Summary
Early Xenopus laevis embryogenesis is a robust system for investigating mechanisms of developmental timing. After a series of rapid cell divisions with concomitant reductions in cell size, the first major developmental transition is the midblastula transition (MBT), when zygotic transcription begins and cell cycles elongate [1-3]. While the maintenance of a constant nuclear-to-cytoplasmic (N/C) volume ratio is a conserved cellular property [4-7], it has long been recognized that the N/C volume ratio changes dramatically during early Xenopus development [8]. We investigated how changes in nuclear size and the N/C volume ratio during early development contribute to the regulation of MBT timing. While previous studies suggested a role for the N/C volume ratio in MBT timing [1, 9-13], none directly tested the effects of altering nuclear size. In this study, we first quantify blastomere and nuclear sizes in X. laevis embryos, demonstrating that the N/C volume ratio increases prior to the MBT. We then manipulate nuclear volume in embryos by microinjecting different nuclear scaling factors, including import proteins, lamins, and reticulons. Using this approach, we show that increasing the N/C volume ratio in pre-MBT embryos leads to premature activation of zygotic gene transcription and early onset of longer cell cycles. Conversely, decreasing the N/C volume ratio delays zygotic transcription and leads to additional rapid cell divisions. While the DNA-to-cytoplasmic ratio has been implicated in MBT timing [1, 9-18], our data show that nuclear size also contributes to the regulation of MBT timing, demonstrating the functional significance of nuclear size during development.
Results
Nuclear and cell volumes become progressively smaller during Xenopus early development with the N/C volume ratio increasing prior to the MBT
The first major developmental transition during early X. laevis embryogenesis is the midblastula transition (MBT). Approximately 1.5 hours after fertilization, twelve rapid synchronous cleavage cell cycles ensue, each about 25-30 minutes long and consisting of alternating S and M phases [1, 2]. Next, cell cycles lengthen with the acquisition of gap phases and major zygotic transcription begins, marking the MBT (Nieuwkoop-Faber stage 8, cleavage 12) [14, 19]. Initially a 1.2 mm single cell, the embryo consists of several thousand 50 μm and smaller blastomeres at the MBT.
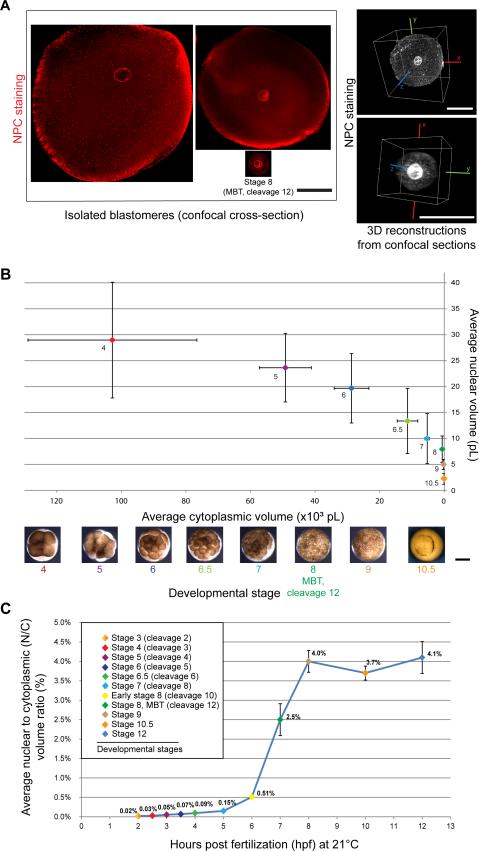
To determine nuclear scaling relationships during X. laevis development, we isolated blastomeres from embryos at different developmental stages (Figure 1A) and quantified cell and nuclear sizes (Figure S1A-B). Average nuclear volume scaled progressively smaller with cytoplasmic volume in all early stages examined (Figure 1B). From stages 4 to 8 (cleavage 12), nuclear volume decreased on average ~ 3-fold while cytoplasmic volume showed a much more dramatic ~ 70-fold reduction in volume (Figure 1B). Within a given stage, we observed large differences in blastomere size [20], and nuclear and cell sizes tended to scale within a given stage (Figure S1A-B).
Figure 1. Nuclear and cell volumes scale smaller during Xenopus early development with the N/C volume ratio increasing prior to the MBT.
