Abstract
Advances in genome sequencing technology have fostered a new era of clinical genomic medicine. Genetic counselors, who have begun to support patients undergoing multi-gene panel testing for hereditary cancer risk, will review brief clinical vignettes, and discuss early experiences with clinical genomic testing. Their experiences will frame a discussion about how current testing may challenge patient understanding and expectations toward the evaluation of cancer risk and downstream preventive behaviors.
I. Introduction
Fast forward to genomic medicine
The advent of next-generation sequencing (NGS) and the ability to sequence millions of DNA base-pairs rapidly and at low-cost has ushered in a new era of genomic medicine that has only just begun to have an impact in medicine. Within only a decade of completion of the Human Genome Project, NGS technologies now permit sequencing of the entire human genome in a matter of days and for only a few thousand dollars. While the early impact of clinical genomics may be most evident in medical sub-specialties such as oncology, the public health implications of genomic medicine are beginning to be recognized [1, 2]. Paralleling the emergence of genomic testing into the clinic, studies in recent years have suggested marked growth in the availability and use of genetic and genomic tests [3–5]. Indeed, expanded indications for genomic testing are fueling innovation and greater demand for cost- and time-efficient approaches to delivery of genomic services [6–11]. However, important unresolved issues related to genomic testing, including standards for informed consent, management of ancillary findings (i.e, off-target or unsolicited), and appropriate formats for results reporting continue to be debated [12–16]. Coupled with this are concerns for the public related to the pace of uptake of complex genetic and genomic medicine. Knowledge and communication gaps among healthcare providers have been highlighted as major weaknesses in an era of increasingly complex pharmacogenomic medicine [17–20]. Indeed, the most recent report from the Secretary’s Advisory Committee on Genetics, Health, and Society (SACGHS) entitled, “The US System of Oversight of Genetic Testing” identified five significant gaps in oversight of genetic testing, including, “Education and training of health professionals to improve the application of genetic testing and the interpretation of genetic test results” and “The level of consumer understanding of genetics and genetic tests”[21].
Clinical cancer genomics
Oncology has emerged as a leader in the diverse application of genomics to the clinical care of the cancer patient. A multitude of genomic tests are now immediately available to cancer care providers (Table 1). Oncologists were among the first clinicians to incorporate tumor pharmacogenomics into the routine management of cancer, including agents targeted to somatic mutations (e.g. the KRAS gene in colorectal cancer; the BRAF gene in melanoma), or therapy guided by multi-gene predictive panels (e.g. OncoTypeDx™ for node negative breast cancer) [22–25]. More recently, NGS has fueled rapid growth in tumor pharmacogenomics, from single- and oligo-gene pharmacogenomic testing to multi-gene somatic panels like Foundation One™. In the evaluation of hereditary cancer risk, multi-gene risk panels have also begun to supplant the standard approach of syndrome-targeted genetic testing of the past (e.g. BRCA1/2 testing for hereditary-breast ovarian cancer). Some newer commercial hereditary panels are designed to test multiple genes associated with cancer in a particular organ system (e.g. Ambry’s Renal Next™ panel), while others have gone the route of including genes associated with a diversity of cancer risks or syndromes in a single panel test, casting a wide net to identify a genetic cause of cancer (e.g. University of Washington’s 49-gene BROCA™ panel). Non-targeted genomic testing, however, adds the additional clinical burden for oncologists and genetic counselors of interpreting mutations and/or variants of uncertain significance (VUS) in genes with an unclear relationship to the patient’s clinical presentation, and attempting to incorporate these results into preventive recommendations [10, 26].
Table 1.
Example clinical genomic testing for cancer patients
| Test type | Gene target(s) tested |
Format | Example(s) |
|---|---|---|---|
|
PHARMACOGENOMIC TESTING |
|||
| SELECT TUMOR | Single gene test | BRAF testing to predict response to vemurafenib therapy for colorectal cancer |
|
| Panel gene test | |||
| Predictive & prognostic test | OncotypeDx test to estimate efficacy of adjuvant chemotherapy for ER/PR positive breast cancer |
||
| Prognostic test only | Mammaprint test to estimate breast cancer prognosis | ||
| Multi-gene test | Foundation One test to identify actionable somatic targets to guide anti-tumor therapies |
||
| LARGE-SCALE SEQUENCING | Whole exome sequencing | Multiple labs | |
| Whole genome sequencing | Multiple labs | ||
| SELECT GERM-LINE | Single-gene/locus test | CYP2D6 test to guide Tamoxifen dosing | |
| Multi-gene test | Myriad On Dose test to guide 5-FU metabolism | ||
|
HEREDITARY CANCER RISK TESTING |
|||
| SELECT GERM-LINE | Single gene test | Many examples; PTEN, VHL, BRCA1 | |
| Panel gene test | |||
| Tumor/syndrome specific | Ambry Renal Next panel, Colorectal Cancer panel | ||
| Tumor/syndrome non-specific | Myriad My Risk; Ambry Cancer Next | ||
| LARGE-SCALE SEQUENCING | Whole exome sequencing | Multiple labs | |
| Whole genome sequencing | Multiple labs | ||
II. Early experiences with counseling for clinical genomic panels
With limited guidance from professional organizations, genetic counselors practicing in high-risk cancer clinics and performing clinical cancer risk assessment have very recently been tasked with incorporating novel genomic tests into patient care. While the frequency of use of gene panels has grown substantially in the high-risk clinic, whole genome and whole exome sequencing remain primarily research modalities. In the following vignettes, we present three clinical scenarios which bring to light several of the most important complexities currently being encountered in the delivery of genomic medicine in the high-risk cancer clinic. In Table 2, an abbreviated summary of the important counseling and communication elements for with each case is provided. Family pedigrees accompanying each vignette may be seen in Figure 1.
Table 2.
