Abstract
In the developing mammalian nervous system, common progenitors integrate both cell extrinsic and intrinsic regulatory programs to produce distinct neuronal and glial cell types as development proceeds. This spatiotemporal restriction of neural progenitor differentiation is enforced, in part, by the dynamic reorganization of chromatin into repressive domains by Polycomb Repressive Complexes, effectively limiting the expression of fate-determining genes. Here, we review distinct roles that the Polycomb Repressive Complexes play during neurogenesis and gliogenesis, while also highlighting recent work describing the molecular mechanisms that govern their dynamic activity in neural development. Further investigation of how Polycomb complexes are regulated in neural development will enable more precise manipulation of neural progenitor differentiation, facilitating the efficient generation of specific neuronal and glial cell types for many biological applications.
Introduction
During development of the nervous system, developmental potential is progressively restricted as pluripotent cells of the early embryo give rise to multi-potent progenitor cells, and as these progenitors differentiate into neurons and glia. By definition, this is an epigenetic phenomenon, whereby cells with the same genome acquire and maintain distinct gene expression patterns that differentiate them in form and function. Mechanisms that reorganize chromatin structure play an essential role in this process. The basic unit of chromatin is the nucleosome, DNA wrapped around core histones, which can be assembled along with non-histone proteins into the complex topology of higher order chromatin structures characteristic of eukaryotic genomes. In its simplest form, the topological arrangement of chromatin partitions the genome into sterically open (euchromatic) and compact (heterochromatic) compartments, respectively, promoting or inhibiting transcriptional initiation and elongation to pattern gene expression in the cell (Armstrong 2012, Olynik and Rastegar 2012, Wutz 2013).
Multipotent stem and progenitor cells have a distinct chromatin structure that facilitates their maintenance of developmental plasticity. In the pluripotent “ground” state of embryonic stem cells (ESCs, derived from the inner cell mass of the early embryo), chromatin is decondensed and histone proteins are loosely bound, exhibiting hyperdynamic exchange rates (Meshorer, Yellajoshula et al. 2006, Meshorer 2007). During differentiation, histone exchange becomes less dynamic and the chromatin becomes more condensed as heterochromatin foci form and spread (Meshorer, Yellajoshula et al. 2006, Meshorer 2007). The exact placement and organization of heterochromatin constrains the competence of a cell by limiting the gene programs available for transcription (Francastel, Schubeler et al. 2000, Arney and Fisher 2004, Bernstein, Meissner et al. 2007, Campos and Reinberg 2009, Zhou, Goren et al. 2011). Understanding how heterochromatin is successively patterned in different progenitors is therefore essential to understanding how cell fate is controlled during development, and how it may be modified ex vivo for experimental and therapeutic purposes.
A number of different regulatory mechanisms have been described that contribute to the formation and dynamic rearrangement of heterochromatin during neural development. These include enzymatic machineries that methylate DNA or covalently modify the amino-terminal tails of histone proteins after translation, alternatively acetylating, ubiquitylating, phosphorylating, or methylating specific residues (Campos and Reinberg 2009, Zhou, Goren et al. 2011). Many of these modifications are well correlated with specific biological functions, including transcriptional activation, repression, and enhancer activity. While the exact consequences of the various post-translation modifications (PTMs) of histone tails is an area of active research, in general these influence transcription by altering nucleosome compaction or mobility, and by modulating the recruitment of non-histone effector proteins (Taverna, Li et al. 2007, Yun, Wu et al. 2011, Zhou, Goren et al. 2011).
Efforts to unravel how chromatin state is regulated during development have been complicated by the fact that many chromatin-modifying proteins are expressed in multiple cell and tissue types. Even within a single cell lineage, these chromatin modifiers can act with temporal specificity, targeting distinct suites of genes during each developmental transition. Therefore, a major current challenge lies in understanding how such spatially and temporally controlled targeting of chromatin modifiers is achieved during development. Here, we will address some of the key histone modification state changes that accompany mammalian neurogenesis and gliogenesis, focusing in particular on temporally distinct roles that the Polycomb Repressor Complexes play in these processes, and on recent advances in research aimed at unraveling the long-standing enigma of how these complexes recognize different genomic targets in different neural cell lineages.
Regulation of Developmental Gene Expression by Polycomb and Trithorax Proteins
Among the most well characterized chromatin modifiers are the Trithorax group (TrxG) and Polycomb group (PcG) proteins. TrxG and PcG proteins were originally discovered in Drosophila melanogaster as multimeric protein complexes that work in opposition to respectively activate or repress Hox gene expression (Schuettengruber, Chourrout et al. 2007). TrxG protein complexes are likewise critical activators of gene expression in mammals, where they catalyze trimethylation of histone 3 lysine 4 (H3K4me3) at promoters to stimulate active transcription. The enzymatic ortholog of Drosophila TrxG in mammals is variable, and can include mixed lineage leukemia (MLL) proteins 1–4, Set1A, and Set1B, all of which bind additional activating proteins to form the multi-subunit MLL1–4 and Set1A/B complexes (Schuettengruber, Martinez et al. 2011).
In contrast, repression of many developmental loci is mediated by the activity of PcG protein complexes. PcG-mediated repression of developmental genes directly antagonizes TrxG-mediated gene activation, preventing ectopic expression of genes associated with alternative lineages (Margueron and Reinberg 2011, Simon and Kingston 2013). The diverse proteins that contribute to PcG-mediated transcriptional regulation are traditionally subdivided into two complexes, Polycomb Repressive Complex 1 (PRC1) and Polycomb Repressive Complex 2 (PRC2), on the basis of their associated enzymatic activity. Mammalian PRC2 consists of three core subunits that are essential for proper catalytic activity and gene repression in vivo: enhancer of zeste 2 (Ezh2) or its homolog Ezh1, embryonic ectoderm development (Eed), and suppressor of zeste 12 (Suz12). Ezh2 and Ezh1 contain a conserved SET domain capable of catalyzing the mono-, di-, and tri-methylation of H3K27 (Margueron, Li et al. 2008, Shen, Liu et al. 2008). H3K27me3 is the most well characterized histone PTM catalyzed by PRC2, and this mark plays a crucial role in the establishment of facultative heterochromatin throughout development (Cao, Wang et al. 2002, Czermin, Melfi et al. 2002, Kuzmichev, Nishioka et al. 2002, Kirmizis, Bartley et al. 2004).
The deposition of H3K27me3 by PRC2 promotes the recruitment of a second Polycomb complex, PRC1, at a subset of targeted loci characterized by longer tracts of GC-rich DNA (Ku, Koche et al. 2008). PRC1 complexes are considerably more heterogeneous in composition than PRC2 (reviewed in (Simon and Kingston 2013) and discussed in some detail below). However, all mammalian PRC1 complexes include a homolog of the Drosophila Ring protein, which catalyzes the mono-ubiquitylation of lysine 119 of histone H2A (H2AK119ub). While the exact consequences of H2AK119ub are not clear, PRC1 is thought to inhibit gene expression through a number of mechanisms, including by impairing transcriptional elongation, increasing chromatin compaction, decreasing nucleosome turnover, and modifying higher order chromatin structure (Simon and Kingston 2013).
Some vertebrates, including mammals and zebrafish, have been shown to utilize an additional mechanism of gene regulation involving combinatorial TrxG and PcG activity (Voigt, Tee et al. 2013). In stem and progenitor cells, genes that promote cell type-specific fate acquisition and differentiation are repressed in a readily reversible manner, through co-modification of their promoters with both the “active” H3K4me3 and “repressive” H3K27me3 modifications (Azuara, Perry et al. 2006, Bernstein, Mikkelsen et al. 2006, Pan, Tian et al. 2007). The promoters of many key developmental regulatory genes are regulated by this ‘bivalent’ histone modification signature, which maintains their expression in a restrained, intermediary state characterized by very low levels of transcription (Bernstein, Mikkelsen et al. 2006, Pan, Tian et al. 2007, Zhao, Han et al. 2007). This bivalent state resolves into sets of loci that are either activated or more stably repressed, depending on the lineage specified for a given progenitor (Mikkelsen, Ku et al. 2007, Alder, Lavial et al. 2010). Thus, bivalent promoters appear to be poised to enable rapid expression once a specific lineage is selected, a process that may involve loading a stalled form of RNA polymerase II that could facilitate swift activation as differentiation is initiated (Brookes, de Santiago et al. 2012).
Dynamic Polycomb Activity Regulates the Differentiation of Neural Stem Cells In Vitro
Bivalency is not restricted to ESCs, but is found in other multipotent cell populations, including multipotent neural and glial progenitor cells (Mohn, Weber et al. 2008, Cui, Zang et al. 2009, Xie, Schultz et al. 2013, Zhu, Adli et al. 2013). Formation of lineage-restricted progenitors during development involves both the loss and de novo acquisition of bivalently-modified loci: some existing PcG targets are resolved to completely active or inactive states, while PcG complexes also relocate to new, progenitor-specific targets that may be expressed in the subsequent stage of differentiation (Mohn, Weber et al. 2008). For instance, as pluripotent ESCs undergo neural fate specification, bivalent developmental genes that must be activated in neural progenitors lose the repressive H3K27me3 modification to become actively transcribed (Burgold, Spreafico et al. 2008, Mohn, Weber et al. 2008). This process is mediated, at least in part, by the Jmjd3 H3K27me3 demethylase, whose ability to facilitate activation of neural progenitor-associated genes like Nestin is indispensable for neural fate acquisition (Burgold, Spreafico et al. 2008). Other activities also contribute to alleviating Polycomb-mediated gene repression during neural specification. For example, Zuotin-related factor 1 (Zrf1) was recently found to be required for chromatin displacement of PRC1 to activate neural genes such as Pax6 during neural cell specification (Aloia, Di Stefano et al. 2014).
Once specified, neural progenitors are restrained from further differentiation by Polycomb-mediated repression of genes associated with neuronal or glial differentiation. This involves both the maintenance of bivalent domains originally established in ES cells as well as the acquisition of new bivalent domains at previously unmarked genes in neural progenitors (Mohn, Weber et al. 2008) (see examples, Figure 1). Successive recruitment of PcG complexes to form new bivalent domains is therefore a recurring process, one which is used by multiple progenitor cell types to establish developmental potential by priming new groups of genes for rapid expression or repression (Figure 1).
Figure 1. Changes in histone modification state accompany cell state transitions during neural development.
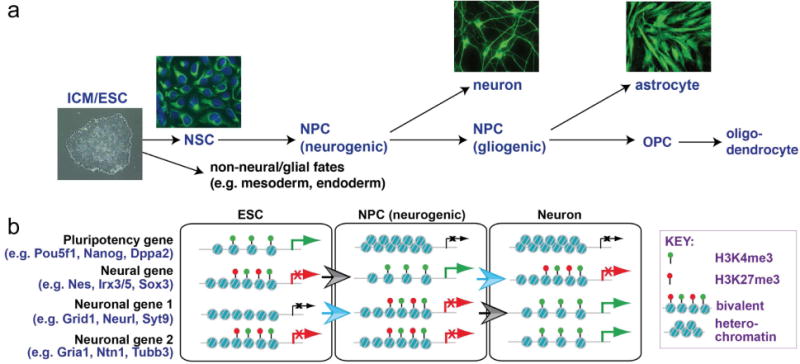
(a) Pluripotent cells of the early embryo (inner cell mass=ICM) and embryonic stem cells (ESCs) can differentiate into multiple neuronal and glial cell types. This process involves cellular transitions through distinct progenitor cell states. Photographs show human ESCs and their directed differentiation into Nestin-expressing neural stem cells (NSCs: green, with DAPI counterstaining of nuclei), Beta-III-tubulin-expressing neurons, and GFAP-expressing astrocytes. (b) Chromatin modification state changes occur at lineage-specific genes during the transition of pluripotent ESCs into differentiated derivatives including neurons. Hallmarks of this process include acquisition of a bivalent modification state by new sets of genes in each progenitor type. Bivalent genes are silent, but primed for rapid expression as differentiation occurs. As a progenitor cell differentiates, bivalent genes associated with the selected lineage resolve into an active chromatin state, while genes associated with alternative lineages adopt more stable heterochromatic configurations. Blue open arrow demarcates transition to a poised bivalent state and gray arrows demarcate transitions from a bivalent to an active (H3K4me3 only) state. Examples for each type of gene are from Mohn et al., 2008.
