Abstract
Understanding the evolution of the neurosensory system of man, able to reflect on its own origin, is one of the major goals of comparative neurobiology. Details of the origin of neurosensory cells, their aggregation into central nervous systems and associated sensory organs, their localized patterning into remarkably different cell types aggregated into variably sized parts of the central nervous system begin to emerge. Insights at the cellular and molecular level begin to shed some light on the evolution of neurosensory cells, partially covered in this review. Molecular evidence suggests that high mobility group (HMG) proteins of pre-metazoans evolved into the definitive Sox [SRY (sex determining region Y)-box] genes used for neurosensory precursor specification in metazoans. Likewise, pre-metazoan basic helix-loop-helix (bHLH) genes evolved in metazoans into the group A bHLH genes dedicated to neurosensory differentiation in bilaterians. Available evidence suggests that the Sox and bHLH genes evolved a cross-regulatory network able to synchronize expansion of precursor populations and their subsequent differentiation into novel parts of the brain or sensory organs. Molecular evidence suggests metazoans evolved patterning gene networks early and not dedicated to neuronal development. Only later in evolution were these patterning gene networks tied into the increasing complexity of diffusible factors, many of which were already present in pre-metazoans, to drive local patterning events. It appears that the evolving molecular basis of neurosensory cell development may have led, in interaction with differentially expressed patterning genes, to local network modifications guiding unique specializations of neurosensory cells into sensory organs and various areas of the central nervous system.
Keywords: basic helix-loop-helix genes, Sox genes, transcription factors, neurosensory, central nervous system, cellular evolution
Introduction
The human brain consists of over 80 billion neurons and similar numbers of glia cells (Herculano-Houzel, 2010), engaged in trillions of synapses to process the information gathered by several sensory organs to respond appropriately to environmental stimuli. While the human brain is now recognized as an organ of extraordinary relative and absolute size and packing density of neurons (Herculano-Houzel, 2012), its origin as a vertebrate adaptation for information processing has led to various ideas mostly revolving around single or multiple origins of the central nervous system (CNS) among metazoans (Nieuwenhuys, 2002, Northcutt, 2012), and subsequent increase and modification of all or parts of the CNS (Nieuwenhuys, et al., 1998, Striedter, 2005). In parallel to this traditional interpretation of macroscopic neuronal evolution, the last 20 years have witnessed the generation of another set of data and associated ideas concentrating on molecular and cellular aspects of neuronal evolution as a variation on the theme of ectodermal cellular diversification (Beccari, et al., 2013, Fritzsch and Glover, 2006, Pani, et al., 2012), integrating neurosensory cellular evolution with the evolution of the major sensory systems, eyes and ears (Fritzsch and Straka, 2014, Lamb, 2013, Patthey, et al., 2014, Schlosser, et al., 2014). More recently, the molecular origin of evolutionary innovations (Wagner, 2011) such as those leading to formation of neurosensory cells and their aggregation into a brain and associated sensory organs begin to direct the deluge of genomic data into a theoretical framework of Darwinian evolution of neurosensory systems (Newman, 2014). Given that only 14 years have passed since the human genome was published, it is clear that a complete understanding of the different levels of neurosensory evolution from genes, to developmental gene regulatory networks, to neurosensory cell and sensory organ development and CNS evolution is beyond the reach of our current understanding as depicted in this short overview. Admittedly, despite tremendous gains, our understanding of gene regulatory networks and their evolution to govern complex macroscopic phenotype changes is only in its infancy (Sauka-Spengler and Bronner-Fraser, 2008, Streit, et al., 2013). As of this writing, our ability to translate mutations at the DNA level into altered phenotypes is not yet deep enough to relate single nucleotide changes causing, for example, a different folding of proteins to macroscopic changes comparable to the morphological alterations of hemoglobin in sickle cell anemia. Despite all the gain in detailed information, we are unable to mechanistically explain how limited sequence differences between protein isoforms can cause major diseases (Forrest, et al., 2014) or how differences of humans and chimpanzees of around 1% can lead to the profound macroscopic differences characterizing these two species.
