Abstract
Neurogenesis, the generation of new neurons, is deregulated in neural stem cell (NSC)- and progenitor-derived murine models of malignant medulloblastoma and glioma, the most common brain tumors of children and adults, respectively. Molecular characterization of human malignant brain tumors, and in particular brain tumor stem cells (BTSCs), has identified neurodevelopmental transcription factors, microRNAs, and epigenetic factors known to inhibit neuronal and glial differentiation. We are starting to understand how these factors are regulated by the major oncogenic drivers in malignant brain tumors. In this review, we will focus on the molecular switches that block normal neuronal differentiation and induce brain tumor formation. Genetic or pharmacological manipulation of these switches in BTSCs has been shown to restore the ability of tumor cells to differentiate. We will discuss potential brain tumor therapies that will promote differentiation in order to reduce treatment-resistance, suppress tumor growth, and prevent recurrence in patients.
Keywords: Brain, Glioma, Medulloblastoma, Neurogenesis, Neural Stem Cell
INTRODUCTION
Pioneering work in the 1960s used radioactive thymidine incorporation to detect ongoing neurogenesis in the adult rodent brain (Altman, 1962). In the late 1990s, a cohort of patients receiving bromodeoxyuridine (BrdU) enabled Eriksson and colleagues to identify hippocampal neurogenesis in the adult human brain (Eriksson, et al., 1998). While a wealth of publications has described rodent neurogenesis, the extent of ongoing adult neurogenesis in humans is debated (Wang, et al., 2014, Yuan, et al., 2014). Neural stem cells (NSCs) and more differentiated precursor cells, capable of producing neurons and glial cells, have been found to give rise to brain tumors in mice. As initiating events during brain tumor formation, we and others have described reduced differentiation and aberrant proliferation of precursor cells. Recent studies have begun to delineate how such cancer-causing events induce a switch from asymmetric to symmetric cell division in transformed cells. We will focus this review on glioma and medulloblastoma (MB), the most common primary malignant brain tumors in adults and children, respectively. Gliomas are classified by grades I–IV (Louis, et al., 2007). Grade I pilocytic astrocytomas are more common in children while grade II–III oligodendrogliomas, astrocytomas, oligoastrocytomas, or ependymomas occur in both children and adults. Genome-wide characterization of both grade IV glioblastomas (GBMs) and grade II–III gliomas has identified molecularly and biologically distinct subgroups in children and adults (Cooper, et al., 2010, Phillips, et al., 2006, Sturm, et al., 2012, Verhaak, et al., 2010). Similarly, extensive profiling of MBs has established multiple subgroups of tumors (Taylor, et al., 2012). Although glioma and MB cells display distinct glial and neuronal phenotypes, both types of brain tumors have been described to originate from neural precursors capable of undergoing neurogenesis. We will first describe regions of forebrain and hindbrain neurogenesis and how the identities of these precursors match distinct subgroups of human gliomas and MBs. We will then summarize findings that characterize neuronal gene expression in these tumors and the ability of brain tumor stem cells (BTSCs) to produce neurons. We will then review factors that make BTSCs pluripotent capable of differentiating into not only neurons and glial phenotypes, but also cells from all three germ layers. Finally, we will suggest that a better understanding of the cell of origin and the initiating events during brain tumor formation can identify new therapeutic targets of the most treatment-resistant BTSCs, ultimately improving the outcome for glioma and MB patients. While the current standard of care includes surgery followed by radio- and chemotherapy, the prognosis for most patients is fatal. The overall survival of MB patients is about 70% but surviving children often succumb to severe “side-effects” due to treatment toxicities, especially from craniospinal radiotherapy (Gilbertson, 2004, Schmidt, et al., 2010). We propose that current advances in neurodevelopmental biology will inform the brain tumor field and may lead to novel therapies that target the root of cancer, rather than individual branches.
Association of forebrain neurogenesis and gliomagenesis
Neurogenesis, the generation of new neurons, is mainly confined to embryonic development of the central nervous system (CNS), but restricted regions continue to produce neurons after birth (Kriegstein and Alvarez-Buylla, 2009). Neuroepithelial stem cells in the ventricles of the forebrain and the spinal canal give rise to radial glial cells (Rowitch and Kriegstein, 2010). The latter in turn are successors that produce mature cells including neurons, oligodendrocytes, astrocytes and ependymal cells (Malatesta, et al., 2000). Radial glia cells express a cytoplasmic protein called brain lipid-binding protein (BLBP) (Feng, et al., 1994, Hartfuss, et al., 2001, Kurtz, et al., 1994). Cellular fate mapping using the BLBP promoter coupled to the Rosa26 reporter shows that most types of neurons in almost all brain regions (Anthony, et al., 2004) originate from BLBP-positive radial glia cells. In the mouse brain, neurogenesis continues throughout life and is restricted to two germinal regions; the subgranular zone (SGZ) of the hippocampus and the subventricular zone (SVZ) lining the lateral ventricles (Alvarez-Buylla and Lim, 2004, Seri, et al., 2001). In the adult mouse SVZ, glial fibrillary acidic protein (GFAP) expressing neural stem cells (NSCs), also called type B1 cells, are thought to undergo asymmetric cell division to generate transit amplifying progenitors (TAPs, type C cells) that further differentiate into immature neuroblasts (type A cells). In mouse brain, type A cells use the rostral migratory stream (RMS) in their migration to the olfactory bulb where they differentiate into olfactory bulb neurons. More recent data demonstrate that cell-intrinsic differences of individual murine SVZ NSCs generate several distinct interneuron subtypes of the olfactory bulb (Merkle, et al., 2007, Merkle, et al., 2004). Although olfactory bulb neurogenesis is not detectable in adult humans, substantial hippocampal neurogenesis with comparable neuronal turnover rates is found in middle-aged humans and mice (Eriksson, et al., 1998, Spalding, et al., 2013). A novel carbon-14 dating approach recently suggested generation of striatal neurons in adult humans, possibly originating from the SVZ (Ernst, et al., 2014). However, another study showed that human and monkey striatal interneurons are derived from the medial ganglionic eminence (Wang C et al., J Neurosci, 2014). Is it possible that oncogenic transformation of forebrain NSCs, neural progenitors or even differentiated neurons can give rise to gliomas?
Similar to normal NSCs, recent findings suggest that treatment-resistant BTSCs in human GBMs possess extensive self-renewal ability, undergo asymmetric cell division, and can differentiate along the three main neural cell lineages, implicating a possible relationship (Hemmati, et al., 2003, Lathia, et al., 2011, Singh, et al., 2003).
Much effort has successfully generated genome-wide characterization of low- and high-grade gliomas into molecularly and biologically distinct subtypes in children and adults (Cooper, et al., 2010, Sturm, et al., 2012, Verhaak, et al., 2010). Recent studies suggest that GBM patients with tumors contacting the SVZ show worse prognosis and increased radiation doses of this region were associated with improved survival in GBM patients (Chen, et al., 2013, Jafri, et al., 2013). In contrast, we have previously shown that human oligodendrogliomas often lack association to the lateral ventricles where NSCs reside and can arise from oligodendrocyte progenitor cells (OPCs) in a murine glioma model (Persson, et al., 2010). Interestingly, oligodendrogliomas and a subset of GBMs display a proneural phenotype associated with improved survival and enriched for genes expressed in OPCs (SOX10, OLIG2, PDGFRA) (Cooper, et al., 2010, Verhaak, et al., 2010). In contrast, the classical and mesenchymal phenotypes of GBMs show worse prognosis and a higher degree of stemness-related genes (HES1, PDPN) (Phillips, et al., 2006, Verhaak, et al., 2010). Studies of several genetically-engineered murine models (GEMMs) found that glioma formation from NSCs leads to reduced neurogenesis, suggesting that initiation of glioma formation from NSCs is associated with a neurogenic-gliogenic shift (Chen, et al., 2012a, Li, et al., 2014a, Zhu, et al., 2005) (Figure 1).
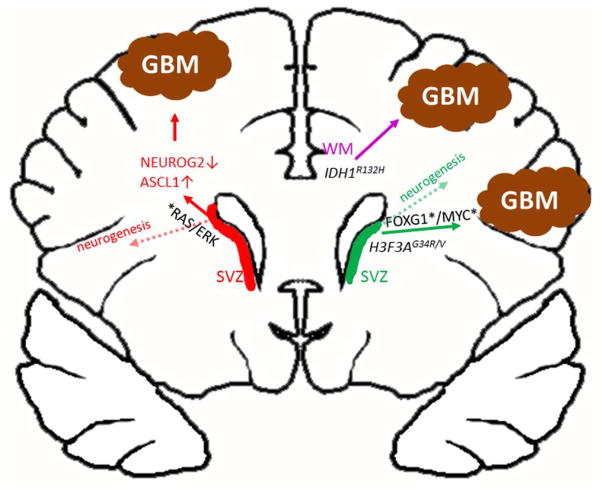
Figure 1. Deregulation of neuronal and glial differentiation as a priming step in GBM formation.
Schematics of frontal lobes and lateral ventricles in the anterior regions of the adult cerebrum. Genetic alterations in SVZ neural precursors in the RAS/ERK pathway induce a NEUROG2-ASCL1 switch that induces gliomagenesis (Li, et al., 2014). The inability of IDH1R132H mutation to transform nestin-expressing NSCs, the high frequency of IDH1R132H mutation in oligodendroglioma (shown to originate from white matter OPCs), and the association of these tumors to the frontal cortex suggest that expression of IDH1R132H mutation in white matter (WM) progenitors inhibits differentiation and drive formation of proneural gliomas (Phillips, et al., 2006, Sturm, et al., 2012). Similarly, childhood GBMs displaying G34R/V mutation in the H3F3A gene express high levels of FOXG1 and MYC, but low/no levels of the OPC-related gene OLIG2, implicating a NSC origin. Misexpression (activation/upregulation (*) or (loss) of important cancer genes is shown in black.
Association of hindbrain neurogenesis and medulloblastoma formation
Neurogenesis is restricted to two germinal zones in the developing cerebellum (small brain) (Hatten and Heintz, 1995). The first is a structure called the rhombic lip (RL) where precursors expressing the mouse homolog of the Drosophila proneural gene atonal (MATH1) reside. They form granule cell neuron precursors (GNPs) that build up the external germinal layer (EGL). GNPs later form glutamatergic granule neurons (Hevner, et al., 2006), the most abundant type of neuron in the brain (Grimmer and Weiss, 2006). Other cerebellar neurons can be formed from a second germinal region, the ventricular zone (VZ) closest to the 4th ventricle. The VZ is similar to the lateral VZ in the forebrain and is also composed of BLBP-positive radial glia cells.
Radial glia cells migrate into the cerebellum and give rise to Purkinje neurons, γ-Aminobutyric acid (GABA) expressing cells as well as interneurons including Basket and Stellate cells (Hoshino, et al., 2005). Release of sonic hedgehog (SHH) from Purkinje neurons stimulates proliferation of GNPs (Dahmane and Ruiz i Altaba, 1999, Wechsler-Reya and Scott, 1999). Bergmann glial cells are derived from radial glia and guide immature GNPs to migrate from EGL to form a new internal granular layer (IGL). Murine postnatal (P7) cerebellum contains multipotent NSCs that are positive for the stem cell marker Prominin (Lee, et al., 2005). The cerebellum starts to mature by 3 weeks of age in mice and at around 2 years of age in humans (Raaf and Kernohan, 1944). Postnatally, Bergmann glia continue to express members of the SRY (sex determining region Y)-box (SOX) family of transcription factors SOX1 and SOX2 together with the stem cell marker Nestin and proliferate upon damage to the cerebellum (Sottile, et al., 2006, Stamatakis, et al., 2004). In contrast to adult forebrain NSCs, there is no clear proof of adult cerebellar NSCs that are still mitotic and that can undergo neurogenesis. Neurogenesis in the brain stem is somewhat different from that in the forebrain and in the cerebellum. The ventricular zone of the medulla and pons generates various brainstem nuclei, including the spinal trigeminal nucleus and the nucleus of the solitary tract (Altman and Bayer, 1980) in which Oligodendrocyte transcription factor 3 (OLIG3) and Pancreas transcription factor 1 subunit alpha (PTF1A) are essential in neuronal fate determination (Storm, et al., 2009). MATH1-positive cells from the lower rhombic lip structure migrate and give rise to mossy fiber neurons in the pontine gray nucleus (PGN) in the pons. In mammals, this process contributes to the greatest number of mossy fibers in the cerebellum where mossy fiber axons extend from PGN, cross the ventral brainstem midline, and enter the cerebellum (Cicirata, et al., 2005). Importantly, Zinc finger of the cerebellum 1 (ZIC1) is involved in the regulation of the pontine neuron cell body position and axonal projections within the pons (Dipietrantonio and Dymecki, 2009).
Medulloblastoma is a grade IV malignant childhood disorder that is capable of disseminating from its primary site of origin. The cells of origin for MB are not clearly identified, and it is likely that more than one type of cell can generate any of the four different molecular subtypes of MB: Wingless (WNT), SHH, Group 3 or Group 4 (Northcott, et al., 2012a). The primary site of human MB is not always intra-cerebellar as suspected when considering MB to be a cerebellar disorder. In fact, only SHH MBs are found as “intra-cerebellar tumors” while WNT, Group 3 and Group 4 tumors are either extra-cerebellar only, or a mix of intra- and extra-cerebellar tumors (Wefers, et al., 2014). Neural stem cells, GNPs or other neural progenitor cells from the lower RL of the developing dorsal spinal cord all give rise to MB in GEMMs (Figure 2) as discussed below. Genetic alterations in MB are often subgroup-specific like mutations in Catenin (cadherin-associated protein), beta 1 (CTNNB1) in the WNT subgroup, aberration in Patched 1 (PTCH1), Smoothened (SMO), Suppressor of fused homolog (SUFU) or GLI family zinc finger 2 (GLI2) in the SHH subgroup, amplification of v-myc avian myelocytomatosis viral oncogene homolog (MYC) in group 3 and amplifications of Orthodenticle homeobox 2 (OTX2) in group 3 and 4 (Jones, et al., 2012, Northcott, et al., 2009b, Northcott, et al., 2012b, Parsons, et al., 2011, Pugh, et al., 2012, Robinson, et al., 2012). On the other hand, some alterations are found across subgroups such as amplifications of v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog (MYCN) (Taylor, et al., 2012a). As will be discussed below, all of these genes have been clearly implicated in various stages of brain and neuronal development, thus further linking MB formation to deregulated neurogenesis.
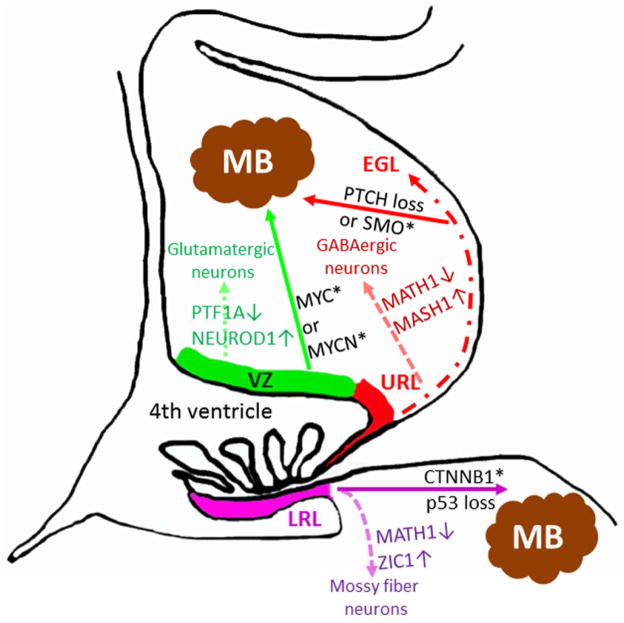
Figure 2. Transformation of neuronal precursors disrupts neurogenesis and drives MB formation.
Schematic figure of the cerebellum and the dorsal brain stem during embryonic development. Various developmental pathways (dotted arrows) drives glutamatergic or GABAergic neurogenesis from stem cells in the ventricular zone (VZ) (green) or from granule neuron precursors in the upper rhombic lip (URL)/external granular layer (EGL) (red), respectively (for further details see (Swartling, et al., 2013)). Similarly, mossy fiber neurons are derived from the lower rhombic lip (LRL) (purple) of the brain stem (Cicirata, et al., 2005). Misexpression (activation/upregulation (*) or (loss) of important cancer genes including CTNNB1 (Gibson, et al., 2010), SMO/Ptch (Schuller, et al., 2008, Yang, et al., 2008), MYC/MYCN (Kawauchi, et al., 2012, Pei, et al., 2012, Swartling, et al., 2010, Swartling, et al., 2012) are shown in black. As described in this review such cancer genes are involved in driving medulloblastoma (MB) in a cell context-dependent fashion from cells within VZ, URL/EGL or LRL.
Disruption of neurogenesis as an initiating event for brain tumor formation
A growth factor-mediated neurogenic-gliogenic switch in forebrain precursor cells
The mechanisms that control cell fate in neocortical NSCs are well understood. Less is known how discrete populations of postnatal NSCs maintain their neurogenic potential, but evidence indicates that both cell-extrinsic signals and cell-intrinsic epigenetic mechanisms are required (Fuentealba, et al., 2012, Ming and Song, 2011). The output of newborn neurons reflects the size of the NSC pool and regulation of cell fate determinants in type B1 NSCs and type C cells. A delicate balance and coordination between neurogenic and gliogenic factors determine the cell fate of NSC progeny and can be disrupted during pathological conditions. Multipotent cortical progenitors are maintained in a proliferative state by basic helix-loop-helix (bHLH) factors from the Inhibitor of differentiation (ID) and hairy and enhancer of split (HES) families (Ross, et al., 2003). The neocortex is derived from the telencephalon, where the proneural gene Neurogenin 2 (NEUROG2) specifies a glutamatergic neuronal fate in dorsal progenitors (Fode, et al., 2000, Mattar, et al., 2008, Parras, et al., 2002, Schuurmans, et al., 2004), while the proneural gene Achaete-scute family bHLH transcription factor 1 (ASCL1) specifies the identities of neocortical GABAergic neurons and embryonic OPCs that are derived from ventral progenitors (Britz, et al., 2006, Casarosa, et al., 1999, Castro, et al., 2011, Parras, et al., 2007). During development, the transition from proliferation to neurogenesis involves a coordinated increase in the activity of proneural bHLH factors (ASCL1, NEUROG1, and NEUROG2) and a decrease in the activity of HES and ID factors. Elegant work from Alvarez-Buylla’s laboratory demonstrates that adult SVZ NSCs in distinct microdomains generate diverse types of olfactory bulb neurons (Merkle, et al., 2014, Merkle, et al., 2007). The transcription factor Paired box 6 (PAX6) promotes the neurogenic potential of cortical radial glia during CNS development (Heins, et al., 2002). In the adult SVZ, expression of PAX6 in TAPs and SVZ neuroblasts is required for generating olfactory bulb neurons of the dopaminergic lineage (Kohwi, et al., 2005). In contrast, expression of OLIG2 specifies the SVZ TAP fate, opposes the neurogenic role of PAX6, and promotes oligodendrogenesis (Hack, et al., 2005, Marshall, et al., 2005). Conversely, PAX6 overexpression inhibits cell-intrinsic expression of OLIG2 (Jang and Goldman, 2011). OLIG2 is expressed by different precursors of glial cells, including OPCs, the most wide-spread population of cycling cells in the adult brain (Geha, et al., 2010, Zhou and Anderson, 2002). Infusion of epidermal growth factor (EGF) or platelet-derived growth factor AA (PDGF-AA) into the lateral ventricles and SVZ of adult mice induces massive expansion of OLIG2+ cells and depletion of neurogenesis (Gonzalez-Perez, et al., 2009, Jackson, et al., 2006, Kuhn, et al., 1997). Establishment of murine embryonic ventricular zone and adult subventricular zone neurosphere cultures with EGF or fibroblast growth factor 2 (FGF-2) results in prominent down-regulation of transcription factors in telencephalic precursors and robust up-regulation of OLIG2 (Hack, et al., 2004). Several members of the SOX family of transcription factors play important roles in regulating neurogenesis (Wegner and Stolt, 2005). SOX9 is a glial fate marker responsible for generating astrocytes and oligodendrocytes in the developing brain and spinal cord (Stolt, et al., 2003). Transcriptionally, SOX9 regulates glial specification in collaboration with the Nuclear factor I family member (NFIA) by targeting the downstream effector Mouse double minute 2 homolog (MDM2) (Kang, et al., 2012). Therefore, high levels of SOX9 in glial precursor cells provide a barrier to neurogenesis. Detailed time-lapse studies monitoring OLIG2, ASCL1, and HES1 expression in NSCs derived from the ventral telencephalon show an oscillatory expression pattern, whereas the differentiated states correlated with sustained expression of single factors (Imayoshi, et al., 2013). In conclusion, this delicate balance of proneurogenic and progliogenic transcription factors is a prerequisite for normal neuronal and glial output in developing and adult germinal regions. Growth factor-mediated stimulation of neural precursor cells favors production of glial progeny, providing an explanation for how downstream signaling effectors can promote gliomagenesis.
