Abstract
Actin functions as a helical polymer, F-actin, but attempts to build an atomic model for this filament have been hampered by the fact that the filament cannot be crystallized and by structural heterogeneity. We have used a direct electron detector, electron cryo-microscopy and the forces imposed on actin filaments in thin films to reconstruct one state of the filament at 4.7 Å resolution, which allows for building the first reliable pseudo-atomic model of F-actin. We also report a different state of the filament where actin protomers adopt a conformation observed in the crystal structure of the G-actin-profilin complex with an open ATP-binding cleft. Comparison of the two structural states provides new insights into ATP-hydrolysis and filament dynamics. The atomic model provides a framework for understanding why every buried residue in actin has been under intense selective pressure.
Actin is one of the most highly conserved as well as abundant eukaryotic proteins. From chickens to humans, an evolutionary separation of ~ 350 million years, there are no amino acid changes in the skeletal muscle isoform of actin (Hennessey et al., 1993). There are at least six different mammalian isoforms that are quite similar to each other, and all appear to have diverged from a common ancestral actin gene (Miwa et al., 1991). In contrast, we now know that bacteria have actin-like proteins which share a common fold (van den Ent et al., 2001; van den Ent et al., 2002), but have vanishingly little sequence similarity both among themselves and to eukaryotic actin (Derman et al., 2009).
While various ideas have been proposed for why every residue in eukaryotic actin has been under intense selection, ideas about allostery have been the most attractive as allosteric networks must place constraints on buried residues (Suel et al., 2003). For example, it has been postulated that there is an allosteric linkage between the N- and C-termini in actin (McKane et al., 2005), which would require a pathway of internal residues to communicate this allosteric information. Similarly, direct allosteric transfer of information has been shown between the DNase I-binding loop in actin and the C-terminus (Crosbie et al., 1994; Khaitlina et al., 1993), which must depend upon a different pathway of buried residues, just as the observed allosteric linkages between other elements in actin must place constraints on yet more buried residues (Egelman, 2001, 2003; Rubenstein and Wen, 2014).
Since the functional form of actin in most instances is a polymer (F-actin), understanding the constraints on actin sequence evolution must involve an understanding of the structure and dynamics of the actin filament. Unfortunately, unless a helical polymer contains exactly two, three, four or six subunits per turn, it cannot be crystallized so that every subunit is in an equivalent crystallographic environment. This has prevented the structural determination of many helical polymers at atomic resolution, and techniques such as electron cryo-microscopy (cryo-EM) (Fujii et al., 2010; Galkin et al., 2010) and x-ray fiber diffraction (Holmes et al., 1990; Oda et al., 2009) have been the main tools available for studying the structure of protein polymers such as F-actin. Recently, the development of direct electron detectors has allowed an unprecedented advance in the ability of cryo-EM to reach near-atomic resolution for many protein polymers and protein complexes (Amunts et al., 2014; Bai et al., 2013; Bammes et al., 2012; Fernandez et al., 2014; Li et al., 2013; Liao et al., 2013; Lu et al., 2014a; Lu et al., 2014b; Voorhees et al., 2014; Wong et al., 2014).
We have used a direct electron detector and have now been able to reconstruct one state of F-actin at 4.7 Å resolution. This has allowed us to build a pseudo-atomic model of this state, which differs in many details from earlier models for F-actin derived from either a substantially lower resolution reconstruction (Fujii et al., 2010) or modeled against x-ray fiber diffraction patterns (Oda et al., 2009). We compare this atomic model with two other distinctly different states that we have determined at ~ 12 Å resolution, and suggest that only by understanding the multiplicity of states possible for F-actin can one understand the selective pressure on many residues and why mutations of some of these residues leads to myopathies and other human disorders (Rubenstein and Wen, 2014).
Results
F-actin structure depends upon applied forces
We recently suggested that the structural state of actin filaments can be modulated by applied forces (Galkin et al., 2012). Samples are prepared for cryo-EM by blotting a drop applied to a grid so that only a thin film remains, which is then rapidly frozen. During the blotting procedure both the thinning of the solvent film and the flux of fluid within this film can introduce significant mechanical forces. This can be seen by the fact that fields of actin filaments are frequently aligned by the fluid flow (Supp. Fig. 1A) showing the highly anisotropic environment, and by the fact that filaments can be observed to break due to these forces (Supp. Fig. 1B). A previous observation using the compression of actin filaments between two thin mica plates observed a stiffening of these filaments under force (Greene et al., 2009), which appears to be the same phenomenon as the rigidification of F-actin observed in thin ice (Galkin et al., 2012). In order to reduce structural heterogeneity within the actin filament to achieve the highest possible resolution we used blotting conditions (see Materials and Methods) which yielded very thin ice (Fig. 1A, Supp. Fig. 2,3).
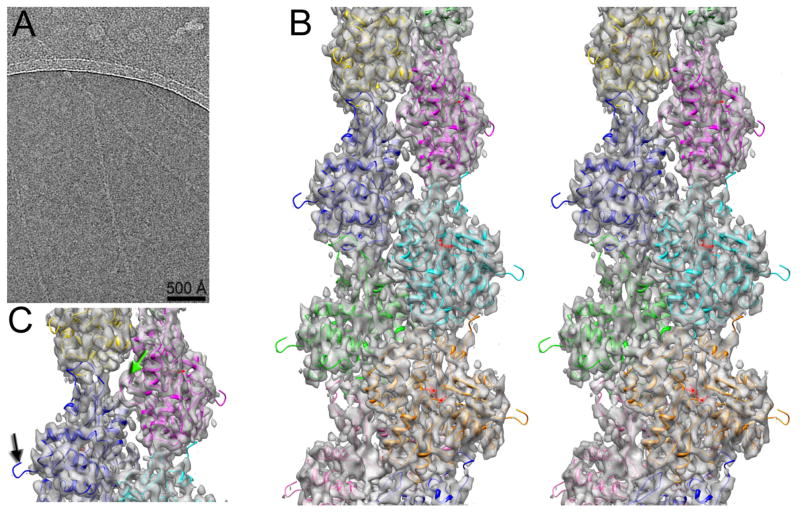
Fig. 1.
3D-reconstruction of frozen hydrated actin filaments. (A) Typical micrograph of actin filaments embedded in thin ice. (B) Stereo view of the 3D-reconstruction at ~ 4.7 Å resolution. (C) The absence of the N-terminal density in the map is indicated with a black arrow, while the hydrophobic plug density is marked with a green arrow.
The relative absence of structural heterogeneity within the thin ice sample allowed us to achieve a high resolution three-dimensional (3D) reconstruction of this one state of F-actin (Fig. 1B) that has been stabilized by mechanical forces without any sorting of the segments by twist or structural state. While the Fourier Shell Correlation (FSC) has been a widely used metric for resolution in cryo-EM, it suffers from the problem that at best it only estimates the internal consistency in the data and not the actual resolution. For example, it is possible to impose the wrong helical symmetry and still measure an FSC that is meaningless (Wu, 2014; Xu et al., 2014). A further problem is that the FSC is based upon comparisons between two half data sets as a way to estimate the resolution of the full data set. Resolution is typically related to the log of the number of images (Stagg et al., 2014) and will therefore tend to become asymptotic as the number of images increases. Thus, with a large data set (where the resolution is near the asymptotic limit) the two half data sets will have a very similar resolution to that of the full data set, while when one has a relatively small number of images the resolution of each half data set will be substantially lower than that of the full data set. We have plotted the FSC as a function of the number of segments in the half data sets (Supp. Fig. 4) and this yields an estimate of 4.5 Å when extrapolating to the full data set. Given the aforementioned problems with the FSC, we estimated the resolution by calculating the FSC between the atomic model and the 3D reconstruction (Supp. Fig. 5) which yielded 4.7 Å using the FSC=0.5 criterion. The best way to judge the resolution, we think, is by actually looking at the map (Fig. 2) and the features seen, such as resolving the individual strands in a β-sheet (Fig. 2A) and visualizing the density from ADP (Fig. 2B), suggest that the 4.7 Å estimate is a reasonable one.
