Abstract
Background
There is emerging evidence suggesting the role of peripheral blood leukocytes in the pathogenesis of obesity and related diseases. However, few studies have taken a genome wide approach to investigating gene expression profiles in peripheral leukocytes between obese and lean individuals with the consideration of obesity related shifts in leukocyte types.
Method
We conducted this study in 95 African Americans of both genders (age 14-20, 46 lean and 49 obese). Complete blood count with differential test (CBC) was performed in whole blood. Genome wide gene expression analysis was obtained using Illumina HumanHT-12 V4 Beadchip with RNA extracted from peripheral leukocytes. Out of the 95 participants, 64 had neutrophils stored. The validation study was based on Real-time polymerase chain reaction with RNA extracted from purified neutrophils.
Results
CBC test suggested that in males, obesity was associated with increased neutrophil percentage (p=0.03). Genome wide gene expression analysis showed that in males, the majority of the most differentially expressed genes were related to neutrophil activation. Validation of the gene expression levels of ELANE (neutrophil elastase) and MPO (myeloperoxidase) in purified neutrophils demonstrated that the expression of these two genes – important biomarkers of neutrophils activation – were significantly elevated in obese males (p=0.01 and p=0.02, respectively).
Conclusion
The identification of increased neutrophil percentage and activation in obese African American males suggests that neutrophils play an essential role in the pathogenesis of obesity related disease. Further functional and mechanistic studies on neutrophils may contribute to the development of novel intervention strategies reducing the burden associated with obesity-related health problems.
Keywords: Obesity, gene expression, African American, neutrophil, leukocyte
Introduction
Obesity is one of the major public health problems worldwide. It significantly increases the risk of various diseases, such as diabetes, cardiovascular diseases and certain types of cancer 1, 2. Low-grade systemic inflammation has been widely shown to accompany obesity and is thought to etiologically contribute to its related comorbidities3. The basis for this view mainly comes from studies on adipose tissues and targeted metabolic organs (e.g., muscle, liver, etc.) in which local productions of cytokines and immune cell infiltrations are discovered 4, 5. However, because of the not-easy availability of intra-abdominal fat or target metabolic organs in human studies, these findings have remained of limited use in clinical practice and development of prevention strategies. Since adipose-infiltrating immune cells in obesity are largely derived from bone marrows, a major gap of knowledge is whether changes in circulating leukocytes or leukocyte populations could provide etiological clues, pharmaceutical targets or as markers of the chronic, low-grade inflammatory response to obesity.
With the rapid development of genome wide technologies, it is of great interest to use genome-wide approaches to investigate obesity induced gene expression changes in circulating leukocytes. To our best knowledge, there have only been two studies exploring the gene expression differences between obese cases and lean controls using genome wide gene expression approaches in peripheral blood cells 6, 7. However, both studies were performed in whole blood rather than leukocytes, for which the influence of the high abundance of globin transcripts cannot be neglected. Moreover, both studies did not consider the potential influence of leukocyte composition changes related to obesity.
In this study, based on the complete blood count with differential counts and the genome-wide gene expression profiles on peripheral leukocytes from 46 obese cases and 49 lean controls, we aimed to identify obesity related gene expression changes after considering alterations in leukocyte composition. Gene ontology analysis was then performed to provide functional interpretations of the differentially expressed genes. Additionally, we conducted further replication in purified neutrophils to demonstrate that the genome wide findings in males were not driven by the increased percentage of neutrophils in obese cases. To our best knowledge, this is the first study investigating obesity induced gene expression profiling using peripheral leukocytes and purified neutrophils in African American youth and young adults with consideration of cell compositions.
Methods
Subjects
We selected 46 African American (AA) male participants (22 obese vs. 24 lean) and 49 AA female participants (24 obese vs. 25 lean) from the EpiGO (EpiGenetic basis of Obesity induced cardiovascular disease and type 2 diabetes) study. The EpiGO study was established in 2011 with the goal of identifying methylation changes involved in the pathogenesis of obesity and its related co-morbidities. Currently it is still ongoing and will in total enroll 400 obese and 400 lean youth aged 14-20 years with roughly equal number of AAs and European Americans (EA) as well as males and females. The detail information of the EpiGO study has been described previously 8.
This study was approved by the Institutional Review Board of Georgia Regents University, and performed under the guide of Declaration of Helsinki. Written informed consent was provided by all participants (if age≥18 years) or by their parents (if age<18 years).
For all participants, height and weight were measured by standard methods using a wall-mounted stadiometer and a scale, respectively. Body mass index (BMI) was calculated as weight/height2, and BMI percentile was calculated according to their age, sex, height and weight 9. The inclusion criteria of the EpiGO study are as followed: (1) age between 14 (with equal) to 21 (less than) years old; (2) both parents of the participants reported being of European or African ancestry; (3) obese participants with BMI ≥ 30 kg/m2 or BMI percentile ≥ 95% if age ≤ 20; (4) lean participants with BMI <25 kg/m2 or BMI percentile ≤ 50% if age ≤ 20; (5) free of any chronic or acute disease; (6) no daily medication controls for illnesses.
Measurements
Metabolic traits
Blood pressure (BP) was obtained at the 11th, 13th, 15th minute during a 15 minute supine relaxation period. The average of the three records was used to represent BP levels. Fasting serum triglyceride (TG), high-density lipoprotein cholesterol (HDLC), insulin and glucose levels were measured in the NIDDK (National Institute of Diabetes and Digestive and Kidney Diseases) supported Clinical Nutrition Research Unit Core Laboratory at the University of Alabama. TG, HDLC and glucoses were measured using colorimetric method on Sirrus analyzer (Stanbio Laboratory, Boerne, TX), while insulin was measured using an immunoenzymatic method on a TOSOH AIA-600 II analyzer (TOSOH Bioscience, S. San Fran, CA). The inter-assay CV from this lab is 3.63%, 1.61% and 2.34%, and the intra-assay CV is 2.53%, 1.99% and 2.93%, for TG, HDLC, and glucose, respectively. The assay sensitivity is 1.0 uU/ml for insulin and the intra-assay CV is 1.49%.