During neurogenesis, a subset of progenitors activates neuronal genes in response to external and internal cues, while other progenitors maintain repression of neuronal genes and have the capacity to contribute to gliogenesis. As neurogenesis is completed and gliogenesis subsequently begins, these gliogenic progenitors resolve bivalent genes with roles in astrocyte or, subsequently, oligodendrocyte differentiation to an actively transcribed state. This process appears to be particularly important for the specification of oligodendrocyte precursor cells (OPCS), which maintain substantially elevated levels of Ezh2 in comparison to differentiating neurons and astrocytes (Sher, Rössler et al. 2008). Ectopic expression of Ezh2 in differentiating NPCs drives the formation of oligodendrocytes, while loss of Ezh2 has the opposite effect. Ezh2 expression remains high until late stages of oligodendrocyte differentiation, implying a role for PRC2, not just in OPCs, but throughout the multistep process of oligodendrocyte maturation (Sher, Boddeke et al. 2012). Indeed, analysis of the genome-wide distribution of Ezh2 in cultured murine neural stem cells and premature oligodendrocytes (pOLs) indicates that pOLs retain Ezh2 at a subset of targets involved in neuronal or astrocyte fate acquisition, supporting the hypothesis that the varying competence of different progenitor cell populations is determined by Polycomb-mediated repression of distinct targets.
Polycomb Complexes Regulate Multiple, Distinct Cell State Transitions in the Developing Nervous System
While the directed differentiation of cultured embryonic stem cells and neural progenitors has provided a tractable experimental model in which to dissect the molecular dynamics of chromatin regulation by PcG protein complexes, in vitro models are necessarily limited in their capacity to recapitulate the in vivo dynamics of mammalian neural development. The mammalian nervous system is among the most complex biological systems in existence. The human brain is composed of billions of neuronal and glial cells of numerous subtypes, all arranged in an intricate three-dimensional topology essential to proper function. The formation of this system requires multiple cell state transitions as progenitor pools proliferate, migrate, differentiate, and integrate to form circuits. Below, we review evidence demonstrating diverse roles for Polycomb complexes throughout this process.
Polycomb Complexes Regulate Cortical Progenitor Renewal and Differentiation
The mammalian nervous system begins its development as a simple neuroepithelial sheet, which will subsequently be organized along the dorso-ventral and rostro-caudal axes through complex interplay between extracellular morphogens with asymmetric spatial distributions. Cells located at the rostral extent of the neural plate will give rise to the brain, including the cortex, while more caudally positioned cells will eventually form the spinal cord.
After neurulation, cells of the rostral neuroepithelium (i.e., the ventricular zone) undergo symmetric self-renewing divisions to expand the neuroepithelial (NE) cell pool. At the beginning of cortical neurogenesis, NE cells transition into multipotent neural progenitors called radial glial (RG) stem cells. RG stem cells will undergo asymmetric divisions to generate additional RG stem cells and neurons, either directly or through production of fate-restricted basal progenitors that leave the apical surface of the ventricular zone and move into the subventricular zone. Basal progenitors usually undergo only a single symmetric division to generate two neurons, although they can also undergo asymmetric divisions to generate a basal progenitor and a neuron. This differentiation program follows a stereotyped inside-to-out pattern to generate the six layers of the cortex, with neurons located in the deep layers of the cortex being produced prior to neurons found in more superficial layers (Kriegstein and Alvarez-Buylla 2009, Martynoga, Drechsel et al. 2012, Greig, Woodworth et al. 2013, MuhChyi, Juliandi et al. 2013).
After its formation, the RG stem cell population must give rise to a large number of different neuronal and glial cell types. In addition to spatial information, which confers a dorsoventral and rostrocaudal identity upon neuronal populations, temporal information contributes to this diversity. Throughout the central nervous system (CNS), neural progenitor cells (NPCs) produce subtypes of neurons in a defined order before astrogliogenesis is initiated, and astrocytes are formed before most oligodendrogenesis is initiated (Walsh and Cepko 1992, Qian, Shen et al. 2000, Hirabayashi and Gotoh 2005, Shen, Wang et al. 2006, Noctor, Martinez-Cerdeno et al. 2008, Costa, Bucholz et al. 2009).
Although secreted extracellular signals guide the patterning of the developing cortex in vivo, clonal analyses of individual murine neural progenitor cells indicate that cell intrinsic mechanisms also play an instrumental role in controlling the differentiation potential of these cells, progressively restricting their competence as development proceeds. Neo-cortical progenitors generate lower-layer neurons after fewer cell divisions than upper-layer neurons, and progenitors from older mice exhibit reduced capacity to generate earlier-born neuronal subtypes (Shen, Wang et al. 2006). Furthermore, experiments using mouse and human neural stem cells demonstrated that the temporal order in which neural progenitors generate subtypes of neocortical neurons in vivo is retained in vitro, implying that the progressive, cell-intrinsic restriction of neural progenitor competence is a general feature of mammalian neural development (Eiraku, Watanabe et al. 2008, Gaspard, Bouschet et al. 2008).
In addition to cortical neuron subtype specification, the switch from neurogenesis to gliogenesis also appears to involve cell-intrinsic mechanisms. At the onset of the neurogenic phase, extracellular Wnt signaling initiates expression of the transcription factors Neurogenin 1 and 2 (Ngn1/2), which activate expression of other neurogenesis-promoting genes (Hirabayashi, Itoh et al. 2004, Israsena, Hu et al. 2004). The transition from neurogenesis to gliogenesis involves the activation of astrocytic genes such as Glial fibrillary acidic protein (GFAP) by the Jak-STAT signaling pathway, signaling which is stimulated by extracellular signals including ciliary neurotrophic factor (CNTF), leukemia inhibitory factor (LIF), and bone morphogenetic protein 2 (BMP2) (Johe, Hazel et al. 1996, Bonni, Sun et al. 1997, Rajan and McKay 1998, Nakashima, Yanagisawa et al. 1999, He, Ge et al. 2005, Hsieh and Gage 2005, Shen, Wang et al. 2006, Yoshimatsu, Kawaguchi et al. 2006). However, despite the presence of gliogenic cytokines during early corticogenesis, early cortical progenitors do not generate glia (Uemura, Takizawa et al. 2002, Derouet, Rousseau et al. 2004), and young cortical progenitors cultured in vitro are less responsive to gliogenic cytokines than older progenitors (He, Ge et al. 2005). Conversely, neurogenic Wnt ligands continue to be expressed during astrogliogenesis, suggesting the existence of a cell-intrinsic mechanism for regulating the switch from neurogenic to gliogenic competence in neural progenitors (Shimogori, VanSant et al. 2004).
The processes that limit cellular competence in cortical progenitors involve negative crosstalk between regulatory pathways. For example, Ngn1/2 blocks gliogenesis by sequestering the coactivators CBP/p300 and Smad1, which are required to promote transcriptional activation of glial genes such as GFAP in response to pro-astrocytic STAT signaling (Sun, Nadal-Vicens et al. 2001). However, a variety of epigenetic mechanisms have also been implicated. In particular, mounting evidence suggests the Polycomb complexes form one of the key regulatory axes responsible for sequential limitation of the gliogenic and neurogenic competence of neural progenitors in the developing cortex. Ezh2 is highly expressed in neural progenitors, but is down-regulated during the differentiation of cortical neurons (Pereira, Sansom et al. 2010). Premature loss of Ezh2 from the start of the neurogenic period (through conditional deletion from embryonic day 9.5 (E9.5) in the mouse) accelerates neurogenesis and neuronal differentiation, exhausting the apical and basal progenitor pools and resulting in a thinned cortex (Pereira, Sansom et al. 2010, Testa 2011). The accelerating effect of Ezh2 deletion is not limited to neurogenic precursors but also includes glial lineages, as evinced by the drastically expedited appearance of mature astrocytes in the mouse E16 ventricular zone (over 4 days ahead of schedule) (Pereira, Sansom et al. 2010, Testa 2011).
In addition to limiting the onset of neural progenitor differentiation, there is evidence that Polycomb activity regulates the developmental transition from neurogenesis to astrogliogenesis. Loss of Ezh2 or the PRC1 component Ring1b at later stages of cortical development prolongs neurogenesis, rather than accelerating it. This protracted neurogenic phase occurs at the expense of astrogliogenesis, the onset of which is delayed (Hirabayashi, Suzki et al. 2009). Control of the transition between the neurogenic versus gliogenic phase of development is exerted by direct recruitment of PcG complexes to the promoters of the Neurogenin 1 and 2 genes, progressively inhibiting their expression as corticogenesis proceeds (Hirabayashi, Suzki et al. 2009). Polycomb-mediated repression of neurogenic genes in turn facilitates activation of astrocytic genes such as Glial fibrillary acidic protein (GFAP), e.g. by alleviating Neurogenin’s sequestration of coactivators of the pro-astrocytic Stat3 transcription factor (Sun, Nadal-Vicens et al. 2001).
There is also some data suggesting that Polycomb activity continues to be important in later stages of cortical gliogenesis, as the production of astrocytes gives way to oligodendrogenesis. In the ventral telencephalon, the Dlx1 and Dlx2 transcription factors are required both to produce inhibitory GABA (gamma-aminobutyric acid)-ergic neurons and to repress oligodendrocyte formation promoted by Olig2 (Petryniak, Potter et al. 2007). Repression of neurogenic Dlx1/2 activity is therefore one requirement for the developmental transition to oligodendrogenesis. In adult neural stem cells, MLL activity maintains Dlx2 expression by antagonizing PcG-mediated repression (Lim, Huang et al. 2009), while in the embryonic cortex loss of Polycomb activity (Ring1b) de-represses Dlx2 (Hirabayashi, Suzki et al. 2009). These findings suggest that cell intrinsic regulatory mechanisms involving PcG also contribute in vivo to the developmental transition to oligodendrogenesis.
Roles for Polycomb function beyond cortical neurogenesis and gliogenesis
Beyond their functions as regulators of progenitor specification in the embryonic cortex, PcG complexes continue to play roles in adult neural progenitors and differentiated neurons. In the adult, a subset of astrocytes in the subventricular zone (SVZ) act as neural stem cells (NSCs), with a neurogenic competence that is lacking in other adult astrocytes. Recent work indicates that continued expression of Ezh2 distinguishes these neurogenic astrocytes from their glialrestricted counterparts. Specifically, Ezh2 controls the neurogenic competence of adult SVZ NSCs by repressing the expression of Olig2 to permit neuronal differentiation, while simultaneously preventing the activation of genes associated with non-SVZ neuronal subtypes (Hwang, Salinas et al. 2014). PRC1 also participates in the regulation of adult NSCs by modulating proliferation and self-renewal. The PRC1 component Bmi1 can maintain the proliferation and self-renewal of adult neural stem and progenitor cells through repression of the cell cycle inhibitors p16Ink4a, p19Arf, and p21 (Molofsky, Pardal et al. 2003, Bruggeman, Valk-Lingbeek et al. 2005, Molofsky, He et al. 2005, Fasano, Dimos et al. 2007, Fasano, Phoenix et al. 2009, Román-Trufero, Méndez-Gómez et al. 2009). Accordingly, depletion of the PRC1 protein Ring1b from cultured adult olfactory bulb NSCs impairs NSC proliferation and self-renewal (Román-Trufero, Méndez-Gómez et al. 2009).