(A) X. laevis embryos that were un-arrested (i.e. not treated with cycloheximide) were cultured from stage 2 in calcium/magnesium-free medium within intact vitelline membranes. At desired stages, dissociated blastomeres were immediately fixed and subjected to whole-mount fluorescence immunocytochemistry staining with mAb414, an antibody against the nuclear pore complex (NPC). Nuclei within intact, roughly spherical blastomeres were visualized by confocal microscopy. Cross-sectional area was quantified for both cells and nuclei (Figure S1A-B). The right side of the panel shows representative 3D reconstructions from cell and nuclear confocal sections. Image acquisition and quantification are detailed in “Supplemental Experimental Procedures.” Direct volume measurements agreed within 9% and 3% of volumes extrapolated from cross-sectional areas for stage 6 and stage 8 (MBT, cleavage 12), respectively, validating our approach of estimating volumes from cross-sectional area measurements. All scale bars, 50 μm.
(B) Nuclear and cell volumes were extrapolated from cross-sectional area measurements (Figure S1A-B). Cytoplasmic volume was determined by subtracting nuclear volume from cell volume. Average values are plotted and error bars represent SD. Scale bar, 500 μm.
(C) Average nuclear-to-cytoplasmic (N/C) volume ratios are plotted as a function of time post-fertilization, and error bars are SE.
See also Figure S1.
Although absolute nuclear size was greater in earlier developmental stages, nuclei in later stage embryos occupied proportionately more of the cell. To quantify this effect, we calculated N/C (nuclear-to-cytoplasmic) volume ratio values on a per cell basis and found that the average N/C volume ratio increased prior to the MBT (stage 8, cleavage 12), reaching a maximum at stage 9 (Figure 1C, 2C, S1H). These data prompted us to test if the N/C volume ratio plays a role in regulating timing of the MBT.
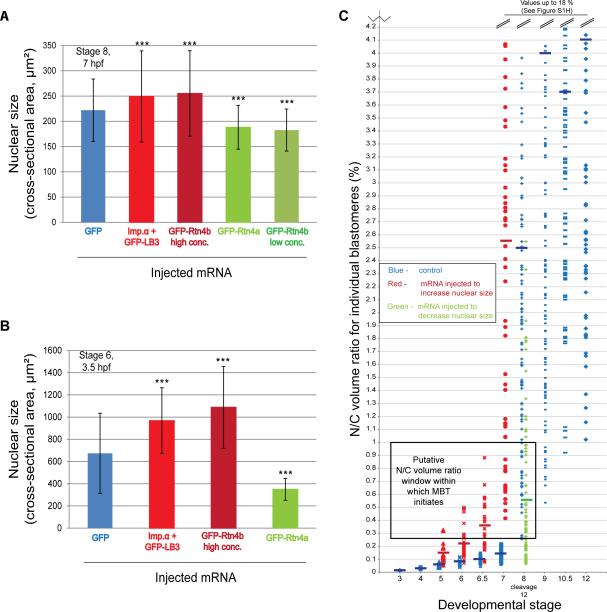
Figure 2. Manipulating nuclear size and the N/C volume ratio in X. laevis embryos.
(A) One-cell embryos were microinjected with the most effective amounts of indicated mRNAs: GFP, 100 pg; Imp.α + GFP-LB3, 1000 pg; GFP-Rtn4b high concentration, 500 pg; GFP-Rtn4a, 350 pg; GFP-Rtn4b low concentration, 150 pg. Nuclei were isolated from stage 8 embryos and visualized by immunofluorescence using mAb414. Nuclear cross-sectional area was quantified for at least 500 nuclei from ~100 embryos for each condition. One representative experiment of three is shown, error bars represent SD, *** P<0.001.
(B) Embryos were injected as described in (A). Nuclei were visualized in isolated stage 6 (3.5 hpf) blastomeres by whole-mount immunocytochemistry as described in Figure 1A. Nuclear cross-sectional area was quantified for at least 50 nuclei for each condition. One representative experiment of two is shown, error bars represent SD, *** P<0.001. (C) Embryos were injected as described in (A). Nuclei were visualized in isolated blastomeres at the indicated stages by whole-mount immunocytochemistry and N/C volume ratios were quantified as in Figure 1. N/C volume ratios for individual blastomeres are plotted. N/C ratios shown in blue represent control blastomeres and were calculated from the data shown in Figure S1A-B. For stages 7-12, N/C ratios greater than 4.2% were excluded for clarity and all of those data are plotted in Figure S1H. N/C ratios shown in red represent blastomeres with increased nuclear size (i.e. injected with importin α + GFP-LB3 or GFP-Rtn4b high concentration). N/C ratios shown in green represent blastomeres with decreased nuclear size (i.e. injected with GFP-Rtn4a or GFP-Rtn4b low concentration). Stage 5 blastomeres with increased nuclear size, n=28; stage 6 blastomeres with increased nuclear size, n=38; stage 6.5 blastomeres with increased nuclear size, n=23; stage 7 blastomeres with increased nuclear size, n=55; stage 8 blastomeres with decreased nuclear size, n=83. Short thick horizontal lines indicate mean values for each stage. The box labeled “Putative N/C volume ratio window within which MBT initiates” denotes the overlap between wild-type stage 8 blastomeres with the smallest N/C volume ratios and stage 5-6.5 blastomeres with increased nuclear size.
