Abstract
Background
Krüpple-like factor 5 (KLF5) is a transcription factor that is highly expressed in the proliferative compartment of the intestinal crypt. There, it is thought to regulate epithelial turnover and homeostasis.
Aim
In this study, we sought to determine the role for Klf5 in the maintenance of cellular proliferation, cytodifferentiation and morphology of the crypt-villus axis.
Methods
Tamoxifen-induced recombination directed by the epithelial-specific Villin promoter (in Villin-CreERT2 transgenic mice) was used to delete Klf5 (in Klf5loxP/loxP mice) from the adult mouse intestine, and analyzed by immunostaining and RT-qPCR. Control mice were tamoxifen-treated Klf5loxP/loxP mice lacking Villin-CreERT2.
Results
Three days after tamoxifen-induced recombination, the mitosis marker phospho-histone H3 was significantly reduced within the Klf5-mutant crypt epithelium, coincident with increased expression of the apoptosis marker cleaved-caspase 3 within the crypt where cell death rarely occurs normally. We also observed a reduction in Chromagranin A expressing enteroendocrine cells, though no significant change was seen in other secretory or absorptive cell types. To examine the long term repercussions of Klf5 loss, we sacrificed mice 5, 14, and 28 days post recombination and found reemerging expression of KLF5. Furthermore, we observed restoration of cellular proliferation, though not to levels seen wildtype intestinal crypts. Reduction of apoptosis to levels comparable to the wildtype intestinal crypt was also observed at later time points. Analysis of cell cycle machinery indicated no significant perturbation upon deletion of Klf5; however a reduction of stem cell markers Ascl2, Lgr5, and Olfm4 was observed at all time points following Klf5 deletion.
Conclusions
These results indicate that Klf5 is necessary to maintain adult intestinal crypt proliferation and proper cellular differentiation. Rapid replacement of Klf5-mutant crypts with wildtype cells and reduction of stem cell markers suggests further that Klf5 is required for self renewal of intestinal stem cells.
Keywords: Krüpple-like factor 5, proliferation, apoptosis, cellular differentiation
Introduction
The intestinal epithelium is a rapidly renewing surface that possesses the ability to regulate nutrient absorption, balance digestive hormone production, and protect the host from noxious lumenal contents [1-4]. The dynamic function of this organ relies on specific coordination between proliferation and differentiation to maintain proper homeostatic regeneration. In the adult intestine, the crypts of Lieberkühn are the niche for epithelial stem cells and contain all proliferative stem and progenitor cells. Differentiating cells exit the cell cycle and migrate out of the crypts and onto the surface epithelium of the intestine, where they perform their physiological roles (e.g., nutrient absorption by enterocytes; mucous secretion by goblet cells) and are eventually shed into the lumen [5, 6]. Homeostatic maintenance of these stem and progenitor cell populations is essential to support the rapid turnover of the intestinal epithelium.
Krüpple-like factors are a family of DNA-binding zinc finger transcription factors that regulate a wide variety cellular and context dependent processes such as proliferation, differentiation, and migration [7-9]. Among these, Krüpple-like factor 5 (KLF5) is a pro-proliferative transcription factor found to be essential in embryogenesis and organogenesis [10-14]. KLF5 is highly expressed in the proliferative compartments of epithelial surfaces such as the skin and gastrointestinal tract [15-17]. Cultured fibroblast and intestinal epithelial cell assays were among the first to show that KLF5 is a positive regulator of cellular proliferation. Furthermore, Klf5 mRNA was found in actively proliferating cells, while transient overexpression of this transcription factor significantly enhanced proliferation, cell growth, and anchorage independent growth [13, 18-20].
Deregulated cellular proliferation can influence life-threatening intestinal pathophysiology such as inflammation or cancer. Studies investigating the role of Klf5 in DSS-induced colitis determined it was requisite for the proliferative and migratory responses required for epithelial repair [21]. Furthermore, it was found to increase the proliferative capacity of the colon after bacterial infection [22]. Interestingly, the KLF5 gene is amplified in colorectal cancers and frequently overexpressed in KRAS mutated colorectal cancers, suggesting a contributory role in tumorigenesis [23, 24]. In vitro analysis showed that KLF5 mediates the hyperproliferative phenotype found in HRAS and KRAS transformed cells through induction of mitogen-activated protein kinase signaling and cell cycle related genes[20, 23, 25]. In vivo studies further concluded that Klf5 mediated tumorigenesis in intestines harboring KRAS mutations, loss of the tumor suppressor adenomatous polyposis coli (APC), or both KRAS and APC mutation: heterozygosity for Klf5 attenuated both number and size of intestinal adenomas in both Apcmin/KRAS and Apcmin induced tumors [26, 27]. These data implicate KLF5 as a key mediator of intestinal health; however, its primary role in regulation of normal epithelial homeostasis remains unclear.
A recent study examined the role of Klf5 in the intestine by generating a mouse model in which conditional deletion of Klf5 was directed by the epithelial-specific Villin promoter. It was concluded that loss of Klf5 in the adult results in the loss of crypt architecture, loss of barrier function, impaired differentiation, migration, and proliferation[28]. However, this study also reported a 66% mortality rate shortly after birth in mice lacking Klf5 in the intestinal epithelium. Subsequently it was determined that Klf5 is integral part of villus formation and cellular differentiation in the embryonic small intestine at the time of villus formation [29]. Based on this result, it was unclear whether the reported adult phenotype of Klf5 mutant mice was due to dysfunction of Klf5 in the adult intestine or a result of maldeveloped tissue thus prompting the need for a novel inducible knockout of this gene. Therefore, we set out to distinguish these possibilities by determining the effect of Klf5 loss in the adult intestinal epithelium utilizing an inducible Klf5 deletion mouse model. In this present study, we found that absence of Klf5 acutely inhibited proliferation and concomitantly increased cell death in the crypts. Long term loss of Klf5 was not sustainable, and strong selection for expansion of the remaining undeleted epithelium allowed repopulation of the crypt with wildtype epithelium along with abatement of cell death. However, loss of stem cell markers, Lgr5, Ascl2, and Olf4 as well as decreased proliferation remained through 28 days after recombination.
Materials and Methods
Animals
VilCreERT2 mice [30] were mated with Klf5loxP/loxP mice [31] to produce an F2 generation of Klf5loxP/loxP; VilCreERT2 experimental mice and Klf5loxP/loxP; VilWT controls. Eight week old mice were given an intraperitoneal injection of 80mg/kg tamoxifen (Sigma-Aldrich, St. Louis, MO) dissolved in sesame oil, for two consecutive days. Animals were sacrificed 3, 5, 14, and28 days after first injection. DNA was extracted from tail clippings and utilized for PCR to determine genotype.