Oncogenic RAS-MEK-ERK signaling as an initiating event during gliomagenesis
As a critical regulator of brain development, the RAS/extracellular-regulated kinase (ERK) signaling cascade influences neural cell fate determination by controlling the expression of transcription factors and bHLH factors (Li, et al., 2014b). Inactivation of Neurofibromin 1 (NF1), a RAS GTPase-activating protein, promotes ERK-dependent expression of OLIG2 specifically in ASCL1+ TAPs, leading to increased gliogenesis at the expense of neurogenesis in neonatal and adult SVZ (Wang, et al., 2012b). In utero transduction found that increased activation of RAS/ERK signaling in neocortical progenitors promoted a NEUROG2-ASCL1 genetic switch that promoted a glial cell fate (Wang, et al., 2012b). As a result, the authors found that activation of the ERK, but not other down-stream effectors of RAS, resulted in a massive expansion of OLIG2+ and SOX9+ progeny. Electroporation with the RASV12 oncogene resulted in glioma formation. Interestingly, this study demonstrated that RAS/ERK signaling regulated ASCL1 transcriptional activity by direct phosphorylation of multiple sites in dose-dependent manner. Moreover, they found that transactivation of the homebox genes Distal-less 1/2 (DLX1/2) and SOX9 reporters required phosphorylation of ASCL1. Their study elegantly demonstrates the importance of RAS/ERK activation for cell lineage determination in NSCs and progenitors. It also provides a basis for glioma initiation as a result of disrupted neurogenesis. In human gliomas, RAS activation is a common pro-proliferative event that is triggered by mutations of upstream receptor tyrosine kinases (RTKs), or downstream signaling components, such as NF1 or the proto-oncogene BRAF (Jones, et al., 2008, Louis, et al., 2007, McLendon, et al., 2008). Reduced NEUROG2 expression was necessary for self-renewal and tumorigenesis from BTSCs (Guichet, et al., 2013). Conversely, ASCL1 expression promoted self-renewal and tumorigenicity (Rheinbay, et al., 2013). We and others have employed GEMMs of glioma based on constitutive activation of the RAS-ERK pathway in NSCs, OPCs, or astrocytes (Acquaviva, et al., 2011, Chen, et al., 2012a, Ding, et al., 2001, Persson, et al., 2010, Zhu, et al., 2005). Aberrant RAS/ERK signaling in NSCs and OPCs blocked differentiation and promoted gliomagenesis (Jackson, et al., 2006, Persson, et al., 2010). To identify a relationship between NSC origin and treatment-resistant BTSCs in developing gliomas, a recent study generated malignant GBMs from SVZ NSCs through conditional inactivation of the tumor suppressors NF1, TP53, and Phosphatase and tensin homolog (PTEN) under the Nestin promoter (Chen, et al., 2012a). They traced a restricted population of Nestin-expressing cells that propagated GBMs in mice and regrowth following treatment with the alkylating agent temozolomide. In conclusion, a developmental neurogenic-gliogenic switch in transcription factor and bHLH expression represents an initiating step for RAS/ERK-driven glioma formation derived from NSCs. In OPC-derived gliomas, RAS-ERK signaling blocks differentiation and promotes a switch from asymmetric to symmetric cell division (Jiang, et al., 2011, Persson, et al., 2010, Sugiarto, et al., 2011). Pharmacological inhibition of RAS-ERK signaling not only rescues cell fate specification in NF1-deficient neural progenitors (Wang, et al., 2012b), but may also block gliomagenesis and restore normal differentiation.
In GBMs, approximately 45% of adult human cases harbor amplifications or activating mutations in the Epidermal growth factor receptor (EGFR) (Frattini, et al., 2013, McLendon, et al., 2008). Increased EGFR signaling promotes self-renewal of NSCs and promotes a neuronal-glial switch (Ayuso-Sacido, et al., 2010, Gonzalez-Perez, et al., 2009, Sun, et al., 2005b). Sensitive fluorescence in situ hybridization (FISH) experiments found that approximately 29% and 21% of pediatric and adult high-grade gliomas, respectively, harbor Platelet-derived growth factor A (PDGFRA) amplifications (Phillips, et al., 2013). Stimulation of PDGFRA expressed on SVZ NSCs or OPCs promotes gliogenesis (Gonzalez-Perez, et al., 2009, Jackson, et al., 2006, Noble, et al., 1988, Phillips, et al., 2013, Richardson, et al., 1988, Woodruff, et al., 2004). Approximately 18% of adult GBMs show mutations or homozygous deletions of the NF1 gene (Frattini, et al., 2013, McLendon, et al., 2008). As a negative regulator of RAS signaling, loss of NF1 in NSCs increased gliogenesis at the expense of neurogenesis in the neonatal and adult SVZ (Wang Y et al., Cell, 2012). The tumor suppressor RB1 also shows frequent deletions or inactivating mutations in GBM (Frattini, et al., 2013, McLendon, et al., 2008), while the normal functioning of RB1 has clearly been linked to neurogenesis and there are indications that loss of its function might lead to increased neural progenitor proliferation and reduced neuronal differentiation (reviewed in (Sage, 2012)). In summary, recent advances have defined oncogenic RAS-MEK-ERK signaling as an initiating event during gliomagenesis, leading to changes in transcription factors controlling cell fate determination and promoting aberrant gliogenesis.
Linking transcriptional regulators of hindbrain neurogenesis and medulloblastoma formation
Proneural genes are involved in early patterning and cell fate specification of the cerebellum. MATH1 is important for the generation of GNPs and for their expansion (Ben-Arie, et al., 1997). Similar to forebrain development, the Neurogenic differentiation 1 (NEUROD1) gene is involved in cerebellar neuronal differentiation and acts as a master gene in granule cell differentiation (Miyata, et al., 1999). Progenitor cells in the ventricular zone (VZ) close to the fourth ventricle express fate markers like PTF1A, NEUROG1 and NEUROG2. GABAergic interneuron precursors are produced from E13.5 to E16.5 from VZ progenitors (Maricich and Herrup, 1999). These interneuron precursors proliferate and express PAX2 until the first postnatal week when they migrate to the deep cerebellar nuclei (DCN). MASH1 suppresses gliogenesis and participates in the generation of GABAergic interneurons and (DCN) neurons. Members of the NOTCH and Bone morphogenic protein (BMP) families regulate cerebellar development in the VZ and in the roof plate, respectively. During embryogenesis BMPs stimulate production of glutamatergic neurons and expression of MATH1 (Alder, et al., 1999, Chizhikov, et al., 2006, Machold, et al., 2007). Depletion of NOTCH1 in the developing cerebellum leads to increased glutamatergic neurogenesis (Lutolf, et al., 2002, Machold, et al., 2007). Specifically, SHH signaling has been identified as a pathway stimulating the expansion and increased proliferation of glutamatergic cerebellar GNPs during their migration through the EGL in normal development of the cerebellum (Gilbertson and Ellison, 2008, Marshall, et al., 2014, Roussel and Hatten, 2011). Aberrant inactivation of SHH signaling antagonists, predominantly PTCH1 and SUFU, or activations of factors such as the GLI2 gene that positively regulate SHH signaling including SMO, have all been found altered in the SHH MB subgroup. Inactivation of SHH antagonists or activation of SHH signaling inhibit cell cycle exit of cerebellar GNPs, reduce differentiation, and lead to abnormal cell growth (Gilbertson and Ellison, 2008, Marshall, et al., 2014, Roussel and Hatten, 2011), suggesting that GNPs may represent the cell of origin of the human SHH MB subgroup. In fact, while GNPs can directly generate SHH MB in mice, hindbrain NSCs adopt a GNP fate upon SHH pathway stimulation by activated SMO (Schuller, et al., 2008) or PTCH1 depletion (Yang, et al., 2008). Binding of SHH ligand to its receptors, PTCH1 and SMO homologue, leads to the activation of GLI transcription factors (Lai, et al., 2003, Palma and Ruiz i Altaba, 2004). Genome sequencing identified mutations in several key genes in the SHH-pathway including PTCH1, SMO and SUFU in as many as 87% of SHH MBs (Kool, et al., 2014). PTCH1 mutations are found evenly distributed among infant, children and adult patients. On the other hand SMO mutations are only found in adult SHH MBs while SUFU mutations were exclusively found in infant SHH MBs. In mice, expression of an activated allele of SMOM2 in multipotent GFAP+/OLIG2+ progenitors or unipotent GNPs both resulted in MB formation (Schuller, et al., 2008). Tumors formed were histologically similar and MATH1 positive despite being generated from different cell types. It is also possible that even late stage GNPs can dedifferentiate in order to give rise to these tumors. However, when instead depleting PTCH1 in various cell types to drive SHH MBs, tumors generated from NSCs develop more rapidly than tumors generated in GNPs (Yang, et al., 2008). Interestingly, two recent reports suggest that also cells from the cochlear nuclei in the spinal cord (Grammel, et al., 2012) or rare quiescent Nestin-positive cells in the EGL that are MATH1 negative (Zhao, et al., 2013) can give rise to SHH MBs. The latter study suggests that MATH1 is not required for generating GNPs or for generating all types of SHH MBs.
The cell of origin for MB subtypes still remains unclear, and it is likely that more than one type of cell can generate any of the four different molecular subtypes of MB: WNT, SHH, Group 3 or Group 4 (Northcott, et al., 2012a) (Table 1). Neural stem cells, GNPs, or other neural progenitor cells from the lower RL of the developing dorsal spinal cord all give rise to MB in mouse models, as further discussed below (Figure 2).
Table 1.
Cell of origin for murine tumors matching human MB subgroups
| MB subgroup | Driver genes* | Pathways | Originating cell** |
|---|---|---|---|
| WNT | CTNNB1, TP53 loss | WNT | LRL progenitor of developing brain stem |
| SHH | SHH, SMO, SUFU, MYCN, GLI2, hTERT promoter mutations, TP53 loss, PTCH loss, | SHH | URL progenitor/EGL cell, Nes+ EGL cerebellar cell or choclear brain stem cell |
| Group 3 | MYC, GFI1/GFI1B, OTX2, LIN28B | Various | Cerebellar NSCs |
| Group 4 | MYCN, GFI1/GFI1B, KDM6A, CDK6, SNCAIP, LIN28B | Various | Unknown/cerebellar NSCs |
a vast selection of the most important cancer driver genes for these brain tumors
potential cellular origin as supported from literature and various brain tumor models
Meanwhile, it has become increasingly clear that brain tumor development shares many characteristics with normal tissue and cell development (Swartling, et al., 2013b) where the cell types in normal brain and neuronal development undergo various changes in proliferation, differentiation and apoptotic capacities (Swartling, et al., 2013a). In MB, a hint at this coupling between normal brain and tumor development can be gathered from the age dependence of tumor subgroups. Additionally it has been proposed that the clustering of adult MBs, in contrast to the four subgroups in pediatric MBs, only contain three subgroups with Group 4 showing also a different appearance and outcome for pediatric and adult cases (Remke, et al., 2011) and similar observations have been made for WNT and SHH groups (Northcott, et al., 2011). Finally, a previous study from our own research group that used an orthotopic mouse model transplanted with NSCs previously transduced with a stabilized form of MYCN has also clearly showed that the cancer type and the subgroup affiliation of the induced tumors is influenced not only by the brain region but also the time point at which NSCs have been dissected and transduced (Swartling, et al., 2012). In summary, it is reasonable to conclude that the intrinsic programs of the respective tumor cells of origins are directly coupled or reflect certain brain or neuronal developmental programs. Tumorigenesis in this case is also restricted to certain developmental time windows, in which the given signaling pathways are susceptible to alterations or mutations of the respective cancer gene (Gilbertson and Ellison, 2008, Liu and Zong, 2012, Marshall, et al., 2014, Sanai, et al., 2005). However, Liu et al. found that genetic mutations resulted in brain tumor formation only as NSCs differentiated into more restricted precursor cells (Liu, et al., 2011). In addition, temozolomide treatment of human gliomas can induce a hypermutated phenotype in recurrent tumors (Johnson, et al., 2014). Moreover, radiotherapy promotes a proneural-mesenchymal transition of high-grade gliomas (Halliday, et al., 2014, Mao, et al., 2013). These studies suggest that, rather than trying to delineate the cell of origin based on the resulting tumor phenotype, it is more informative to perform careful analyses of GEMMs to delineate the cells that are most susceptible to defined genetic mutations.
Mutational analysis and transcriptomal profiling of human medulloblastoma subgroups imply distinct developmental origins
Development of GEMMs for MB have identified spatially and temporally defined neural precursors that generate tumors with gene expression signatures for different human MB subgroups. (Gilbertson and Ellison, 2008, Hatten and Roussel, 2011, Liu and Zong, 2012, Marino, 2005, Marshall, et al., 2014, Roussel and Hatten, 2011). Specifically, SHH signaling has been identified as a pathway stimulating the expansion and increased proliferation of cerebellar GNPs and granule cell progenitors (GCPs) during their migration through the EGL in normal development of the cerebellum (Gilbertson and Ellison, 2008, Marshall, et al., 2014, Roussel and Hatten, 2011). GNPs might be likely cell of origin candidates of human SHH tumors and while GCPs can directly generate SHH MB in mice, hindbrain NSCs turn on a GCP fate upon SHH pathway stimulation by activated SMO (Schuller, et al., 2008) or PTCH1 depletion (Yang, et al., 2008). The tumorigenic properties of MB cells might be brought about by the loss of other signals antagonizing SHH activity (Fan and Eberhart, 2008, Hatten and Roussel, 2011, Roussel and Hatten, 2011). One example is the Potassium channel tetramerisation domain containing 11 (KCTD11, REN) gene, often deleted in MB due to loss of chromosome 17p, expression of which is thought to antagonize SHH-induced proliferation and thus stimulating differentiation of GCPs (Argenti, et al., 2005, Di Marcotullio, et al., 2004). Another example is given by the BMP pathway, effectors of which are often found transcriptionally down-regulated in MB, and that has been found to facilitate GNP differentiation by antagonizing SHH-induced proliferation (Zhao, et al., 2008).
Similar to the SHH signaling pathway, WNT signaling has also been implicated in neural development and neural cell differentiation and proliferation (Ciani and Salinas, 2005, Lange, et al., 2006, Pei, et al., 2012a). Specifically, active WNT signaling is thought to interfere with the formation of a multi protein complex including Adenomatous polyposis coli (APC), AXIN2 and β-Catenin, thus abrogating β-Catenin phosphorylation and subsequent degradation and instead leading to an increased accumulation of β-Catenin proteins in the cell, which then causes downstream activation of other WNT targets and eventually down-regulates neuronal differentiation and up-regulates cell proliferation (Gilbertson and Ellison, 2008, Marino, 2005, Pei, et al., 2012a). In addition to activation of WNT signaling, deletion of exon 3 in the β-Catenin encoding gene CTNNB1 is sufficient for β-Catenin accumulation in the cell due to evasion from the protein degradation machinery (Harada, et al., 1999, Zechner, et al., 2003). Since mutations activating WNT signaling (in particular deletions of exon 3 in the CTNNB1 gene) are frequent in the WNT subgroup, it is reasonable to conclude that this is one factor determining the aberrant proliferation of this MB subgroup (Marshall, et al., 2014), although these alterations alone appear not sufficient for MB development (Pei, et al., 2012a). These WNT MBs have been suggested to derive from lower rhombic lip or embryonic dorsal brainstem progenitors (Gibson, et al., 2010).
Notch signaling plays a major role coordinating proliferation and differentiation in NSCs, GNPs and Bergmann glia in forebrain and hindbrain germinal regions (Helgesson, et al., 2007, Kool, et al., 2014, Machold, et al., 2007, Mathieu, et al., 2010, Rousseau-Merck, et al., 1985). Specifically, it has been shown that NOTCH1 is required for GNP differentiation (Kang, et al., 2012), while NOTCH2 stimulates GNP proliferation (Kool, et al., 2014). Since NOTCH1 expression levels are often found low and Notch2 expression is found upregulated in MB and other brain tumors (Fan, et al., 2004, Wefers, et al., 2014), deregulated Notch signaling might resemble another pathway that is contributing to tumorigenic proliferation in these cancers.
Hindbrain NSCs are suggested to represent the cell of origin for Group 3 and Group 4 MBs in GEMMs driven by MYC proteins (Kawauchi, et al., 2012, Pei, et al., 2012b, Swartling, et al., 2010, Swartling, et al., 2012). However, less is known about the intrinsic program of Group 3 and Group 4 MB, although there exist several hints at pathways or genes, which are ordinarly contributing to normal neural differentiation and proliferation but are deregulated in these tumors.
Aberrations in the MYC family genes, especially characterized by amplification of MYC and MYCN, are common in group 3 and 4 MB. It is well known that both MYC and MYCN transcription factors play roles in normal brain development, including cerebellar development, as well as in determining the fate, proliferation and differentiation of neural progenitors (Knoepfler, et al., 2002, Wey and Knoepfler, 2010, Wey, et al., 2010, Zinin, et al., 2014). Additionally, MYC proteins have been widely implicated in various aspects of brain tumor biology (reviewed in (Swartling, 2012)). It has been shown that MYCN is a downstream target of SHH signaling and is often required for the induction of SHH induced proliferation discussed above (Hatton, et al., 2006, Kenney, et al., 2003, Oliver, et al., 2003), explaining why amplification or upregulated expression of MYCN is often seen in SHH-dependent MBs. However, amplification and aberrant expression of MYC as well as amplification of MYCN are also recognized as some of the most recurrent tumorigenic alterations in MB subgroup 3 and 4, respectively, and generally correlate with a poor clinical prognosis (Kool, et al., 2012, Korshunov, et al., 2012). Thus it is clear that SHH-independent aberrant expression of MYC and MYCN might be key factors determining the deregulated proliferation and differentiation properties of neural progenitors in these tumor subtypes, which is also underscored by the fact that current mouse models of these subgroups all depend on either MYC (Kawauchi, et al., 2012, Pei, et al., 2012b) or MYCN (Swartling, et al., 2010, Swartling, et al., 2012).
There is a possible role for members of the OTX gene family in MB tumorigenesis (de Haas, et al., 2006) and in driving and defining especially the group 3 and group 4 subclasses (Marshall, et al., 2014). Specific patterns of OTX1 and OTX2 have been found delineating borders and layers between and within hindbrain, midbrain and forebrain and regulated expression of these transcription factors has been assumed important for controlling neurogenesis, neural progenitor identity and fate as well as local brain regionalization (Pennartz, et al., 2004, Rajendran, et al., 2008, Su, et al., 2009, Sun, et al., 2005a, Zupanc, et al., 2005). Importantly, OTX2 has been shown to induce proliferation and impede differentiation of cancer cells in MB (Adamson, et al., 2010, Lachyankar, et al., 2000, Ross, et al., 2001), and since it is often found amplified in group 3 and group 4 MBs (Adamson, et al., 2010, Northcott, et al., 2012b), it might be argued to be a driver of those subgroups. Most recently, growth factor independent 1 (GFI1) family proto-oncogenes were found as common drivers of Group 3 and Group 4 MB (Northcott, et al., 2014). Interestingly, in tumor cells GFI1 genes are altered and directed proximal to active enhancer elements in the genome where they promote tumorigenesis. In conclusion, studies in mice have shown that genetic alterations and expression of transcription factors found in human MB subgroups lead to changes in developmental programs, known to play important roles for cell homeostasis and cell fate determination (Table 1).