Summary of vignettes
| Vignette subject | Major theme | Communication challenges |
|---|---|---|
| 1. An African American patient with several variants in multiple genes |
Risk of VUS with panel tests; risk of VUS in non-Caucasian populations; risk of VUS misinterpretation; risk of ancillary findings in genes unrelated to the family history |
-Explaining VUS vs disease causing mutations -Race/ethnicity differences in VUS rates -Management of ancillary VUS in clinically relevant and non-relevant genes |
| 2. A patient with cancer and a single (heterozygous) mutation in the NBN gene |
Communicating risks of cancer in the carrier state for recessive gene mutations; testing of family members for recessive genes |
-Impact of heterozygous mutations on hereditary cancer risk and prevention -Approach to the family for testing for heterozygous mutations |
| 3. Genetic testing to guide treatment decisions for breast cancer discovers a mutation in the RAD51D gene |
Interpreting cancer risks for gene mutations unrelated to family history; interpreting cancer risks for clinically less studied genes |
-Interpreting genetic mutations in the context of clinical history -Management of mutations in genes with limited clinical information |
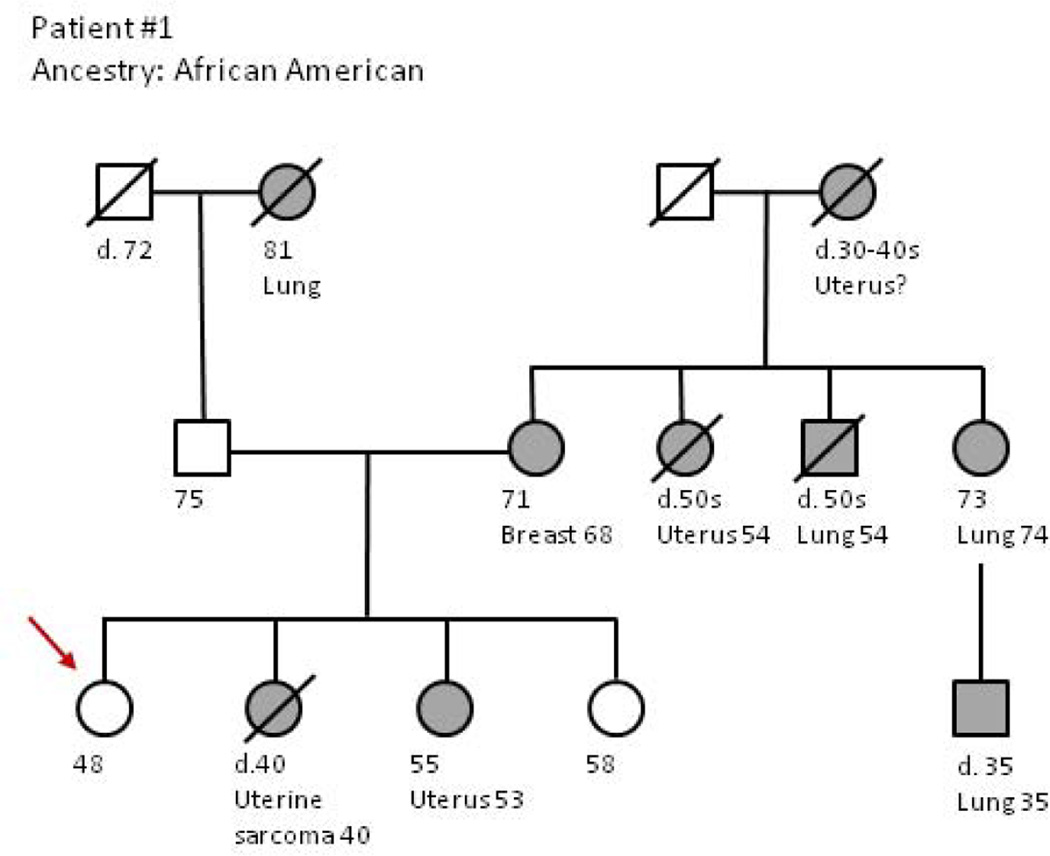
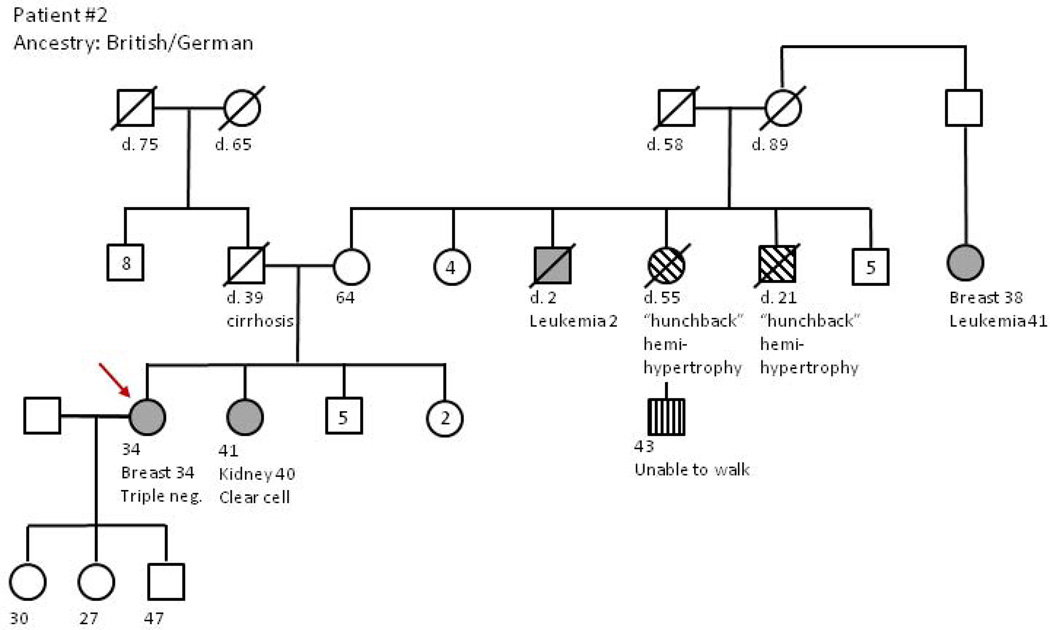
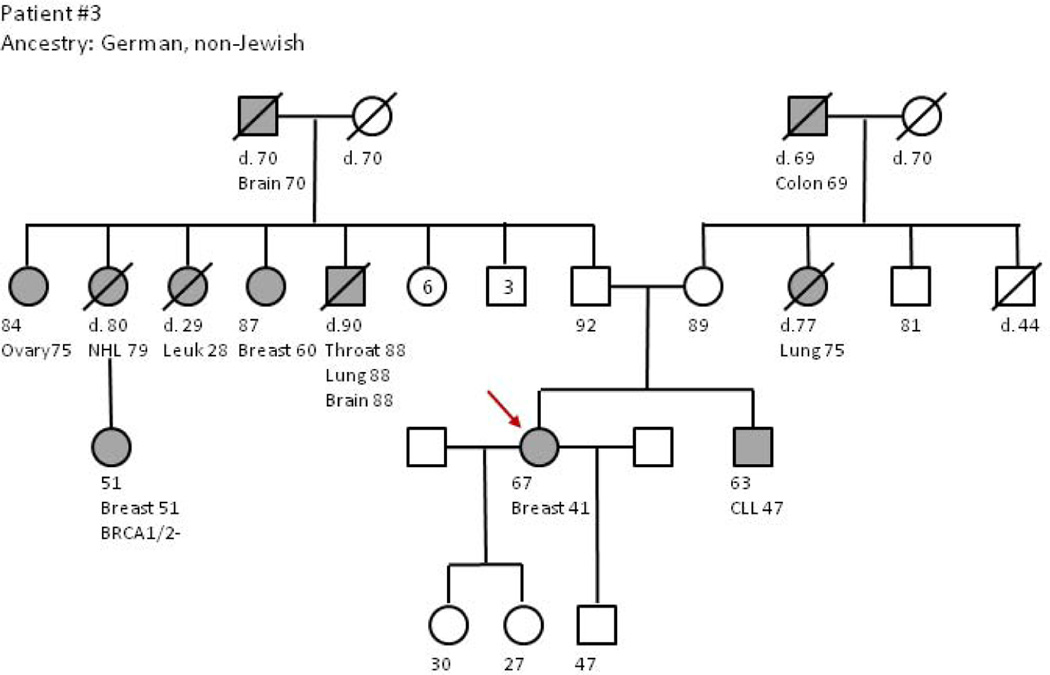
Figure 1.