Finally, recent research expands the contexts in which PcG complexes function to include terminally differentiated neurons. During tangential migration of precerebellar neurons and formation of connections with the cortex, Ezh2-dependent regulation of transcriptional programs is required to maintain positional information to control topographic neuronal guidance and connectivity (Di Meglio, Kratochwil et al. 2013). Mouse knockout studies of Jmjd3, the histone demethylase primarily responsible for the removal of H3K27me3, further demonstrate the importance of precise regulation of H3K27me3-dependent gene repression for neuronal maintenance. Inactivation of Jmjd3 in the mouse leads to perinatal lethality as a result of disrupted maintenance of the pre-Botzinger complex (PBC), the pacemaker of the respiratory rhythm generator (RRG) (Burgold, Voituron et al. 2012). Specifically, while the early formation of the RRG was not affected by loss of Jmjd3, maintenance of the PBC is perturbed due to aberrant silencing of PBC-related genes. Among the genes dysregulated by loss of Jmjd3 are reelin, a glycoprotein involved in neuronal migration, and Neph2, a transmembrane protein with roles in synaptogenesis (Burgold, Voituron et al. 2012). In differentiated neurons, PRC2 also modulates neuronal activity-dependent processes including dendritic arborization (Qi, Liu et al. 2014). Together, these results support a continuing role for PcG complexes as regulators of neural circuit formation and maintenance, above and beyond their developmental functions as regulators of neuronal and glial fate specification.
Polycomb Complexes Regulate Motor Neuron Subtype Specification in the Spinal Cord
During the period of embryogenesis, when patterning of the rostral neural tube into the brain occurs, the caudal neuroepithelium is also patterned as the neural plate closes to form the spinal neural tube. The ventral region of the spinal neural tube is colonized by a progenitor population that will give rise to diverse subtypes of neurons, including motor neurons (MN). Spinal motor neuron progenitors are grouped into columns on the basis of their rosto-caudal location along the neuraxis, which is established by the expression of region-specific Hox transcription factors (Philippidou and Dasen 2013, Davis-Dusenbery, Williams et al. 2014). MN columns are comprised of sets of MNs arranged longitudinally along the rostro-caudal axis of the spinal cord, and neurons of each column project to distinct regions in the periphery. For example, the preganglionic motor column (PGC) is located at the thoracic level of the neuraxis and contains visceral MNs that innervate sympathetic ganglia, while MNs of the lateral motor column (LMC) span both the brachial and lumbar levels to innervate the limbs.
Hox gene expression in the spinal neural tube is patterned in two phases. First, extracellular signals secreted from the adjacent rostral somitic mesoderm and caudal presomitic mesoderm create opposing morphogen gradients that establish broad territories of Hox gene expression in proliferating spinal progenitors (Philippidou and Dasen 2013, Davis-Dusenbery, Williams et al. 2014). After spinal progenitors undergo their final mitosis, the boundaries of posterior Hox domains are further refined by cross-repressive interactions between the transcriptional programs originally induced by the various extracellular signals, a process that is required to sharpen and maintain expression borders.
Recent work has demonstrated the involvement of Polycomb Repressive Complexes at both steps of spinal progenitor differentiation. In one study, PRC2 was found to be essential for the initial repression of Hox gene expression prior to regionalization of spinal motor neurons by gradients of retinoic acid (RA), Wnt, and fibroblast growth factor (FGF), as well as maintaining the repression of alternative Hox codes after regionalization (Mazzoni, Mahony et al. 2013). Treatment of differentiating mouse neural progenitors with RA resulted in the binding of retinoic acid receptors (RARs) to the Hox1-Hox5 genes, triggering the rapid, domain-wide clearance of PcG-dependent H3K27me3 repression to enable Hox gene activation and acquisition of cervical spinal identity. Wnt and fibroblast growth factor (FGF) signals instead activated the expression of the Cdx2 transcription factor, whose subsequent binding to Hox1-Hox9 genes cleared H3K27me3 from these domains to specify brachial or thoracic spinal identity.
Polycomb also appears to be critical for the cross-repressive refinement of Hox expression boundaries in post-mitotic spinal neurons, with PRC1 exhibiting a dose-dependent regulation of MN subtype differentiation (Golden and Dasen 2012). Depletion of Bmi1 from the developing spinal cord results in the de-repression of more posterior Hox genes and alters MN fate, converting forelimb lateral motor column (LMC) MNs to a thoracic preganglionic (PGC) identity. Intriguingly, ectopic expression of Bmi1 at thoracic levels has the opposite effect, converting PGC MNs to an LMC identity. The dose-dependent roles of PRC1 in the developing spinal cord imply that absolute levels of Polycomb activity may be an important determinant of its regulatory targets in this context.
Molecular Mechanisms Governing PcG Protein Activity in Neural Development
The previous examples emphasize the multiple, temporally distinct roles played by the PcG complexes in neural development. Polycomb functions differ, not just between stem cells of different types, but also within the same cell type at different developmental stages. These observations imply the existence of molecular machinery capable of modifying the cohort of genes targeted by Polycomb in response to intrinsic and environmental cues. In Drosophila, specific DNA sequences called Polycomb Response Elements (PREs) recruit PcG protein complexes to their targets. However, in vertebrates, isolating response elements with similar functionality to fly PREs has proven to be exceedingly difficult, and no consensus motif for a DNA element capable of recruiting PRC2 or PRC1 has been identified. In the absence of a clear association between transcription factors and Polycomb binding, several alternative mechanisms contributing to Polycomb complex recruitment have been proposed. The complex biochemistry governing the general recruitment of mammalian Polycomb complexes is outside the scope of this review, and has been described elsewhere (see (Simon and Kingston 2013) for an excellent review). Instead, we will focus on the progress that has been made toward uncovering context-specific mechanisms that govern Polycomb recruitment specifically in the nervous system.
Several types of mechanisms for Polycomb recruitment have been proposed. In general, these involve: 1. the role of the chromatin environment and histone modification state in Polycomb recruitment, 2. the ability of several proteins to interact with either PRC2 or PRC1 complexes to mediate their recruitment to specific target sites, or 3. the role of interactions between Polycomb complexes and long non-coding RNAs in Polycomb recruitment. In the sections below, we discuss each of these potential recruitment mechanisms in detail and describe their known or likely roles in PcG complex recruitment during nervous system development.
The Local Chromatin Environment Influences the Activity of PRC2
Recently, the chromatin environment and histone modification state have been shown to influence recruitment of PcG complexes to and activity on chromatin. In addition to the enzymatic subunit Ezh2, PRC2 complexes include the proteins Eed and Suz12 in stoichiometric ratios, and both proteins are required for effective enzymatic activity (Figure 2A and 2B). The latest research suggests that both Suz12 and Eed function as adaptors that bind to and modify Ezh2 catalysis in response to local chromatin cues.
Figure 2. Polycomb complex diversity.
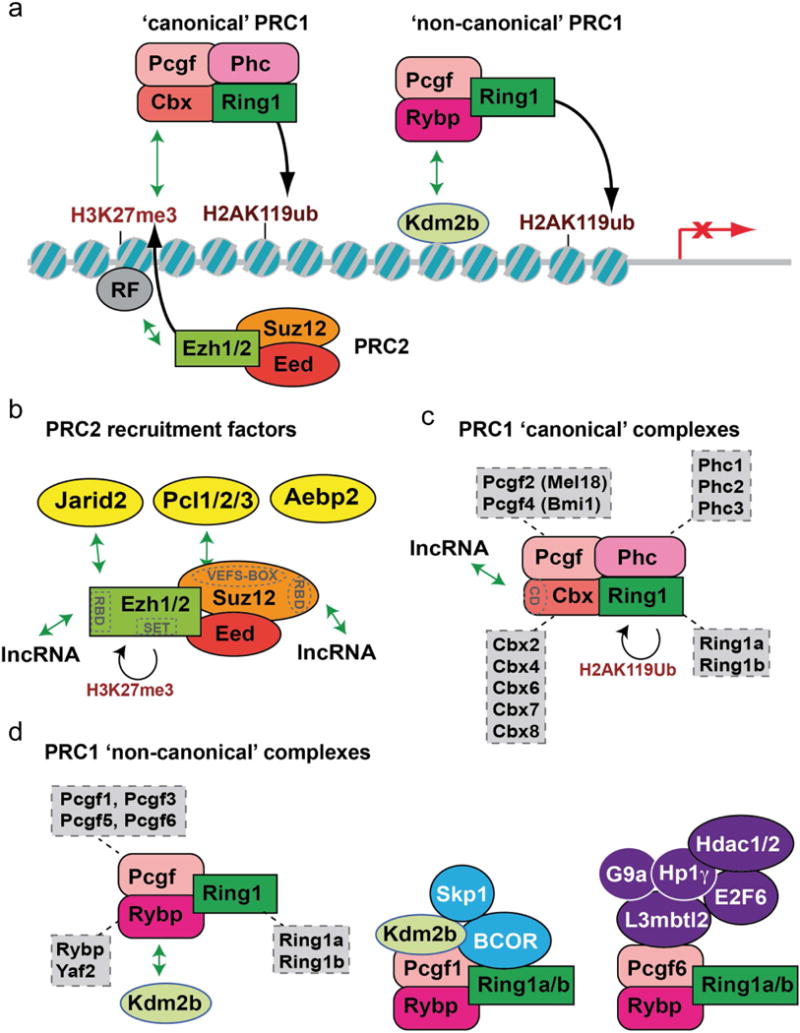
(a) H3K27me3 modification of chromatin by the EZH1/2 subunit of PRC2 promotes recruitment of the ‘canonical’ PRC1 complex, which catalyzes ubiquitylation of H2AK119 to repress gene expression. Additional ‘non-canonical’ forms of the PRC1 complex can be recruited to chromatin in a PRC2-independent manner. (b) PRC2 associates with several non-stoichiometric proteins, shown in yellow. PRC2 subunits EZH2 and SUZ12 also have functional domains described in text (shown in gray), including RNA binding domains (RBDs) that can associate with long non-coding RNAs (lncRNA). Both protein and lncRNA associations have the potential to facilitate context-dependent recruitment to chromatin. (c) ‘Canonical’ PRC1 complexes have multiple forms, through inclusion of specific subunit variants. The chromodomain (CD) of the Cbx subunit can also associate with lncRNAs. (d) Non-canonical PRC1 complexes have an even more diverse potential subunit composition, with some complex variants shown here (see text for further information).