With this caveat in mind, this overview aims to define some major steps relevant for the evolution of neurosensory systems by casting data on both macroscopic and molecular evolution into a framework of developmental gene regulatory network evolution acting at the cellular level. The presentation adopts the perspective of Wagner (Wagner, 2011) that an intermediate level of abstraction is paramount for human understanding of otherwise hopelessly entangled innovations across all molecules and levels of analysis. The level of abstraction attempted here will provide pa ‘neurocentric’ view of the evolution and development of neurosensory systems of bilaterians. To achieve this, we will progress from the evolution of general cell fate decision making networks to special cases of limited complexity of lineage related differential cell fate decisions (inner ear neurons and hair cells) to overall patterning with and without mesodermal induction. Another layer of complexity, related to the gene expression regulation (Davidson, 2010) and differential splicing in neurosensory cells (Nakano, et al., 2012, Raj, et al., 2014), is beyond the scope of this review.
1. Evolving single cells to multicellular organisms with dedicated neurosensory cells
The origin and molecular repertoire of single and multicellular organisms has been worked out over the last 10 years (Fig. 1): it is commonly agreed that all multicellular animals, including their brains, have evolved from single-celled ancestors comparable to choanoflagellates (Fairclough, et al., 2013, King, 2004) and other related single-celled organisms (Sebé-Pedrós, et al., 2013, Suga, et al., 2013). These single-celled organisms can form transient aggregates with complex life cycle changes (Levin, et al., 2014, Sebé-Pedrós, et al., 2013) and already contain many genes previously speculated to be unique to metazoans. While single cells, by definition, do not have specialized neurosensory cells, it seems plausible that some gene regulatory network present only in unicellular organisms leading to metazoans, but not those leading to plants or fungi, should have evolved into the gene regulatory network driving the neurosensory cell development of metazoans. In short, we speculate here that the cellular basis of brain and sensory organ evolution lies in the multiplication, diversification and specialization of gene networks (Wagner, 2011) that evolved in single-celled precursors of metazoans to guide temporal changes in transient multicellular organization associated with reproduction related life cycles. In essence, metazoans may have evolved through incompletely understood innovations of the single-celled metazoan ancestors the molecular repertoire to develop sophisticated developmental fate decision processes (Lai, et al., 2013, Stergachis, et al., 2013) that generate specific cells capable of collecting and processing information. Whatever the details of this process are, it turned the single-celled metazoan ancestors, fully capable of sensing their environment and responding to it in an appropriate way to ensure survival of the species (Dayel, et al., 2011, Fairclough, et al., 2013, Sebé-Pedrós, et al., 2013, Suga, et al., 2013), into an aggregate of cells with a segregation of the sensory and information processing aspect to morphologically distinct neurosensory cell types. Ultimately, such neurosensory cells grouped into the CNS and associated sensory organs (eyes, ears, nose). The evolution of gene networks past single-celled ancestors now governs the transformation of the single fertilized egg at the start of each metazoans life cycle into multiple distinct neurosensory cell types through the topological and temporal precise expression of gene regulatory networks that govern neuronal (Beccari, et al., 2013, Puelles, et al., 2013) and sensory organ evolution (Fortunato, et al., 2014, Fritzsch and Straka, 2013).
Fig. 1.