Complete blood count with differential test (CBC test)
Peripheral blood was collected and brought to the clinical pathology core lab within 2 hours for the complete blood count with differential count, which includes the total leukocyte count and percentages of peripheral blood cell types including neutrophils, lymphocytes, monocytes, eosinophils and basophils.
Leukocyte isolation and RNA extraction
Leukocytes were obtained after removal of plasma and lysis of all the red blood cells. The cell pellet was dissolved in RNA protect Cell Reagent (QIAGEN, Inc.) immediately and stored at −80°C. For the RNA extraction, the frozen cells were brought back to the room temperature and repelleted. After removal of the RNA protect Cell Reagent, RNA was extracted using QIAamp RNA mini Kit (QIAGEN, Inc.). RNA concentration and purity were evaluated on NanoDrop spectrophotometer (Thermo Scientific Inc.). RNA integrity (RIN) was evaluated on Bioanalyzer 2100 (Agilent Technologies, Inc.). The RIN score of all these 95 samples were greater than 8, indicating good RNA qualities.
Neutrophil isolation and RNA extraction
We used the approach developed by De et al 10. With a high purity of ≥97%, this method is simple for rapid isolation of functional neutrophils using commercially available cell preparation tube (CPT). This tube is produced by BD Biosciences Corp (San Jose, CA), which contains a gel lock to maintain the gradient between the Ficoll-hypaque and the blood. The red blood cells/neutrophil mixture was collected. After lysis of the red blood cells, the neutrophil cells were pelleted and washed with ice cold phosphate buffered saline (PBS). Similarly to the leukocytes, the neutrophil cell pellet was dissolved in RNA protect Cell Reagent (QIAGEN, Inc.) immediately and stored at −80°C. Neutrophil RNA was extracted using the same approach as the one used for leukocytes.
Genome wide gene expression profiling in leukocyte
The Illumina HumanHT-12 v4 Expression BeadChip (Illumina, Inc.), a direct hybridization assay, was used for genome wide gene expression analysis with 1 μg total RNA. This chip targets more than 48,000 probes that provide genome-wide coverage of well-characterized genes, gene candidates and splice variants. All of these 95 samples were tested in one batch at the Genomics Core Facility of the University of Chicago.
Real-Time polymerase chain reaction (RT-PCR) in neutrophils
Out of the 95 participants, 64 had frozen neutrophils stored (33 obese vs. 31 lean, 27 male vs. 37 female, details see online table S1). To validate the microarray findings, the expression levels of ELANE and MPO genes in purified neutrophils were quantified relative to endogenous control gene, beta actin (β-actin) using pre-designed BIO-RAD gene expression assays (BIO-RAD, Hercules, CA) following the instructions.
Statistical analysis
All the statistical analyses, with the exception of the genome wide gene expression analysis, were conducted using Stata SE Version 12 (StataCorp). Linear regression was performed to test whether the percentages of cell types in peripheral leukocyte were different between obese and lean subjects with age and sex included as covariates. The percentage of monocyte was log-transformed and the percentage of eosinophil was square root transformed to obtain normal distribution. For the percentage of basophil which cannot be transformed to normal distribution, the nonparametric Wilcoxon rank-sum test was further conducted. Similar results were obtained, so results for the regression analysis were reported here. We further tested whether this difference was dependent on sex by including the group (obese vs. lean) × sex interaction term. This is, leukocyte subtype percentages were used as dependent variable, group, sex and group × sex interaction as independent variables, and age as covariate. Linear regression was also used to test whether the expression levels of ELANE and MPO genes in purified neutrophils were different between obese cases and lean controls in each sex with age adjusted. Log transformation was applied to the expression levels of these two genes to obtain normal distribution.
For the genome wide gene expression analysis, the background subtracted signals were imported into R-environment. Probes with detection P-value less than 0.05 in more than 50% of the samples were defined as “present” and a total of 19,066 presented probes harbored 11,776 known genes from all the 95 subjects were selected for data analysis. Quartile normalization and log transform was performed before the analysis. The Limma package 11 was used to test the mean difference and to evaluate obesity-related differentially expressed genes in males and females separately with age adjustment. Raw P-values were assigned based on the empirical Bayes shrinkage from the designed linear model. To correct for multiple testing, the set of raw p values were converted to false discovery rates (FDR) according to Benjamini and Hochberg 12.
Gene ontology analysis was performed using DAVID (the Database for annotation visualization and Integrated Discovery v6.7) with GOTERM biological progress selected (http://david.abcc.ncifcrf.gov). The human genome was used as background, and the enrichment P-values were derived from a modified Fisher's exact test. The most significant probe was selected to present the gene if one transcript has more than one probe. The genes with p-value<0.01 and absolute log2 fold change>0.5 were selected for both genders and imported into the analysis. The top ten enriched pathways were exported from the output.
Results
The general characteristic of the 95 subjects are presented by group and sex in Table 1. The average age of all participants was around 17 years, and there were no significant age differences between males and females or between obese cases and lean controls. In both males and females, obesity was associated with higher systolic blood pressure (p<0.001) higher fasting insulin (p<0.001) levels and lower high-density lipoprotein cholesterol levels.
Table 1.