Suz12 is a zinc-finger protein, whose cumulative interactions with Ezh2 and neighboring histones (via an amino-terminal VEFS-BOX domain) can positively and negatively regulate PRC2 activity (Schmitges, Prusty et al. 2011, Chan, Chen et al. 2012, Ciferri, Lander et al. 2012, Yuan, Wu et al. 2012). The ability of Suz12 to bind neighboring histone H3 confers upon PRC2 a substantial preference for densely packed polynucleosomes, relative to dispersed substrates. Indeed, increased polynucleosome density augments PRC2 catalysis up to 30-fold over controls in vitro (Chan, Chen et al. 2012, Yuan, Wu et al. 2012). Eed instead seems to primarily be involved in feed-forward of PRC2 activity, via the ability of its WD40 β-propeller to recognize H3K27me3 and positively stimulate Ezh2 (Margueron, Justin et al. 2009, Xu, Bian et al. 2010). The addition of K27me3-containing peptide to purified PRC2 complexes in vitro results in an up to seven-fold increase in PRC2 catalytic activity, consistent with experiments demonstrating the essential requirement of Eed for proper PRC2 function (Montgomery, Yee et al. 2005, Margueron, Justin et al. 2009). The potent allosteric activation of Ezh2 catalysis induced by EED binding to H3K27me3 may explain why acetylation of H3K27 inhibits PRC2 activity, and accordingly, why the removal of H3K27ac by the nucleosome remodeling and deacetylation (NuRD) complex is essential for PRC2 recruitment during ES differentiation (Tie, Banerjee et al. 2009, Pasini, Malatesta et al. 2010, Reynolds, Salmon-Divon et al. 2012).
Given the considerable influence of the noncatalytic subunits of PRC2 on methyltransferase activity, PRC2 appears to function as a complex holoenzyme, integrating the regulatory inputs from the core subunits and other cofactors to sense the local chromatin environment and adjust activity accordingly (Margueron and Reinberg 2011, Ciferri, Lander et al. 2012). PRC2’s ability to sense the local chromatin environment may also link PRC2 target selection in neural development to the activities of other chromatin regulators with well-described functions in neural differentiation, such as the ATP-dependent BAF chromatin remodeling complexes (Ronan, Wu et al. 2013, Narayanan and Tuoc 2014).
PRC1 Subunit Composition Confers Cell-State Specific Activity
Compared to PRC2, PRC1 complexes are highly heterogeneous in composition. ‘Canonical’ PRC1 complexes have four core subunit types, each of which can be represented by several different PcG proteins: Cbx2/4/6/7/8, Ring1a/b, Pcgf1/2/3/4/5/6, and Ph1/2/3 (Figure 2C) (Levine, Weiss et al. 2002). Canonical PRC1 complexes can be recruited to chromatin through Cbx protein binding to PRC2-deposited H3K27me3 (Wang, Brown et al. 2004), although H3K27me3-independent targeting of PRC1 complexes has also been documented. Once chromatin bound, the Ring1a/b subunit ubiquitylates H2A lysine 119 (H2AK119Ub) (de Napoles, Mermoud et al. 2004, Cao, Tsukada et al. 2005) (Figure 2A). This ubiquitylation event promotes gene repression (Endoh, Endo et al. 2012), and can impair transcriptional elongation (Stock, Giadrossi et al. 2007). PRC1 can also repress gene expression through Ring1b/Rnf2-mediated chromatin compaction (Francis, Kingston et al. 2004, Eskeland, Leeb et al. 2010, Endoh, Endo et al. 2012).
Evidence suggests that subunit variants are not redundant, but rather function in distinct complexes. For example, mutation of the PRC1 protein Ring1b results in embryonic lethality during gastrulation (Voncken, Roelen et al. 2003), while Ring1a mice are viable (de Napoles, Mermoud et al. 2004). Although the roles of many of the PRC1 subunit variants in development remain uncharacterized, it seems likely that they will contribute to context-specific functions of PRC1 (Turner and Bracken 2013). For instance, replacement of Cbx7-containing PRC1 complexes by Cbx2/4-containing PRC1 complexes was recently shown to mediate a transition from pluripotency to fate acquisition (Morey, Pascual et al. 2012, O’Loghlen, Munoz-Cabello et al. 2012). This example illustrates the potential for variant subunits to endow PRC1 with cell-state specific functions.
PRC1 complexes also exist in non-canonical forms in which the Cbx subunit is substituted for alternative proteins (Figure 2D) (García, Marcos-Gutiérrez et al. 1999, Sánchez, Sánchez et al. 2007, Gao, Zhang et al. 2012, Hisada, Sanchez et al. 2012, Junco, Wang et al. 2013). Notably, Pcgf-Ring1a/b complexes lacking a Cbx subunit can instead associate with Rybp or its homolog Yaf2, in which case Rybp stimulates Ring1b-mediated ubiquitylation of H2AK119 in a PRC2/H3K27me3-independent manner (Gao, Zhang et al. 2012, Tavares, Dimitrova et al. 2012).
Many PRC2 and PRC1 core subunits, variant subunits, and associated recruitment factors or accessory proteins are expressed in the developing nervous system. The Gene Expression Database at the Mouse Genome Informatics Resource catalogs published temporal and spatial expression patterns of Polycomb protein expression reported to date for the mouse nervous system (Figure 3). Some Polycomb subunits are expressed in the central nervous system from the onset of neural plate formation (embryonic day 8–8.5), with other subunits being detected from the onset of neurogenesis (around embryonic day 10.5) and through post-natal stages. Within the nervous system, expression of many subunits is reported in the fore-, mid-, and hindbrain regions, with core PcG subunits frequently showing enrichment in the ventricular zone of the cortex. Expression of some subunits has also been reported in the spinal cord. These data support potential roles for both canonical and non-canonical Polycomb complexes in neural development.
Figure 3. Expression of Polycomb complex proteins and recruitment factors in the developing nervous system.
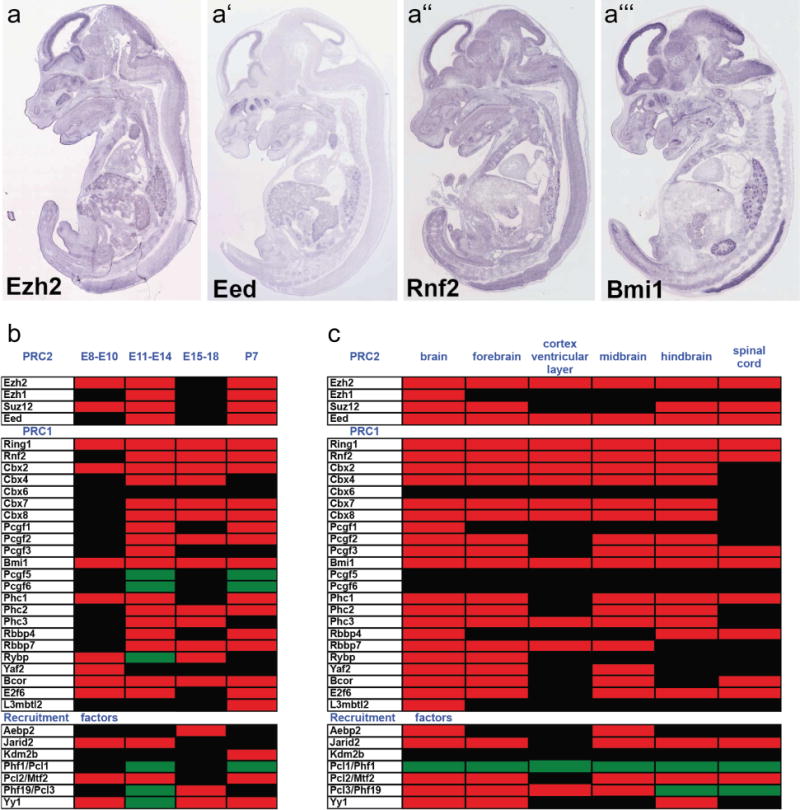
(a–a′″) In situ hybridization data for four core Polycomb proteins at embryonic day 14.5 (Genepaint database) shows robust expression in the ventricular zone of the cortex and other CNS locations. (b–c) The Gene Expression Database at the Mouse Genome Informatics Resource was used to catalog (b) temporal and (c) regional expression of Polycomb subunits that has been reported for the mouse central nervous system between embryonic day 8 (E8) and post-natal day 7 (P7). Heat maps indicate developmental time windows (b) or CNS regions (c) where expression is documented as present (red), as absent (green), or where no data is reported (black). CNS expression of some Polycomb core subunits is reported from the onset of neural plate formation, with expression of other subunits being detected during neurogenesis and through post-natal stages. Within the CNS, expression of many subunits is reported in the fore-, mid-, and hindbrain regions, with core PcG subunits in particular showing enrichment in the ventricular zone of the cortex. Expression of some subunits has also been reported in the spinal cord.
Evidence from mouse knockout models suggests that non-canonical PRC1 complexes may play specific roles in neural development. While constitutive knockout of Rybp results in embryonic lethality at early post-implantation stages, chimeric embryos show myriad phenotypes indicative of aberrant neural development, including forebrain overgrowth and localized regions of disrupted neural tube closure (Pirity, Locker et al. 2005).
Protein Recruitment Factors Regulate Polycomb Complex Targeting in Neural Development
Both PRC2 and PRC1 have been shown to directly interact with proteins that can facilitate their recruitment to chromatin through distinct mechanisms. Below, we discuss how some of these proteins may regulate the recruitment of Polycomb complexes to specific target subsets in neural development.
Jarid2 May Regulate Context-Dependent PRC2 Activity During Neural Development
Among the most prominent of the non-core PRC2-associated proteins is Jarid2, a catalytically inactive member of the jumonji family of histone demethylases that directly interacts with PRC2 in nearly stoichiometric ratios in ES cells (Peng, Valouev et al. 2009, Shen, Kim et al. 2009, Li, Margueron et al. 2010, Pasini, Cloos et al. 2010). Jarid2 has well established roles in neurulation in mouse models and in directed differentiation of ESCs, including differentiation toward neuronal lineages: Jarid2 (also called Jumonji) was initially identified based upon a gene trap mutation in the mouse that resulted in defects in neural tube closure in the midbrain region, demonstrating strain-dependent requirements for neurulation (Takeuchi, Yamazaki et al. 1995, Takeuchi, Kojima et al. 1999) Jarid2 was further shown to be required to repress cyclin D1 expression to coordinate cell cycle exit and neuronal migration during neurogenesis in the mouse hindbrain (Takahashi, Kojima et al. 2007). In addition, several studies have examined the role of Jarid2 during directed differentiation of mouse ESCs. In this context, Jarid2 mapped to many PcG target genes (Pasini, Cloos et al. 2010) and its activity was required for induction of differentiation-related genes (Peng, Valouev et al. 2009, Shen, Kim et al. 2009, Pasini, Cloos et al. 2010), including expression of the neuronal marker Sox11 (Shen, Kim et al. 2009). While both Jarid2 and PRC2 are required for neural development, it is not known whether Jarid2-PRC2 interaction is required for PcG recruitment or activity in the developmental contexts described above.
Several studies do suggest that Jarid2 is important for the recruitment of PRC2 to its targets in ESCs, although it is not clear whether this involves the direct binding of Jarid2 to DNA via its zinc finger or ARID domains, or whether some other mechanism is involved. However, it seems unlikely that Jarid2 is the sole factor responsible for PRC2 recruitment in ESCs: loss of Jarid2 does not result in the extensive re-expression of PRC2 target genes, as is observed when ablating a core PRC2 subunit, and the overall impact of Jarid2 loss upon H3K27me3 levels in cells is decidedly mild (Landeira, Sauer et al. 2010, Margueron and Reinberg 2011).
