Peripheral T-cell lymphoma not otherwise specified (PTCL-NOS) is a diagnosis of exclusion making up the largest fraction (25–30%) of PTCL. Although traditionally considered a ‘wastebasket' diagnosis, recent gene-expression results suggest the disease comprises two biologic sub-entities characterized by expression of the transcription factors GATA3 or TBX21 and their target genes.1 The mutational landscape of PTCL-NOS remains largely undefined.
We sought a better understanding the disease using a targeted deep-sequencing approach to identify pathogenic mechanisms and potential therapeutic targets that might fuel further studies. There is a substantial need for new therapies for PTCL-NOS, which leads to the death of more than two-thirds of patients within 5 years of diagnosis.2 The median age of onset for PTCL-NOS is 60, two-thirds of patients are male, and 69% have advanced-stage at diagnosis. Front line treatment remains CHOP (cyclophosphamide, doxorubicin, vincristine and prednisone) or other CHOP-based combinations optimized for use in B-cell lymphomas. Efforts to address the substantial unmet clinical need of PTCL-NOS patients are hampered by poor understanding of its biology, thwarting the development of specific therapies.
We collected 61 formalin-fixed paraffin embedded (FFPE) tumor samples from patients seen at Memorial Sloan-Kettering Cancer Center (MSKCC) with original diagnosis of PTCL-NOS, anaplastic large-cell lymphoma (ALCL) or angioimmunoblastic T-cell lymphoma (AITL). After re-review (JTF) of pathology and clinical factors, 31 cases met criteria for inclusion in this study of PTCL-NOS, lacking features indicative of other PTCL types. Pathologic details including morphology and immunophenotype are provided in Supplementary Table 1. In particular, we excluded cases with features of AITL because several studies have illuminated its mutational landscape,3, 4, 5, 6, 7 while our interest was in PTCL-NOS, for which few disease-specific recurrent mutational targets have been reported. We chose 237 genes for deep sequencing that have been reported as recurrent mutational targets in other hematologic cancers (Supplementary Table 2).
Analyzed tumor samples came from patients who consented to institutional tissue banking and analysis protocols, approved by the MSKCC Institutional Review Board and in compliance with the Declaration of Helsinki. Specific authorization for use and collection of de-identified clinical data came from the Human Biospecimen Use Committee. We isolated DNA from FFPE scrolls using the Formapure kit from Beckman Coulter Genomics in a semi-automated fashion on a Biomek NX liquid Handler. Illumina-compatible libraries were prepared from ~250 ng of sheared DNA (~150 bp in size) on a Biomek SPRI-Works HT robot using the Kapa Biosystems High Throughput library preparation kit with SPRI solution (magnetic beads) and amplified using the Kapa Standard PCR Library Amplification/Illumina series. During library preparation, adapters with barcodes were added to the DNA fragments for sample identification. All exons of the 237 genes were captured using the Nimblegen system (Roche SeqCap EZ Custom bait hybridization probes). The samples were then pooled and run on an Illumina HiSeq sequencer.
Reads were aligned to the hg19 build of the human genome using BWA 0.6.2-r126 followed by duplicate removal using Picard-Tools-1.55. The Genome Analysis Toolkit (GATK-2.6–3-gdee51c4) was used to perform local realignment around known indels and base quality score recalibration. Variant detection was performed using the GATK Unified Genotyper. Quality settings in the GATK HaplotypeCaller resulted in the elimination of candidate variants at very low allele frequency, which while improving the overall confidence of reported mutations likely also excluded some tumor-specific sub-clonal variants. Variants were annotated with the SNPeff annotation program to identify protein-coding changes and cross-referenced against the dbSNP132, 1000 Genomes and Catalog of Somatic Mutations in Cancer (COSMIC) databases. We eliminated variants listed in dbSNP132 or 1000 genomes and reviewed all remaining variants manually in IGV 2.3 browser, resulting in the elimination of additional mutation calls based on sequencing quality, allele frequency (if similar to known single-nucleotide polymorphisms (SNPs) in the same sample) and by searching the internet to identify additional SNPs. Mean sequencing depth was 232X (range 6–701). Cases with mean sequencing depth <100X (7 of 31) were included only if mutations were confirmed by targeting validation sequencing (see below), resulting in inclusion of four and exclusion of three such cases. This left 28 total cases for which we report mutations. Targeted validation sequencing of all mutations was performed with Illumina miSeq after re-amplification of DNA from the FFPE tumor samples, again using the Nimblegen capture system.