General characteristics of all participants in genome-wide gene expression analysis using mRNA from peripheral leukocytes (n=95)
| General character | Male participants |
Female participants |
||||
|---|---|---|---|---|---|---|
| Lean (n=24) | Obese (n=22) | P-value* | Lean (n=25) | Obese (n=24) | P-value* | |
| Age (years) | 17.42±1.91 | 17.58±1.96 | 0.78 | 17.91±1.72 | 17.35±1.49 | 0.23 |
| BMI (kg/m2) | 19.34±1.34 | 39.11±6.95 | <0.001 | 18.57±1.16 | 44.43±7.52 | <0.001 |
| BMI percentile (%) | 22.47±12.57 | 99.15±0.73 | <0.001 | 19.05±9.84 | 99.07±0.46 | <0.001 |
| SBP (mmHg) | 110.78±9.63 | 127.24±14.02 | <0.001 | 103.69±5.81 | 129.16±19.69 | <0.001 |
| DBP (mmHg) | 64.54±7.74 | 63.88±6.57 | 0.73 | 63.27±5.09 | 67.19±10.77 | 0.06 |
| TG (mg/dl) | 60.57±26.94 | 67.76±24.54 | 0.19 | 54.60±22.99 | 61.54±19.40 | 0.15 |
| HDLC (mg/dl) | 56.09±13.17 | 44.67±11.53 | <0.001 | 61.24±15.01 | 47.67±8.84 | <0.001 |
| Fasting insulin (uU/ml) | 8.23±3.40 | 19.44±9.94 | <0.001 | 12.71±7.25 | 30.52±14.95 | <0.001 |
| Fasting glucose (mg/dl) | 88.83±7.70 | 90.48±7.71 | 0.60 | 83.80±6.99 | 88.08±9.07 | 0.11 |
BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; TG, triglycerides; HDLC, high-density lipoprotein cholesterol.
P-value adjusted for age (if applicable)
In terms of the percentages of the leukocyte subtypes, we observed a significant sex × group (obese vs. lean) interaction for percentage of neutrophils (p=0.037), therefore, we conducted the stratified analyses in each gender. As shown in Table 2, in the male participants, obesity was associated with significantly increase in the percentage of neutrophils (52.95±9.74 vs. 46.45±11.15, p=0.03). In females, none of the percentages of these major leukocyte types showed significant differences between obese cases and lean controls. In terms of the cell counts, there was a significant increase in the total number of white blood cells in male obese subjects (6.03±1.62 vs. 4.97±1.57, p=0.05). Similar trend was observed in females but did not reach significance (6.20±1.47 vs.5.51±1.41, p=0.13). There were also significant increases of lymphocytes counts in female obese subjects (2.13±1.34 vs. 1.80±0.55, p=0.03) and significant increases of neutrophil counts in male obese subjects (3.33±1.62 vs. 2.39±1.14, p=0.04), which were due to the increases of the total number of white blood cells in obese cases.
Table 2.
Complete blood count with differential (mean ± standard deviation)
| Blood cell composition | Male participants |
Female participants |
||||
|---|---|---|---|---|---|---|
| Lean | Obese | P-value* | Lean | Obese | P-value* | |
| Neutrophils (%) | 46.45±11.15 | 52.95±9.74 | 0.03 | 55.92±10.77 | 54.18±10.30 | 0.63 |
| Lymphocytes (%) | 40.63±10.70 | 35.72±9.32 | 0.11 | 33.72±9.47 | 35.68±8.65 | 0.50 |
| Monocytes (%) | 9.29±2.79 | 7.95±1.53 | 0.06 | 7.80±3.20 | 7.64±2.11 | 0.81 |
| Eosinophils (%) | 3.29±1.78 | 2.55±1.79 | 0.18 | 1.96±1.90 | 2.27±2.43 | 0.55 |
| Basophils (%) | 0.63±0.49 | 0.81±0.66 | 0.29 | 0.60±0.58 | 0.55±0.60 | 0.85 |
| White blood cell (1000/mm3) | 4.97±1.57 | 6.03±1.62 | 0.05 | 5.51±1.41 | 6.20±1.47 | 0.13 |
| Neutrophils (1000/mm3) | 2.39±1.14 | 3.33±1.62 | 0.04 | 3.18±1.19 | 3.48±1.34 | 0.46 |
| Lymphocytes (1000/mm3) | 1.95±0.71 | 2.03±0.59 | 0.51 | 1.80±0.55 | 2.13±0.33 | 0.03 |
| Monocytes (1000/mm3) | 0.45±0.16 | 0.46±0.12 | 0.80 | 0.40±0.16 | 0.48±0.16 | 0.15 |
| Eosinophils (1000/mm3) | 0.19±0.15 | 0.17±0.13 | 0.81 | 0.11±0.10 | 0.13±0.11 | 0.34 |
| Basophils (1000/mm3) | 0.01±0.04 | 0.05±0.08 | 0.08 | 0.03±0.05 | 0.03±0.05 | 0.86 |
P-value adjusted for age
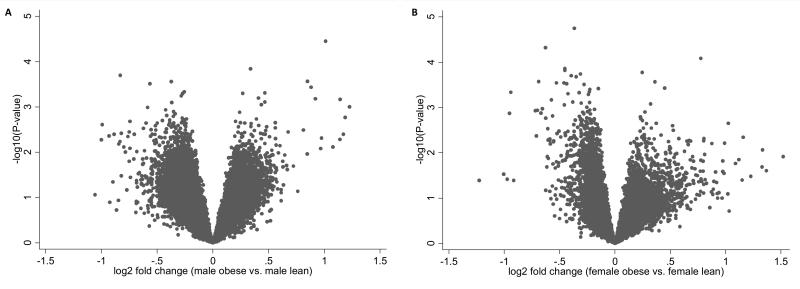
Because of the different effects of obesity on peripheral leukocyte cell compositions in males and females, we performed the following gene expression analysis in males and females separately. Figure 1 is the volcano plot showing the raw p-values versus mean gene expression difference for all genes between the case and the control group in males and females separately. The most significant gene was LCN2 in males showing a raw p value of 3.43×10−5 and a FDR of 0.42 and the most significant gene was FAAH in females showing a raw p value of 1.79×10−5 and a FDR of 0.34. Among the 11,776 unique genes, we identified in total 226 genes as obesity related differentially expressed genes in males and 222 genes in females with p-value <0.01. Out of the 226 genes, 73 (32.30%) genes were up-regulated and 153 (67.70%) were down-regulated in obese males. For females, there were 57 (25.68%) up-regulated genes and 165 (74.32%) down-regulated genes in obese.