See also Figure S2.
Manipulating nuclear size and the N/C volume ratio in Xenopus embryos
To determine how nuclear size and the N/C volume ratio might impact developmental progression, we sought multiple approaches employing different mechanisms to manipulate nuclear size in the embryo, utilizing factors known to regulate nuclear size: importins, lamins, and reticulons [21, 22]. Previous work in Xenopus egg extracts and early embryos demonstrated that rates of nuclear import influence nuclear size, with the levels of importin α being particularly important [23]. An importin α cargo essential for nuclear envelope (NE) growth and for regulating nuclear size is lamin B3 (LB3), the primary lamin present in the egg and early embryo that is a major constituent of the nuclear lamina [24-26]. Since the NE and endoplasmic reticulum (ER) are continuous, the structure of the ER also impacts nuclear size. Proteins in the reticulon (Rtn) family mediate ER tubule formation [27, 28], and Rtn4 overexpression converts ER sheets to tubules and decreases nuclear size in tissue culture cells [29, 30].
We first tested if increasing the levels of importin α and LB3 would affect nuclear size in the embryo. Fertilized one-cell X. laevis embryos were microinjected with a range of mRNA amounts to titrate the expression levels of importin α and GFP-LB3, and nuclear size was quantified in stage 8 embryos (cleavage 12; 7 hours post fertilization, hpf). While ectopic expression of either importin α or GFP-LB3 increased nuclear size, co-expression had a greater effect (Figure 2A, S1C-E). Effects of importin α and LB3 expression on nuclear size were more dramatic when quantified in embryos at stage 6 (3.5 hpf) (Figure 2B) compared to stage 8 (7 hpf) (Figure 2A).
To identify an approach to altering nuclear size that does not directly involve nucleocytoplasmic transport or lamins, we focused on ER structure and reticulons. Ectopic expression of GFP-Rtn4a in X. laevis embryos decreased nuclear size at all concentrations tested, consistent with results in mammalian tissue culture [30] (Figure 2A-B, S1F). We also tested the smaller isoform Rtn4b. While low concentrations of GFP-Rtn4b decreased nuclear size, as expected, high expression levels increased nuclear size (Figure 2A-B, S1G). A possible explanation for this observation is that high GFP-Rtn4b concentrations led to the formation of reticulon aggregates (Figure S2) that potentially act in a dominant negative fashion by extracting reticulons from the ER, promoting ER sheet formation and increased nuclear size. For our studies, an advantage of this result is that we were able to increase or decrease nuclear size by manipulating the levels of the same factor.
Having identified multiple conditions that increase or decrease nuclear size in the embryo, we next determined how these manipulations affected the N/C volume ratios of individual cells. In stage 5 and 6 embryos with increased nuclear size, the average N/C volume ratio increased to a value larger than that found in wild-type stage 7 embryos, and the spread of these values also became greater (Figure 2C). This effect was more pronounced in stage 6.5 and 7 embryos, with injected stage 7 embryos having the same average and spread of N/C volume ratios as untreated stage 8 embryos (Figure 2C, S1H). Even though the average N/C volume ratio in stage 5-6.5 embryos with increased nuclear size did not reach that of MBT embryos, we noted that many individual cells had N/C volume ratios that overlapped with the lower range found in stage 8 MBT blastomeres (Figure 2C). These observations led us to test if the MBT might be initiated earlier in these blastomeres upon reaching a threshold N/C volume ratio. Conversely, in stage 8 embryos with reduced nuclear size, the average N/C volume ratio decreased ~5-fold. Many individual blastomeres with decreased nuclear size had N/C volume ratios characteristic of blastomeres at stage 7 and earlier (Figure 2C, S1H), prompting us to test if MBT initiation might be delayed in these blastomeres.