Epigenetics, neurogenesis, and brain tumor formation
Epigenetics, heritable changes in gene activity that are not caused by changes in the DNA sequence, governs fine-tuning and precision of gene expression programs that define the molecular basis of differentiation and reprogramming in NSCs and more restricted progenitors. As NSCs differentiate, the genome becomes more transcriptionally restrained, due to chromatin condensation and maturation of heterochromatin. Epigenetic mechanisms include post-translational modifications of histones, incorporation of histone variants, changes in DNA methylation, chromatin remodeling, and implementation of RNAi pathways and non-protein coding RNAs. Recent molecular characterization of malignant gliomas and MBs show that genetic alterations in cancer-causing genes produce broad epigenetic changes that play important roles in tumor initiation and progression.
Regulation of neurogenesis through post-translational modification of histones and DNA
Acetylation of the core histones H3, H4, H2A, and H2B is normally associated with a more relaxed chromatin conformation and thus activation of gene transcription. Opposite actions of histone acetyltransferases (HATs) and histone deacetylases (HDACs) on histone acetylation in NSCs regulate self-renewal and differentiation (Balasubramaniyan, et al., 2006). HDACs can form complexes with the DNA binding protein REST (RE1 silencing transcription factor; also called NRSF) and its co-repressor CoREST to repress expression of neuronal genes (Bruce, et al., 2004, Lunyak, et al., 2002). As NSCs differentiate into neurons, REST and its co-repressors dissociate from the REST binding site (RE1), triggering activation of neuronal genes (Ballas, et al., 2005).
In addition to post-translational modifications of histones, the methylation status of DNA plays critical roles in the regulation of gene expression during development (Bird, 2002). DNA methylation predominantly occurs at the cytosine residue of CpG dinucleotides to generate methylation of the 5′ site (5-mC) on the pyrimidine ring. DNA methyltransferases (DNMTs) and members of the methyl-CpG-binding domain (MBD) family are responsible for the generation and maintenance of methylation patterns in the DNA sequence (Nan, et al., 1998, Smith and Meissner, 2013). Although previously thought to be an irreversible process, recent studies suggest that reversible DNA methylation occurs (Popp, et al., 2010, Rai, et al., 2008).
In addition to 5-mC methylation of DNA, histone lysine methylation regulates transcription and modulates other chromatin-related processes such as replication (Zhang and Reinberg, 2001). The lysine (K) residues of histones H3 and H4 can be mono-, di-, and tri-methylated, whereas arginine (R) can be mono- or dimethylated. Specific histone methylation marks are associated with transcriptional activation and/or repression. In contrast to differentiated cells, members of the Ten-11 translocation family TET1-3 have the capacity to oxidate 5-mC to 5-hydroxymethyl-cytosine (5-hmC), thought to be an intermediary toward 5-mC demethylation (Tahiliani, et al., 2009). Loss of or TET2-3 leads to defective neuronal differentiation, suggesting that formation of 5-hmc and loss of H3K27me3 promotes brain development (Hahn, et al., 2013). In fact, increased 5-hmc levels during embryonic mouse brain development parallel neuronal differentiation where it preferentially associates with gene bodies of activated neuronal function-related genes (Hahn, et al., 2013). H3K4me3 and H3K27me3 mark genes that are actively transcribed or repressed, respectively. The gain of 5-hmC DNA methylation is often accompanied by the loss of H3K27me3 during neuronal differentiation (Hahn, et al., 2013). Histone methylation is also regulated by repressive Polycomb group (PcG) and activating Trithorax (TrxG) chromatin modifying complexes. The PcG gene Polycomb repressive complex 2 (PRC2) acts to stabilize repressive chromatin structure through the function of chromatin modifiers, all of which are Histone methyltransferases (HMTs) responsible for depositing H3K27me2 and H3K27me3 marks on chromatin (Cao, et al., 2002). PRC2 contains enhancer of zeste 2 (EZH2), a catalytically active core component that trimethylates H3K27 (Pereira, et al., 2010). A recent study found that expression of EZH2 in SVZ NSCs was required for NSC proliferation and neuronal differentiation through direct repression of cyclin-dependent kinase inhibitor 2A (CDK2A; Ink4a) and OLIG2 transcription, respectively (Hwang WW et al., Elife, 2014). In contrast to PcG genes, the TRXG group of histone modifiers mediates post-translational modifications of active transcription such as H3K4me3. PcG-mediated gene silencing can be antagonized by mixed-lineage leukemia (MLL) HMTs that catalyze methylation of H3K4 or the Jumonji-family of proteins that removes methylation of H3K27me3. The TRXG member MLL1 is required for neurogenesis in the postnatal mouse brain (Lim, et al., 2009). MLL1-deficient SVZ NSCs progeny express the early proneural transcription factor ASCL1 and gliogenic OLIG2, but not DLX2, a key downstream regulator of neurogenesis. MLL1 deficiency leads to a switch from high levels of H3K4me3 to bivalent expression of H3K4me3 and H3K27me3 at the DLX2 loci (Lim, et al., 2009).
Dynamic DNA methylation of bHLHs and members of the SOXC family have been implicated for both embryonic and adult neurogenesis ((see review (Covic, et al., 2010)). A closed conformation by H3K27me marks of NEUROG1, NEUROG2, and NeuroD2 promoters and an open conformation (no H3K27me3) of SOX4 and SOX11 promoters suggest that SOXC family members in NSCs may control proneuronal gene expression (Mikkelsen, et al., 2007, Mohn, et al., 2008). Other PcG and TRXG chromatin remodeling genes that regulate NSC self-renewal and neuronal differentiation includes polycomb complex protein BMI1 and the Chromodomain-helicase-DNA-binding 7 (CHD7) protein (Fasano, et al., 2009, Feng, et al., 2013, Molofsky, et al., 2005, Molofsky, et al., 2003). In recent years, extensive literature has demonstrated the many roles that post-translational modifications of histones and DNA play in NSC biology.
miRNAs function as epigenetic modifiers and regulate neurogenesis
As 98% of the transcriptional output in mammalians consist of non-protein coding RNAs (ncRNAs), they regulate chromosome dynamics, chromatin modification, and epigenetic memory (Mattick, 2003). For example, small 21-nucleotide long microRNAs (miRNAs) regulate the stability and translation of target mRNAs. Knockouts of the miRNA processing enzymes Dicer and DGCR8 abolish the ability of embryonic stem cells to silence their self-renewal program and lead to severe defects in their ability to differentiate (Murchison, et al., 2005, Wang, et al., 2007). Down-regulation of REST removes the repression on miR9/9* and miR124 promoters, leading to a switch in the chromatin remodeling complex mSWI/SNF subunit composition and differentiation into neurons (Yoo, et al., 2009). Up-regulation of miR-184 and miR137 is mediated by the MBD proteins MBD1 and Methyl-CpG-binding protein 2 (MeCP2), respectively, and promotes neuronal differentiation during development (Liu, et al., 2010, Szulwach, et al., 2010). A regulatory circuit between let-7 miRNA and the RNA-binding protein Lin28 regulates neurogenesis in NSCs. As cells undergo neuronal differentiation, let-7 down-regulates expression of genes important for self-renewal including ASCL1, NEUROG1, HMGA2, Lin28, MYC, MYCN, TLX (Rehfeld, et al., 2014). Let-7-mediated down-regulation of TLX indirectly increases expression of cell cycle inhibitors and miR-9 (Zhao, et al., 2013). Let-7 also targets the oncofetal mRNA-binding protein Imp1, necessary for transition from proliferative fetal NSCs to more quiescent adult NSCs (Nishino, et al., 2013). In conclusion, we have described examples where both a regulatory circuit of let-7 and lin28 miRNAs, or functions of miRNAs as epigenetic modifiers, can regulate neurogenesis and proliferation of neural precursors.
Distinct mutations are associated with subgroup-specific methylation signatures in human GBMs
Deregulation of chromatin structure by DNA methylation, acetylation and methylation of histones, and nucleosome repositioning, have all been linked to tumor suppressor gene silencing, cancer initiation and progression of brain tumors (Spyropoulou, et al., 2013). Genetic alterations that impact epigenetic regulation of genes may block differentiation and initiate brain tumor formation. Mutation of the Isocitrate dehydrogenase (IDH1) or IDH2 genes in diffuse and high-grade gliomas lead to global hypermethylation of CpG islands (Noushmehr, et al., 2010, Parsons, et al., 2008). Diffuse gliomas and GBMs displaying IDH1/2 mutations were classified into the proneural transcriptomal subgroup (Cooper, et al., 2010, Verhaak, et al., 2010, Yan, et al., 2009). Although the IDH1R132H mutation is present in astrocytic and oligodendrocytic tumors, the mutation is often co-expressed with TP53 mutation in astrocytoma while it is associated with combined loss of chromosome 1p and 19q in oligodendroglioma. Despite a transcriptional signature similar to other proneural tumors, IDH1R132H mutant gliomas show accumulation of (2-HG) and a hypermethylated phenotype (Chaumeil, et al., 2013, Turcan, et al., 2012). Introduction of the IDH1R132H mutation increased expression of the NSC marker Nestin in immortalized human astrocytes and promoted a glial-neuronal switch in murine forebrain NSCs (Lu, et al., 2012, Turcan, et al., 2012) (Figure 1). The inability of IDH1R132H-transduced NSCs to demethylate histones was proposed to block differentiation. Exome sequencing of high-grade pediatric gliomas, including supratentorial GBM and diffuse intrinsic pontine gliomas (DIPG) identified missense mutations Lys27Met (K27M) and Gly34Arg/Val (G34R/V) in genes encoding histone H3.3 (H3F3A) and H3.1 (HIST3H1B) (Schwartzentruber, et al., 2012, Wu, et al., 2012). Interestingly, Sturm and colleagues found IDH mutations occurred in a distinct group of patient samples from H3F3A mutants, and the K27 and G34 mutations were mutually exclusive and clustered into their own subgroup when examining gene expression and methylation profile of tumors harboring these mutations (Sturm, et al., 2012). Anatomically, the K27-mutant tumors arise in pontine and more rostral midline brain structures, whereas the G34-mutant tumors are usually hemispheric and found in the forebrain. K27 mutations occur in children while G34 mutations occur in adolescents and IDH mutations were found in young adults (Khuong-Quang, et al., 2012, Sturm, et al., 2012). G34 mutations correlated with hypermethylation and silencing of oligodendrocyte lineage genes such as OLIG1 and OLIG2, while expressing the forebrain marker FOXG1. In contrast, K27 mutant tumors express OLIG1 and OLIG2, but not FOXG1 (Sturm, et al., 2012). Mutation of the chromatin remodeling gene Alpha thalassemia/mental retardation syndrome X-linked (ATRX) gene is more tightly associated with older patients and found in all tumors with G34 mutations in the Schwartzentruber et al study (Schwartzentruber, et al., 2012, Sturm, et al., 2012). Hypomethylation in G34 mutant tumors is especially prominent at chromosome ends, which may provide a link with alternative lengthening of telomeres (ALT), a phenomenon commonly observed with ATRX mutations (Sturm et al. 2012; Schwartzentruber et al. 2012). Overexpression of the G34 mutation induces expression of MYCN in normal human astrocytes and fetal glial cells (Khuong-Quang et al. 2012; Bjerke et al. 2013) (Figure 1). In summary, cohorts of human GBMs show IDH1/2 and H3F3A mutations that produce distinct methylation signatures, correlate with age of patients, and can be associated to certain brain regions. It is therefore plausible that these tumors derive from different populations of precursor cells.
Subgroup-specific epigenetic regulators in human medulloblastoma
In recent years, several lines of evidence have emerged implicating epigenetic deregulation in the formation of MB (Batora, et al., 2014, Dubuc, et al., 2012, Hovestadt, et al., 2014, Jones, et al., 2013). The events in the underlying epigenome are exemplified mainly by aberrant patterns of DNA methylation, genomic alterations of the chromatin modeling and modifying machinery resulting also in deregulated patterns of histone modifications, as well as aberrant expression of numerous miRNAs. Initial studies on the role of these deregulations appear to converge on a common scheme by which these epigenetic aberrations might ablate tumor cell differentiation and enforce the maintenance of progenitor and proliferating cellular states (Batora, et al., 2014, Dubuc, et al., 2012, Jones, et al., 2013). A high degree of methylation in MB patients has been found to associate with a worsened clinical prognosis (Pfister, et al., 2007), while DNA methylation profiles in general have recently been shown to effectively reinforce the robustness of four distinct molecular subtypes of MB: Wingless (WNT), Sonic Hedgehog (SHH), Group 3 and Group 4 (Hovestadt, et al., 2013, Northcott, et al., 2012a, Schwalbe, et al., 2013). Targeted screens for aberrantly methylated gene loci revealed hypermethylation induced silencing of tumor suppressor genes such as the cell growth regulator Hypermethylated in cancer 1 (HIC1) (Rood, et al., 2002), the cell cycle regulator Cyclin-dependent kinase inhibitor 2A (CDKN2A) (Fruhwald, et al., 2001) or the SHH signaling and WNT signaling antagonists PTCH1 (Diede, et al., 2010) and SFRP family members (Kongkham, et al., 2010). A recent study by Hovestadt et al. has further employed whole genome bisulfite sequencing coupled to RNA sequencing data to detect regions of DNA methylation and expression downstream of promoters in 34 human MBs (Hovestadt, et al., 2014). The study showed that overall DNA methylation and expression of genes displayed clear differences between MB subgroups, exemplified by a novel promoter of Lin-28 Homolog B (LIN28B), which was hypomethylated and thus leading to upregulated gene expression in almost all Group 3 and Group 4 MBs. LIN28B plays a role in downregulating the expression of tumor suppressors in the LET-7 miRNA family and the study showed that increased expression of LIN28B correlated clearly with a worsened prognosis in these MB subgroups. Further efforts in detecting deregulated epigenetic mechanisms in MB formation have also revealed numerous miRNAs with up- and downregulated expression levels and potential implications in the onset of this disease (reviewed by (Batora, et al., 2014)). Downregulation of miRNAs in MB affects the tumor suppressor miR-218 (Venkataraman, et al., 2013), a cluster of miRNAs (miR-125b, miR-126, miR-324-5p and miR-326) that regulate proliferation by antagonizing SHH signaling (Ferretti, et al., 2008). Induced expression of miR-128a inhibits MB cell growth by downregulating the polycomb complex protein and oncogene BMI-1 (Venkataraman, et al., 2010). Furthermore, overexpression of miR-34a reduces MB cell proliferation and stimulates neural differentiation (de Antonellis, et al., 2011, Weeraratne, et al., 2011). MiR-17/92 is another important miRNA highly expressed in MB that operates downstream of the SHH pathway and has been suggested to act in concert with SHH signaling to promote MB cell proliferation (Northcott, et al., 2009a, Uziel, et al., 2009). Finally, methylation of miR-9, the inhibitor of WNT signaling Dickkopf 3 (DKK3), and Kruppel-like factor 4 (KLF4) genes in human MB represents additional examples of epigenetic silencing (Fiaschetti G et al., Br J Cancer, 2014; Valdora F et al., Int J Mol Sci, 2013; Nakahara Y et al., Neoplasia, 2010).
The most intriguing implication of epigenetics in MB formation is via deregulated histone modifying and chromatin remodeling functions. Several large-scale genomic screens performed on MB patients, have consistently reported on alterations, predominantly somatic mutations but also copy number aberrations, in various HATs, HDACs, HMTs and Histone demethylases (HDMs) (Jones, et al., 2012, Northcott, et al., 2009b, Parsons, et al., 2008, Pugh, et al., 2012, Robinson, et al., 2012). A comprehensive list of the most prominent, recurrently mutated histone modifiers can be found in (Batora, et al., 2014, Jones, et al., 2013, Northcott, et al., 2012a). Specifically, the Lysine (K)-specific demethylase 6A (KDM6A) gene represents the most frequently mutated gene in MB subgroup 4 and these events comprise largely non-sense mutations, suggesting a loss of its H3K27me3/2 demethylating capacity (Northcott, et al., 2012a). Due to its ability to remove inactivating H3K27me3 marks, KDM6A has been implicated in facilitating cellular differentiation (Agger, et al., 2007) and promoting ES cell-derived neuronal differentiation (Shahhoseini, et al., 2013). In contrast, MLL2 and MLL3 are the most frequently mutated H3K4 methylases in MB, occurring with mutation rates of 5.8% (Northcott, et al., 2012a)/8% (Dubuc, et al., 2013) and 2.6% (Northcott, et al., 2012a), respectively. A majority of such mutations has been interpreted as loss-of-function events (Parsons, et al., 2011), suggesting an ablation of H3K4me3 operability, which is needed for induction of cell differentiation. Interestingly, mutations in MLL2 and KDM6A were found to be mutually exclusive, suggesting alternative but converging ways of maintaining a K27+ phenotype mainly in MB subgroups 3 and 4, a picture that is further supported when considering additional alterations affecting the H3K27/H3K4 machinery (Dubuc, et al., 2013). This scheme is also supportive of a previous finding by Robinson et al. (2012), documenting a mutual exclusivity between EZH2 overexpression and the abrogated function of KDM6A or other epigenetic effectors such as Zinc finger, MYM-type 3 (ZMYM3) in group 4 or CHD7 in group 3 and 4 (Robinson, et al., 2012). Northcott et al. (2009) further showed a loss of the heterochromatic H3K9me2 mark in 41% of investigated MB patients (Northcott, et al., 2009b), suggesting a second avenue of epigenetic events with a converging phenotype in MB (Dubuc, et al., 2013). Finally, recurrent mutations also frequently effect chromatin remodelers (Batora, et al., 2014, Jones, et al., 2013, Northcott, et al., 2012a), the most prominent of which is the SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 (SMARCA4), which is enriched in WNT and group 3 tumors (Jones, et al., 2012, Pugh, et al., 2012, Robinson, et al., 2012). Interestingly, the chromatin remodeling complex SMARCA4, has previously been proposed to function as a neuronal-glial cell fate switch in NSCs (Matsumoto, et al., 2006). Further work is needed to better understand how genetic alterations in MBs influence epigenetics as a function of subgroup, with the goal of developing epigenetic-based therapies (for examples see later section of this review).
Neuronal differentiation of malignant brain tumors
Post-mitotic neurons display growth arrest and share few similarities with malignant brain tumors. Although a recent study found that lentiviral transformation of post-mitotic neurons can generate GBMs in mice (Friedmann-Morvinski, et al., 2012), glial precursor cells and GNPs, both able to differentiate into neuronal progeny, represent more likely origins for gliomas and MB. As described in a previous section, cancer-inducing genes can block neuronal differentiation and promote a glial phenotype as malignant gliomas develop. Still, human malignant glioma cells express proteins associated with an immature neuronal phenotype, including Class III beta-tubulin (TujI) and Doublecortin (DCX) (Yan, et al., 2011). In contrast, the majority of MB cells express proteins that are normally abundant in immature or mature neurons (Lastowska, et al., 2013, Maraziotis, et al., 1992). It raises the question of whether the extent of neuronal differentiation in gliomas and MBs is associated with a more differentiated phenotype and reduced tumorigenicity. As described earlier, proneural transcription factors promote neurogenesis during CNS development and in adult NSCs. However, the proneural genes PAX6, NEUROG2, and ASCL1 can individually function as master regulators and induce functional neurons when introduced into cultured primary astroglial cells from the early postnatal cerebral cortex (Berninger, et al., 2007, Heins, et al., 2002). Expression of NEUROD1 in reactive astrocytes and NG2+ OPCs induced glutaminergic neurons, or a combination of glutaminergic and GABAergic neurons, respectively. Even mouse and human fibroblasts can be reprogrammed to transdifferentiate into neurons following expression of ASCL1 and two other neurogenic transcription factors (Torper, et al., 2013, Vierbuchen, et al., 2010). In human pediatric and adult brain tumors, subpopulations of stem-like BTSCs can differentiate into an immature neuronal phenotype (Hemmati, et al., 2003, Singh, et al., 2003).