Figure 1. Volcano plot of obesity related differentially expressed genes in males and females.
(A) Volcano plot of the log2 fold change of gene expression levels (X-axis) against the –log10 P-value (Y-axis) comparing obese and lean males.
(B) Volcano plot of the log2 fold change of gene expression levels (X-axis) against the –log10 P-value (Y-axis) comparing obese and lean females.
The top twenty differentially expressed genes ranked by the smallest p-value or the largest log2 fold change were presented in Table 3. Among the top lists in males, LCN2 (lipocalin-2), CEACAM6 (carcinoembryonic antigen-related cell adhesion molecule 6, CD66c), ELANE and AZU1 (azurocidin 1) were directly related with neutrophil activation. Interestingly, there were more genes associated with neutrophil activations in the top list ranked by the largest fold change. Not only LCN2, CEACAM6 and ELANE gene, another 8 genes (LYZ-lysozyme, OLFM4-olfactomedin 4, DEFA1B-defensin alpha 1b, DEFA3-defensin alpha 3, DEFA4-defensin alpha 4, DEFA1 defensin alpha 1, CTSG-cathepsin G and CEACAM8- carcinoembryonic antigen-related cell adhesion molecule 8) all have functions closely linked to neutrophil activation. Different from the findings in males, several of the genes in the top lists in females are involved in the activation of natural killer cells including KIR3DL1, KIR3DL3, KIR2DL1, KIR2DL3, KIR2DL4 and KIR2DS5 (killer cell immunoglobulin-like receptor, three domains long cytoplasmic tail 1; three domains long cytoplasmic tail 3; two domains long cytoplasmic tail 1; two domains long cytoplasmic tail 3; two domains long cytoplasmic tail 4; two domains short cytoplasmic tail 5) and KLRC1 (killer cell lectin-like receptor subfamily C, member 1).
Table 3.
Top 20 differentially expressed genes sorted by P-value or Log2 fold changes
| Sorted by P-value |
Sorted by Log2 fold changes |
||||
|---|---|---|---|---|---|
| Gene Symbol | Log2 fold change | P-value* | Gene Symbol | Log2 fold change | P-value* |
| Obese (n=22) vs. lean (n=24) males | |||||
| LCN2** | 1.01 | 3.43E-05 | DEFA1B | 1.19 | 1.66E-03 |
| PLEKHJ1 | 0.34 | 1.40E-04 | ALAS2 | 1.14 | 6.58E-04 |
| PID1 | −0.83 | 2.95E-04 | DEFA3 | 1.14 | 5.10E-03 |
| CEACAM6 | 0.85 | 2.65E-04 | LCN2 | 1.01 | 3.43E-05 |
| C6orf97 | −0.37 | 2.68E-04 | IGJ | −1.00 | 5.16E-03 |
| HSD17B12 | −0.56 | 2.96E-04 | LYZ | −0.99 | 2.39E-03 |
| ELANE | 0.88 | 3.59E-04 | DEFA4 | 0.98 | 4.70E-03 |
| SLC7A6OS | −0.26 | 4.51E-04 | DEFA1 | 0.97 | 8.13E-03 |
| AZU1 | 0.47 | 4.77E-04 | OLFM4 | 0.92 | 6.47E-04 |
| FGF18 | 0.27 | 4.88E-04 | ELANE | 0.88 | 3.59E-04 |
| MRPL45 | −0.27 | 4.94E-04 | CEACAM6 | 0.85 | 2.65E-04 |
| TESSP5 | −0.28 | 5.56E-04 | PID1 | −0.83 | 1.95E-04 |
| WASH1 | 0.41 | 6.21E-04 | TNFRSF17 | −0.82 | 3.71E-03 |
| OLFM4 | 0.92 | 6.47E-04 | CEACAM8 | 0.81 | 3.16E-03 |
| ALAS2 | 1.14 | 6.58E-04 | TXNDC5 | −0.80 | 7.41E-03 |
| C17orf67 | 0.47 | 7.51E-04 | RPS23 | −0.78 | 9.65E-03 |
| BRCC3 | −0.37 | 7.73E-04 | GLDC | −0.74 | 8.03E-03 |
| NCRNA00092 | 0.44 | 8.64E-04 | RPL12P6 | −0.68 | 9.43E-03 |
| CEP68 | −0.42 | 1.14E-03 | CTSG | 0.68 | 3.50E-03 |
| ANKRD37 | −0.34 | 1.37E-03 | KIAA0101 | −0.61 | 9.05E-03 |
| Obese (n=24) vs. lean (n=25) females | |||||
| FAAH | −0.37 | 1.79E-05 | SLPI | 1.34 | 8.66E-03 |
| KIR3DL1 | −0.63 | 4.76E-05 | IL1B | 1.16 | 4.58E-03 |
| NRG1 | 0.78 | 8.22E-05 | CASP5 | 1.02 | 2.23E-03 |
| FLJ42627 | −0.45 | 1.38E-04 | TREML3 | 0.99 | 6.11E-03 |
| ITIH4 | −0.45 | 1.50E-04 | KIR2DL3 | −0.95 | 1.34E-03 |
| SYTL3 | −0.40 | 1.99E-04 | KIR2DL4 | −0.94 | 4.62E-04 |
| LIMD2 | −0.35 | 2.09E-04 | NAIP | 0.88 | 6.24E-03 |
| KLHL5 | 0.36 | 2.71E-04 | CCR1 | 0.80 | 4.83E-03 |
| ABCA2 | −0.46 | 2.88E-04 | DSC2 | 0.80 | 5.38E-03 |
| NCAM1 | −0.44 | 2.97E-04 | NRG1 | 0.78 | 8.22E-05 |