Altering the N/C volume ratio changes the timing of the onset of zygotic gene transcription
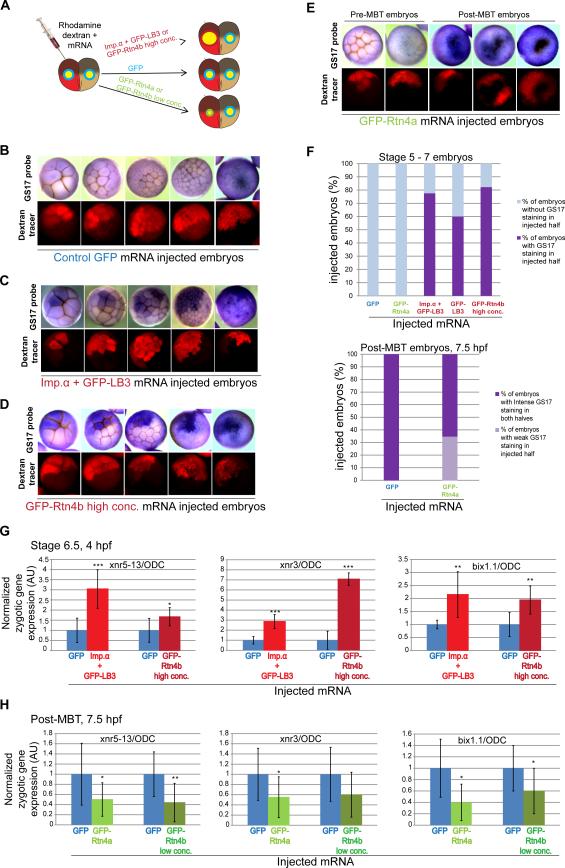
To test if altering nuclear size in early embryos affects MBT timing we monitored a key molecular hallmark of the MBT, onset of zygotic gene expression. We first used whole-mount in situ hybridization to detect the GS17 transcript, one of the first abundantly transcribed zygotic genes and a classical molecular marker of the MBT [31, 32]. Our approach was to alter nuclear size in half of each embryo by microinjecting one blastomere at the two-cell stage and allowing the embryo to develop (Figure 3A). Differential GS17 staining in the two halves of each embryo indicated altered timing for the onset of zygotic transcription. Control GFP-injected embryos showed no GS17 staining in pre-MBT stages, and at stage 8 both halves stained equally for GS17, indicating no effect on MBT timing (Figure 3B, F). Premature expression of GS17 was detected in stage 5-7 embryos when nuclear size was increased by i) co-injection of importin α and GFP-LB3 (Figure 3C, F), ii) injection of the high concentration of GFPRtn4b (Figure 3D, F), or iii) injection of GFP-LB3 alone (Figure 3F). Furthermore, stage 8 embryos exhibited more intense GS17 staining in the embryo halves with increased nuclear size. We noted that not all cells in injected embryo halves showed differential staining. We quantified these cells and found that blastomeres expressing GS17 had larger N/C volume ratios than both un-injected cells and injected cells with weak or absent GS17 staining (Figure S3A-D). These data are consistent with the hypothesis that GS17 transcription initiates upon cells reaching a threshold N/C volume ratio. Conversely, decreasing nuclear size by GFP-Rtn4a mRNA microinjection led to weaker GS17 staining in late stage 8 embryos (post-MBT, 7.5 hpf), suggestive of a delay in MBT timing (Figure 3E, F).
Figure 3. Altering the N/C volume ratio changes the timing of the onset of zygotic transcription.
(A) The experimental scheme is depicted in which one blastomere of a two-cell embryo is co-injected with rhodamine-labeled dextran and mRNA to alter nuclear size in half of the embryo.
(B-E) Embryos at the indicated stages and microinjected with the indicated mRNAs were subjected to in situ hybridization to detect the GS17 transcript. The top panels are bright field images of embryos stained for GS17 (purple). The bottom panels are the corresponding rhodamine fluorescence images indicating cells in the embryo that received the indicated mRNA. Representative embryos are shown.
(F) The top graph shows the percentage of stage 5-7 embryos in which the injected half showed GS17 staining while the un-injected half did not (dark purple bars). GFP, n=57; GFP-Rtn4a, n=42; Imp.α + GFP-LB3, n=119; GFP-LB3, n=124; GFP-Rtn4b high conc., n=139. The bottom graph shows the percentage of post-MBT (7.5 hpf) embryos in which the injected half showed weak GS17 staining relative to the un-injected half (light purple bar). GFP, n=38; GFP-Rtn4a, n=70. (G,H) One-cell embryos were microinjected with the indicated mRNAs to increase (G) or decrease (H) nuclear size and allowed to develop to stage 6.5 (G) or post-MBT 7.5 hpf (H). Total RNA was isolated from 12 embryos for each condition and converted to cDNA. Expression levels of three zygotic genes (xnr5-13, xnr3 and bix1.1) were determined by qPCR, normalized to ODC. Gene expression levels are plotted in arbitrary units (AU) relative to GFP mRNA injected control embryos. The means from two independent experiments are shown, error bars represent SD, *** P<0.001, ** P<0.01, * P<0.05.
