.addr.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Castranova V, Schulte PA, Zumwalde RD. Acc. Chem. Res. 2012;46(3):642–649. doi: 10.1021/ar300004a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang K, Liu Z. Curr Drug Metab. 2012;13:1057–1067. doi: 10.2174/138920012802850029. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Zhao Y, Sun B, Chen C. Acc. Chem. Res. 2012;46(3):702–713. doi: 10.1021/ar300028m. [DOI] [PubMed] [Google Scholar]
- 9.Ren L, Yan D, Zhong W. Carbon. 2012;50(3):1303–1310. doi: 10.1016/j.carbon.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuo G, Huang Q, Wei G, Zhou R, Fang H. Acs Nano. 2010;4(12):7508–7514. doi: 10.1021/nn101762b. [DOI] [PubMed] [Google Scholar]
- 11.Park KH, Chhowalla M, Iqbal Z, Sesti F. J. Biol. Chem. 2003;278(50):50212–50216. doi: 10.1074/jbc.M310216200. [DOI] [PubMed] [Google Scholar]
- 12.(a) Mu Q, Liu W, Xing Y, Zhou H, Li Z, Zhang Y, Ji L, Wang F, Si Z, Zhang B, Yan B. J. Phys. Chem. C. 2008;112(9):3300–3307. [Google Scholar]; (b) Sun Y-P, Fu K, Lin Y, Huang W. Acc. Chem. Res. 2002;35(12):1096–1104. doi: 10.1021/ar010160v. [DOI] [PubMed] [Google Scholar]
- 13.Sayes CM, Liang F, Hudson JL, Mendez J, Guo W, Beach JM, Moore VC, Doyle CD, West JL, Billups WE, Ausman KD, Colvin VL. Toxicol. Lett. 2006;161(2):135–142. doi: 10.1016/j.toxlet.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 14.(a) Zheng M, Jagota A, Semke ED, Diner BA, McLean RS, Lustig SR, Richardson RE, Tassi NG. Nat. Mater. 2003;2(5):338–342. doi: 10.1038/nmat877. [DOI] [PubMed] [Google Scholar]; (b) Nepal D, Geckeler KE. Small. 2006;2(3):406–412. doi: 10.1002/smll.200500351. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Humphreys ES, Chung S-Y, Delduco DF, Lustig SR, Wang H, Parker KN, Rizzo NW, Subramoney S, Chiang Y-M, Jagota A. Nat. Mater. 2003;2(3):196–200. doi: 10.1038/nmat833. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X, Liu R, Chi Z, Teng Y, Qin P. J. Phys. Chem. B. 2010;114(16):5625–5631. doi: 10.1021/jp100903x. [DOI] [PubMed] [Google Scholar]
- 17.Ge C, Du J, Zhao L, Wang L, Liu Y, Li D, Yang Y, Zhou R, Zhao Y, Chai Z, Chen C. Proc. Natl. Acad. Sci. USA. 2011;108(41):16968–16973. doi: 10.1073/pnas.1105270108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt BD, Dahl KN, Islam MF. Small. 2011;7(16):2348–2355. doi: 10.1002/smll.201100437. [DOI] [PubMed] [Google Scholar]
- 19.Dutta D, Sundaram SK, Teeguarden JG, Riley BJ, Fifield LS, Jacobs JM, Addleman SR, Kaysen GA, Moudgil BM, Weber TJ. Toxicol. Sci. 2007;100(1):303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- 20.Dominguez-Medina S, Blankenburg J, Olson J, Landes CF, Link S. ACS Sustainable Chem. Eng. 2013 doi: 10.1021/sc400042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Johnston HJ, Hutchison GR, Christensen FM, Peters S, Hankin S, Aschberger K, Stone V. Nanotoxicology. 2010;4(2):207–246. doi: 10.3109/17435390903569639. [DOI] [PubMed] [Google Scholar]; (b) Jacobsen NR, Pojana G, White P, Møller P, Cohn CA, Smith Korsholm K, Vogel U, Marcomini A, Loft S, Wallin H. Environ. Mol. Mutagen. 2008;49(6):476–487. doi: 10.1002/em.20406. [DOI] [PubMed] [Google Scholar]
