Abstract
Notch signaling induced by canonical Notch ligands is critical for normal embryonic development and tissue homeostasis through the regulation of a variety of cell fate decisions and cellular processes. Activation of Notch signaling is normally tightly controlled by direct interactions with ligand-expressing cells and dysregulated Notch signaling is associated with developmental abnormalities and cancer. While canonical Notch ligands are responsible for the majority of Notch signaling, a diverse group of structurally unrelated non-canonical ligands has also been identified that activate Notch and likely contribute to the pleiotropic effects of Notch signaling. Soluble forms of both canonical and non-canonical ligands have been isolated, some of which block Notch signaling and could serve as natural inhibitors of this pathway. Ligand activity can also be indirectly regulated by other signaling pathways at the level of ligand expression, serving to spatio-temporally compartmentalize Notch signaling activity and integrate Notch signaling into a molecular network that orchestrates developmental events. Here, we review the molecular mechanisms underlying the dual role of Notch ligands as activators and inhibitors of Notch signaling. Additionally, evidence that Notch ligands function independent of Notch are presented. We also discuss how ligand post-translational modification, endocytosis, proteolysis and spatio-temporal expression regulate their signaling activity.
1. INTRODUCTION
The Notch pathway functions as a core signaling system during embryonic development and is also required for the regulation of tissue homeostasis and stem cell maintenance in the adult (Artavanis-Tsakonas et al., 1999; Gridley, 1997, 2003). Ligand-induced Notch signaling directs the specification of a variety of cell types and contributes to tissue patterning and morphogenesis through effects on cellular differentiation, proliferation, survival and apoptosis (Bray, 2006; Fiuza and Arias, 2007). Given the widespread usage of the Notch pathway in different cell types and cellular processes, it is not surprising that defects in Notch ligands are associated with hereditary diseases such as Alagille syndrome and spondylocostal dysostosis and that aberrant ligand expression is detected in several cancers (Koch and Radtke, 2007; Leong and Karsan, 2006; Piccoli and Spinner, 2001; Turnpenny et al., 2007).
The canonical ligands that bind and activate Notch receptors are integral cell surface proteins, and thus activation of Notch signaling is dependent on direct cell-to-cell interactions. The transmembrane nature of Notch ligands serves to limit signaling to local cell interactions and additionally provides a signaling system for cells to communicate directly with their neighbors. Interestingly, during certain developmental processes, ligands have been found to activate Notch expressed on the surface of distantly located cells. Such long range signaling may utilize actin-based cellular projections to deliver activating signals to Notch at distant sites (de Joussineau et al., 2003). In support of such a model, the ligand Delta appears to concentrate in filopodia-like projections, possibly inducing and stabilizing these structures to facilitate long-range signaling (de Joussineau et al., 2003; Renaud and Simpson, 2001). Similarly, the C. elegans distal tip cell has long cellular processes that contain the ligand Lag2 and appear to extend all the way to the mitotic/meiotic border where they regulate proliferation of the germ line through activation of the Notch homolog Glp1 (Fitzgerald and Greenwald, 1995).
Signaling induced by Notch cells following engagement with ligand cells involves a series of proteolytic cleavages in Notch to release the intracellular domain that functions directly as the biologically active signal transducer (Kopan and Ilagan, 2009). During maturation and trafficking to the cell surface, the Notch receptor is processed by a furin-like protease to produce an intramolecular heterodimer that predisposes Notch to proteolytic activation by ligand. Interactions with ligand cells results in an extracellular juxtamembrane cleavage in Notch catalyzed by an A-Disintegrin-And-Metalloprotease (ADAM), which is followed by an intramembrane cleavage by γ-secretase to release the Notch intracellular domain (NICD) from the membrane (Fig. 1). NICD translocates to the nucleus where it functions directly in signal transduction through complexing with the CSL (CBF1, Su(H), LAG1) DNA binding protein and transcriptional coactivators to switch on expression of Notch target genes such as hairy and enhancer of split (HES) family. The mechanism and details of Notch transcriptional activation are covered extensively in the Chapter by Bray. In addition to the well-characterized role for activation of Notch signaling through cell-cell interactions (trans-interactions), ligands can also interact with Notch cell autonomously (cis-interactions) leading to inhibition of Notch signaling. The nature and mechanisms underlying the inhibitory role of Notch ligands will be discussed in section 3 of this review. Additional characteristics of canonical and non-canonical Notch ligands required to activate signaling are discussed below.
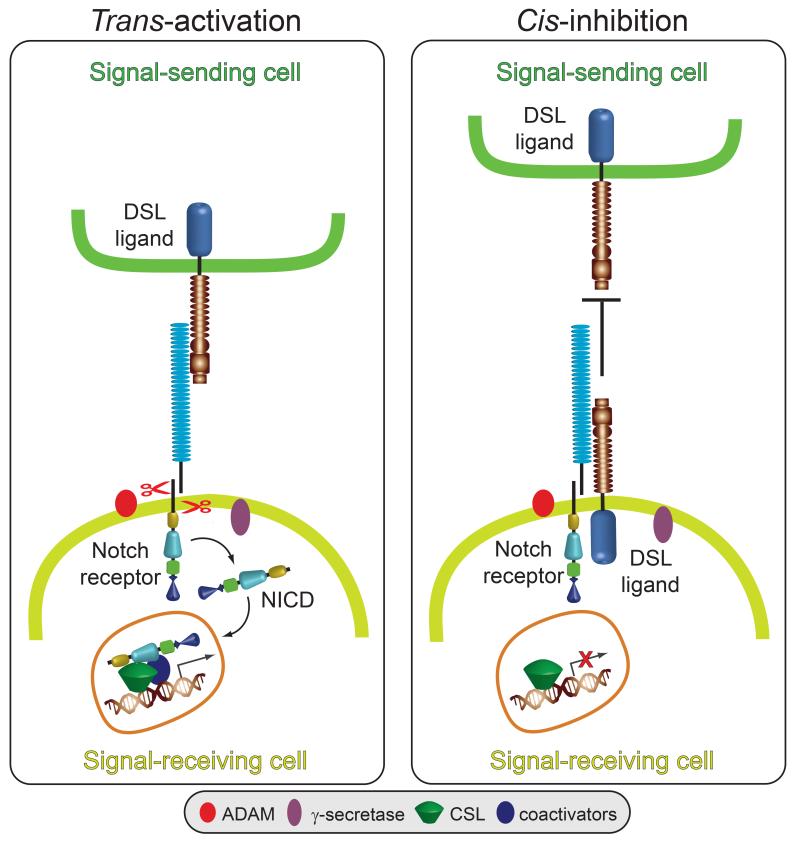
Fig. 1. Models for DSL ligand trans-activation and cis-inhibition in Notch signaling.
Ligand expressed on the surface of the signal-sending cell binds to Notch expressed on the surface of the signal-receiving cell (trans-interactions) and induces sequential cleavages by A-Disintegrin-And-Metalloprotease (ADAM) and -secretase in Notch releasing the Notch intracellular domain (NICD) from the membrane. NICD translocates to the nucleus where it directly interacts with the CSL (CBF1, Su(H), LAG1) transcription factor and recruits coactivators to induce Notch target gene expression. Ligand binding to Notch expressed in the same cell (cis-interactions) prevents Notch activation by trans- ligand by competing with trans-ligand for Notch binding.
2. CANONICAL NOTCH LIGAND STRUCTURE
The majority of Notch signaling is induced by a family of DSL ligands that are characterized by the presence of a DSL (Delta, Serrate, and Lag2) domain (Henderson et al., 1994; Tax et al., 1994). The mammalian DSL ligands are classified as either Delta-like (Dll1, Dll3 and Dll4) or Serrate (Jagged)-like (Jagged1 and Jagged2) based on homology to their Drosophila prototypes Delta and Serrate (Kopan and Ilagan, 2009). DSL ligands are type1 transmembrane proteins that share a common modular arrangement in their extracellular domains comprising an N-terminal (NT) domain followed by the DSL domain and multiple tandemly arrayed Epidermal Growth Factor (EGF)-like repeats (both calcium binding and non-calcium binding, see Fig. 2).
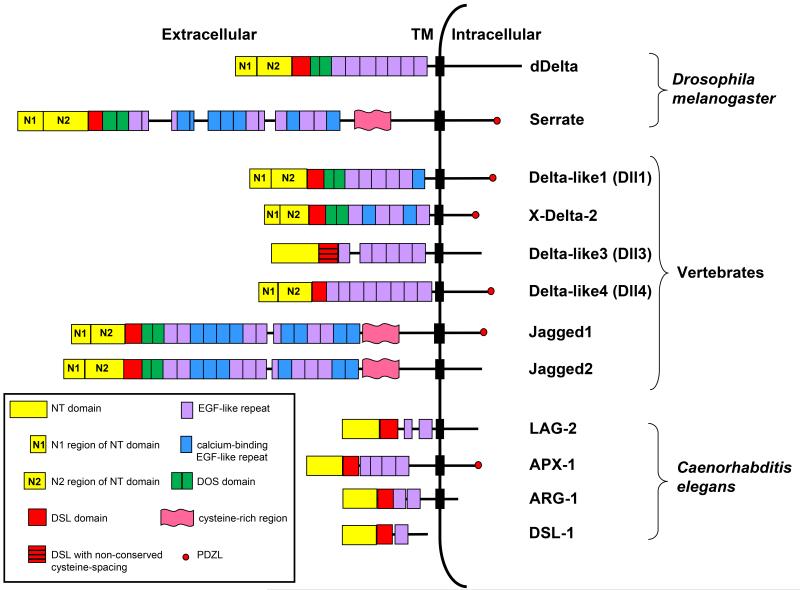
Fig. 2. Structural domains of canonical ligands.
The extracellular domains of canonical ligands are characterized by the presence of an N-terminal (NT) domain followed by a Delta/Serrate/LAG-2 (DSL) domain and multiple tandemly arranged Epidermal Growth Factor (EGF)-like repeats (see text for details). The DSL domain together with the flanking NT domain and the first two EGF repeats containing the Delta and OSM-11-like proteins (DOS) motif are required for canonical ligands to bind Notch. The NT domain of vertebrate and Drosophila ligands is subdivided into a region containing six conserved cysteine residues, N1 and a cysteine- free region, N2. Serrate/Jagged ligands contain an additional cysteine-rich region not present in Delta-like ligands. The intracellular domains of some canonical ligands contain a carboxy-terminal PSD-95/Dlg/ZO-1-ligand (PDZL) motif that plays a role independent of Notch signaling. C. elegans DSL ligands lack a DOS motif but have been proposed to cooperate with DOS-only containing ligands (not depicted) to activate Notch signaling. Dll3 is the most structurally divergent vertebrate DSL ligand and lacks structural features required by other DSL ligands to bind and activate Notch.
The DSL is a degenerate EGF-like repeat that is necessary but not sufficient for interactions with Notch (Shimizu et al., 1999). Mutations in conserved residues within the DSL domain are associated with losses in Notch signaling in both vertebrate and invertebrates (Henderson et al., 1997; Henderson et al., 1994; Morrissette et al., 2001; Parks et al., 2006; Tax et al., 1994; Warthen et al., 2006). In particular, DSL mutations in Jagged1 are linked to Alagille syndrome. In addition, a conserved motif called DOS (Delta and OSM-11-like proteins) has been identified within the first two EGF-like repeats that is proposed to cooperate with the DSL domain (Komatsu et al., 2008). Mutational and structural studies indicate a contributory role for the DOS domain in Notch binding and signaling distinguishing them from the remaining EGF-like repeats (Cordle et al., 2008; Komatsu et al., 2008; Parks et al., 2006; Shimizu et al., 1999). In particular, the sequence and spacing within the DOS are important for signaling (Geffers et al., 2007). Furthermore, mutations associated with Alagille syndrome and the congenital disorder Tetralogy of Fallot map to the DOS motif of Jagged1, highlighting the importance of this region in Notch signaling (Eldadah et al., 2001; Guarnaccia et al., 2009; Warthen et al., 2006). Surprisingly, Dll4 and Dll3 and all C. elegans DSL ligands lack a DOS motif and it has been proposed that optimal activation of Notch signaling by DSL domain-only containing ligands requires cooperative Notch binding by DOS domain containing non-canonical ligands (Komatsu et al., 2008).
In addition to the DSL and DOS domains, sequences N-terminal to the DSL are also conserved among the canonical ligands that appear important for function (Fleming, 1998; Henderson et al., 1997; Parks et al., 2006). The NT domain can be subdivided into two distinct regions based on differential cysteine content: N1 is cysteine-rich while N2 is cysteine-free (Parks et al., 2006), and Alagille mutations map to the N1 and N2 regions of Jagged1 (Morrissette et al., 2001; Warthen et al., 2006). More recently, a conserved glycosphingolipid (GSL)-binding motif (GBM) has been identified within the N2 region that may regulate ligand membrane association and endocytosis (Hamel et al., 2010).
Despite the similarity in the overall modular organization of the extracellular domains (Fig. 2), some structural differences exist among the DSL ligands. For example, the number of EGF-like repeats varies as does the spacing between this motif. Moreover, the Serrate-like Jagged ligands have a cysteine-rich region sharing partial homology with the von Willebrand factor type C domain (VWFC) that is absent from Delta ligands (Vitt et al., 2001). Although the non-DOS containing EGF-like repeats have not been reported to regulate signaling activity (Henderson et al., 1997; Parks et al., 2006), mutations in some of these repeats in Jagged1 are associated with Alagille syndrome (Morrissette et al., 2001; Warthen et al., 2006).
The intracellular domains of DSL ligands exhibit the lowest level of overall sequence homology (Pintar et al., 2007). With the exception of Dll3, they contain multiple lysine residues that are potential sites for modification by distinct E3 ubiquitin ligases (outlined in section 4). Although ubiquitination is critical for ligands to activate Notch signaling, the role that this modification plays is poorly defined. In addition, most, but not all DSL ligands have a C-terminal PDZ (PSD-95/Dlg/ZO-1)-ligand motif that promotes interactions with the actin cytoskeleton that appear to play a role independent of Notch signaling (Pintar et al., 2007). Although PDZ-ligand motifs are predicted for some invertebrate DSL ligands, including Drosophila Serrate and C. elegans APX-1 (Sheng and Sala, 2001), the functional relevance remains to be determined.
Finally, Dll3 is the most structurally divergent DSL ligand having a degenerate DSL domain (Dunwoodie et al., 1997) and lacking both a DOS motif (Komatsu et al., 2008) and intracellular domain lysine residues (Pintar et al., 2007). Although these structural features are critical for ligand signaling activity, losses in Dll3 are associated with vertebral segmentation and rib malformations similar to those caused by defects in Notch signaling (Dunwoodie, 2009). Dll3, however, does not bind Notch in trans or activate Notch signaling (Ladi et al., 2005), and the majority of Dll3 is detected in the Golgi, with relatively little, if any, cell surface expression (Geffers et al., 2007). Gene replacement studies in mice clearly show that Dll3 cannot substitute for the loss of Dll1 (Geffers et al., 2007), indicating that these DSL ligands are not functionally equivalent. In contrast to Dll1 that both activates and inhibits Notch signaling, Dll3 functions exclusively as a Notch antagonist (Ladi et al., 2005). Despite these findings, it is still unclear how Dll3 functions in Notch signaling, and while the Dll3 structural differences are predicted to perturb ligand signaling activity, it is difficult to reconcile how Dll3 in the Golgi would participate in Notch signaling.
3. CANONICAL LIGANDS AS INHIBITORS OF NOTCH SIGNALING
The Notch receptors and DSL ligands are widely expressed during development, and in many cases, interacting cells express both ligands and receptors. Cells take on distinct fates because Notch signaling is consistently activated in only one of the two interacting cells, indicating that the signaling polarity must be highly regulated. The relative levels of Notch and its ligands present on interacting cells are thought to establish the signaling polarity necessary to ensure that the correct cell fates are generated at the right time in development. In fact, developmental processes are sensitive to Notch ligand and receptor gene dosage, underscoring the importance of Notch ligand and receptor expression levels for normal signaling. In humans, haploinsufficiency of either Jagged1 or Notch2 is associated with Alagille syndrome (McDaniell et al., 2006), while Notch1 haploinsufficiency is implicated in a subtype of inherited aortic disease (Garg et al., 2005).
Studies in flies and worms have identified positive and negative transcriptional feedback mechanisms that amplify small differences in Notch and DSL ligand expression that could introduce a bias for which of the interacting cells sends or receives the Notch signal (Greenwald and Rubin, 1992; Seugnet et al., 1997b). If this were the case, cells competent to send a signal would be expected to display higher DSL ligand levels then cells receiving the Notch signal; however, Delta expression appears uniform among cells undergoing lateral inhibition during selection of the neural fate (Kooh et al., 1993; Kopczynski and Muskavitch, 1989). Therefore, mechanisms in addition to transcription must exist to ensure fidelity in cell fate decisions regulated by Notch signaling. In this regard, interactions between Notch and its ligand in the same cell may provide additional mechanisms to regulate the cell’s potential to send or receive a Notch signal.
3.1. Cis-interactions between ligand and Notch inhibit signaling by trans-ligand
In contrast to the trans-interactions between Notch ligand and receptor cells that activate signaling (Fig. 1), interactions between Notch ligands and receptors in the same cell result in inhibition of signaling through a poorly defined process of cis-inhibition (Glittenberg et al., 2006; Jacobsen et al., 1998; Klein and Arias, 1998; Klein et al., 1997; Ladi et al., 2005; Micchelli et al., 1997; Sakamoto et al., 2002a). Nonetheless, cis-inhibition appears to be a particularly important mechanism to establish and maintain the signaling polarity required for specific Notch-dependent cell fate determinations (Becam et al., 2010; de Celis and Bray, 1997; Jacobsen et al., 1998; Klein and Arias, 1998; Klein et al., 1997; Matsuda and Chitnis, 2009; Miller et al., 2009; Sprinzak et al., 2010). Ectopic expression of truncated ligands lacking most of the intracellular domain function cell autonomously to block Notch signaling and promote retinal neurogenesis and neurite outgrowth as well as inhibit keratinocyte differentiation within the epidermal stem cell niche (Dorsky et al., 1997; Franklin et al., 1999; Henrique et al., 1997; Lowell et al., 2000; Lowell and Watt, 2001). Although these studies have relied on overexpression of DSL ligands, loss of function studies have also demonstrated that endogenous ligands can function in a cis-inhibitory manner (Micchelli et al., 1997; Miller et al., 2009).
3.2. Cis-interactions between ligand and Notch determine signal polarity
A recent study using mammalian cell culture to manipulate ligand expression levels in Notch expressing cells has provided insight into understanding how cis versus trans - ligand expression could influence the Notch signaling response (Sprinzak et al., 2010). Specifically, cells adopt mutually exclusive signaling states so that depending on their relative levels of Notch ligand and receptor they either ‘send’ or ‘receive’, but not both. According to this model, an ‘ultrasensitive switch’ between these states is capable of amplifying small differences between interacting cells even in the absence of transcriptional feedback. One way to set up this signaling asymmetry is to control levels of both ligand and receptor on the cell surface such that the signal-sending cell maintains high ligand surface expression while the signal-receiving cell has high surface Notch. Previously this asymmetry has been explained solely by a feedback mechanism through which activation of Notch down regulates ligand expression at the level of transcription (Greenwald and Rubin, 1992; Seugnet et al., 1997b). Changes in signal sending and receiving potential, however, have been observed that do not involve overall changes in ligand or receptor transcription (Becam et al., 2010; Sprinzak et al., 2010). These studies suggest that cis-interactions between ligands and receptors would mutually inhibit the potential of ligands to signal as well as restrict Notch activation to the receiving cell. Studies in the developing fly eye have provided additional support for ligand cis-inhibition in establishing unidirectional signaling, and have also suggested a role for maintaining signaling polarity once cell fates have been determined (Miller et al., 2009).