See also Figure S3.
To corroborate our in situ hybridization data, we quantified additional zygotic transcripts by qPCR [3, 33]. Stage 6.5 (4 hpf) embryos with increased nuclear size showed increased expression of three additional MBT transcripts compared to GFP control-injected embryos, up to seven times greater in some cases (Figure 3G). These changes in gene expression were comparable to those reported for other studies in which MBT timing was altered [34-36]. Importantly, non-zygotic transcripts did not show this trend, suggesting that the effect of nuclear size is specific to MBT transcription (Figure S3E). In late stage 8 embryos (7.5 hpf) with decreased nuclear size, we observed less expression of zygotic transcripts compared to control embryos (Figure 3H), but not reduced expression of non-zygotic transcripts (Figure S3F). Taken together, these results demonstrate that increasing the N/C volume ratio in pre-MBT embryos is sufficient to cause premature onset of zygotic transcription, while decreasing the N/C volume ratio delays zygotic gene transcription.
Altering the N/C volume ratio changes the timing of cellular hallmarks of the MBT
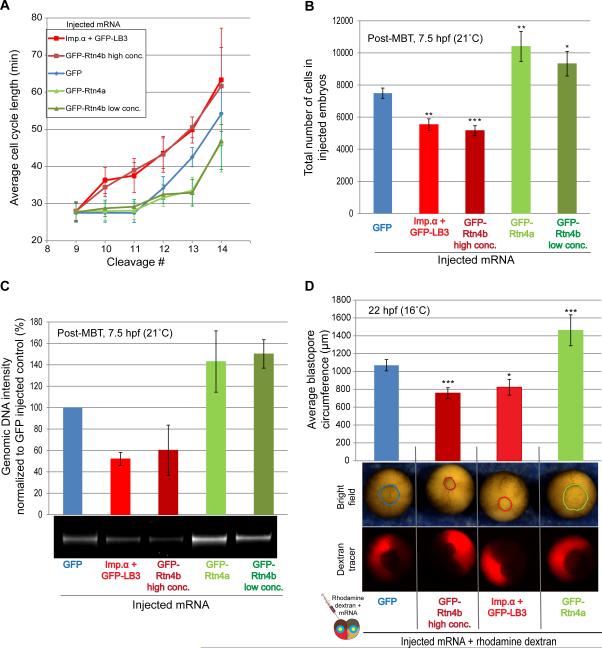
Changes in the cell cycle are another key feature of the MBT. To test if altering nuclear size in early embryos affects the timing of cellular hallmarks of the MBT, we analyzed time-lapse movies of microinjected embryos (Figure 4A, S4, Movie S1). Generally, control GFP-injected embryos cleaved synchronously until the 12th cleavage (i.e. stage 8, the normal MBT), at which time cell cycles lengthened (Figure 4A, S4C). Embryos with decreased nuclear size cleaved rapidly until, on average, the 14th cell cycle, while embryos with increased nuclear size exhibited longer cell cycles after only the 10th cleavage (Figure 4A, S4C). To further support these observations, we quantified the number of cells and amount of genomic DNA per embryo, as a proxy for the number of cell divisions. Consistent with our time-lapse movies, 7.5 hpf embryos with increased nuclear size had considerably fewer cells and less total genomic DNA than control embryos, while embryos with decreased nuclear size had greater numbers of cells and more DNA at the same post-fertilization time point (Figure 4B-C, S4A). In addition, as a result of altered cell cycle timing, cell sizes differed in embryos with altered nuclear size when analyzed at 7.5 hpf (Figure S4A-B). Furthermore, blastopore closure rates were also affected in embryos with altered nuclear size (Figure 4D, Movie S2).
Figure 4. Altering the N/C volume ratio changes the timing of cellular hallmarks of the MBT.
(A) One-cell embryos were microinjected with the indicated mRNAs. Bright field time-lapse imaging was performed on the animal pole at 5 min intervals. Cell cycle lengths were measured for at least four cells per embryo starting at the ninth cell division, for six embryos per condition. Red squares, blue diamonds, and green triangles represent average cycle lengths for embryos with increased, control, and decreased nuclear size, respectively. Data from two independent experiments are shown, error bars represent SD.