- 22.(a) Yang Z, Zhang Y, Yang Y, Sun L, Han D, Li H, Wang C. Nanomed: Nanotechnol. 2010;6(3):427–441. [Google Scholar]; (b) Manna SK, Sarkar S, Barr J, Wise K, Barrera EV, Jejelowo O, Rice-Ficht AC, Ramesh GT. Nano. Lett. 2005;5(9):1676–1684. doi: 10.1021/nl0507966. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Shvedova AA, Pietroiusti A, Fadeel B, Kagan VE. Toxicol. Appl. Pharm. 2012;261(2):121–133. doi: 10.1016/j.taap.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright MRTAK. Biophys. 1975;15(137–141) doi: 10.1016/s0006-3495(75)85797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Birch ME, Ruda-Eberenz TA, Chai M, Andrews R, Hatfield RL. Ann. Occup. Hyg. 2013 doi: 10.1093/annhyg/met042. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fujiwara A, Ishii K, Suematsu H, Kataura H, Maniwa Y, Suzuki S, Achiba Y. Chem. Phys. Lett. 2001;336(3–4):205–211. [Google Scholar]
- 25.Li X, Chen W, Zhan Q, Dai L, Sowards L, Pender M, Naik RR. J. Phys. Chem. B. 2006;110(25):12621–12625. doi: 10.1021/jp061518d. [DOI] [PubMed] [Google Scholar]
- 26.Yaron PN, Holt BD, Short PA, Losche M, Islam MF, Dahl KN. J. Nanobiotechnol. 2011;9:45. doi: 10.1186/1477-3155-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.(a) Aggarwal P, Hall JB, McLeland CB, Dobrovolskaia MA, McNeil SE. Adv. Drug Deliv. Rev. 2009;61(6):428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lesniak A, Fenaroli F, Monopoli MP, Åberg C, Dawson KA, Salvati A. Acs Nano. 2012;6(7):5845–5857. doi: 10.1021/nn300223w. [DOI] [PubMed] [Google Scholar]
- 28.Chen R, Choudhary P, Schurr RN, Bhattacharya P, Brown JM, Ke PC. Appl. Phys. Lett. 2012;100(1) doi: 10.1063/1.3672035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walczyk D, Bombelli FB, Monopoli MP, Lynch I, Dawson KA. J. Am. Chem. Soc. 2010;132(16):5761–5768. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 30.Ren L, Zhong W. Environ. Sci. Technol. 2010;44(18):6954–6958. doi: 10.1021/es101821m. [DOI] [PubMed] [Google Scholar]
- 31.Kyte JD, Russell F. J. Mol. Biol. 1982;157(1):27. [Google Scholar]
- 32.Cedervall T, Lynch I, Lindman S, Berggård T, Thulin E, Nilsson H, Dawson KA, Linse S. P. Natl. Acad. Sci. USA. 2007;104(7):2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(a) Shams H, Holt BD, Mahboobi SH, Jahed Z, Islam MF, Dahl KN, Mofrad MRK. Acs Nano. 2013 doi: 10.1021/nn402865e. [DOI] [PubMed] [Google Scholar]; (b) Dinu CZ, Bale SS, Zhu G, Dordick JS. Small. 2009;5(3):310–315. doi: 10.1002/smll.200801434. [DOI] [PubMed] [Google Scholar]
- 34.(a) Holt BD, Short PA, Rape AD, Wang Y-l, Islam MF, Dahl KN. Acs Nano. 2010;4(8):4872–4878. doi: 10.1021/nn101151x. [DOI] [PubMed] [Google Scholar]; (b) Sargent LM, Hubbs AF, Young SH, Kashon ML, Dinu CZ, Salisbury JL, Benkovic SA, Lowry DT, Murray AR, Kisin ER, Siegrist KJ, Battelli L, Mastovich J, Sturgeon JL, Bunker KL, Shvedova AA, Reynolds SH. Mutat. Res-Gen. Tox. En. 2012;745(1–2):28–37. doi: 10.1016/j.mrgentox.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kam NWS, Liu Z, Dai H. Angew. Chem. Int. Ed. 2006;118(4):591–595. [Google Scholar]
- 36.Li A-L, Li H-Y, Jin B-F, Ye Q-N, Zhou T, Yu X-D, Pan X, Man J-H, He K, Yu M, Hu M-R, Wang J, Yang S-C, Shen B-F, Zhang X-M. J. Biol. Chem. 2004;279(47):49251–49258. doi: 10.1074/jbc.M407165200. [DOI] [PubMed] [Google Scholar]
- 37.Neumann CA, Cao J, Manevich Y. Cell Cycle. 2009;8(24):4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.