Fig. 2.
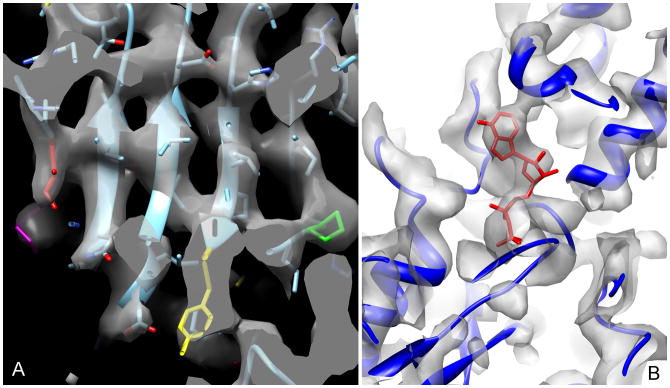
Visual inspection of the map supports the resolution estimated. (A) A close-up view of the β-sheet in SD3 shows that the strands are resolved. ILE330 (red), Pro172 (green), Tyr166 (yellow) and Lys328 (magenta) are highlighted. (B) The region surrounding the ADP (red) is shown.
The structure of the actin filament in canonical state
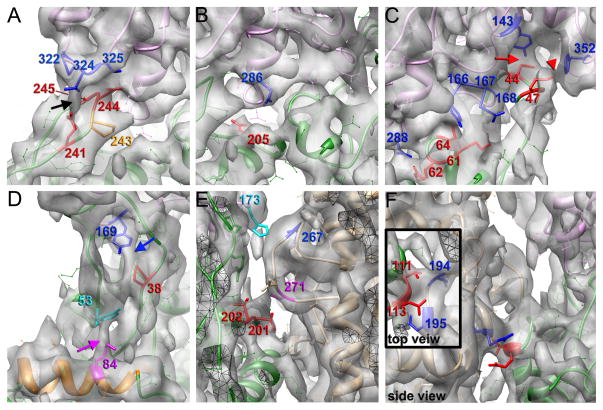
Two surprising observations are that the N-terminus of actin is disordered, and not visualized in the 3D reconstruction (Fig. 1C, black arrow) in contrast to a previous reconstruction where it was quite rigid (Fujii et al., 2010), and that the hydrophobic plug (Fig. 1C, green arrow) originally suggested to maintain the lateral contacts between the two strands of the actin filaments (Holmes et al., 1990) in fact does not appear to make any prominent contacts with the opposite strand. At 4.7 Å resolution the density due to many large side chains can be visualized, and thus a reliable pseudo-atomic model of the filament can be built. Fig. 3 shows details of the longitudinal and lateral interactions that are observed. Actin contains two major domains, with the nucleotide bound in the cleft between them. The outer domain in the actin filament contains two subdomains, subdomain-1 (SD1) and subdomain-2 (SD2), while the inner domain contains subdomain-3 (SD3) and subdomain-4 (SD4). Longitudinal contacts between subunits include the SD3/SD4 and SD2/SD1 interfaces. In agreement with the previous report (Fujii et al., 2010) we found that Asp241 and Glu245 (Fig. 3A, shown in red) interact with Thr324 and Pro322, respectively (Fig. 3A, shown in blue). At the same time Asp244 makes a contact with Met325 instead of Arg290 (Fig. 3A, black arrow). Residues Glu205 (Fig. 3B, shown in red), Asp286 and Asp292 (Fig. 3B, shown in blue) suggested to be a part of this interface (Fujii et al., 2010) are well-resolved in the map and apparently do not make any interactions with residues in the adjacent protomer. Interestingly, we found that Ala204 does not participate in intrafilament interactions, as well, suggesting that the non-polymerizability of the double A204E/P243K mutation (Joel et al., 2004; Rould et al., 2006) may be mainly due to the P243K single mutation. In our map Pro243 can be positioned (Fig. 3A, orange) and forms a kink in the 240–245 loop. We believe that this kink is crucial for a proper positioning of Asp241 and Asp244 in the vicinity of the Thr324 and Met325 in a neighboring subunit, and thus any substitution that changes the local geometry of the loop will result in non-polymerizable actin. We confirm the previous suggestion (Fujii et al., 2010) that Ile64, Lys61, and Arg62 (Fig. 2, marked in red) make interactions with Tyr166, Glu167, and Asp288 (Fig. 3C, marked in blue), respectively. Analysis of our atomic model with PISA (Krissinel and Henrick, 2007) predicts formation of salt bridges between the Lys61-Glu167 and Arg62-Asp288 pairs. At the same time we found that Met44, but not Val45 interacts with Tyr143 (Fig. 3C, red arrow), while Tyr169 is in proximity to Pro38 (Fig. 3D, blue arrow) rather than Pro40, which does not make any intrafilament interactions. The backbone nitrogen of Met44 also makes a hydrogen bond with Gly168 (Fig. 3C, red and blue, respectively). Our filament model thus explains the destabilization of F-actin upon Met44 oxidation (Hung et al., 2011). In addition, Met47 makes a contact with Phe352 (Fig. 3C, red arrowhead) which stabilizes the top of the D-loop. Also, our model explains the increased critical concentration and reduced rates of nucleation after phosphorylation of Tyr53 (Baek et al., 2008; Liu et al., 2006), sinceTyr53 makes a contact with Lys84 (Fig 3D, magenta arrow) and thus staples the D-loop to the body of SD1. A detailed comparison between the interactions present in our model and those predicted in both Fujii et al. (2010) and Oda et al. (2009) is provided in Supp. Table I.
Fig. 3.
Interface between actin protomers in the canonical structural state of F-actin. (A) Interface between SD3 and SD4 is composed of three pairs of interacting residues: 245–322, 324–241, and 244–325. Residues in SD4 of the lower protomer are in red, while residues in SD3 of the upper protomer are in blue. Black arrow indicates a bridge of density between residues 241 and 324. Pro243 that forms a kink in the top of the loop and is believed to position Asp241 and Asp244 is shown in orange. (B) Side-chains of residues 205 (in red) and 286 (in blue) are seen in the map and are not involved in the SD3/SD4 interactions. (C) Interface between SD2 of the lower protomer (red residues) and SD1/3 of the upper protomer (blue residues) includes contacts between residues 44 and 143, 47 and 352, 61 and 167, 62 and 288, and finally 64 and166. Red arrow indicates a contact between residues 44 and 143, while the red arrowhead marks a contact between residues 47 and 352. (D) Pro38 (red) makes a bridge of density with Tyr169 in blue (blue arrow), while Tyr53 (cyan) makes an interaction with Lys84 which is in magenta (magenta arrow). (E) Hydrophobic plug does not make any strong specific interactions with protomers in the opposite strand. There are hydrogen bonds between Val201/Thr202 (in red) and Ser271 (in magenta), along with a hydrophobic interaction between His173 (in cyan) and Ile267 (in blue) (F) Lateral contacts between the two strands contain two pairs of interacting residues: 113–195, and 111–194. Residues in SD1 are in blue, while residues in SD4 are in red.
Buried surface area calculations using PISA (Krissinel and Henrick, 2007) showed that the longitudinal interface buries 1,150 Å2 of surface area while the lateral interface only buries 405 Å2. Surprisingly, the hydrophobic plug, which is nicely resolved in our map and accounts for approximately half of the buried surface area in the lateral interface, does not make any prominent interactions with residues within protomers on the opposite strand. The lateral contacts at this interface are hydrogen bonds between Val201/Thr202 (Fig. 3E, in red) and Ser271 (Fig. 3E, in magenta), along with a hydrophobic interaction between His173 (Fig. 3E, in cyan) and Ile267 (Fig. 3E, in blue). The absence of any prominent interactions between the hydrophobic plug and the adjacent protomers is consistent with cross-linking experiments which suggested that the hydrophobic plug is mobile within the mature filament (Scoville et al., 2006; Shvetsov et al., 2008; Shvetsov et al., 2006). Nevertheless, the hydrophobic plug is crucial for actin polymerization (Shvetsov et al., 2002), it certainly contributes to the lateral interface, and we suggest that it may be more important to the lateral interactions in the other structural states associated with actin polymerization (see below).