Tissue Staining
All intestinal specimens were fixed in 4% paraformaldehyde overnight at 4°C, embedded in paraffin, and cut into 5μm sections. For immunofluorescence, antigen retrieval was performed in 10mM sodium citrate solution (pH 6.0). Tissue was blocked in 4% normal donkey serum and then stained with antibodies raised against KLF5 (a gift from Dr. Jeffrey Whitsett, Cincinnati Children's Hospital, OH; 1:500), Ki67 (Leica Biosystems, New Castle, UK; 1:1000), Chromagranin A (CHGA) (Immuostar, Hudson, WI; 1:5000), Mucin2 (MUC2) (Santa Cruz, Dallas, TX; 1:1000), Lysozyme (LYZ) (Zymed Laboratories, San Francisco, CA; 1:5000), or phospho-Histone H3 (PHH3) (Cell Signaling, Boston, MA; 1:1000). The sections were then mounted with Vectashield containing DAPI stain. For immunohistochemistry, antigen retrieval was performed in 10mM Tris, 1mM EDTA, 0.05% Tween solution (pH 9.0). Tissues were blocked with 0.3% H2O2 in Methanol, then Avidin/Biotin(Vector Laboratories, Burlingame, CA), and finally in 4% normal goat serum. Staining was performed for cleaved-caspase 3 (CC3) (Cell Signaling, Boston, MA; 1:100) using the Vector ABC kit (Vector Laboratories, Burlingame, CA). Development was completed utilizing liquid DAB (Vector Laboratories, Burlingame, CA) and counterstained with hemotoxylin (Leica Biosystems,New Castle, UK) . Intestinal tissue was evaluated from at least three animals per time point.
RNA Isolation and qRT-PCR
Jejunal segments of approximately 1cm were excised from the small intestine and immediately placed in TRIzol reagent (Life Technologies, Carlsbad, CA). RNA was isolated as described in the manufacturer's instructions and subsequently purified with the RNeasy kit (QIAGEN, Valencia, CA) with DNase treatment (QIAGEN, Valencia, CA). Five micrograms of RNA were used to synthesize of cDNA using Superscript III First Strand Synthesis System (Invitrogen, Carlsbad, CA) according to manufacturer's protocol. Taqman assays were carried out utilizing ABsolute Blue QPCR Low ROX Mix (Thermo Scientific, Waltham, MA) with commercially available probe and primer sets were used for Klf5 (Mm00456521_m1), Ccnd1 (Catalog # Mm00432359_m1), Ccnb1 (Catalog # Mm03053893_gH), and Cdk1 (Catalog # Mm00772472_m1) (Life Technologies, Carlsbad, CA) with 3 to 6 animals per group. Relative RNA expression was found by absolute quantification with standard curves created from pooled cDNA, normalized to Actb (Mm00607939_s1), and reported expression relative to the control group. SYBR green assays were carried out utilizing Brilliant III Ultra Fast SYBR Green Master Mix (Agilent Technologies, Santa Clara, CA) with ChgA (F: 5′-CCCACTGCAGCATCCAGTT-3′ , R: 5′- AGTCCGACTGACCATCATCTTTC-3′ ), Muc2 (F: 5′– ATGCCCACCTCCTCAAAGAC-3′, R: 5′–GTAGTTTCCGTTGGAACAGTGAA-3′), Fabp1 (F: 5′–ATGAACTTCTCCGGCAAGTACC-3′, R: 5′–CTGACACCCCCTTGATGTCC-3′), Gata4 (F: 5′–CCCTACCCAGCCTACATGG-3′, R: 5′–ACATATCGAGATTGGGGTGTCT-3′), Lgr5 (F – ACATTCCCAAGGGAGCGTTC, R – ATGTGGTTGGCATCTAGGCG), Ascl2 (F – AAGCACACCTTGACTGGTACG, R-AAGCACACCTTGACTGGTACG), and Olfm4 (F – CAGCCACTTTCCAATTTCACTG, R – GCTGGACATACTCCTTCACCTTA) primers. Relative RNA expression was found by absolute quantification with standard curves created from pooled cDNA, normalized to Gapdh (F: 5′-AGGTCGGTGTGAACGGATTTG-3′, R: 5′-TGTAGACCATGTAGTTGAGGTCA-3′), and reported as expression relative to the control group.
Cell Quantification
Proliferation was calculated as an average of PHH3 positive cells per total crypt cell numbers for at least 20 crypts per region per mouse. Apoptosis was calculated as an average number of CC3 positive cells per crypt. The final numbers were averaged per mouse group (wildtype or Klf5-mutant at 3, 5, 14, or 28 days following tamoxifen treatment) to give the average proliferation or apoptosis rate for each segment of the intestine. These rates were then compared for each segment of the intestine, with significance assessed by Student's t-test. Data is reported as mean ± SD.
Results
KLF5 expression overlaps with proliferative cells in the small and large intestine