A recent study suggests that the conflicting reports regarding the importance of Jarid2 to PRC2 function might be reconciled by the newly-discovered ability of Ezh1 to compensate for Jarid2: both Ezh1 and Jarid2 have innate nucleosome binding capacity, and PRC2 recruitment and enzymatic activity in Jarid2-deficient myoblasts was shown to depend on expression of Ezh1 but not Ezh2 (Son, Shen et al. 2013). Jarid2 and Ezh1 also display reciprocal expression patterns: Jarid2 is expressed most highly in pluripotent and early lineage-committed cells, while Ezh1 is most highly expressed in later, more differentiated cell types. Thus, it may be that Ezh1 has functions that are non-redundant with Ezh2, including promoting the access of Ezh2-containing PRC2 complexes to chromatin in committed cell types that lack Jarid2 (Son, Shen et al. 2013). It will be important to test this hypothesis in models of lineage commitment other than myoblast differentiation. In the particular context of neural development, it will be interesting to determine whether there is a developmental time at which PRC2’s dependence on Jarid2 is supplanted by Ezh1, and whether this switch may be associated with progressive restriction of neural progenitor competence.
Chd4 and Chd5 Regulate PRC2 Recruitment to Promote Neurogenesis in Cortical Progenitors
Effector proteins endowed with H3K27me3-“reader” domains can impinge on PRC2 activity in a cell type-specific manner. For example, chromodomain helicase DNA-binding protein 5 (Chd5) is a protein with the ability to remodel nucleosomes that is frequently deleted in aggressive neuroblastoma (Koyama, Zhuang et al. 2012). Chd5 is characterized by two chromodomains that bind H3K27me3 and are essential for its function as a regulator of cortical neurogenesis (Egan, Nyman et al. 2013). Depletion of Chd5 in differentiating neural progenitors leads to de-repression of a subset of PRC2 targets, as well as the failure to activate expression of key neuronal genes (Egan, Nyman et al. 2013).
Another chromodomain helicase DNA-binding protein, Chd4, has also recently been shown to be a critical interaction partner of Ezh2 in cortical progenitors, where it is required specifically for PRC2-mediated suppression of the astrogliogenic marker gene GFAP. Accordingly, experimentally depleting Chd4 RNA from cortical progenitors in the developing neocortex promotes astrogliogenesis in vivo. While Chd4 is a frequent component of the NuRD complex, depletion of other NuRD components did not result in increased astrogliogenesis, suggesting that Chd4 may function independently of the NuRD complex to regulate neural progenitor competence. Together, these data suggest that target gene-specific mechanisms involving cross talk with other chromatin “readers” can influence Polycomb activity to control neural cell fate transitions.
AEBP2 is a Co-Activator of PRC2 that May Regulate Recruitment in Neural Development
Aebp2 is a Gli-type zinc finger that is frequently found in association with PRC2, where it appears to enhance PRC2 enzymatic activity (Cao, Wang et al. 2002, Cao and Zhang 2004, Ciferri, Lander et al. 2012). While homozygous loss of Aebp2 in mouse models is embryonic lethal, heterozygous animals have phenotypes suggesting a role for Aebp2 in regulation of neural crest cell development (Kim, Kang et al. 2011). At present, it is not clear whether Aebp2 primarily influences PRC2 enzymatic activity or recruits PRC2 to some of its targets through its ability to bind DNA. Analysis of Aebp2 target sites in brains of one month old mice revealed considerable overlap between Aebp2 and PRC2 target genes, supportive of a role for Aebp2 in recruitment of PRC2 complexes during neural development (Kim, Kang et al. 2009). However, because this study relied on cloning of DNA fragments isolated by ChIP, combined with Sanger sequencing, it will be necessary to perform this analysis on a genome-wide scale to validate the extent of Aebp2 and PRC2 overlap in bound chromatin locations. Moreover, if Aebp2 is required for PRC2 recruitment, loss of Aebp2 should abrogate PRC2 recruitment specifically at common target sites, without affecting sites targeted only by PRC2 and this remains to be tested. Therefore, while not definitive, the current evidence is supportive of a role for Aebp2 in targeting of PRC2 during development of the brain and neural crest.
Kdm2b Regulates the Recruitment of Non-Canonical PRC1 Complexes
Kdm2b (also known as Fbxl10 or Jhdm1b) is an H3K36-specific histone demethylase that was initially identified as a factor controlling cell proliferation and senescence by regulating the Ink4a-ARF-Ink4b locus (He, Kallin et al. 2008). Recent work has revealed that Kdm2b also facilitates recruitment of PRC1 in some contexts (Wu, Johansen et al. 2013). Kdm2b-containing PRC1 complexes constitute a distinct type of non-canonical complex containing Ring1b, Pcgf1 (Nspc1), and Rybp, but not Cbx proteins (Wu, Johansen et al. 2013)(Figure 2A,D). Kdm2b binds non-methylated CpG island sequences (CGIs) via its CxxC-type zinc-finger domain, promoting PRC1-mediated H2AK119 ubiquitylation at a subset of its target sites (Koyama-Nasu, David et al. 2007, Farcas, Blackledge et al. 2012, He, Shen et al. 2013, Wu, Johansen et al. 2013). Although Kdm2b does bind the CGIs of Polycomb-repressed genes, it also binds CGIs throughout the genome, a large fraction of which correspond to actively transcribed genes that are not PRC1 targets. These observations indicate that the presence of Kdm2b at unmethylated CGIs is not sufficient for stable PRC1 recruitment.
Interestingly, while the process of PRC1 recruitment to target sites has been generally considered a hierarchical event that depends on prior PRC2-mediated H3K27me3, Kdm2b was recently shown to recruit a Pcgf1-containing variant PRC1 complex to CpG islands, with PRC1 chromatin binding leading to subsequent PRC2 recruitment and H3K27 methylation (Blackledge, Farcas et al. 2014). Another recent study also found that PRC1 recruitment to and H2A ubiquitylation at unmethylated CpG rich chromatin regions in ES cells was sufficient to recruit PRC2 to chromatin (Cooper, Dienstbier et al. 2014). This work supports a role for PRC1 recruitment in directing PRC2-mediated H3K27me3 and indicates that non-canonical complexes that associate with Kdm2b may act in this manner.
Kdm2b appears to play required roles in neural development. While loss of Kdm2b in cultured ESCs can cause de-repression of lineage-specific genes and precocious differentiation (He, Shen et al. 2013), Kdm2b mutant embryos die perinatally with defects specifically in neural development, including incomplete neural tube closure, exencephaly, and increased proliferation and apoptosis of neural progenitor cells (Fukuda, Tokunaga et al. 2011). However, at a mechanistic level, the role of Kdm2b in non-canonical PRC1 targeting in the context of neural and glial development has yet to be elucidated.
Methylation of CpG Islands Flanking Promoters Inhibits PRC2 in Neural Progenitors
Another aspect of chromatin state that can influence Polycomb recruitment is methylation of CpG islands (CGIs). Highly methylated CGIs are strongly anti-correlated with H3K27me3 and PRC2 binding, a finding supported by multiple studies that could partially explain the restricted repertoire of PRC2 targets in different lineages (Mohn, Weber et al. 2008, Brinkman, Gu et al. 2012, Lynch, Smith et al. 2012, Xie, Schultz et al. 2013). While sequential chromatin immunoprecipitation (ChIP) and bisulfite sequencing experiments confirm that H3K27me3 and DNA methylation can indeed co-localize in the genome, this co-localization is never seen in regions with high CpG density, implying that high levels of DNA methylation somehow inhibit the local activity of PRC2 (Brinkman, Gu et al. 2012, Statham, Robinson et al. 2012).
Loss-of-function studies of Dnmt3a and Dnmt3b also support the link between DNA methylation and PRC2 recruitment, particularly in neural development. In murine ESCs, simultaneous knockout of Dnmt3a/b results in depletion of DNA methylation from intragenic and intergenic CpG islands, concomitant with an increase of H3K27me3 (Hagarman, Motley et al. 2013). Consistent with these results, a study examining the role of Dnmt3a in murine neural stem cells found that, while Dnmt3a is dispensable for their renewal and proliferation, knockout of Dnmt3a causes stunted neurogenic potential both in vitro and in vivo (Wu, Coskun et al. 2010). Loss of Dnmt3a significantly decreased the expression of neurogenic genes while genes involved in gliogenesis were significantly upregulated, even though both classes of targets experienced a decrease in DNA methylation.
The results above are inconsistent with the straightforward interpretation of DNA methylation as a repressive mark. However, they might be explained by the striking observation that most genes that were down-regulated after Dnmt3a ablation had H3K4me-rich CGIs in their proximal promoters, while Dnmt3a binding (and associated DNA methylation) was present only in the inter- and intragenic regions flanking their promoter CGIs (Wu, Coskun et al. 2010). In contrast, genes that were up-regulated after loss of Dnmt3a featured CpG-poor proximal promoters, with low expression and little modification by H3K4me3. This led the authors to hypothesize that DNA methylation in regions flanking CpG-rich proximal promoters acted as an activating signal, preventing repression by some alternative mechanism (Wu, Coskun et al. 2010). Genome-wide profiling of H3K27me3 in Dnmt3a-knockout NSCs confirmed that genes that were down-regulated in response to DNA hypomethylation exhibited increased PRC2 activity. These results suggest that DNA methylation in gene bodies and in intragenic regions flanking the proximal promoter may activate gene expression by inhibiting the enzymatic activity of PRC2 (Figure 4). Future experiments exploring the dynamics of genome-wide DNA methylation in models of neural development will be required to determine the extent to which changing patterns of DNA methylation can explain the cell state-specific selection of targets by otherwise ubiquitous chromatin modifiers like PRC2.
Figure 4. Relationship between DNA methylation and Polycomb recruitment.
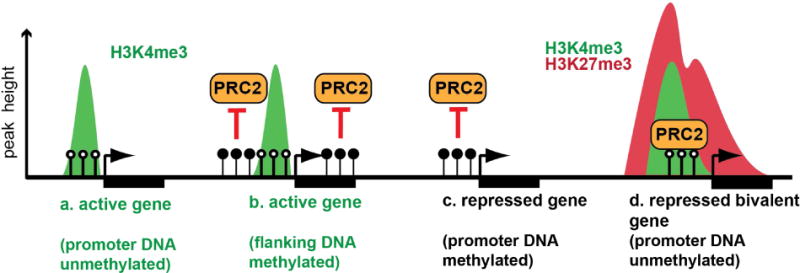
(a–b) Promoters of active genes are frequently (a) unmethylated (open circles), although (b) DNA methylation (closed circles) flanking the promoter can also facilitate the active state by preventing PRC2-mediated repression. Repressed genes may have methylated promoter DNA (c), although demethylation allows PRC2-mediated silencing (d). Arrows indicate the transcription start site and rectangles indicate gene bodies.
Long non-coding RNAs in neural development
A host of studies have also implicated long, non-coding RNAs (lncRNAs) in the targeting of PRC2. While the investigation of lncRNAs in the CNS is still in its infancy, studies have already emerged implicating lncRNAs in neurodegenerative, neuro-oncological, and psychiatric disorders (Johnson 2012, Qureshi and Mehler 2013, Tushir and Akbarian 2013, Ziats and Rennert 2013, Zhang and Leung 2014). Therefore, it is likely that many additional connections between lncRNAs and Polycomb recruitment in the context of neural development will be defined. Below, we first highlight roles of lncRNAs in neural development and nascent connections to Polycomb recruitment in this context. We then discuss lncRNA-dependent Polycomb PRC2 recruitment mechanisms. For more comprehensive information regarding the roles of lncRNAs in CNS development, we refer the reader to several recent reviews (Qureshi and Mehler 2012, Ng, Lin et al. 2013, Fatica and Bozzoni 2014).