The second lateral interface, composed of interactions between SD1 and SD4 of actin protomers on two different strands, is also quite modest in our map and includes two observed pairs of interacting residues: Asn111 is in proximity to Thr194, while Lys113 is next to Glu195 (Fig. 3F).
How does the observed interface explain mutations in actin linked to human diseases? Substitution of Lys241, which makes a contact with Thr324, causes hearing impairment (Hennessey et al., 1993), while mutation of Ile64 causes typical nemaline myopathy (NM) (Feng and Marston, 2009). Similarly, mutation of His40, which hydrogen bonds with Ala170, leads to severe NM (Feng and Marston, 2009). But the majority of the residues that we found to be involved in the intrafilament interactions have not been linked to any diseases. We believe that substitutions of these residues are probably lethal. Our model of the actin filament raises questions about the role of several residues linked to diseases in humans, but not involved in the inter-subunit interactions. For example, substitution of Glu205 causes congenital fiber type disproportion (CFTD) and in vitro results in a 42% decrease in the length of filaments (Guo et al., 2007). Mutations of Asp286 and Asp292 have no effects on actin polymerization in vitro, but cause severe NM and CFTD, respectively (Feng and Marston, 2009). Yet these three amino acids do not participate in F-actin contacts (Fig. 3B). Mutations of residues Asn115, Val370 (Feng and Marston, 2009) and Glu270 (Costa et al., 2004), which are also absent in the interface between protomers, cause typical or severe NM in humans. We suggest that the role of these residues may only be understood by looking at other structural states of F-actin (Galkin et al., 2010). In the next section we will show that T-actin contacts may explain the effect of all of these mutations.
Atomic model of T-actin
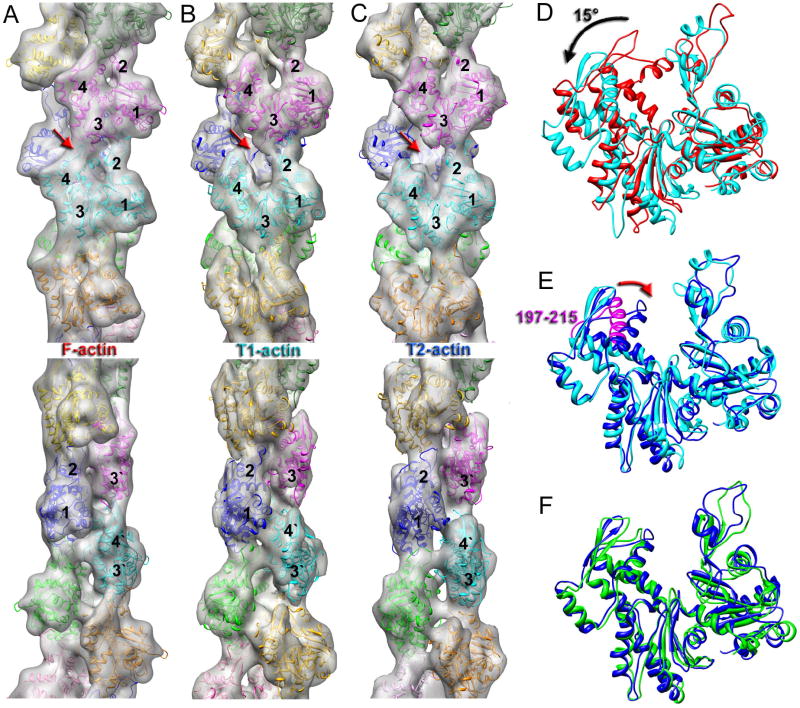
In previous work (Galkin et al., 2010) we reported a 3D reconstruction of the tilted structural state of actin (T-actin) which had a resolution of ~16 Å. Even at that low resolution we were not able to dock in a crystal structure of actin reasonably, since several loops and helices were protruding out of the density envelope even after allowing domain-domain rotations within the actin subunit. This suggested that the imperfections of that reconstruction were caused by the heterogeneity of the T-actin set. To improve the T-actin 3D reconstruction we used images of frozen-hydrated actin filaments over holes (rather than on carbon as was done previously) to enhance the signal-to noise ratio. Next, extensive sorting was used (see Materials and Methods) to divide the T-actin set into two more structurally homogeneous classes. Since models were used for the sorting, concerns about model bias might exist. However, we have previously shown (Galkin et al., 2010) that with the IHRSR approach, where solid cylinders are used as the initial references in all reconstructions, the details of the models used for the sorting never enter into the reconstruction process and thus cannot bias the final results. 3D reconstructions of the two classes, which we call T1 and T2 (Fig. 4), are at a resolution of ~12 Å (Supp. Fig. 6), which allows for flexible fitting of actin crystal structures. Both T1 and T2 maps possess a significant opening of the ATP-binding cleft which results from the rotation by ~15° of subdomains 3 and 4 (the inner domain) of actin around the hinge region (Fig. 4D, black arrow). The main difference between the two tilted actins is not the extent of the opening of the cleft, but rather a movement of the 197–215 region of SD4 towards SD2 (Fig. 4E, red arrow). Importantly, protomers in T2 are similar in conformation with respect to the domain positions with the crystal structure of G-actin in the open state (Chik et al., 1996) (Fig 4F). The flattening of the actin subunits (Oda et al., 2009) is reduced in the tilted state, making the protomers more G-like. Since the tilted state of the actin filament has been linked to early stages of actin polymerization (Galkin et al., 2001; Orlova et al., 2004), it is not surprising that the flattening of actin protomers in the T2 state is reduced.
Fig. 4.
Comparison of canonical F-actin (A) with T1-actin (B) and T2-actin (C). The four subdomains of actin (SD1-SD4) are labeled 1–4. The bottom panels in A–C are the same 3D-reconstructions rotated by 90 degrees. Actin protomers are shown in different colors. All 3D-reconstructions are filtered to 12 Å resolution. (D) Transition from canonical actin (red) to T1-actin (cyan) involves a rotation of SD4/SD3 by ~15° around the hinge region (black arrow). (E) Transition from T1-actin (cyan) to T2-actin (blue) mainly involves transition of the 197–215 region of SD4 (magenta) towards SD2 (red arrow). (F) Alignment of T2-protomer (blue) with the crystal structure of actin in the open state (Chik et al., 1996).
At ~12 Å resolution it is impossible to determine the interface between the protomers in the T1 and T2 states accurately, but we can predict regions involved in these interfaces (Fig. 5). The main difference in the inter-protomer contacts between the canonical and tilted actins is the absence of a contact between SD4 of the lower protomer and SD3 of the upper protomer within the same strand (Fig. 5A,C red arrows). At the same time the difference between the two tilted states is a lack of interaction of the top of SD4 (Fig 5B,D, residues 195–205 in magenta) of one protomer with SD1 (Fig. 5B, residues 112–118 in orange) of the adjacent protomer on the opposite strand (Fig. 5A,C red arrow) due to the movement of the 197–215 region of SD4 towards SD2 in the T2-actin (Fig. 4E, red arrow). Pseudo-atomic models of the tilted structural states of actin place all of the residues involved in human diseases discussed earlier (40, 115, 205, 270, 286, and 370) at the interface between the protomers (Fig. 5E).
Fig. 5.