Of 28 patients, 25 with available demographic data were an average age of 52 years at diagnosis (range 9–76), with 11/25 age⩾60 and 13/25 male. Treatment and survival data were available for 23 patients followed long term at MSKCC. The majority of these (16) received CHOP or CHOP-like chemotherapy (Supplementary Figure 1A), whereas three received more intensive chemotherapy. Median event-free survival was 11.5 months, whereas median overall survival (OS) was 40.2 months (Supplementary Figure 1B). Subjects showed somewhat lower average age and less male predominance than is typical.2 There was no OS difference between cases with nodal or extranodal presentation (Supplementary Figure 1C). Twenty-four of 28 samples were pretreatment and 4 were relapsed.
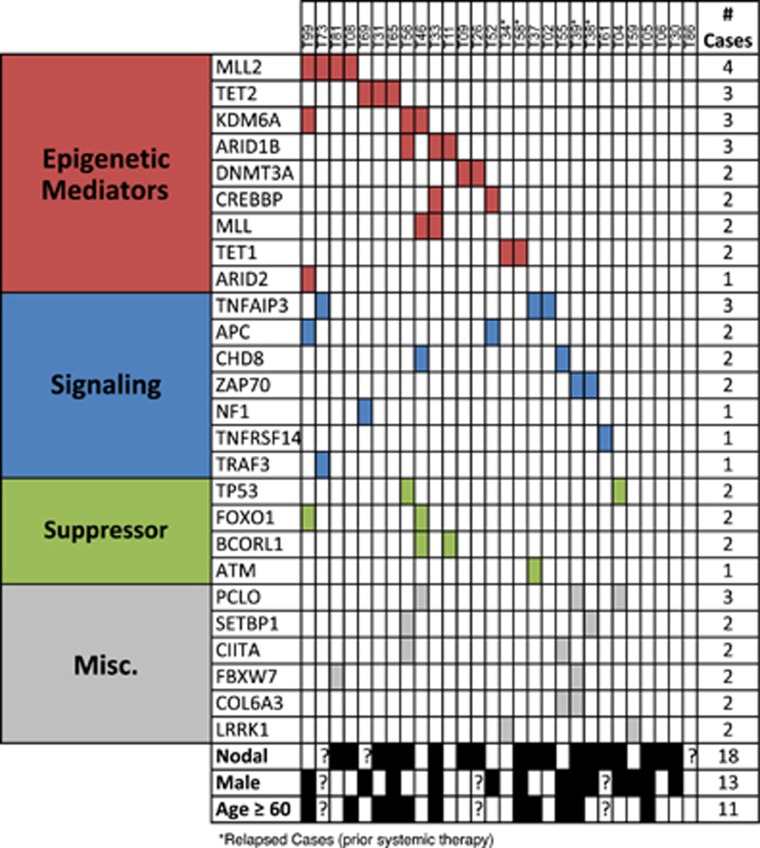
Table 1 shows 89 protein-coding mutations found in the 28 cases, affecting 59 genes, including 74 single-nucleotide variants and 15 indels. There was a mean of 3.0 mutations per case (range 0–11). There was no significant difference between the mutational load in the four relapsed samples and others (P=0.283), but we can't exclude the possibility some mutations detected in these four samples were not present at diagnosis. Lack of germ-line DNA to confirm the somatic nature of mutations introduces the possibility that some mutations in Table 1 are SNPs that are not reported in dbSNP132 or detected in the 1000 genomes project. We therefore limited further analysis to genes either recurrently mutated or containing mutations previously shown to be tumor specific in other studies. Figure 1 shows breakdown of genes affected by such mutations by functional category and whether cases had a nodal or extranodal presentation.