In the intestine, expression of KLF5 is predominantly localized in the proliferative crypt cells [32] extending up to the lower 1/3 of the villi in the small intestine [27, 28, 32]. This was confirmed by examining the co-expression of KLF5 with the proliferative marker Ki67. Along the epithelium, KLF5 expression is highest in the top two-thirds of the crypt with expression fading towards the bottom third. The expression of Ki67 is confined to the proliferative population of the crypts in both the large and small bowel. Immunoflourescent staining showed extensive overlap between Ki67 and KLF5 indicating that proliferative cells express this transcription factor (Figure 1A-C).
Figure 1. KLF5 is expressed in proliferative crypts of the small and large intestine.
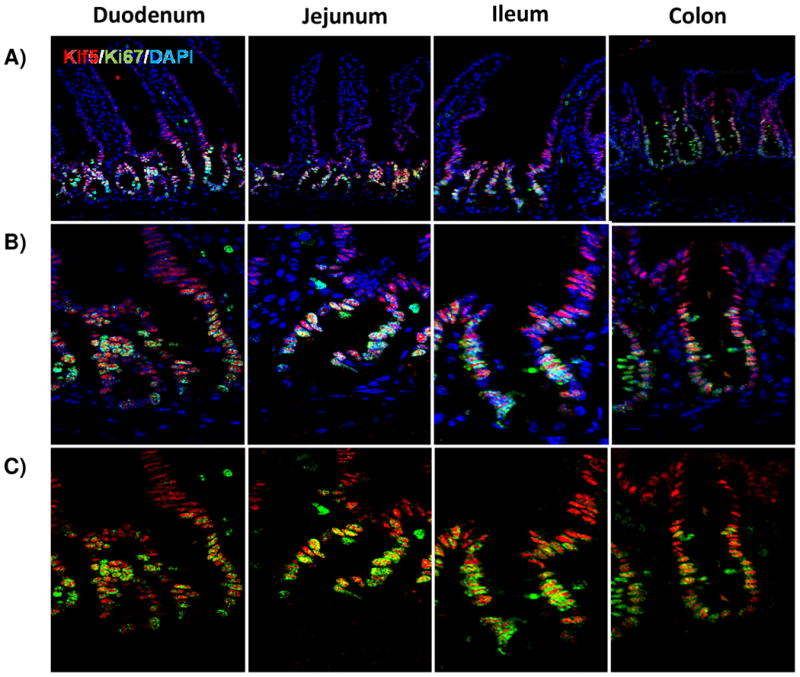
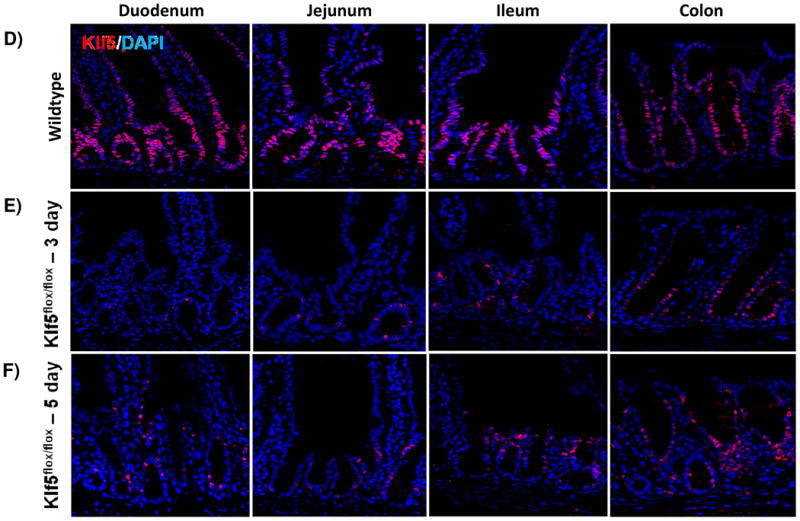
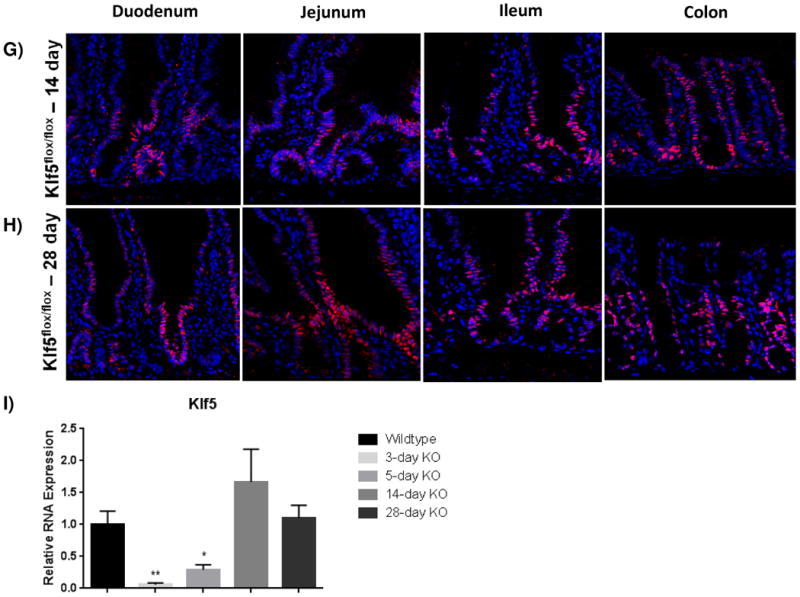
(A) Immunofluorescence staining for coexpression of KLF5 (red) and Ki67 (green). DAPI (blue) was utilized as a nuclear stain.(B) Immunofluorescence staining of the crypt with DAPI (B) and without DAPI (C) show regions of Klf5 and Ki67 expression overlap. Colocalization between these two proteins is seen in yellow. (D-H) Regional histological analysis of tamoxifen-induced recombination in Klf5loxP/loxP; VilWT control mice (D) and Klf5loxP/loxP; VilCreERT2 experimental mice 3 (E), 5 (F), 14 (G), and 28 days (H). (I) Comparison of Klf5 mRNA expression analysis by RT-qPCR from jejunal segments of the small intestine. Student's t-test was performed to determine p-values; expression was normalized to Actb. (Mean ± SEM: P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n = 3-6).
To determine the role of KLF5 in the adult intestine, we utilized conditional Klf5 knockout mice that contain loxP sites flanking exons 2 and 3, as previously described [31]. Immunofluorescence was used to determine the efficiency of recombination in the adult mouse. Three days after the first intraperitoneal injection, histological staining showed efficient depletion of KLF5 throughout the proximal small intestine and a large decrease in KLF5 positive cells in the ileum and colon (Figure 1D-E). As Klf5 loss was previously reported to be detrimental to barrier function and overall intestinal maintenance [28], we wanted to examine the long-term effects of Klf5 absence. Interestingly, we found that Klf5 loss was not maintained for 28 days for any experimental mice (Figure 1B). Intermediate time points were examined to determine when KLF5 reemerged. KLF5 remained low 5 days after tamoxifen treatment, whereas extensive reemergence of KLF5 could be seen by 14 days after recombination (Figure 1A, 1G-H). The results were confirmed by RT-qPCR on isolated jejunal segments which showed almost complete loss of Klf5 mRNA at 3 and 5 days after recombination and restoration of Klf5 expression at days 14 and 28 (Figure 1I).
Loss of Klf5 affects cellular differentiation
It was previously reported that loss of Klf5 in the adult intestine has a significant effect on villus formation and inhibits cytodifferentiation [28, 33]. To examine these effects, we utilized immunofluorescence analysis to visualize cellular differentiation within the intestine 3 and 5 days after tamoxifen treatment coincident with Klf5 depletion. Furthermore, it is at these time points that turnover of the villus epithelium is nearly complete. The overall crypt-villus architecture of the small and large intestines was preserved in Klf5-mutant mice (Figure 2). Immunofluorescence indicated a decrease in the number of enteroendocrine cells (CHGA-positive cells) within the small intestine of the 3 and 5 day animals (Figure 2A-C). RNA analysis indicated a significant reduction in ChgA was maintained for 14 days, but returned to baseline levels by 28 days (Figure 2M). Normal numbers and placement of goblet cells (MUC2-positive) (Figure D-F) and enterocytes (DPP4-positive cells) (Figure G-I) were seen within the small and large intestines of Klf5-mutant mice (Figure 2D-F). RNA expression analysis confirmed normal expression of goblet cell marker, Muc2, and enterocyte markers, Gata4 and Fabp1, 3 days post tamoxifen treatment (Figure M).
Figure 2. Loss of KLF5 affects cytodifferentiation.