| GTF3C1 | −0.28 | 3.09E-04 | KLRC1 | −0.71 | 4.23E-03 |
| DACT1 | 0.45 | 3.73E-04 | THBD | 0.68 | 5.56E-03 |
| SNX1 | −0.23 | 4.01E-04 | MPL | 0.66 | 6.58E-03 |
| KIAA2026 | −0.23 | 4.51E-04 | C17orf97 | 0.66 | 6.51E-03 |
| KIR2DL4 | −0.94 | 4.61E-04 | PROS1 | 0.65 | 5.52E-03 |
| STAG3L4 | −0.32 | 4.66E-04 | EMR2 | 0.63 | 9.26E-03 |
| ZCCHC14 | −0.24 | 5.15E-04 | KIR3DL1 | −0.63 | 4.76E-05 |
| ANKRD20A1 | −0.57 | 6.77E-04 | KIR2DS5 | −0.63 | 1.70E-03 |
| GSTM2 | −0.38 | 7.56E-04 | KIR2DL1 | −0.61 | 2.94E-03 |
| CLK2 | −0.23 | 7.69E-04 | KIR3DL3 | −0.60 | 5.19E-03 |
P-value adjusted age
Bold genes were closely related to neutrophil activation
Gene ontology analysis using DAVID showed the similar results with innate immune system including defense response to fungus and defense response to bacterium among the top GO categories in males (Table 4). Similarly, defense response is also one of the top GO categories in females (Table 4).
Table 4.
Pathway analysis of obesity related differentially expressed genes*
| Terms | Genes | P-value |
|---|---|---|
| Obese (n=22) vs. lean (n=24) males | ||
| Defense response to bacterium | CTSG, DEFA1B, DEFA3, DEFA4, LYZ | 1.0E-04 |
| Defense response to fungus | DEFA1B, DEFA3, DEFA4 | 3.6E-04 |
| Response to other organism | CTSG, DEFA1B, DEFA3, DEFA4, LCN2, LYZ | 3.9E-04 |
| Killing of cells of another organism | DEFA1B, DEFA3, DEFA4 | 4.2E-04 |
| Response to bacterium | CTSG, DEFA1B, DEFA3, DEFA4, LYZ | 8.0E-04 |
| Immune system process | ALAS2, CEACAM8, CTSG, DEFA1B, ELANE, IGJ, IL4R, TNFRSF17 | 1.1E-03 |
| Response to fungus | DEFA1B, DEFA3, DEFA4 | 1.3E-03 |
| Cell killing | DEFA1B, DEFA3, DEFA4 | 1.3E-03 |
| Response to biotic stimulus | CTSG, DEFA1B, DEFA3, DEFA4, LCN2, LYZ | 1.4E-03 |
| Translational elongation | RPL12P6, RPL21, RPS21, RPS23 | 1.4E-03 |
| Obese (n=24) vs. lean (n=25) females | ||
| Immune response | BMP6, CCR1, IL1B, KIR3DL1, KIR2DL1, KIR2DL3, KIR2DS5, SPON2 | 4.1E-05 |
| Immune system process | BMP6, CCR1, IL1B, KIR3DL1, KIR2DL1, KIR2DL3, KIR2DS5, SPON2 | 4.2E-04 |
| Response to wounding | BMP6, CCR1, IL1B, NRG1, PROS1, THBD | 9.0E-04 |
| Response to stimulus | BMP6, CCR1, IL1B, KIR3DL2, KIR3DL1, KIR2DL1, KIR2DL3, KIR2DL4, KIR2DS5, NRG1, PROS1, SPON2, THBD | 1.6E-03 |
| Defense response | BMP6, CCR1, IL1B, KIR3DL2, KIR2DL4, KIR2DS5 | 1.8E-03 |
| Response to stress | BMP6, CCR1, IL1B, KIR3DL2, KIR2DL4, KIR2DS5, NRG1, PROS1, THBD | 2.0E-03 |
| Wound healing | IL1B, NRG1, PROS1, THBD | 2.7E-03 |
| Cellular defense response | KIR3DL2, KIR2DL4, KIR2DS5 | 3.7E-03 |
| Response to external stimulus | BMP6, CCR1, IL1B, NRG1, PROS1, THBD | 9.6E-03 |
| Anti-apoptosis | NAIP, IL1B, NRG1 | 3.7E-02 |
Genes were selected with p-value<0.01 and absolute log2 fold change > 0.5, and the human genome was chosen as background in the analysis
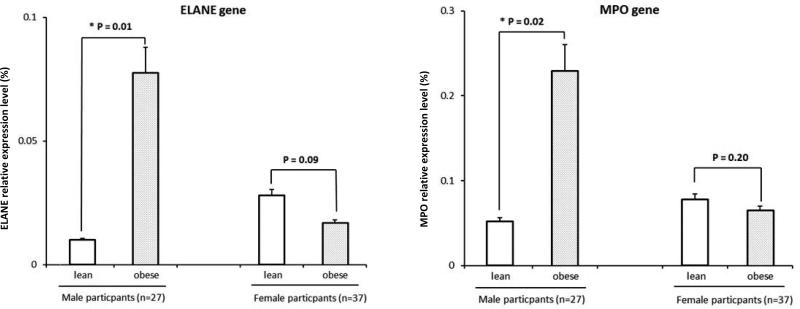
Due to the fact that in this study obesity was only associated with increased percentage of neutrophils in males, we conducted further replication in purified neutrophils to exclude the possibility that the findings in males were driven by the increased percentage of neutrophils in obese cases. We performed RT-PCR for two genes closely related with neutrophil activation and extensively studied in the literature (ELANE, one of the genes in our top list, and MPO, also showing significantly increased expression levels in our study [LogFC=0.44, P=0.005]) in 64 out of the 95 subjects for whom we have purified neutrophil stored (obese group: 15M/18F; lean group: 12M/19F; online table S1). Results were shown in Figure 2. Similar to the results in the microarray analysis using the leukocytes, obesity was associated with significantly higher expression levels of both genes in neutrophils in males (ELANE, p=0.01; MPO, p=0.02) but not in females.