3.3. Molecular mechanisms for ligand cis-inhibition of Notch signaling
The molecular mechanism underlying cis-inhibition is poorly understood, and has remained highly controversial. Competition between trans- and cis-ligand binding to Notch is likely to underlie the ability of ligands to activate or inhibit Notch signaling. This hypothesis assumes that the ligand-Notch binding interfaces overlap. Consistent with this idea, the Jagged1 DSL domain has been proposed to contain a highly conserved binding site for both trans- and cis-interactions with Notch (Cordle et al., 2008). At odds with the competition model, the binding sites in Notch for cis- and trans-interactions might not overlap. Extensive data indicate that the 11th and 12th EGF repeats in Notch are critical for trans-ligand binding and signaling activity (see Blacklow Chapter for details), however, early studies in flies implicated EGF-like repeats 24-29 in cis-inhibition (de Celis and Bray, 2000). More recent studies report a requirement for the 11th and 12th EGF repeats in cis-inhibition (Becam et al., 2010; Cordle et al., 2008; Fiuza et al., 2010), suggesting that the cis- and trans-ligand binding sites in Notch do overlap. Together these findings support a competitive mechanism for ligand-Notch interactions that ultimately results in either trans-activation or cis-inhibition of Notch signaling. Interestingly, the switch from an active to inhibited signaling state requires a high threshold of cis-ligand, while signaling responses over a range of trans-ligand are linear (Sprinzak et al., 2010). Since low levels of activated Notch are sufficient to induce Notch target gene expression (Schroeter et al., 1998), it seems likely that most, if not all, Notch receptors would need to interact with ligand in cis for signaling by trans-ligand to be suppressed.
Even though both cis- and trans-interactions with Notch may involve overlapping binding sites, only trans-ligand interactions activate Notch. Based on structural studies discussed below, trans-ligand is thought to induce conformational changes in Notch that facilitate proteolytic activation required for downstream signaling. Since cis-interactions do not lead to Notch activation, ligand-Notch interactions formed within the plane of the same membrane must not be able to induce the conformational changes required to activate Notch. In support of this idea, a recent study has suggested that ligand cis-interactions with Notch prevent proteolytic activation (Fiuza et al., 2010).
Although the majority of findings are consistent with cis-inhibition involving ligand-receptor interactions at the cell surface, inhibitory cis-interactions formed in the secretory pathway have been proposed to prevent Notch receptors from reaching the cell surface to account for losses in signaling (Sakamoto et al., 2002a). At odds with this notion, ligands retained within the biosynthetic pathway are defective in cis-inhibition, providing indirect support that ligand-Notch cis-interactions occur at the cell surface (Glittenberg et al., 2006; Ladi et al., 2005). Consistent with this, defects in ligand endocytosis that promote accumulation of ligand on the cell surface diminish trans-activation yet potentiate cis-inhibition (Glittenberg et al., 2006). Together these findings suggest that mechanisms must exist to coordinate the trans and cis activities mediated by ligands.
In addition to ligand-receptor cis-interactions inhibiting the ability of Notch cells to receive a signal, similar cis-interactions also inhibit the ability of ligand cells to send signals (Becam et al., 2010; Matsuda and Chitnis, 2009; Miller et al., 2009; Sprinzak et al., 2010), indicating that ligand-receptor interactions in the same cell can be mutually inactivating for sending or receiving signals. Although these studies did not detect losses in protein expression, Notch stimulated endocytosis has been reported to result in a decrease of cell surface ligand available for activation of signaling in adjacent cells (Becam et al., 2010; Matsuda and Chitnis, 2009). Specifically, studies in both zebrafish and flies report that under Notch knockdown conditions that the ligands DeltaD and Serrate accumulate on the cell surface, suggesting that Notch cis-interactions result in removal of cell surface ligand through endocytosis (Becam et al., 2010; Matsuda and Chitnis, 2009). Further, DeltaD-Notch cis-interactions have been proposed to inhibit Notch signaling through removing both the ligand and receptor from the cell surface (Matsuda and Chitnis, 2009). Studies in flies have found that a truncated form of Notch lacking intracellular domain sequences accumulates at the cell surface without increasing levels of Serrate (Becam et al., 2010). This suggests that interaction with the extracellular domain of Notch is sufficient to promote clearance of cell surface Serrate alone without simultaneous receptor internalization. Importantly, the 11th and 12th EGF-like repeats that function in trans-ligand binding are also required for this clearance and inhibitory affect on Serrate signaling. Interestingly, not all DSL ligands are susceptible to down regulation by Notch cis-interactions, and the molecular basis and biological relevance of these findings are unclear.
Even though cis-inhibition has been proposed to involve intercellular ligand-ligand interactions leading to a decrease in ligand available for trans-activation of Notch, a recent report has challenged this view by demonstrating that cells coexpressing both Notch and Delta form cell aggregates with Delta cells even though these same cell-cell interactions do not activate Notch signaling (Fiuza et al., 2010). Together these findings support the idea that cis-inhibition involves ligand-receptor interactions at the surface of the same cell to restrict signaling to one of the two interacting cells.
4. REGULATION OF LIGAND-INDUCED NOTCH SIGNALING BY POST-TRANSLATIONAL MODIFICATION
4.1. Glycosylation
The Notch ligands and receptors undergo O- and N-linked glycan modifications at conserved sequences within specific EGF repeats; however, only O-fucose and O-glucose additions to Notch have so far been reported to affect signaling. N-glycan-modifications of Notch, on the other hand, do not appear to alter Notch-dependent development in mice (Haltiwanger and Lowe, 2004). Glycosylation of Notch both positively and negatively regulates signaling induced by ligands, presumably through modulating the strength of the ligand-receptor interactions. Although DSL ligands are glycosylated as reported for Notch (Panin et al., 2002), affects on ligand signaling activity have so far not been detected. Roles for glycosylation in Notch signaling are the subject of the chapter by Stanley and Okajima and the reader is encouraged to consult the indicated chapter for further details.
4.2. Ubiquitination
Post-translational modification of Notch ligands by ubiquitination regulates cell surface levels and is an absolute requirement for ligand signaling activity (Chitnis, 2006; Le Borgne, 2006; Le Borgne and Schweisguth, 2003a; Nichols et al., 2007b). As found for Drosophila Delta and Serrate, the intracellular domains of Dll1, Dll4, Jagged1 and Jagged2 contain multiple lysine residues that can serve as potential sites for the addition of ubiquitin by E3 ligases. Two structurally distinct RING-containing E3 ligases, Neuralized (Neur) and Mind bomb (Mib), influence Notch signaling through interacting with and ubiquitinating DSL ligands to enhance their endocytosis. Initial studies in Drosophila and Xenopus reported that Neur had intrinsic ubiquitin ligase activity and interacted with Delta to promote its internalization and degradation through ubiquitination (Deblandre et al., 2001; Lai et al., 2001; Pavlopoulos et al., 2001; Yeh et al., 2001). Given that Neur is required for Notch signaling these findings are difficult to reconcile; however, based on the cell autonomous activity identified for Neur (Lai and Rubin, 2001a, b; Yeh et al., 2000) a model was suggested in which the loss of cell surface Delta induced by Neur might indirectly enhance Notch signaling through relieving cis-inhibition imposed by Delta (Deblandre et al., 2001). More recent studies, however, have clearly shown that cis-inhibition does not require ligand ubiquitination (Glittenberg et al., 2006). Moreover, Neur expression is enhanced in signal-sending cells where it is asymmetrically localized and functions to direct cell fate decisions regulated by Notch signaling (Bardin and Schweisguth, 2006; Le Borgne and Schweisguth, 2003b; Morel et al., 2003; Pavlopoulos et al., 2001), providing support for the idea that Neur-induced endocytosis functions to stimulate ligand signaling activity.
Although studies in flies and frogs support a role for Neur in regulating cell surface levels and generating a productive signal, mice lacking the mammalian Neur homolog do not display any obvious Notch developmental phenotypes (Ruan et al., 2001; Vollrath et al., 2001). This surprising finding suggested that mammalian Neur might not be an essential component of the Notch signaling pathway. Alternatively, additional E3 ubiquitin ligases could exist to modify DSL ligands and facilitate Notch activation. Supporting the latter idea, a structurally distinct E3 ligase was subsequently identified as the target of the Mib neurogenic mutant in zebrafish (Chen and Casey Corliss, 2004; Itoh et al., 2003). Mib binds and ubiquitinates Delta and upregulates Delta endocytosis as reported for Neur, but in contrast to Neur, Mib functions exclusively in the ligand cell to activate Notch signaling and is unable to reverse the cis-inhibitory effects of Delta on Notch reception (Itoh et al., 2003; Koo et al., 2005a).
Neur and Mib homologs have been isolated from a number of different species and despite being conserved and having similar molecular activities, Neur and Mib genes may have evolved to serve different roles in vertebrate Notch signaling. Drosophila has a single Neur gene (dNeur) and two related Mib genes (dMib1 and dMib2) that regulate distinct Notch-dependent developmental events (Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005), apparently through differential expression. Both Neur and Mib ubiquitinate the Drosophila ligands, Delta and Serrate and stimulate ligand endocytosis and signaling activities. Importantly, gene rescue experiments indicate that for the most part these structurally distinct E3 ligases are functionally redundant. In contrast to these findings in flies, studies in mice indicate the surprising findings that the mammalian Neur1 and Neur2 genes are dispensable for normal development. Additionally, animals defective in Neur1, Neur2 and Mib2 gene expression do not display any Notch-dependent phenotypes, while additional removal of Mib1 produces embryonic lethal pheontypes associated with losses in Notch signaling (Koo et al., 2007). Importantly, disruption of only the Mib1 gene produces the known constellation of Notch mutant phenotypes in developing mouse embryos (Barsi et al., 2005; Koo et al., 2005a). Although Mib1 and Mib2 appear functionally redundant (Zhang et al., 2007a; Zhang et al., 2007b), Mib2 is not strongly expressed during embryonic development accounting for the absolute requirement for Mib1 in Notch-dependent developmental processes (Koo et al., 2007). In contrast to findings reported for the functionally redundant E3 ligases in flies, Mib2 but not Neur1 or Neur2 can rescue the Mib1 mutant neurogenic phenotype in zebrafish (Koo et al., 2005b). Further, while both Neur1 and Neur2 are dispensable for normal neurogenesis in mice, Mib1 mutant embryos display strong neurogenic phenotypes in the developing brain and neural tube (Koo et al., 2005b; Koo et al., 2007). Therefore, while Neur and Mib appear to perform similar roles in Notch signaling in flies, the vertebrate Neur and Mib proteins do not appear to be functionally equivalent.
Findings from mammalian cells have suggested that Mib, not Neur is the E3 ligase responsible for DSL ligand endocytosis that activates Notch signaling, while Neur functions downstream of Mib to direct lysosomal degradation of internalized ligands and thereby regulate the level of ligand available for Notch activation (Song et al., 2006). Consistent with this idea, overexpression of Neur1 monoubiqutinates Jagged1 leading to degradation and attenuation of Jagged1-induced Notch signaling (Koutelou et al., 2008); however, Mib2 (skeletrophin) ubiquitination of Jagged2 is associated with activation of Notch signaling (Takeuchi et al., 2005). The different functional roles for Neur and Mib ligases in Notch signaling might reflect different ubiquitin states of DSL ligands mediated by these structurally distinct E3 ligases. Notch ligands have been reported to be mono- and/or polyubiquitinated; however, the functional consequences of these types of ubiquitination to Notch signaling are not well documented. Polyubiquitination is associated with proteasome degradation, while both mono and multi-mono ubiqutination can signal endocytosis of membrane proteins from the cell surface and further influence intracellular trafficking (Staub and Rotin, 2006). Trafficking events that degrade internalized DSL ligands could function to downregulate Notch signaling, while recognition of ubiquitinated ligands by specific adaptor/sorting molecules might promote signaling.
In addition to inducing different types of ubiquitination, Mib and Neur could potentially regulate ligand activity by modifying distinct lysine residues. Ligand intracellular domains contain multiple lysine residues that could potentially be modified by the addition of ubiquitin. Mutation of two intracellular lysine residues in Serrate produces signaling defects that are associated with losses in endocytosis and accumulation of ligand at the cell surface (Glittenberg et al., 2006). In contrast, mutation of all seventeen intracellular lysine residues in Dll1 did not prevent internalization or promote accumulation of this lysine-less mutant at the cell surface (Heuss et al., 2008). Nonetheless, the internalized lysine-less mutant was unable to recycle or activate Notch signaling, and these defects were associated with decreased fractionation to detergent resistant lipid microdomains compared to wild-type Dll1. In addition, the inability of the lysine-less mutant to recycle also correlated with defects in binding a soluble form of Notch. In contrast, a Dll1/Dll3 chimeric ligand containing the extracellular domain of Dll1 and the transmembrane and intracellular domains of Dll3, internalized, recycled, and displayed high affinity binding to Notch despite lacking lysines required for ubiquitination (Heuss et al., 2008). Underscoring the importance of ligand ubiquitination in signaling activity, the Dll1/Dll3 chimera did not activate Notch signaling and this also correlated with a loss in fractionation to lipid microdomains. Based on these findings, the authors concluded that ubiquitination is not required for ligand endocytosis but rather functions to direct ligand to a specific recycling pathway where it acquires high affinity binding to Notch. As exciting as these findings are, the authors failed to unravel the connections between ubiquitination, recycling, lipid microdomain fractionation and high affinity binding in the generation of an active ligand. Importantly, this study did not determine the signaling activity of wild-type Dll1 when either protein recycling or lipid raft formation are disrupted.
Studies in flies indicate that Neur may play additional roles in Notch ligand endocytosis to enhance signaling activity beyond ubiquitination (Pitsouli and Delidakis, 2005; Skwarek et al., 2007). Specifically, a phosphoinositide-binding domain was identified in Neur that is necessary for its interactions with the plasma membrane. Although Neur membrane localization is not required for Neur to interact with or ubiquitinate Delta, membrane association of Neur is required for Delta endocytosis (Skwarek et al., 2007). In this regard, a recent study identified a link between the glycosphingolipid (GSL) content of the plasma membrane and Mib-dependent endocytosis of Delta that is required to activate Notch signaling (Hamel et al., 2010). A conserved GSL-binding motif (GBM) was identified in the N-terminal region of Delta and Serrate that conferred binding to specific GSLs, which is proposed to modulate ligand membrane association and in turn ligand endocytosis. Together these studies underscore the importance of membrane lipids in modulating ligand endocytosis and signaling activity.
5. LIGAND ENDOCYTOSIS IN ACTIVATION OF NOTCH SIGNALING
A requirement for direct cell-to-cell interactions is a hallmark of Notch signaling, however, the transmembrane property of the ligands may underlie the basic mechanism of Notch activation that is dependent on ligand endocytosis. Specifically, in the absence of endocytosis, ligands accumulate at the cell surface but fail to activate signaling (Itoh et al., 2003; Nichols et al., 2007a; Parks et al., 2000). That ligands need to be internalized by the signal-sending cell to activate Notch on the signal-receiving cell represents a fundamentally new paradigm for endocytic activation of a signaling pathway. Nonetheless, the exact mechanism by which ligand endocytosis triggers Notch signaling has remained a mystery.
5.1. Identifying the endocytic machinery required for ligand cells to activate Notch
The majority of cell surface proteins are internalized via clathrin-mediated endocytosis (CME); however, additional portals of entry exist that do not involve clathrin (Conner and Schmid, 2003; Doherty and McMahon, 2009). The specific endocytic pathways used by Notch ligands are poorly characterized, but what is certain is that only ubiquitinated ligands internalized in an epsin-dependent manner are competent to signal (Chen and Casey Corliss, 2004; Deblandre et al., 2001; Glittenberg et al., 2006; Haltiwanger and Lowe, 2004; Itoh et al., 2003; Koo et al., 2005a; Lai et al., 2001; Overstreet et al., 2004; Pavlopoulos et al., 2001; Wang and Struhl, 2005; Yeh et al., 2001). Genetic and cellular studies indicate that ligand cells require the key endocytic factor dynamin to activate Notch (Nichols et al., 2007a; Parks et al., 2000; Seugnet et al., 1997a), however, dynamin functions to release endocytic vesicles from the plasma membrane during both clathrin-dependent and -independent endocytosis (Conner and Schmid, 2003), so either or both pathways could function in ligand activity. In addition, the clathrin adaptor epsin that is critical for ligand activity has also been implicated in endocytosis independent of clathrin (Chen and De Camilli, 2005; Sigismund et al., 2005).
Indirect support for CME in ligand signaling activity has come from genetic studies indicating that Notch dependent developmental events require auxilin and the ubiquitious cyclin G-associated kinase (GAK) that function at multiple steps in clathrin-coated pit formation and un-coating of clathrin-coated vesicles (Eisenberg and Greene, 2007; Yim et al., 2010). Moreover, the Notch signaling defects identified with auxilin mutants can be partially rescued by ectopic clathrin expression (Eun et al., 2008), suggesting that losses in auxilin produce un-coating defects that limit clathrin availability for ligand endocytosis. Together with findings from mammalian cell culture indicating that blockade of CME in ligand cells inhibits Notch signaling (Nichols et al., 2007a), it seems likely that ligand endocytosis required for activation of Notch is clathrin dependent. Nonetheless, a role for ligand endocytosis independent of clathrin for signaling activity in specific cellular contexts cannot be ruled out.
Although it is clear that endocytosis by the ligand cell is critical for activation of signaling in the Notch cell, the exact role that ligand endocytosis serves in signaling has remained poorly defined. Studies in flies and mammalian cells have suggested that ligands undergo two distinct endocytic events to activate Notch (Fig. 3). The first ligand endocytic event occurs prior to engagement of Notch and is proposed to facilitate recycling to generate an active ligand. Following interactions with Notch on adjacent cells, a second ligand endocytic event is proposed to generate a pulling force to allow activating Notch proteolysis. It is important to note that whether the first, second, or both endocytic events are necessary for ligand activation of Notch is controversial.
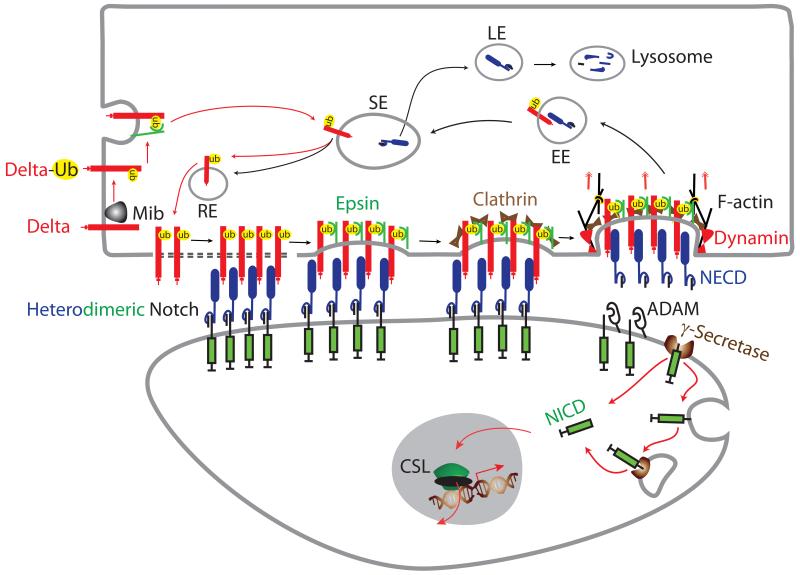
Fig. 3. Models for distinct endocytic events by the ligand cell to activate signaling in the Notch cell.
Prior to Notch engagement, endocytosis allows ligand to enter the sorting endosome (SE) or recycling endosome (RE) where it is processed into an active ligand and returned to the cell surface to activate Notch. Ligand ubiquitination by Mib may facilitate interactions with epsin that direct the required endocytosis and/or trafficking. Alternatively, ligand binding to Notch may induce ligand ubiquitination for recruitment of epsin to orchestrate the formation of a clathrin- coated endocytic structure specialized in force generation to pull the non- covalent heterodimeric Notch apart. Heterodimer dissociation would account for the observed uptake of the Notch extracellular domain (NECD) by ligand cells. In the early endosome (EE), internalized NECD dissociates from the ligand and trafficks to the late endsome (LE) where it is targeted for lysosomal degradation. Ligand dissociated in the EE traffics to the SE or RE for return to the cell surface where it is available to activate Notch on adjacent cells. Removal of the NECD exposes the ADAM site in the membrane-bound heterodimer subunit to facilitate γ-secretase cleavage and release of the Notch intracellular domain (NICD) from the membrane. Released NICD translocates to nucleus where it interacts with CSL to activate Notch target gene transcription. As discussed in the text, these models that account for the critical requirement for endocytosis by the ligand cell to activate signaling in the Notch cell may not be mutually exclusive.