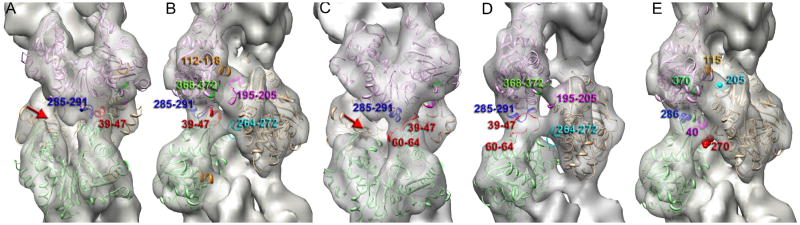
Interface between actin protomers in T1- and T2- actins. 3D-reconstructions are front (A and C) and side (B, D and E) views. (A) Longitudinal contacts in T1-actin reside in the 39–47 region located in SD2 of the lower protomer (red) and region 285–291 (blue) in SD3 of the upper protomer. (B) Lateral interactions are likely to involve residues 112–118 (orange) and 368–371 (green) in SD1 of the upper protomer, and 195–205 (magenta) in SD4 along with the hydrophobic plug (264–272 in cyan) in the lower protomer. (C) Longitudinal contacts in T2-actin are similar to the T1-actin, but also may include residue 60–64 (marked in red). (D) lateral interactions in T2-actin map to the same regions as in T1-actin except for the112–118 region. (E) Residues linked to human diseases are shown as spheres in different colors. Note that these residues are at the interfaces between protomers in the tilted actin.
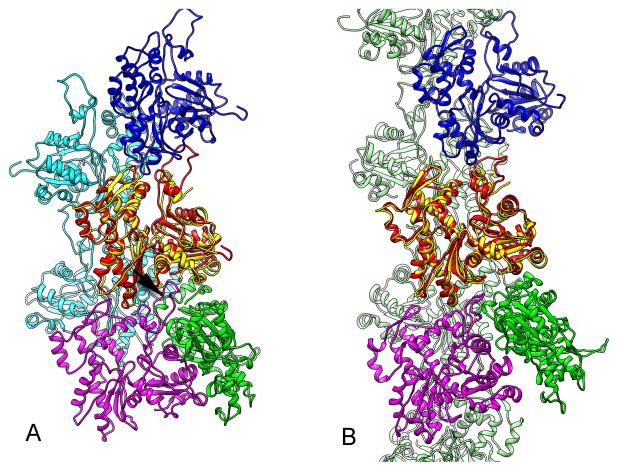
A model for T-actin allows an interesting observation about the relationship between an actin dimer (Bubb et al., 2002; Millonig et al., 1988; Silvan et al., 2012) and the branched and ragged actin filaments that have been observed at short times after the initiation of polymerization (Steinmetz et al., 1997). In solution, monomeric actin can be readily crosslinked into an antiparallel dimer that has been called the lower-dimer (Bubb et al., 2002; Millonig et al., 1988). Surprisingly, since every subunit in an actin filament has the same polarity, this dimer nucleates actin filament formation (Bubb et al., 2002). If one subunit of the crystallographic dimer (Bubb et al., 2002) is fit into a model for canonical F-actin, the antiparallel subunit of the dimer makes large clashes with another F-actin protomer (Fig. 6A). In contrast, there are no significant clashes when the same antiparallel dimer is docked into our model for T-actin (Fig. 6B), allowing the second subunit of the dimer to serve as a nucleus for a branched actin filament.
Fig. 6.
A comparison between the antiparallel actin dimer 1LCU (Bubb et al., 2002) and two states of the actin filament. A) When chain A of the dimer (in yellow) is superimposed upon one subunit (red) in our model for canonical F-actin (1.1 A rmsd, over 236 Cα pairs), the second chain of the dimer (in green) makes extensive clashes with another protomer in the filament (arrow). B) When the same chain A of 1LCU (in yellow) is superimposed upon a protomer in the T1 actin (in red) (1.3 A rmsd, over 242 Cα pairs), the second chain of the dimer (in green) makes no significant clashes with the filament.
Position of the nucleotide in the canonical and tilted actin
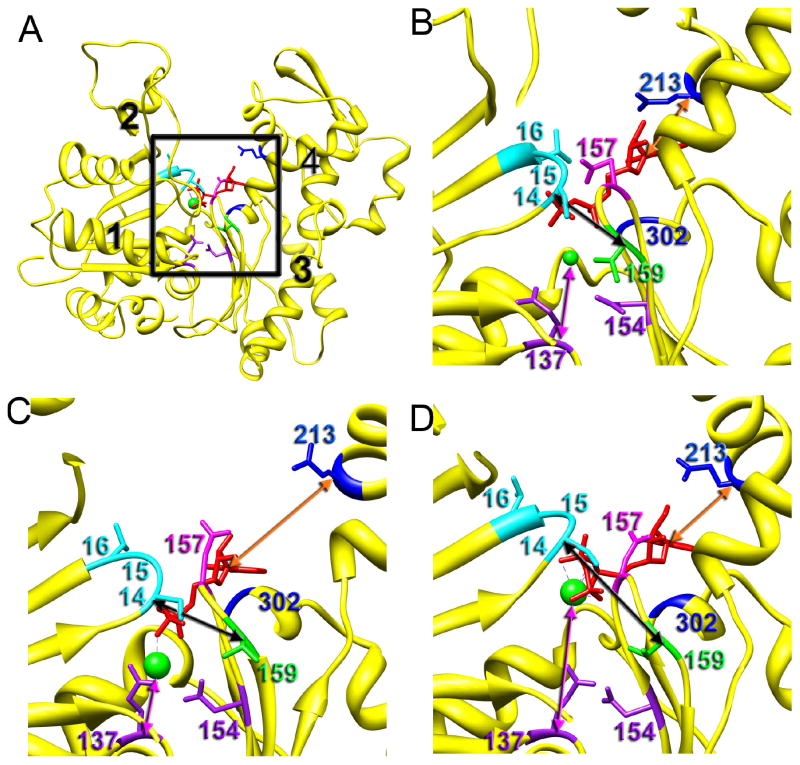
Polymerization of G-actin into its filamentous form triggers the hydrolysis of ATP located in the binding cleft located between the two major domains. At 4.7 Å resolution we can visualize the ADP molecule in the cleft (Fig. 2B). The base of the ADP makes contacts with Lys213 and Glu302 (Fig. 7B, in blue). In our model, Asp157 (Fig. 7B, in magenta) forms a hydrogen bond with the β-phosphate which also makes hydrogen bonds with the backbone nitrogens of Ser14, Gly15 and Leu16 (Fig. 7B, in cyan). While the Mg2+ ion is not seen at the resolution of our map, it is presumably coordinated by the ADP oxygens on the top and residues Gln137 and Asp154 (Fig. 7D, in purple) on the bottom. We do not observe interaction of Lys18 and Lys336 with the nucleotide as was observed in a crystal structure of non-polymerizable ADP-actin (Rould et al., 2006). To predict the position of the nucleotide and the Mg2+ ion in the cleft of the T1 state we aligned the T1 protomer with the crystal structures of actin in the closed (Rould et al., 2006) and open states (Chik et al., 1996). We found a significantly better match of the cleft region of the T1 protomer with actin in the closed state (Rould et al., 2006), which was surprising since the cleft in the T1 state is wide open. Because of the opening of the cleft the distance between Lys213 and the ADP base (from the Cα to C4 of the adenine base) increases from ~ 5 Å in the canonical state to ~ 10 Å in T1 (Fig. 5B and C, orange arrow). At the same time the distance between Asp157 and Glu15, which reflects the tightness of the packing of the phosphate groups, is similar to the canonical state. Importantly, the distance between Ser14 (Fig. 7, in cyan) and Val159 (Fig. 7, in green) that coordinate γ-phosphate in the crystal structure of the ATP-bound G-actin (Rould et al., 2006) is ~ 7 Å (all distances between Cα atoms) in both canonical and T1 (Fig. 7B,C, black arrows). The distance between Glu137 and the divalent cation is also similar to the canonical state (Fig. 7B,C, magenta arrow). Thus, the only difference between the canonical and the T1 states in the cleft region seems to be a movement of Lys213 away from the base of the nucleotide. As mentioned earlier (Fig. 4F), the T2-protomer yields a better match with the crystal structure of actin in the open state (Chik et al., 1996). Due to the backshift of the 197–215 region of SD4 in the T2 actin (Fig. 4E, red arrow) the distance between the nucleotide base and Lys213 is reduced to ~ 7 Å, but is still larger than that in canonical actin (Fig. 7B,D, orange arrows). The divalent cation is coordinated only by the terminal phosphates in this model since the distance between Glu137 and Asp154 is twice that in the canonical or T1 protomers (Fig. 7B,D, magenta arrows). The distance between Ser14 and Val159 is increased from ~ 7 Å in T1 to ~ 10 Å in T2-actin, which suggests that the terminal phosphate is less coordinated in the cleft of T2. Overall, our data suggest that in both T1 and T2 protomers interaction of the ADP base with the actin molecule is compromised, while in T2 the terminal phosphate group interaction with actin via Ser14 and Val159 is weakened. Thus, in the T2 state dissociation of the γ-phosphate should be favored because of the weaker nucleotide-actin interaction. This is consistent with our previous results which showed an elevated level of T-actin in early stages of actin polymerization (Galkin et al., 2001; Orlova et al., 2004) when ATP is actively hydrolyzed and the γ-phosphate group is released.