Gene panel testing and the risk and interpretation of variants of uncertain significance (VUS) Andrea Forman, MS, LCGC
Patient #1 is a 45 year old woman of African American ancestry who was referred for risk assessment due to a strong family history of breast and uterine cancer. Cancer diagnoses in the family history included a sister who died (age 40) from uterine cancer, suspected to be a sarcoma; a second sister (age 53) was actively undergoing treatment for advanced uterine cancer; a maternal aunt died from uterine cancer (age 54), the patient’s maternal grandmother died (age 30–40) from possible uterine cancer; the patient’s mother carried a diagnosis of breast cancer (age 68); three additional maternal relatives had lung cancer, including a maternal aunt, (age 74), a maternal uncle (age 54) and a maternal 1st cousin (age 35). Hereditary cancer risk syndromes considered during risk assessment based on the family history included Lynch syndrome due to the uterine cancer history, Cowden syndrome due to the uterine and breast cancer history, and Li-Fraumeni syndrome due to the reported uterine sarcoma. Of note, the proband’s sister and mother had previously declined testing for Lynch syndrome or other cancer risks syndromes on multiple occasions.
As part of the cancer risk consultation, a multi-gene panel was offered to the proband due the varied and overlapping syndrome risks. The result identified no deleterious mutations in any of the 25 genes tested as part of a non-targeted multi-gene panel. However, 6 variants of uncertain significance (VUS) were found within four different genes tested: BRIP1, NBN, PALB2, and SMAD4. Three of these four genes have been associated with familial breast cancer, while the fourth, SMAD4, is associated with a rare cancer risk syndrome (juvenile polyposis syndrome) primarily involving the gastrointestinal tract that appeared unrelated to the patient’s history. Despite 6 findings in 4 genes, these VUS are not helpful in quantifying the genetic risk in this family.
Studies have shown that patients can have difficulty understanding genetic results including the difference between a deleterious and an uncertain genetic result, and are at risk of making inappropriate prevention decisions due to misunderstanding [27]. Some patient’s perceive their cancer risk to be elevated based on these results, leading to confusion over utilization of risk reducing strategies or screening [28]. Providers have expressed concern over increased fear and uncertainty for patients with uncertain results [29]. Reclassification of uncertain variants is complex [30] and may take years. The burden to re-contact patients with updates is a challenge and a responsibility of both the patient and the providers [31, 32]. Here, the gene panel results could not be used in formulating medical management recommendations for the family or helping unaffected family members understand their cancer risks, because most VUS will ultimately be reclassified to benign polymorphisms [33]. Rather, recommendations were tailored to the patient’s personal and family history [34]. Nonetheless, explaining the results to the patient was extremely challenging, primarily due to the large number of VUS discovered and the fact that they were found within 3 genes linked with breast cancer about which clinical testing experience is more limited, and one gene seemingly unrelated to the family history. Notably, higher than average rates of VUS have been recognized in African American populations undergoing genetic testing due to less testing in this population [33, 35, 36]. Moreover, in data examining the first 87 clinical test results from a hereditary risk multi-gene panel at our center, 26.5% of Caucasians (18/68) compared to 57.9% (11/19) of minority patients (African American, Latin American/Caribbean, Asian, or Filipino) were found to carry at least one VUS [37]. Thus panel tests may present a greater clinical challenge to counselors and patients from racial/ethnic groups where overall testing experience is lower.
Interpretation and impact of a deleterious mutation in a recessively inherited cancer risk gene Sue Montgomery, BSN, OCN, GCN
Patient #2 is a 34 year old woman diagnosed with triple negative (ER-/PR-, Her2-) invasive ductal carcinoma of the breast. Given the association of BRCA1/2 mutations with early-onset, triple negative breast cancer, she was referred for genetic risk assessment. The patient’s family history was notable for a sister with a clear cell kidney cancer (age 40), a maternal uncle who died of leukemia (age 2), and a maternal second cousin with breast cancer (age 38) followed by leukemia (age 41). Interestingly, a maternal aunt, uncle, and cousin were all reported to have short stature and scoliosis—the cousin is 43 and is unable to care for himself, though further details of his limitations were not known by the patient. The paternal family history was negative for cancer and the patient’s father was deceased. Because of the diversity of cancers in the family, a 25 gene multi-gene panel was selected for testing. The patient was found to be heterozygous for a deleterious mutation in the NBN gene. Individuals homozygous for NBN mutations have Nijmegen breakage syndrome (NBS), a rare autosomal recessive syndrome characterized by microcephaly at birth, growth retardation, mild-to-moderate intellectual disability and predisposition to malignancies. Some studies have found an increased risk of cancer in heterozygous NBN mutation carriers, including a 2–3 fold increased risk of breast cancer [38, 39] Other studies have found variable penetrance of this mutation in high risk breast cancer families [40].
The patient and her sister (with kidney cancer) came together for the discussion of the genetic test results. We began our discussion by disclosing the uncertainty of whether the heterozygous NBN mutation was causally related to the patient’s breast cancer. A careful literature review had not revealed an increased risk for kidney cancer in NBN heterozygotes, so we were unable to correlate that diagnosis with the test result. The patient has 8 siblings total who could potentially benefit from the discovery of an inherited cancer predisposition gene, but our lack of consistent evidence regarding cancer risk and this NBN mutation suggests that additional testing in the family for this gene mutation would not be beneficial for quantifying cancer risks in unaffected family members. The patient decided to proceed with bilateral mastectomy as her definitive surgical treatment following neoadjuvant chemotherapy, independent of her genetic test results. Given her kidney cancer diagnosis, the patient’s sister requested site-specific testing for the NBN mutation, with awareness of the caveat that current clinical recommendations based on a positive result would be very limited.
A second challenge to our discussion with the patient and her sister was the possibility that Nijmegen breakage syndrome (NBS) had affected some members of the family, suggesting that a second NBN mutation may also be present in this family. Three individuals had physical anomalies which could be associated with NBS. We recommended genetic testing for the all the patient’s siblings and their spouses to be certain none were NBN mutation carriers. If both parents were found to have an NBN mutation, they would have a 25% chance of having a child with NBS. The commercial genetics lab has initially offered site-specific testing for the NBN mutation in the patient’s mother, in an attempt to ascertain whether this mutation has been inherited through the maternal lineage.
Interpreting mutations in genes unrelated to the familial cancer risks Kim Rainey, MS, M.Ed, LCGC
Patient #3 is a 66-year-old woman with a personal history of ER- breast cancer at age 41 managed with mastectomy and chemotherapy. Recently, she was diagnosed with ER+/PR+ ductal carcinoma-in-situ (DCIS) of the left breast. Her new diagnosis provided incentive for genetic evaluation, primarily to inform personal surgical decisions and to help better quantify breast cancer risk for her daughters. The patient’s family history includes: a brother with chronic lymphocytic leukemia (diagnosed age 50) and melanoma; a maternal aunt (smoker) with lung cancer (age 78); a maternal grandfather with colon cancer (age 70); a father (non-smoker) with lung cancer (age 80); a paternal aunt with leukemia (age 30); a paternal uncle with multiple cancers in his 90s; a paternal aunt with bilateral breast cancer; a paternal aunt with ovarian cancer (age 82), and a paternal grandfather with brain cancer (age 71); a paternal cousin with breast cancer in her 50s was reportedly BRCA1/2 negative. The patient chose to pursue gene panel testing in an effort to have a more comprehensive assessment of her cancer risk, and a deleterious mutation was detected in the RAD51D gene. RAD51D mutations are only known to affect ovarian cancer risk, and the relative risk of ovarian cancer for RAD51D mutation carriers is estimated to be 6.30 (95% CI 2.86–13.85, P= 4.8 × 10−6). [41] When we reviewed the test results with the patient, we discussed that there are no known additional cancer risks associated with a RAD51D gene mutation other than ovarian cancer, and that this gene mutation is likely, but not certain to be, unrelated to the patient’s strong breast cancer history. Further, we attempted to explain that RAD51D mutations have been seen in families with both breast and ovarian cancers, however the studies did not find a statistically significantly increased breast cancer risk. Finally, we recommended that testing the daughters for the RAD51D mutation would not impact our assessment of their breast cancer risk or screening. After this discussion, the patient was inclined to have her ovaries and fallopian tubes (BSO) removed for risk reduction, and she further elected to have a left segmental mastectomy and radiation therapy rather than mastectomy.