5.2. Recycling to generate an active ligand
The recycling model assumes that newly synthesized ligand delivered to the cell surface cannot activate Notch and requires endocytosis, trafficking and recycling back to the cell surface to gain signaling activity (Heuss et al., 2008; Rajan et al., 2009; Wang and Struhl, 2004). To account for the absolute requirements for epsin and ligand ubiquitination in signaling activity, this model further proposes that epsin selectively promotes endocytosis and/or trafficking of a sub-population of ubiquitinated ligand for conversion in the recycling endosome into an active ligand. The changes conferred by recycling to obtain signaling activity are completely unknown, however concentration, clustering, and proteolytic processing of ligand, as well as localization of ligand to a specific microdomain or recruitment of co-factors have all been suggested as possible modifications (Chitnis, 2006; Le Borgne, 2006; Nichols et al., 2007b).
Even though Notch ligands are known to recycle (Heuss et al., 2008; Rajan et al., 2009), the role that ligand recycling plays, if any, in activating Notch is poorly defined. In addition to returning internalized proteins and membrane to the cell surface, recycling is used to establish distinct apical and basolateral membranes in polarized cells (Grant and Donaldson, 2009; Maxfield and McGraw, 2004). Therefore, it may not be surprising that the strongest support for ligand recycling in activation of Notch signaling comes from studies on cell fates derived from sensory organ precursors (SOP) that involve polarized cells (see Bellen Chapter). Specifically, SOP progeny that activate Notch signaling in neighboring cells are enriched in Rab11 recycling endosomes that concentrate Delta and apically internalized Delta must traffic from the basolateral membrane to an apical actin-rich structure for SOP progeny to acquire signaling activity (Emery et al., 2005; Jafar-Nejad et al., 2005; Rajan et al., 2009). However, Sec15 that functions with Rab11 in the recycling endosome to regulate SOP derived cell fates is not required in every developmental event regulated by Notch (Jafar-Nejad et al., 2005; Windler and Bilder, 2010). Additionally, loss of Rab11 activity does not perturb Delta signaling in the germline (Windler and Bilder, 2010), and Rab11 mutants do not display Notch eye phenotypes (Li et al., 2007a) as expected if ligand recycling is an absolute requirement for Notch signaling. Moreover, Rab11 does not overlap with Delta in the morphogenetic furrow (Hagedorn et al., 2006) where Notch signaling directs normal eye development. If recycling is absolutely required to generate an active ligand, then Rab5 that is a prerequisite for entry into the Rab11 recycling pathway should also be required, however, defects in Rab5 do not perturb Delta signaling activity (Windler and Bilder, 2010). Together these findings suggest that ligand recycling, at least that dependent on Rab11 and Sec15, is not a general requirement of Notch signaling.
5.3. Ligand endocytosis in force generation to activate Notch
A general requirement for ligand endocytosis has been proposed to reflect the need for Notch to undergo conformational changes to effect activating proteolysis that ligand binding alone would not induce (Gordon et al., 2008a; Gordon et al., 2008b). Proteolytic activation of Notch signaling involves the specific uptake of the Notch extracellular domain (NECD) by the ligand cell (Nichols et al., 2007a; Parks et al., 2000), and although ligand cells defective in endocytosis bind and cluster Notch they do not internalize NECD or activate signaling (Nichols et al., 2007a). These findings first suggested a role for ligand endocytosis in activation of signaling that involved a mechanical force to dissociate the NECD from intact Notch. The force produced by ligand endocytosis is thought to induce conformational changes that destabilize the non-covalent interactions that keep the Notch heterodimer intact and inactive in the absence of ligand. The identification and characterization of a negative regulatory region (NRR) in the Notch ectodomain that stabilizes the Notch heterodimer and prevents activating proteolysis provide additional support for the ligand endocytosis pulling-force model (see Blacklow Chapter). Specifically, structural analyses of the NRR confirm that multiple non-covalent interactions stabilize the structure and serve to occlude the ADAM cleavage site that is required to initiate activating Notch proteolysis (Gordon et al., 2007). Moreover, these findings have suggested that ligand binding alone would not be sufficient to induce the required global conformational changes, but rather, endocytosis of ligand-bound Notch would be necessary to produce a force to pull on Notch and expose the ADAM cleavage site for activating proteolysis. Although endocytosis is a good force-generating candidate, it is not known if force is produced during the process of ligand endocytosis, or if such a force could destabilize the NRR structure and expose the ADAM cleavage site.
Endocytosis of ligand-bound Notch by the ligand cell may be mechanistically different than constitutive ligand endocytosis. Specifically, cells may experience a resistance to ligand internalization of Notch attached to the surface of an adjacent cell. To overcome such a resistance, ligand cells may need to recruit specific cellular factors to form an endocytic structure specialized in force generation to effect ligand endocytosis of cell surface Notch. Specifically, ligand endocytic force induced conformational changes in Notch could physically release NECD by dissociating the heterodimer subunits or unmasking the ADAM cleavage site and both mechanisms have been proposed (discussed in Blacklow Chapter). In any event, removal of NECD from the intact Notch heterodimer would be necessary for activating proteolysis of the remaining membrane-bound Notch for downstream signaling (Fig. 3).
The requirement for epsin in ligand signaling activity (Overstreet et al., 2003; Overstreet et al., 2004; Tian et al., 2004) has been proposed to reflect a role for epsin in ligand endocytosis and/or trafficking to allow access to a specific recycling pathway for conversion into an active ligand (Wang and Struhl, 2004). Nonetheless, epsin is not known to regulate protein recycling (Vanden Broeck and De Wolf, 2006) and data are lacking to show that losses in epsin actually perturb ligand recycling. Although it is clear that epsin is required for ligand signaling activity, it is possible that this does not involve ligand recycling prior to engagement with Notch. Rather, we propose that epsin may function downstream of ligand binding to Notch to induce the formation of a force-producing endocytic structure. Notch binding may induce ligand ubiquitination and/or clustering to amass multiple ubiquitin-binding sites for epsin. By assembling multiple low affinity mono-ubiquitin interactions, strong epsin-UIM/ubiquitinated-DSL interactions could be generated (Barriere et al., 2006; Hawryluk et al., 2006), and this may be necessary for ligand to overcome resistance to internalization when bound to cell surface Notch. In fact, replacement of the Delta intracellular domain with a single ubiquitin motif that can undergo polyubiquitination promotes internalization and signaling activity in zebrafish (Itoh et al., 2003). However, a non-extendable ubiquitin only weakly signals even though it promotes endocytosis (Wang and Struhl, 2004), supporting the idea that multiple ubiquitin interaction sites are required for ligands to activate Notch, possibly through providing stable associations with epsin-containing endocytic vesicles.
Consistent with these ideas, ligand cells require epsin, dynamin and the actin cytoskeleton to activate signaling in Notch cells, and all of these cellular factors have been implicated in inducing membrane constriction and tension that could contribute to force generation during the process of endocytosis (Itoh et al., 2005; Roux et al., 2006). Therefore, it is tempting to speculate that ligand cells require epsin to orchestrate the formation of a molecularly distinct clathrin-coated endocytic structure specialized in force generation. In addition to membrane bending, epsin has also been reported to regulate the actin cytoskeleton during endocytosis (Horvath et al., 2007; Maldonado-Baez and Wendland, 2006), which together could endow cells with sufficient endocytic force to induce conformational changes in ligand-bound Notch required to initiate activating proteolysis. Although epsin participates in endocytosis through simultaneous binding to the plasma membrane, clathrin endocytic vesicles, and ubiquitinated cargo (Horvath et al., 2007), interactions between ligands and epsin have yet to be reported and it is still unclear how epsin and ubiquitinated ligands contribute to Notch activation.
Implicit in the pulling-force model is the need for ligand-Notch interactions to survive the endocytic force that induces conformational changes required for NECD transendocytosis and activating Notch proteolysis. That NECD transendocytosis by ligand cells is required for activation of Notch (Heuss et al., 2008; Nichols et al., 2007a; Parks et al., 2000) indicates that ligand-Notch interactions do indeed survive the putative endocytic force required for global conformational changes in Notch to expose the ADAM cleavage site. In this regard, reported atomic force microscopy (AFM) measurements for Delta cells binding to uncleaved Notch are stronger than those detected for furin-cleaved Notch (Ahimou et al., 2004), suggesting that Delta-Notch interactions are indeed stronger than the non-covalent interactions that hold the heterodimer subunits together (see Blacklow Chapter for further discussion). Therefore, ligand endocytosis could function first to allow recycling to produce a high affinity ligand for avid binding to Notch, and this in turn would enable ligand-Notch interactions to survive the pulling force produced by ligand endocytosis of Notch bound to adjacent cells (Fig. 3).
Recycling has been suggested to generate a high affinity ligand by directing ligand to a specific membrane microdomain (Heuss et al., 2008) and this could provide a mechanism to produce strong ligand-Notch interactions. While it is attractive to propose that ligand endocytosis regulates recycling to generate a high affinity ligand, the fact that soluble ligands that have never recycled can signal when attached to surfaces (Varnum-Finney et al., 2000) argues against a requirement for endosomal processing to generate an active ligand. Additionally, the dependence of soluble ligands on surface attachment to activate signaling is consistent with the proposed role for force in exposing Notch to activating proteolysis; however, in this case the Notch cell would provide the force to disrupt the NRR structure through cell migration. Finally, in contrast to the absolute requirement for ligand endocytosis in signaling, studies have failed to establish a firm correlation between ligand recycling and signaling activity (Glittenberg et al., 2006; Heuss et al., 2008), implying that endocytosis rather than recycling is a general requirement for ligands to activate Notch signaling.
Confirmation of endocytic force in ligand signaling activity awaits biophysical analyses to establish that ligand cells can indeed produce a mechanical force following interactions with Notch. AFM studies have provided support that Delta binds Notch with high avidity (Ahimou et al., 2004), but whether ligands need to recycle to acquire strong binding potential is unknown. To confirm a requirement for ligand recycling in non-polarized cells it will be necessary to show that losses in ligand recycling produce signaling defects. Moreover, elucidating how epsin functions to regulate signaling activity of ubiquitinated ligands is critical to understanding the molecular, cellular and physical basis of ligand endocytosis in Notch activation. Biophysical studies will also be required to determine if activating Notch proteolysis and downstream signaling are regulated by mechanical force; however, the ultimate challenge will be to obtain evidence for endocytic force in regulating Notch signaling in whole animals.
6. REGULATION OF DSL LIGAND ACTIVITY BY PROTEOLYSIS
DSL ligands undergo proteolytic cleavage by ADAMs and γ-secretase as described for Notch, however, in contrast to signaling induced by Notch proteolysis, proteolytic removal of cell surface ligand can either inhibit or enhance Notch signaling. Although Notch proteolysis generates an intracellular fragment that acts as the signal transducer, it is less clear if the cleavage products generated by ligand proteolysis have intrinsic activity (Fig. 4). A detailed review describing the proteases that cleave DSL ligands and the biological significance has been previously published (Zolkiewska, 2008); here we discuss possible mechanisms by which ligand proteolysis could affect Notch signaling. While mammalian DSL ligands are cleaved by several ADAMs (ADAM9, ADAM10, ADAM12, ADAM17), Drosophila ligands have been reported to be cleaved by only the homologs of ADAM10 (Kuzbanian/Kuz and Kuzbanian-like/Kul) and ADAM17 (DTACE).
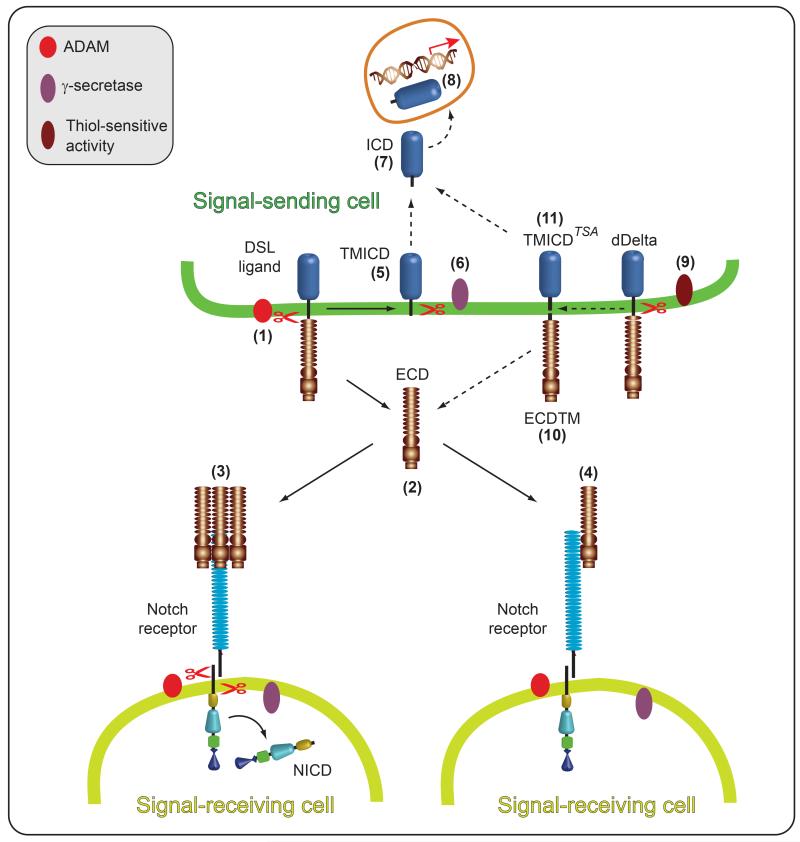
Fig. 4. Regulation of DSL ligand signaling activity by proteolysis.
Mammalian and Drosophila DSL ligands undergo proteolytic cleavages within the juxtamembrane and intramembrane regions. A-Disintegrin-And-Metalloprotease (ADAM) mediated cleavage (1) of mammalian and Drosophila DSL (Delta/Serrate/LAG-2) ligands within the juxtamembrane region results in shedding of the extracellular domain (2, ECD). The shed ECD requires clustering to activate Notch signaling (3). Although unclustered soluble ECD can bind Notch, it may antagonize Notch signaling (4). In mammalian cells, the remaining membrane-tethered ADAM cleavage product, the membrane-tethered fragment containing the intracellular domain (TMICD, 5) may undergo further cleavage by -secretase (6) to release the intracellular domain (ICD) from the membrane (7) allowing it translocate to the nucleus and activate gene transcription (8) (see text for details). However, the Drosophila Delta (dDelta) TMICD (5) is not further processed and could antagonize Notch signaling (see text for details). Like mammalian DSL ligands, dDelta also undergoes intramembrane cleavage, however, this event does not require prior ADAM cleavage and is catalyzed by a thiol-sensitive activity (TSA, 9). It is unclear if the resulting cleavage products remain membrane-tethered. If the ECD containing fragment (ECDTM) remains membrane-tethered (10), it could antagonize Notch signaling, but if released from the membrane, ECDTM could function as proposed for soluble ECD (2, 3, 4) (see text for details). If the ICD-containing intramembrane cleavage product TMICDTSA remains membrane-bound (11), it could antagonize Notch signaling, but if released from the membrane (7), TMICDTSA could translocate to the nucleus and activate gene transcription (8) (see text for details).
6.1. ADAM ectodomain shedding of DSL ligands as regulators of Notch signaling
One of the consequences of ADAM cleavage of DSL ligands is shedding of the ectodomain that contains the Notch binding site. Accordingly, ADAM-shedding of ligands would decrease ligand-Notch interactions both in trans and in cis, however, these scenarios would produce opposing outcomes on Notch signaling. Specifically, losses in trans-interactions would lead to losses in Notch signaling while losses in cis-interactions would relieve cis-inhibition and thereby enhance signaling. Therefore, in addition to transcriptional feedback loops and endocytosis discussed in section 3, ligand shedding provides an additional mechanism to regulate the cell’s potential to send or receive a Notch signal. Furthermore, in addition to regulating signal polarity, ligand shedding could also determine the intensity and duration of Notch signaling.
Several lines of evidence suggest a role for ADAM-mediated ligand ectodomain shedding in establishing and/or maintaining an asymmetric distribution of cell surface ligand between signal-sending and signal-receiving cells. In flies, this has been best demonstrated for the ADAM Kul, where both losses and gains in Kul activity produce wing vein defects characteristic of aberrant Notch signaling (Lieber et al., 2002; Sapir et al., 2005). These studies suggest that Kul, which exclusively cleaves ligands and not Notch, is required to maintain asymmetric distribution of Delta in the developing wing to facilitate unidirectional signaling. In the signal-receiving cell, Kul acts as a positive regulator of Notch signaling by maintaining low levels of ligand at the cell surface to prevent cis-inhibition and ensure efficient signal reception necessary for normal wing margin formation (Sapir et al., 2005). Similar to the requirement for Kul in the signal-receiving cell, ectopic expression of ADAM12 (an ADAM that cleaves Dll1 but not Notch) results in Dll1 shedding and enhanced Notch signaling in mammalian cells again, presumably by relieving cis-inhibition (Dyczynska et al., 2007; Sun et al., 2008a). Dll1 shedding is also thought to deplete ligand available for activation of Notch signaling that would result in decreases in signaling. Such asymmetry in Notch signaling among initially equivalent myogenic progenitors, created through Dll1 shedding, is proposed to maintain the balance between self-renewal and differentiation (Sun et al., 2008a).
ADAM expression and activity could regulate ligand ectodomain shedding and thus, Notch signaling. In this regard, transforming growth factor (TGF)-β3 downregulates ADAM10 expression and correlates with activation of Notch signaling in cultured chick leg bud mesenchymal cells (Jin et al., 2007). In this scenario, TGF-β3-induced downregulation of ADAM10 prevents Dll1 ectodomain shedding, and this correlates with an inhibition in cell proliferation and subsequent precartilage condensation through increases in Notch signaling. The glycosylphosphatidyl-anchored cell-surface protein, RECK (reversion-inducing cysteine-rich protein with kazal motifs), specifically inhibits ADAM10 activity leading to inhibition of ectodomain shedding of DSL ligands and activation of Notch signaling (Muraguchi et al., 2007). Consistent with this role, RECK-deficient mouse embryos exhibit a loss in Notch target gene expression and display some Notch-dependent developmental defects, presumably due to loss of cell surface ligand available for interaction with Notch in trans (Muraguchi et al., 2007).