Given the high complexity of the vertebrate nervous system, it may not be surprising that developing neural cells exhibit intense transcriptional activity, including expression of a large number of lncRNAs. Initial attempts to characterize the breadth and specificity of lncRNA expression in the brain include high-throughput in situ hybridization data from the Allen Brain Atlas, which found regional, cell-type, and subcellular specificity in the expression patterns of over 849 lncRNAs from diverse locations in the genome (Mercer, Dinger et al. 2008). Similarly, microarray analysis of lncRNA expression in human brain samples detected thousands of lncRNA transcripts with regionally-restricted expression, many of which are primate-specific (Derrien, Johnson et al. 2012). The enormous lncRNA transcriptome of the mammalian CNS, as well as the preponderance of primate-specific lncRNAs expressed in the human brain, suggests that functionalization of non-coding RNAs may be essential to the evolution of higher brain function. This hypothesis is consistent with an apparent correlation between the proportion of non-coding DNAs in the genome and organismal complexity (Taft, Hawkins et al. 2011). The mammalian brain is therefore an interesting context for investigating how lncRNAs might function.
In one of the earliest studies to assess the neural lncRNA transcriptome, researchers used custom microarrays to evaluate expression of a large set of lncRNAs in embryonic forebrain-derived murine neural stem cells, and in NSCs induced in vitro to become Nkx2.1-expressing bipotent neuronal-glial progenitors, which can produce both cortical GABAergic neurons and oligodendrocytes (Mercer, Qureshi et al. 2010). Interestingly, unique subsets of lncRNAs were differentially expressed throughout the process of neuro-gliogenesis, including during bipotent progenitor specification, GABAergic neurogenesis, the switch to oligodendrogenesis, and the maturation of myelin-producing oligodendrocytes. While none of the transcripts identified in this study were functionally characterized, extensive differential expression of lncRNAs during neural fate specification was later confirmed in human cells (Ng, Johnson et al. 2012). In this work, custom microarrays were used to examine the expression of a set of previously identified lncRNAs in human embryonic stem cell-derived neural progenitor cells (NPCs), both in the process of in vitro NPC specification and during the differentiation of NPCs to produce dopaminergic neurons (Ng, Johnson et al. 2012). In this study, a subset of four intergenic lncRNAs that were preferentially enriched in differentiated neurons was also selected for transient depletion in neural progenitors using siRNAs. Depleting each of these four intergenic lncRNAs inhibited neurogenesis, creating an average of five-fold reduction in the numbers of Tuj1-positive neurons after differentiation, compared to non-targeting siRNA controls. Subsequent expression analysis of lncRNA-depleted NPCs by quantitative PCR revealed an apparent switch from a neurogenic to a gliogenic program, as indicated by reduced expression of neurogenic markers like Neurog2 and concomitant increase in expression of gliogenic marker genes like PDGRalpha and Myelin Basic Protein.
In addition to establishing that lncRNAs can indeed act as critical regulators of neurogenic and gliogenic homeostasis, at least in vitro, Ng et al. (2012) also demonstrated that neurogenic lncRNAs identified in this study bind selectively to the Suz12 subunit of PRC2 and to a subunit of the REST transcriptional repressor complex, implying a role in the epigenetic control of transcription (Ng, Johnson et al. 2012). A follow-up study by the same group revealed that one of the previously identified neurogenic lncRNAs that did not bind to either PRC2 or REST, previously dubbed RMST for rhabdomyosarcoma 2-associated transcript, nevertheless regulates neurogenesis at the transcriptional level by binding to the Sox2 transcription factor (Ng, Bogu et al. 2013). Like previous examples of lncRNAs that contribute to transcriptional regulation, RMST was found to act as a physical bridge between Sox2 and chromatin, effectively functioning as a molecular scaffold that facilitated the activation of key neurogenic genes. Taken together, these data reinforce the notion that lncRNAs can act as key transcriptional regulators in neural development, and that this regulation involves the recruitment of nuclear proteins to their genomic targets.
Finally, recent experiments using mouse models have provided insight into the neural lncRNA transcriptome and roles for lncRNAs in the nervous system in vivo (Aprea, Prenninger et al. 2013, Ramos, Diaz et al. 2013, Sauvageau, Goff et al. 2013). In one study, researchers combined laser capture microdissection with RNA-sequencing to profile the expression of lncRNAs in the adult mouse subventricular zone (SVZ), uncovering over 3,000 completely novel lncRNAs and identifying complex isoforms of many others (Ramos, Diaz et al. 2013). By combining expression data with chromatin state maps, these researchers were also able to identify a subset of approximately 100 lncRNAs with putative roles as regulators of neural development. These lncRNAs exhibit the bivalent (H3K27me3 and H3K4me3, repressed but poised) chromatin state in embryonic stem cells that resolves to H3K4me3-only (active expression) specifically in SVZ-derived neural stem cells (SVZ-NSCs) (Ramos, Diaz et al. 2013). The set of genes that are bivalent in embryonic stem cells is highly enriched for lineage-determining factors, which has led to the hypothesis that bivalency pre-patterns lineage specification. Indeed, chromatin state maps have been used successfully to identify novel proteins that act as regulators of cell fate in a variety of tissue contexts (Paige, Thomas et al. 2012, Wamstad, Alexander et al. 2012, Xu and Zaret 2012, Xie, Schultz et al. 2013). Application of this chromatin state model to predict functional lncRNAs in the adult SVZ also seems fruitful. For example, two lncRNAs identified as putative regulators of neural cell fate on the basis of chromatin state transitions during neural development, Six3os and Dlx1as, were found to increase the production of astrocytes at the expense of neurons when depleted from cultured SVZ-NSCs (Ramos, Diaz et al. 2013).
Long, non-coding RNAs may influence recruitment of PRC2 in neural development
Although investigations are ongoing, the section above highlights roles that have already been defined for lncRNAs in neural and glial development. Evidence in multiple contexts also indicates that some lncRNAs can control gene expression by recruiting transcription factors and complexes that modify chromatin, including Polycomb complexes, to their genomic targets. The original inspiration for a lncRNA-based model of Polycomb complex recruitment is the lncRNA Xist, which directly recruits PRC2 in cis to initiate the process of X chromosome inactivation in mammals (Engreitz, Pandya-Jones et al. 2013, Froberg, Yang et al. 2013). The idea that lncRNAs might contribute to epigenetic regulation by PcG complexes subsequently led to the identification of the lncRNA Hotair, which is transcribed from the HoxD locus and directly binds PRC2 to regulate the HoxC locus in trans (Rinn, Kertesz et al. 2007, Chu, Qu et al. 2011, Li, Liu et al. 2013). These findings stimulated additional work investigating the entire population of RNAs bound by PRC2. RNA immunoprecipitation followed by microarray analysis (RIP-chip) or sequencing (RIP-seq) of PRC2 components including Ezh2 suggest that as many as 20% of the known lncRNAs bind to PRC2 (Khalil, Guttman et al. 2009, Zhao, Ohsumi et al. 2010). These observations, combined with the fact that lncRNAs exhibit highly tissue and cell-type specific expression patterns, have led researchers to propose an attractive model, whereby the changing expression of cell type-specific lncRNAs as development progresses alters the recruitment of PRC2 and other chromatin modifiers to their targets (Lee 2012, Nakagawa and Kageyama 2014).
The role of lncRNAs as general recruiters of PRC2 remains controversial, primarily due to the limited availability of experimental evidence directly supporting the model. The overwhelming number of putative RNA-PRC2 interactions predicted by RIP has led some to question the specificity of the assay. Indeed, native RIP (RIP performed without crosslinking, as was performed in (Khalil, Guttman et al. 2009, Zhao, Ohsumi et al. 2010)) suffers from a notoriously high background signal (Mili and Steitz 2004, Brockdorff 2013). While these caveats and limitations must be kept in mind, high-stringency RIP of PRC2 from cells fixed by a variety of strategies (including formaldehyde fixation and UV crosslinking) recapitulates the large number of PRC2-interacting RNAs found by prior studies (Guttman, Donaghey et al. 2011, Davidovich, Zheng et al., Kaneko, Bonasio et al. 2013). A substantial fraction of the identified RNAs are therefore likely to be bona fide interactors with Polycomb, regardless of the functional relevance of this interaction or lack thereof.
Nevertheless, the overwhelming number of RNAs that bind PRC2 poses a significant challenge to experimentalists. No single RNA motif has been identified that mediates PRC2 binding. Although e.g. Xist and Hotair have specific sequence tracts that mediate binding (Cui, Zang et al. 2009, Engreitz, Pandya-Jones et al. 2013, Wu, Murat et al. 2013), these tracts are not particularly similar at the level of primary sequence. In the absence of a defined RNA motif, how does PRC2 recognize such a vast repertoire of RNAs, and how is the necessary specificity for a subset of functionally relevant RNAs achieved? Recent work from two different groups illustrates that PRC2 may have generic affinity for RNA that is important for its function (Davidovich, Zheng et al. 2013, Kaneko, Bonasio et al. 2013). In one study, it was demonstrated that mammalian PRC2 binds RNA promiscuously and with submicromolar affinity, including exogenous RNA from ciliates and bacteria (Davidovich, Zheng et al. 2013). While sequence appears to be largely irrelevant, size matters: longer RNAs are bound with higher affinity than shorter RNAs in a salt independent-manner, illustrating that the interaction with PRC2 is not merely electrostatic and may involve a more complex mechanism, such as base stacking with aromatic amino acid side chains (Davidovich, Zheng et al. 2013).
These data demonstrate the relatively high affinity of PRC2 for RNA and suggest its biological significance, but they also challenge the simplistic model of lncRNAs as a specialized class of molecular recruiter for PRC2. ChIP-seq verifies that most promoters with Ezh2 are enriched for H3K27me3 and correspond to repressed genes, while RIP-seq indicates that Ezh2 tends to bind nascent RNAs from actively transcribed genes, where H3K27me3 is absent (Davidovich, Zheng et al. 2013, Kaneko, Bonasio et al. 2013). These observations suggest that PRC2 may globally survey transcription by interacting with nascent RNAs – including protein coding and noncoding transcripts – while actual repression only occurs where other factors (e.g., the local chromatin context) are conducive to PRC2 transferring from RNA to chromatin.
On top of this basic level of regulation by transcriptional surveillance, it is clear that some lncRNAs – e.g., Xist and Hotair – have evolved to achieve higher affinity and specificity for PRC2. Both Xist and Hotair have specific domains that are required for their interaction with PRC2, suggesting that their structures are optimized to preferentially recruit PRC2 with higher affinity and thereby modify the distribution of PRC2 across the genome in biologically significant ways (Cui, Zang et al. 2009, Engreitz, Pandya-Jones et al. 2013, Wu, Murat et al. 2013). Given the diversity of processes regulated by these two transcripts alone, as well as the sheer abundance of lncRNAs that bind PRC2, it is highly possible that other RNAs have evolved a similar, uniquely potent affinity for PRC2 that may modulate its activity in other contexts. Such a mechanism might be particularly important in the nervous system, where lncRNA transcription is exceptionally abundant and complex, and where chromatin-interacting lncRNAs that are essential for neurogenesis have already been identified in vitro and in vivo (Dinger, Amaral et al. 2008, Derrien, Johnson et al. 2012, Ng, Bogu et al. 2013, Ramos, Diaz et al. 2013, Sauvageau, Goff et al. 2013).