(B) One-cell embryos were microinjected with the indicated mRNAs. Stage four embryos were cultured in calcium/magnesium-free medium within intact vitelline membranes. 7.5 hpf embryos were fixed, cells from individual embryos were dissociated in 2.5 ml of fixative, and the number of cells in 25 μl aliquots were counted and corrected for the total volume. For each condition, cells from 10 embryos were counted. Means from two independent experiments are shown, error bars represent SE, *** P<0.001, ** P<0.01, * P<0.05.
(C) One-cell embryos were microinjected with the indicated mRNAs and allowed to develop to 7.5 hpf. Genomic DNA was extracted from 10 embryos for each condition, and 0.4 embryo equivalents of genomic DNA were run on a 1.2% agarose gel stained with ethidium bromide. DNA amounts were quantified using imageJ and normalized to the GFP control. Means from three independent experiments are shown, error bars represent SD, one representative gel is shown.
(D) One blastomere of a two-cell embryo was co-injected with rhodamine-labeled dextran and the indicated mRNAs to alter nuclear size. Bright field time-lapse imaging was performed on the vegetal pole of gastrulating embryos at 5 min intervals. Blastopore circumference was measured for twelve embryos per condition. Means from three independent experiments are shown, error bars represent SE, *** P<0.001, * P<0.05.
Discussion
Our data show that nuclear and cell volumes scale progressively smaller throughout early embryogenesis, consistent with data from previous studies [8, 23], that the N/C volume ratio increases rapidly prior to the MBT, and that altering the N/C volume ratio leads to changes in the timing of both zygotic gene transcription and cell cycle lengthening. We manipulated nuclear size in embryos by altering the levels of a variety of different factors, strengthening our conclusion that nuclear size impacts MBT timing and minimizing concerns about pleiotropic effects of mRNA microinjections. In particular, our reticulon injection experiments likely alter nuclear size independently of nucleocytoplasmic transport.
Stage 5-7 embryos with increased N/C volume ratios exhibited premature zygotic transcription, however not all injected embryos and not all cells in injected embryo halves stained positive for GS17 and the average increase in zygotic gene expression was less than that occurring at the normal MBT (Figure 3). One explanation is that nuclear size changes in a subset of cells were insufficient to increase the N/C volume ratio to that associated with the MBT (Figure 2C). Variability in the amount of mRNA received by individual cells as well as variation in cell sizes within a given embryo (Figure S1A-B) might account for why only some cells possessed a sufficiently increased N/C volume ratio to trigger zygotic gene expression. In support of this idea, we observed a striking correlation between GS17 expression and increased N/C volume ratios when examined on a cell-by-cell basis (Figure S3A-D). Taken together, our data suggest that the initiation of MBT zygotic transcription is triggered within a threshold N/C volume ratio window (Figure 2C).
One mechanism that regulates MBT timing is the DNA-to-cytoplasmic ratio [1, 9-18]. The proposed model is that maternally-derived MBT inhibitors are present in a fixed volume of embryonic cytoplasm, and once a critical genomic DNA amount is reached, titration of these factors by DNA induces the MBT [1, 14]. Recently, a number of potential limiting components have been identified relevant to the Xenopus MBT [34, 37, 38]. Importantly, none of these factors appear to fully account for the abrupt changes in gene expression and cell behavior that occur at the MBT, and it seems likely that redundant mechanisms regulate this critical developmental transition [39, 40]. We show that altering the N/C volume ratio independently of ploidy affects MBT timing, thus demonstrating that nuclear size, as well as DNA amount, regulates the MBT. Interestingly, we find that increasing nuclear size affects zygotic transcription earlier than cell cycle length. It is possible that transcription is more sensitive to nuclear size while DNA amount is more critical to cell cycle changes, and there is precedence for uncoupling of these two events during the MBT [20, 41-45].
Future experiments will address the relative contributions of nuclear size and DNA amount to MBT timing, and it seems likely that the two mechanisms are related. One possibility is that the effects of DNA amount on MBT timing in classical experiments [1, 14] were mediated through changes in nuclear size. Altering ploidy in Xenopus can subtly affect nuclear size. For example, haploids have smaller nuclei than diploids in early stages of development, and nuclei assembled de novo in egg extract are slightly smaller when assembled with less DNA [23]. Furthermore, previously-demonstrated activation of MBT transcription by cycloheximide treatment [16, 42] may have been mediated by nuclear size changes, as nuclei grow continuously in early embryos treated with cycloheximide [23]. Early MBT induction by microinjection of plasmid DNA into embryos [14] might also have been mediated through the N/C volume ratio as this injected DNA was subsequently shown to assemble into nuclei [46, 47]. Another possibility is that nuclear volume is relevant to models of MBT timing that invoke titration of an inhibitor by DNA binding, and a notable property of the putative limiting components identified to date is that they all act within the nucleus [34, 37, 38]. Perhaps the MBT is regulated not only by the absolute amounts of key DNA-binding factors that are maternally-derived and titrated by DNA to trigger the MBT, but by their nuclear concentrations, determined by changes in total embryonic nuclear volume during development.