6.2. Activity of the ADAM-shed ectodomain of DSL ligands in Notch signaling
ADAM proteolysis of DSL ligands generates several cleavage products that could potentially affect Notch signaling (Fig. 4-1). The putative activity of soluble ligand extracellular domains (ECD) (Fig. 4-2) has best been examined through the use of recombinant ligands containing ECD sequences. While some studies have suggested that the ECDs are inactive others have suggested that they can either activate or inhibit Notch signaling depending on the cellular context. Nonetheless, soluble forms of Delta have been detected in Drosophila embryos (Klueg et al., 1998; Qi et al., 1999) and ectopic expression of Delta or Serrate ECDs antagonize Notch signaling (Hukriede et al., 1997; Sun and Artavanis-Tsakonas, 1997). A requirement for ADAM10/Kuzbanian in Notch signaling was initially interpreted to reflect shedding of Delta to produce an active ligand (Qi et al., 1999), however, subsequent findings from this same group has questioned this idea (Mishra-Gorur et al., 2002). The agonistic activity of soluble ligands is not easy to reconcile given the strict requirement for ligand endocytosis in Notch activation. Providing insight into this paradox, pre-fixed Delta cells that are presumably endocytosis-defective can activate Notch target genes (Delwig and Rand, 2008; Mishra-Gorur et al., 2002), suggesting that a physical force required to dissociate the Notch heterodimer may be provided by other mechanisms. Perhaps movement of Notch cells away from soluble ligand attached to the extracellular matrix or cell surface could produce the required force for heterodimer dissociation. In support of this idea, several studies have demonstrated that recombinant soluble ligands need to be pre-clustered or immobilized to activate Notch signaling and induce biological responses (Hicks et al., 2002; Karanu et al., 2000; Morrison et al., 2000; Shimizu et al., 2002; Varnum-Finney et al., 2000; Vas et al., 2004) (Fig. 4-3). Additionally, while unclustered soluble ligands can bind Notch, they are unable to activate signaling but rather appear to antagonize signaling induced by trans-ligands (Hicks et al., 2002; Shimizu et al., 2002; Varnum-Finney et al., 2000; Vas et al., 2004) (Fig. 4-4). In these cases, soluble ligands may compete with membrane-bound ligands for Notch binding, providing a mechanistic basis for the antagonistic activities identified for putative soluble forms of Drosophila (Hukriede et al., 1997; Sun and Artavanis-Tsakonas, 1997) and mammalian DSL ligands (Li et al., 2007b; Lobov et al., 2007; Noguera-Troise et al., 2006; Small et al., 2001; Trifonova et al., 2004).
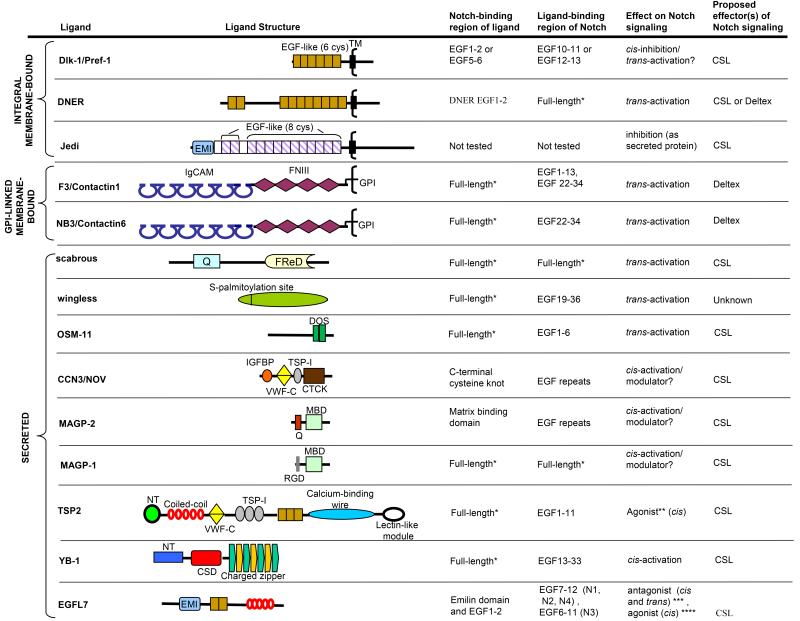
Naturally occurring soluble DSL ligands that function as Notch agonists have been identified in C. elegans and mammalian cells (Aho, 2004; Chen and Greenwald, 2004; Komatsu et al., 2008). In fact five of the ten C. elegans DSL ligands are soluble which represents the highest proportion of soluble DSL ligands identified for any phylum. Interestingly, neither the soluble nor membrane-bound C. elegans DSL ligands have a DOS motif, which is present in most, but not all, DSL ligands from other phyla (Komatsu et al., 2008). In flies and mammalian systems, both the DSL and DOS domains are required for high affinity binding to Notch receptors and activation of signaling (Parks et al., 2006; Shimizu et al., 1999). Genetic studies in C. elegans have identified the existence of soluble proteins that contain a DOS domain that are required in some developmental contexts for DSL ligands to activate the Notch related LIN-12 receptor by (Komatsu et al., 2008). To account for the biological activity observed for the DOS-containing proteins in DSL ligand activation of Notch signaling, the authors propose that optimal signaling requires the formation of a bipartite ligand system comprising distinct DSL and DOS domain-containing ligands. These findings emphasize the cooperative action of DSL and DOS domains for optimal Notch signaling, irrespective of whether these domains are present collinearly (as in the case of Drosophila Delta and Serrate and vertebrate ligands Dll1, Jagged1 and Jagged2) or within distinct proteins (as in the case of C. elegans ligands). Of the vertebrate DSL ligands, only Dll4 and Dll3 lack DOS domains (Komatsu et al., 2008) and similar to C. elegans DSL ligands their signaling activity may be dependent on collaboration with DOS domain-containing ligands (Kopan and Ilagan, 2009). At odds with this idea, Dll4 has been reported to be the most avid DSL ligand (Funahashi et al., 2008; Karanu et al., 2001; Sun et al., 2008b) and Dll3 is unable to bind or activate Notch (Ladi et al., 2005). While DSL and DOS domains may cooperate to activate Notch signaling, it is possible that on their own they function to antagonize Notch signaling as discussed in section 9.1.
6.3. Activity of the ADAM-cleaved membrane-tethered fragment in signaling
ADAM cleavage of DSL ligands also produces a membrane-tethered fragment containing the intracellular domain (TMICD, Fig. 4-5), which in mammalian cells undergoes further cleavage by γ-secretase (Ikeuchi and Sisodia, 2003; LaVoie and Selkoe, 2003; Six et al., 2003) (Fig. 4-6). Several studies have indicated that the released ligand ICD translocates to the nucleus (Hiratochi et al., 2007; Ikeuchi and Sisodia, 2003; Kolev et al., 2005; LaVoie and Selkoe, 2003; Six et al., 2003) and activates gene transcription (Hiratochi et al., 2007; Kolev et al., 2005; LaVoie and Selkoe, 2003 6) (Fig. 4-7, -8), similar to that identified for cleaved Notch. In support of this idea, the ICDs contain positively charged amino acids that when mutated prevent nuclear translocation and transcriptional activation (Kolev et al., 2005; LaVoie and Selkoe, 2003). Although these studies provide some support for the idea that DSL ligands undergo reverse signaling it is important to note that this has mostly relied on the use of engineered fragments, rather than physiological proteolytic cleavage of full-length ligands. Nonetheless, the possibility that DSL ligand-Notch signaling is bi-directional is exciting and awaits a clear demonstration of signaling events triggered in both DSL ligand and Notch cells following ligand-Notch interactions as established for the prototypic EphB/ephrinB bi-directional signaling system (Aoto and Chen, 2007; Dravis et al., 2004; Holland et al., 1996), that also involves transmembrane ligands and receptors.
Unlike mammalian DSL ligands, the TMICD fragment produced by ADAM cleavage of Drosophila Delta does not appear to undergo further processing and likely remains membrane-bound (Bland et al., 2003; Delwig et al., 2006) (Fig. 4-5). Although this fragment lacks a Notch binding domain, it could potentially compete with full-length ligands for the ubiquitination and/or endocytic machinery and thus antagonize ligand signaling activity. Another distinguishing feature of the proteolytic cleavage of Drosophila Delta is that although intramembrane cleavage occurs, this event does not require prior ADAM cleavage and does not involve γ-secretase (Delwig et al., 2006). Rather, this cleavage is induced by a thiol-sensitive activity (TSA) and occurs close to the extracellular face of the membrane (Fig. 4-9). Hence, it is uncertain whether the ICD would be readily released as proposed for ligand ICDs generated by γ-secretase (Delwig et al., 2006). If the ECD containing fragment (ECDTM) remains membrane-tethered (Fig. 4-10), it could function like ICD truncated ligands, which are endocytosis-defective and unable to activate signaling but are efficient cis-inhibitors (Chitnis et al., 1995; Henrique et al., 1997; Nichols et al., 2007a; Shimizu et al., 2002), but, if released, the ECDTM could function as proposed for soluble DSL ligands (Figure 4-2, -3, -4). The corresponding ICD-containing intramembrane cleavage product (TMICDTSA, Fig. 4-11) would be expected to function similarly to the Drosophila Delta TMICD (Fig. 4-5) if it remained membrane-bound; however, if released (Fig. 4-7), it could translocate to the nucleus and activate gene transcription (Fig. 4-8). In this regard, nuclear staining of Delta has only been detected using engineered ICD forms (Bland et al., 2003; Sun and Artavanis-Tsakonas, 1996), and hence, it is unclear whether the ICD is in fact released from full-length Delta and moves to the nucleus. Like Delta, Serrate also undergoes ADAM cleavage (Sapir et al., 2005); however, intramembrane cleavage of Serrate has not been reported to date.
6.4. Regulation of ligand proteolysis
Compared to the proteolytic activation of Notch that is tightly regulated by ligand, it is less clear if or how ligand proteolysis is induced or regulated. DSL ligands are actively cleaved in cell culture (Bland et al., 2003; Delwig et al., 2006; Dyczynska et al., 2007; LaVoie and Selkoe, 2003; Six et al., 2003; Yang et al., 2005), however, this proteolysis could be induced by signaling pathways trigged by serum components (Seals and Courtneidge, 2003). In fact, phorbol esters are known to activate intracellular signaling as well as ADAMs, both of which can induce DSL ligand proteolysis (Seals and Courtneidge, 2003). The extracellular matrix protein MAGP2 has also been reported to regulate DSL ligand proteolysis (Nehring et al., 2005). Interestingly, MAGP2 interacts with several DSL ligands, yet only the Jagged1 ectodomain is shed in a metalloprotease-dependent manner following interactions with MAGP2. Direct cell-cell interactions also contribute to ADAM cleavage of DSL ligands and both homotypic ligand-ligand and ligand-Notch interactions have been implicated (Bland et al., 2003; Delwig et al., 2006; Dyczynska et al., 2007; Hiratochi et al., 2007; LaVoie and Selkoe, 2003). Finally, gains and losses in Neur activity have been shown to be associated with Delta proteolytic processing in flies (Delwig et al., 2006; Haltiwanger and Lowe, 2004; Pavlopoulos et al., 2001), raising the possibility that ligand cleavage may occur intracellularly and involve endocytosis.
7. DSL LIGAND INTERACTIONS WITH PDZ-DOMAIN CONTAINING PROTEINS
The vertebrate DSL ligands Dll1, Dll4 and Jagged1 have PDZ-binding motifs at their carboxy termini (Pintar et al., 2007), which mediate interactions with PDZ-containing scaffold/adaptor proteins (Ascano et al., 2003; Estrach et al., 2007; Mizuhara et al., 2005; Pfister et al., 2003; Six et al., 2004; Wright et al., 2004). While being dispensable for both ligand activation (Ascano et al., 2003; Mizuhara et al., 2005; Six et al., 2004; Wright et al., 2004) and inhibition of Notch signaling (Glittenberg et al., 2006), the PDZ-binding sequences are required to mediate the effects of ligands on cell adhesion (Estrach et al., 2007; Mizuhara et al., 2005), migration (Six et al., 2004; Wright et al., 2004), and oncogenic transformation (Ascano et al., 2003). DSL ligands exhibit some preference for binding specific PDZ-containing proteins, most likely a reflection of the sequence differences in their PDZ-binding motifs (Pintar et al., 2007). For example, Jagged1 is unable to bind the PDZ domain proteins, MAGI-1 (membrane-associated guanylate kinase with inverted domain arrangement-1) and Dlg1 (human homolog of Drosophila discs large 1) (Mizuhara et al., 2005; Six et al., 2004), while the closely related Dll1 and Dll4 proteins both bind Dlg1 (Six et al., 2004). Although PDZ interactions do not mediate activation of Notch signaling, loss of the PDZ motif enhances the signaling activity of Delta (Estrach et al., 2007). These findings raise the intriguing possibility that PDZ-based interactions may restrict access of ligands to specific endocytic pathways necessary for their signaling activity.
PDZ-containing proteins play an important role in organizing specialized sites of cell-cell contact at adherens junctions as well as facilitating the cytoskeletal attachment of membrane proteins (Brone and Eggermont, 2005; Harris and Lim, 2001; Jelen et al., 2003). In fact, DSL ligands co-localize with actin (Lowell and Watt, 2001) and their specific PDZ-domain partners at regions of cell-cell contact (Estrach et al., 2007; Mizuhara et al., 2005; Six et al., 2004; Wright et al., 2004), consistent with the proposed role for DSL ligands in promoting cell adhesion and inhibiting cell motility. Additionally, Jagged1-PDZ interactions may produce changes in gene expression that promote oncogenic transformation (Ascano et al., 2003). How such interactions at the cell surface lead to transcriptional events in the nucleus is unknown, but PDZ-domain proteins such as calcium/calmodulin-dependent serine protein kinase (CASK), Bridge-1 or glutamate receptor interacting protein (GRIP)-tau are known to directly act as transcriptional activators (Hsueh et al., 2000; Lee et al., 2005; Nakata et al., 2004) whereas others such as the Dll1 interacting PDZ domain protein Acvrinp1 and the Jagged1 PDZ domain partner afadin/AF6 could indirectly effect gene transcription by binding the signal transducers Smad3 (Pfister et al., 2003; Shoji et al., 2000) or Ras (Ascano et al., 2003; Quilliam et al., 1999). Finally, that the cellular responses associated with DSL-PDZ interactions require both the extracellular and intracellular domains of DSL ligands suggests that homotypic ligand-ligand interactions could activate ligand signaling (Lowell et al., 2000; Lowell and Watt, 2001), while ligand-Notch interactions could induce bi-directional signaling (Ascano et al., 2003). Interestingly, a model in which fringe could block Jagged1-induced Notch1 signaling yet allow Jagged1 to mediate PDZ-dependent intracellular signaling has been proposed (Ascano et al., 2003).
8. REGULATION OF DSL LIGAND EXPRESSION PATTERNS
Notch signaling can both positively and negatively regulate DSL ligand expression, such that defects in Notch signaling are associated with increased expression of Dll1 (Barrantes et al., 1999; de la Pompa et al., 1997) or Dll4 (Suchting et al., 2007). On the other hand, Notch inductive signals upregulate DSL ligand expression, which is necessary for proper wing margin formation in flies (Doherty et al., 1996) as well as somite formation and patterning in vertebrates (Barrantes et al., 1999; Cheng et al., 2007; Cheng et al., 2003; de la Pompa et al., 1997; Doherty et al., 1996; Takahashi et al., 2003).
8.1. Cellular factors that regulate Notch ligand expression
In addition to Notch, other signaling systems are thought to intersect with the Notch pathway at the level of ligand expression (Hurlbut et al., 2007). In particular, the signaling pathways outlined in Table I are known to regulate ligand expression and produce specific cellular responses. These include, vascular endothelial growth factor (VEGF) (Benedito et al., 2009; Hellstrom et al., 2007; Limbourg et al., 2007; Liu et al., 2003; Lobov et al., 2007; Patel et al., 2005; Seo et al., 2006; Williams et al., 2006), tumor necrosis factor alpha (TNFα) (Benedito et al., 2009), fibroblast growth factor (Akai et al., 2005; Faux et al., 2001; Limbourg et al., 2007), platelet derived growth factor (PDGF) (Campos et al., 2002), TGFβ (Zavadil et al., 2004), lipopolysaccharide (LPS) (Amsen et al., 2004; Liotta et al., 2008), interleukin-6 (IL6) (Sansone et al., 2007; Studebaker et al., 2008), Hedgehog (McGlinn et al., 2005), Drosophila epidermal growth factor receptor (Carmena et al., 2002; Tsuda et al., 2002) and Wnt (Estrach et al., 2006; Hofmann et al., 2004; Pannequin et al., 2009; Rodilla et al., 2009).
Table I.
Cellular factors that regulate DSL ligand expression
| Effector of DSL ligand expression |
DSL ligand |
Effect on ligand expression: Upregulation (+) Downregulation (−) |
Cell type | Biological effect | References |
|---|---|---|---|---|---|
| VEGFa | Dll4 b | + | Endothelial | Inhibition of angiogenic sprouting; arterial specification |
Hellstrom et al., 2007; Liu et al., 2003; Lobov et al., 2007; Patel et al., 2005; Seo et al., 2006; Williams et al., 2006 |
| TNFαc | Jagged1 | + | Endothelial | Promotion of angiogenic sprouting |
Benedito et al., 2009; Sainson et al., 2008 |
| FGFd | Dll1b | + | Neural stem cells |
Maintenance of spinal cord stem cells |
Akai et al., 2005 |
| LPSe | Dll4b | + | Dendritic cells | CD4+ Th1k polarization |
Amsen et al., 2004 |
| LPSe / PGE2f | Jagged1 | + | Dendritic cells | CD4+ Th2k polarization |
Amsen et al., 2004 |
| IL6g | Jagged1 | + | Mammary epithelial cells |
Proliferation and invasion |
Sansone et al., 2007; Studebaker et al., 2008 |
| Hedgehog | Jagged1 | + | Mesenchymal cells |
Limb development | McGlinn et al., 2005 |
| VEGFa+ FGF2d | Dll1b | + | Endothelial cells | Postnatal Arteriogenesis |
Limbourg et al., 2007 |
| Wnt | Jagged1 | + | Hair follicle precortex |
Hair follicle differentiation |
Estrach et al., 2006 |
| Wnt | Jagged1 | + | Intestinal epithelial cells |
Proliferation (tumorigenesis) |
Rodilla et al., 2009; Pannequin et al., 2009 |
| Wnt | Dll1b | + | Presomitic mesoderm |
Somitogenesis | Hofmann et al., 2004 |
| DERh and/or Heartless | Drosophila Delta |
+ | Embryonic mesoderm |
Specification of muscle and heart progenitors, photo- receptor and non- neuronal cone cells |
Carmena et al., 2002; Tsuda et al., 2002 |
| TGFβi | Jagged1 | + | Epithelial cells | Epithelial - mesenchymal transformation |
Zavadil et al., 2004 |
| FGF1d / FGF2d | Dll1b | - | Neuroepithelium | Maintenance of neuroepithelial precursors |
Faux et al., 2001 |
| PDGFi / angiotensin II | Jagged1 | - | Vascular smooth muscle cells |
Growth retardation | Campos et al., 2002 |
| LPSe | Jagged1 | - | Bone-marrow mesenchymal stem cells |
Proliferation of CD4+ T cells |
Liotta et al., 2008 |
Footnotes:
Vascular Endothelial Growth Factor
Dll: Delta-like
Tumor Necrosis Factor α
FGF: Fibroblast Growth Factor
Lipopolysaccharide
Prostaglandin E2
Interleukin 6
Drosophila Epidermal Growth Factor Receptor
Transforming Growth Factor β
Platelet-derived Growth Factor
Th: T helper cell
The majority of these signaling pathways enhance ligand expression, such as canonical Wnt signaling that activates Jagged1 transcription during hair follicle differentiation (Hofmann et al., 2004). In the angiogenic vasculature, VEGF induces Dll4 expression in endothelial cells to prevent sprouting angiogenesis (Roca and Adams, 2007; Sainson and Harris, 2008; Thurston et al., 2007; Yan and Plowman, 2007) while TNFα-induced Jagged1 expression has the opposite effect (Benedito et al., 2009; Sainson et al., 2008). The differential regulation of expression of Dll4 and Jagged1 with opposing roles in angiogenesis has been proposed to guide the specification of tip cells and stalk cells to regulate the number of sprouting vessels (see Chapter by Gridley). In the immune system, specific inflammatory responses upregulate expression of either Delta-like or Jagged1 ligands in dendritic cells to guide activated CD4+ T cells toward either a T-helper (Th)-1 or Th-2 response, respectively (Amsen et al., 2004; Maekawa et al., 2003). However, more recent findings have questioned the role of Notch signaling in T cell fate acquisition (Ong et al., 2008). Additionally, ligand-specific effects of Notch signaling have also been reported in non-small cell lung cancer cells and hematopoietic progenitors (Choi et al., 2009; de La Coste and Freitas, 2006). Fringe-mediated modulation of the sensitivity of Notch for different ligands as well as interaction of different ligands with distinct Notch receptors have been proposed to regulate some of these ligand-dependent effects (Amsen et al., 2004; Cheng and Gabrilovich, 2007; de La Coste and Freitas, 2006; Maekawa et al., 2003; Raymond et al., 2007).