The greatest difficulty with communicating this test result involved formulating clinical recommendations for the patient’s daughters (both in their late 20s without children). If they are found to have the RAD51D mutation, their risk for ovarian cancer is increased, but not currently thought to be at the same high level of risk as a BRCA1/2 mutation. Would a lifetime ovarian cancer risk of 7–10% be sufficient to warrant prophylactic bilateral salpingo-oophorectomy (BSO)? If so, at what age should this be done? The daughters would still be considered at increased risk for breast cancer based on the patient’s early onset and bilateral breast cancer diagnoses, regardless of their RAD51D carrier status. Thus, formulating evidence-based medical management recommendations may be an enormous challenge for unaffected relatives who are found to carry mutations in less clinically studied genes like RAD51D.
III. Health care providers: translators of genomic findings into clinical decisions
Providers’ positive attitudes toward genomic medicine
A growing body of research has examined the role of primary and specialty health care providers in the delivery of genomic medicine. Within this literature are studies examining providers’ views on the advantages and disadvantages to genomic testing in the delivery of medical care, and their perceived preparedness to effectively integrate genomic medicine into their practice. Among the new challenges presented by genomic medicine includes understanding and conveying the value of various genomic tests to patients, the ability to assess, interpret, and make clinical decisions using the complex data that may be generated by genomic tests, management of ancillary information that may be produced by a test, and how to communicate the range of potential risks of genomic testing to the patient in a clear and timely manner [10, 12, 16, 26, 42–47]. Today’s providers may also be faced with interpreting direct-to-consumer tests that patients access via the Internet.
Despite the complexity of genomic medicine, providers have expressed generally positive attitudes toward genetic testing, yet demonstrate modest experience with and knowledge of clinical genetics and genomics. Providers recognize personal limitations in delivering genomic medicine [48–54], and may be further disadvantaged in this area by numeracy and health literacy deficiencies among patients that create barriers to communication of genetic information [46]. In one study, only 15% of family medicine and internal medicine providers felt prepared to answer questions about genomic tests available to patients [49]. Additionally, providers have expressed frustration with what they see as unrealistically high expectations among patients that have accompanied genomic medicine [52, 53, 55, 56]. Many feel the public remain unaware of the scarcity of evidence supporting clinical utility for many tests [46], and the lack of guidelines for using newer tests and managing uncertain findings or ancillary information [46, 57].
Are providers ready for genomic medicine?
Recently several studies have examined preparedness and confidence toward the application of genomic testing in the clinic among healthcare providers. A study examining the potential use of genomic testing to assess prostate cancer risk found participating urologists to have low levels of confidence in their genomics skill-set, and to be highly reluctant to use genomic testing in practice [58]. Confidence in genomic skills and know-how has been noted in several studies to impact provider behaviors towards genomic tests, and has been associated with greater likelihood of ordering tumor-based genomic testing, knowledge, and being an academic clinician[59–63]. Recently, Grey at al found highly variable levels of genomic confidence among oncologists at a tertiary care cancer center related to the incorporation of a novel multi-gene genomic test into clinical practice. Confidence was positively associated with several behaviors related to adapting genomics into practice, including using test results to guide therapy and disclosing results to patients [59]. These findings and others highlight how experience and perceived competence can impact physician behaviors towards new genomic testing technologies. Above all else, it would appear that healthcare providers’ positive attitudes towards genomics may be tempered by perceptions of ill-preparedness and lack of confidence to effectively translate genomic medicine into clinical care. It further seems likely that before providers are ready to fully embrace genomic medicine, substantial progress to demonstrate the utility of genomics is needed, as well as focused education, interpretation and communication skills training for providers to be able to respond to the informational needs of patients. Training may be particularly important among primary care and community providers, who to date may have less practical experience with genomic testing [19, 48, 50, 54, 59, 60, 64]. Unquestionably, the delivery of germ-line and somatic genomic medicine is rapidly changing at this time, and even current established knowledge levels, attitudes, and behaviors are likely to change. Interestingly, genetic counselors appear to be less involved in the delivery of multi-gene tumor or somatic testing in the oncology clinic, and data from our group suggest many counselors do not perceive a strong role for their services in this arena [Hall, Boland, Ruth][62]. Nonetheless, increased uptake of multi-gene somatic panels by the oncology community has continued to uncover somatic mutations in tumors that, in the context of personal/family history, may be suspicious for also being germ-line mutations and thus may warrant further investigation.
IV. Patient perceptions and expectations of genomics in medical and cancer care
Patients are highly optimistic about genomic medicine and its potential for improving health. Expectations are diverse, and reflect largely positive attitudes towards genetics, a high regard for the power of medicine, but also concerns about personal privacy. For example, cancer patients are now facing complicated shared decisions related to new germ-line and somatic genomic tests—these decisions are both challenging for providers to facilitate as well as difficult for the patient to understand. In the setting of poor understanding of genetics and potentially incomplete clinical information about some of the genes examined in newer panel tests, many of the basic elements of informed consent (e.g. pros and cons of a test, potential therapeutic implications), are challenging to address. Public attitudes towards genomic medicine, while largely positive, may in fact be founded in incomplete understanding. The complexity of new tests is compounded by providers with insufficient skills to communicate about these tests and effectively translate genomic technologies into appropriate clinical care.
Patient views of genomics: Positive attitudes yet limited understanding
Patients perceive high value to genetic information, and many view genomic testing as a novel route to better understand and manage health risks [53, 58, 65, 66]. However, an abundance of research has demonstrated that patients have limited knowledge of genetic principles and limited understanding of the current genetic and genomic tests available to them [67, 68]. In the prevention and treatment of cancer, patients have been shown to have a limited understanding of what “personalized medicine” means [68]. They may not understand the differences between a hereditary versus a somatic genomic test, or a diagnostic test versus one examining a disease association. Despite acknowledging that environmental risks of disease are on par with genetic risks, patients place high importance on the results of genetic tests [69], and anticipate that tests are accurate and replicable. Futhermore, substantial limitations in genetic health literacy and numeracy skill in the general population makes the integration of genomic testing into practice even more challenging [70–72].
Impact of genomics on patient behaviors
Genomics will impact the delivery of medical care in a variety of formats, several of which will rely on patient comprehension of a genomic test result and subsequent behaviors like screening. Currently, the informed patient is integral to long-term outcomes of diagnostic tests for hereditary cancer risk genes, genes that may be important in environmental health risks, and disease association tests that do not identify causal genomic determinants of disease, but may quantify health risks through genomic markers. Evidence suggests that genomic test results can effectively motivate behavioral changes in patients, whether to seek prevention or a novel therapeutic agent, but it may be mediated by number of factors. The behavioral health impact of genomic information may vary by the degree of risk identified and the health options following testing [73–76]. For example, research from Graves et al found that patients’ interest in SNP tests for cancer risk increased dependent on the degree of risk conveyed if the test was positive (e.g. higher interest in a test that detects a higher risk) and the type of preventive options available [e.g. lower interest when options to mitigate risk were behavioral (exercise) versus chemopreventive (take a pill)] [29].