Table 1. Protein-affecting variants by gene and case.
| Gene | Case | CHR | POS | REF | ALT | Mutant Allele Fraction | Type | Effect | Previous Report |
|---|---|---|---|---|---|---|---|---|---|
| ALMS1 | T06 | chr2 | 73 676 742 | T | A | 0.39444 | Missense | p.S1029T | None |
| ALPK2 | T46 | chr18 | 56 203 629 | C | T | 0.35000 | Missense | p.G1264S | None |
| APC | 99–31720 | chr5 | 112 164 629 | G | A | 0.50131 | Missense | p.S568N | COSMIC |
| APC | T52 | chr5 | 112 176 308 | G | A | 0.42678 | Missense | p.E1673K | COSMIC |
| ARID1B | T11 | chr6 | 157 099 420 | G | GCAGCAA | 0.33333 | Codon insertion | p.119_120insQQ | None |
| ARID1B | T33 | chr6 | 157 431 662 | G | A | 0.42798 | Missense | p.A709T | None |
| ARID1B | T56 | chr6 | 157 528 066 | CTG | C | 0.44118 | Frameshift | p.C1932fs | None |
| ARID2 | 99–31720 | chr12 | 46 125 011 | GA | G | 0.28737 | Frameshift | p.N67fs | COSMIC |
| ATM | T37 | chr11 | 108 160 480 | T | G | 0.44118 | Missense | p.F1463C | COSMIC |
| BCL6 | T34 | chr3 | 187 447 027 | T | C | 0.41648 | Missense | p.N389S | None |
| BCL9 | T55 | chr1 | 147 095 762 | C | T | 0.41615 | Missense | p.P1095S | None |
| BCORL1 | T11 | chrX | 129 150 080 | C | T | 0.53977 | Missense | p.T1111M | COSMIC |
| BCORL1 | T46 | chrX | 129 147 806 | C | T | 0.47740 | Missense | p.P353L | None |
| BRCA2 | T39 | chr13 | 32 906 921 | A | G | 0.40000 | Missense | p.K436E | None |
| BRD4 | T37 | chr19 | 15 376 223 | G | A | 0.44444 | Missense | p.A264V | None |
| BRIP1 | T81 | chr17 | 59 885 858 | C | G | 0.42308 | Missense | p.E296D | None |
| CD58 | T39 | chr1 | 117 061 887 | T | C | 0.85185 | Missense | p.I237V | None |
| CDH23 | T34 | chr10 | 73 501 454 | G | A | 0.40785 | Missense | p.V1541M | None |
| CHD8 | T46 | chr14 | 21 894 360 | G | T | 0.46903 | Missense | p.T269N | None |
| CHD8 | T55 | chr14 | 21 859 651 | C | T | 0.48592 | Missense | p.E2067K | None |
| CIITA | T55 | chr16 | 11 004 047 | C | T | 0.44654 | Missense | p.T940M | None |
| CIITA | T56 | chr16 | 11 000 940 | G | A | 0.43501 | Missense | p.G531S | None |
| CMYA5 | T33 | chr5 | 79 034 658 | G | C | 0.36957 | Missense | p.S3357T | None |
| COL6A3 | T39 | chr2 | 238 296 329 | G | A | 0.42345 | Missense | p.P403L | COSMIC |
| COL6A3 | T55 | chr2 | 238 277 596 | G | A | 0.38728 | Missense | p.R1504W | COSMIC |
| CREBBP | T33 | chr16 | 3 824 628 | C | G | 0.40741 | Missense | p.R704P | None |
| CREBBP | T52 | chr16 | 3 778 708 | C | T | 0.42241 | Missense | p.G2076S | None |
| CUL9 | T34 | chr6 | 43 154 017 | C | G | 0.51064 | Missense | p.Q359E | Ref. 15 |