Contrasting with the positive regulatory effects of signaling pathways on DSL ligand expression, downregulation of Jagged1 expression by PDGF and angiotensin II restricts vascular smooth muscle cell growth in vitro (Campos et al., 2002), while LPS-mediated downregulation of Jagged1 expression in bone marrow mesenchymal stem cells inhibits proliferation of CD4+ T cells (Liotta et al., 2008). The regulation of ligand expression not only plays a role in coordinating normal cellular responses but also in promoting cancer. In fact, upregulation of Jagged1 expression has recently emerged as a ‘pathological link’ between Wnt and IL6 signaling pathways and Notch activation in colon (Pannequin et al., 2009; Rodilla et al., 2009) and breast (Sansone et al., 2007; Studebaker et al., 2008) cancer.
8.2. Spatio-temporal regulation of Notch ligand expression
The existence of mechanisms to regulate ligand expression provides a means to temporally and/or spatially compartmentalize Notch signaling activity and coordinate specific Notch-dependent responses. In fact, the establishment of developmental boundaries and the segmentation of limbs and appendages is dependent on Notch signaling and coordination of these processes can be regulated by the spatio-temporal distribution of ligand expression (Bishop et al., 1999; de Celis et al., 1998; Klein and Arias, 1998; Panin et al., 1997; Rauskolb and Irvine, 1999). For instance, in the developing wing disc of flies, Serrate is expressed dorsally while higher Delta expression occurs ventrally and Notch signaling directs this ligand expression pattern (Blair, 2000; Doherty et al., 1996). The coexpression of Fringe in the dorsal compartment ensures that Serrate can only signal to adjacent ventral cells that lack Fringe, while ventral Delta signals preferentially to adjacent dorsal cells. In this manner, reciprocal Notch signaling between dorsal and ventral cells restricts Notch activation to cells along the dorsoventral boundary required for proper wing margin formation. Further, establishment of leg segments in Drosophila which is dependent on Notch signaling is regulated by the ‘leg gap genes’ homothorax, dachshund and Distal-less that temporally control the segmental pattern of Notch ligand expression as well as the glycosyltransferase Fringe (Bishop et al., 1999; Rauskolb, 2001). Together these findings indicate that the regulation of DSL ligand expression by other signaling pathways serves to spatio-temporally compartmentalize Notch signaling activity. This allows Notch signaling to be integrated into a highly ordered and complex molecular network (Hurlbut et al., 2007), which could regulate embryonic development, the induction of immune and vascular responses as well as contribute to disease states such as cancer in the adult.
9. NON-CANONICAL LIGANDS
In contrast to other signaling systems that employ a large number of activating ligands, there are only 4 mammalian ligands known to activate Notch receptors. It is difficult to account for the pleiotropic affects of Notch given this limited number of DSL ligands; however, the identification of non-canonical ligands expands the repertoire of ligands reported to activate signaling. Unlike the activating canonical ligands that contain a DSL domain required to interact with Notch (Fig. 2), non-canonical ligands lack this essential motif and comprise a group of structurally diverse proteins that include integral and glycosylphosphatidylinositol (GPI)-linked membrane as well as secreted proteins outlined in Fig. 5.
Fig. 5. Non-canonical ligand structure and proposed effects on Notch signaling.
Non-canonical ligands lack a DSL domain (Delta/Serrate/LAG-2), are structurally diverse and include integral- and GPI-linked membrane proteins as well as secreted proteins (see text for details). EGF-like (6 cys), 6-cysteine epidermal growth factor-like repeat as found in canonical ligands; cys, cysteine; TM, transmembrane domain, CSL (CBF1, Su(H), LAG1); EMI, emilin-like domain; EGF-like (8 cys), EGF-like motif with 8 cysteines that is not laminin-like; Ig-CAM, immunoglobulin-containing cell adhesion molecule domain; FNIII, fibronectin type III domain; GPI, glycosylphosphatidylinositol; Q, glutamine-rich region; FReD, fibrinogen-related domain; DOS, Delta and OSM-11-like proteins; IGFBP, insulin-like growth factor-binding protein-like domain; VWF-C, von Willebrand factor type C-like domain; TSP-1, thrombospondin type 1-like domain; CTCK, C-terminal cysteine knot domain; MBD, matrix binding domain; RGD, integrin binding motif; NT, N-terminal domain; CSD, cold shock domain, N1, Notch1; N2, Notch2; N3, Notch3; N4, Notch4. *Only full-length constructs were tested for binding. **Agonist of Jagged1 signaling ***Antagonist of Jagged1 signaling. **** Agonist of Dll4 (Delta-like 4) signaling.
9.1. Membrane-tethered non-canonical ligands
Delta-like 1 (Dlk-1), also known as Pref-1, or FA-1 is one of the first reported non-canonical ligands for Notch (Bachmann et al., 1996; Laborda et al., 1993; Smas and Sul, 1993) and is best known for its role in preventing adipogenesis (Wang et al., 2006). While lacking a DSL domain, Dlk-1 is otherwise structurally similar to Delta-like proteins. Dlk1 is also cleaved by ADAMs and is negatively regulated by Notch signaling (Ross et al., 2004; Wang and Sul, 2006). Most evidence support the idea that Dlk-1 and Notch only interact in cis and the affects of Dlk-1 overexpression on Notch target gene expression and phenotype are consistent with Dlk-1 functioning as a cis inhibitor of Notch signaling (Baladron et al., 2005; Bray et al., 2008; Nueda et al., 2007). Interestingly, an ADAM-resistant, membrane-bound form of Dlk-1 is more potent than wild-type or soluble forms in functioning in cis-inhibition, suggesting that Dlk-1-mediated antagonism of Notch signaling may require low cellular ADAM activity to maintain membrane-bound Dlk-1 (Bray et al., 2008). The molecular basis for Dlk-1 mediated Notch antagonism is unclear, but given the overlap in the binding sites for Dlk-1 and DSL ligands on Notch (Baladron et al., 2005), it seems plausible that Dlk-1 antagonizes Notch signaling by competing with DSL ligands for Notch binding. Although Dlk-1 and Notch have been shown to interact by yeast-two hybrid analysis (Baladron et al., 2005; Komatsu et al., 2008), interactions between these proteins has not been demonstrated for endogenous or ectopic proteins. Neither is there a consensus on whether Dlk-1-induced loss of Hes-1 expression directly involves Notch since, Hes-1 is regulated by other signaling pathways (Hatakeyama et al., 2004; Kluppel and Wrana, 2005; Ross et al., 2004).
More recently, the identification of a DOS domain in Dlk-1 and Dlk-2/EGFL9 has led to the proposal that these proteins may also function as co-activating non-canonical Notch ligands (Komatsu et al., 2008). In fact, genetic studies have shown that Dlk-1 can functionally substitute for the C.elegans DOS-only containing ligand, OSM-11, in cooperating with the DSL-only ligand, DSL-1, to activate Notch signaling, suggesting that the role of DOS-motif in Notch signaling may be conserved across species (Komatsu et al., 2008). However, these findings are difficult to reconcile given that Dlk-1 has been suggested to antagonize Jagged1-induced Notch signaling (Baladron et al., 2005). In light of the fact that Jagged1 contains both a DSL domain and a DOS motif, it has been proposed that the DOS-only containing ligand Dlk-1 competes with Jagged1 for Notch binding and thus antagonizes Jagged1 signaling (Komatsu et al., 2008). Thus, DOS domain proteins may function as Notch agonists in cooperation with DSL domain-only containing ligands but antagonize signaling by ligands containing both DSL and DOS domains.
Another integral membrane-bound Notch ligand lacking a DSL domain is Delta/Notch-like EGF-related receptor (DNER) that like Dlk-1 also contains extracellular tandem EGF repeats (Eiraku et al., 2002). In contrast to Dlk-1, DNER binds Notch when presented in trans and DNER-expressing cells activate a CSL reporter in cocultured cells (Eiraku et al., 2005). Both in vitro and in vivo studies support DNER’s function as a trans-ligand to effect glial morphological changes through activation of Notch. DNER, however, does not affect glial cell number in vivo, suggesting that it functions at later stages of differentiation. Consistent with the expression of DNER in Purkinje cells and Notch in the adjacent Bergmann glia, DNER mutant mice exhibit morphological defects in Bergmann glia (Eiraku et al., 2005). A soluble recombinant form of DNER can also affect Bergmann glia morphology in vitro in a γ-secretase-dependent but CSL-independent manner. Instead of CSL, the E3 ubiquitin ligase Deltex has been implicated as an alternative downstream effector of Notch in DNER-induced glial morphological changes. Deltex can bind directly to the Notch intracellular domain, and mediate a trimeric complex between itself, full-length Notch, and β-arrestin (Mukherjee et al., 2005), making it possible that Notch could activate signaling through β-arrestin that would require Deltex but not CSL. Whether the effects of DNER are dependent on Notch receptor expression in Bergmann glia have yet to be determined.
A putative DSL ligand-like protein called Jagged and Delta protein (Jedi) has been identified based on sequence data (Krivtsov et al., 2007). However, the putative DSL and EGF repeats of Jedi lack the conserved cysteine spacing common to either the signature motif of canonical ligands or EGF repeats present in DNER and Dlk-1. Instead, the Jedi ECD contains an N-terminal emilin domain followed by multiple tandem repeats of an 8-cysteine variation of the EGF domain interspersed with two single 6-cysteine EGF repeats (Krivtsov et al., 2007; Nanda et al., 2005). In fact, Jedi has not been reported to interact with any of the Notch receptors and lacks trans-activating or cis-inhibitory activity. Although soluble Jedi added to Notch-expressing cells weakly inhibits a Notch reporter, there is currently no strong evidence linking Jedi to Notch signaling. The closely related Jedi family member, multiple EGF-like domains 10 protein (MEGF10) (Krivtsov et al., 2007) has also been proposed to interact with the Notch signaling pathway (Holterman et al., 2007); however, like Jedi, there has been no formal demonstration that MEGF10 can directly interact with Notch receptors.
9.2. GPI-linked non-canonical ligands
Structurally distinct from the integral membrane non-canonical ligands are the GPI-linked neural cell adhesion molecules, F3/contactin1 and NB3/contactin6 that activate Notch signaling to induce oligodendrocyte (OL) differentiation (Cui et al., 2004; Hu et al., 2003). Although binding and fractionation studies have indicated that these contactins interact with Notch in trans, cis interactions cannot be ruled out since both endogenous F3 and NB3 coimmunoprecipitate with Notch. Both contactins interact with Notch EGF repeats distal to the DSL binding site; however, F3 can also interact with Notch EGF repeats 1-13 that includes the DSL ligand-binding site at EGF 11-12. While this interaction would initially suggest that F3 competes for the DSL ligand-binding site, further studies are required to determine whether the F3 and DSL ligand-binding sites actually overlap.
As found for DSL ligand treatment, soluble forms of either contactin induce γ-secretase-dependent NICD production in OL cells. However, F3-Notch signaling does not activate Hes-1 transcription, and there is no evidence of NB3 activating canonical CSL-induced Notch signaling (Hu et al., 2003; Lu et al., 2008). Instead of CSL, both contactins utilize Deltex as an effector of Notch signaling to induce glial maturation. An interesting conundrum is raised in these in vitro assays since the same cells (that presumably utilize the same Notch receptors) differentiate in response to contactins but remain progenitors in response to DSL ligand or NICD expression. It is thought that differences in the temporal expression of DSL ligands and contactins could dictate which effect takes precedence in vivo since DSL ligands and contactins are expressed at distinct developmental time points. Therefore, like DNER, the contactins appear to utilize Notch to effect changes late in differentiation as opposed to DSL ligands that can impact early cell fate decisions (Hu et al., 2003).
The important role for contactin-induced Notch signaling in OL differentiation, has led to speculation that in multiple sclerosis lesions, contactin expression may be lost in demyelinated axons that would normally activate Notch in neighboring OL precursor cells. Recent findings, however have demonstrated that not only is contactin expression maintained in demyelinated axons but also that NICD is generated in the OL precursor cells within these lesions (Nakahara et al., 2009). Instead, translocation of NICD to the nucleus was inhibited in these cells and the proapototic factor TAT-interacting protein 30 that prevents nuclear transport has been implicated in this process. These findings identify a novel mechanism for regulation of Notch activity downstream of NICD generation.
9.3. Secreted non-canonical ligands
Two secreted non-DSL ligands have been identified in Drosophila. The first, Scabrous (Sca), plays a role in Notch-dependent patterning of eye ommatidia and sensory bristles (Baker et al., 1990; Mlodzik et al., 1990). Sca binds to Notch in trans and activates transcription of the Notch target gene E(spl)C m3 (Mok et al., 2005; Powell et al., 2001). It is not known however, if the effects of Sca require γ-secretase cleavage of Notch, the Notch downstream effector Su(H), or indeed activation of some other signaling pathway. Another reported Drosophila secreted non-DSL ligand for Notch is Wingless (Wg), the fly ortholog of mammalian Wnt proteins. Wg was identified as a Notch-binding protein in a screen of a phage display library expressing Drosophila embryo transcripts and immunoprecipitation of endogenous Notch and Wg in fly embryos supports such an interaction in vivo (Wesley, 1999). Although the gene shaggy can be transcriptionally activated in a Wg- and Notch-dependent manner, it is not clear if binding of Wg to Notch is required for its transcription or which Notch downstream effectors are required. While many vertebrate Wnt proteins exist, none have been reported to bind Notch as demonstrated for Drosophila Wg.
In C. elegans, five secreted putative Notch ligands lacking DSL domains have been identified: OSM11, OSM7, DOS1, DOS2 and DOS3 (Komatsu et al., 2008). Interestingly, all five proteins contain conserved amino acids within a common motif called DOS and although this motif is lacking in all C. elegans DSL ligands, it is present in most DSL ligands from other phyla. The best characterized of these C. elegans DOS-containing ligands, OSM11, interacts with the extracellular domain of LIN-12 in yeast two-hybrid assays and genetic analyses suggest that OSM-11 enhances LIN-12 signaling during vulval development by acting upstream of or during LIN12 receptor activation. While losses of osm11 produce defects in vulval precursor cell specification associated with losses in Notch signaling, loss of the DSL domain-containing ligand dsl-1 potentiates osm11 loss-of-function defects, suggesting that DOS domain-only and DSL domain-only containing ligands cooperate to activate signaling in some developmental contexts in C. elegans. Although these results suggest the existence of a bipartite ligand system for activating Notch in C. elegans, biochemical studies are necessary to confirm that OSM11 directly interacts with endogenous LIN12 to activate signaling. Furthermore, it is not clear from these studies whether the effects of OSM11 on Notch signaling are indeed mediated by the DOS motif. In this regard, a role for the DOS motif in Notch binding and signaling has been extrapolated (Komatsu et al., 2008; Kopan and Ilagan, 2009) from mutational and structural studies of Drosophila and mammalian DSL ligands (Cordle et al., 2008; Parks et al., 2006; Shimizu et al., 1999). Significantly, mutations that map to the DOS motif of Jagged1 are associated with human syndromes (Eldadah et al., 2001; Guarnaccia et al., 2009; Warthen et al., 2006) and genetic malformations in mice (Kiernan et al., 2001; Tsai et al., 2001). Although X-ray crystallography and NMR-based binding studies have suggested that the binding interface between Jagged1 and Notch1 includes amino acid residues from not only the DSL domain but also the DOS domain, the topography of this interface is not known (Cordle et al., 2008). Understanding how the DOS and DSL domains cooperatively bind Notch will require crystal structure studies of the ligand-Notch complex.
Secreted Notch ligands lacking a DSL domain have also been identified in vertebrates. One of these is the Connective Tissue Growth Factor/cysteine-rich 61/Nephroblastoma Overexpressed Gene family member, CCN3. When coexpressed, CCN3 interacts with Notch via the CCN3 C-terminal cysteine knot that appears to be a general tandem EGF repeat-binding domain (Sakamoto et al., 2002b; Thibout et al., 2003). Co-expression of CCN3 potentiates endogenous CSL-dependent Notch signaling in reporter assays and losses in CCN3 reduce trans-DSL ligand induced activation of a CSL reporter (Gupta et al., 2007; Minamizato et al., 2007; Sakamoto et al., 2002b). Further supporting a role for CCN3 as an activating cofactor for canonical ligand-induced signaling is the observation that soluble CCN3 can enhance hematopoietic precursor cell colony formation induced by Jagged-1 (Gupta et al., 2007). Additionally, gains and losses in CCN3 expression produce corresponding changes in Hes-1 expression, suggesting that CCN3 may activate Notch in an autocrine fashion (Gupta et al., 2007; Minamizato et al., 2007; Sakamoto et al., 2002b). Autocrine Notch signaling by CCN3 may be relevant to cell types such as chondrocytes and vascular smooth muscle cells that secrete extracellular matrix and consequently are isolated and unable to undergo juxtacrine signaling by canonical ligands. Consistent with this notion, both chondrocytes and vascular smooth muscle cells express CCN3 (Ellis et al., 2000; Perbal, 2004).
A second secreted, non-DSL vertebrate protein that can activate Notch signaling is the microfibril-associated glycoprotein family, MAGP-1 and MAGP-2 (Gibson et al., 1996; Gibson et al., 1991). Both MAGP proteins can interact with Notch leading to γ-secretase-dependent NICD generation and activation of CSL-dependent reporter constructs (Miyamoto et al., 2006). Like CCN3, MAGP-2 only activates Notch when co-expressed in the same cell, and vascular smooth muscle cells that express MAGP2 may use this non-canonical ligand to activate Notch signaling in an autocrine manner (Albig et al., 2008; Miyamoto et al., 2006). Interestingly, similar to DSL ligands, MAGP-2 can activate Notch by inducing ADAM-independent dissociation of the Notch heterodimer and in fact, MAGP-2 is the only non-canonical ligand that has so far been demonstrated to cause non-enzymatic dissociation of Notch (Miyamoto et al., 2006). The biological significance of MAGP-2-induced Notch signaling is as yet unclear and it appears that MAGP-2 can also inhibit Notch signaling in certain cell types; however, the molecular basis for these cell-type differences are not understood (Albig et al., 2008).
In addition to CCN3/NOV and MAGP proteins, a third vertebrate matrix protein, thrombospondin2 (TSP2), has been implicated as a non-canonical Notch ligand (Meng et al., 2009). As found for CCN3/NOV, TSP2 enhances signaling induced by trans-DSL ligands either when coexpressed or when exposed to Notch cells as a soluble recombinant protein (Meng et al., 2009). The effect of TSP2 on Notch signaling is γ-secretase-dependent and requires the Notch extracellular sequences. Consistent with these findings, coimmunoprecipitation studies suggest that TSP2 interacts with the Notch3 extracellular domain at the cell surface. In vitro binding assays using recombinant proteins further suggest that TSP2 can directly interact with the first 11 EGF-like repeats of Notch3. It is surprising that TSP2 enhances rather than inhibits ligand-induced Notch signaling given that the region of Notch3 includes the DSL ligand-binding domain; however, it is not known if the TSP2 and DSL ligand binding sites actually overlap. Interestingly, TSP2 can also interact with Jagged1 and enhance binding to Notch3 EGF repeats suggesting a molecular basis for increased Notch signaling by TSP2. Further supporting interactions between TSP2 and Notch is the observation that arterial tissue from TSP2 knockout mice exhibit significant reduction in Notch target expression. Although the closely related thrombospondin, TSP1, also interacts with Notch3 and Jagged1 (Meng et al., 2009) it is neither able to enhance binding between the ligand-receptor pair nor enhance Notch signaling - subtle structural differences between these TSP family members may account for their differential effects on Notch signaling.