Expectations of genomic medicine: the patient’s perspective
Patients anticipate that genomic medicine will improve their health through the selection of more appropriate treatments and through side effect reduction, better prognostication, earlier detection of disease, better identification of the causes of serious diseases like cancer, and improved cure rates [65, 68]. However, public enthusiasm is tempered by concerns of genetic discrimination, including provider discrimination about who merits testing or does not. Concern about the loss of employment or insurance, costs of testing, privacy concerns, and psychological harm are additional concerns raised. For other patients, genomic medicine simply remains unattractive and something they don’t want to know about. Many fear that tests will lead to “information overload” during the course of treatment for an illness like cancer. Interestingly, studies have found that the type of test and how the public understands it can influence attitudes toward testing. For example, patients express more negative attitudes about whole genome testing versus testing of individual genes, anticipating and fearing greater breaches of privacy if the entire genome is examined [68]. Other research has shown that patients may view tests with multiple levels of meaning or those producing ancillary information in a paradoxically positive manner, viewing multiple levels of information in an optimistic “two-for-one” light [65].
For patients, genomic tests are largely indiscernible from other forms of medical information, and thus their expectation is that a genomic test result is both reproducible and accurate. Moreover, information from a genomic test is expected to be interpretable, and that the test result’s health relevance should be clear [68]. Further, patients feel that genomic information should be presented in a way that they may understand, and that is non-overwhelming [14, 16, 77–79]. From their providers, patients additionally expect competence in ordering the correct tests and interpreting them in the context of their personal and family medical history. Patients overall demand a high level of privacy in the treatment and protection of genomic information, and believe that it should not be used to determine employment eligibility or insurability [15]. Finally, patients prefer to maintain personal control over how their genomic information is accessed and used, including the release of information to their own family members [16, 65, 80, 81].
Practical limitations of genomic testing
Increasing availability and reduced costs of clinical genomic tests, coupled with increasing public interest and easier access to these tests, risk fueling an environment where genomic medicine could be improperly delivered in the clinic. Most concerning, patients risk being tested inappropriately with new genomic tests and receiving poorly interpreted and/or contextualized information, and thus risk adopting behaviors that are non-beneficial to their health. Low confidence and knowledge towards genomic testing among providers only magnifies these risks. Many facets of the complex genetic and genomic testing process increase the potential for misguided use of genomic medicine, and it is at these junctures that genetics professionals are most critical to be certain that genomic tests are selected appropriately, that the rationale for testing is communicated properly to the patient, and that test results are interpreted and contextualized correctly. Interest and enthusiasm for novel tests to better diagnose risks or guide therapy may lead to use of tests of uncertain validity. Studies have shown early adopters of novel genomic tests may be less savvy about genomics than counterparts who remain skeptical about these tests and use them later [64, 66, 69]. Patients, in the context of trying to find answers for risks or to improve health outcomes, may also focus on personal needs and immediate payoffs over familial or socio-psychological implications of genomic tests and the information they generate (e.g. on the ability to get life insurance if a mutation is found)[82]. Patients may fail to fully understand how one immediately available test is different from another that may be more appropriate for them but that is less readily available [68].
Even more concerning, studies suggest patients and their providers may have little awareness of the evidence-base associated with various genomic tests or the likelihood of test findings having clear clinical benefits or interpretable implications [53, 64, 68, 71]. On a more granular level, research has shown patients and providers are at risk of misinterpreting genetic results due to a generally poor understanding of genetic principles. For example, errors in understanding the differences between variants of uncertain significance and deleterious mutations, low versus high penetrance genes, autosomal dominant versus recessive inheritance, and associations versus causative mutations have all been documented in the literature [27, 64]. Risks to patients are magnified further when the issues above lead to a flawed interpretation of genetic risk within the context of a strong family history [83]. It is common in the high-risk clinic to consult with patients with a very strong family history of cancer (but in whom no mutation was identified by testing) who understood their result as meaning they do not have an increased risk of cancer. On the other hand, it is equally common to see patients unaffected by cancer who have been found to have an uncertain variant in a high-risk gene that has been interpreted as being disease causing.
Powerful tests but uncertain results—managing ancillary information from genomic tests
In the coming years, patients will be subjected to increased use of genomic tests and a concurrent increased risk that tests will generate information that is very difficult to interpret in the context of their personal/family history, or in the context of current medical knowledge. Research has shown that patients are largely unaware that ancillary or off-target results may be an undesired outcome of genomic testing [13, 65, 81, 84]. Important practical and ethical questions that remain unresolved in this area are whether patients, at the time of consent for a clinical genomic test, should have the right to refuse to receive certain ancillary results of a genomic test if they choose, whether they should have additional controls over how genomic information is accessed and mined for research and quality improvement through clinical databases, and, perhaps most importantly, the responsibilities of healthcare providers toward family members when target and off-target genomic information is generated in a patient who may be reluctant or unable to communicate these findings to family members [84].
V. Conclusion
Genomic testing is rapidly entering routine clinical care, most notably in the practice of oncology. New and more powerful testing technologies are reducing the time and costs associated with large scale gene sequencing, but patients and providers must be aware of the limitations and risks associated with these tests. Conversations between patients and providers should outline the potential benefits of genetic evaluation with new multi-gene panels and other technologies, but must also carefully address the technical and clinical limitations of these technologies. Expert recommendations and procedures for clinical testing using multi-gene panels are needed to guide the appropriate use of powerful genetic and genomic technologies in the evaluation of hereditary cancer risk and the molecular evaluation of tumors for treatment.