Identifying the subset of “high affinity” PRC2-recruiting lncRNAs, should they exist, is an ongoing challenge. To truly demonstrate a direct role for a given RNA in the recruitment of a chromatin complex like PRC2, it is not sufficient to simply illustrate that a given RNA binds to PRC2 or that its depletion affects deposition of H3K27me3. For example, some lncRNAs have been shown to antagonize DNA methylation by inhibiting the activity of DNMT1, and lncRNA binding has been also been suggested to modulate the function of other chromatin complexes, underscoring the myriad ways that expression of a given lncRNA might be linked to PRC2 recruitment through any of several alternative models discussed in this review (Wang, Yang et al. 2011, Di Ruscio, Ebralidze et al. 2013). Distinguishing between direct recruitment of PRC2 and the other complex ways in which lncRNAs can regulate chromatin structure will necessitate experimental evidence that a putative RNA recruitment factor is located in physical proximity to the supposed sites of recruitment, as has been demonstrated for the lncRNAs Xist and Hotair (Chu, Qu et al. 2011, Engreitz, Pandya-Jones et al. 2013). Many groups have reported technical advancements that will facilitate future research, including a variety of techniques involving the use of tiled, biotinylated oligonucleotide probes to immunoprecipitate cross-linked lncRNA-chromatin complexes, which can be combined with high throughput DNA sequencing to characterize the genome-wide localization of a lncRNA (Simon, Wang et al. 2011, Chu, Quinn et al. 2012, Engreitz, Pandya-Jones et al. 2013). Annotation of the cistrome of PRC2-interacting lncRNAs in neural and glial progenitors and their derivatives will provide invaluable information for modeling how and to what extent lncRNAs contribute to PcG complex function in neural development, and may shed some light on the rich complexity of the neural lncRNA transcriptome.
Summary
The multiplicity of Polycomb activities in the developing nervous system is readily apparent, even from the relatively limited data available. To date, these activities include maintaining the balance between neural progenitor cell self-renewal and the onset of neurogenesis in the cortex, promoting the transition from neurogenesis to gliogenesis, directing the astrocyte versus oligodendrocyte precursor fate switch, and even regulating transcriptional events in terminally differentiated neurons. Although more work is required to tease apart the exact functions of individual PcG proteins at different stages of neural development, it is clear that the dynamic activity of Polycomb complexes contributes to the regulation of neural progenitor fate specification at nearly every step. Exactly how Polycomb complexes are recruited to regulate unique targets across the spectrum of neural fates is among the most important, unanswered questions in the field.
In recent years, a number of potential mechanisms have emerged that can influence the dynamic recruitment of Polycomb to chromatin. These mechanisms include facilitators of Polycomb recruitment, such as unmethylated CGI sequences and the proteins that maintain their unmethylated status, protein recruitment factors that can sense the local chromatin environment to influence the recruitment of PcG complexes in a context-dependent manner, and long non-coding RNAs. Additionally, the targeting of Polycomb occurs in a milieu of multiple chromatin modifications that can directly or indirectly modulate the recruitment process, consistent with the increasingly appreciated multidimensional complexity of chromatin structure. However, much of what is known about Polycomb recruitment is derived from in vitro studies of embryonic stem cells, an extremely useful model that nevertheless cannot fully recapitulate the complexity of Polycomb activity during the generation of complex tissues like the nervous system.
Recent work, such as the finding that Chd4 promotes neurogenesis by specifically recruiting PRC2 to silence gliogenic target genes, underscores the fact that unique PcG recruitment mechanisms are used to specify distinct neural cell fates. Continued research to identify these unique methods of PcG recruitment is a necessary precursor to the design of experimental and therapeutic approaches that can target specific aspects of PcG recruitment, rather than globally affecting PcG activity. For instance, the outcome of cortical progenitor differentiation is not dictated solely by the absolute and relative concentrations of instructive extracellular signals, but also by an intrinsic mechanism that selectively recruits Polycomb to specific targets in order to modify the response of the cells to such signals. Targeted manipulation of PcG recruitment should therefore enable more precise control of the timing and efficiency of neural progenitor differentiation into therapeutically relevant cell-types than would be achievable by manipulating extracellular signaling pathways alone.
As an example, current protocols for the production of OPCs from human neural progenitors can take months, with relatively low efficiency yields. Manipulating PcG recruitment to accelerate the developmental ‘clock’ could potentially permit more rapid production of homogenous populations of OPCs for use in applications like in vitro disease modeling, or the development of cell transplantation therapies. Therefore, it seems likely that expanding our knowledge of how PcG proteins are regulated during neural fate acquisition will simultaneously expand the molecular toolkit available for this research and its application to regenerative medicine.
Acknowledgments
We thank Kesavan Meganathan for providing photographs of hESCs undergoing directed differentiation (Figure 1) and Ethan Patterson, Bryan Teets, Laura Waller, and Kesavan Meganathan for critical reading of the manuscript. This work was funded by grants from the NIH (GM66815), the March of Dimes (FY13-413), and the Association for Research on Childhood Cancer to K.K. and the Cell and Molecular Biology Training Grant (GM007067) to M.C.
References
- Alder O, Lavial F, Helness A, Brookes E, Pinho S, Chandrashekran A, Arnaud P, Pombo A, O’Neill L, Azuara V. Ring1B and Suv39h1 delineate distinct chromatin states at bivalent genes during early mouse lineage commitment. Development. 2010;137(15):2483–2492. doi: 10.1242/dev.048363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloia L, Di Stefano B, Sessa A, Morey L, Santanach A, Gutierrez A, Cozzuto L, Benitah SA, Graf T, Broccoli V, Di Croce L. Zrf1 is required to establish and maintain neural progenitor identity. Genes Dev. 2014;28(2):182–197. doi: 10.1101/gad.228510.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprea J, Prenninger S, Dori M, Ghosh T, Monasor LS, Wessendorf E, Zocher S, Massalini S, Alexopoulou D, Lesche M, Dahl A, Groszer M, Hiller M, Calegari F. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32(24):3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L. Epigenetic control of embryonic stem cell differentiation. Stem Cell Rev. 2012;8(1):67–77. doi: 10.1007/s12015-011-9300-4. [DOI] [PubMed] [Google Scholar]
- Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004;117(Pt 19):4355–4363. doi: 10.1242/jcs.01390. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8(5):532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157(6):1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278(5337):477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Brinkman AB, Gu H, Bartels SJ, Zhang Y, Matarese F, Simmer F, Marks H, Bock C, Gnirke A, Meissner A, Stunnenberg HG. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22(6):1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, Kimura H, Ragoussis J, Teichmann SA, Pombo A. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10(2):157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SWM, Valk-Lingbeek ME, van der Stoop PPM, Jacobs JJL, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 2005;19(12):1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS ONE. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgold T, Voituron N, Caganova M, Tripathi PP, Menuet C, Tusi BK, Spreafico F, Bevengut M, Gestreau C, Buontempo S, Simeone A, Kruidenier L, Natoli G, Casola S, Hilaire G, Testa G. The H3K27 demethylase JMJD3 is required for maintenance of the embryonic respiratory neuronal network, neonatal breathing, and survival. Cell Rep. 2012;2(5):1244–1258. doi: 10.1016/j.celrep.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y-I, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Chan THM, Chen L, Liu M, Hu L, Zheng B-j, Poon VK-M, Huang P, Yuan Y-F, Huang J-d, Yang J, Tsao GS-w, Guan X-Y. Translationally controlled tumor protein induces mitotic defects and chromosome missegregation in hepatocellular carcinoma development. Hepatology. 2012;55:491–505. doi: 10.1002/hep.24709. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012;(61) doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. Elife. 2012;1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, Heger A, Brockdorff N. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014;7(5):1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MR, Bucholz O, Schroeder T, Gotz M. Late origin of glia-restricted progenitors in the developing mouse cerebral cortex. Cereb Cortex. 2009;19(Suppl 1):i135–143. doi: 10.1093/cercor/bhp046. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4(1):80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Dusenbery BN, Williams LA, Klim JR, Eggan K. How to make spinal motor neurons. Development. 2014;141(3):491–501. doi: 10.1242/dev.097410. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Derouet D, Rousseau F, Alfonsi F, Froger J, Hermann J, Barbier F, Perret D, Diveu C, Guillet C, Preisser L, Dumont A, Barbado M, Morel A, deLapeyriere O, Gascan H, Chevalier S. Neuropoietin, a new IL-6-related cytokine signaling through the ciliary neurotrophic factor receptor. Proc Natl Acad Sci U S A. 2004;101(14):4827–4832. doi: 10.1073/pnas.0306178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meglio T, Kratochwil CF, Vilain N, Loche A, Vitobello A, Yonehara K, Hrycaj SM, Roska B, Peters AH, Eichmann A, Wellik D, Ducret S, Rijli FM. Ezh2 orchestrates topographic migration and connectivity of mouse precerebellar neurons. Science. 2013;339(6116):204–207. doi: 10.1126/science.1229326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alo F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan CM, Nyman U, Skotte J, Streubel G, Turner S, O’Connell DJ, Rraklli V, Dolan MJ, Chadderton N, Hansen K, Farrar GJ, Helin K, Holmberg J, Bracken AP. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev Cell. 2013;26:223–236. doi: 10.1016/j.devcel.2013.07.008. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, Vidal M, Bernstein BE, Koseki H. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8(7):e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farcas AM, Blackledge NP, Sudbery I, Long HK, McGouran JF, Rose NR, Lee S, Sims D, Cerase A, Sheahan TW, Koseki H, Brockdorff N, Ponting CP, Kessler BM, Klose RJ. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. Elife. 2012;1:e00205. doi: 10.7554/eLife.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell stem cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 2009;23:561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1(2):137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306(5701):1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Froberg JE, Yang L, Lee JT. Guided by RNAs: X-Inactivation as a Model for lncRNA Function. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Tokunaga A, Sakamoto R, Yoshida N. Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol Cell Neurosci. 2011;46:614–624. doi: 10.1016/j.mcn.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45(3):344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García E, Marcos-Gutiérrez C, del Mar Lorente M, Moreno JC, Vidal M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999;18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspard N, Bouschet T, Hourez R, Dimidschstein J, Naeije G, van den Ameele J, Espuny-Camacho I, Herpoel A, Passante L, Schiffmann SN, Gaillard A, Vanderhaeghen P. An intrinsic mechanism of corticogenesis from embryonic stem cells. Nature. 2008;455(7211):351–357. doi: 10.1038/nature07287. [DOI] [PubMed] [Google Scholar]
- Golden MG, Dasen JS. Polycomb repressive complex 1 activities determine the columnar organization of motor neurons. Genes Dev. 2012;26(19):2236–2250. doi: 10.1101/gad.199133.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14(11):755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagarman JA, Motley MP, Kristjansdottir K, Soloway PD. Coordinate regulation of DNA methylation and H3K27me3 in mouse embryonic stem cells. PLoS One. 2013;8(1):e53880. doi: 10.1371/journal.pone.0053880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Ge W, Martinowich K, Becker-Catania S, Coskun V, Zhu W, Wu H, Castro D, Guillemot F, Fan G, de Vellis J, Sun YE. A positive autoregulatory loop of Jak-STAT signaling controls the onset of astrogliogenesis. Nat Neurosci. 2005;8(5):616–625. doi: 10.1038/nn1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada YI, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008 doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Shen L, Wan M, Taranova O, Wu H, Zhang Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nat Cell Biol. 2013;15(4):373–384. doi: 10.1038/ncb2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y. Stage-dependent fate determination of neural precursor cells in mouse forebrain. Neurosci Res. 2005;51(4):331–336. doi: 10.1016/j.neures.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131(12):2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hisada K, Sanchez C, Endo TA, Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol Cell Biol. 2012;32(6):1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Current Opinion in Cell Biology. 2005;17(6):664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Hwang WW, Salinas RD, Siu JJ, Kelley KW, Delgado RN, Paredes MF, Alvarez-Buylla A, Oldham MC, Lim DA. Distinct and separable roles for EZH2 in neurogenic astroglia. Elife. 2014;3:e02439. doi: 10.7554/eLife.02439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israsena N, Hu M, Fu W, Kan L, Kessler JA. The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev Biol. 2004;268:220–231. doi: 10.1016/j.ydbio.2003.12.024. [DOI] [PubMed] [Google Scholar]