Fig. 7.
Comparison of the nucleotide position in the ATP-binding cleft of the canonical, T1 and T2 actins. (A) Back view of the actin molecule with domains numbered in black. Black square marks the ATP-binding cleft magnified in B–D. (B) Atomic model of the actin-ADP interaction derived from the 4.7 Å 3D-reconstruction of the canonical F-actin. (C) Modeling of the nucleotide interaction with the T1 protomer. (D) Modeling of the nucleotide interaction with the T2 protomer. (B–C) The base of the nucleotide interacts with Lys213 and Gln302 marked in blue. Phosphate groups are coordinated through the interaction with Ser14, Glu15, and Leu16 of the SD1 (in cyan), and Asp157 (magenta) and Val159 (green) located in SD3. Divalent metal ion interacts with phosphate groups on the top, and Gln137 and Asp154 on the bottom (in purple). The distances between the nucleotide base and Lys213 are shown as orange arrows, distances between the Mg2+ ion and Gln137 are shown as magenta arrows, while the distances between Ser14 and Val159 that coordinate terminal phosphate are marked with black arrows.
Discussion
Given the multiplicity of structural states possible for F-actin (Galkin et al., 2010) we do not claim to have generated an improved model for the only state that can exist. Rather, we have taken advantage of new developments in cryo-EM, such as a direct electron detector (Li et al., 2013), to generate a model for only one state, and this model differs in many important details (Supp. Table I) from models generated with lower resolution data (Fujii et al., 2010; Oda et al., 2009). Given the polymorphisms observed in vitro, what is the biological significance of these different states? A naturally occurring actin bundle, the acrosomal process of the Limulus sperm, provides some interesting insights into how cells may actually use the structural polymorphisms that are inherent in F-actin. Within the acrosomal process a multiplicity of actin states were observed, which differed not only in helical twist but in tertiary structure (Cong et al., 2008; Schmid et al., 2004). When actin filaments are decorated with cofilin (Galkin et al., 2011), which changes the twist of the filament by ~ 5° per subunit (McGough et al., 1997), the conformational changes in the actin subunit in the filament are greater than the conformational changes associated with the monomer to polymer transition (Oda et al., 2009). Given that intracellular bundles of actin decorated with cofilin have been observed in neurodegenerative diseases (Bamburg et al., 2010), and given that cofilin decoration of actin requires very large conformational changes in actin (Galkin et al., 2011), actin’s structural plasticity cannot be considered as being an in vitro artifact.
T-actin and phosphate hydrolysis
Polymerization of G-actin into filaments involves hydrolysis of the nucleotide tightly bound in the cleft of the actin molecule. Although ATP hydrolysis is not required for actin polymerization per se (De La Cruz et al., 2000; Kasai et al., 1965) it has been shown that the stability of the actin filament and the rate of polymerization both depend upon the bound nucleotide. Thus, a great deal of attention has been devoted to the structural consequences of ATP hydrolysis and dissociation of the inorganic phosphate. Early EM studies noted that the gross morphology of filaments changes over time after initial polymerization (Steinmetz et al., 1997). Dominguez and colleagues suggested from a crystal structure that the D-loop goes from a loop to helix upon nucleotide hydrolysis (Otterbein et al., 2001), but this conclusion was undercut by subsequent crystallographic studies where the main difference seen between ADP- and ATP-bound G-actin was a small movement of the sensor loop in the back of the ATP-binding cleft (Rould et al., 2006). Thus, crystallographic studies failed to find the difference between the ATP- and ADP-bound actin that could explain large changes in actin morphology after ATP hydrolysis during the early stages of polymerization. Image analysis of negatively stained actin filaments suggested that ATP-bound F-actin has its cleft tightly closed and ATP hydrolysis results in the opening of the cleft (Belmont et al., 1999), but subsequent data obtained from frozen-hydrated actin filaments showed that in mature F-actin, where all ATP has been hydrolyzed to ADP, the ATP-binding cleft is tightly closed (Fujii et al., 2010; Galkin et al., 2010). However, after short times of polymerization F-actin is enriched with an unconventional structural state of the filament which we called T-actin (Orlova et al., 2004). In this structural state the longitudinal contacts, which play the largest role in the stability of the filament (as opposed to the weaker lateral contacts), are weakened since the interaction between SD4 and SD3 of two adjacent actin protomers along the same strand is broken (Fig. 4B,C, red arrow). Such a weakening of the contacts within T-actin is consistent with the elevated instability of the filament at the region of attachment of a cap containing actin-ATP to the body of the filament (which is actin-ADP) and has been associated with “minor catastrophe” events upon actin polymerization (Fujiwara et al., 2002). Importantly, in the presence of proteins that disrupt actin filaments and enhance ATP hydrolysis (ADF and cofilins) the frequency of T-actin was also elevated (Galkin et al., 2002), suggesting a mechanism of depolymerization by means of a time reversal of polymerization (Orlova et al., 2004).
Our model for T-actin can now serve to reconcile a number of seemingly paradoxical observations, such as how an antiparallel actin dimer that readily forms in solution (Millonig et al., 1988) can serve as a nucleator for actin filaments (Bubb et al., 2002) where every subunit has a parallel orientation. The fact that the dimer is compatible with T-actin and incompatible with canonical actin (Fig. 6), and the fact that T-actin is greatly enriched at early times after the initiation of polymerization (Orlova et al., 2004), are consistent with the appearance of branched filaments observed at early stages of polymerization (Steinmetz et al., 1997), exactly what would be expected if the second subunit within the dimer were nucleating a second actin filament.
Materials and Methods
Reconstruction volumes have been deposited for the canonical F-actin, T1 and T2 states with accession numbers EMDB-6179, EMDB-6180 and EMDB-6181, with the corresponding atomic models 3J8I.pdb, 3J8J.pdb and 3J8K.pdb, respectively.
Actin
Rabbit skeletal actin was prepared as previously described (Galkin et al., 2010).