To assess neuronal differentiation, most studies rely on expression of TujI, a marker of immature neurons (Katsetos, et al., 2001, Singh, et al., 2003). As a marker of migrating type A neuroblasts of the SVZ and SGZ, DCX is also expressed in invasive malignant gliomas (Daou, et al., 2005). Ectopic expression of DCX in human GBM cells effectively inhibited invasion, reduced self-renewal, inhibited tumorigenicity, and induced terminal differentiation into neuron-like cells (Santra, et al., 2011, Santra, et al., 2009). Introduction of NEUROG2 or NEUROD1 in GBM BTSCs resulted in massive cell death, proliferation arrest, and the few surviving cells differentiated into neuron-like cells (Guichet, et al., 2013). Overexpression of PAX6 reduced tumorigenicity of human GBM cells and in a platelet-derived growth factor B (PDGFB)-driven murine glioma model (Appolloni, et al., 2012, Zhou, et al., 2005). SOX9 is a glial fate marker responsible for generating astrocytes and oligodendrocytes in the developing brain and spinal cord (Stolt, et al., 2003). Transcriptionally, SOX9 regulates glial specification in collaboration with the nuclear factor I family member, NFIA by targeting the downstream effector Mmd2 (Kang, et al., 2012). SOX9 is further under control of miR-124 which is essential for neurogenesis driven from adult stem cells residing in the lateral ventricles. SOX9 is required for NSC maintenance and multipotency. In NSCs, SOX9 is a direct target of miR-124a, the most highly expressed miRNA in the adult brain (Cheng, et al., 2009). The absent/low levels of miR-124a in NSCs increase as NSCs become activated and differentiate into neurons. Overexpression of miR-124a effectively increased neuronal differentiation in mouse SVZ (Akerblom, et al., 2012, Silber, et al., 2008). We have previously shown that miR-124a, and to some extent miR-137 induction, increased neuronal differentiation in GBM tumorsphere cultures (Silber, et al., 2008). More surprisingly, miR-124a induction generated neuronal progeny from OPC-derived murine oligodendroglioma cells (Silber, et al., 2008). It is possible that activated RAS/MEK signaling in OPC-derived oligodendroglioma cells regulate other SOX genes (such as SOX10) that function as barriers to oligodendrocyte differentiation. However, SOX9 might be a target for miR-124a and enable aberrant neuronal differentiation in both normal OPCs and OPC-derived gliomas (Persson, et al., 2010). Hypermethylation of miR-124a was found in 82% of human GBMs and regulated by activity of REST (Tivnan, et al., 2014). Therefore, REST inhibitors or demethylating agents may represent effective therapies in GBM. In support of this notion, a recent report demonstrated that BTSCs in human GBMs are highly sensitive to Lysine (K)-specific demethylase 1 (LSD1) suppression, leading to a collapse of the stem cell phenotype and reduced tumorigenicity (Suva, et al., 2014). Altogether, these studies suggest that neurogenic factors promote neuronal differentiation and reduce tumorigenicity in human malignant gliomas. It remains unclear if malignant brain tumor cells expressing neuronal markers show properties of normal neurons.
Both diffusible and membrane-bound factors from hippocampal astrocytes can promote proliferation and neuronal fate commitment of hippocampal NSCs (Song, et al., 2002). In hippocampal co-culture experiments from mice lacking the intermediate filament proteins GFAP and Vimentin, reduced endocytosis of the NOTCH1 ligand Jagged1 and decreased Notch1 signaling in primary astrocytes resulted in increased neuronal differentiation of co-cultured neurospheres (Wilhelmsson, et al., 2012). High expression of GFAP and Vimentin in astrocytic gliomas may therefore inhibit neuronal differentiation. Several studies suggest incomplete neuronal differentiation in malignant gliomas and MBs (Varghese, et al., 2008). Differentiation protocols show that neuronal differentiation in GBM seems to be inhibited at the stage of the neuronal intermediate phenotype (Wolanczyk, et al., 2010). As a result of imperfect differentiation arrest differentiated human GBMs co-express neural and mesenchymal markers (Rieske, et al., 2009). However, differentiation of GBM BTSCs into a neuronal phenotype showed that differentiated cells displayed high electrical membrane resistance and the ability to generate action potentials, consistent with more mature neurons (Varghese, et al., 2008). Furthermore, neuronal differentiation was only observed following short-term passaging of human GBMs xenografts (Higgins, et al., 2013). In a later section, we will discuss the therapeutic potential of differentiating brain tumors into neurons and other differentiated phenotypes. Current studies suggest that neuronal differentiation in human brain tumors only reaches an intermediate stage, potentially avoiding aberrant integration with surrounding normal brain and seizures.
Pluripotent brain tumors - beyond neural differentiation
Initial studies of human primary GBM cells demonstrated these cells to be multipotent; capable of differentiating along neuronal and glial cell lineages. Recent studies have shown that subpopulations of stem-like human GBM cells can differentiate into endothelial cells, pericytes, and into multiple mesenchymal lineages (Cheng, et al., 2013, Ricci-Vitiani, et al., 2010, Ricci-Vitiani, et al., 2008, Tso, et al., 2006, Wang, et al., 2010). How can GBMs originating from a neuroectodermal cell differentiate along mesenchymal cell lineages? Introduction of the two transcription factors CCAAT/enhancer-binding protein β (C/EBPβ) and Signal transducer and activator of transcription 3 (STAT3) reprogramed fetal human NSCs towards a mesenchymal phenotype and blocked differentiation along neural cell lineages (Carro, et al., 2010). Recent studies found that radiation induces a proneural-mesenchymal shift in a murine proneural glioma model and human GBMs (Halliday, et al., 2014, Mao, et al., 2013). Interestingly, the authors found that transcriptional targets down-stream of C/EBPβ and STAT3 signaling were induced in the murine gliomas following radiation (Halliday, et al., 2014). In human GBMs, depletion of C/EBPβ and STAT3 collapsed the mesenchymal signature and reduced aggressiveness (Carro, et al., 2010). These data suggest that transformation of multipotent NSCs or more differentiated progeny can give rise to GBM cells that are pluripotent. Such plasticity of brain tumor cells should provide a survival benefit to changes in the tumor microenvironment following therapy.
Pluripotency of somatic cells was elegantly demonstrated by Yamanaka and colleagues in a landmark study showing that four factors (OCT4, SOX2, MYC and KLF4) can convert human skin fibroblasts into induced pluripotent stem (iPS) cells (Takahashi and Yamanaka, 2006). More recent studies found that other factors (like LIN28 and NANOG) can also promote generation of iPS cells (Park, et al., 2008). Conversion of adult multipotent NSCs into iPS cells required even fewer factors (excluding MYC and SOX2) (Kim, et al., 2008). High levels of these reprogramming factors are expressed in aggressive human cancers, including malignant gliomas (Ben-Porath, et al., 2008). Low- and high-grade human gliomas express NANOG, KLF4, SOX2, and MYC, where NANOG expression was found as a predictor of survival (Elsir, et al., 2014). In this section, we will review the effects of these reprogramming factors on pluripotency and tumorigenicity in malignant brain tumors. The three transcriptional factors NANOG, OCT4 and SOX2 are thought to play central roles in cancer stem cells and embryonic stem (ES) cells (Kashyap, et al., 2009). The theory was supported in glioma by a study of Guo et al. (Guo, et al., 2011), in which they demonstrate expression of NANOG, OCT4 and SOX2 in the majority of GBM BTSCs. Interestingly, expression of those transcription factors was not only associated with Nestin-positive NSCs, but they were also dependent of the grade in gliomas, suggesting their involvement in glioma progression (Guo, et al., 2011, Yang, et al., 2013) and increased malignancy (Holmberg, et al., 2011). Over-expression of NANOG, OCT4 and SOX2 in cultured human glioma samples further demonstrates their importance in stemness maintenance (Clement, et al., 2007). Similar to GBMs, expression of the cell-surface antigen CD133 (AC133) has been suggested to label a subpopulation of BTSCs in human MB (Hemmati, et al., 2003, Singh, et al., 2003). However, CD15 rather than CD133 seems to be a tumor propagating marker for SHH-driven MB (Read, et al., 2009, Ward, et al., 2009). Most studies have investigated BTSCs in GBMs while reprogramming ability and multi-lineage potential is less known for MB BTSCs.
NANOG is a transcription factor involved in maintaining self-renewal capacity of ES cells (Cavaleri and Schöler, 2003). NANOG is often found to be enriched in subset of glioma cells positive for certain stemness markers (Clement, et al., 2007, Niu, et al., 2011, Zbinden, et al., 2010). GLI1 is an important effector of the SHH signaling pathway and is up-regulated together with NANOG in human glioma (Clement, et al., 2007, Zbinden, et al., 2010). Indeed, NANOG is activated through the SHH mediator GLI1 in both NSCs and mouse and human MB stem cells (Po, et al., 2010), suggesting NANOG involvement in SHH-driven stemness of brain tumors. The tumor suppressor TP53 is often mutated in human glioma patients (England, et al., 2013) and seems to be an important negative regulator of GLI1/NANOG signaling (Zbinden, et al., 2010). Another study found that self-renewal in NSCs is promoted by NANOG (Garg, et al., 2013). NANOG was found to increases the transcription of the microRNA cluster 17–92, resulting in inhibition of Trp53inp1, a down-stream component of the TP53 pathway, promoting self-renewal. Up-stream SHH-GLI1 signaling was suggested to regulate NSC self-renewal either through direct inhibition of the TP53 pathway or indirect regulation of transcriptional activation of NANOG.
OCT4 is expressed in many human gliomas (Du, et al., 2009). In this study, authors showed that OCT4 expression in C6 rat glioma cells increase the self-renewal capacity and promotes an undifferentiated phenotype. In addition, RNA interference of OCT4 reduced tumor-forming capacity and increased sensitivity to temozolomide treatment in cultured glioma cells (Ikushima, et al., 2011).
SOX2 levels are reduced as NSCs and progenitors differentiate along neural cell lineages (Uwanogho, et al., 1995). High-grade gliomas express high levels of SOX2, varying between 50–100% nuclear staining (Annovazzi, et al., 2011). Human MBs also express SOX2, especially desmoplastic and nodular tumors that fall into the SHH subgroup (Ahlfeld, et al., 2013). Interestingly, SOX2 positive MB cells are the likely BTSCs for the SHH subtype (Vanner, et al., 2014). These tumor cells are slowly dividing and produce rapidly cycling doublecortin-positive neuronal cells that will comprise tumor bulk. SOX2 positive tumor cells further show resistance to chemotherapy, suggesting that this population is responsible for recurrences. SOX2 is an important factor responsible for brain tumor progression as depletion of SOX2 by ectopic expression of SOX21 diminishes capability of GBM cells to form in vitro spheres and additionally induces aberrant differentiation and apoptosis (Caglayan, et al., 2013, Ferletta, et al., 2011). However, elevated SOX2 levels reduced proliferation and CD133 expression in GBM and MB cell lines, suggesting that the threshold level of SOX2 expression regulate stemness and cell fate in brain tumor cells (Cox, et al., 2012).
MYC markedly promotes generation of iPS cells, but also increases tumor formation. Accordingly, the expression levels in reprogrammed cells and MYC-transformed cells are quite similar when comparing differentially expressed (Riggs, et al., 2013). However, different functional moieties on the MYC proteins are involved in the transformation and promotion of directed reprogramming (Nakagawa, et al., 2010).
Malignant primary brain tumor cells often express pluripotency genes and stemness, suggesting that the high degree of tumor plasticity is driven by oncogenic drivers. However, recent studies show that also the tumor microenvironment and therapy can have profound effects on brain tumor plasticity and promote a mesenchymal tumor phenotype. For example, GBMs displaying a high degree of necrosis or angiogenesis express higher levels of the master transcriptional regulators CEPB and STAT3 that drive a mesenchymal phenotype (Cooper, et al., 2012). Other studies found that hypoxia and tumor-associated macrophages promote stemness in human GBMs (Heddleston, et al., 2009, Seidel, et al., 2010, Wang, et al., 2012a, Yi, et al., 2011). In addition, temozolomide treatment induced expression of OCT4 and SOX2 in cultured human GBM cell lines and primary GBM cells (Auffinger, et al., 2014). The acquired stem cell phenotype showed increased self-renewal capacity and increased tumorigenicity following xenografting into recipient mice. In summary, reprogramming factors are found at high levels in brain tumors (Ben-Porath, et al., 2008, Clement, et al., 2007). The mechanisms involved in stemness and dedifferentiation are extensively studied and there is an increasing amount of reports on how transformation and reprogramming are driven by common transcriptional networks (reviewed in (Semi, et al., 2013, Swartling, et al., 2013a)). A core set of four neurodevelopmental transcription factors (SOX2, OLIG2, Sal-like protein 2 (SALL2), and POU domain, class 3, transcription factor 2 (POU3F2)) was recently shown to be essential for GBM propagation and maintenance of BTSCs (Suva, et al., 2014). Expression of these factors in differentiated GBM cells “induced” a BTSC phenotype by reprogramming the epigenetic cell state, providing an epigenetic basis for a developmental hierarchy in GBMs that underscores the cellular plasticity.
Potential therapeutic avenues to induce neuronal differentiation in brain tumors
The presence of specific BTSCs is associated with unfavorable outcome in many brain cancer types (Piccirillo and Vescovi, 2007, Visvader, 2011). Current cancer therapies effectively target the tumor bulk of brain tumor cells but leave behind BTSCs that underlie regrowth and recurrence (Chen, et al., 2012a). Development of differentiation regimens of cancers (Table 2), rather than cytotoxic therapies, may be an effective way to reduce treatment-resistance and prevent regrowth of brain tumors.
Table 2.
Potential differentiation therapies for brain tumors
| Compound | Cancer type | Phase | Targeted pathway |
|---|---|---|---|
| Retinoic acid | Acute promyelocytic leukemia Glioma Medulloblastoma |
Clinical practice (Dos Santos, et al., 2013) Preclinical trials (Patties, et al., 2013, Shi, et al., 2013, Spiller, et al., 2008) Clinical trials (Defer, et al., 1997, Grauer, et al., 2011, Jaeckle, et al., 2003, Levin, et al., 2006, Phuphanich, et al., 1997) |
MAPK/ERK, JNK |
| NGF | Prostate cancer Neuroblastoma Medulloblastoma Glioma |
Preclinical trials (Chen, et al., 2012b, Kimura, et al., 2002, Lavoie, et al., 2005, Muragaki, et al., 1997, Zhu, et al., 2013) | NGF/Trk |
| Farnesyl-transferase inhibitors | Glioma Medulloblastoma |
Preclinical trials (Lackner, et al., 2005, Wang and Macaulay, 1999) Clinical trials (Fouladi, et al., 2007) |
NGF/Rab7 |
| BMP4/Vaccinia virus expressing BMP4 | Medulloblastoma Glioblastoma multiforme |
Preclinical trials (Zhao, et al., 2008), (Duggal, et al., 2013, Piccirillo, et al., 2006) | BMP pathway |
| Isoxazole | Glioblastoma multiforme (astrocytoma) | Preclinical trials (Zhang, et al., 2011) | G1 blockade |
| AICAR | Glioma Other cancers |
Clinical and preclinical trials (Liu, et al., 2014) | mTOR pathway |
| Metformin | Glioma Other cancers |
Clinical and preclinical trials (Liu, et al., 2014) | mTOR pathway |
| HDAC inhibitors | Glioma Medulloblastoma |
Clinical and preclinical trials (Galanis, et al., 2009, Lee, et al., 2013, Nor, et al., 2013, Weller, et al., 2011) | HDAC inhibition |
| Decitabine | Glioma | Preclinical and clinical trials (Turcan, et al., 2013) | DNMT |
| IDH1R132H | Glioma | Preclinical trials (Rohle, et al., 2013) | IDH1R132H |
Retinoic acid
Retinoic acids have for many years been known for their anti-tumor effect, predominantly causing growth arrest and differentiation of target cells (Connolly, et al., 2013). All-trans retinoic acid (ATRA) induces arrest in G1 phase and promotes apoptosis in human squamous cell carcinoma (Zhang, et al., 2014). The authors found that RAS-ERK signaling pathway is a direct target of ATRA that acts by decreasing levels of activated ERK and c-Jun NH2-terminal kinase 1/2 (JNK1/2). Effects of ATRA have been well characterized in acute pro-myelocytic leukemia, with important roles in targeting transcription, proteolysis and differentiation (Dos Santos, et al., 2013). The differentiation potential observed in case of ATRA is rather promising not only in case of leukemia, but also in other cancer types such as glioma (Table 2). A recent study showed proof of concept results where ATRA treatment reduced proliferation and induced apoptosis in U87 GBM cells (Shi, et al., 2013). Differentiation potential and anti-tumor effect of ATRA have also been evaluated in preclinical MB models, where growth arrest and extensive apoptosis have been observed both in vitro on human MB cell lines and mouse xenografts (Patties, et al., 2013, Spiller, et al., 2008). Potential of ATRA to differentiate BTSCs has been evaluated in several clinical trials alone (Phuphanich, et al., 1997) or in combination with temozolomide (Grauer, et al., 2011, Jaeckle, et al., 2003), celecoxib (Levin, et al., 2006) or cytosine arabinosine (Defer, et al., 1997) (Table 2). Therapeutic effect was either lost after six months of treatment (Defer, et al., 1997) or there was no difference in progression-free survival (PFS) (Grauer, et al., 2011). Modest effects of ATRA on PFS were only observed in a small subset of patients at low-dose ATRA treatment (Jaeckle, et al., 2003, Levin, et al., 2006, Phuphanich, et al., 1997). As the authors proposed, better understanding of ATRA response mechanism is crucial for therapy success, as well as increase of dosage, which could be beneficial to proven modest responders to low-dose ATRA treatment. Although ATRA has been proven for its differentiation potential in brain tumor cells, the current clinical evaluation does not provide conclusive results, asking for other treatment modalities aiming to differentiate BTSCs, some of which we will further describe below.
Nerve growth factor
Nerve growth factor (NGF) treatment induces differentiation of cancer stem cells of multiple cancers. One study shows remarkable effects of NGF on reducing the number of stem cells present in androgen-independent prostate cancer, where authors showed decreased proliferative capacity both in vitro and in vivo (Chen, et al., 2012b). Neuroblastoma, a childhood nerve-sheet tumor originating from the peripheral nervous system, is highly responsive to NGF signaling (Verdi and Anderson, 1994), showing significant neuronal differentiation (Matsumoto, et al., 1995, Nakagawara, et al., 1993) and apoptosis (Lavoie, et al., 2005). Similarly, rat C6 glioma cells respond to NGF treatment both in vitro and in vivo by reducing proliferative potential and inducing neuronal differentiation (Kimura, et al., 2002). NGF signaling is crucial in differentiation of cancer stem cells, predominantly through Tropomyosin receptor kinase A (TRKA) and TRKB receptor activation. High expression of these TRK receptors is found in low-grade gliomas, while highly aggressive and malignant gliomas have almost completely suppressed TRK expression (Wadhwa, et al., 2003), demonstrating the importance of the NGF pathway in glioma progression and clinical outcome. Success of NGF treatment is hence dependent on TRK receptor expression. In attempt to differentiate MB cell lines DAOY, Muragaki et al. overexpressed TRK receptors and subsequently evaluated effects of NGF on cell survival (Muragaki, et al., 1997). As expected, authors indicated reduced metabolism and increased apoptosis in cells with upregulated TRK, thus bringing up another potential therapeutic approach in fighting MB. A small GTPase Rab7 seems to suppress TRK levels in gliomas (Saxena, et al., 2005) and it was recently identified as a target for farnesyl-transferase inhibitors (Lackner, et al., 2005), suggesting such drugs could work in differentiating brain tumors cells. Farnesyl-transferase inhibitor Manumycin A was also shown to be effective against MB cell lines in preclinical trials by inducing G1 arrest and apoptosis (Wang and Macaulay, 1999). Another farnesyl-transferase inhibitor Tipifarnib was recently evaluated in a clinical setting, where GBM and MB patients were evaluated for response (Fouladi, et al., 2007). Although none of the MB patients showed response, 10% of evaluated GBM patients showed either partial response or stable disease within four-year trial.
BMP
Bone morphogenetic proteins (BMPs) form a group of proteins with multiple functions in the adult brain stem cell niche, favoring the astroglial fate of cells. In attempt to differentiate cells, Piccirillo et al. (Piccirillo, et al., 2006) showed arrest in proliferation of GBM cells after BMP4 exposure. They observed accumulation of cells in G0/G1 phase and reduction in the number of cells with active DNA synthesis. The effect was not only observed in vitro, but also in vivo, where tumor-bearing animals survived significantly longer when treated with BMP4-releasing carrier beads. In a recent study, overexpression of BMP4 showed a robust antitumor effect on GBM cells (Duggal, et al., 2013). BMP2 and BMP4 also inhibit MB proliferation by inducing neuronal differentiation. This inhibition is caused by a rapid proteasome-mediated degradation of MATH1 (Zhao, et al., 2008). Despite the increasing number of preclinical studies evaluating therapeutic effects on BTSCs, BMP targeted therapies have still not entered clinical trials. BMPs are currently approved modes of treatment for several bone-related malformations and injuries including open fractures of long bones (Gautschi, et al., 2007), proving the safety of those agents for patients.