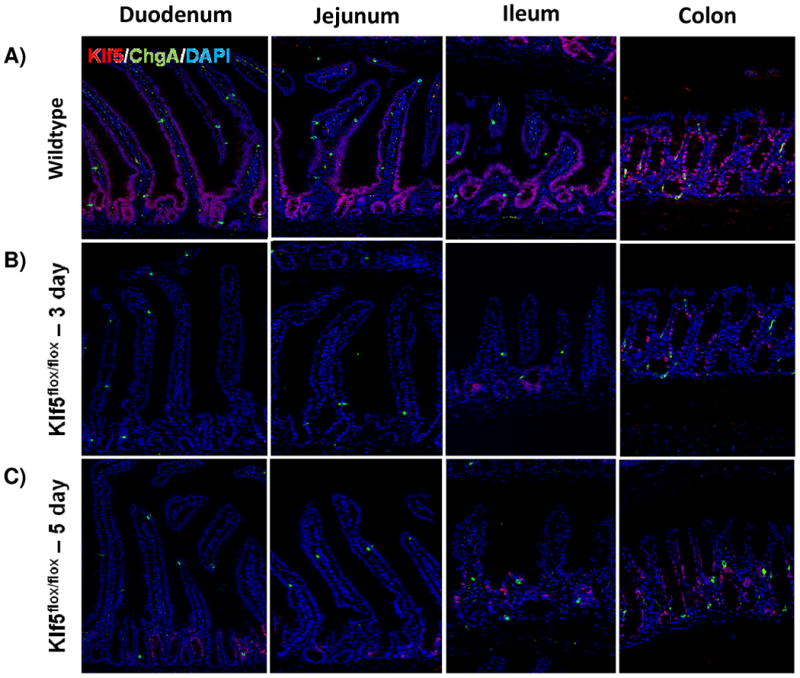
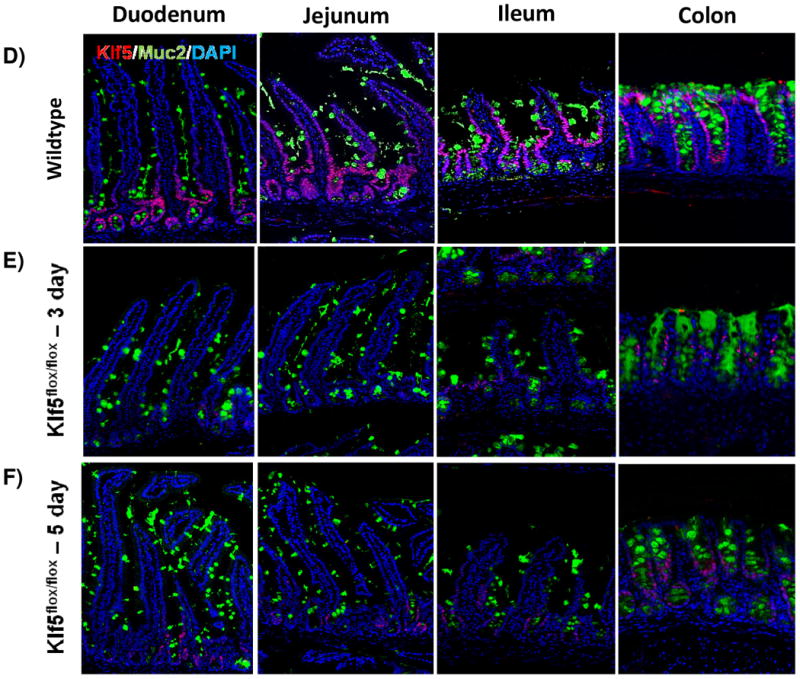
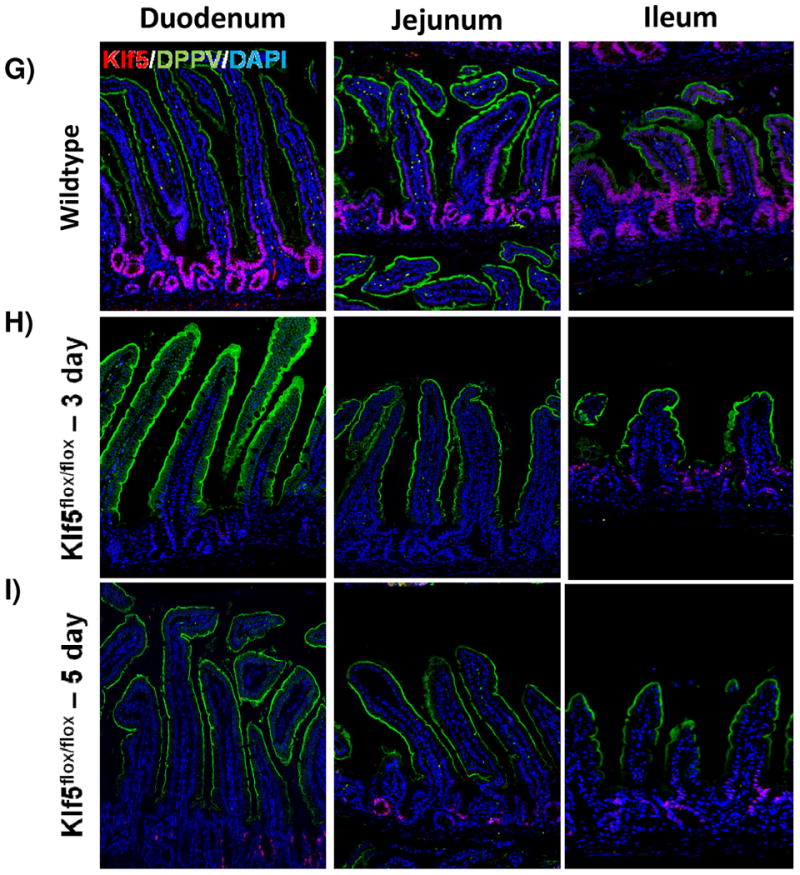
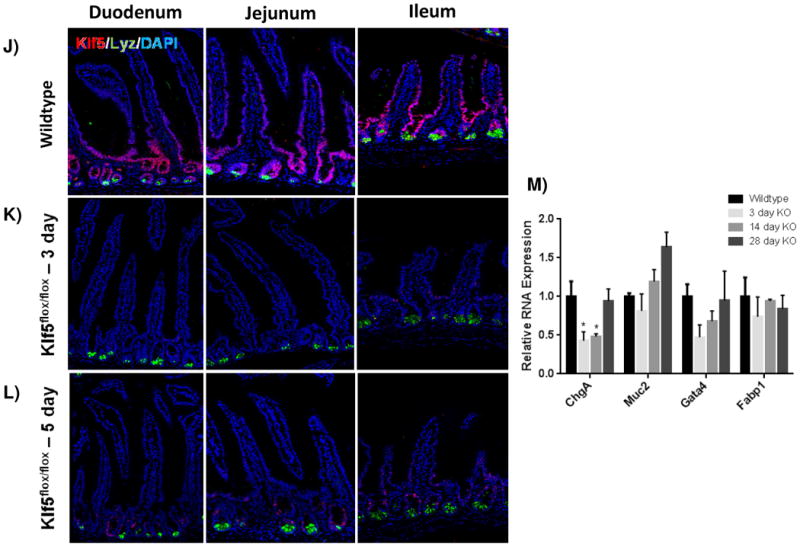
Histological comparison of Klf5loxP/loxP; VilWT (WT) and Klf5loxP/loxP; VilCreERT2 (Klf5-mutant) intestinal tissues for cellular differentiation utilizing immunofluorescent staining for chromagranin A (green; enteroendocrine cells) (A-C),mucin2 (green; goblet cells) (D-F), lysozyme (green; Paneth cells) (G-I), and dipeptidyl peptidase-4 (green; enterocytes) (J-L). Nuclear stain is DAPI (blue). (M) Comparison of ChgA, Muc2, Gata4, and Fabp1 mRNA expression analysis by RT-qPCR from jejunal segments of the small intestine. Student's t-test was performed to determine p-values; expression was normalized to Gapdh. (Mean ± SEM: P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n = 3-6).
Paneth cell lifespans (approximately 60 days) are longer than cells migrating up the villi and thus deletion of Klf5 for 3-5 days is not expected to perturb the census of these long-lived cells. However, it was previously reported that Klf5 loss interfered with proper cell migration, specifically in Paneth cell placement [28]. Therefore we examined their localization in our knockout mice. Paneth cells (LYZ-positive) remained localized along the bottom of the small intestinal crypts of Klf5-mutant mice (Figure 2J-L). Together these data suggest that loss of Klf5 results in enteroendocrine cell loss within the small intestine, but does not significantly affect the other cell types during the time periods examined.