| DDX3X | T46 | chrX | 41 204 494 | A | T | 0.48918 | Nonsense | p.R363* | None |
| DNMT3A | T09 | chr2 | 25 463 248 | G | A | 0.30313 | Missense | p.R749C | COSMIC |
| DNMT3A | T26 | chr2 | 25 467 432 | CAT | C | 0.19303 | Frameshift | p.M548fs | None |
| FBXW7 | T39 | chr4 | 153 332 910 | C | CAGG | 0.42920 | Codon insertion | p.15_16insP | COSMIC |
| FBXW7 | T81 | chr4 | 153 268 155 | TG | T | 0.17647 | Frameshift | p.Q100fs | COSMIC |
| FOXO1 | 99–31720 | chr13 | 41 240 039 | C | G | 0.31250 | Missense | p.G104A | None |
| FOXO1 | T46 | chr13 | 41 240 273 | G | A | 0.25547 | Missense | p.P26L | None |
| FYB | T59 | chr5 | 39 202 971 | C | A | 0.37037 | Missense | p.G31V | None |
| IDH2 | T06 | chr15 | 90 645 600 | A | G | 0.41176 | Missense | p.V8A | None |
| IL7R | T39 | chr5 | 35 876 541 | C | T | 0.45918 | Nonsense | p.Q445* | None |
| IRF4 | T39 | chr6 | 394 888 | C | G | 0.37700 | Missense | p.T95R | None |
| IRF8 | T39 | chr16 | 85 936 739 | T | A | 0.38928 | Missense | p.W40R | None |
| JAK3 | T52 | chr19 | 17 937 710 | G | A | 0.44845 | Missense | p.L1073F | None |
| KDM4C | T46 | chr9 | 7 046 915 | T | A | 0.30758 | Missense | p.N771K | None |
| KDM6A | 99–31720 | chrX | 44 941 837 | G | GT | 0.54369 | Frameshift | p.R1054fs | None |
| KDM6A | T46 | chrX | 44 733 220 | C | T | 0.42655 | Missense | p.A71V | None |
| KDM6A | T56 | chrX | 44 913 193 | C | CT | 0.41379 | Frameshift | p.G291fs | None |
| KIAA1618 | T52 | chr17 | 78 264 463 | AGAG | A | 0.42010 | Codon deletion | p.G404del | None |
| LRRK1 | T34 | chr15 | 101 514 110 | C | T | 0.36364 | Missense | p.R67C | None |
| LRRK1 | T34 | chr15 | 101 549 251 | C | G | 0.34553 | Missense | p.D324E | None |
| LRRK1 | T59 | chr15 | 101 567 909 | G | A | 0.41379 | Missense | p.D865N | None |
| MLL | T33 | chr11 | 118 366 578 | C | T | 0.32051 | Missense | p.P1840S | None |
| MLL | T46 | chr11 | 118 373 835 | A | G | 0.43956 | Missense | p.M2407V | None |
| MLL2 | 99–31720 | chr12 | 49 434 709 | G | A | 0.51190 | Missense | p.R2282W | None |
| MLL2 | T08 | chr12 | 49 445 392 | G | T | 0.51471 | Missense | p.P692T | Ref. 8 |
| MLL2 | T73 | chr12 | 49 433 883 | G | A | 0.44056 | Missense | p.P2557L | None |
| MLL2 | T81 | chr12 | 49 448 530 | C | G | 0.32143 | Missense | p.G61R | None |
| MPDZ | T39 | chr9 | 13 192 237 | C | A | 0.67901 | Nonsense | p.E621* | None |
| NF1 | T69 | chr17 | 29 553 477 | A | AC | 0.30303 | Frameshift | p.P678fs | COSMIC |
| PASD1 | T34 | chrX | 150 844 560 | C | T | 0.39912 | Missense | p.A756V | None |
| PASK | T06 | chr2 | 242 080 117 | C | T | 0.41535 | Missense | p.C83Y | None |