AMPK and mTOR
In an attempt to differentiate malignant astrocytoma cells and overcome reprogramming induced by TP53 and Ink4a mutations, Zhang et al. successfully delayed G1 phase and M phase entry by activating AMP-activated protein kinase (AMPK) by Isoxazole treatment (Zhang, et al., 2011). Moreover, cells treated with Isoxazole show re-expression of neuronal markers, e.g. TuJ1. It is suggested that BTSCs reprogram their metabolism for optimal growth and proliferation, where AMP-activated protein kinase (AMPK) has an important role. AMPK is activated in both human and mouse gliomas and its activity can be suppressed by using AMPK-agonists AICAR and metformin (Liu, et al., 2014). Both drugs are nowadays evaluated in more than 50 different clinical trials and have recently been identified as potential therapeutics against gliomas (Liu, et al., 2014). Both drugs were able to induce G2M arrest previously correlated with cell differentiation (Xiang, et al., 2008). Several mTOR inhibitors including sirolimus (rapamycin), everolimus (RAD-001) and temsirolimus (CCI-779) are currently evaluated in approximately 40 clinical trials involving patients with diagnosed gliomas and GBMs (https://clinicaltrials.gov). Recently completed phase II clinical trial involving temsirolimus treatment in combination with temozolomide and bevacizumab however did not show any partial response in all 13 GBM patients, providing only two patients with stable disease (Lassen, et al., 2013). Similarly, none of the 18 recurrent GBM patients involved in phase II study of sorafenib in combination with temsirolimus remained progression free at six months, with only two partial responses reported (Lee, et al., 2012). Everolimus on the other hand seems to be promising in GBM patients in combination with bevacizumab, temozolomide and radiation (Hainsworth, et al., 2012). More than half of the patients enrolled in this trial (61%) had objective response with a median progression-free survival 11.3 months and overall survival of 13.9 months. Combined, AMPK and mTOR inhibitors show promise in GBM. Despite strong therapeutic effect in preclinical studies, failing to provide benefits in clinical settings require more advanced inhibitors that specifically target the mTOR pathway.
HDAC inhibition
Use of HDAC inhibitors has emerged as a potential strategy to reverse aberrant chromatin changes and induce cell differentiation in cancer. The HDAC inhibitor valproic acid is FDA-approved for use in several neurological disorders. Several classes of HDAC inhibitors reduce GBM cell proliferation in vitro and tumor growth in GBM xenograft models (Asklund, et al., 2012, Benitez, et al., 2008, Yin, et al., 2007). In GBM patients, valproic acid treatment during radiotherapy improved overall survival (Barker, et al., 2013). Furthermore, dual inhibition of LSD1 and HDAC cooperated to induce cell death in GBM cell lines (Singh, et al., 2011). In MB, HDAC inhibition reversed GNP proliferation in a SHH-driven GEMM of MB (Lee, et al., 2013). In human MB cell lines, the incubation with the HDAC inhibitor sodium butyrate promoted cell death and induced neuronal differentiation (Nor, et al., 2013). Clinical trials suggest that the HDAC inhibitors vorinostat and valproic acid are well-tolerated in GBM patients (Galanis, et al., 2009, Weller, et al., 2011). Valproic acid treatment is well-tolerated in GBM patients (Weller, et al., 2011). These studies validate HDAC inhibition as a potential target in human GBM and MB patients.
DNMT, EZH2, and IDH1R132H inhibition
Widespread resetting of DNA methylation pattern in GBM BTSCs inhibited malignant properties (Stricker, et al., 2013). Inhibition of the PRC2 complexing protein EZH2 in human GBMs reduced tumor growth by both H3K27me3-dependent and STAT3-dependent pathways (Kim, et al., 2013). In another study, pharmacological inhibition of EZH2 impaired invasion into surrounding brain tissue of xenografted GBMs (Natsume, et al., 2013). Furthermore, the DNMT1 inhibitor Decitabine successfully inhibited differentiation and suppressed tumor growth of IDH1R132H mutant human glioma cells (Turcan, et al., 2013). A more personalized approach include the successful use of a small molecule IDH1R132H inhibitor AGI-5198 that increased glial differentiation in murine NSC cultures and delayed growth of human IDH1R132H mutant glioma xenografts (Rohle, et al., 2013).
MYC/MYCN
G34R or G34V H3F3A mutations are also found in children and young adults, but these do not result in inhibition of PRC2. Instead, G34R/V mutations result in altered binding of proteins that recognize trimethylated H3K36 (H3K36me3) (Bjerke, et al., 2013). This results in a gene expression signature that is more commonly seen in the developing forebrain and that is associated with increased MYCN transcription. This is in line with data that we demonstrated and which showed that constitutively activated T58A MYCN can drive gliomagenesis in forebrain murine NSCs (Swartling, et al., 2012). Glial differentiation by the transcriptional co-activator p300 was blocked by overexpression of MYC that instead promoted a stem cell-phenotype (Panicker, et al., 2010). Interestingly, MYC was found to be a direct target of EZH2 and essential for maintenance of the stem cell-phenotype (Suva, et al., 2009). These studies show that H3F3A mutations in GBMs converge on MYC and MYCN. As a major engine and driver of an undifferentiated state in GBMs, future therapies targeting MYC are expected to induce differentiation and apoptosis. However, aberrant recruitment of the PRC2 complex to K27M mutant H3.3 and enzymatic inhibition of EZH2 was found to produce a global reduction of the repressive histone mark H3K27me3 (Bender, et al., 2013), suggesting that EZH2 inhibition might accelerate tumor growth in K27M mutant tumors (Lewis, et al., 2013). Alternative pathways to inhibit MYC-driven MBs include cell cycle arrest following epigenetic regulation using BET bromodomain inhibition or Aurora kinase A inhibition (Bandopadhayay, et al., 2014, Bjerke, et al., 2013).
Use of location and time to delineate a relationship between cell of origin and brain tumor type
Refined transcriptomal profiling of gliomas and MB have enabled researchers to associate tumor subgroups with genetic alterations and methylation signatures. Interestingly, several studies show that subgroups of primary malignant brain tumors associate with the patient age and the location in the brain, implicating that temporally and regionally restricted populations of neural precursors give rise to distinct subsets of tumors. As the tumor phenotype change and accumulate new driver mutations, it becomes even more difficult following therapy to delineate a relationship between the origin and tumor phenotype. Therefore this discussion will only focus on primary non-recurrent brain tumors.
The methylation profiles of pediatric and adult GBMs were found to correlate with mutational status and cytogenetic aberrations (Sturm D et al. Cancer Cell, 2012). Young children are diagnosed with GBMs that often display H3F3A K27M mutations and a hypomethylated phenotype. In contrast, GBMs in older children are associated with H3F3A G34R/V mutations that display a hypermethylated phenotype. While most H3F3AK27M tumors are found in brainstem regions, H3F3AG34R/V tumors are instead observed in cortical forebrain lobes. Interestingly, H3F3AG34R/V tumors express low levels of OLIG2, expressed at high levels in other GBM subgroups, but are still aggressive tumors. In young adults, IDH1R132H mutated GBMs are highly hypermethylated and associated with a better prognosis. Despite the different methylation signatures, H3F3A and IDH1R132H mutated tumors are highly associated with TP53 mutations. It has been suggested that a population of SHH-responsive glial progenitors expressing OLIG2 and Nestin peaks in the ventral pons when most H3F3AK27M GBMs and diffuse intrinsic pontine gliomas (DIPGs) are diagnosed in patients (Monje M et al. PNAS, 2011). It is possible that cooperation of TP53 and H3F3AG34R/V mutations in SVZ NSCs of young children produce GBMs during late childhood (Figure 1). Development of IDH1R132H mutation in young adults is consistent with transformation of white matter OPCs as the major population of cycling cells. In childhood GBMs displaying PDGFRA amplifications, SVZ NSCs and OPCs are two populations susceptible to transformation. In adults, PDGFRA amplified tumors are more likely to originate from OPCs, known to be highly dependent of PDGF signaling. During late adulthood, it remains unclear if remnants of transformed NSCs, OPCs, or dedifferentiated astrocytes produce tumors displaying a mesenchymal gene expression signature or GBMs displaying EGFR amplifications. Interestingly, studies of a GEMM of pediatric low-grade glioma found that tumors arise from NSC lining the third ventricle, and not from SVZ NSCs lining the lateral ventricles (Lee DY et al. Cancer Cell, 2012).
For MB, SHH tumors are unique since they occur in infants but not in older children (Taylor MD et al., Acta Neuropathol, 2012). Similar to WNT tumors, they can also occur in adults. Since several populations of cerebellar and brain stem precursors expand around birth and during early childhood, it has been difficult to assign a unique cell of origin for each MB subgroup. However, development of novel GEMMs for MB has identified specific cell populations that can generate tumors matching gene expression profiles of human MB subgroups (Poschl J et al. Acta Neuropathol, 2014). To identify the cell of origin, most studies have relied on GEMMs that rely on oncogenic expression under a specific gene promoter. A better spatial and temporal resolution can be attained by using a cross-species analysis approach where murine tumors, generated from temporally defined precursors, are compared to cohorts of human tumors. This approach was elegantly employed to define the cell of origin for different ependymoma subtypes (Johnson RA et al. Nature, 2010). However, this approach is more difficult to use for gliomas and MBs where multiple candidates of precursor populations can produce tumors. Given that there are major differences in the development of neocortex and postnatal neurogenesis between mouse and humans, future work should validate that tumor-susceptible precursor populations expand during corresponding time windows also in humans.
CONCLUSION
Advances in NSC biology have identified genes and pathways that regulate neurogenesis in animals and humans. In parallel, genome-wide characterization of human gliomas and MBs has identified genetically and epigenetically distinct subgroups. Of interest, genetic alterations in pathways regulating normal neurogenesis are often perturbed in human brain tumors, and when reversed, often promote neuronal differentiation. Newly developed GEMM of gliomas and MBs have demonstrated that subgroup-specific genomic alterations in precursor cells often disrupt normal differentiation and promote tumorigenesis in a cell context-dependent manner. Studies of GEMMs and human brain tumors serve complimentary roles and are both important to identify driver mutations that maintain tumor growth and as pre-clinical tools for targeted therapy. Direct pharmacological targeting of driver mutations or down-stream effectors, manipulation of the tumor microenvironment, or cell-based therapies all represent novel approaches to reduce levels of reprogramming genes, induce differentiation, and reduce treatment-resistance in brain tumors.
Acknowledgments
AIP was supported by the TDC Foundation, the Guggenhime Endowment Fund, the National Brain Tumor Society, and National Institute of Health (U54CA163155 and R21NS088114). FJS was supported by the Swedish Childhood Cancer Foundation, the Swedish Cancer Society, the Swedish Research Council, the Ragnar Söderberg Foundation, the Swedish Society of Medicine, the Åke Wiberg Foundation and the Association for International Cancer Research/Worldwide Cancer Research.
References
- Acquaviva J, Jun HJ, Lessard J, Ruiz R, Zhu H, Donovan M, Woolfenden S, Boskovitz A, Raval A, Bronson RT, Pfannl R, Whittaker CA, Housman DE, Charest A. Chronic activation of wild-type epidermal growth factor receptor and loss of Cdkn2a cause mouse glioblastoma formation. Cancer Res. 2011;71:7198–7206. doi: 10.1158/0008-5472.CAN-11-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson DC, Shi Q, Wortham M, Northcott PA, Di C, Duncan CG, Li J, McLendon RE, Bigner DD, Taylor MD, Yan H. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Ahlfeld J, Favaro R, Pagella P, Kretzschmar HA, Nicolis S, Schuller U. Sox2 requirement in sonic hedgehog-associated medulloblastoma. Cancer Res. 2013;73:3796–3807. doi: 10.1158/0008-5472.CAN-13-0238. [DOI] [PubMed] [Google Scholar]
- Akerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879–8889. doi: 10.1523/JNEUROSCI.0558-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder J, Lee KJ, Jessell TM, Hatten ME. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- Altman J. Are new neurons formed in the brains of adult mammals? Science. 1962;135:1127–1128. doi: 10.1126/science.135.3509.1127. [DOI] [PubMed] [Google Scholar]
- Altman J, Bayer SA. Development of the brain stem in the rat. I. Thymidine-radiographic study of the time of origin of neurons of the lower medulla. J Comp Neurol. 1980;194:1–35. doi: 10.1002/cne.901940102. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41:683–686. doi: 10.1016/s0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 Expression and Amplification in Gliomas and Glioma Cell Lines. Cancer Genomics - Proteomics. 2011;8:139–147. [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Appolloni I, Calzolari F, Barilari M, Terrile M, Daga A, Malatesta P. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. Int J Cancer. 2012;131:E1078–1087. doi: 10.1002/ijc.27606. [DOI] [PubMed] [Google Scholar]
- Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S, De Smaele E, Greco A, Fiorenza MT, Maroder M, Screpanti I, Alesse E, Gulino A. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci. 2005;25:8338–8346. doi: 10.1523/JNEUROSCI.2438-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asklund T, Kvarnbrink S, Holmlund C, Wibom C, Bergenheim T, Henriksson R, Hedman H. Synergistic killing of glioblastoma stem-like cells by bortezomib and HDAC inhibitors. Anticancer Res. 2012;32:2407–2413. [PubMed] [Google Scholar]
- Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso-Sacido A, Moliterno JA, Kratovac S, Kapoor GS, O’Rourke DM, Holland EC, Garcia-Verdugo JM, Roy NS, Boockvar JA. Activated EGFR signaling increases proliferation, survival, and migration and blocks neuronal differentiation in post-natal neural stem cells. J Neurooncol. 2010;97:323–337. doi: 10.1007/s11060-009-0035-x. [DOI] [PubMed] [Google Scholar]
- Balasubramaniyan V, Boddeke E, Bakels R, Kust B, Kooistra S, Veneman A, Copray S. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–951. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, Bolin S, Schumacher SE, Zeid R, Masoud S, Yu F, Vue N, Gibson WJ, Paolella BR, Mitra SS, Cheshier SH, Qi J, Liu KW, Wechsler-Reya R, Weiss WA, Swartling FJ, Kieran MW, Bradner JE, Beroukhim R, Cho YJ. BET Bromodomain Inhibition of MYC-Amplified Medulloblastoma. Clin Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CA, Bishop AJ, Chang M, Beal K, Chan TA. Valproic acid use during radiation therapy for glioblastoma associated with improved survival. Int J Radiat Oncol Biol Phys. 2013;86:504–509. doi: 10.1016/j.ijrobp.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batora NV, Sturm D, Jones DT, Kool M, Pfister SM, Northcott PA. Transitioning from genotypes to epigenotypes: why the time has come for medulloblastoma epigenomics. Neuroscience. 2014;264:171–185. doi: 10.1016/j.neuroscience.2013.07.030. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Hojfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Seker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu CM, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho YJ, Pfister SM. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Benitez JA, Arregui L, Cabrera G, Segovia J. Valproic acid induces polarization, neuronal-like differentiation of a subpopulation of C6 glioma cells and selectively regulates transgene expression. Neuroscience. 2008;156:911–920. doi: 10.1016/j.neuroscience.2008.07.065. [DOI] [PubMed] [Google Scholar]
- Berninger B, Costa MR, Koch U, Schroeder T, Sutor B, Grothe B, Gotz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, Bax DA, Carvalho D, Taylor KR, Vinci M, Bajrami I, McGonnell IM, Lord CJ, Reis RM, Hargrave D, Ashworth A, Workman P, Jones C. Histone H3.3 Mutations Drive Pediatric Glioblastoma through Upregulation of MYCN. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britz O, Mattar P, Nguyen L, Langevin LM, Zimmer C, Alam S, Guillemot F, Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16(Suppl 1):i138–151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- Bruce AW, Donaldson IJ, Wood IC, Yerbury SA, Sadowski MI, Chapman M, Gottgens B, Buckley NJ. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc Natl Acad Sci U S A. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan D, Lundin E, Kastemar M, Westermark B, Ferletta M. Sox21 inhibits glioma progression in vivo by forming complexes with Sox2 and stimulating aberrant differentiation. International Journal of Cancer. 2013;133:1345–1356. doi: 10.1002/ijc.28147. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, Dolle D, Bithell A, Ettwiller L, Buckley N, Guillemot F. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25:930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaleri F, Schöler HR. Nanog: A New Recruit to the Embryonic Stem Cell Orchestra. Cell. 2003;113:551–552. doi: 10.1016/s0092-8674(03)00394-5. [DOI] [PubMed] [Google Scholar]
- Chaumeil MM, Larson PE, Yoshihara HA, Danforth OM, Vigneron DB, Nelson SJ, Pieper RO, Phillips JJ, Ronen SM. Non-invasive in vivo assessment of IDH1 mutational status in glioma. Nat Commun. 2013;4:2429. doi: 10.1038/ncomms3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012a;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Guerrero-Cazares H, Ye X, Ford E, McNutt T, Kleinberg L, Lim M, Chaichana K, Quinones-Hinojosa A, Redmond K. Increased subventricular zone radiation dose correlates with survival in glioblastoma patients after gross total resection. Int J Radiat Oncol Biol Phys. 2013;86:616–622. doi: 10.1016/j.ijrobp.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Wang GM, Guo JM, Sun LA, Wang H. NGF/gamma-IFN Inhibits Androgen-Independent Prostate Cancer and Reverses Androgen Receptor Function Through Downregulation of FGFR2 and Decrease in Cancer Stem Cells. Stem Cells and Development. 2012b;21:3372–3380. doi: 10.1089/scd.2012.0121. [DOI] [PubMed] [Google Scholar]
- Cheng L, Huang Z, Zhou W, Wu Q, Donnola S, Liu JK, Fang X, Sloan AE, Mao Y, Lathia JD, Min W, McLendon RE, Rich JN, Bao S. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature neuroscience. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhikov VV, Lindgren AG, Currle DS, Rose MF, Monuki ES, Millen KJ. The roof plate regulates cerebellar cell-type specification and proliferation. Development. 2006;133:2793–2804. doi: 10.1242/dev.02441. [DOI] [PubMed] [Google Scholar]
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Cicirata F, Zappala A, Serapide MF, Parenti R, Panto MR, Paz C. Different pontine projections to the two sides of the cerebellum. Brain Res Brain Res Rev. 2005;49:280–294. doi: 10.1016/j.brainresrev.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly RM, Nguyen NK, Sukumar S. Molecular pathways: current role and future directions of the retinoic acid pathway in cancer prevention and treatment. Clin Cancer Res. 2013;19:1651–1659. doi: 10.1158/1078-0432.CCR-12-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y, Kurc T, Van Meir EG, Saltz JH, Moreno CS, Brat DJ. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180:2108–2119. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper LA, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, Saltz JH, Brat DJ, Moreno CS. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS One. 2010;5:e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covic M, Karaca E, Lie DC. Epigenetic regulation of neurogenesis in the adult hippocampus. Heredity (Edinb) 2010;105:122–134. doi: 10.1038/hdy.2010.27. [DOI] [PubMed] [Google Scholar]