Ice thickness and structural heterogeneity
In our hands the thinnest ice was achieved using lacey carbon grids, while quite thick ice was observed when C-flat grids were used. Since it is not possible to determine the thickness of the ice in individual images, searches are done for regions of a grid where the ice appears thin based upon the reduced attenuation of the electron beam. We have been able to correlate the perceived thickness of the ice with the amount of out-of-plane tilt (Supp. Fig. 2), since the thinner the ice the more filaments will be constrained to be in a plane perpendicular to the beam. To evaluate the degree of heterogeneity in each sample we looked at both the variability in the twist and the frequency of multiple structural states that we previously reported as being present in thicker ice (Galkin et al., 2010). In thin ice actin filaments have reduced variability of twist (Supp. Fig. 3), but nevertheless variable twist is still observable in these filaments. Power spectra of the 166° and 167° classes (Supp. Fig 3A, insert) unambiguously show shifts of the n=4 layer line, demonstrating that the sorting is working as expected, and filaments are not being misclassified due to a poor signal to noise ratio or other effects. Variability of the twist in the thicker ice sample was twice that of the thinner ice, since Gaussian fits of the two twist histograms yielded σ=0.73° for the thinner ice and σ=1.5° for the thicker ice. Using the formalism of cumulative random angular disorder (Egelman and DeRosier, 1982; Egelman et al., 1982; Galkin et al., 2012) these values correspond to random deviations per actin subunit of 2.9° and 6.0° for the thinner and thicker ice, respectively. As with the thinner ice, power spectra from two classes of the thicker ice sample validated the sorting (Supp. Fig. 3B, insert). The degree of structural heterogeneity paralleled the variability in the twist in the two sets. While 95% of the segments from the thin ice set were classified as being canonical F-actin, we found only 70% of such segments in the thick ice set. The remaining 30% were comprised of segments with subdomain 2 (SD2) of actin disordered (~12%) and in a tilted (T-actin) state (Galkin et al., 2010; Galkin et al., 2002) (~18%). We found that the T-actin set was itself heterogeneous and two modes of that state can be sorted out.
3D-reconstruction of canonical F-actin
Samples (2.5 μL) were applied to lacey carbon grids and vitrified in a Vitrobot Mark IV (FEI, Inc.). They were imaged with a Titan Krios at 300 keV using the Falcon 2 direct electron detector. Images were acquired using the EPU software for automated control of the microscope. The SPIDER software package (Frank et al., 1996) was used for most image processing, but the CTFFIND3 software (Mindell and Grigorieff, 2003) was used to determine the defocus values in the micrographs while helixboxer in the EMAN package (Ludtke et al., 1999) was used to extract filament images from micrographs. We used 850 micrographs possessing a defocus range from 0.5 to 3.0 μm at a raster of 1.05 Å per pixel. Initial correction for the contrast transfer function (CTF) was made by multiplying each image by its theoretical CTF. From these CTF-corrected images, 55,052 overlapping segments (460 pixels long) were extracted and reconstructed with the IHRSR method (Egelman, 2000). After 10 iterations the set yielded a stable solution of 166.7°/27.6 Å. While out-of-plane tilt can be ignored in negatively-stained images (due to adsorption to a carbon substrate as well as limited resolution), it cannot be ignored at higher resolution in cryo-EM. The sensitivity to out-of-plane tilt is the same as the sensitivity to azimuthal rotations, and is given by d/D where d is the resolution and D is the diameter. So for an actin filament with an approximate diameter of 90 Å and with an anticipated resolution of 4.5 Å, the sampling angle needed for both azimuthal rotations and out-of-plane tilt is ~ 3°. We treated out-of-plane tilt by including reference projections with an out-of-plane tilt range from −9° to +9° for all IHRSR cycles done at high resolution.
3D-reconstruction of T1- and T2- actins
Samples were applied to C-flat grids, vitrified in a lab-built plunger, and imaged on a Tecnai F20 at 200 keV using film. A Nikon COOLPIX 8000 scanner was used to digitize 437 micrographs having a defocus range from 0.9 to 3.0 μm at a raster of 1.25 Å per pixel. Initial correction for the contrast transfer function (CTF) was made by multiplying each image by its theoretical CTF. From these images we extracted 89,160 overlapping segments (400 px long). Three model volumes were created using a model of F-actin in the canonical state (Fujii et al., 2010), the same model with missing SD2, and one in the “tilted” state (Galkin et al., 2010). These volumes were scaled to 5.0 Å per pixel and projected onto 100 × 100 px images with an azimuthal rotational increment of 4°, generating 270 reference projections (3 × 90). The F-actin segments were down-sampled to 5 Å per pixel and cross-correlated with the 270 reference projections. A set of 62,550 segments (70%) was selected as similar to the “canonical” state of F-actin, 10,304 segments were assigned to the SD2-disordered class (12%), and 16,306 segments (18%) yielded the best match with the tilted reference. The T-actin set was sorted out into two classes based upon the opening of the ATP-binding cleft using two model volumes generated using either a crystal structure of the G-actin-profilin complex in the open state (Chik et al., 1996), or actin molecules having a partially open cleft where the two major domains of actin were moved apart half way between the canonical actin and the open structural state found in the G-actin-profilin complex. Both sets were reconstructed using the IHRSR approach. T1-set (n=8,747) converged to a symmetry of −166.8°/28.3 Å, while T2-set (n=7,559) yielded a symmetry of −168°/29.8Å. A test of whether models might always return segments that resemble the model was done using a bogus model, one lacking SD3, in addition to the models above. Very few segments showed the highest correlation with this bogus model, and a reconstruction from these segments (starting with a solid cylinder) did not look like anything recognizable.
Supplementary Material
Supplementary Figure 1, related to Figure 1. Preparation of grids for cryo-EM involves large forces on actin filaments. A) An image of a field of filaments showing a common orientation, almost certainly due to fluid flow in one direction during the blotting that occurs prior to freezing. B) Breaks (black arrows) can occasionally be observed in actin filaments due to the mechanical forces that are placed on these filaments from the fluid flow. The sharp bend (white arrow) is inconsistent with the known rigidity of the actin filament and thermal flexing, and must also arise from mechanical forces.
Supplementary Figure 2, related to Figure 1. Out-of-plane tilt of segments correlates with the perceived thickness of the ice. Typical images of actin in what were perceived as thin ice (A) and thick ice (C), both using lacey carbon grids imaged with a Titan Krios. In the thin ice, actin filaments (A, arrows) can be frequently found that are anomalously straight (Galkin et al., 2012) due to the large mechanical forces that are present. Histograms for out-of-plane tilt from thin ice (B) and thick ice (D) have been fitted with Gaussians, and the distributions show that the amount of out-of-plane tilt is increased when the ice is thicker, consistent with the fact that filaments have a greater freedom to deviate away from the perpendicular to the beam when the film containing the filaments is thicker prior to freezing.
Supplementary Figure 3, related to Figure 1. Comparison of twist distributions of F-actin in thinner ice (A) and thicker ice (B) were calculated using cross-correlation of images with projections from a set of models having a range of twist from 162° to 170° with a step of 0.5°. The reliability of the sorting was established by computing averaged power spectra from segments having smaller (red bins) or larger (blue bins) angles. A) Twist distribution based on a sorting of 55,052 segments in thinner ice, with the two classes used to calculate power spectra marked in red (n=8,727) and blue (n=11,900). Comparison of the two power spectra is shown in insert where positions of the n=4 layer line are marked with white arrows. B) Twist distribution based on a sorting of 89,160 segments in thicker ice, with the two classes used to calculate power spectra in red (n=6,770) and blue (n=8,995). Comparison of the two power spectra is shown in insert where positions of the n=4 layer line are marked with white arrows.
Supplementary Figure 4, related to Figure 2. (A) The Fourier Shell Correlation (FSC) between reconstructions from half data sets, with each half set containing from 6,250 to 25,000 segments. Completely independent data sets were generated, so that there was no overlap of boxes (as opposed to the extensive overlap that would exist if one took odd/even images to create the two sets). (B) The resolution (using the FSC=0.143) found in (A) is plotted versus the number of segments, using a log10 scale. This can be fit nicely by a straight line (dotted red line) as observed by others (Stagg et al., 2014). The estimate for the resolution of 50,000 segments (approximately the number of segments in the full data set) is 0.223 (vertical black line) or 1/(4.5 Å).