A fourth secreted vertebrate non-DSL ligand that activates Notch signaling, Y-box (YB) protein-1, belongs to the cold shock protein family (Frye et al., 2009; Rauen et al., 2009). Yeast-two hybrid and coimmunoprecipitation studies have demonstrated that YB1 interacts with EGF repeats 13-33 of Notch3 (Rauen et al., 2009), a region distinct from that required for DSL ligand binding. Furthermore, confocal microscopy and fluorescence activated cell sorter analyses suggest that YB1-Notch3 interactions occur at the cell surface and soluble YB1-Notch3 interactions activate CSL-dependent reporter constructs in a γ-secretase dependent manner. Interestingly, YB1 does not bind Notch1 and although interactions with Notch2 and Notch4 have not been examined, it is tempting to speculate that YB1 interactions may be specific for Notch3. YB1-Notch3 interactions may be relevant to kidney disease since in a mouse model of mesanglioproliferative disease in which YB1 and Notch3 expression are coordinately upregulated during the course of the disease, both the cleaved extracellular domain of Notch3 and YB-1 are detected in urine samples of diseased animals. The molecular basis of these finding are unclear, however, they could reflect YB-1 induced dissociation of the Notch3 heterodimer.
More recently, a fifth vertebrate secreted non-DSL Notch ligand, Epidermal growth factor-like domain 7 (EGFL7) was identified in yeast-two-hybrid screens (Schmidt et al., 2009). EGFL7 interacts with a set of EGF-like repeats that includes the DSL binding sites of all four human Notch receptors and antagonizes Notch signaling induced by Jagged1-type ligands. The inhibitory effects of EGFL7 on Jagged1-induced Notch signaling were demonstrated both in cis and in trans using biochemical studies as well as a neurosphere model and appear to result from competition with Jagged1 for Notch binding. Expression of EGFL7 prevents self-renewal of neural stem cells cultured as neurospheres, a process dependent on Jagged1-Notch1 interactions. Further supporting a role for EGFL7 as a Notch antagonist, EGFL7 expression promotes differentiation of NSCs into neurons and oligodendrocytes at the expense of astrocytes. Surprisingly, the inhibitory effects of EGFL7 seem specific to Jagged-type ligands. Furthermore, ablation of EGFL7 reduces Dll4-induced activation of a CSL-dependent reporter suggesting that EGFL7 enhances Dll4-induced Notch signaling. Although it is difficult to reconcile the differential DSL ligand-dependent effects of EGFL7 on Notch signaling, yeast two-hybrid studies have shown that EGFL7 can also interact with Dll4 but not with Jagged1 or Jagged2. It is possible that as proposed for TSP2, EGFL7 enhances ligand-Notch interactions accounting for its ability to potentiate Dll4-induced Notch signaling. It is important to note that the opposing DSL ligand-specific effects of EGFL7 were demonstrated in different cell contexts and thus could reflect cell type-specific differences for EGFL7 on Notch signaling as found for MAGP2.
In summary, non-canonical ligands represent a subset of Notch ligands that despite lacking a DSL domain can activate Notch signaling. Compared to the canonical ligands that all require binding to Notch EGF repeats 11 and 12 to activate signaling, non-canonical ligands do not appear to have a consensus Notch binding site, yet some activate Notch γ-secretase cleavage and CSL-dependent transcription. Given that non-enzymatic dissociation of Notch leads to signaling, it would appear that any protein that can bind Notch and dissociate the heterodimeric structure activates Notch signaling. Indeed, binding of MAGP2 can cause non-enzymatic dissociation of Notch and activation of Notch signaling, and it remains to be demonstrated if other non-canonical ligands also follow a similar mechanism for Notch activation. Interestingly, like the membrane-bound DSL ligands, all type-1 transmembrane non-canonical ligands contain lysines in their intracellular domains that could serve as ubiquitination sites to facilitate transendocytosis; however, it is not known if endocytosis is required for activity of these non-canonical ligands. Even less clear is how Notch binding to secreted non-canonical ligands like MAGP2 could produce force for heterodimer dissociation, but perhaps cooperative binding with membrane-bound ligands or tethering to the extracellular matrix would induce a pulling force on Notch.
While non-canonical ligands may contribute for the pleiotropic nature of Notch signaling, the effects of many have only been demonstrated using in vitro assays and need to be confirmed in vivo. In this regard, it is noteworthy that while DSL ligands are crucial for embryonic development and viability in the mouse, none of the reported non-canonical ligands are similarly required. It thus appears that if non-canonical ligands function in vivo, they may do so as modulators of Notch signaling in the adult animal.
10. CONCLUSIONS AND FUTURE DIRECTIONS
Although unique ligand-receptor combinations have been identified that induce specific cellular responses, the molecular mechanisms underlying ligand-specific signaling remains an outstanding question in the field. Moreover, given the direct and somewhat simple signaling mechanism ascribed to Notch, it is unclear how different Notch ligands could induce distinct signaling responses. It will be important to determine if different ligand-Notch complexes recruit unique signaling effectors and whether the distinct responses involve activation of cytoplasmic and/or nuclear signaling pathways. In this regard, identification of the endocytic machinery used by ligand cells to activate Notch signaling and the potential role for endocytosis in force generation are critical avenues that remain to be tested. That ligands have intrinsic signaling activity independent of Notch as well as their potential to participate in bi-directional signaling, are exciting but relatively unexplored areas of ligand biology that warrant further investigation. The importance of Notch ligands in cancer and other pathological states involving aberrant angiogenesis have identified Notch ligands as potential and promising therapeutic targets (Roca and Adams, 2007; Sainson and Harris, 2008; Thurston et al., 2007; Yan and Plowman, 2007). Finally, the use of Notch ligands in the expansion and maintenance of stem cells for tissue regeneration and replacement underscores their fundamental biological importance (Dallas et al., 2005; Delaney et al., 2005).
Acknowledgements
We thank Abdiwahab Musse and Jason Tchieu for help with the illustrations and Alison Miyamoto for contributions to the material previously published in Oncogene (2008) 27, 5148–5167. The authors acknowledge the NIH R01-GM0850032 (GW), JCCF and NIH Training Grant-GM007185 (LMK) and AICR (BD) for financial support.
REFERENCES
- Ahimou F, Mok LP, Bardot B, Wesley C. The adhesion force of Notch with Delta and the rate of Notch signaling. J Cell Biol. 2004;167:1217–1229. doi: 10.1083/jcb.200407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aho S. Soluble form of Jagged1: unique product of epithelial keratinocytes and a regulator of keratinocyte differentiation. J Cell Biochem. 2004;92:1271–1281. doi: 10.1002/jcb.20125. [DOI] [PubMed] [Google Scholar]
- Akai J, Halley PA, Storey KG. FGF-dependent Notch signaling maintains the spinal cord stem zone. Genes Dev. 2005;19:2877–2887. doi: 10.1101/gad.357705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albig AR, Becenti DJ, Roy TG, Schiemann WP. Microfibril-associate glycoprotein-2 (MAGP-2) promotes angiogenic cell sprouting by blocking notch signaling in endothelial cells. Microvasc Res. 2008 doi: 10.1016/j.mvr.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–526. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- Aoto J, Chen L. Bidirectional ephrin/Eph signaling in synaptic functions. Brain Res. 2007;1184:72–80. doi: 10.1016/j.brainres.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Ascano JM, Beverly LJ, Capobianco AJ. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J Biol Chem. 2003;278:8771–8779. doi: 10.1074/jbc.M211427200. [DOI] [PubMed] [Google Scholar]
- Bachmann E, Krogh TN, Hojrup P, Skjodt K, Teisner B. Mouse fetal antigen 1 (mFA1), the circulating gene product of mdlk, pref-1 and SCP-1: isolation, characterization and biology. J Reprod Fertil. 1996;107:279–285. doi: 10.1530/jrf.0.1070279. [DOI] [PubMed] [Google Scholar]
- Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250:1370–1377. doi: 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- Baladron V, Ruiz-Hidalgo MJ, Nueda ML, Diaz-Guerra MJ, Garcia-Ramirez JJ, Bonvini E, Gubina E, Laborda J. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp Cell Res. 2005;303:343–359. doi: 10.1016/j.yexcr.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Schweisguth F. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev Cell. 2006;10:245–255. doi: 10.1016/j.devcel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Barrantes IB, Elia AJ, Wunsch K, Hrabe de Angelis MH, Mak TW, Rossant J, Conlon RA, Gossler A, de la Pompa JL. Interaction between Notch signalling and Lunatic fringe during somite boundary formation in the mouse. Curr Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- Barriere H, Nemes C, Lechardeur D, Khan-Mohammad M, Fruh K, Lukacs GL. Molecular basis of oligoubiquitin-dependent internalization of membrane proteins in Mammalian cells. Traffic. 2006;7:282–297. doi: 10.1111/j.1600-0854.2006.00384.x. [DOI] [PubMed] [Google Scholar]
- Barsi JC, Rajendra R, Wu JI, Artzt K. Mind bomb1 is a ubiquitin ligase essential for mouse embryonic development and Notch signaling. Mech Dev. 2005;122:1106–1117. doi: 10.1016/j.mod.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Becam I, Fiuza UM, Arias AM, Milan M. A Role of Receptor Notch in Ligand cis-Inhibition in Drosophila. Curr Biol. 2010;20:554–560. doi: 10.1016/j.cub.2010.01.058. [DOI] [PubMed] [Google Scholar]
- Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- Bishop SA, Klein T, Arias AM, Couso JP. Composite signalling from Serrate and Delta establishes leg segments in Drosophila through Notch. Development. 1999;126:2993–3003. doi: 10.1242/dev.126.13.2993. [DOI] [PubMed] [Google Scholar]
- Blair SS. Notch signaling: Fringe really is a glycosyltransferase. Curr Biol. 2000;10:R608–612. doi: 10.1016/s0960-9822(00)00633-3. [DOI] [PubMed] [Google Scholar]
- Bland CE, Kimberly P, Rand MD. Notch induced proteolysis and nuclear localization of the Delta ligand. J Biol Chem. 2003;278:13607–13610. doi: 10.1074/jbc.C300016200. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Bray SJ, Takada S, Harrison E, Shen SC, Ferguson-Smith AC. The atypical mammalian ligand Delta-like homologue 1 (Dlk1) can regulate Notch signalling in Drosophila. BMC Dev Biol. 2008;8:11. doi: 10.1186/1471-213X-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brone B, Eggermont J. PDZ proteins retain and regulate membrane transporters in polarized epithelial cell membranes. Am J Physiol Cell Physiol. 2005;288:C20–29. doi: 10.1152/ajpcell.00368.2004. [DOI] [PubMed] [Google Scholar]
- Campos AH, Wang W, Pollman MJ, Gibbons GH. Determinants of Notch-3 receptor expression and signaling in vascular smooth muscle cells: implications in cell-cycle regulation. Circ Res. 2002;91:999–1006. doi: 10.1161/01.res.0000044944.99984.25. [DOI] [PubMed] [Google Scholar]
- Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, Michelson AM. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- Chen H, De Camilli P. The association of epsin with ubiquitinated cargo along the endocytic pathway is negatively regulated by its interaction with clathrin. Proc Natl Acad Sci U S A. 2005;102:2766–2771. doi: 10.1073/pnas.0409719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Greenwald I. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev Cell. 2004;6:183–192. doi: 10.1016/s1534-5807(04)00021-8. [DOI] [PubMed] [Google Scholar]
- Chen W, Casey Corliss D. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev Biol. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HT, Miner JH, Lin M, Tansey MG, Roth K, Kopan R. Gamma-secretase activity is dispensable for mesenchyme-to-epithelium transition but required for podocyte and proximal tubule formation in developing mouse kidney. Development. 2003;130:5031–5042. doi: 10.1242/dev.00697. [DOI] [PubMed] [Google Scholar]
- Cheng P, Gabrilovich D. Notch signaling in differentiation and function of dendritic cells. Immunol Res. 2007 doi: 10.1007/s12026-007-8011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A. Why is delta endocytosis required for effective activation of notch? Dev Dyn. 2006;235:886–894. doi: 10.1002/dvdy.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta [see comments] Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Choi K, Ahn YH, Gibbons DL, Tran HT, Creighton CJ, Girard L, Minna JD, Qin FX, Kurie JM. Distinct biological roles for the notch ligands Jagged-1 and Jagged-2. J Biol Chem. 2009;284:17766–17774. doi: 10.1074/jbc.M109.003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Cordle J, Johnson S, Tay JZ, Roversi P, Wilkin MB, de Madrid BH, Shimizu H, Jensen S, Whiteman P, Jin B, et al. A conserved face of the Jagged/Serrate DSL domain is involved in Notch trans-activation and cis-inhibition. Nat Struct Mol Biol. 2008;15:849–857. doi: 10.1038/nsmb.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XY, Hu QD, Tekaya M, Shimoda Y, Ang BT, Nie DY, Sun L, Hu WP, Karsak M, Duka T, et al. NB-3/Notch1 pathway via Deltex1 promotes neural progenitor cell differentiation into oligodendrocytes. J Biol Chem. 2004;279:25858–25865. doi: 10.1074/jbc.M313505200. [DOI] [PubMed] [Google Scholar]
- Dallas MH, Varnum-Finney B, Delaney C, Kato K, Bernstein ID. Density of the Notch ligand Delta1 determines generation of B and T cell precursors from hematopoietic stem cells. J Exp Med. 2005;201:1361–1366. doi: 10.1084/jem.20042450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis CF, Bray SJ. The Abruptex domain of Notch regulates negative interactions between Notch, its ligands and Fringe. Development. 2000;127:1291–1302. doi: 10.1242/dev.127.6.1291. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124:3241–3251. doi: 10.1242/dev.124.17.3241. [DOI] [PubMed] [Google Scholar]
- de Celis JF, Tyler DM, de Celis J, Bray SJ. Notch signalling mediates segmentation of the Drosophila leg. Development. 1998;125:4617–4626. doi: 10.1242/dev.125.23.4617. [DOI] [PubMed] [Google Scholar]
- de Joussineau C, Soule J, Martin M, Anguille C, Montcourrier P, Alexandre D. Delta-promoted filopodia mediate long-range lateral inhibition in Drosophila. Nature. 2003;426:555–559. doi: 10.1038/nature02157. [DOI] [PubMed] [Google Scholar]
- de La Coste A, Freitas AA. Notch signaling: distinct ligands induce specific signals during lymphocyte development and maturation. Immunol Lett. 2006;102:1–9. doi: 10.1016/j.imlet.2005.06.014. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- Deblandre GA, Lai EC, Kintner C. Xenopus neuralized is a ubiquitin ligase that interacts with XDelta1 and regulates Notch signaling. Dev Cell. 2001;1:795–806. doi: 10.1016/s1534-5807(01)00091-0. [DOI] [PubMed] [Google Scholar]
- Delaney C, Varnum-Finney B, Aoyama K, Brashem-Stein C, Bernstein ID. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106:2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwig A, Bland C, Beem-Miller M, Kimberly P, Rand MD. Endocytosis-independent mechanisms of Delta ligand proteolysis. Exp Cell Res. 2006;312:1345–1360. doi: 10.1016/j.yexcr.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Delwig A, Rand MD. Kuz and TACE can activate Notch independent of ligand. Cell Mol Life Sci. 2008;65:2232–2243. doi: 10.1007/s00018-008-8127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty D, Feger G, Younger-Shepherd S, Jan LY, Jan YN. Delta is a ventral to dorsal signal complementary to Serrate, another Notch ligand, in Drosophila wing formation. Genes Dev. 1996;10:421–434. doi: 10.1101/gad.10.4.421. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Chang WS, Rapaport DH, Harris WA. Regulation of neuronal diversity in the Xenopus retina by Delta signalling. Nature. 1997;385:67–70. doi: 10.1038/385067a0. [DOI] [PubMed] [Google Scholar]
- Dravis C, Yokoyama N, Chumley MJ, Cowan CA, Silvany RE, Shay J, Baker LA, Henkemeyer M. Bidirectional signaling mediated by ephrin-B2 and EphB2 controls urorectal development. Dev Biol. 2004;271:272–290. doi: 10.1016/j.ydbio.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL. The role of Notch in patterning the human vertebral column. Curr Opin Genet Dev. 2009;19:329–337. doi: 10.1016/j.gde.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL, Henrique D, Harrison SM, Beddington RS. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development. 1997;124:3065–3076. doi: 10.1242/dev.124.16.3065. [DOI] [PubMed] [Google Scholar]
- Dyczynska E, Sun D, Yi H, Sehara-Fujisawa A, Blobel CP, Zolkiewska A. Proteolytic processing of delta-like 1 by ADAM proteases. J Biol Chem. 2007;282:436–444. doi: 10.1074/jbc.M605451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Hirata Y, Takeshima H, Hirano T, Kengaku M. Delta/notch-like epidermal growth factor (EGF)-related receptor, a novel EGF-like repeat-containing protein targeted to dendrites of developing and adult central nervous system neurons. J Biol Chem. 2002;277:25400–25407. doi: 10.1074/jbc.M110793200. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, Kengaku M. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873–880. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Greene LE. Multiple roles of auxilin and hsc70 in clathrin-mediated endocytosis. Traffic. 2007;8:640–646. doi: 10.1111/j.1600-0854.2007.00568.x. [DOI] [PubMed] [Google Scholar]
- Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, Elkins R, Dietz HC. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- Ellis PD, Chen Q, Barker PJ, Metcalfe JC, Kemp PR. Nov gene encodes adhesion factor for vascular smooth muscle cells and is dynamically regulated in response to vascular injury. Arterioscler Thromb Vasc Biol. 2000;20:1912–1919. doi: 10.1161/01.atv.20.8.1912. [DOI] [PubMed] [Google Scholar]
- Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 2005;122:763–773. doi: 10.1016/j.cell.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Estrach S, Ambler CA, Lo Celso C, Hozumi K, Watt FM. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development. 2006;133:4427–4438. doi: 10.1242/dev.02644. [DOI] [PubMed] [Google Scholar]
- Estrach S, Legg J, Watt FM. Syntenin mediates Delta1-induced cohesiveness of epidermal stem cells in culture. J Cell Sci. 2007;120:2944–2952. doi: 10.1242/jcs.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun SH, Banks SM, Fischer JA. Auxilin is essential for Delta signaling. Development. 2008;135:1089–1095. doi: 10.1242/dev.009530. [DOI] [PubMed] [Google Scholar]
- Faux CH, Turnley AM, Epa R, Cappai R, Bartlett PF. Interactions between fibroblast growth factors and Notch regulate neuronal differentiation. J Neurosci. 2001;21:5587–5596. doi: 10.1523/JNEUROSCI.21-15-05587.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K, Greenwald I. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development. 1995;121:4275–4282. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- Fiuza UM, Arias AM. Cell and molecular biology of Notch. J Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- Fiuza UM, Klein T, Martinez Arias A, Hayward P. Mechanisms of ligand-mediated inhibition in Notch signaling activity in Drosophila. Dev Dyn. 2010;239:798–805. doi: 10.1002/dvdy.22207. [DOI] [PubMed] [Google Scholar]
- Fleming RJ. Structural conservation of Notch receptors and ligands. Semin Cell Dev Biol. 1998;9:599–607. doi: 10.1006/scdb.1998.0260. [DOI] [PubMed] [Google Scholar]