Figure 2. Real time PCR replication of ELANE and MPO in neutrophils.
(A) Histogram showing the gene expression levels of ELANE relative to β-actin (Y-axis, %) comparing obese and lean males (p=0.01 with age adjusted) and females (p=0.09 with age adjusted), respectively
(B) Histogram showing the gene expression levels of MPO relative to β-actin (Y-axis, %) comparing obese and lean males (p=0.02 with age adjusted) and females (p=0.20 with age adjusted), respectively
Discussion
In this study, we not only demonstrated the gene expression differences in peripheral leukocytes from African American obese and lean subjects, in both males and females, but also observed that in African American males obesity was associated with increased percentage of neutrophils and increased expression of neutrophil activation related genes.
Obesity has been found to be associated with high leukocyte count, severing as one piece of evidence of the involvement of inflammation13, 14. The major interests on circulating leukocyte subtypes are monocytes and T cells since the discovery of enhanced adipose tissue macrophage and T cells infiltration in obesity15, 16. While several studies suggest a mild increase in the total circulating numbers of these subtypes, this has not been a universal finding in other studies with even reports of negative association between the percentage of these cell types and BMI17, 18. In contrast with the inconsistent finding in these leukocyte subtypes, obesity is consistently associated with persistent neutrophilia, manifesting not only by elevated neutrophil counts but also increased percentages14, 17, 19, 20. All of these studies were conducted in Caucasians. In this study, we observed the same pattern in African American males, that is, obese cases have significantly higher peripheral neutrophil percentages and total cell numbers.
The importance of neutrophils in the pathogenesis of obesity related disease was further emphasized recently. In 2012 and 2013 a series of studies were published demonstrating the importance of neutrophil infiltration in obesity induced immune dysfunction21-23. For example, Talukdar et al 22 found that the neutrophil was the first immune cell responding to inflammation and its infiltration into the adipose tissue can occur as early as after 3 days of high fat feeding and can be maintained for up to 90 days on a high fat diet. Treatment with neutrophil elastase (ELANE, a key enzyme secreted by activated neutrophil) causes increased cellular insulin resistance and the deletion of ELANE in high fat diet induced obese mice leads to less tissue inflammation associated with lower adipose tissue neutrophil and macrophage infiltrations. Using an unbiased proteomic approach in a mouse model, Mansuy-Aubert et al 21 observed that obesity was associated with an imbalance between ELANE and its inhibitor α1-Antitrypin and this imbalance was linked with insulin sensitivity and inflammation in target organs. Previous studies in humans also observed that serum ELANE activity was positively and significantly correlated with BMI24-27. In consistent with these two animal studies and previous human studies on ELANE, in the current study we also observed that the expression of ELANE was significantly increased in the peripheral leukocytes of obese AA males. To exclude the possibility that this finding was driven by the different cell compositions observed in obese and lean AA males, we further validated the findings of two genes, ELANE and MPO in purified neutrophils. The MPO gene encodes a peroxidase enzyme which is a major component of the azurophilic granules of neutrophils. MPO is one of the keys in the initiation and progression of both acute and chronic inflammatory diseases, including cardiovascular disease, and has been identified as a marker of neutrophil infiltration into tissues. A recent study 28 in 446 Caucasian prepubertal children (age 6-12 years, 223 normal weight vs. 223 obese) found that plasma MPO level was not only elevated in obese cases but also associated with pro-inflammatory and cardiovascular risk biomarkers. In the current study, the expression of MPO was also significantly increased in the peripheral leukocytes of obese males. The results from our validation study in purified neutrophils are consistent with our genome wide microarray findings and with these previous studies. In males, the obese participants showed increased expression levels of both the MPO and ELANE genes in purified neutrophils.
The other top signals related to neutrophil activation in males included LCN2, CEACAM6, CEACAM8, LYZ, OLFM4, AZU1 and alpha defensin families. LCN2, lipocalin-2, also known as neutrophil gelatinase-associated lipocalin, is involved in innate immunity and mainly expressed by neutrophils [3]. CEACAM6 and CEACAM8, also known as CD66c and CD66b (Cluster of Differentiation 66c and 66b), come from the same family which encode the subunits of carcinoembryonic antigen-related cell adhesion molecule 6 and are cell-adhesion proteins on neutrophils [4]. LYZ, also known as muramidase or N-acetylmuramide glycanhydrolase, is mainly present in cytoplasmic granules of neutrophils with the major function of damaging bacterial cell walls. A recent study suggested that OLFM4 was associated with neutrophil-specific granules and defines a subset of human neutrophils [5]. AZU1 gene, azurocidin 1, is mainly expressed in azurophil granules which is one of the specialized lysosomes of the neutrophils [6]. The alpha defensin families, including DEFA1B, DEFA3, DEFA4, DEFA1, are particularly abundant in neutrophils with the activations of against bacteria, fungi and viruses.