References
- 1.EGAPP website. [ http://www.egappreviews.org/]
- 2.Healthy people website. [ http://www.healthypeople.gov/2020/default.aspx]
- 3.Hudson KL, Murphy JA, Kaufman DJ, Javitt GH, Katsanis SH, Scott J. Oversight of US genetic testing laboratories. Nat Biotechnol. 2006;24:1083–1090. doi: 10.1038/nbt0906-1083. [DOI] [PubMed] [Google Scholar]
- 4.Frank TS, Deffenbaugh AM, Reid JE, Hulick M, Ward BE, Lingenfelter B, Gumpper KL, Scholl T, Tavtigian SV, Pruss DR, Critchfield GC. Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol. 2002;20:1480–1490. doi: 10.1200/JCO.2002.20.6.1480. [DOI] [PubMed] [Google Scholar]
- 5.Beamer LC, Grant ML, Espenschied CR, Blazer KR, Hampel HL, Weitzel JN, MacDonald DJ. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol. 2012;30:1058–1063. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwartz F, Valdimarsdottir HB, Peshkin BN, Mandelblatt J, Nusbaum R, Huang AT, Chang Y, Graves K, Isaacs C, Wood M, et al. Randomized noninferiority trial of telephone versus in-person genetic counseling for hereditary breast and ovarian cancer. J Clin Oncol. 2014;32:618–626. doi: 10.1200/JCO.2013.51.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patrick-Miller L, Egleston BL, Daly M, Stevens E, Fetzer D, Forman A, Bealin L, Rybak C, Peterson C, Corbman M, Bradbury AR. Implementation and outcomes of telephone disclosure of clinical BRCA1/2 test results. Patient Educ Couns. 2013;93:413–419. doi: 10.1016/j.pec.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall MJ, Herda MM, Handorf EA, Rybak CC, Keleher CA, Siemon M, Daly MB. Direct-to-patient disclosure of results of mismatch repair (MMR) screening for Lynch syndrome via electronic personal health record (ePHR): A feasibility study. Genet Med. 2014 doi: 10.1038/gim.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swanson A, Ramos E, Snyder H. Next Generation Sequencing is the impetus for the next generation of laboratory-based genetic counselors. J Genet Couns. 2014 doi: 10.1007/s10897-013-9684-1. [DOI] [PubMed] [Google Scholar]
- 10.Mauer CB, Pirzadeh-Miller SM, Robinson LD, Euhus DM. The integration of next-generation sequencing panels in the clinical cancer genetics practice an institutional experience. Genet Med. 2013 doi: 10.1038/gim.2013.160. [DOI] [PubMed] [Google Scholar]
- 11.Meder B, Haas J, Keller A, Heid C, Just S, Borries A, Boisguerin V, Scharfenberger-Schmeer M, Stahler P, Beier M, et al. Targeted next-generation sequencing for the molecular genetic diagnostics of cardiomyopathies. Circ Cardiovasc Genet. 2011;4:110–122. doi: 10.1161/CIRCGENETICS.110.958322. [DOI] [PubMed] [Google Scholar]
- 12.Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health meeting the challenge one bin at a time. Genet Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- 13.Netzer C, Klein C, Kohlhase J, Kubisch C. New challenges for informed consent through whole genome array testing. Journal of Medical Genetics. 2009;46:495–496. doi: 10.1136/jmg.2009.068015. [DOI] [PubMed] [Google Scholar]
- 14.Ormond KE, Wheeler MT, Hudgins L, Klein TE, Butte AJ, Altman RB, Ashley EA, Greely HT. Challenges in the clinical application of whole-genome sequencing. Lancet. 2010;375:1749–1751. doi: 10.1016/S0140-6736(10)60599-5. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong K, Putt M, Halbert CH, Grande D, Schwartz JS, Liao KJ, Marcus N, Demeter MB, Shea J. The Influence of Health Care Policies and Health Care System Distrust on Willingness to Undergo Genetic Testing. Medical Care. 2012;50:381–387. doi: 10.1097/MLR.0b013e31824d748b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigter T, Henneman L, Kristoffersson U, Hall A, Yntema HG, Borry P, Tonnies H, Waisfisz Q, Elting MW, Dondorp WJ, Cornel MC. Reflecting on earlier experiences with unsolicited findings points to consider for next-generation sequencing and informed consent in diagnostics. Hum Mutat. 2013;34:1322–1328. doi: 10.1002/humu.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Latif DA. Pharmacogenetics and pharmacogenomics instruction in schools of pharmacy in the USA: is it adequate? Pharmacogenomics. 2005;6:317–319. doi: 10.1517/14622416.6.4.317. [DOI] [PubMed] [Google Scholar]
- 18.Gurwitz D, Lunshof JE, Dedoussis G, Flordellis CS, Fuhr U, Kirchheiner J, Licinio J, Llerena A, Manolopoulos V, Sheffield LJ, Pharmacogenomics education, et al. International Society of Pharmacogenomics recommendations for medical, pharmaceutical, and health schools deans of education. Pharmacogenomics Journal. 2005;5:221–225. doi: 10.1038/sj.tpj.6500312. [DOI] [PubMed] [Google Scholar]
- 19.Frueh FW, Gurwitz D. From pharmacogenetics to personalized medicine a vital need for educating health professionals and the community. Pharmacogenomics. 2004;5:571–579. doi: 10.1517/14622416.5.5.571. [DOI] [PubMed] [Google Scholar]
- 20.Shah J. Recent developments - Criteria influencing the clinical uptake of pharmacogenomic strategies. British Medical Journal. 2004;328:1482–1486. doi: 10.1136/bmj.328.7454.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.U.S. System of Oversight of Genetic Testing: A Response to the Charge of the Secretary of Health and Human Services Report of the Secretary’s Advisory Committee on Genetics, Health, Society; Department of Health , Human Services. Bethesda ,Maryland,Secretary’s Advisory Committee on Genetics, Health Society, April 2008. Book US System of Oversight of Genetic Testing A Response to the Charge of the Secretary of Health and Human Services, Report of the Secretary’s Advisory Committee on Genetics, Health, and Society; Department of Health and Human Services. Bethesda, Maryland, Secretary’s Advisory Committee on Genetics, Health, and Society,2008(Editor ed.^eds.) 2008 Apr; City. [Google Scholar]
- 22.Ratain MJ, Innocenti F. Individualizing dosing of irinotecan. Clin Cancer Res. 2010;16:371–372. doi: 10.1158/1078-0432.CCR-09-2936. [DOI] [PubMed] [Google Scholar]
- 23.Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 24.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchionni L, Wilson RF, Wolff AC, Marinopoulos S, Parmigiani G, Bass EB, Goodman SN. Systematic review gene expression profiling assays in early-stage breast cancer. Ann Intern Med. 2008;148:358–369. doi: 10.7326/0003-4819-148-5-200803040-00208. [DOI] [PubMed] [Google Scholar]
- 26.Hiraki S, Rinella ES, Schnabel F, Oratz R, Ostrer H. Cancer Risk Assessment Using Genetic Panel Testing Considerations for Clinical Application. J Genet Couns. 2014 doi: 10.1007/s10897-014-9695-6. [DOI] [PubMed] [Google Scholar]
- 27.Grover S, Stoffel EM, Mercado RC, Ford BM, Kohlman WK, Shannon KM, Conrad PG, Blanco AM, Terdiman JP, Gruber SB, et al. Colorectal cancer risk perception on the basis of genetic test results in individuals at risk for Lynch syndrome. J Clin Oncol. 2009;27:3981–3986. doi: 10.1200/JCO.2008.18.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon I, Hooker G, Erby L, Klein W, Giardiello F, Axilbund F, Blanco A, Hampel H, Semotiuk K, Leonard L. Annual Conference of the National Society of Genetic Counselors; 2013. 32nd. Anaheim, CA: 2013. Living in Lynch syndrome limbo: Understanding the meaning of uncertain genetic test results. [Google Scholar]