| PCLO | T04 | chr7 | 82 763 889 | T | A | 0.31897 | Missense | p.S993C | None |
| PCLO | T39 | chr7 | 82 546 098 | C | T | 0.41736 | Missense | p.G3735E | None |
| PCLO | T39 | chr7 | 82 583 972 | G | T | 0.40136 | Missense | p.D2099E | None |
| PCLO | T46 | chr7 | 82 595 148 | T | G | 0.25290 | Missense | p.E1319A | None |
| PHLPP | T04 | chr18 | 60 645 819 | G | A | 0.47619 | Missense | p.G925S | None |
| PLCG2 | T55 | chr16 | 81 902 872 | G | A | 0.48000 | Missense | p.S178N | None |
| RELN | T37 | chr7 | 103 136 199 | T | C | 0.48413 | Missense | p.I3114V | None |
| SAMD9 | T55 | chr7 | 92 731 734 | C | A | 0.38095 | Missense | p.R1226I | None |
| SETBP1 | T38 | chr18 | 42 456 670 | C | CTCTT | 0.19608 | Frameshift | p.T228fs | None |
| SETBP1 | T56 | chr18 | 42 456 691 | A | C | 0.25641 | Missense | p.E234D | None |
| SMARCA2 | T73 | chr9 | 2 039 844 | A | T | 0.41176 | Missense | p.Q245L | None |
| STAT5B | T81 | chr17 | 40 375 521 | C | G | 0.38710 | Missense | p.Q143H | None |
| TET1 | T34 | chr10 | 70 333 197 | G | C | 0.42177 | Missense | p.A368P | None |
| TET1 | T58 | chr10 | 70 426 857 | C | T | 0.38255 | Missense | p.T1506I | None |
| TET2 | T31 | chr4 | 106 193 809 | CT | C | 0.36364 | Frameshift | p.S1424fs | COSMIC |
| TET2 | T65 | chr4 | 106 157 694 | GCAATATTT | G | 0.30000 | Frameshift | p.Q866fs | COSMIC |
| TET2 | T69 | chr4 | 106 164 733 | C | T | 0.28571 | Missense | p.R1201C | None |
| TNFAIP3 | T02 | chr6 | 138 195 991 | A | G | 0.39706 | Missense | p.N102S | COSMIC |
| TNFAIP3 | T37 | chr6 | 138 201 240 | A | C | 0.48916 | Missense | p.T647P | COSMIC |
| TNFAIP3 | T73 | chr6 | 138 201 240 | A | C | 0.37647 | Missense | p.T647P | COSMIC |
| TNFRSF14 | T61 | chr1 | 2 488 104 | A | G | 0.16413 | Missense; Start codon | p.M1V | COSMIC |
| TP53 | T04 | chr17 | 7 579 492 | TCTGGGAGCTTCATCTGGAC | T | 0.31169 | Frameshift | p.G59fs | COSMIC |
| TP53 | T56 | chr17 | 7 578 190 | T | C | 0.86620 | Missense | p.Y220C | COSMIC |
| TRAF3 | T73 | chr14 | 103 363 658 | C | T | 0.52830 | Nonsense | p.Q294* | COSMIC |
| ULK4 | T81 | chr3 | 41 860 984 | C | CT | 0.21053 | Frameshift | p.N594fs | None |
| ZAP70 | T38 | chr2 | 98 351 166 | G | C | 0.21287 | Missense | p.R358P | None |
| ZAP70 | T39 | chr2 | 98 354 531 | C | T | 0.36923 | Missense | p.P566L | None |
| ZFHX3 | T34 | chr16 | 72 831 834 | C | A | 0.37500 | Missense | p.G1583C | None |
| ZMYM3 | T38 | chrX | 70 469 934 | G | C | 0.33333 | Missense | p.P398R | None |
| ZNF608 | T46 | chr5 | 123 985 372 | A | G | 0.39334 | Missense | p.V394A | None |
Figure 1.