Our results were also consistent with a recently study of bariatric surgery 29. Berisha et al performed a genome-wide gene expression analysis using whole blood samples from 11 obese subjects prior to and 6-12 months after bariatric surgery. The LCN2 mRNA level decreased with -46% changes following bariatric surgery. Besides, several other genes, including DEFA1, DEFA3, CEACAM8, which were increased in obese subjects in our study, were found to be significantly decreased after bariatric surgery. These consistent findings suggested that these differentially expressed genes are closely related to obesity status and their expression levels can be reversed following weight change.
Different from the findings in males, we did not observe that obesity was associated with increased percentage of neutrophil, nor did the gene expression analysis show increased expression of neutrophil activation related genes in females. On the contrary, several genes in the top lists of the gene expression analysis in females were involved in the activation of nature killer cells. Interestingly, in Ilavska et al's study in which the potential sex difference was explored, a significantly negative correlation between the percentage of NK cells and BMI was observed only in females but not in males17. The other two studies which observed a decreased percentage of NK cells in obese cases vs. lean controls also have samples dominantly by females (65%-70%)30, 31. In consistent with the lower percentage of NK cells in obese females, the NK cell activation related genes are down-regulated in our gene expression analysis in females. It will be of great interest to know whether this observation is due to changes in NK cell percentages or obesity is indeed associated with impaired NK functions including decreased percentage and decreased activation in females. Unfortunately, neither the percentage of NK cells was measured, nor was purified NK cell kept in the current study. Nevertheless, both of neutrophils and nature killer cells are essential parts of innate immune system, which is the first line of defense response in a non-specific manner. This is consistent with the results of gene ontology analysis in the current study. This is, defense response is the top GO term in both males and females.
The strength of current study includes: 1) the focus on youth, which reduces the probability that gene expression changes are hidden or biased by the coexistence of obesity related co-morbidities and the use of various medications, which are commonly required by obese adults. Youth are a population frequently showing obesity related metabolic risks but not yet manifesting overt clinical disease; 2) the availability of complete blood counts with differentials, which enabled us to consider the potential influence of major leukocyte composition changes; 3) the availability of purified neutrophils, which enabled us to exclude the possibility that the findings in males were driven by the increased percentage of neutrophils in obese cases.
Nevertheless, several limitations of this study need to be recognized. First, genome-wide gene expression analysis on purified neutrophils is needed to provide further confirmation of the relationship between obesity and neutrophil activation related genes. Second, this study was based on African American populations. Further replication in Caucasians as well as other ethnicities will be needed.
In conclusion, we found that in young AA males, obese status was not only associated with increased percentage of peripheral neutrophil, but also with increased neutrophil gene activation. Further functional and mechanistic studies on neutrophils could contribute to the development of novel intervention strategies reducing the burden associated with obesity-related health problems.
Supplementary Material
Acknowledgement
The current study is funded by NIH HL105689. We would like to extend a special thanks to all the participants for involving in our study. We are especially grateful to the staffs in our departments for their help in recruitment, data collection and lab experiments.
Footnotes
Conflicts
There is no conflict of interest.
Supplementary information is available at International Journal of Obesity's website.
References
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365(9455):217–23. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Poirier P, Giles TD, Bray GA, Hong Y, Stern JS, Pi-Sunyer FX, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on Obesity and Heart Disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation 2006. 113(6):898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 3.O'Rourke RW. Inflammation in obesity-related diseases. Surgery. 2009;145(3):255–9. doi: 10.1016/j.surg.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bullo M, Garcia-Lorda P, Megias I, Salas-Salvado J. Systemic inflammation, adipose tissue tumor necrosis factor, and leptin expression. Obesity research. 2003;11(4):525–31. doi: 10.1038/oby.2003.74. [DOI] [PubMed] [Google Scholar]
- 5.Festa A, D'Agostino R, Jr., Williams K, Karter AJ, Mayer-Davis EJ, Tracy RP, et al. The relation of body fat mass and distribution to markers of chronic inflammation. International journal of obesity and related metabolic disorders : journal of the International Association for the Study of Obesity. 2001;25(10):1407–15. doi: 10.1038/sj.ijo.0801792. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh S, Dent R, Harper ME, Gorman SA, Stuart JS, McPherson R. Gene expression profiling in whole blood identifies distinct biological pathways associated with obesity. BMC medical genomics. 2010;3:56. doi: 10.1186/1755-8794-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hindle AK, Koury J, McCaffrey T, Fu SW, Brody F. Dysregulation of gene expression within the peroxisome proliferator activated receptor pathway in morbidly obese patients. Surgical endoscopy. 2009;23(6):1292–7. doi: 10.1007/s00464-008-0152-1. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Su S, Barnes VA, De Miguel C, Pollock J, Ownby D, et al. A genome-wide methylation study on obesity: Differential variability and differential methylation. Epigenetics : official journal of the DNA Methylation Society. 2013;8(5) doi: 10.4161/epi.24506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Body Mass Index: BMI for Children and Teens. Center for Disease Control; 2013. [Google Scholar]