- 29.Graves KD, Christopher J, Harrison TM, Peshkin BN, Isaacs C, Sheppard VB. Providers’ Perceptions and Practices Regarding BRCA1/2 Genetic Counseling and Testing in African American Women. Journal of Genetic Counseling. 2011;20:674–689. doi: 10.1007/s10897-011-9396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindor NM, Goldgar DE, Tavtigian SV, Plon SE, Couch FJ. BRCA1/2 sequence variants of uncertain significance a primer for providers to assist in discussions and in medical management. Oncologist. 2013;18:518–524. doi: 10.1634/theoncologist.2012-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pyeritz RE. The coming explosion in genetic testing--is there a duty to recontact? Engl J Med. 2011;365:1367–1369. doi: 10.1056/NEJMp1107564. [DOI] [PubMed] [Google Scholar]
- 32.Hunter AG, Sharpe N, Mullen M, Meschino WS. Ethical Legal practical concerns about recontacting patients to inform them of new information the case in medical genetics. Am J Med Genet. 2001;103:265–276. [PubMed] [Google Scholar]
- 33.Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, Wenstrup RJ, Ward BE, Scholl TA, Noll WW. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009;115(Suppl 1):S56–S62. doi: 10.1002/cncr.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast and Ovarian. Book National Comprehensive Cancer Network. Genetic/Familial High-Risk Assessment: Breast and Ovarian (Editor ed.^eds.) (Version 1.2014) City. [Google Scholar]
- 35.Forman AD, Hall MJ. Influence of race/ethnicity on genetic counseling and testing for hereditary breast and ovarian cancer. Breast J. 2009;(Suppl 1):S56–S62. doi: 10.1111/j.1524-4741.2009.00798.x. [DOI] [PubMed] [Google Scholar]
- 36.Eggington JM, Burbidge LA, Roa BBea. ACMG Annual Clinical Genetics Meeting. Charlotte, NC: 2012. Current variant of uncertain significance rates in BRCA1/2 and Lynch Syndrome testing (MLH1, MSH2, MSH6, PMS2, EPCAM) [Google Scholar]
- 37.Obeid E, Forman AJ, Hall MJ, Daly MB. Multigene Panel Genetic Testing - a Pilot Program’s Experience at Fox Chase Cancer Center (FCCC) Proceedings of the American Society of Clinical Oncology. 2014 [Google Scholar]
- 38.Hsu HM, Wang HC, Chen ST, Hsu GC, Shen CY, Yu JC. Breast Cancer Risk Is Associated with the Genes Encoding the DNA Double-Strand Break Repair Mre11/Rad50/Nbs1 Complex. Biomarkers Prev. 2007;16:2024–2032. doi: 10.1158/1055-9965.EPI-07-0116. [DOI] [PubMed] [Google Scholar]
- 39.Bogdanova N, Feshchenko S, Schürmann P, Waltes R, Wieland B, Hillemanns P, Rogov YI, Dammann O, Bremer M, Karstens JH, et al. Nijmegen Breakage Syndrome mutations and risk of breast cancer. Int J Cancer. 2008;122:802–806. doi: 10.1002/ijc.23168. [DOI] [PubMed] [Google Scholar]
- 40.Desjardins S, Beauparlant JC, Labrie Y, Ouellette G, BRCAs I, Durocher F. Variations in the NBN/NBS1 gene and the risk of breast cancer in non-BRCA1/2 French Canadian families with high risk of breast cancer. BMC Cancer. 2009;9:1–17. doi: 10.1186/1471-2407-9-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loveday CCT, Ramsay E, Hughes D, Ruark E, Frankum JR, Bowden G, Kalmyrzaev B, Warren-Perry M, Snape K, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:873–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Institute of Medicine—Value of genetic and genomic. [ http://www.iom.edu/Reports/2010/The-Value-of-Genetic-and-Genomic-Technologies.aspx]
- 43.Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a Personalized Medicine Infrastructure at a Major Cancer Center. Journal of Clinical Oncology. 2013;31:1849–1857. doi: 10.1200/JCO.2012.45.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dienstmann R, Rodon J, Barretina J, Tabernero J. Genomic medicine frontier in human solid tumors prospects and challenges. J Clin Oncol. 2013;31:1874–1884. doi: 10.1200/JCO.2012.45.2268. [DOI] [PubMed] [Google Scholar]
- 45.Henrikson NB, Burke W, Veenstra DL. Ancillary risk information pharmacogenetic tests social and policy implications. Pharmacogenomics J. 2008;8:85–89. doi: 10.1038/sj.tpj.6500457. [DOI] [PubMed] [Google Scholar]
- 46.Haga SB, Tindall G, O’Daniel JM. Professional Perspectives About Pharmacogenetic Testing and Managing Ancillary Findings. Genetic Testing and Molecular Biomarkers. 2012;16:21–24. doi: 10.1089/gtmb.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lohn Z, Adam S, Birch P, Townsend A, Friedman J. Genetics professionals’ perspectives on reporting incidental findings from clinical genome-wide sequencing. Am J Med Genet A. 2013;161A:542–549. doi: 10.1002/ajmg.a.35794. [DOI] [PubMed] [Google Scholar]
- 48.Chen LS, Goodson P. Public health genomics knowledge and attitudes a survey of public health educators in the United States. Genet Med. 2007;9:496–503. doi: 10.1097/gim.0b013e31812e95b5. [DOI] [PubMed] [Google Scholar]
- 49.Powell KP, Cogswell WA, Christianson CA, Dave G, Verma A, Eubanks S, Henrich VC. Primary care physicians’ awareness, experience and opinions of direct-to-consumer genetic testing. J Genet Couns. 2012;21:113–126. doi: 10.1007/s10897-011-9390-9. [DOI] [PubMed] [Google Scholar]
- 50.Chen LS, Kim M. Needs Assessment in Genomic Education A Survey of Health Educators in the United States. Health Promot Pract. 2013 doi: 10.1177/1524839913483470. [DOI] [PubMed] [Google Scholar]
- 51.Guttmacher AE, Porteous ME, McInerney JD. Educating healthcare professionals about genetics and genomics. Nat Rev Genet. 2007;8:151–157. doi: 10.1038/nrg2007. [DOI] [PubMed] [Google Scholar]
- 52.O’Daniel JM, Haga SB, Willard HF. Considerations for the Impact of Personal Genome Information A Study of Genomic Profiling among Genetics and Genomics Professionals. Journal of Genetic Counseling. 2010;19:387–401. doi: 10.1007/s10897-010-9297-x. [DOI] [PubMed] [Google Scholar]
- 53.Fargher EA, Eddy C, Newman W, Qasim F, Tricker K, Elliott RA, Payne K. Patients’ and healthcare professionals’ views on pharmacogenetic testing and its future delivery in the NHS. Pharmacogenomics. 2007;8:1511–1519. doi: 10.2217/14622416.8.11.1511. [DOI] [PubMed] [Google Scholar]
- 54.Nelson EA, McGuire AL. The need for medical education reform: genomics and the changing nature of health information. Genome Med. 2010;2:18. doi: 10.1186/gm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Rourke PP. Genomic medicine: too great expectations? Clin Pharmacol Ther. 2013;94:188–190. doi: 10.1038/clpt.2013.44. [DOI] [PubMed] [Google Scholar]
- 56.Hall MJPML, McCully KC, Bradbury AR. American Society of Preventive Oncology. Washington DC: Annual Meeting; Mar, 2012. Genomic tests in cancer care Attitudes, informed consent needs, and preferences for information sharing; pp. 4–6. 2012. [Google Scholar]
- 57.Haga SB, Burke W. Pharmacogenetic testing not as simple as it seems. Genet Med. 2008;10:391–395. doi: 10.1097/GIM.0b013e31817701d4. [DOI] [PubMed] [Google Scholar]