Distribution of mutations in 28 diagnostic peripheral T-cell lymphoma not otherwise specified (PTCL-NOS) cases. Included are all genes affected in multiple cases, or those affected in single cases with mutations listed in COSMIC or other reports as indicated in Table 1. Nodal: original presentation as nodal disease (black boxes) vs original extranodal presentation (white boxes).
As seen in other hematologic cancers, epigenetic regulation is the most mutated category overall. Regulators of histone methylation were mutated in 25% of cases, including MLL28 (4/28 cases), KDM6A (3/28) and MLL (2/28). Regulators of DNA methylation also were affected in 25% of cases. TET2 showed previously reported frameshifts in two cases and a missense mutation in a third, whereas DNMT3A had a frameshift in one case and a previously reported missense mutation in a second. The significance of two previously unreported TET1 missense mutations is less clear. There was no overlap between cases with histone methylation and DNA methylation alterations (Supplementary Figure 2A). Chromatin remodeling mediated by SWI/SNF complex activity is affected in 18%, specifically, ARID1B (3/28 cases), ARID2 (1/28) and SMARCA2 (1/28). These frequencies are similar to a recent meta-analysis of 44 cancer-sequencing studies.9 Overall, epigenetic regulators emerge as recurrent targets of somatic mutations in PTCL-NOS.
Activation of T-cell receptor (TCR) signaling is a known pathogenic mechanism in PTCL-NOS containing t(5;9)(q33;q22), found in <10 percent of cases.10 The resulting ITK-SYK fusion kinase localizes to lipid-rafts and mimics constitutive TCR activation.11 Our data highlight additional mechanisms activating TCR and downstream signaling. TNFAIP3, encoding the A20-negative regulator of NF-kB activation, had missense mutations in 11% (3/28) of cases, all of which are reported in the COSMIC. A20 is known to be a key regulator of NF-kB activation in T cells after TCR stimulation.12 WNT/β-Catenin negative regulators APC and CHD8 were affected in two cases each, or 14% (4/28) overall. Three additional genes with known suppressive roles in TCR activation had mutations previously reported in COSMIC: NF1 (frameshift), TNFRSF14 (missense affecting the start codon) and TRAF3 (nonsense). Overall, 46% (13/28) had at least one mutation in TCR or downstream mediators, expanding the role for these processes in PTCL-NOS pathogenesis.
The TP53 tumor suppressor gene had loss-of-function alterations in two cases, consistent with prior reports showing it is not mutated at a high rate in PTCL.13,14 Additional affected suppressors include the ATM DNA-repair kinase (one case) and the transcription factors FOXO1 and BCORL1 (two cases each).
Examination of survival effects (Supplementary Figure 2B) showed cases with alterations in histone methylation (MLL2, KDM6A, or MLL; P=0.0198) had worse OS than unaffected cases, whereas there was no such effect for either DNA methylation (TET2, DNMT3A, or TET1; P=0.2694) or signaling (TNFAIP3, APC, CHD8, ZAP70, NF1, TNFRSF14, or TRAF3; P=0.6695). We also examined differences in mutational patterns between cases with nodal or extranodal presentation (Supplementary Table 3). Although there was no significant difference in the above categories, interestingly all four cases affected by WNT/β-Catenin alterations were in the extranodal category (P=0.003).