The maintenance of a constant N/C volume ratio is a highly conserved cellular feature [4-7], and aberrations in this ratio and nuclear size are associated with certain disease states, most notably cancer [48]. Our study identifies a functional role for nuclear size in regulating developmental timing and paves the way for future research into the functional significance of nuclear size in the context of development, differentiation, and disease.
Experimental Procedures
Supplementary Material
Acknowledgments
We thank members of the Levy and Gatlin labs as well as our colleagues in the Department of Molecular Biology for helpful advice and discussions, Ana Milunović-Jevtić for assistance with confocal microscopy, and David Fay, Rebecca Heald, Dan Starr, and Karen White for constructive comments on the manuscript. We also thank Rebecca Heald for support in the early stages of this research. P.J. is supported by a graduate assistantship from the University of Wyoming Agricultural Experiment Station. Research in the Levy lab is supported by the NIH/NIGMS (R15GM106318).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 2.Newport JW, Kirschner MW. Regulation of the cell cycle during early Xenopus development. Cell. 1984;37:731–742. doi: 10.1016/0092-8674(84)90409-4. [DOI] [PubMed] [Google Scholar]
- 3.Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, Sesay AK, Smith JN, Smith JC, Gilchrist MJ. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141:1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson EB. The Cell in Development and Heredity. Third Edition. The Macmillan Company; New York: 1925. The karyoplasmic ratio. pp. 727–733. [Google Scholar]
- 5.Conklin E. Cell size and nuclear size. J. Exp. Embryol. 1912;12:1–98. [Google Scholar]
- 6.Jorgensen P, Edgington NP, Schneider BL, Rupes I, Tyers M, Futcher B. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann FR, Nurse P. Nuclear size control in fission yeast. J Cell Biol. 2007;179:593–600. doi: 10.1083/jcb.200708054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerhart JC. Mechanisms regulating pattern formation in the amphibian egg and early embryo. In: Goldberger RF, editor. Biological Regulation and Development. Vol. 2. Plenum; New York: 1980. pp. 133–316. [Google Scholar]
- 9.Clute P, Masui Y. Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Dev Biol. 1995;171:273–285. doi: 10.1006/dbio.1995.1280. [DOI] [PubMed] [Google Scholar]
- 10.Kobayakawa Y, Kubota HY. Temporal pattern of cleavage and the onset of gastrulation in amphibian embryos developed from eggs with the reduced cytoplasm. J Embryol Exp Morphol. 1981;62:83–94. [PubMed] [Google Scholar]
- 11.Edgar BA, Kiehle CP, Schubiger G. Cell cycle control by the nucleo-cytoplasmic ratio in early Drosophila development. Cell. 1986;44:365–372. doi: 10.1016/0092-8674(86)90771-3. [DOI] [PubMed] [Google Scholar]
- 12.Kane DA, Kimmel CB. The zebrafish midblastula transition. Development. 1993;119:447–456. doi: 10.1242/dev.119.2.447. [DOI] [PubMed] [Google Scholar]
- 13.Mita I. Studies on Factors Affecting the Timing of Early Morphogenetic Events During Starfish Embryogenesis. The Journal of Experimental Zoology. 1983;225:293–299. [Google Scholar]
- 14.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 15.Clute P, Masui Y. Microtubule dependence of chromosome cycles in Xenopus laevis blastomeres under the influence of a DNA synthesis inhibitor, aphidicolin. Dev Biol. 1997;185:1–13. doi: 10.1006/dbio.1997.8540. [DOI] [PubMed] [Google Scholar]
- 16.Edgar BA, Schubiger G. Parameters controlling transcriptional activation during early Drosophila development. Cell. 1986;44:871–877. doi: 10.1016/0092-8674(86)90009-7. [DOI] [PubMed] [Google Scholar]
- 17.Signoret J, Lefresne J. Contribution a l'etude de la segmentation de l'oeuf d'axolotl. Annales d'Embryologie et de Morphogenese. 1973;6:299–307. [Google Scholar]
- 18.Mita I, Obata C. Timing of Early Morphogenetic Events in Tetraploid Starfish Embryos. The Journal of Experimental Zoology. 1984;229:215–222. [Google Scholar]