Loss of Klf5 reduces proliferation
KLF5 is a well-known pro-proliferative transcription factor, and its loss is associated with decreased proliferation in other epithelial cell types[11, 13]. As the expression of KLF5 overlaps with the proliferative transient amplifying population of cells, we hypothesized that loss of Klf5 would have an effect on proliferation. Tissue analyzed three days after recombination showed a significant decrease in proliferating crypt cells as measured by the mitotic marker, PHH3. After 3 days, the number of cells in mitosis dropped significantly in all regions of the intestine. This drop was nearly three-fold in the duodenum (9.13±0.97% to 3.22±0.79%), jejunum (8.86±0.83% to 3.28±0.63%), and colon (8.82±0.67% to 3.68±0.12%). While magnitude of effect was slightly less, probably due to a less robust loss of Klf5, the loss in proliferation was also significant in the ileum (7.05±0.35% to 3.22±0.34%) (Figure 3A-B, 3F). Reduced proliferation was similar in the intestines after loss of Klf5 for 5 days with proliferation rates also reduced nearly three-fold in (3.75±1.26% in the duodenum, 2.78±0.65% in the jejunum, 2.64±0.29% in the ileum, and 3.73±0.38% in the colon) (Figure 3C, 3F).
Figure 3. Loss of KLF5 attenuates cellular proliferation in the adult intestine.
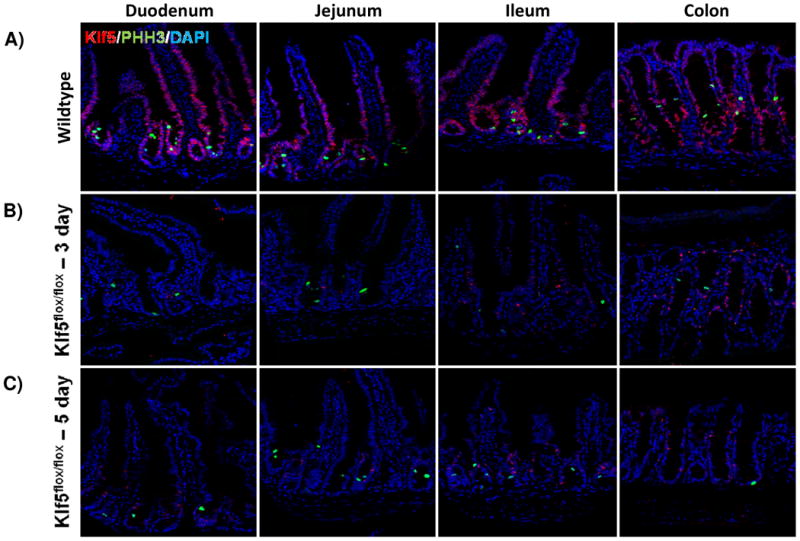
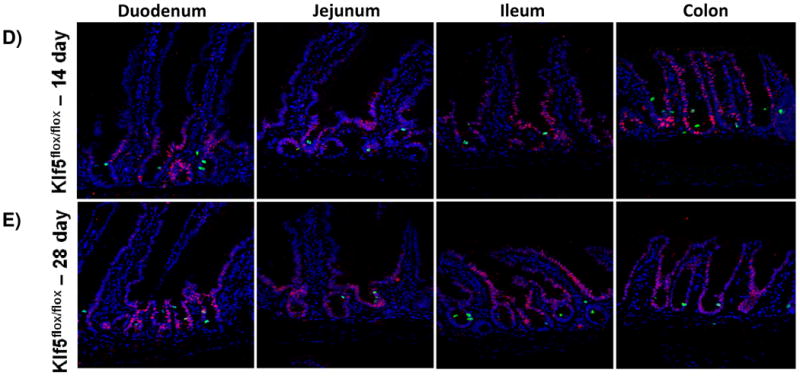
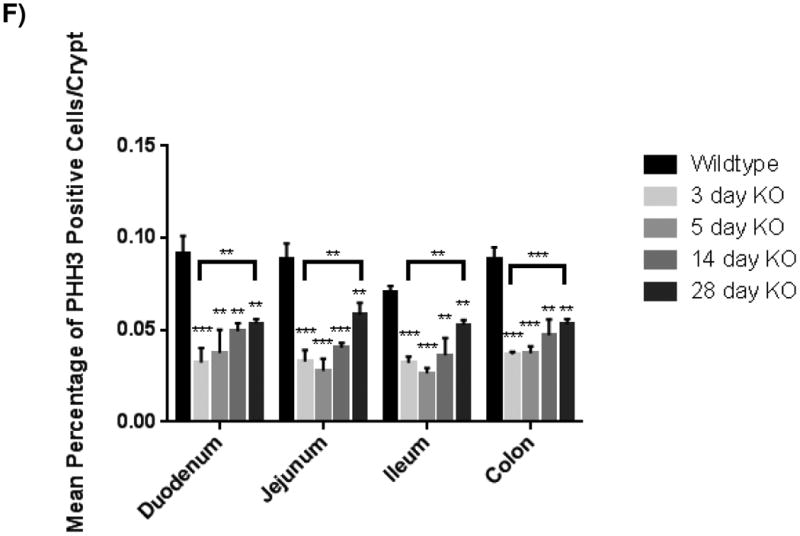
(A) Histological analysis for the comparison of cellular proliferation utilizing immunofluorescent staining for mitotic marker, phosphohistoneH3 (green) in wildtype and Klf5 loss-of-function mice (3, 5, 14, and 28 days post tamoxifen-induced recombination). Nuclear stain is DAPI (blue). (B) Quantification of average percentage of phosphohistoneH3 positive cells per crypt. Significance determined between groups of two (wildtype vs. loss of Klf5 for 3 days, wildtype vs. loss of Klf5 for 5 days, wildtype vs. loss of Klf5 for 14 days, wildtype vs. loss of Klf5 for 28 days, and loss of Klf5 for 3 vs. 28 days). (Mean ± SD, T-test: * P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n = 3)
Because KLF5 expression returned between 14 and 28 days after tamoxifen-induced recombination, we hypothesized that proliferation would increase accordingly. While there were increased numbers of PHH3+ mitotic figures found after 14 days (4.92±0.45% in the duodenum, 4.04±0.26% in the jejunum, 3.62±0.94% in the ileum, and 4.72±0.86% in the colon) (Figure 3D, 3F), the rate of proliferation, as measured by this marker, was significantly improved only 28 days after Klf5 deletion. The number of PHH3 positive cells rose in all regions of the epithelium: to 5.34±0.25% in the duodenum, 5.82±0.66% in the jejunum, 5.26±0.28 in the ileum, and 5.31±0.28% in the colon (Figure 3E-F). Although a restoration in the level of proliferation was seen 28 days after deletion of Klf5, this level of proliferation remained less than wildtype (Figure 3F). Altogether, these data indicate that disruption of Klf5 has an immediate and perdurant effect on normal intestinal proliferation.
Loss of Klf5 increases apoptosis in the crypt
In the uninjured intestine, cell death within the epithelium is rare and typically observed only at the villus tips. After cells migrate up the villi, they detach from the basement membrane and are shed into the luminal cavity, where they undergo detachment-induced apoptosis, called anoikis [34, 35]. Due to the decrease in proliferation and the lack of change in tissue morphology, we sought to determine the fate of cells within the crypt. Interestingly, the loss of Klf5 was correlated with an acute increase in apoptosis in the crypts as visualized by CC3. On average, the number of apoptotic cells was about 1.5 cells per crypt three days after ablation of Klf5, with the number of dying cells per crypt ranging from 1-5 (Figure 4A-B, 4F). Apoptosis was still apparent in the crypt 5 days after loss of Klf5, but did was not significantly increased as seen in 3 days (Figure 4A, 4C, and 4F). Intestinal tissues examined 14 and 28 days after deletion of Klf5 had negligible amounts of crypt cell death comparable to the wildtype controls (Figure 4A, 4D-F). This may be explained by the reemergence of the KLF5 and restoration of normal homeostatic function. Collectively, these data indicate that acute loss of KLF5 expression correlates with increased apoptosis.