- Cox JL, Wilder PJ, Desler M, Rizzino A. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PLoS One. 2012;7:e44087. doi: 10.1371/journal.pone.0044087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Daou MC, Smith TW, Litofsky NS, Hsieh CC, Ross AH. Doublecortin is preferentially expressed in invasive human brain tumors. Acta Neuropathol. 2005;110:472–480. doi: 10.1007/s00401-005-1070-0. [DOI] [PubMed] [Google Scholar]
- de Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, De Vita G, Carotenuto M, Bello A, Formiggini F, Galeone A, De Rosa G, Virgilio A, Scognamiglio I, Sciro M, Basso G, Schulte JH, Cinalli G, Iolascon A, Zollo M. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS One. 2011;6:e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas T, Oussoren E, Grajkowska W, Perek-Polnik M, Popovic M, Zadravec-Zaletel L, Perera M, Corte G, Wirths O, van Sluis P, Pietsch T, Troost D, Baas F, Versteeg R, Kool M. OTX1 and OTX2 expression correlates with the clinicopathologic classification of medulloblastomas. J Neuropathol Exp Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- Defer GL, Adle-Biassette H, Ricolfi F, Martin L, Authier FJ, Chomienne C, Degos L, Degos JD. All-trans retinoic acid in relapsing malignant gliomas: clinical and radiological stabilization associated with the appearance of intratumoral calcifications. J Neurooncol. 1997;34:169–177. doi: 10.1023/a:1005701507111. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, De Smaele E, Argenti B, Mincione C, Zazzeroni F, Gallo R, Masuelli L, Napolitano M, Maroder M, Modesti A, Giangaspero F, Screpanti I, Alesse E, Gulino A. REN(KCTD11) is a suppressor of Hedgehog signaling and is deleted in human medulloblastoma. Proc Natl Acad Sci U S A. 2004;101:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Guenthoer J, Geng LN, Mahoney SE, Marotta M, Olson JM, Tanaka H, Tapscott SJ. DNA methylation of developmental genes in pediatric medulloblastomas identified by denaturation analysis of methylation differences. Proc Natl Acad Sci U S A. 2010;107:234–239. doi: 10.1073/pnas.0907606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Roncari L, Shannon P, Wu X, Lau N, Karaskova J, Gutmann DH, Squire JA, Nagy A, Guha A. Astrocyte-specific expression of activated p21-ras results in malignant astrocytoma formation in a transgenic mouse model of human gliomas. Cancer Res. 2001;61:3826–3836. [PubMed] [Google Scholar]
- Dipietrantonio HJ, Dymecki SM. Zic1 levels regulate mossy fiber neuron position and axon laterality choice in the ventral brain stem. Neuroscience. 2009;162:560–573. doi: 10.1016/j.neuroscience.2009.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos GA, Kats L, Pandolfi PP. Synergy against PML-RARa: targeting transcription, proteolysis, differentiation, and self-renewal in acute promyelocytic leukemia. J Exp Med. 2013;210:2793–2802. doi: 10.1084/jem.20131121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y, Cao X, Ling E-A, Hao A. Oct4 is expressed in human gliomas and promotes colony formation in glioma cells. Glia. 2009;57:724–733. doi: 10.1002/glia.20800. [DOI] [PubMed] [Google Scholar]
- Dubuc A, Mack S, Unterberger A, Northcott P, Taylor M. The Epigenetics of Brain Tumors. In: Dumitrescu RG, Verma M, editors. Cancer Epigenetics, vol 863. Methods in Molecular Biology. Humana Press; 2012. pp. 139–153. [DOI] [PubMed] [Google Scholar]
- Dubuc AM, Remke M, Korshunov A, Northcott PA, Zhan SH, Mendez-Lago M, Kool M, Jones DT, Unterberger A, Morrissy AS, Shih D, Peacock J, Ramaswamy V, Rolider A, Wang X, Witt H, Hielscher T, Hawkins C, Vibhakar R, Croul S, Rutka JT, Weiss WA, Jones SJ, Eberhart CG, Marra MA, Pfister SM, Taylor MD. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 2013;125:373–384. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal R, Geissinger U, Zhang Q, Aguilar J, Chen NHG, Binda E, Vescovi AL, Szalay AA. Vaccinia virus expressing bone morphogenetic protein-4 in novel glioblastoma orthotopic models facilitates enhanced tumor regression and long-term survival. Journal of Translational Medicine. 2013:11. doi: 10.1186/1479-5876-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsir T, Edqvist PH, Carlson J, Ribom D, Bergqvist M, Ekman S, Popova SN, Alafuzoff I, Ponten F, Nister M, Smits A. A study of embryonic stem cell-related proteins in human astrocytomas: identification of Nanog as a predictor of survival. Int J Cancer. 2014;134:1123–1131. doi: 10.1002/ijc.28441. [DOI] [PubMed] [Google Scholar]
- England B, Huang T, Karsy M. Current understanding of the role and targeting of tumor suppressor p53 in glioblastoma multiforme. Tumor Biology. 2013;34:2063–2074. doi: 10.1007/s13277-013-0871-3. [DOI] [PubMed] [Google Scholar]
- Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, Gage FH. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- Ernst A, Alkass K, Bernard S, Salehpour M, Perl S, Tisdale J, Possnert G, Druid H, Frisen J. Neurogenesis in the striatum of the adult human brain. Cell. 2014;156:1072–1083. doi: 10.1016/j.cell.2014.01.044. [DOI] [PubMed] [Google Scholar]
- Fan X, Eberhart CG. Medulloblastoma stem cells. J Clin Oncol. 2008;26:2821–2827. doi: 10.1200/JCO.2007.15.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 2009;23:561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Feng W, Khan MA, Bellvis P, Zhu Z, Bernhardt O, Herold-Mende C, Liu HK. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, Westermark B. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. International Journal of Cancer. 2011;129:45–60. doi: 10.1002/ijc.25647. [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, Bozzoni I, Gulino A. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Fouladi M, Nicholson HS, Zhou T, Laningham F, Helton KJ, Holmes E, Cohen K, Speights RA, Wright J, Pollack IF. A phase II study of the farnesyl transferase inhibitor, tipifarnib, in children with recurrent or progressive high-grade glioma, medulloblastoma/primitive neuroectodermal tumor, or brainstem glioma: A children’s oncology group study. Cancer. 2007;110:2535–2541. doi: 10.1002/cncr.23078. [DOI] [PubMed] [Google Scholar]
- Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, Danussi C, Dolgalev I, Porrati P, Pellegatta S, Heguy A, Gupta G, Pisapia DJ, Canoll P, Bruce JN, McLendon RE, Yan H, Aldape K, Finocchiaro G, Mikkelsen T, Prive GG, Bigner DD, Lasorella A, Rabadan R, Iavarone A. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of Neurons and Astrocytes by Oncogenes Can Induce Gliomas in Mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruhwald MC, O’Dorisio MS, Dai Z, Tanner SM, Balster DA, Gao X, Wright FA, Plass C. Aberrant promoter methylation of previously unidentified target genes is a common abnormality in medulloblastomas--implications for tumor biology and potential clinical utility. Oncogene. 2001;20:5033–5042. doi: 10.1038/sj.onc.1204613. [DOI] [PubMed] [Google Scholar]
- Fuentealba LC, Obernier K, Alvarez-Buylla A. Adult neural stem cells bridge their niche. Cell Stem Cell. 2012;10:698–708. doi: 10.1016/j.stem.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanis E, Jaeckle KA, Maurer MJ, Reid JM, Ames MM, Hardwick JS, Reilly JF, Loboda A, Nebozhyn M, Fantin VR, Richon VM, Scheithauer B, Giannini C, Flynn PJ, Moore DF, Jr, Zwiebel J, Buckner JC. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2009;27:2052–2058. doi: 10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Po A, Miele E, Campese AF, Begalli F, Silvano M, Infante P, Capalbo C, De Smaele E, Canettieri G, Di Marcotullio L, Screpanti I, Ferretti E, Gulino A. microRNA-17–92 cluster is a direct Nanog target and controls neural stem cell through Trp53inp1. EMBO J. 2013;32:2819–2832. doi: 10.1038/emboj.2013.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi OP, Frey SP, Zellweger R. BONE MORPHOGENETIC PROTEINS IN CLINICAL APPLICATIONS. ANZ Journal of Surgery. 2007;77:626–631. doi: 10.1111/j.1445-2197.2007.04175.x. [DOI] [PubMed] [Google Scholar]
- Geha S, Pallud J, Junier MP, Devaux B, Leonard N, Chassoux F, Chneiweiss H, Daumas-Duport C, Varlet P. NG2+/Olig2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol. 2010;20:399–411. doi: 10.1111/j.1750-3639.2009.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson M, Currle D, Eden C, Kranenburg T, Hogg T, Poppleton H, Martin J. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson RJ. Medulloblastoma: signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu Rev pathmechdis Mech Dis. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez O, Romero-Rodriguez R, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Epidermal growth factor induces the progeny of subventricular zone type B cells to migrate and differentiate into oligodendrocytes. Stem Cells. 2009;27:2032–2043. doi: 10.1002/stem.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammel D, Warmuth-Metz M, von Bueren AO, Kool M, Pietsch T, Kretzschmar HA, Rowitch DH, Rutkowski S, Pfister SM, Schuller U. Sonic hedgehog-associated medulloblastoma arising from the cochlear nuclei of the brainstem. Acta Neuropathol. 2012;123:601–614. doi: 10.1007/s00401-012-0961-0. [DOI] [PubMed] [Google Scholar]
- Grauer O, Pascher C, Hartmann C, Zeman F, Weller M, Proescholdt M, Brawanski A, Pietsch T, Wick W, Bogdahn U, Hau P. Temozolomide and 13-cis retinoic acid in patients with anaplastic gliomas: a prospective single-arm monocentric phase-II study (RNOP-05) J Neurooncol. 2011;104:801–809. doi: 10.1007/s11060-011-0548-y. [DOI] [PubMed] [Google Scholar]
- Grimmer MR, Weiss WA. Childhood tumors of the nervous system as disorders of normal development. Curr Opin Pediatr. 2006;18:634–638. doi: 10.1097/MOP.0b013e32801080fe. [DOI] [PubMed] [Google Scholar]
- Guichet PO, Bieche I, Teigell M, Serguera C, Rothhut B, Rigau V, Scamps F, Ripoll C, Vacher S, Taviaux S, Chevassus H, Duffau H, Mallet J, Susini A, Joubert D, Bauchet L, Hugnot JP. Cell death and neuronal differentiation of glioblastoma stem-like cells induced by neurogenic transcription factors. Glia. 2013;61:225–239. doi: 10.1002/glia.22429. [DOI] [PubMed] [Google Scholar]
- Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing L, Zhang Y, Ling E-A, Gao J, Hao A. Expression profile of embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human gliomas. Histopathology. 2011;59:763–775. doi: 10.1111/j.1365-2559.2011.03993.x. [DOI] [PubMed] [Google Scholar]
- Hack MA, Saghatelyan A, de Chevigny A, Pfeifer A, Ashery-Padan R, Lledo PM, Gotz M. Neuronal fate determinants of adult olfactory bulb neurogenesis. Nat Neurosci. 2005;8:865–872. doi: 10.1038/nn1479. [DOI] [PubMed] [Google Scholar]
- Hack MA, Sugimori M, Lundberg C, Nakafuku M, Gotz M. Regionalization and fate specification in neurospheres: the role of Olig2 and Pax6. Mol Cell Neurosci. 2004;25:664–678. doi: 10.1016/j.mcn.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291–300. doi: 10.1016/j.celrep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainsworth JD, Shih KC, Shepard GC, Tillinghast GW, Brinker BT, Spigel DR. Phase II study of concurrent radiation therapy, temozolomide, and bevacizumab followed by bevacizumab/everolimus as first-line treatment for patients with glioblastoma. Clin Adv Hematol Oncol. 2012;10:240–246. [PubMed] [Google Scholar]
- Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, Holland EC. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111:5248–5253. doi: 10.1073/pnas.1321014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton BA, Knoepfler PS, Kenney AM, Rowitch DH, de Alboran IM, Olson JM, Eisenman RN. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 2006;66:8655–8661. doi: 10.1158/0008-5472.CAN-06-1621. [DOI] [PubMed] [Google Scholar]
- Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Helgesson G, Eriksson S, Swartling U. Limited relevance of the right not to know--reflections on a screening study. Account Res. 2007;14:197–209. doi: 10.1080/08989620701456322. [DOI] [PubMed] [Google Scholar]
- Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RA, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Higgins DM, Wang R, Milligan B, Schroeder M, Carlson B, Pokorny J, Cheshier SH, Meyer FB, Weissman IL, Sarkaria JN, Henley JR. Brain tumor stem cell multipotency correlates with nanog expression and extent of passaging in human glioblastoma xenografts. Oncotarget. 2013;4:792–801. doi: 10.18632/oncotarget.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg J, He X, Peredo I, Orrego A, Hesselager G, Ericsson C, Hovatta O, Oba-Shinjo SM, Marie SK, Nister M, Muhr J. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS One. 2011;6:e18454. doi: 10.1371/journal.pone.0018454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, Terashima T, Wright CV, Kawaguchi Y, Nakao K, Nabeshima Y. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Hovestadt V, Jones DT, Picelli S, Wang W, Kool M, Northcott PA, Sultan M, Stachurski K, Ryzhova M, Warnatz HJ, Ralser M, Brun S, Bunt J, Jager N, Kleinheinz K, Erkek S, Weber UD, Bartholomae CC, von Kalle C, Lawerenz C, Eils J, Koster J, Versteeg R, Milde T, Witt O, Schmidt S, Wolf S, Pietsch T, Rutkowski S, Scheurlen W, Taylor MD, Brors B, Felsberg J, Reifenberger G, Borkhardt A, Lehrach H, Wechsler-Reya RJ, Eils R, Yaspo ML, Landgraf P, Korshunov A, Zapatka M, Radlwimmer B, Pfister SM, Lichter P. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- Hovestadt V, Remke M, Kool M, Pietsch T, Northcott PA, Fischer R, Cavalli FM, Ramaswamy V, Zapatka M, Reifenberger G, Rutkowski S, Schick M, Bewerunge-Hudler M, Korshunov A, Lichter P, Taylor MD, Pfister SM, Jones DT. Robust molecular subgrouping and copy-number profiling of medulloblastoma from small amounts of archival tumour material using high-density DNA methylation arrays. Acta Neuropathol. 2013;125:913–916. doi: 10.1007/s00401-013-1126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, Miyazono K. Glioma-initiating Cells Retain Their Tumorigenicity through Integration of the Sox Axis and Oct4 Protein. Journal of Biological Chemistry. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, Kageyama R. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science. 2013;342:1203–1208. doi: 10.1126/science.1242366. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Garcia-Verdugo JM, Gil-Perotin S, Roy M, Quinones-Hinojosa A, VandenBerg S, Alvarez-Buylla A. PDGFR alpha-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron. 2006;51:187–199. doi: 10.1016/j.neuron.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Jaeckle KA, Hess KR, Yung WK, Greenberg H, Fine H, Schiff D, Pollack IF, Kuhn J, Fink K, Mehta M, Cloughesy T, Nicholas MK, Chang S, Prados M North American Brain Tumor C. Phase II evaluation of temozolomide and 13-cis-retinoic acid for the treatment of recurrent and progressive malignant glioma: a North American Brain Tumor Consortium study. J Clin Oncol. 2003;21:2305–2311. doi: 10.1200/JCO.2003.12.097. [DOI] [PubMed] [Google Scholar]
- Jafri NF, Clarke JL, Weinberg V, Barani IJ, Cha S. Relationship of glioblastoma multiforme to the subventricular zone is associated with survival. Neuro Oncol. 2013;15:91–96. doi: 10.1093/neuonc/nos268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang ES, Goldman JE. Pax6 expression is sufficient to induce a neurogenic fate in glial progenitors of the neonatal subventricular zone. PLoS One. 2011;6:e20894. doi: 10.1371/journal.pone.0020894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Boije M, Westermark B, Uhrbom L. PDGF-B Can sustain self-renewal and tumorigenicity of experimental glioma-derived cancer-initiating cells by preventing oligodendrocyte differentiation. Neoplasia. 2011;13:492–503. doi: 10.1593/neo.11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJ, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, Cho Y-J, Pugh TJ, Hovestadt V, Stütz AM. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, Collins VP. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Northcott PA, Kool M, Pfister SM. The role of chromatin remodeling in medulloblastoma. Brain Pathol. 2013;23:193–199. doi: 10.1111/bpa.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, Deneen B. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsetos CD, Del Valle L, Geddes JF, Assimakopoulou M, Legido A, Boyd JC, Balin B, Parikh NA, Maraziotis T, de Chadarevian JP, Varakis JN, Matsas R, Spano A, Frankfurter A, Herman MM, Khalili K. Aberrant localization of the neuronal class III beta-tubulin in astrocytomas. Arch Pathol Lab Med. 2001;125:613–624. doi: 10.5858/2001-125-0613-ALOTNC. [DOI] [PubMed] [Google Scholar]
- Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison DW. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer cell. 2012;21:168–180. doi: 10.1016/j.ccr.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, Bourgey M, Bourque G, Montpetit A, Bourret G, Lepage P, Fleming A, Lichter P, Kool M, von Deimling A, Sturm D, Korshunov A, Faury D, Jones DT, Majewski J, Pfister SM, Jabado N, Hawkins C. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam DH, Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Kimura S, Yoshino A, Katayama Y, Watanabe T, Fukushima T. Growth control of C6 glioma in vivo by nerve growth factor. Journal of Neuro-Oncology. 2002;59:199–205. doi: 10.1023/a:1019919019497. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Osumi N, Rubenstein JL, Alvarez-Buylla A. Pax6 is required for making specific subpopulations of granule and periglomerular neurons in the olfactory bulb. J Neurosci. 2005;25:6997–7003. doi: 10.1523/JNEUROSCI.1435-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongkham PN, Northcott PA, Croul SE, Smith CA, Taylor MD, Rutka JT. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene. 2010;29:3017–3024. doi: 10.1038/onc.2010.32. [DOI] [PubMed] [Google Scholar]
- Kool M, Jones DT, Jager N, Northcott PA, Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, Milde T, Bourdeaut F, Ryzhova M, Sturm D, Pfaff E, Stark S, Hutter S, Seker-Cin H, Johann P, Bender S, Schmidt C, Rausch T, Shih D, Reimand J, Sieber L, Wittmann A, Linke L, Witt H, Weber UD, Zapatka M, Konig R, Beroukhim R, Bergthold G, van Sluis P, Volckmann R, Koster J, Versteeg R, Schmidt S, Wolf S, Lawerenz C, Bartholomae CC, von Kalle C, Unterberg A, Herold-Mende C, Hofer S, Kulozik AE, von Deimling A, Scheurlen W, Felsberg J, Reifenberger G, Hasselblatt M, Crawford JR, Grant GA, Jabado N, Perry A, Cowdrey C, Croul S, Zadeh G, Korbel JO, Doz F, Delattre O, Bader GD, McCabe MG, Collins VP, Kieran MW, Cho YJ, Pomeroy SL, Witt O, Brors B, Taylor MD, Schuller U, Korshunov A, Eils R, Wechsler-Reya RJ, Lichter P, Pfister SM, Project IPT. Genome Sequencing of SHH Medulloblastoma Predicts Genotype-Related Response to Smoothened Inhibition. Cancer Cell. 2014;25:393–405. doi: 10.1016/j.ccr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van Meeteren A, van Vuurden D, Clifford SC, Pietsch T, von Bueren AO, Rutkowski S, McCabe M, Collins VP, Backlund ML, Haberler C, Bourdeaut F, Delattre O, Doz F, Ellison DW, Gilbertson RJ, Pomeroy SL, Taylor MD, Lichter P, Pfister SM. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunov A, Remke M, Kool M, Hielscher T, Northcott PA, Williamson D, Pfaff E, Witt H, Jones DT, Ryzhova M, Cho YJ, Wittmann A, Benner A, Weiss WA, von Deimling A, Scheurlen W, Kulozik AE, Clifford SC, Peter Collins V, Westermann F, Taylor MD, Lichter P, Pfister SM. Biological and clinical heterogeneity of MYCN-amplified medulloblastoma. Acta Neuropathol. 2012;123:515–527. doi: 10.1007/s00401-011-0918-8. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn HG, Winkler J, Kempermann G, Thal LJ, Gage FH. Epidermal growth factor and fibroblast growth factor-2 have different effects on neural progenitors in the adult rat brain. J Neurosci. 1997;17:5820–5829. doi: 10.1523/JNEUROSCI.17-15-05820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A, Zimmer A, Schnutgen F, Bruning G, Spener F, Muller T. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- Lachyankar MB, Sultana N, Schonhoff CM, Mitra P, Poluha W, Lambert S, Quesenberry PJ, Litofsky NS, Recht LD, Nabi R, Miller SJ, Ohta S, Neel BG, Ross AH. A role for nuclear PTEN in neuronal differentiation. J Neurosci. 2000;20:1404–1413. doi: 10.1523/JNEUROSCI.20-04-01404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Kindt RM, Carroll PM, Brown K, Cancilla MR, Chen CY, de Silva H, Franke Y, Guan B, Heuer T, Hung T, Keegan K, Lee JM, Manne V, O’Brien C, Parry D, Perez-Villar JJ, Reddy RK, Xiao HJ, Zhan HJ, Cockett M, Plowman G, Fitzgerald K, Costa M, Ross-Macdonald P. Chemical genetics identifies Rab geranylgeranyl transferase as an apoptotic target of farnesyl transferase inhibitors. Cancer Cell. 2005;7:325–336. doi: 10.1016/j.ccr.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH, Schaffer DV. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci. 2003;6:21–27. doi: 10.1038/nn983. [DOI] [PubMed] [Google Scholar]