- 10.De AK, Roach SE, De M, Minielly RC, Laudanski K, Miller-Graziano CL, et al. Development of a simple method for rapid isolation of polymorphonuclear leukocytes from human blood. Journal of immunoassay & immunochemistry. 2005;26(1):35–42. doi: 10.1081/ias-200041157. [DOI] [PubMed] [Google Scholar]
- 11.Smthy GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 2005 [Google Scholar]
- 12.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society, Series B. 1995;57(1):289–300. [Google Scholar]
- 13.Fisch IR, Freedman SH. Smoking, oral contraceptives, and obesity. Effects on white blood cell count. JAMA : the journal of the American Medical Association. 1975;234(5):500–6. [PubMed] [Google Scholar]
- 14.Dixon JB, O'Brien PE. Obesity and the white blood cell count: changes with sustained weight loss. Obesity surgery. 2006;16(3):251–7. doi: 10.1381/096089206776116453. [DOI] [PubMed] [Google Scholar]
- 15.Kintscher U, Hartge M, Hess K, Foryst-Ludwig A, Clemenz M, Wabitsch M, et al. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arteriosclerosis, thrombosis, and vascular biology. 2008;28(7):1304–10. doi: 10.1161/ATVBAHA.108.165100. [DOI] [PubMed] [Google Scholar]
- 16.Subramanian V, Ferrante AW., Jr. Obesity, inflammation, and macrophages. Nestle Nutrition workshop series. Paediatric programme. 2009;63:151–9. doi: 10.1159/000209979. discussion 159-62, 259-68. [DOI] [PubMed] [Google Scholar]
- 17.Ilavska S, Horvathova M, Szabova M, Nemessanyi T, Jahnova E, Tulinska J, et al. Association between the human immune response and body mass index. Human immunology. 2012;73(5):480–5. doi: 10.1016/j.humimm.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 18.Viardot A, Heilbronn LK, Samocha-Bonet D, Mackay F, Campbell LV, Samaras K. Obesity is associated with activated and insulin resistant immune cells. Diabetes/metabolism research and reviews. 2012;28(5):447–54. doi: 10.1002/dmrr.2302. [DOI] [PubMed] [Google Scholar]
- 19.Al-Sufyani AA, Mahassni SH. Obesity and immune cells in Saudi females. Innate immunity. 2011;17(5):439–50. doi: 10.1177/1753425910372536. [DOI] [PubMed] [Google Scholar]
- 20.Herishanu Y, Rogowski O, Polliack A, Marilus R. Leukocytosis in obese individuals: possible link in patients with unexplained persistent neutrophilia. European journal of haematology. 2006;76(6):516–20. doi: 10.1111/j.1600-0609.2006.00658.x. [DOI] [PubMed] [Google Scholar]
- 21.Mansuy-Aubert V, Zhou QL, Xie X, Gong Z, Huang JY, Khan AR, et al. Imbalance between neutrophil elastase and its inhibitor alpha1-antitrypsin in obesity alters insulin sensitivity, inflammation, and energy expenditure. Cell metabolism. 2013;17(4):534–48. doi: 10.1016/j.cmet.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talukdar S, Oh da Y, Bandyopadhyay G, Li D, Xu J, McNelis J, et al. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nature medicine. 2012;18(9):1407–12. doi: 10.1038/nm.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadad N, Burgazliev O, Elgazar-Carmon V, Solomonov Y, Wueest S, Item F, et al. Induction of cytosolic phospholipase a2alpha is required for adipose neutrophil infiltration and hepatic insulin resistance early in the course of high-fat feeding. Diabetes. 2013;62(9):3053–63. doi: 10.2337/db12-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paczek L, Michalska W, Bartlomiejczyk I. Trypsin, elastase, plasmin and MMP-9 activity in the serum during the human ageing process. Age and ageing. 2008;37(3):318–23. doi: 10.1093/ageing/afn039. [DOI] [PubMed] [Google Scholar]
- 25.Bonithon-Kopp C, Touboul PJ, Berr C, Magne C, Ducimetiere P. Factors of carotid arterial enlargement in a population aged 59 to 71 years: the EVA study. Stroke; a journal of cerebral circulation. 1996;27(4):654–60. doi: 10.1161/01.str.27.4.654. [DOI] [PubMed] [Google Scholar]
- 26.Bizbiz L, Bonithon-Kopp C, Ducimetiere P, Berr C, Alperovitch A, Robert L. Relation of serum elastase activity to ultrasonographically assessed carotid artery wall lesions and cardiovascular risk factors. The EVA study. Atherosclerosis. 1996;120(1-2):47–55. doi: 10.1016/0021-9150(95)05676-9. [DOI] [PubMed] [Google Scholar]
- 27.Piwowar A, Knapik-Kordecka M, Warwas M. Concentration of leukocyte elastase in plasma and polymorphonuclear neutrophil extracts in type 2 diabetes. Clinical chemistry and laboratory medicine : CCLM / FESCC. 2000;38(12):1257–61. doi: 10.1515/CCLM.2000.198. [DOI] [PubMed] [Google Scholar]
- 28.Olza J, Aguilera CM, Gil-Campos M, Leis R, Bueno G, Martinez-Jimenez MD, et al. Myeloperoxidase is an early biomarker of inflammation and cardiovascular risk in prepubertal obese children. Diabetes care. 2012;35(11):2373–6. doi: 10.2337/dc12-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berisha SZ, Serre D, Schauer P, Kashyap SR, Smith JD. Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PloS one. 2011;6(3):e16729. doi: 10.1371/journal.pone.0016729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Shea D, Cawood TJ, O'Farrelly C, Lynch L. Natural killer cells in obesity: impaired function and increased susceptibility to the effects of cigarette smoke. PloS one. 2010;5(1):e8660. doi: 10.1371/journal.pone.0008660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch LA, O'Connell JM, Kwasnik AK, Cawood TJ, O'Farrelly C, O'Shea DB. Are natural killer cells protecting the metabolically healthy obese patient? Obesity (Silver Spring) 2009;17(3):601–5. doi: 10.1038/oby.2008.565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.