- 58.Birmingham WC, Agarwal N, Kohlmann W, Aspinwall LG, Wang M, Bishoff J, Dechet C, Kinney AY. Patient and provider attitudes toward genomic testing for prostate cancer susceptibility: a mixed method study. BMC Health Serv Res. 2013;13:279. doi: 10.1186/1472-6963-13-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gray SW, Hicks-Courant K, Cronin A, Rollins BJ, Weeks JC. Physicians’ Attitudes About Multiplex Tumor Genomic Testing. J Clin Oncol. 2014 doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woods JIRK, Boland PM, Rainey KL, Fang CY, Cohen SJ, Matro JM, Chertock Y, Wong YN, Daly MB, Hall MJ. Academic (AO) and community (CO) oncologists’ knowledge, understanding, and preparedness for clinical next-generation sequencing genomic testing (NGSGT) Proceedings of the American Society of Clinical Oncology. 2014 [Google Scholar]
- 61.Haga SB, Carrig MM, O’Daniel JM, Orlando LA, Killeya-Jones LA, Ginsburg GS, Cho A. Genomic Risk Profiling Attitudes and Use in Personal and Clinical Care of Primary Care Physicians Who Offer Risk Profiling. Journal of General Internal Medicine. 2011;26:834–840. doi: 10.1007/s11606-011-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hall MJ, Boland PM, Ruth K, Matro JM, Rainey KL, Fang CY, Wong YN, Daly MB. Incorporating genomic testing using next-generation sequencing (NGS) into clinical practice:Genetic counselors’ (GC) experience, knowledge and perceived competence. Book Incorporating genomic testing using next-generation sequencing (NGS) into clinical practice:Genetic counselors’ (GC) experience, knowledge and perceived competence (Editor ed.^eds.) 2014 City. [Google Scholar]
- 63.Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases a systematic review. JAMA. 2008;299:1320–1334. doi: 10.1001/jama.299.11.1320. [DOI] [PubMed] [Google Scholar]
- 64.Sturm AC, Manickam K. Direct-to-consumer personal genomic testing a case study and practical recommendations for “genomic counseling. J Genet Couns. 2012;21:402–412. doi: 10.1007/s10897-012-9489-7. [DOI] [PubMed] [Google Scholar]
- 65.Hall MJ, Patrick Miller L, McCully KC, Bradbury AR. American Society of Preventive Oncology, Annual Meeting. Washington DC: Mar, 2012. Genomic tests in cancer care Attitudes, informed consent needs, and preferences for information sharing; pp. 4–6. [Google Scholar]
- 66.Gollust SE, Gordon ES, Zayac C, Griffin G, Christman MF, Pyeritz RE, Wawak L, Bernhardt BA. Motivations and perceptions of early adopters of personalized genomics perspectives from research participants. Public Health Genomics. 2012;15:22–30. doi: 10.1159/000327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kolor K, Duquette D, Zlot A, Foland J, Anderson B, Giles R, Wrathall J, Khoury MJ. Public awareness and use of direct-to-consumer personal genomic tests from four state population-based surveys, and implications for clinical and public health practice. Genetics in Medicine. 2012;14:860–867. doi: 10.1038/gim.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gray SW, Hicks-Courant K, Lathan CS, Garraway L, Park ER, Weeks JC. Attitudes of patients with cancer about personalized medicine and somatic genetic testing. J Oncol Pract. 2012;8:329–335. doi: 10.1200/JOP.2012.000626. 322 p following 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Characteristics of users of online personalized genomic risk assessments implications for physician-patient interactions. Genet Med. 2009;11:582–587. doi: 10.1097/GIM.0b013e3181b22c3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaphingst KA, McBride CM, Wade C, Alford SH, Brody LC, Baxevanis AD. Consumers’ Use of Web-Based Information and Their Decisions About Multiplex Genetic Susceptibility Testing. Journal of Medical Internet Research. 2010;12 doi: 10.2196/jmir.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leighton JW, Valverde K, Bernhardt BA. The general public’s understanding and perception of direct-to-consumer genetic test results. Public Health Genomics. 2012;15:11–21. doi: 10.1159/000327159. [DOI] [PubMed] [Google Scholar]
- 72.Kelly KM, Graves KD, Harper FWK, Schmidt JE, Dickinson SL, Andrykowski MA. Assessing perceptions of cancer risk: Does mode of assessment or numeracy matter? Cancer Detection and Prevention. 2007;31:465–473. doi: 10.1016/j.cdp.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 73.Chao S, Roberts JS, Marteau TM, Silliman R, Cupples LA, Green RC. Health behavior changes after genetic risk assessment for Alzheimer disease The REVEAL Study. Alzheimer Dis Assoc Disord. 2008;22:94–97. doi: 10.1097/WAD.0b013e31815a9dcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McBride CM, Bepler G, Lipkus IM, Lyna P, Samsa G, Albright J, Datta S, Rimer BK. Incorporating genetic susceptibility feedback into a smoking cessation program for African-American smokers with low income. Cancer Epidemiol Biomarkers Prev. 2002;11:521–528. [PubMed] [Google Scholar]
- 75.Ito H, Matsuo K, Wakai K, Saito T, Kumimoto H, Okuma K, Tajima K, Hamajima N. An intervention study of smoking cessation with feedback on genetic cancer susceptibility in Japan. Prev Med. 2006;42:102–108. doi: 10.1016/j.ypmed.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 76.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. N Engl J Med. 2011;364:524–534. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaphingst KA, Facio FM, Cheng MR, Brooks S, Eidem H, Linn A, Biesecker BB, Biesecker LG. Effects of informed consent for individual genome sequencing on relevant knowledge. Clin Genet. 2012;82:408–415. doi: 10.1111/j.1399-0004.2012.01909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ayuso C, Millán JM, Mancheño M, Dal-Ré R. Informed consent for whole-genome sequencing studies in the clinical setting. Proposed recommendations on essential content and process. Eur J Hum Genet. 2013;21:1054–1059. doi: 10.1038/ejhg.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ormond KE, Iris M, Banuvar S, Minogue J, Annas GJ, Elias S. What do patients prefer informed consent models for genetic carrier testing. J Genet Couns. 2007;16:539–550. doi: 10.1007/s10897-007-9094-3. [DOI] [PubMed] [Google Scholar]
- 80.Thorogood A, Knoppers BM, Dondorp WJ, de Wert GM. Whole-genome sequencing and the physician. Clin Genet. 2012;81:511–513. doi: 10.1111/j.1399-0004.2012.01868.x. [DOI] [PubMed] [Google Scholar]
- 81.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire AL, Nussbaum RL, O’Daniel JM, Ormond KE, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, Beskow LM, Cho MK, Christman MF, Green RC, Hall R, et al. Managing incidental findings and research results in genomic research involving biobanks and archived data sets. Genet Med. 2012;14:361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giardiello FM, Brensinger JD, Petersen GM, Luce MC, Hylind LM, Bacon JA, Booker SV, Parker RD, Hamilton SR. The use and interpretation of commercial APC gene testing for familial adenomatous polyposis. New England Journal of Medicine. 1997;336:823–827. doi: 10.1056/NEJM199703203361202. [DOI] [PubMed] [Google Scholar]
- 84.Bennette CS, Trinidad SB, Fullerton SM, Patrick D, Amendola L, Burke W, Hisama FM, Jarvik GP, Regier DA, Veenstra DL. Return of incidental findings in genomic medicine measuring what patients value--development of an instrument to measure preferences for information from next-generation testing (IMPRINT) Genet Med. 2013;15:873–881. doi: 10.1038/gim.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]