- Lange C, Mix E, Rateitschak K, Rolfs A. Wnt signal pathways and neural stem cell differentiation. Neurodegener Dis. 2006;3:76–86. doi: 10.1159/000092097. [DOI] [PubMed] [Google Scholar]
- Lassen U, Sorensen M, Gaziel TB, Hasselbalch B, Poulsen HS. Phase II study of bevacizumab and temsirolimus combination therapy for recurrent glioblastoma multiforme. Anticancer Res. 2013;33:1657–1660. [PubMed] [Google Scholar]
- Lastowska M, Al-Afghani H, Al-Balool HH, Sheth H, Mercer E, Coxhead JM, Redfern CP, Peters H, Burt AD, Santibanez-Koref M, Bacon CM, Chesler L, Rust AG, Adams DJ, Williamson D, Clifford SC, Jackson MS. Identification of a neuronal transcription factor network involved in medulloblastoma development. Acta Neuropathol Commun. 2013;1:35. doi: 10.1186/2051-5960-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Hitomi M, Gallagher J, Gadani SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, Minhas S, Soeda A, Hoeppner DJ, Ravin R, McKay RD, McLendon RE, Corbeil D, Chenn A, Hjelmeland AB, Park DM, Rich JN. Distribution of CD133 reveals glioma stem cells self-renew through symmetric and asymmetric cell divisions. Cell Death Dis. 2011;2:e200. doi: 10.1038/cddis.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie J-F, LeSauteur L, Kohn J, Wong J, Furtoss O, Thiele CJ, Miller FD, Kaplan DR. TrkA Induces Apoptosis of Neuroblastoma Cells and Does So via a p53-dependent Mechanism. Journal of Biological Chemistry. 2005;280:29199–29207. doi: 10.1074/jbc.M502364200. [DOI] [PubMed] [Google Scholar]
- Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, Johnson JE, Wechsler-Reya RJ. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci. 2005;8:723–729. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EQ, Kuhn J, Lamborn KR, Abrey L, DeAngelis LM, Lieberman F, Robins HI, Chang SM, Yung WK, Drappatz J, Mehta MP, Levin VA, Aldape K, Dancey JE, Wright JJ, Prados MD, Cloughesy TF, Gilbert MR, Wen PY. Phase I/II study of sorafenib in combination with temsirolimus for recurrent glioblastoma or gliosarcoma: North American Brain Tumor Consortium study 05–02. Neuro Oncol. 2012;14:1511–1518. doi: 10.1093/neuonc/nos264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Lindsey S, Graves B, Yoo S, Olson JM, Langhans SA. Sonic hedgehog-induced histone deacetylase activation is required for cerebellar granule precursor hyperplasia in medulloblastoma. PLoS One. 2013;8:e71455. doi: 10.1371/journal.pone.0071455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin VA, Giglio P, Puduvalli VK, Jochec J, Groves MD, Yung WK, Hess K. Combination chemotherapy with 13-cis-retinoic acid and celecoxib in the treatment of glioblastoma multiforme. J Neurooncol. 2006;78:85–90. doi: 10.1007/s11060-005-9062-4. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mattar P, Dixit R, Lawn SO, Wilkinson G, Kinch C, Eisenstat D, Kurrasch DM, Chan JA, Schuurmans C. RAS/ERK signaling controls proneural genetic programs in cortical development and gliomagenesis. J Neurosci. 2014a;34:2169–2190. doi: 10.1523/JNEUROSCI.4077-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Mattar P, Dixit R, Lawn SO, Wilkinson G, Kinch C, Eisenstat D, Kurrasch DM, Chan JA, Schuurmans C. RAS/ERK Signaling Controls Proneural Genetic Programs in Cortical Development and Gliomagenesis. The Journal of Neuroscience. 2014b;34:2169–2190. doi: 10.1523/JNEUROSCI.4077-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Teng ZQ, Santistevan NJ, Szulwach KE, Guo W, Jin P, Zhao X. Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell. 2010;6:433–444. doi: 10.1016/j.stem.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zong H. Developmental origins of brain tumors. Curr Opin Neurobiol. 2012;22:844–849. doi: 10.1016/j.conb.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chhipa RR, Pooya S, Wortman M, Yachyshin S, Chow LML, Kumar A, Zhou X, Sun Y, Quinn B, McPherson C, Warnick RE, Kendler A, Giri S, Poels J, Norga K, Viollet B, Grabowski GA, Dasgupta B. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proceedings of the National Academy of Sciences. 2014;111:E435–E444. doi: 10.1073/pnas.1311121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O’Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G, Rosenfeld MG. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Lutolf S, Radtke F, Aguet M, Suter U, Taylor V. Notch1 is required for neuronal and glial differentiation in the cerebellum. Development. 2002;129:373–385. doi: 10.1242/dev.129.2.373. [DOI] [PubMed] [Google Scholar]
- Machold RP, Kittell DJ, Fishell GJ. Antagonism between Notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2007;2:5. doi: 10.1186/1749-8104-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, Wang M, Hu B, Cheng SY, Sobol RW, Nakano I. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraziotis T, Perentes E, Karamitopoulou E, Nakagawa Y, Gessaga EC, Probst A, Frankfurter A. Neuron-associated class III beta-tubulin isotype, retinal S-antigen, synaptophysin, and glial fibrillary acidic protein in human medulloblastomas: a clinicopathological analysis of 36 cases. Acta Neuropathol. 1992;84:355–363. doi: 10.1007/BF00227661. [DOI] [PubMed] [Google Scholar]
- Maricich SM, Herrup K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J Neurobiol. 1999;41:281–294. doi: 10.1002/(sici)1097-4695(19991105)41:2<281::aid-neu10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Marino S. Medulloblastoma: developmental mechanisms out of control. Trends Mol Med. 2005;11:17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Marshall CA, Novitch BG, Goldman JE. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J Neurosci. 2005;25:7289–7298. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall GM, Carter DR, Cheung BB, Liu T, Mateos MK, Meyerowitz JG, Weiss WA. The prenatal origins of cancer. Nat Rev Cancer. 2014;14:277–289. doi: 10.1038/nrc3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu P, Battista D, Depino A, Roca V, Graciarena M, Pitossi F. The more you have, the less you get: the functional role of inflammation on neuronal differentiation of endogenous and transplanted neural stem cells in the adult brain. J Neurochem. 2010;112:1368–1385. doi: 10.1111/j.1471-4159.2009.06548.x. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Wada RK, Yamashiro JM, Kaplan DR, Thiele CJ. Expression of brain-derived neurotrophic factor and p145TrkB affects survival, differentiation, and invasiveness of human neuroblastoma cells. Cancer Res. 1995;55:1798–1806. [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, Metzger D, Chambon P, Rao MS, Sherman LS. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Developmental Biology. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Mattar P, Langevin LM, Markham K, Klenin N, Shivji S, Zinyk D, Schuurmans C. Basic helixloop-helix transcription factors cooperate to specify a cortical projection neuron identity. Mol Cell Biol. 2008;28:1456–1469. doi: 10.1128/MCB.01510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays. 2003;25:930–939. doi: 10.1002/bies.10332. [DOI] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, Olson JJ, Mikkelsen T, Lehman N, Aldape K. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Fuentealba LC, Sanders TA, Magno L, Kessaris N, Alvarez-Buylla A. Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat Neurosci. 2014;17:207–214. doi: 10.1038/nn.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, Garcia-Verdugo JM, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci U S A. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muragaki Y, Chou TT, Kaplan DR, Trojanowski JQ, Lee VM-Y. Nerve Growth Factor Induces Apoptosis in Human Medulloblastoma Cell Lines that Express TrkA Receptors. The Journal of Neuroscience. 1997;17:530–542. doi: 10.1523/JNEUROSCI.17-02-00530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicerdeficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M, Takizawa N, Narita M, Ichisaka T, Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc Natl Acad Sci U S A. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawara A, Arima-Nakagawara M, Scavarda NJ, Azar CG, Cantor AB, Brodeur GM. Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N Engl J Med. 1993;328:847–854. doi: 10.1056/NEJM199303253281205. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Natsume A, Ito M, Katsushima K, Ohka F, Hatanaka A, Shinjo K, Sato S, Takahashi S, Ishikawa Y, Takeuchi I, Shimogawa H, Uesugi M, Okano H, Kim SU, Wakabayashi T, Issa JP, Sekido Y, Kondo Y. Chromatin regulator PRC2 is a key regulator of epigenetic plasticity in glioblastoma. Cancer Res. 2013;73:4559–4570. doi: 10.1158/0008-5472.CAN-13-0109. [DOI] [PubMed] [Google Scholar]
- Nishino J, Kim S, Zhu Y, Zhu H, Morrison SJ. A network of heterochronic genes including Imp1 regulates temporal changes in stem cell properties. Elife. 2013;2:e00924. doi: 10.7554/eLife.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu CS, Li DX, Liu YH, Fu XM, Tang SF, Li J. Expression of NANOG in human gliomas and its relationship with undifferentiated glioma cells. Oncol Rep. 2011;26:593–601. doi: 10.3892/or.2011.1308. [DOI] [PubMed] [Google Scholar]
- Noble M, Murray K, Stroobant P, Waterfield MD, Riddle P. Platelet-derived growth factor promotes division and motility and inhibits premature differentiation of the oligodendrocyte/type-2 astrocyte progenitor cell. Nature. 1988;333:560–562. doi: 10.1038/333560a0. [DOI] [PubMed] [Google Scholar]
- Nor C, Sassi FA, de Farias CB, Schwartsmann G, Abujamra AL, Lenz G, Brunetto AL, Roesler R. The histone deacetylase inhibitor sodium butyrate promotes cell death and differentiation and reduces neurosphere formation in human medulloblastoma cells. Mol Neurobiol. 2013;48:533–543. doi: 10.1007/s12035-013-8441-7. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009a;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Hielscher T, Dubuc A, Mack S, Shih D, Remke M, Al-Halabi H, Albrecht S, Jabado N, Eberhart CG. Pediatric and adult sonic hedgehog medulloblastomas are clinically and molecularly distinct. Acta neuropathologica. 2011;122:231–240. doi: 10.1007/s00401-011-0846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Jones DTW, Kool M, Robinson GW, Gilbertson RJ, Cho Y-J, Pomeroy SL, Korshunov A, Lichter P, Taylor MD, Pfister SM. Medulloblastomics: the end of the beginning. Nat Rev Cancer. 2012a;12:818–834. doi: 10.1038/nrc3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Lee C, Zichner T, Stutz AM, Erkek S, Kawauchi D, Shih DJ, Hovestadt V, Zapatka M, Sturm D, Jones DT, Kool M, Remke M, Cavalli FM, Zuyderduyn S, Bader GD, VandenBerg S, Esparza LA, Ryzhova M, Wang W, Wittmann A, Stark S, Sieber L, Seker-Cin H, Linke L, Kratochwil F, Jager N, Buchhalter I, Imbusch CD, Zipprich G, Raeder B, Schmidt S, Diessl N, Wolf S, Wiemann S, Brors B, Lawerenz C, Eils J, Warnatz HJ, Risch T, Yaspo ML, Weber UD, Bartholomae CC, von Kalle C, Turanyi E, Hauser P, Sanden E, Darabi A, Siesjo P, Sterba J, Zitterbart K, Sumerauer D, van Sluis P, Versteeg R, Volckmann R, Koster J, Schuhmann MU, Ebinger M, Grimes HL, Robinson GW, Gajjar A, Mynarek M, von Hoff K, Rutkowski S, Pietsch T, Scheurlen W, Felsberg J, Reifenberger G, Kulozik AE, von Deimling A, Witt O, Eils R, Gilbertson RJ, Korshunov A, Taylor MD, Lichter P, Korbel JO, Wechsler-Reya RJ, Pfister SM. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, Mack S, Kongkham PN, Peacock J, Dubuc A, Ra Y-S, Zilberberg K, McLeod J, Scherer SW, Sunil Rao J, Eberhart CG, Grajkowska W, Gillespie Y, Lach B, Grundy R, Pollack IF, Hamilton RL, Van Meter T, Carlotti CG, Boop F, Bigner D, Gilbertson RJ, Rutka JT, Taylor MD. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009b;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stütz AM, Korshunov A, Reimand J, Schumacher SE. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012b doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM, Wickramasinghe R, Scott MP, Wechsler-Reya RJ. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma V, Ruiz i Altaba A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development. 2004;131:337–345. doi: 10.1242/dev.00930. [DOI] [PubMed] [Google Scholar]
- Panicker SP, Raychaudhuri B, Sharma P, Tipps R, Mazumdar T, Mal AK, Palomo JM, Vogelbaum MA, Haque SJ. p300-and Myc-mediated regulation of glioblastoma multiforme cell differentiation. Oncotarget. 2010;1:289–303. doi: 10.18632/oncotarget.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Parras CM, Hunt C, Sugimori M, Nakafuku M, Rowitch D, Guillemot F. The proneural gene Mash1 specifies an early population of telencephalic oligodendrocytes. J Neurosci. 2007;27:4233–4242. doi: 10.1523/JNEUROSCI.0126-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patties I, Kortmann R-D, Glasow A. Inhibitory effects of epigenetic modulators and differentiation inducers on human medulloblastoma cell lines. Journal of Experimental & Clinical Cancer Research. 2013;32:27. doi: 10.1186/1756-9966-32-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Brun SN, Markant SL, Lento W, Gibson P, Taketo MM, Giovannini M, Gilbertson RJ, Wechsler-Reya RJ. WNT signaling increases proliferation and impairs differentiation of stem cells in the developing cerebellum. Development. 2012a;139:1724–1733. doi: 10.1242/dev.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, Witt H, Korshunov A, Read TA, Sun JL, Schmitt EM, Miller CR, Buckley AF, McLendon RE, Westbrook TF, Northcott PA, Taylor MD, Pfister SM, Febbo PG, Wechsler-Reya RJ. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012b;21:155–167. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennartz S, Belvindrah R, Tomiuk S, Zimmer C, Hofmann K, Conradt M, Bosio A, Cremer H. Purification of neuronal precursors from the adult mouse brain: comprehensive gene expression analysis provides new insights into the control of cell migration, differentiation, and homeostasis. Mol Cell Neurosci. 2004;25:692–706. doi: 10.1016/j.mcn.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S, Ayers-Ringler J, Nishiyama A, Stallcup WB, Berger MS, Bergers G, McKnight TR, Goldman SA, Weiss WA. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S, Schlaeger C, Mendrzyk F, Wittmann A, Benner A, Kulozik A, Scheurlen W, Radlwimmer B, Lichter P. Array-based profiling of reference-independent methylation status (aPRIMES) identifies frequent promoter methylation and consecutive downregulation of ZIC2 in pediatric medulloblastoma. Nucleic Acids Res. 2007;35:e51. doi: 10.1093/nar/gkm094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Phillips JJ, Aranda D, Ellison DW, Judkins AR, Croul SE, Brat DJ, Ligon KL, Horbinski C, Venneti S, Zadeh G, Santi M, Zhou S, Appin CL, Sioletic S, Sullivan LM, Martinez-Lage M, Robinson AE, Yong WH, Cloughesy T, Lai A, Phillips HS, Marshall R, Mueller S, Haas-Kogan DA, Molinaro AM, Perry A. PDGFRA amplification is common in pediatric and adult high-grade astrocytomas and identifies a poor prognostic group in IDH1 mutant glioblastoma. Brain Pathol. 2013;23:565–573. doi: 10.1111/bpa.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phuphanich S, Scott C, Fischbach AJ, Langer C, Yung WK. All-trans-retinoic acid: a phase II Radiation Therapy Oncology Group study (RTOG 91–13) in patients with recurrent malignant astrocytoma. J Neurooncol. 1997;34:193–200. doi: 10.1023/a:1005765915288. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Vescovi AL. Brain tumour stem cells: possibilities of new therapeutic strategies. Expert Opinion on Biological Therapy. 2007;7:1129–1135. doi: 10.1517/14712598.7.8.1129. [DOI] [PubMed] [Google Scholar]
- Po A, Ferretti E, Miele E, De Smaele E, Paganelli A, Canettieri G, Coni S, Di Marcotullio L, Biffoni M, Massimi L, Di Rocco C, Screpanti I, Gulino A. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. The EMBO Journal. 2010;29:2646–2658. doi: 10.1038/emboj.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh TJ, Weeraratne SD, Archer TC, Krummel DAP, Auclair D, Bochicchio J, Carneiro MO, Carter SL, Cibulskis K, Erlich RL. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012 doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaf J, Kernohan J. A study of the external granular layer in the cerebellum. Am J Anat. 1944:151–172. [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran RS, Wellbrock UM, Zupanc GK. Apoptotic cell death, long-term persistence, and neuronal differentiation of aneuploid cells generated in the adult brain of teleost fish. Dev Neurobiol. 2008;68:1257–1268. doi: 10.1002/dneu.20656. [DOI] [PubMed] [Google Scholar]
- Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135–147. doi: 10.1016/j.ccr.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeld F, Rohde AM, Nguyen DT, Wulczyn FG. Lin28 and let-7: ancient milestones on the road from pluripotencyto neurogenesis. Cell Tissue Res. 2014 doi: 10.1007/s00441-014-1872-2. [DOI] [PubMed] [Google Scholar]
- Remke M, Hielscher T, Northcott PA, Witt H, Ryzhova M, Wittmann A, Benner A, von Deimling A, Scheurlen W, Perry A. Adult medulloblastoma comprises three major molecular variants. Journal of Clinical Oncology. 2011;29:2717–2723. doi: 10.1200/JCO.2011.34.9373. [DOI] [PubMed] [Google Scholar]
- Rheinbay E, Suva ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, Louis DN, Kasif S, Chi AS, Bernstein BE. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3:1567–1579. doi: 10.1016/j.celrep.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Biffoni M, Todaro M, Invernici G, Cenci T, Maira G, Parati EA, Stassi G, Larocca LM, De Maria R. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Pallini R, Larocca LM, Lombardi DG, Signore M, Pierconti F, Petrucci G, Montano N, Maira G, De Maria R. Mesenchymal differentiation of glioblastoma stem cells. Cell Death Differ. 2008;15:1491–1498. doi: 10.1038/cdd.2008.72. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Pringle N, Mosley MJ, Westermark B, Dubois-Dalcq M. A role for platelet-derived growth factor in normal gliogenesis in the central nervous system. Cell. 1988;53:309–319. doi: 10.1016/0092-8674(88)90392-3. [DOI] [PubMed] [Google Scholar]