- Franklin JL, Berechid BE, Cutting FB, Presente A, Chambers CB, Foltz DR, Ferreira A, Nye JS. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr Biol. 1999;9:1448–1457. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- Frye BC, Halfter S, Djudjaj S, Muehlenberg P, Weber S, Raffetseder U, En-Nia A, Knott H, Baron JM, Dooley S, et al. Y-box protein-1 is actively secreted through a non-classical pathway and acts as an extracellular mitogen. EMBO Rep. 2009;10:783–789. doi: 10.1038/embor.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi Y, Hernandez SL, Das I, Ahn A, Huang J, Vorontchikhina M, Sharma A, Kanamaru E, Borisenko V, Desilva DM, et al. A notch1 ectodomain construct inhibits endothelial notch signaling, tumor growth, and angiogenesis. Cancer Res. 2008;68:4727–4735. doi: 10.1158/0008-5472.CAN-07-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Geffers I, Serth K, Chapman G, Jaekel R, Schuster-Gossler K, Cordes R, Sparrow DB, Kremmer E, Dunwoodie SL, Klein T, et al. Divergent functions and distinct localization of the Notch ligands DLL1 and DLL3 in vivo. J Cell Biol. 2007;178:465–476. doi: 10.1083/jcb.200702009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MA, Hatzinikolas G, Kumaratilake JS, Sandberg LB, Nicholl JK, Sutherland GR, Cleary EG. Further characterization of proteins associated with elastic fiber microfibrils including the molecular cloning of MAGP-2 (MP25) J Biol Chem. 1996;271:1096–1103. doi: 10.1074/jbc.271.2.1096. [DOI] [PubMed] [Google Scholar]
- Gibson MA, Sandberg LB, Grosso LE, Cleary EG. Complementary DNA cloning establishes microfibril-associated glycoprotein (MAGP) to be a discrete component of the elastin-associated microfibrils. J Biol Chem. 1991;266:7596–7601. [PubMed] [Google Scholar]
- Glittenberg M, Pitsouli C, Garvey C, Delidakis C, Bray S. Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. Embo J. 2006;25:4697–4706. doi: 10.1038/sj.emboj.7601337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling--a structural and biochemical perspective. J Cell Sci. 2008a;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1 negative regulatory region: Implications for normal activation and pathogenic signaling in T-ALL. Blood. 2008b doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I, Rubin GM. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell. 1992;68:271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling in vertebrate development and disease. Mol Cell Neurosci. 1997;9:103–108. doi: 10.1006/mcne.1997.0610. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Spec No 1):R9–13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- Guarnaccia C, Dhir S, Pintar A, Pongor S. The tetralogy of Fallot-associated G274D mutation impairs folding of the second epidermal growth factor repeat in Jagged-1. FEBS J. 2009;276:6247–6257. doi: 10.1111/j.1742-4658.2009.07333.x. [DOI] [PubMed] [Google Scholar]
- Gupta R, Hong D, Iborra F, Sarno S, Enver T. NOV (CCN3) functions as a regulator of human hematopoietic stem or progenitor cells. Science. 2007;316:590–593. doi: 10.1126/science.1136031. [DOI] [PubMed] [Google Scholar]
- Hagedorn EJ, Bayraktar JL, Kandachar VR, Bai T, Englert DM, Chang HC. Drosophila melanogaster auxilin regulates the internalization of Delta to control activity of the Notch signaling pathway. J Cell Biol. 2006;173:443–452. doi: 10.1083/jcb.200602054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Lowe JB. Role of glycosylation in development. Annu Rev Biochem. 2004;73:491–537. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- Hamel S, Fantini J, Schweisguth F. Notch ligand activity is modulated by glycosphingolipid membrane composition in Drosophila melanogaster. J Cell Biol. 2010;188:581–594. doi: 10.1083/jcb.200907116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris BZ, Lim WA. Mechanism and role of PDZ domains in signaling complex assembly. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hawryluk MJ, Keyel PA, Mishra SK, Watkins SC, Heuser JE, Traub LM. Epsin 1 is a polyubiquitin-selective clathrin-associated sorting protein. Traffic. 2006;7:262–281. doi: 10.1111/j.1600-0854.2006.00383.x. [DOI] [PubMed] [Google Scholar]
- Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Christensen S, Kimble J. Functional domains of LAG-2, a putative signaling ligand for LIN-12 and GLP-1 receptors in Caenorhabditis elegans. Mol Biol Cell. 1997;8:1751–1762. doi: 10.1091/mbc.8.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signalling in the embryonic chick retina. Curr Biol. 1997;7:661–670. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Heuss SF, Ndiaye-Lobry D, Six EM, Israel A, Logeat F. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc Natl Acad Sci U S A. 2008;105:11212–11217. doi: 10.1073/pnas.0800695105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks C, Ladi E, Lindsell C, Hsieh J, Hayward S, Collazo A, Weinmaster G. A secreted Delta1-Fc fusion protein functions both as an activator and inhibitor of Notch1 signaling. Journal of Neuroscience Research. 2002;69:60–71. doi: 10.1002/jnr.10263. [DOI] [PubMed] [Google Scholar]
- Hiratochi M, Nagase H, Kuramochi Y, Koh CS, Ohkawara T, Nakayama K. The Delta intracellular domain mediates TGF-beta/Activin signaling through binding to Smads and has an important bi-directional function in the Notch-Delta signaling pathway. Nucleic Acids Res. 2007;35:912–922. doi: 10.1093/nar/gkl1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M, Schuster-Gossler K, Watabe-Rudolph M, Aulehla A, Herrmann BG, Gossler A. WNT signaling, in synergy with T/TBX6, controls Notch signaling by regulating Dll1 expression in the presomitic mesoderm of mouse embryos. Genes Dev. 2004;18:2712–2717. doi: 10.1101/gad.1248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SJ, GAle NW, Mbamalu G, Yancopoulos G, Henkemeyer M, Pawson T. Bidirectional signaling through the EPH family receptor Nuk and its transmembrane ligands. Nature. 1996;383:722–725. doi: 10.1038/383722a0. [DOI] [PubMed] [Google Scholar]
- Holterman CE, Le Grand F, Kuang S, Seale P, Rudnicki MA. Megf10 regulates the progression of the satellite cell myogenic program. J Cell Biol. 2007;179:911–922. doi: 10.1083/jcb.200709083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Epsin: inducing membrane curvature. Int J Biochem Cell Biol. 2007;39:1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Hsueh YP, Wang TF, Yang FC, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, Takeda Y, Chia W, Sankar N, Ng YK, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–175. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Hukriede NA, Gu Y, Fleming RJ. A dominant-negative form of Serrate acts as a general antagonist of Notch activation. Development. 1997;124:3427–3437. doi: 10.1242/dev.124.17.3427. [DOI] [PubMed] [Google Scholar]
- Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr Opin Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, Sisodia SS. The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent “gamma-secretase” cleavage. J Biol Chem. 2003;278:7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Jacobsen TL, Brennan K, Arias AM, Muskavitch MA. Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development. 1998;125:4531–4540. doi: 10.1242/dev.125.22.4531. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Jelen F, Oleksy A, Smietana K, Otlewski J. PDZ domains - common players in the cell signaling. Acta Biochim Pol. 2003;50:985–1017. [PubMed] [Google Scholar]
- Jin EJ, Choi YA, Sonn JK, Kang SS. Suppression of ADAM 10-induced Delta-1 shedding inhibits cell proliferation during the chondro-inhibitory action of TGF-beta3. Mol Cells. 2007;24:139–147. [PubMed] [Google Scholar]
- Karanu FN, Murdoch B, Gallacher L, Wu DM, Koremoto M, Sakano S, Bhatia M. The notch ligand jagged-1 represents a novel growth factor of human hematopoietic stem cells. J Exp Med. 2000;192:1365–1372. doi: 10.1084/jem.192.9.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karanu FN, Murdoch B, Miyabayashi T, Ohno M, Koremoto M, Gallacher L, Wu D, Itoh A, Sakano S, Bhatia M. Human homologues of Delta-1 and Delta-4 function as mitogenic regulators of primitive human hematopoietic cells. Blood. 2001;97:1960–1967. doi: 10.1182/blood.v97.7.1960. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Ahituv N, Fuchs H, Balling R, Avraham KB, Steel KP, Hrabe de Angelis M. The Notch ligand Jagged1 is required for inner ear sensory development. Proc Natl Acad Sci U S A. 2001;98:3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T, Arias AM. Interactions among Delta, Serrate and Fringe modulate Notch activity during Drosophila wing development. Development. 1998;125:2951–2962. doi: 10.1242/dev.125.15.2951. [DOI] [PubMed] [Google Scholar]
- Klein T, Brennan K, Arias AM. An intrinsic dominant negative activity of serrate that is modulated during wing development in Drosophila. Dev Biol. 1997;189:123–134. doi: 10.1006/dbio.1997.8564. [DOI] [PubMed] [Google Scholar]
- Klueg KM, Parody TR, Muskavitch MA. Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol Biol Cell. 1998;9:1709–1723. doi: 10.1091/mbc.9.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluppel M, Wrana JL. Turning it up a Notch: cross-talk between TGF beta and Notch signaling. Bioessays. 2005;27:115–118. doi: 10.1002/bies.20187. [DOI] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell Mol Life Sci. 2007;64:2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev V, Kacer D, Trifonova R, Small D, Duarte M, Soldi R, Graziani I, Sideleva O, Larman B, Maciag T, et al. The intracellular domain of Notch ligand Delta1 induces cell growth arrest. FEBS Lett. 2005;579:5798–5802. doi: 10.1016/j.febslet.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Komatsu H, Chao MY, Larkins-Ford J, Corkins ME, Somers GA, Tucey T, Dionne HM, White JQ, Wani K, Boxem M, et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008;6:e196. doi: 10.1371/journal.pbio.0060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005a;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon KJ, Yoo KW, Lim HS, Song R, So JH, Kim CH, Kong YY. Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem. 2005b;280:22335–22342. doi: 10.1074/jbc.M501631200. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon MJ, Yoon KJ, Im SK, Kim YY, Kim CH, Suh PG, Jan YN, Kong YY. An obligatory role of mind bomb-1 in notch signaling of Mammalian development. PLoS ONE. 2007;2:e1221. doi: 10.1371/journal.pone.0001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooh PJ, Fehon RG, Muskavitch MA. Implications of dynamic patterns of Delta and Notch expression for cellular interactions during Drosophila development. Development. 1993;117:493–507. doi: 10.1242/dev.117.2.493. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski CC, Muskavitch MA. Complex spatio-temporal accumulation of alternative transcripts from the neurogenic gene Delta during Drosophila embryogenesis. Development. 1989;107:623–636. doi: 10.1242/dev.107.3.623. [DOI] [PubMed] [Google Scholar]
- Koutelou E, Sato S, Tomomori-Sato C, Florens L, Swanson SK, Washburn MP, Kokkinaki M, Conaway RC, Conaway JW, Moschonas NK. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Rozov FN, Zinovyeva MV, Hendrikx PJ, Jiang Y, Visser JW, Belyavsky AV. Jedi--a novel transmembrane protein expressed in early hematopoietic cells. J Cell Biochem. 2007;101:767–784. doi: 10.1002/jcb.21232. [DOI] [PubMed] [Google Scholar]
- Laborda J, Sausville EA, Hoffman T, Notario V. dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Deblandre GA, Kintner C, Rubin GM. Drosophila neuralized is a ubiquitin ligase that promotes the internalization and degradation of delta. Dev Cell. 2001;1:783–794. doi: 10.1016/s1534-5807(01)00092-2. [DOI] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Lai EC, Rubin GM. neuralized functions cell-autonomously to regulate a subset of notch-dependent processes during adult Drosophila development. Dev Biol. 2001a;231:217–233. doi: 10.1006/dbio.2000.0124. [DOI] [PubMed] [Google Scholar]
- Lai EC, Rubin GM. Neuralized is essential for a subset of Notch pathway-dependent cell fate decisions during Drosophila eye development. Proc Natl Acad Sci U S A. 2001b;98:5637–5642. doi: 10.1073/pnas.101135498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- Le Borgne R. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr Opin Cell Biol. 2006;18:213–222. doi: 10.1016/j.ceb.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Notch signaling: endocytosis makes delta signal better. Curr Biol. 2003a;13:R273–275. doi: 10.1016/s0960-9822(03)00199-4. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev Cell. 2003b;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Lee JH, Volinic JL, Banz C, Yao KM, Thomas MK. Interactions with p300 enhance transcriptional activation by the PDZ-domain coactivator Bridge-1. J Endocrinol. 2005;187:283–292. doi: 10.1677/joe.1.06305. [DOI] [PubMed] [Google Scholar]
- Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- Li BX, Satoh AK, Ready DF. Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J Cell Biol. 2007a;177:659–669. doi: 10.1083/jcb.200610157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JL, Sainson RC, Shi W, Leek R, Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E, Hainfellner JA, et al. Delta-like 4 Notch ligand regulates tumor angiogenesis, improves tumor vascular function, and promotes tumor growth in vivo. Cancer Res. 2007b;67:11244–11253. doi: 10.1158/0008-5472.CAN-07-0969. [DOI] [PubMed] [Google Scholar]
- Lieber T, Kidd S, Young MW. kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbourg A, Ploom M, Elligsen D, Sorensen I, Ziegelhoeffer T, Gossler A, Drexler H, Limbourg FP. Notch ligand Delta-like 1 is essential for postnatal arteriogenesis. Circ Res. 2007;100:363–371. doi: 10.1161/01.RES.0000258174.77370.2c. [DOI] [PubMed] [Google Scholar]
- Liotta F, Angeli R, Cosmi L, Fili L, Manuelli C, Frosali F, Mazzinghi B, Maggi L, Pasini A, Lisi V, et al. Toll-like receptors 3 and 4 are expressed by human bone marrow-derived mesenchymal stem cells and can inhibit their T-cell modulatory activity by impairing Notch signaling. Stem Cells. 2008;26:279–289. doi: 10.1634/stemcells.2007-0454. [DOI] [PubMed] [Google Scholar]
- Liu ZJ, Shirakawa T, Li Y, Soma A, Oka M, Dotto GP, Fairman RM, Velazquez OC, Herlyn M. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobov IB, Renard RA, Papadopoulos N, Gale NW, Thurston G, Yancopoulos GD, Wiegand SJ. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci U S A. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- Lowell S, Watt FM. Delta regulates keratinocyte spreading and motility independently of differentiation. Mech Dev. 2001;107:133–140. doi: 10.1016/s0925-4773(01)00459-2. [DOI] [PubMed] [Google Scholar]
- Lu L, Chen X, Zhang CW, Yang WL, Wu YJ, Sun L, Bai LM, Gu XS, Ahmed S, Dawe GS, et al. Morphological and functional characterization of predifferentiation of myelinating glia-like cells from human bone marrow stromal cells through activation of F3/Notch signaling in mouse retina. Stem Cells. 2008;26:580–590. doi: 10.1634/stemcells.2007-0106. [DOI] [PubMed] [Google Scholar]
- Maekawa Y, Tsukumo S, Chiba S, Hirai H, Hayashi Y, Okada H, Kishihara K, Yasutomo K. Delta1-Notch3 interactions bias the functional differentiation of activated CD4+ T cells. Immunity. 2003;19:549–559. doi: 10.1016/s1074-7613(03)00270-x. [DOI] [PubMed] [Google Scholar]
- Maldonado-Baez L, Wendland B. Endocytic adaptors: recruiters, coordinators and regulators. Trends Cell Biol. 2006;16:505–513. doi: 10.1016/j.tcb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Chitnis AB. Interaction with Notch determines endocytosis of specific Delta ligands in zebrafish neural tissue. Development. 2009;136:197–206. doi: 10.1242/dev.027938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinn E, van Bueren KL, Fiorenza S, Mo R, Poh AM, Forrest A, Soares MB, Bonaldo Mde F, Grimmond S, Hui CC, et al. Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech Dev. 2005;122:1218–1233. doi: 10.1016/j.mod.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Meng H, Zhang X, Hankenson KD, Wang MM. Thrombospondin 2 potentiates notch3/jagged1 signaling. J Biol Chem. 2009;284:7866–7874. doi: 10.1074/jbc.M803650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: Notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol. 2009;19:1378–1383. doi: 10.1016/j.cub.2009.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamizato T, Sakamoto K, Liu T, Kokubo H, Katsube K, Perbal B, Nakamura S, Yamaguchi A. CCN3/NOV inhibits BMP-2-induced osteoblast differentiation by interacting with BMP and Notch signaling pathways. Biochem Biophys Res Commun. 2007;354:567–573. doi: 10.1016/j.bbrc.2007.01.029. [DOI] [PubMed] [Google Scholar]
- Mishra-Gorur K, Rand MD, Perez-Villamil B, Artavanis-Tsakonas S. Down-regulation of Delta by proteolytic processing. J Cell Biol. 2002;159:313–324. doi: 10.1083/jcb.200203117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Lau R, Hein PW, Shipley JM, Weinmaster G. Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation. J Biol Chem. 2006;281:10089–10097. doi: 10.1074/jbc.M600298200. [DOI] [PubMed] [Google Scholar]
- Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y, Takai Y. MAGI1 recruits Dll1 to cadherin-based adherens junctions and stabilizes it on the cell surface. J Biol Chem. 2005;280:26499–26507. doi: 10.1074/jbc.M500375200. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4:1848–1861. doi: 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- Mok LP, Qin T, Bardot B, LeComte M, Homayouni A, Ahimou F, Wesley C. Delta activity independent of its activity as a ligand of Notch. BMC Dev Biol. 2005;5:6. doi: 10.1186/1471-213X-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel V, Le Borgne R, Schweisguth F. Snail is required for Delta endocytosis and Notch-dependent activation of single-minded expression. Dev Genes Evol. 2003;213:65–72. doi: 10.1007/s00427-003-0296-x. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Perez SE, Qiao Z, Verdi JM, Hicks C, Weinmaster G, Anderson DJ. Transient Notch activation initiates an irreversible switch from neurogenesis to gliogenesis by neural crest stem cells. Cell. 2000;101:499–510. doi: 10.1016/s0092-8674(00)80860-0. [DOI] [PubMed] [Google Scholar]
- Morrissette JD, Colliton RP, Spinner NB. Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet. 2001;10:405–413. doi: 10.1093/hmg/10.4.405. [DOI] [PubMed] [Google Scholar]
- Mukherjee A, Veraksa A, Bauer A, Rosse C, Camonis J, Artavanis-Tsakonas S. Regulation of Notch signalling by non-visual beta-arrestin. Nat Cell Biol. 2005;7:1191–1201. doi: 10.1038/ncb1327. [DOI] [PubMed] [Google Scholar]
- Muraguchi T, Takegami Y, Ohtsuka T, Kitajima S, Chandana EP, Omura A, Miki T, Takahashi R, Matsumoto N, Ludwig A, et al. RECK modulates Notch signaling during cortical neurogenesis by regulating ADAM10 activity. Nat Neurosci. 2007;10:838–845. doi: 10.1038/nn1922. [DOI] [PubMed] [Google Scholar]
- Nakahara J, Kanekura K, Nawa M, Aiso S, Suzuki N. Abnormal expression of TIP30 and arrested nucleocytoplasmic transport within oligodendrocyte precursor cells in multiple sclerosis. J Clin Invest. 2009;119:169–181. doi: 10.1172/JCI35440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata A, Ito T, Nagata M, Hori S, Sekimizu K. GRIP1tau, a novel PDZ domain-containing transcriptional activator, cooperates with the testis-specific transcription elongation factor SII-T1. Genes Cells. 2004;9:1125–1135. doi: 10.1111/j.1365-2443.2004.00795.x. [DOI] [PubMed] [Google Scholar]
- Nanda N, Bao M, Lin H, Clauser K, Komuves L, Quertermous T, Conley PB, Phillips DR, Hart MJ. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J Biol Chem. 2005;280:24680–24689. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- Nehring LC, Miyamoto A, Hein PW, Weinmaster G, Shipley JM. The extracellular matrix protein MAGP-2 interacts with Jagged1 and induces its shedding from the cell surface. J Biol Chem. 2005;280:20349–20355. doi: 10.1074/jbc.M500273200. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Olsen SL, D’Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007a;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Weinmaster G. Notch signaling--constantly on the move. Traffic. 2007b;8:959–969. doi: 10.1111/j.1600-0854.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- Noguera-Troise I, Daly C, Papadopoulos NJ, Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD, Thurston G. Blockade of Dll4 inhibits tumour growth by promoting non-productive angiogenesis. Nature. 2006;444:1032–1037. doi: 10.1038/nature05355. [DOI] [PubMed] [Google Scholar]