- 19.Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) 2nd Edition North-Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- 20.Wang P, Hayden S, Masui Y. Transition of the blastomere cell cycle from cell size-independent to size-dependent control at the midblastula stage in Xenopus laevis. J Exp Zool. 2000;287:128–144. doi: 10.1002/1097-010x(20000701)287:2<128::aid-jez3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 21.Edens LJ, White KH, Jevtic P, Li X, Levy DL. Nuclear size regulation: from single cells to development and disease. Trends Cell Biol. 2013;23:151–159. doi: 10.1016/j.tcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Jevtic P, Edens LJ, Vukovic LD, Levy DL. Sizing and shaping the nucleus: mechanisms and significance. Curr Opin Cell Biol. 2014;28C:16–27. doi: 10.1016/j.ceb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy DL, Heald R. Nuclear size is regulated by importin alpha and Ntf2 in Xenopus. Cell. 2010;143:288–298. doi: 10.1016/j.cell.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106(Pt 1):275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- 26.Misteli T, Spector DL. The Nucleus. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2011. [Google Scholar]
- 27.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 28.Friedman JR, Voeltz GK. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 2011;21:709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson DJ, Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol. 2007;9:1160–1166. doi: 10.1038/ncb1636. [DOI] [PubMed] [Google Scholar]
- 30.Anderson DJ, Hetzer MW. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol. 2008;182:911–924. doi: 10.1083/jcb.200805140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieg PA, Melton DA. Developmental regulation of a gastrula-specific gene injected into fertilized Xenopus eggs. Embo J. 1985;4:3463–3471. doi: 10.1002/j.1460-2075.1985.tb04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvey RP, Tabin CJ, Melton DA. Embryonic expression and nuclear localization of Xenopus homeobox (Xhox) gene products. Embo J. 1986;5:1237–1244. doi: 10.1002/j.1460-2075.1986.tb04352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013;341:893–896. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altmann CR, Bell E, Sczyrba A, Pun J, Bekiranov S, Gaasterland T, Brivanlou AH. Microarray-based analysis of early development in Xenopus laevis. Dev Biol. 2001;236:64–75. doi: 10.1006/dbio.2001.0298. [DOI] [PubMed] [Google Scholar]
- 36.Kerns SL, Schultz KM, Barry KA, Thorne TM, McGarry TJ. Geminin is required for zygotic gene expression at the Xenopus mid-blastula transition. PLoS One. 2012;7:e38009. doi: 10.1371/journal.pone.0038009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy CM, Michael WM. Control of DNA replication by the nucleus/cytoplasm ratio in Xenopus. J Biol Chem. 2013;288:29382–29393. doi: 10.1074/jbc.M113.499012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vastag L, Jorgensen P, Peshkin L, Wei R, Rabinowitz JD, Kirschner MW. Remodeling of the metabolome during early frog development. PLoS One. 2011;6:e16881. doi: 10.1371/journal.pone.0016881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veenstra GJ. Early Embryonic Gene Transcription in Xenopus. Advances in Developmental Biology and Biochemistry. 2002;12:85–105. [Google Scholar]
- 40.Almouzni G, Wolffe AP. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. Embo J. 1995;14:1752–1765. doi: 10.1002/j.1460-2075.1995.tb07164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeichi T, Satoh N, Tashiro K, Shiokawa K. Temporal control of rRNA synthesis in cleavage-arrested embryos of Xenopus laevis. Developmental Biology. 1985;112:443–450. [Google Scholar]
- 42.Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- 43.Yasuda GK, Schubiger G. Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet. 1992;8:124–127. doi: 10.1016/0168-9525(92)90369-F. [DOI] [PubMed] [Google Scholar]
- 44.Lund E, Dahlberg JE. Control of 4-8S RNA transcription at the midblastula transition in Xenopus laevis embryos. Genes Dev. 1992;6:1097–1106. doi: 10.1101/gad.6.6.1097. [DOI] [PubMed] [Google Scholar]
- 45.Lu X, Li JM, Elemento O, Tavazoie S, Wieschaus EF. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development. 2009;136:2101–2110. doi: 10.1242/dev.034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forbes DJ, Kirschner MW, Newport JW. Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell. 1983;34:13–23. doi: 10.1016/0092-8674(83)90132-0. [DOI] [PubMed] [Google Scholar]
- 47.Dreyer C. Differential accumulation of oocyte nuclear proteins by embryonic nuclei of Xenopus. Development. 1987;101:829–846. doi: 10.1242/dev.101.4.829. [DOI] [PubMed] [Google Scholar]
- 48.Jevtic P, Levy DL. Mechanisms of nuclear size regulation in model systems and cancer. Adv Exp Med Biol. 2014;773:537–569. doi: 10.1007/978-1-4899-8032-8_25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.