Figure 4. Intestinal crypt cell death is increased early after loss of KLF5, but disappears over time.
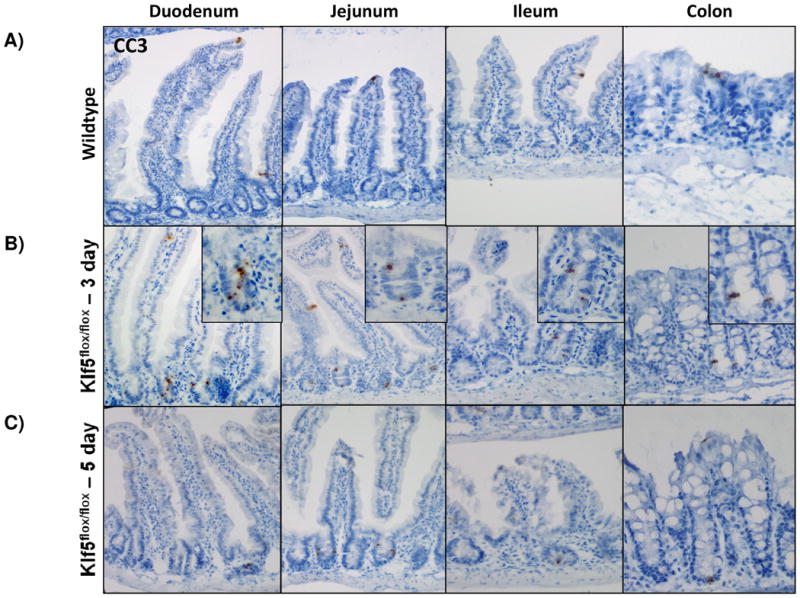
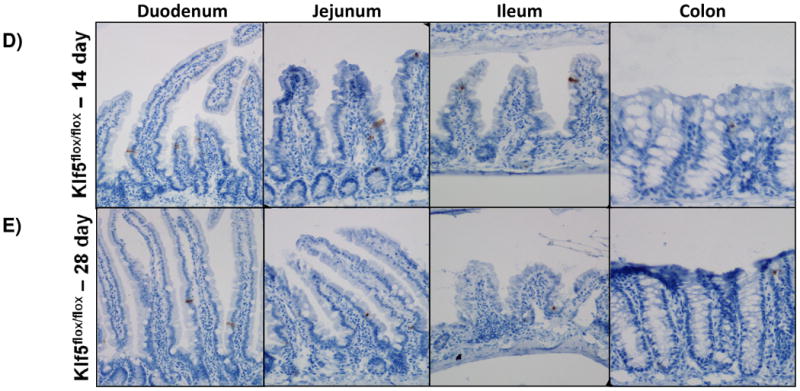
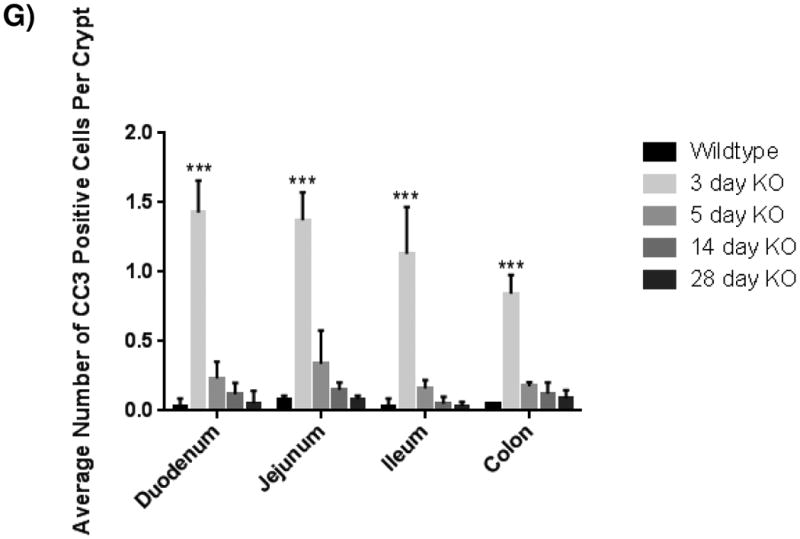
(A-E) Immunohistochemical analysis of apoposis is shown using cleaved-caspase 3 (brown) counterstained with hemotoxylin (blue) in wildtype and Klf5 loss-of-function mice (3, 5, 14, 28 days post tamoxifen-induced recombination). (F) Quantification of the number of cleaved-caspase 3 positive cells per crypt. Significance determined between groups of two (wildtype vs. loss of Klf5 for 3 days, wildtype vs. loss of Klf5 for 5 days, wildtype vs. loss of Klf5 for 14 days, wildtype vs. loss of Klf5 for 28 days, and loss of Klf5 for 3 vs. 28 days). (Mean ± SD, T-test: * P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n = 3)
Loss of Klf5 does not globally affect cell cycle machinery in vivo, but does affect stem cell marker expression
As KLF5 has been shown to play a role in the regulation of cell cycle machinery such as CyclinD1 (Ccnd1), CyclinB1 (Ccnb1), and Cdc2 (Cdk1) [10, 20, 25, 36], we examined the RNA expression levels of these proteins from jejunal segments where deletion of Klf5 was most robust (Figure 1). In contrast to the previous reports, no significant change was found in the expression of any of these genes (Figure 5A).
Figure 5. Loss of Klf5 results in loss of active stem cell marker expression in the small intestine.
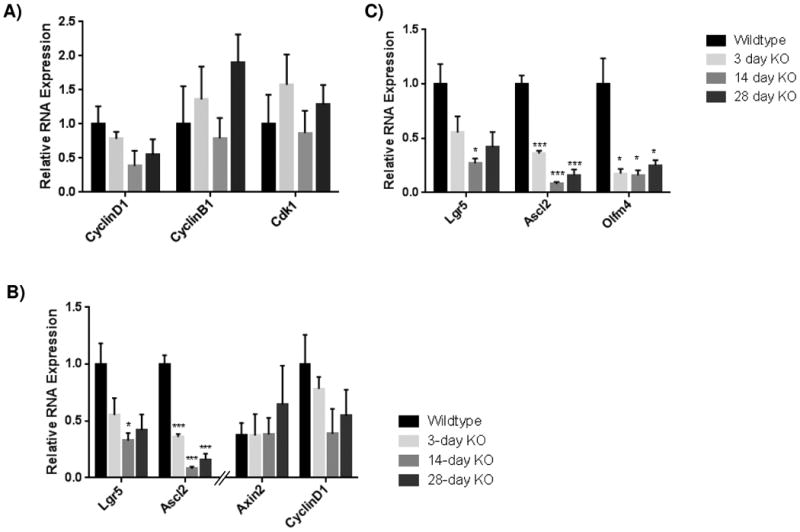
Results from of mRNA of expression for (A) cell cycle proteins ( CyclinD1, CyclinB1, and Cdk1) , (B) downstream targets of canonical Wnt signaling (Lgr5, Ascl2, Axin2, and CyclinD1), and (C) active intestinal stem cell markers (Lgr5, Ascl2, Axin2, CyclinD1) by RT-qPCR from jejunal segments of the small intestine. Student's t-test was performed to determine p-values; expression was normalized to Gapdh (SYBR) or Actb (Taqman). (Mean ± SEM: P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, n = 3-6). Note that some qRT-PCR results for relative RNA expression appear on two graphs for comparison purposes.