- Nueda ML, Baladron V, Sanchez-Solana B, Ballesteros MA, Laborda J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J Mol Biol. 2007;367:1281–1293. doi: 10.1016/j.jmb.2006.10.043. [DOI] [PubMed] [Google Scholar]
- Ong CT, Sedy JR, Murphy KM, Kopan R. Notch and presenilin regulate cellular expansion and cytokine secretion but cannot instruct Th1/Th2 fate acquisition. PLoS One. 2008;3:e2823. doi: 10.1371/journal.pone.0002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet E, Chen X, Wendland B, Fischer JA. Either part of a Drosophila epsin protein, divided after the ENTH domain, functions in endocytosis of delta in the developing eye. Curr Biol. 2003;13:854–860. doi: 10.1016/s0960-9822(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Overstreet E, Fitch E, Fischer JA. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 2004;131:5355–5366. doi: 10.1242/dev.01434. [DOI] [PubMed] [Google Scholar]
- Panin VM, Papayannopoulos V, Wilson R, Irvine KD. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- Panin VM, Shao L, Lei L, Moloney DJ, Irvine KD, Haltiwanger RS. Notch ligands are substrates for EGF protein O-fucosyltransferase and Fringe. J Biol Chem. 2002;29:29. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- Pannequin J, Bonnans C, Delaunay N, Ryan J, Bourgaux JF, Joubert D, Hollande F. The wnt target jagged-1 mediates the activation of notch signaling by progastrin in human colorectal cancer cells. Cancer Res. 2009;69:6065–6073. doi: 10.1158/0008-5472.CAN-08-2409. [DOI] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Parks AL, Stout JR, Shepard SB, Klueg KM, Dos Santos AA, Parody TR, Vaskova M, Muskavitch MA. Structure-function analysis of delta trafficking, receptor binding and signaling in Drosophila. Genetics. 2006;174:1947–1961. doi: 10.1534/genetics.106.061630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NS, Li JL, Generali D, Poulsom R, Cranston DW, Harris AL. Up-regulation of delta-like 4 ligand in human tumor vasculature and the role of basal expression in endothelial cell function. Cancer Res. 2005;65:8690–8697. doi: 10.1158/0008-5472.CAN-05-1208. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Pitsouli C, Klueg KM, Muskavitch MA, Moschonas NK, Delidakis C. neuralized Encodes a peripheral membrane protein involved in delta signaling and endocytosis. Dev Cell. 2001;1:807–816. doi: 10.1016/s1534-5807(01)00093-4. [DOI] [PubMed] [Google Scholar]
- Perbal B. CCN proteins: multifunctional signalling regulators. Lancet. 2004;363:62–64. doi: 10.1016/S0140-6736(03)15172-0. [DOI] [PubMed] [Google Scholar]
- Pfister S, Przemeck GK, Gerber JK, Beckers J, Adamski J, Hrabe de Angelis M. Interaction of the MAGUK family member Acvrinp1 and the cytoplasmic domain of the Notch ligand Delta1. J Mol Biol. 2003;333:229–235. doi: 10.1016/j.jmb.2003.08.043. [DOI] [PubMed] [Google Scholar]
- Piccoli DA, Spinner NB. Alagille syndrome and the Jagged1 gene. Semin Liver Dis. 2001;21:525–534. doi: 10.1055/s-2001-19036. [DOI] [PubMed] [Google Scholar]
- Pintar A, De Biasio A, Popovic M, Ivanova N, Pongor S. The intracellular region of Notch ligands: does the tail make the difference? Biol Direct. 2007;2:19. doi: 10.1186/1745-6150-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitsouli C, Delidakis C. The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development. 2005;132:4041–4050. doi: 10.1242/dev.01979. [DOI] [PubMed] [Google Scholar]
- Powell PA, Wesley C, Spencer S, Cagan RL. Scabrous complexes with Notch to mediate boundary formation. Nature. 2001;409:626–630. doi: 10.1038/35054566. [DOI] [PubMed] [Google Scholar]
- Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the notch ligand delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- Quilliam LA, Castro AF, Rogers-Graham KS, Martin CB, Der CJ, Bi C. M-Ras/R-Ras3, a transforming ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6. J Biol Chem. 1999;274:23850–23857. doi: 10.1074/jbc.274.34.23850. [DOI] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen T, Raffetseder U, Frye BC, Djudjaj S, Muhlenberg PJ, Eitner F, Lendahl U, Bernhagen J, Dooley S, Mertens PR. YB-1 acts as a ligand for Notch-3 receptors and modulates receptor activation. J Biol Chem. 2009;284:26928–26940. doi: 10.1074/jbc.M109.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauskolb C. The establishment of segmentation in the Drosophila leg. Development. 2001;128:4511–4521. doi: 10.1242/dev.128.22.4511. [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Irvine KD. Notch-mediated segmentation and growth control of the Drosophila leg. Dev Biol. 1999;210:339–350. doi: 10.1006/dbio.1999.9273. [DOI] [PubMed] [Google Scholar]
- Raymond T, Schaller M, Hogaboam CM, Lukacs NW, Rochford R, Kunkel SL. Toll-like receptors, Notch ligands, and cytokines drive the chronicity of lung inflammation. Proc Am Thorac Soc. 2007;4:635–641. doi: 10.1513/pats.200706-067TH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud O, Simpson P. scabrous modifies epithelial cell adhesion and extends the range of lateral signalling during development of the spaced bristle pattern in Drosophila. Dev Biol. 2001;240:361–376. doi: 10.1006/dbio.2001.0482. [DOI] [PubMed] [Google Scholar]
- Roca C, Adams RH. Regulation of vascular morphogenesis by Notch signaling. Genes Dev. 2007;21:2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- Rodilla V, Villanueva A, Obrador-Hevia A, Robert-Moreno A, Fernandez-Majada V, Grilli A, Lopez-Bigas N, Bellora N, Alba MM, Torres F, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A. 2009;106:6315–6320. doi: 10.1073/pnas.0813221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DA, Rao PK, Kadesch T. Dual roles for the Notch target gene Hes-1 in the differentiation of 3T3-L1 preadipocytes. Mol Cell Biol. 2004;24:3505–3513. doi: 10.1128/MCB.24.8.3505-3513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- Ruan Y, Tecott L, Jiang MM, Jan LY, Jan YN. Ethanol hypersensitivity and olfactory discrimination defect in mice lacking a homolog of Drosophila neuralized. Proc Natl Acad Sci U S A. 2001;98:9907–9912. doi: 10.1073/pnas.171321098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainson RC, Harris AL. Regulation of angiogenesis by homotypic and heterotypic notch signalling in endothelial cells and pericytes: from basic research to potential therapies. Angiogenesis. 2008;11:41–51. doi: 10.1007/s10456-008-9098-0. [DOI] [PubMed] [Google Scholar]
- Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, Davis J, Conn E, Hughes CC. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood. 2008;111:4997–5007. doi: 10.1182/blood-2007-08-108597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Ohara O, Takagi M, Takeda S, Katsube K. Intracellular cell-autonomous association of Notch and its ligands: a novel mechanism of Notch signal modification. Dev Biol. 2002a;241:313–326. doi: 10.1006/dbio.2001.0517. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Yamaguchi S, Ando R, Miyawaki A, Kabasawa Y, Takagi M, Li CL, Perbal B, Katsube K. The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J Biol Chem. 2002b;277:29399–29405. doi: 10.1074/jbc.M203727200. [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapir A, Assa-Kunik E, Tsruya R, Schejter E, Shilo BZ. Unidirectional Notch signaling depends on continuous cleavage of Delta. Development. 2005;132:123–132. doi: 10.1242/dev.01546. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Bicker F, Nikolic I, Meister J, Babuke T, Picuric S, Muller-Esterl W, Plate KH, Dikic I. Epidermal growth factor-like domain 7 (EGFL7) modulates Notch signalling and affects neural stem cell renewal. Nat Cell Biol. 2009;11:873–880. doi: 10.1038/ncb1896. [DOI] [PubMed] [Google Scholar]
- Schroeter E, Kisslinger J, Kopan R. Notch1 signalling requires ligand-induced proteolytic release of the intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Seo S, Fujita H, Nakano A, Kang M, Duarte A, Kume T. The forkhead transcription factors, Foxc1 and Foxc2, are required for arterial specification and lymphatic sprouting during vascular development. Dev Biol. 2006;294:458–470. doi: 10.1016/j.ydbio.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Simpson P, Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997a;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- Seugnet L, Simpson P, Haenlin M. Transcriptional regulation of Notch and Delta: requirement for neuroblast segregation in Drosophila. Development. 1997b;124:2015–2025. doi: 10.1242/dev.124.10.2015. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, Kanda Y, Hamada Y, Yazaki Y, Hirai H. Mouse Jagged1 Physically Interacts with Notch2 and Other Notch Receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Takahashi T, Kumano K, Hamada Y, Hirai H. Integrity of intracellular domain of Notch ligand is indispensable for cleavage required for release of the Notch2 intracellular domain. Embo J. 2002;21:294–302. doi: 10.1093/emboj/21.3.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji H, Tsuchida K, Kishi H, Yamakawa N, Matsuzaki T, Liu Z, Nakamura T, Sugino H. Identification and characterization of a PDZ protein that interacts with activin type II receptors. J Biol Chem. 2000;275:5485–5492. doi: 10.1074/jbc.275.8.5485. [DOI] [PubMed] [Google Scholar]
- Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, Transidico P, Di Fiore PP, Polo S. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and gamma-secretase. Proc Natl Acad Sci U S A. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Six EM, Ndiaye D, Sauer G, Laabi Y, Athman R, Cumano A, Brou C, Israel A, Logeat F. The notch ligand Delta1 recruits Dlg1 at cell-cell contacts and regulates cell migration. J Biol Chem. 2004;279:55818–55826. doi: 10.1074/jbc.M408022200. [DOI] [PubMed] [Google Scholar]
- Skwarek LC, Garroni MK, Commisso C, Boulianne GL. Neuralized contains a phosphoinositide-binding motif required downstream of ubiquitination for delta endocytosis and notch signaling. Dev Cell. 2007;13:783–795. doi: 10.1016/j.devcel.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Small D, Kovalenko D, Kacer D, Liaw L, Landriscina M, Di Serio C, Prudovsky I, Maciag T. Soluble Jagged 1 represses the function of its transmembrane form to induce the formation of the Src-dependent chord-like phenotype. J Biol Chem. 2001;276:32022–32030. doi: 10.1074/jbc.M100933200. [DOI] [PubMed] [Google Scholar]
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- Song R, Koo BK, Yoon KJ, Yoon MJ, Yoo KW, Kim HT, Oh HJ, Kim YY, Han JK, Kim CH, et al. Neuralized-2 regulates a Notch ligand in cooperation with Mind bomb-1. J Biol Chem. 2006;281:36391–36400. doi: 10.1074/jbc.M606601200. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz ME. Cis Interactions beween Notch and Delta generate mutally exclusive signaling states. Nature. 2010 doi: 10.1038/nature08959. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86:669–707. doi: 10.1152/physrev.00020.2005. [DOI] [PubMed] [Google Scholar]
- Studebaker AW, Storci G, Werbeck JL, Sansone P, Sasser AK, Tavolari S, Huang T, Chan MW, Marini FC, Rosol TJ, et al. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68:9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- Suchting S, Freitas C, le Noble F, Benedito R, Breant C, Duarte A, Eichmann A. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci U S A. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Li H, Zolkiewska A. The role of Delta-like 1 shedding in muscle cell self-renewal and differentiation. J Cell Sci. 2008a;121:3815–3823. doi: 10.1242/jcs.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Krawczyk CJ, Pearce EJ. Suppression of Th2 cell development by Notch ligands Delta1 and Delta4. J Immunol. 2008b;180:1655–1661. doi: 10.4049/jimmunol.180.3.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S. The intracellular deletions of Delta and Serrate define dominant negative forms of the Drosophila Notch ligands. Development. 1996;122:2465–2474. doi: 10.1242/dev.122.8.2465. [DOI] [PubMed] [Google Scholar]
- Sun X, Artavanis-Tsakonas S. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development. 1997;124:3439–3448. doi: 10.1242/dev.124.17.3439. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Inoue T, Gossler A, Saga Y. Feedback loops comprising Dll1, Dll3 and Mesp2, and differential involvement of Psen1 are essential for rostrocaudal patterning of somites. Development. 2003;130:4259–4268. doi: 10.1242/dev.00629. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Adachi Y, Ohtsuki Y. Skeletrophin, a novel ubiquitin ligase to the intracellular region of Jagged-2, is aberrantly expressed in multiple myeloma. Am J Pathol. 2005;166:1817–1826. doi: 10.1016/S0002-9440(10)62491-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tax FE, Yeargers JJ, Thomas JH. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature. 1994;368:150–154. doi: 10.1038/368150a0. [DOI] [PubMed] [Google Scholar]
- Thibout H, Martinerie C, Creminon C, Godeau F, Boudou P, Le Bouc Y, Laurent M. Characterization of human NOV in biological fluids: an enzyme immunoassay for the quantification of human NOV in sera from patients with diseases of the adrenal gland and of the nervous system. J Clin Endocrinol Metab. 2003;88:327–336. doi: 10.1210/jc.2002-020304. [DOI] [PubMed] [Google Scholar]
- Thurston G, Noguera-Troise I, Yancopoulos GD. The Delta paradox: DLL4 blockade leads to more tumour vessels but less tumour growth. Nat Rev Cancer. 2007;7:327–331. doi: 10.1038/nrc2130. [DOI] [PubMed] [Google Scholar]
- Tian X, Hansen D, Schedl T, Skeath JB. Epsin potentiates Notch pathway activity in Drosophila and C. elegans. Development. 2004;131:5807–5815. doi: 10.1242/dev.01459. [DOI] [PubMed] [Google Scholar]
- Trifonova R, Small D, Kacer D, Kovalenko D, Kolev V, Mandinova A, Soldi R, Liaw L, Prudovsky I, Maciag T. The non-transmembrane form of Delta1, but not of Jagged1, induces normal migratory behavior accompanied by fibroblast growth factor receptor 1-dependent transformation. J Biol Chem. 2004;279:13285–13288. doi: 10.1074/jbc.C300564200. [DOI] [PubMed] [Google Scholar]
- Tsai H, Hardisty RE, Rhodes C, Kiernan AE, Roby P, Tymowska-Lalanne Z, Mburu P, Rastan S, Hunter AJ, Brown SD, et al. The mouse slalom mutant demonstrates a role for Jagged1 in neuroepithelial patterning in the organ of Corti. Hum Mol Genet. 2001;10:507–512. doi: 10.1093/hmg/10.5.507. [DOI] [PubMed] [Google Scholar]
- Tsuda L, Nagaraj R, Zipursky SL, Banerjee U. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell. 2002;110:625–637. doi: 10.1016/s0092-8674(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Turnpenny PD, Alman B, Cornier AS, Giampietro PF, Offiah A, Tassy O, Pourquie O, Kusumi K, Dunwoodie S. Abnormal vertebral segmentation and the notch signaling pathway in man. Dev Dyn. 2007;236:1456–1474. doi: 10.1002/dvdy.21182. [DOI] [PubMed] [Google Scholar]
- Vanden Broeck D, De Wolf MJ. Selective blocking of clathrin-mediated endocytosis by RNA interference: epsin as target protein. Biotechniques. 2006;41:475–484. doi: 10.2144/000112265. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney B, Wu L, Yu M, Brashem-Stein C, Staats S, Flowers D, Griffin JD, Bernstein ID. Immobilization of Notch ligand, Delta-1, is required for induction of notch signaling. J Cell Sci. 2000;113(Pt 23):4313–4318. doi: 10.1242/jcs.113.23.4313. [DOI] [PubMed] [Google Scholar]
- Vas V, Szilagyi L, Paloczi K, Uher F. Soluble Jagged-1 is able to inhibit the function of its multivalent form to induce hematopoietic stem cell self-renewal in a surrogate in vitro assay. J Leukoc Biol. 2004;75:714–720. doi: 10.1189/jlb.1003462. [DOI] [PubMed] [Google Scholar]
- Vitt UA, Hsu SY, Hsueh AJ. Evolution and classification of cystine knot-containing hormones and related extracellular signaling molecules. Mol Endocrinol. 2001;15:681–694. doi: 10.1210/mend.15.5.0639. [DOI] [PubMed] [Google Scholar]
- Vollrath B, Pudney J, Asa S, Leder P, Fitzgerald K. Isolation of a murine homologue of the Drosophila neuralized gene, a gene required for axonemal integrity in spermatozoa and terminal maturation of the mammary gland. Mol Cell Biol. 2001;21:7481–7494. doi: 10.1128/MCB.21.21.7481-7494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Struhl G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development. 2004;131:5367–5380. doi: 10.1242/dev.01413. [DOI] [PubMed] [Google Scholar]
- Wang W, Struhl G. Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development. 2005;132:2883–2894. doi: 10.1242/dev.01860. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim KA, Kim JH, Sul HS. Pref-1, a preadipocyte secreted factor that inhibits adipogenesis. J Nutr. 2006;136:2953–2956. doi: 10.1093/jn/136.12.2953. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sul HS. Ectodomain shedding of preadipocyte factor 1 (Pref-1) by tumor necrosis factor alpha converting enzyme (TACE) and inhibition of adipocyte differentiation. Mol Cell Biol. 2006;26:5421–5435. doi: 10.1128/MCB.02437-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthen DM, Moore EC, Kamath BM, Morrissette JJ, Sanchez P, Piccoli DA, Krantz ID, Spinner NB. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27:436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- Wesley CS. Notch and wingless regulate expression of cuticle patterning genes. Mol Cell Biol. 1999;19:5743–5758. doi: 10.1128/mcb.19.8.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CK, Li JL, Murga M, Harris AL, Tosato G. Up-regulation of the Notch ligand Delta-like 4 inhibits VEGF-induced endothelial cell function. Blood. 2006;107:931–939. doi: 10.1182/blood-2005-03-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windler SL, Bilder D. Endocytic Internalization Routes Required for Delta/Notch Signaling. Curr Biol. 2010 doi: 10.1016/j.cub.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GJ, Leslie JD, Ariza-McNaughton L, Lewis J. Delta proteins and MAGI proteins: an interaction of Notch ligands with intracellular scaffolding molecules and its significance for zebrafish development. Development. 2004;131:5659–5669. doi: 10.1242/dev.01417. [DOI] [PubMed] [Google Scholar]
- Yan M, Plowman GD. Delta-like 4/Notch signaling and its therapeutic implications. Clin Cancer Res. 2007;13:7243–7246. doi: 10.1158/1078-0432.CCR-07-1393. [DOI] [PubMed] [Google Scholar]
- Yang LT, Nichols JT, Yao C, Manilay JO, Robey EA, Weinmaster G. Fringe glycosyltransferases differentially modulate Notch1 proteolysis induced by Delta1 and Jagged1. Mol Biol Cell. 2005;16:927–942. doi: 10.1091/mbc.E04-07-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Dermer M, Commisso C, Zhou L, McGlade CJ, Boulianne GL. Neuralized functions as an E3 ubiquitin ligase during Drosophila development. Curr Biol. 2001;11:1675–1679. doi: 10.1016/s0960-9822(01)00527-9. [DOI] [PubMed] [Google Scholar]
- Yeh E, Zhou L, Rudzik N, Boulianne GL. Neuralized functions cell autonomously to regulate Drosophila sense organ development. Embo J. 2000;19:4827–4837. doi: 10.1093/emboj/19.17.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim YI, Sun T, Wu LG, Raimondi A, De Camilli P, Eisenberg E, Greene LE. Endocytosis and clathrin-uncoating defects at synapses of auxilin knockout mice. Proc Natl Acad Sci U S A. 2010;107:4412–4417. doi: 10.1073/pnas.1000738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. Embo J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Li Q, Jiang YJ. Zebrafish Mib and Mib2 are mutual E3 ubiquitin ligases with common and specific delta substrates. J Mol Biol. 2007a;366:1115–1128. doi: 10.1016/j.jmb.2006.11.096. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li Q, Lim CH, Qiu X, Jiang YJ. The characterization of zebrafish antimorphic mib alleles reveals that Mib and Mind bomb-2 (Mib2) function redundantly. Dev Biol. 2007b;305:14–27. doi: 10.1016/j.ydbio.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Zolkiewska A. ADAM proteases: ligand processing and modulation of the Notch pathway. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-7586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]