Supplementary Figure 5, related to Figure 2. The FSC between the atomic model for the canonical state of F-actin and the actual reconstruction. The FSC is 0.5 at 4.66 Å.
Supplementary Figure 6, related to Figure 4. The FSC for the T2 reconstruction. The data were split into two halves with almost no overlap of boxes. Each subset was iterated independently for five cycles, and then aligned to generate the FSC curve.
Supplementary Figure 7, related to Experimental Procedures. Rfree value during the final refinement and model building cycles. Simulated annealing refinements were performed with CNS using DEN restraints. Structure factors between 4.4–4.5 Å were chosen for the calculation of Rfree.
Supplementary Table I, related to Figure 3
The residues predicted to be involved in the subunit-subunit interface are shown for the fiber diffraction model of Oda et al. (Oda et al., 2009), the cryo-EM model of Fujii et al. (Fujii et al., 2010) and this paper. Differences between our model and that of Fujii et al. are shown in red.
Supplementary Table II, related to Supplementary Experimental Procedures
Acknowledgments
We thank Kelly Dryden for assistance with the microscopy at UVa. This work was supported by grant GM081303 (to E.H.E.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K, Liu X, Ferron F, Shu S, Korn ED, Dominguez R. Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding. Proc Natl Acad Sci U S A. 2008;105:11748–11753. doi: 10.1073/pnas.0805852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Bernstein BW, Davis RC, Flynn KC, Goldsbury C, Jensen JR, Maloney MT, Marsden IT, Minamide LS, Pak CW, et al. ADF/Cofilin-actin rods in neurodegenerative diseases. Curr Alzheimer Res. 2010;7:241–250. doi: 10.2174/156720510791050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bammes BE, Rochat RH, Jakana J, Chen DH, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. Journal of Structural Biology. 2012;177:589–601. doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont LD, Orlova A, Drubin DG, Egelman EH. A change in actin conformation associated with filament instability after Pi release. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubb MR, Govindasamy L, Yarmola EG, Vorobiev SM, Almo SC, Somasundaram T, Chapman MS, Agbandje-McKenna M, McKenna R. Polylysine induces an antiparallel actin dimer that nucleates filament assembly: crystal structure at 3.5-A resolution. Journal of Biological Chemistry. 2002;277:20999–21006. doi: 10.1074/jbc.M201371200. [DOI] [PubMed] [Google Scholar]
- Chik JK, Lindberg U, Schutt CE. The structure of an open state of β-actin at 2.65 èngstrom resolution. Journal of Molecular Biology. 1996;263:607–623. doi: 10.1006/jmbi.1996.0602. [DOI] [PubMed] [Google Scholar]
- Cong Y, Topf M, Sali A, Matsudaira P, Dougherty M, Chiu W, Schmid MF. Crystallographic conformers of actin in a biologically active bundle of filaments. Journal of Molecular Biology. 2008;375:331–336. doi: 10.1016/j.jmb.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa CF, Rommelaere H, Waterschoot D, Sethi KK, Nowak KJ, Laing NG, Ampe C, Machesky LM. Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J Cell Sci. 2004;117:3367–3377. doi: 10.1242/jcs.01172. [DOI] [PubMed] [Google Scholar]
- Crosbie RH, Miller C, Cheung P, Goodnight T, Muhlrad A, Reisler E. Structural connectivity in actin: effect of C-terminal modifications on the properties of actin. Biophysical Journal. 1994;67:1957–1964. doi: 10.1016/S0006-3495(94)80678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, Mandinova A, Steinmetz MO, Stoffler D, Aebi U, Pollard TD. Polymerization and structure of nucleotide-free actin filaments. J Mol Biol. 2000;295:517–526. doi: 10.1006/jmbi.1999.3390. [DOI] [PubMed] [Google Scholar]
- Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol. 2009;73:534–552. doi: 10.1111/j.1365-2958.2009.06771.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Actin allostery again? Nat Struct Biol. 2001;8:735–736. doi: 10.1038/nsb0901-735. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Actin’s prokaryotic homologs. Curr Opin Struct Biol. 2003;13:244–248. doi: 10.1016/s0959-440x(03)00027-7. [DOI] [PubMed] [Google Scholar]
- Egelman EH, DeRosier DJ. The Fourier transform of actin and other helical systems with cumulative random angular disorder. Acta Crystallographica. 1982;A38:796–799. [Google Scholar]
- Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature. 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Feng JJ, Marston S. Genotype-phenotype correlations in ACTA1 mutations that cause congenital myopathies. Neuromuscul Disord. 2009;19:6–16. doi: 10.1016/j.nmd.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Takahashi S, Tadakuma H, Funatsu T, Ishiwata S. Microscopic analysis of polymerization dynamics with individual actin filaments. Nat Cell Biol. 2002;4:666–673. doi: 10.1038/ncb841. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Egelman EH. Actin filaments as tension sensors. Current Biology. 2012;22:R96–R101. doi: 10.1016/j.cub.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Lukoyanova N, Wriggers W, Egelman EH. Actin Depolymerizing Factor Stabilizes an Existing State of F-Actin and Can Change the Tilt of F-Actin Subunits. Journal of Cell Biology. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Schrîder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17:1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, VanLoock MS, Orlova A, Egelman EH. A new internal mode in F-actin helps explain the remarkable evolutionary conservation of actin’s sequence and structure. Current Biology. 2002;12:570–575. doi: 10.1016/s0960-9822(02)00742-x. [DOI] [PubMed] [Google Scholar]
- Greene GW, Anderson TH, Zeng H, Zappone B, Israelachvili JN. Force amplification response of actin filaments under confined compression. Proc Natl Acad Sci USA. 2009;106:445–449. doi: 10.1073/pnas.0812064106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo DC, Pannu H, Tran-Fadulu V, Papke CL, Yu RK, Avidan N, Bourgeois S, Estrera AL, Safi HJ, Sparks E, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- Hennessey ES, Drummond DR, Sparrow JC. Molecular genetics of actin function. The Biochemical journal. 1993;291(Pt 3):657–671. doi: 10.1042/bj2910657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joel PB, Fagnant PM, Trybus KM. Expression of a nonpolymerizable actin mutant in Sf9 cells. Biochemistry. 2004;43:11554–11559. doi: 10.1021/bi048899a. [DOI] [PubMed] [Google Scholar]
- Kasai M, Nakano E, Oosawa F. Polymerization Of Actin Free From Nucleotides And Divalent Cations. Biochim Biophys Acta. 1965;94:494–503. doi: 10.1016/0926-6585(65)90058-0. [DOI] [PubMed] [Google Scholar]
- Khaitlina SY, Moraczewska J, Strzelecka-Golaszewska H. The actin/actin interactions involving the N-terminus of the DNase-I-binding loop are crucial for stabilization of the actin filament. European Journal of Biochemistry. 1993;218:911–920. doi: 10.1111/j.1432-1033.1993.tb18447.x. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Shu S, Hong MS, Levine RL, Korn ED. Phosphorylation of actin Tyr-53 inhibits filament nucleation and elongation and destabilizes filaments. Proc Natl Acad Sci U S A. 2006;103:13694–13699. doi: 10.1073/pnas.0606321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, Schroder GF, Fitzgerald KA, Wu H, Egelman EH. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014a;156:1193–1206. doi: 10.1016/j.cell.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Bai XC, Ma D, Xie T, Yan C, Sun L, Yang G, Zhao Y, Zhou R, Scheres SH, et al. Three-dimensional structure of human gamma-secretase. Nature. 2014b;512:166–170. doi: 10.1038/nature13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of Structural Biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. Journal of Cell Biology. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKane M, Wen KK, Boldogh IR, Ramcharan S, Pon LA, Rubenstein PA. A mammalian actin substitution in yeast actin (H372R) causes a suppressible mitochondria/vacuole phenotype. Journal of Biological Chemistry. 2005;280:36494–36501. doi: 10.1074/jbc.M506970200. [DOI] [PubMed] [Google Scholar]