KLF5 has also been shown to affect canonical Wnt signaling, a positive regulator of intestinal cellular proliferation, as reduction of Klf5 reduced nuclear localization of β-catenin in the context of Apcmin mice [26]. Therefore we examined RNA expression of well-known downstream targets of Wnt signaling in the intestine, Ccnd1, Axin2, Lgr5 and Ascl2. No significant decrease was seen in Ccnd1 or Axin2 levels; however loss of stem cell markers Lgr5 and Ascl2 were found to be significantly decreased (Figure 5B-C). Loss of Lgr5 expression dropped to 55.2±15.0% of wildtype levels after 3 days, and continued to drop to 26.8±4.6% of wildtype levels 14 days after Klf5 loss. Twenty-eight days after recombination, Lgr5 expression remained below half of wildtype expression at 41.9±13.8%. Ascl2 exhibited a more dramatic effect after recombination as expression dropped to 35.8±2.7% in 3 days, and continued to lower to 8.0±1.9% in 14 days. After 28 days, mRNA levels were still low at 16.0±5.1% of wildtype. This prompted us to examine another well-known stem cell marker Olfm4, which is thought to depend on Notch activity rather that beta-catenin transactivation [37] (Figure 5C). Expression of Olfm4 plummeted to 17.5±4.4% of wildtype levels after 3 days, and continued to fall slightly to 15.8±4.8% after 14 days. As seen with the other two stem cell markers, there was some restoration of expression after 28 days (25%±5%), coincident with restored KLF5 protein expression, but this still remained significantly less than wildtype expression. Overall these data indicate that loss of Klf5 does not globally disrupt cell cycle machinery nor canonical beta-catenin transactivation, but rather points to a role for this transcription factor in the maintenance of stem cells.
Discussion
The goal of this study was to determine the effects of Klf5 loss in the adult intestine. Previously loss of Klf5 was reported to have adverse effects on barrier function, cytodifferentiation, and migration. However, that prior study used the Villin-Cre driver to delete Klf5 in the intestinal epithelium, resulting in perinatal death in two-thirds of the mutant offspring [28]. It was later discovered that this transcription factor plays an important role in intestinal morphogenesis wherein deletion of Klf5 from the embryonic intestine disrupted villus emergence and cellular differentiation [29]. As these phenotypes emerged at the onset of villus outgrowth, we hypothesized that the effects of acute loss of Klf5 in the adult would be distinct from those phenotypes reported in the previous study using the continuously active Villin-Cre transgenic line, which we suggest may have developmental origins. To test this hypothesis, we used the tamoxifen-inducible Villin-CreERT2 driver to delete Klf5 in the adult intestine as a means to test its function in this context.
We found that loss of Klf5 significantly decreased the number of proliferating cells per crypt along the entire region of the small and large intestines. Simultaneously, we observed an acute increase the number of apoptotic cells in Klf5-deficient crypts, and a decreased number of enteroendocrine cells in the small intestine. The diminished number of these cell types may indicate a role for Klf5 specific to enteroendocrine cell differentiation. Additionally, we saw no change in intestinal morphology, suggesting that production of new epithelial cells continues in Klf5-mutant intestines. In contrast to the minimal changes in intestinal cellular differentiation, we observed decreased proliferation coincident with increased apoptosis in Klf5 mutant intestines. We suggest that the lack of cell cycle progression potentiated the onset of cell death early after loss of Klf5. Caspase-3 is known to induce apoptosis after G1 phase cell cycle arrest [38]. As KLF5 is known to regulate expression of CyclinD1 [10, 36], which is required for G1/S phase transition, it is possible that loss of this transcription factor could result in a G1 phase arrest and activation of caspase activity and thus cellular apoptosis. However we did not see a significant change in CyclinD1 mRNA expression. We note that RNA was isolated from whole jejunum, and therefore, these results measure the entire tissue including mesenchyme, and may have reduced the sensitivity of our assay to detect subtle changes in CyclinD1 expression. As an alternative mechanism underlying the observed increased cell death, Klf5 deletion may have caused a loss of a prosurvival gene such as survivin, which has previously been linked to KLF5 in leukemia [39], and thus may play a similar role in the intestine. Spontaneous crypt cell apoptosis is generally a sign of stress or toxicity in the epithelium [40] therefore we interpret our results to suggest that lack of KLF5 triggered a stress response within the intestinal epithelium.
Interesting, Klf5-deficient cells were rapidly replaced with wildtype cells, such that crypts had restored KLF5 expression within 14 to 28 days of induced deletion. While this is probably the result of the lack of 100% efficient knock out of Klf5, we suggest that, given the increased rate of apoptosis, there could be selection against Klf5-mutant cells that is required to reestablish the proliferative capacity of the crypt to maintain proper epithelial turnover. Our results are consistent with the previous studies of Klf5-deficient intestinal epithelium, in which surviving mice showed re-expression of KLF5 in tissues examined 8 weeks after birth [28] and, very recently, in inducible loss of Klf5 in the colon [41]. Regardless of its origin, the reemergence of this transcription factor and incomplete loss of proliferation could explain the lack of disruption in cellular differentiation and intestinal morphology in Klf5-mutant mice.
The main conclusion of this study is that KLF5's primary function in the intestine is to maintain epithelial proliferation. Due to extensive turnover of the epithelium, three to five days is sufficient to see potential differentiation and morphological defects caused by acute KLF5 loss; however, loss of proliferation is the prominent phenotype observed in Klf5-mutant mice. Increased apoptosis is likely coupled to this effect early after recombination, but the number of PHH3 positive cells remains significantly lower than wildtype 28 days post tamoxifen treatment suggesting a lasting effect from the mutation.
KLF5 has been linked to both the Wnt and MAPK pathways [17, 20, 23, 26], and has been reported to regulate cell cycle components such as Cyclin D1, Cyclin B1, and Cdc2 [25, 36]. However, we did not see a significant decrease in the expression of any of these genes. As previously mentioned, analysis of whole jejunal tissue may have diminished the sensitivity of our assay to detect cell-specific changes in gene expression, for example specifically in transient amplifying or stem cells. It is interesting that in the developing intestine, Klf5 deletion causes a profound loss of CyclinD1 expression, however loss of these genes had no effect on proliferation of the embryonic intestine [29]. Therefore the reduction in proliferation seen here may suggest that KLF5 regulates cell cycle in the adult intestine via mechanisms distinct from identified in tumors and cell lines..
Reemergence of Klf5 coupled with the lasting effects of reduced proliferation indicates that the initial loss of Klf5 may have an effect on another important element of homeostasis. . While it is thought that expression is rare or weak in adult stem cells [10], loss of Klf5 in Lgr5 positive stem cells has resulted in suppression of oncogenic transformation [42] indicating a prominent role for this transcription factor in this cell type. Indeed we saw a significant reduction in active intestinal stem cell markers, Lgr5, Ascl2, and Olfm4 which can explain a decreased proliferative response in the intestine in the absence of an overt change in cell cycle machinery or canonical Wnt signaling. In contract to what we expected, there seems to be no lack of crypt cell numbers in these intestines which is possibly due to an incomplete loss of stem cells entirely. In any instance, it is clear that given its potential roles in the initiation of tumorigenesis and in stress induced epithelial repair, understanding the molecular mechanisms by which KLF5 controls homeostatic cellular proliferation and stem cell maintenance could provide valuable answers for future therapies of intestinal diseases.