Our study sheds new light on pathogenesis of a poorly understood clinical entity in need of better therapeutic options and for which poor sample availability has limited interrogation of the mutational landscape to date. Although some findings are confirmatory, others highlight novel disease mechanisms or better define frequency or prognostic implications. In particular, histone methylation alterations were present in a quarter of cases and associated with a worse OS. We believe studies in additional case series are warranted for elaboration of this result. Frequent mutations in regulators of TCR signaling meanwhile highlight mechanisms of activation, further extending the importance of this pathway beyond cases containing the previously identified ITK-SYK fusion kinase. The clustering of all mutations affecting WNT/β-Catenin mediators APC or CHD8 in cases with an extranodal presentation represented a significant difference that should be explored in additional cases and could shed new light on extranodal PTCL-NOS. Therapeutic opportunities from some results are limited. Loss of function of A20, for example, does not easily lend itself to targeted treatment, as NF-kB Inducing Kinase inhibitors have not made their way to clinical evaluation. Low frequency of TP53 mutations, however, highlights a potential for MDM2 inhibition in PTCL-NOS. In sum, we identify promising candidates for evaluation in additional cases and functional studies and to aid the development of better model systems for one of the least well understood hematologic malignancies.
Acknowledgments
This research was supported by Cycle for Survival (JS and all MSKCC authors), the Gabrielle's Angel Foundation (JS), the Lymphoma Research Foundation (JS), NCI R01-CA142798-01 (HGW), the Leukemia Research Foundation (HGW), the Experimental Therapeutics Center at MSKCC (HGW), the American Cancer Society 10284 (HGW), the Geoffrey Beene Cancer Center (HGW), an American Cancer Society Research Scholar Grant (DW) and Nonna's Garden Fund (SH).
Author Contributions
JS designed the research and wrote the manuscript. HGW, SH and DW designed the research and edited the manuscript. ML, AV, AH and KH designed the research. JT-F designed the research and reviewed the pathology of included cases. NL, ID, NS and MP performed bioinformatic analyses. JM collected and helped analyze clinical data.15
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biologic and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123:2915–2923. doi: 10.1182/blood-2013-11-536359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose J, Armitage J, Weisenburger D. International T-Cell Lymphoma Project. International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol. 2008;26:4124–4130. doi: 10.1200/JCO.2008.16.4558. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119:1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Couronné L, Khiabanian H, Kim M-Y, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46:166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120:1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T cell lymphoma. Blood. 2013;123:1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D, Pasqualucci L, Trifonov V, Fangazio M, Bruscaggin A, Rasi S, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streubel B, Vinatzer U, Willheim M, Raderer M, Chott A. Novel t(5;9)(q33;q22) fuses ITK to SYK in unspecified peripheral T-cell lymphoma. Leukemia. 2005;20:313–318. doi: 10.1038/sj.leu.2404045. [DOI] [PubMed] [Google Scholar]
- Pechloff K, Holch J, Ferch U, Schweneker M, Brunner K, Kremer M, et al. The fusion kinase ITK-SYK mimics a T cell receptor signal and drives oncogenesis in conditional mouse models of peripheral T cell lymphoma. J Exp Med. 2010;207:1031–1044. doi: 10.1084/jem.20092042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M. Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima AY, Cesarman E, Chadburn A, Knowles DM. Post-thymic T cell lymphomas frequently overexpress p53 protein but infrequently exhibit p53 gene mutations. Am J Pathol. 1994;144:573–584. [PMC free article] [PubMed] [Google Scholar]
- Petit B, Leroy K, Kanavaros P, Boulland ML, Druet-Cabanac M, Haioun C, et al. Expression of p53 protein in T- and natural killer-cell lymphomas is associated with some clinicopathologic entities but rarely related to p53 mutations. Hum Pathol. 2001;32:196–204. doi: 10.1053/hupa.2001.21569. [DOI] [PubMed] [Google Scholar]
- Chang H, Jackson DG, Kayne PS, Ross-Macdonald PB, Ryseck R-P, Siemers NO. Exome sequencing reveals comprehensive genomic alterations across eight cancer cell lines. PLoS One. 2011;6:e21097. doi: 10.1371/journal.pone.0021097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.