- Millonig R, Salvo H, Aebi U. Probing actin polymerization by intermolecular cross-linking. The Journal of cell biology. 1988;106:785–796. doi: 10.1083/jcb.106.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Miwa T, Manabe Y, Kurokawa K, Kamada S, Kanda N, Bruns G, Ueyama H, Kakunaga T. Structure, chromosome location, and expression of the human smooth muscle (enteric type) gamma-actin gene: evolution of six human actin genes. Mol Cell Biol. 1991;11:3296–3306. doi: 10.1128/mcb.11.6.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Orlova A, Shvetsov A, Galkin VE, Kudryashov DS, Rubenstein PA, Egelman EH, Reisler E. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc Natl Acad Sci USA. 2004;101:17664–17668. doi: 10.1073/pnas.0407525102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterbein LR, Graceffa P, Dominguez R. The crystal structure of uncomplexed actin in the ADP state. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- Rould MA, Wan Q, Joel PB, Lowey S, Trybus KM. Crystal structures of expressed non-polymerizable monomeric actin in the ADP and ATP states. Journal of Biological Chemistry. 2006;281:31909–31919. doi: 10.1074/jbc.M601973200. [DOI] [PubMed] [Google Scholar]
- Rubenstein PA, Wen KK. Insights into the effects of disease-causing mutations in human actins. Cytoskeleton. 2014;71:211–229. doi: 10.1002/cm.21169. [DOI] [PubMed] [Google Scholar]
- Schmid MF, Sherman MB, Matsudaira P, Chiu W. Structure of the acrosomal bundle. Nature. 2004;431:104–107. doi: 10.1038/nature02881. [DOI] [PubMed] [Google Scholar]
- Scoville D, Stamm JD, Toledo-Warshaviak D, Altenbach C, Phillips M, Shvetsov A, Rubenstein PA, Hubbell WL, Reisler E. Hydrophobic loop dynamics and actin filament stability. Biochemistry. 2006;45:13576–13584. doi: 10.1021/bi061229f. [DOI] [PubMed] [Google Scholar]
- Shvetsov A, Galkin VE, Orlova A, Phillips M, Bergeron SE, Rubenstein PA, Egelman EH, Reisler E. Actin hydrophobic loop 262–274 and filament nucleation and elongation. J Mol Biol. 2008;375:793–801. doi: 10.1016/j.jmb.2007.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvetsov A, Musib R, Phillips M, Rubenstein PA, Reisler E. Locking the hydrophobic loop 262–274 to G-actin surface by a disulfide bridge prevents filament formation. Biochemistry. 2002;41:10787–10793. doi: 10.1021/bi020205f. [DOI] [PubMed] [Google Scholar]
- Shvetsov A, Stamm JD, Phillips M, Warshaviak D, Altenbach C, Rubenstein PA, Hideg K, Hubbell WL, Reisler E. Conformational dynamics of loop 262–274 in G- and F-actin. Biochemistry. 2006;45:6541–6549. doi: 10.1021/bi052558v. [DOI] [PubMed] [Google Scholar]
- Silvan U, Boiteux C, Sutterlin R, Schroeder U, Mannherz HG, Jockusch BM, Berneche S, Aebi U, Schoenenberger CA. An antiparallel actin dimer is associated with the endocytic pathway in mammalian cells. J Struct Biol. 2012;177:70–80. doi: 10.1016/j.jsb.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Noble AJ, Spilman M, Chapman MS. ResLog plots as an empirical metric of the quality of cryo-EM reconstructions. J Struct Biol. 2014;185:418–426. doi: 10.1016/j.jsb.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz MO, Goldie KN, Aebi U. A correlative analysis of actin filament assembly, structure and dynamics. Journal of Cell Biology. 1997;138:559–574. doi: 10.1083/jcb.138.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lîwe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO Journal. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Fernandez IS, Scheres SH, Hegde RS. Structure of the mammalian ribosome-Sec61 complex to 3.4 A resolution. Cell. 2014;157:1632–1643. doi: 10.1016/j.cell.2014.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, Scheres SH. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife. 2014:e03080. doi: 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BPA, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular imprinting as a signal activation mechanism of the viral RNA sensor RIG-I. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.06.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ, et al. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. eLife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1, related to Figure 1. Preparation of grids for cryo-EM involves large forces on actin filaments. A) An image of a field of filaments showing a common orientation, almost certainly due to fluid flow in one direction during the blotting that occurs prior to freezing. B) Breaks (black arrows) can occasionally be observed in actin filaments due to the mechanical forces that are placed on these filaments from the fluid flow. The sharp bend (white arrow) is inconsistent with the known rigidity of the actin filament and thermal flexing, and must also arise from mechanical forces.
Supplementary Figure 2, related to Figure 1. Out-of-plane tilt of segments correlates with the perceived thickness of the ice. Typical images of actin in what were perceived as thin ice (A) and thick ice (C), both using lacey carbon grids imaged with a Titan Krios. In the thin ice, actin filaments (A, arrows) can be frequently found that are anomalously straight (Galkin et al., 2012) due to the large mechanical forces that are present. Histograms for out-of-plane tilt from thin ice (B) and thick ice (D) have been fitted with Gaussians, and the distributions show that the amount of out-of-plane tilt is increased when the ice is thicker, consistent with the fact that filaments have a greater freedom to deviate away from the perpendicular to the beam when the film containing the filaments is thicker prior to freezing.
Supplementary Figure 3, related to Figure 1. Comparison of twist distributions of F-actin in thinner ice (A) and thicker ice (B) were calculated using cross-correlation of images with projections from a set of models having a range of twist from 162° to 170° with a step of 0.5°. The reliability of the sorting was established by computing averaged power spectra from segments having smaller (red bins) or larger (blue bins) angles. A) Twist distribution based on a sorting of 55,052 segments in thinner ice, with the two classes used to calculate power spectra marked in red (n=8,727) and blue (n=11,900). Comparison of the two power spectra is shown in insert where positions of the n=4 layer line are marked with white arrows. B) Twist distribution based on a sorting of 89,160 segments in thicker ice, with the two classes used to calculate power spectra in red (n=6,770) and blue (n=8,995). Comparison of the two power spectra is shown in insert where positions of the n=4 layer line are marked with white arrows.
Supplementary Figure 4, related to Figure 2. (A) The Fourier Shell Correlation (FSC) between reconstructions from half data sets, with each half set containing from 6,250 to 25,000 segments. Completely independent data sets were generated, so that there was no overlap of boxes (as opposed to the extensive overlap that would exist if one took odd/even images to create the two sets). (B) The resolution (using the FSC=0.143) found in (A) is plotted versus the number of segments, using a log10 scale. This can be fit nicely by a straight line (dotted red line) as observed by others (Stagg et al., 2014). The estimate for the resolution of 50,000 segments (approximately the number of segments in the full data set) is 0.223 (vertical black line) or 1/(4.5 Å).
Supplementary Figure 5, related to Figure 2. The FSC between the atomic model for the canonical state of F-actin and the actual reconstruction. The FSC is 0.5 at 4.66 Å.
Supplementary Figure 6, related to Figure 4. The FSC for the T2 reconstruction. The data were split into two halves with almost no overlap of boxes. Each subset was iterated independently for five cycles, and then aligned to generate the FSC curve.
Supplementary Figure 7, related to Experimental Procedures. Rfree value during the final refinement and model building cycles. Simulated annealing refinements were performed with CNS using DEN restraints. Structure factors between 4.4–4.5 Å were chosen for the calculation of Rfree.
Supplementary Table I, related to Figure 3
The residues predicted to be involved in the subunit-subunit interface are shown for the fiber diffraction model of Oda et al. (Oda et al., 2009), the cryo-EM model of Fujii et al. (Fujii et al., 2010) and this paper. Differences between our model and that of Fujii et al. are shown in red.
Supplementary Table II, related to Supplementary Experimental Procedures