- Johe KK, Hazel TG, Muller T, Dugich-Djordjevic MM, McKay RD. Single factors direct the differentiation of stem cells from the fetal and adult central nervous system. Genes Dev. 1996;10(24):3129–3140. doi: 10.1101/gad.10.24.3129. [DOI] [PubMed] [Google Scholar]
- Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Junco SE, Wang R, Gaipa JC, Taylor AB, Schirf V, Gearhart MD, Bardwell VJ, Demeler B, Hart PJ, Kim CA. Structure of the polycomb group protein PCGF1 in complex with BCOR reveals basis for binding selectivity of PCGF homologs. Structure. 2013;21(4):665–671. doi: 10.1016/j.str.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and Noncoding RNAs Regulate PRC2 Recruitment to Chromatin. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Ekram MB, Roh T-Y, Kim J. Aebp2 as an epigenetic regulator for neural crest cells. PLoS ONE. 2011;6:e25174. doi: 10.1371/journal.pone.0025174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama H, Zhuang T, Light JE, Kolla V, Higashi M, McGrady PW, London WB, Brodeur GM. Mechanisms of CHD5 Inactivation in neuroblastomas. Clin Cancer Res. 2012;18(6):1588–1597. doi: 10.1158/1078-0432.CCR-11-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Nasu R, David G, Tanese N. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat Cell Biol. 2007;9(9):1074–1080. doi: 10.1038/ncb1628. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, Brookes E, Pombo A, Fisher C, Skarnes WC, Snoek T, Bezstarosti K, Demmers J, Klose RJ, Casanova M, Tavares L, Brockdorff N, Merkenschlager M, Fisher AG. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12(6):618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Liu B, Wapinski OL, Tsai M-C, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458(7237):529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch MD, Smith AJ, De Gobbi M, Flenley M, Hughes JR, Vernimmen D, Ayyub H, Sharpe JA, Sloane-Stanley JA, Sutherland L, Meek S, Burdon T, Gibbons RJ, Garrick D, Higgs DR. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2012;31(2):317–329. doi: 10.1038/emboj.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, Pirrotta V, Reinberg D, Gamblin SJ. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461(7265):762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynoga B, Drechsel D, Guillemot F. Molecular control of neurogenesis: a view from the mammalian cerebral cortex. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Mahony S, Peljto M, Patel T, Thornton SR, McCuine S, Reeder C, Boyer LA, Young RA, Gifford DK, Wichterle H. Saltatory remodeling of Hox chromatin in response to rostrocaudal patterning signals. Nat Neurosci. 2013;16(9):1191–1198. doi: 10.1038/nn.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E. Chromatin in embryonic stem cell neuronal differentiation. Histol Histopathol. 2007;22:311–319. doi: 10.14670/HH-22.311. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10(1):105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10(11):1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park I-K, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15(10):942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- MuhChyi C, Juliandi B, Matsuda T, Nakashima K. Epigenetic regulation of neural stem cell fate during corticogenesis. Int J Dev Neurosci. 2013;31:424–433. doi: 10.1016/j.ijdevneu.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Kageyama Y. Nuclear lncRNAs as epigenetic regulators-Beyond skepticism. Biochim Biophys Acta. 2014;1839(3):215–222. doi: 10.1016/j.bbagrm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284(5413):479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Narayanan R, Tuoc TC. Roles of chromatin remodeling BAF complex in neural differentiation and reprogramming. Cell Tissue Res. 2014;356(3):575–584. doi: 10.1007/s00441-013-1791-7. [DOI] [PubMed] [Google Scholar]
- Ng S-Y, Bogu GK, Soh B-S, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Ng S-Y, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S-Y, Lin L, Soh B-S, Stanton LW. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet. 2013;29:461–468. doi: 10.1016/j.tig.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508(1):28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loghlen A, Munoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, Masui O, Vermeulen M, Carroll T, Graumann J, Heard E, Dillon N, Azuara V, Snijders AP, Peters G, Bernstein E, Gil J. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell. 2012;10(1):33–46. doi: 10.1016/j.stem.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olynik BM, Rastegar M. The genetic and epigenetic journey of embryonic stem cells into mature neural cells. Front Genet. 2012;3:81. doi: 10.3389/fgene.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, Moon RT, Stamatoyannopoulos J, Murry CE. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151(1):221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, Helin K. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JD, Sansom SN, Smith J, Dobenecker M-W, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55(3):417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80(1):12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirity MK, Locker J, Schreiber-Agus N. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol Cell Biol. 2005;25(16):7193–7202. doi: 10.1128/MCB.25.16.7193-7202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C, Liu S, Qin R, Zhang Y, Wang G, Shang Y, Wang Y, Liang J. Coordinated Regulation of Dendrite Arborization by Epigenetic Factors CDYL and EZH2. J Neurosci. 2014;34(13):4494–4508. doi: 10.1523/JNEUROSCI.3647-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Long Non-coding RNAs: Novel Targets for Nervous System Disease Diagnosis and Therapy. Neurotherapeutics. 2013 doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, McKay RD. Multiple routes to astrocytic differentiation in the CNS. J Neurosci. 1998;18(10):3620–3629. doi: 10.1523/JNEUROSCI.18-10-03620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AD, Diaz A, Nellore A, Delgado RN, Park K-Y, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of Genome-wide Approaches Identifies lncRNAs of Adult Neural Stem Cells and Their Progeny In Vivo. Cell Stem Cell. 2013;12:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012;31(3):593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román-Trufero M, Méndez-Gómez H, Pérez C, Hijikata A, Fujimura Y, Endo T, Koseki H, Vicario-Abejón C, Vidal M. Maintenance of Undifferentiated State and Self-Renewal of Embryonic Neural Stem Cells By Polycomb Protein Ring1B. Stem Cells. 2009 doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C, Sánchez I, Demmers JAA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, Liapis SC, Mallard W, Morse M, Swerdel MR, D’Ecclessis MF, Moore JC, Lai V, Gong G, Yancopoulos GD, Frendewey D, Kellis M, Hart RP, Valenzuela DM, Arlotta P, Rinn JL. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W, Müller J, Thomä NH. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan G-C, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Boddeke E, Olah M, Copray S. Dynamic changes in Ezh2 gene occupancy underlie its involvement in neural stem cell self-renewal and differentiation towards oligodendrocytes. PLoS ONE. 2012;7:e40399. doi: 10.1371/journal.pone.0040399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Rössler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- Shimogori T, VanSant J, Paik E, Grove EA. Members of the Wnt, Fz, and Frp gene families expressed in postnatal mouse cerebral cortex. J Comp Neurol. 2004;473(4):496–510. doi: 10.1002/cne.20135. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proceedings of the National Academy of Sciences. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27(24):2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statham AL, Robinson MD, Song JZ, Coolen MW, Stirzaker C, Clark SJ. Bisulfite sequencing of chromatin immunoprecipitated DNA (BisChIP-seq) directly informs methylation status of histone-modified DNA. Genome Res. 2012;22(6):1120–1127. doi: 10.1101/gr.132076.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104(3):365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Hawkins PG, Mattick JS, Morris KV. The relationship between transcription initiation RNAs and CCCTC-binding factor (CTCF) localization. Epigenetics Chromatin. 2011;4:13. doi: 10.1186/1756-8935-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Kojima M, Nakajima K, Suzuki-Migishima R, Takeuchi T. Functions of a jumonji-cyclin D1 pathway in the coordination of cell cycle exit and migration during neurogenesis in the mouse hindbrain. Dev Biol. 2007;303(2):549–560. doi: 10.1016/j.ydbio.2006.11.031. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Kojima M, Nakajima K, Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech Dev. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148(4):664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G. The time of timing: how Polycomb proteins regulate neurogenesis. Bioessays. 2011;33:519–528. doi: 10.1002/bies.201100021. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136(18):3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SA, Bracken AP. A “complex” issue: deciphering the role of variant PRC1 in ESCs. Cell Stem Cell. 2013;12(2):145–146. doi: 10.1016/j.stem.2013.01.014. [DOI] [PubMed] [Google Scholar]
- Tushir JS, Akbarian S. Chromatin-bound RNA and the neurobiology of psychiatric disease. Neuroscience. 2013 doi: 10.1016/j.neuroscience.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura A, Takizawa T, Ochiai W, Yanagisawa M, Nakashima K, Taga T. Cardiotrophin-like cytokine induces astrocyte differentiation of fetal neuroepithelial cells via activation of STAT3. Cytokine. 2002;18(1):1–7. doi: 10.1006/cyto.2002.1006. [DOI] [PubMed] [Google Scholar]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27(12):1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C, Cepko CL. Widespread dispersion of neuronal clones across functional regions of the cerebral cortex. Science. 1992;255(5043):434–440. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, Erwin G, Kattman SJ, Keller GM, Srivastava D, Levine SS, Pollard KS, Holloway AK, Boyer LA, Bruneau BG. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151(1):206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14(5):637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52(52):9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Johansen JV, Helin K. Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol Cell. 2013;49(6):1134–1146. doi: 10.1016/j.molcel.2013.01.016. [DOI] [PubMed] [Google Scholar]
- Wutz A. Epigenetic regulation of stem cells: the role of chromatin in cell differentiation. Adv Exp Med Biol. 2013;786:307–328. doi: 10.1007/978-94-007-6621-1_17. [DOI] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen C-a, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung W-Y, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell, Elsevier Inc. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, Yang H, Wang T, Lee AY, Swanson SA, Zhang J, Zhu Y, Kim A, Nery JR, Urich MA, Kuan S, Yen CA, Klugman S, Yu P, Suknuntha K, Propson NE, Chen H, Edsall LE, Wagner U, Li Y, Ye Z, Kulkarni A, Xuan Z, Chung WY, Chi NC, Antosiewicz-Bourget JE, Slukvin I, Stewart R, Zhang MQ, Wang W, Thomson JA, Ecker JR, Ren B. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153(5):1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Yang W, Galka M, Ouyang H, Chen C, Qiu W, Liu H, Jones AE, MacKenzie F, Pan P, Li SS-C, Wang H, Min J. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci USA. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CR, Zaret KS. Chromatin “pre-pattern” and epigenetic modulation in the cell fate choice of liver over pancreas in the endoderm. Nucleus. 2012;3(2):150–154. doi: 10.4161/nucl.19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu T, Kawaguchi D, Oishi K, Takeda K, Akira S, Masuyama N, Gotoh Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development. 2006;133(13):2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, Han Z, Chai J, Zhou XJ, Gao S, Zhu B. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science. 2012;337(6097):971–975. doi: 10.1126/science.1225237. [DOI] [PubMed] [Google Scholar]
- Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XQ, Leung GK. Long non-coding RNAs in glioma: Functional roles and clinical perspectives. Neurochem Int. 2014 doi: 10.1016/j.neuint.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1(3):286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zhu J, Adli M, Zou JY, Verstappen G, Coyne M, Zhang X, Durham T, Miri M, Deshpande V, De Jager PL, Bennett DA, Houmard JA, Muoio DM, Onder TT, Camahort R, Cowan CA, Meissner A, Epstein CB, Shoresh N, Bernstein BE. Cell. Vol. 152. Elsevier Inc; 2013. Genome-wide Chromatin State Transitions Associated with Developmental and Environmental Cues; pp. 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziats MN, Rennert OM. Aberrant expression of long noncoding RNAs in autistic brain. J Mol Neurosci. 2013;49(3):589–593. doi: 10.1007/s12031-012-9880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]