- Rieske P, Golanska E, Zakrzewska M, Piaskowski S, Hulas-Bigoszewska K, Wolanczyk M, Szybka M, Witusik-Perkowska M, Jaskolski DJ, Zakrzewski K, Biernat W, Krynska B, Liberski PP. Arrested neural and advanced mesenchymal differentiation of glioblastoma cells-comparative study with neural progenitors. BMC Cancer. 2009;9:54. doi: 10.1186/1471-2407-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs JW, Barrilleaux BL, Varlakhanova N, Bush KM, Chan V, Knoepfler PS. Induced pluripotency and oncogenic transformation are related processes. Stem Cells Dev. 2013;22:37–50. doi: 10.1089/scd.2012.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, Chalhoub N, Baker SJ, Huether R, Kriwacki R, Curley N, Thiruvenkatam R, Wang J, Wu G, Rusch M, Hong X, Becksfort J, Gupta P, Ma J, Easton J, Vadodaria B, Onar-Thomas A, Lin T, Li S, Pounds S, Paugh S, Zhao D, Kawauchi D, Roussel MF, Finkelstein D, Ellison DW, Lau CC, Bouffet E, Hassall T, Gururangan S, Cohn R, Fulton RS, Fulton LL, Dooling DJ, Ochoa K, Gajjar A, Mardis ER, Wilson RK, Downing JR, Zhang J, Gilbertson RJ. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- Ross AH, Lachyankar MB, Recht LD. PTEN: a newly identified regulator of neuronal differentiation. Neuroscientist. 2001;7:278–281. doi: 10.1177/107385840100700404. [DOI] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Rousseau-Merck MF, Nogues C, Roth A, Nezelof C, Bourdeau A, Leblanc A, Trincard MD. Hypercalcemic infantile renal tumors: morphological, clinical, and biological heterogeneity. Pediatr Pathol. 1985;3:155–164. doi: 10.3109/15513818509078781. [DOI] [PubMed] [Google Scholar]
- Roussel MF, Hatten ME. Cerebellum development and medulloblastoma. Curr Top Dev Biol. 2011;94:235–282. doi: 10.1016/B978-0-12-380916-2.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 2012;26:1409–1420. doi: 10.1101/gad.193730.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- Santra M, Santra S, Buller B, Santra K, Nallani A, Chopp M. Effect of doublecortin on self-renewal and differentiation in brain tumor stem cells. Cancer Sci. 2011;102:1350–1357. doi: 10.1111/j.1349-7006.2011.01952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santra M, Santra S, Roberts C, Zhang RL, Chopp M. Doublecortin induces mitotic microtubule catastrophe and inhibits glioma cell invasion. J Neurochem. 2009;108:231–245. doi: 10.1111/j.1471-4159.2008.05758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Bucci C, Weis J, Kruttgen A. The small GTPase Rab7 controls the endosomal trafficking and neuritogenic signaling of the nerve growth factor receptor TrkA. Journal of Neuroscience. 2005;25:10930–10940. doi: 10.1523/JNEUROSCI.2029-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AL, Brunetto AL, Schwartsmann G, Roesler R, Abujamra AL. Recent therapeutic advances for treating medulloblastoma: focus on new molecular targets. CNS Neurol Disord Drug Targets. 2010;9:335–348. doi: 10.2174/187152710791292602. [DOI] [PubMed] [Google Scholar]
- Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and - independent pathways. Embo J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalbe EC, Williamson D, Lindsey JC, Hamilton D, Ryan SL, Megahed H, Garami M, Hauser P, Dembowska-Baginska B, Perek D, Northcott PA, Taylor MD, Taylor RE, Ellison DW, Bailey S, Clifford SC. DNA methylation profiling of medulloblastoma allows robust subclassification and improved outcome prediction using formalin-fixed biopsies. Acta Neuropathol. 2013;125:359–371. doi: 10.1007/s00401-012-1077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, Jabado N. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- Seidel S, Garvalov BK, Wirta V, von Stechow L, Schanzer A, Meletis K, Wolter M, Sommerlad D, Henze AT, Nister M, Reifenberger G, Lundeberg J, Frisen J, Acker T. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133:983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- Semi K, Matsuda Y, Ohnishi K, Yamada Y. Cellular reprogramming and cancer development. Int J Cancer. 2013;132:1240–1248. doi: 10.1002/ijc.27963. [DOI] [PubMed] [Google Scholar]
- Seri B, Garcia-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahhoseini M, Taghizadeh Z, Hatami M, Baharvand H. Retinoic acid dependent histone 3 demethylation of the clustered HOX genes during neural differentiation of human embryonic stem cells. Biochem Cell Biol. 2013;91:116–122. doi: 10.1139/bcb-2012-0049. [DOI] [PubMed] [Google Scholar]
- Shi Z, Lou M, Zhao Y, Zhang Q, Cui D, Wang K. Effect of All-Trans Retinoic Acid on the Differentiation of U87 Glioma Stem/Progenitor Cells. Cellular and Molecular Neurobiology. 2013;33:943–951. doi: 10.1007/s10571-013-9960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MM, Manton CA, Bhat KP, Tsai WW, Aldape K, Barton MC, Chandra J. Inhibition of LSD1 sensitizes glioblastoma cells to histone deacetylase inhibitors. Neuro Oncol. 2011;13:894–903. doi: 10.1093/neuonc/nor049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417:39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- Sottile V, Li M, Scotting PJ. Stem cell marker expression in the Bergmann glia population of the adult mouse brain. Brain research. 2006;1099:8–17. doi: 10.1016/j.brainres.2006.04.127. [DOI] [PubMed] [Google Scholar]
- Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, Bostrom E, Westerlund I, Vial C, Buchholz BA, Possnert G, Mash DC, Druid H, Frisen J. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–1227. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S, Ditzler S, Pullar B, Olson J. Response of preclinical medulloblastoma models to combination therapy with 13-cis retinoic acid and suberoylanilide hydroxamic acid (SAHA) Journal of Neuro-Oncology. 2008;87:133–141. doi: 10.1007/s11060-007-9505-1. [DOI] [PubMed] [Google Scholar]
- Spyropoulou A, Piperi C, Adamopoulos C, Papavassiliou AG. Deregulated chromatin remodeling in the pathobiology of brain tumors. Neuromolecular Med. 2013;15:1–24. doi: 10.1007/s12017-012-8205-y. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Barbas H, Dermon CR. Late granule cell genesis in quail cerebellum. J Comp Neurol. 2004;474:173–189. doi: 10.1002/cne.20066. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm R, Cholewa-Waclaw J, Reuter K, Brohl D, Sieber M, Treier M, Muller T, Birchmeier C. The bHLH transcription factor Olig3 marks the dorsal neuroepithelium of the hindbrain and is essential for the development of brainstem nuclei. Development. 2009;136:295–305. doi: 10.1242/dev.027193. [DOI] [PubMed] [Google Scholar]
- Stricker SH, Feber A, Engström PG, Carén H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P, Smith A, Beck S, Pollard SM. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes & Development. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tonjes M, Sill M, Bender S, Kool M, Zapatka M, Becker N, Zucknick M, Hielscher T, Liu XY, Fontebasso AM, Ryzhova M, Albrecht S, Jacob K, Wolter M, Ebinger M, Schuhmann MU, van Meter T, Fruhwald MC, Hauch H, Pekrun A, Radlwimmer B, Niehues T, von Komorowski G, Durken M, Kulozik AE, Madden J, Donson A, Foreman NK, Drissi R, Fouladi M, Scheurlen W, von Deimling A, Monoranu C, Roggendorf W, Herold-Mende C, Unterberg A, Kramm CM, Felsberg J, Hartmann C, Wiestler B, Wick W, Milde T, Witt O, Lindroth AM, Schwartzentruber J, Faury D, Fleming A, Zakrzewska M, Liberski PP, Zakrzewski K, Hauser P, Garami M, Klekner A, Bognar L, Morrissy S, Cavalli F, Taylor MD, van Sluis P, Koster J, Versteeg R, Volckmann R, Mikkelsen T, Aldape K, Reifenberger G, Collins VP, Majewski J, Korshunov A, Lichter P, Plass C, Jabado N, Pfister SM. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Su H, Zhang W, Guo J, Guo A, Yuan Q, Wu W. Lithium enhances the neuronal differentiation of neural progenitor cells in vitro and after transplantation into the avulsed ventral horn of adult rats through the secretion of brain-derived neurotrophic factor. J Neurochem. 2009;108:1385–1398. doi: 10.1111/j.1471-4159.2009.05902.x. [DOI] [PubMed] [Google Scholar]
- Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, Lim DA, Vandenberg S, Stallcup W, Berger MS, Bergers G, Weiss WA, Petritsch C. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328–340. doi: 10.1016/j.ccr.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Colello RJ, Daugherty WP, Kwon TH, McGinn MJ, Harvey HB, Bullock MR. Cell proliferation and neuronal differentiation in the dentate gyrus in juvenile and adult rats following traumatic brain injury. J Neurotrauma. 2005a;22:95–105. doi: 10.1089/neu.2005.22.95. [DOI] [PubMed] [Google Scholar]
- Sun Y, Goderie SK, Temple S. Asymmetric distribution of EGFR receptor during mitosis generates diverse CNS progenitor cells. Neuron. 2005b;45:873–886. doi: 10.1016/j.neuron.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva ML, Riggi N, Janiszewska M, Radovanovic I, Provero P, Stehle JC, Baumer K, Le Bitoux MA, Marino D, Cironi L, Marquez VE, Clement V, Stamenkovic I. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- Swartling FJ. Myc proteins in brain tumor development and maintenance. Upsala journal of medical sciences. 2012;117:122–131. doi: 10.3109/03009734.2012.658975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Bolin S, Phillips JJ, Persson AI. Signals that regulate the oncogenic fate of neural stem cells and progenitors. Experimental neurology. 2013a doi: 10.1016/j.expneurol.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, Lau J, Masic S, Nguyen K, Yakovenko S, Zhe XN, Gilmer HC, Collins R, Nagaoka M, Phillips JJ, Jenkins RB, Tihan T, Vandenberg SR, James CD, Tanaka K, Taylor MD, Weiss WA, Chesler L. Pleiotropic role for MYCN in medulloblastoma. Genes & development. 2010;24:1059–1072. doi: 10.1101/gad.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Hede SM, Weiss WA. What underlies the diversity of brain tumors? Cancer metastasis reviews. 2013b;32:5–24. doi: 10.1007/s10555-012-9407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, Perry A. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, Eberhart CG, Parsons DW, Rutkowski S, Gajjar A, Ellison DW, Lichter P, Gilbertson RJ, Pomeroy SL, Kool M, Pfister SM. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivnan A, Zhao J, Johns TG, Day BW, Stringer BW, Boyd AW, Tiwari S, Giles KM, Teo C, McDonald KL. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumour Biol. 2014;35:1459–1465. doi: 10.1007/s13277-013-1200-6. [DOI] [PubMed] [Google Scholar]
- Torper O, Pfisterer U, Wolf DA, Pereira M, Lau S, Jakobsson J, Bjorklund A, Grealish S, Parmar M. Generation of induced neurons via direct conversion in vivo. Proc Natl Acad Sci U S A. 2013;110:7038–7043. doi: 10.1073/pnas.1303829110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tso CL, Freije WA, Day A, Chen Z, Merriman B, Perlina A, Lee Y, Dia EQ, Yoshimoto K, Mischel PS, Liau LM, Cloughesy TF, Nelson SF. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- Turcan S, Fabius AW, Borodovsky A, Pedraza A, Brennan C, Huse J, Viale A, Riggins GJ, Chan TA. Efficient induction of differentiation and growth inhibition in IDH1 mutant glioma cells by the DNMT Inhibitor Decitabine. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwanogho D, Rex M, Cartwright EJ, Pearl G, Healy C, Scotting PJ, Sharpe PT. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci U S A. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, Kushida M, Head R, Morrissy S, Zhu X, Aviv T, Voisin V, Clarke ID, Li Y, Mungall AJ, Moore RA, Ma Y, Jones SJ, Marra MA, Malkin D, Northcott PA, Kool M, Pfister SM, Bader G, Hochedlinger K, Korshunov A, Taylor MD, Dirks PB. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26:33–47. doi: 10.1016/j.ccr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese M, Olstorn H, Sandberg C, Vik-Mo EO, Noordhuis P, Nister M, Berg-Johnsen J, Moe MC, Langmoen IA. A comparison between stem cells from the adult human brain and from brain tumors. Neurosurgery. 2008;63:1022–1033. doi: 10.1227/01.NEU.0000335792.85142.B0. discussion 1033–1024. [DOI] [PubMed] [Google Scholar]
- Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5:e10748. doi: 10.1371/journal.pone.0010748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman S, Birks DK, Balakrishnan I, Alimova I, Harris PS, Patel PR, Handler MH, Dubuc A, Taylor MD, Foreman NK, Vibhakar R. MicroRNA 218 Acts as a Tumor Suppressor by Targeting Multiple Cancer Phenotype-associated Genes in Medulloblastoma. Journal of Biological Chemistry. 2013;288:1918–1928. doi: 10.1074/jbc.M112.396762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdi JM, Anderson DJ. Neurotrophins regulate sequential changes in neurotrophin receptor expression by sympathetic neuroblasts. Neuron. 1994;13:1359–1372. doi: 10.1016/0896-6273(94)90421-9. [DOI] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- Wadhwa S, Nag TC, Jindal A, Kushwaha R, Mahapatra AK, Sarkar C. Expression of the neurotrophin receptors Trk A and Trk B in adult human astrocytoma and glioblastoma. Journal of Biosciences. 2003;28:181–188. doi: 10.1007/BF02706217. [DOI] [PubMed] [Google Scholar]
- Wang C, You Y, Qi D, Zhou X, Wang L, Wei S, Zhang Z, Huang W, Liu Z, Liu F, Ma L, Yang Z. Human and monkey striatal interneurons are derived from the medial ganglionic eminence but not from the adult subventricular zone. J Neurosci. 2014;34:10906–10923. doi: 10.1523/JNEUROSCI.1758-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468:829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- Wang S-C, Hong J-H, Hsueh C, Chiang C-S. Tumor-secreted SDF-1 promotes glioma invasiveness and TAM tropism toward hypoxia in a murine astrocytoma model. Lab Invest. 2012a;92:151–162. doi: 10.1038/labinvest.2011.128. [DOI] [PubMed] [Google Scholar]
- Wang W, Macaulay RJ. Apoptosis of medulloblastoma cells in vitro follows inhibition of farnesylation using manumycin A. Int J Cancer. 1999;82:430–434. doi: 10.1002/(sici)1097-0215(19990730)82:3<430::aid-ijc17>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kim E, Wang X, Novitch BG, Yoshikawa K, Chang LS, Zhu Y. ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell. 2012b;150:816–830. doi: 10.1016/j.cell.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RJ, Lee L, Graham K, Satkunendran T, Yoshikawa K, Ling E, Harper L, Austin R, Nieuwenhuis E, Clarke ID, Hui CC, Dirks PB. Multipotent CD15+ cancer stem cells in patched-1-deficient mouse medulloblastoma. Cancer Res. 2009;69:4682–4690. doi: 10.1158/0008-5472.CAN-09-0342. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Weeraratne SD, Amani V, Neiss A, Teider N, Scott DK, Pomeroy SL, Cho YJ. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13:165–175. doi: 10.1093/neuonc/noq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers AK, Warmuth-Metz M, Poschl J, von Bueren AO, Monoranu CM, Seelos K, Peraud A, Tonn JC, Koch A, Pietsch T, Herold-Mende C, Mawrin C, Schouten-van Meeteren A, van Vuurden D, von Hoff K, Rutkowski S, Pfister SM, Kool M, Schuller U. Subgroupspecific localization of human medulloblastoma based on pre-operative MRI. Acta Neuropathol. 2014 doi: 10.1007/s00401-014-1271-5. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Weller M, Gorlia T, Cairncross JG, van den Bent MJ, Mason W, Belanger K, Brandes AA, Bogdahn U, Macdonald DR, Forsyth P, Rossetti AO, Lacombe D, Mirimanoff RO, Vecht CJ, Stupp R. Prolonged survival with valproic acid use in the EORTC/NCIC temozolomide trial for glioblastoma. Neurology. 2011;77:1156–1164. doi: 10.1212/WNL.0b013e31822f02e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey A, Knoepfler PS. c-myc and N-myc promote active stem cell metabolism and cycling as architects of the developing brain. Oncotarget. 2010;1:120–130. doi: 10.18632/oncotarget.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c-and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum. 2010;9:537–547. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsson U, Faiz M, de Pablo Y, Sjoqvist M, Andersson D, Widestrand A, Potokar M, Stenovec M, Smith PL, Shinjyo N, Pekny T, Zorec R, Stahlberg A, Pekna M, Sahlgren C, Pekny M. Astrocytes negatively regulate neurogenesis through the Jagged1-mediated Notch pathway. Stem Cells. 2012;30:2320–2329. doi: 10.1002/stem.1196. [DOI] [PubMed] [Google Scholar]
- Wolanczyk M, Hulas-Bigoszewska K, Witusik-Perkowska M, Papierz W, Jaskolski D, Liberski PP, Rieske P. Imperfect oligodendrocytic and neuronal differentiation of glioblastoma cells. Folia Neuropathol. 2010;48:27–34. [PubMed] [Google Scholar]
- Woodruff RH, Fruttiger M, Richardson WD, Franklin RJ. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol Cell Neurosci. 2004;25:252–262. doi: 10.1016/j.mcn.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Baker SJ. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang AC, Xie J, Zhang XJ. Acetylcholinesterase in intestinal cell differentiation involves G2/M cell cycle arrest. Cellular and Molecular Life Sciences. 2008;65:1768–1779. doi: 10.1007/s00018-008-8016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Skaftnesmo KO, Leiss L, Sleire L, Wang J, Li X, Enger PO. Neuronal markers are expressed in human gliomas and NSE knockdown sensitizes glioblastoma cells to radiotherapy and temozolomide. BMC Cancer. 2011;11:524. doi: 10.1186/1471-2407-11-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Niu CS, Cheng CD. Pin1-Nanog expression in human glioma is correlated with advanced tumor progression. Oncol Rep. 2013;30:560–566. doi: 10.3892/or.2013.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schuller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Xiao H, Xu M, Ye X, Hu J, Li F, Li M, Luo C, Yu S, Bian X, Feng H. Glioma-initiating cells: A predominant role in microglia/macrophages tropism to glioma. Journal of Neuroimmunology. 2011;232:75–82. doi: 10.1016/j.jneuroim.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Yin D, Ong JM, Hu J, Desmond JC, Kawamata N, Konda BM, Black KL, Koeffler HP. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor: effects on gene expression and growth of glioma cells in vitro and in vivo. Clin Cancer Res. 2007;13:1045–1052. doi: 10.1158/1078-0432.CCR-06-1261. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TF, Li J, Ding F, Arias-Carrion O. Evidence of adult neurogenesis in non-human primates and human. Cell Tissue Res. 2014 doi: 10.1007/s00441-014-1980-z. [DOI] [PubMed] [Google Scholar]
- Zbinden M, Duquet A, Lorente-Trigos A, Ngwabyt SN, Borges I, Ruiz i Altaba A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010;29:2659–2674. doi: 10.1038/emboj.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner D, Fujita Y, Hulsken J, Muller T, Walther I, Taketo MM, Crenshaw EB, 3rd, Birchmeier W, Birchmeier C. beta-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–418. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li P, Hsu T, Aguilar HR, Frantz DE, Schneider JW, Bachoo RM, Hsieh J. Small-molecule blocks malignant astrocyte proliferation and induces neuronal gene expression. Differentiation. 2011;81:233–242. doi: 10.1016/j.diff.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Ml, Tao Y, Zhou Wq, Ma Pc, Cao Yp, He Cd, Wei J, Li Lj. All-trans retinoic acid induces cell-cycle arrest in human cutaneous squamous carcinoma cells by inhibiting the mitogen-activated protein kinase–activated protein 1 pathway. Clinical and Experimental Dermatology. 2014;39:354–360. doi: 10.1111/ced.12227. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Ye P, Li S, Shi Y. MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci Rep. 2013;3:1329. doi: 10.1038/srep01329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Wu X, Tan F, Shi YX, Glass T, Liu TJ, Wathen K, Hess KR, Gumin J, Lang F, Yung WK. PAX6 suppresses growth of human glioblastoma cells. J Neurooncol. 2005;71:223–229. doi: 10.1007/s11060-004-1720-4. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YY, Li YY, Haraguchi S, Yu M, Ohira M, Ozaki T, Nakagawa A, Ushijima T, Isogai E, Koseki H, Nakamura Y, Kong CZ, Mehlen P, Arakawa H, Nakagawara A. Dependence receptor UNC5D mediates nerve growth factor depletion-induced neuroblastoma regression. Journal of Clinical Investigation. 2013;123:2935–2947. doi: 10.1172/JCI65988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinin N, Adameyko I, Wilhelm M, Fritz N, Uhlen P, Ernfors P, Henriksson MA. MYC proteins promote neuronal differentiation by controlling the mode of progenitor cell division. EMBO Rep. 2014;15:383–391. doi: 10.1002/embr.201337424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupanc GK, Hinsch K, Gage FH. Proliferation, migration, neuronal differentiation, and long-term survival of new cells in the adult zebrafish brain. J Comp Neurol. 2005;488:290–319. doi: 10.1002/cne.20571. [DOI] [PubMed] [Google Scholar]