Acknowledgments
We thank Drs. Jeffrey Whitsett and Yi Zheng of Cincinnati Children's Hospital for providing various materials to complete this work. Thank you to Dr. Taeko Noah for technical advice and Yuan-Hung Lo and Kate Maurer for additional support throughout this study.
Grant acknowledgements: This work is supported by the National Institute of Health grant DK092306, NIH grant P30 DK078392 for the pathology core of the Digestive Disease Research Core Center in Cincinnati at Cincinnati Children's Medical Center, and by the Integrated Microscopy Core at Baylor College of Medicine with funding from the NIH (HD007495, DK56338, and CA125123), the Dan L. Duncan Cancer Center, and the John S. Dunn Gulf Coast Consortium for Chemical Genomic.
References
- 1.Buddington RK, Diamond JM. Ontogenetic development of intestinal nutrient transporters. Annual Review of Physiology. 1989;1989(51):601–619. doi: 10.1146/annurev.ph.51.030189.003125. [DOI] [PubMed] [Google Scholar]
- 2.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. American Journal of Clincal Nutrition. 2001;73:1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 3.Shimizu M. Modulation of intestinal functions by food substances. Die Nahrung. 1999;43(3):154–158. doi: 10.1002/(SICI)1521-3803(19990601)43:3<154::AID-FOOD154>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 4.Strader AD, Woods SC. Gastrointestinal hormones and food intake. Gastroenterology. 2005;128(1):175–191. doi: 10.1053/j.gastro.2004.10.043. [DOI] [PubMed] [Google Scholar]
- 5.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. The American Journal of Anatomy. 1974;141(4):537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 6.Noah TK, Donahue B, Shroyer N. Intestinal development and differentiation. Experimental Cell Research. 2011;317(19):2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276(37):34355–8. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- 8.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biology. 2003;4(2):206.1–206.8. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–81. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66(16):2691–706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghaleb AM, et al. Krupple-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Research. 2005;15(2):92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parisi S, Russo T. Regulatory role of Klf5 in early mouse development and in embryonic stem cells. Vitam Horm. 2011;87:381–97. doi: 10.1016/B978-0-12-386015-6.00037-8. [DOI] [PubMed] [Google Scholar]
- 13.Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like Factor 5) is a positive regulator of cellular proliferation. Journal of Biological Chemistry. 2001;276(10):6897–6900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin SC, et al. Klf5 regulates lineage formation in the pre-implantation mouse embryo. Development. 2010;137(23):3953–63. doi: 10.1242/dev.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein BG, et al. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. American Journal of Gastrointestinal Liver Physiology. 2007;292:G1784–G1792. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- 16.Sur I. Human Kruppel-like factor5/KLF5: Synergy with NF-?B/Rel factors and expression in human skin and hair follicles. European Journal of Cell Biology. 2002;81(6):323–334. doi: 10.1078/0171-9335-00257. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, et al. Kruppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J. 2007;21(2):543–50. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 18.Bateman NW, et al. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279(13):12093–101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 19.Chanchevalap S, et al. All-trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Krüppel-like factor 5. FeBS Letter. 2004;578(1-2):99–105. doi: 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nandan MO, et al. Krupple-like factor mediates the transforming activing of oncogenic H-Ras. Oncogene. 2004;23(19):3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConnell BB, et al. Kruppel-like factor 5 protects against dextran sulfate sodium-induced colonic injury in mice by promoting epithelial repair. Gastroenterology. 2011;140(2):540–549 e2. doi: 10.1053/j.gastro.2010.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McConnell BB, et al. Kruppel-like factor 5 mediates transmissible murine colonic hyperplasia caused by Citrobacter rodentium infection. Gastroenterology. 2008;134(4):1007–16. doi: 10.1053/j.gastro.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nandan MO, et al. Krupple-like Factor 5 Mediates Cellular Transformation during Oncogenic KRAS-Induced Intestinal Tumorigenesis. Gastroenterology. 2008;134(1):120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nandan MO, et al. Kruppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 2005;579(21):4757–62. doi: 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McConnell BB, et al. Haploinsufficiency of Kruppel-like factor 5 rescues the tumor-initiating effect of the Apc(Min) mutation in the intestine. Cancer Res. 2009;69(10):4125–33. doi: 10.1158/0008-5472.CAN-08-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandan MO, et al. Kruppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMin and KRASV12 mutations. Mol Cancer. 2010;9:63. doi: 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McConnell BB, et al. Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011;141(4):1302-13–1313 e1-6. doi: 10.1053/j.gastro.2011.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell SM, et al. Kruppel-like factor 5 controls villus formation and initiation of cytodifferentiation in the embryonic intestinal epithelium. Dev Biol. 2013;375(2):128–39. doi: 10.1016/j.ydbio.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.el Marjou F, et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–93. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 31.Wan H, et al. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135(15):2563–72. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conckright MD, et al. A gene encoding an intestinal-enriched member of the Krüppel-like factor family expressed in intestinal epithelial cells. Nucleic Acids Res. 1999;27(5):1263–1270. doi: 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shindo T, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8(8):856–63. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 34.Grossman J. Molecular mechanisms of “detachment-induced apoptosis—Anoikis”. Apoptosis. 2002;7:247–260. doi: 10.1023/a:1015312119693. [DOI] [PubMed] [Google Scholar]
- 35.Rosen K, et al. Cell detachment triggers p38 mitogen-activated protein kinase-dependent overexpression of Fas ligand. A novel mechanism of Anoikis of intestinal epithelial cells. J Biol Chem. 2002;277(48):46123–30. doi: 10.1074/jbc.M207883200. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, et al. Kruppel-like factor (KLF) 5 mediates cyclin D1 expression and cell proliferation via interaction with c-Jun in Ang II-induced VSMCs. Acta Pharmacol Sin. 2010;31(1):10–8. doi: 10.1038/aps.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.VanDussen KL, et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2011;139(3):488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alenzi FQB. Links between apoptosis, proliferation, and the cell cycle. British Journal of Biomedical Sceince. 2004;61(2):99–102. doi: 10.1080/09674845.2004.11732652. [DOI] [PubMed] [Google Scholar]
- 39.Zhu N, et al. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281(21):14711–8. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]
- 40.Watson AJ, Pritchard DM. Lessons From Genetically Engineered Animal Models VII. Apoptosis in intestinal epithelium: lessons from transgenic and knockout mice. American Journal of Physiology, Gastrointestinal and Liver Physiolgoy. 2000;278:G1–G5. doi: 10.1152/ajpgi.2000.278.1.G1. [DOI] [PubMed] [Google Scholar]
- 41.Nandan MO, et al. Inducible intestine-specific deletion of Krüppel-like factor 5 is characterized by a regenerative response in adult mouse colon. Dev Biol. 2014;387(2):191–202. doi: 10.1016/j.ydbio.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakaya T, et al. KLF5 Regulates the Integrity and Oncogenicity of Intestinal Stem Cells. Cancer Res. 2014;74(10):2882–2891. doi: 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]