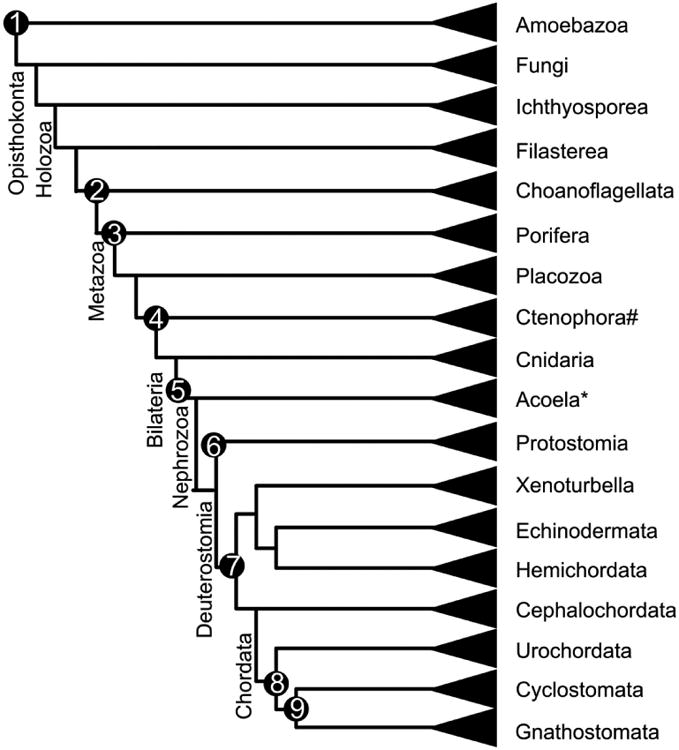
This figure displays the relationship of single-celled and multicellular organisms and some critical events concerning the evolution of neurons and sensory systems. 1) Complicated life cycle with transient multicellularity; 2) most cellular communication signals are present, of Sox like and bHLH genes are present 3) class A bHLH genes and SoxB genes that can induce neurons are present. 4) epithelial nerve nets and sensory organs evolve; 5) miR-124 specific for neurons and miR-183 specific for sensory cells appear; 6) ventral central nervous system evolves; 7) ‘skin brains’ with chordate like patterning evolve; 8) neurons are concentrated in a dorsal neural tube and composite, organ like sensory cell groups appear. 9) Vertebrate sensory organs and nervous system appear. # note that the position of Ctenophora is disputed. *Acoela are sometimes combined with Xenoturbella, indicating perhaps limited molecular distinction between basic bilaterians and basic deuterostomes. Compiled after (Gyoja, 2014, Osigus, et al., 2013, Peterson, et al., 2013, Schnitzler, et al., 2014, Suga, et al., 2013, Swalla and Smith, 2008)
Integrated into the topological expression regulation is the sequential gene activation that governs coordinated transitions between different stages of cell fate commitment, a process dominated by two closely interacting partners, the (SRY (sex determining region Y) box (Sox) genes (Guth and Wegner, 2008) and the bHLH genes (Degnan, et al., 2009, Simionato, et al., 2007). Metazoan Sox and bHLH genes associated with neurosensory development seem to have no precursors in single-celled metazoan ancestors (Guth and Wegner, 2008, Neriec and Desplan, 2014). However, both Sox gene-related and bHLH gene-related precursors with similarities in DNA binding sites are well known in single cell organisms (Albert, et al., 2013, Gordan, et al., 2013) and are also found in plants (Ikeda, et al., 2012, Xia, et al., 2014). In fact, single bHLH genes such as Tcf25 (Nulp1) are extremely conserved http://www.ensembl.org/Mus_musculus/Gene/Compara_Tree?collapse=6638243%2C6638340%2C6638209%2C6638328%2C6638179%2C6638173%2C6638249%2C6638233%2C6638296;db=core;g=ENSMUSG00000001472;r=8:123373824-123404173 and play a significant role in sensory function (Wolber, et al., 2014). Combined these data suggest that indeed transcription factors that may have signaled in single-celled organism and evolved into a gene regulatory network dedicated to metazoan neurosensory cell development. We propose that gene regulatory networks of metazoan neurosensory development could have originated in DNA binding transcription factors that may already form an interactive network in single cells to regulate specific vegetative or generative states of a cell.
1.A. Evolving bHLH and Sox gene networks to specify neurosensory cell types
Single celled organisms have the molecular machinery to transit through vegetative and generative stages of their life cycle, using molecular cascades to progress into or forgo mitosis, or change cellular communication to engage in multicellularity with or without sexual reproduction. Such intracellular molecular interactions, once evolved in single-celled organisms to define temporal progression of specific stages in the life of one cell, may have evolved to form the basis for the equally fundamental decision of any given cell during development of multicellular organism: to proliferate or to differentiate. It appears that evolution picked bHLH and Sox-related transcription factors to translate such cellular decision processes into action already in single-celled ancestors (Sebé-Pedrós, et al., 2013). Consistent with this assumption is that during evolution, bHLH and Sox proteins and their DNA binding sites have been mostly multiplied and diversified (Degnan, et al., 2009, Guth and Wegner, 2008, Pan, et al., 2012) and in particular, bHLH genes govern in extant metazoans the progression of individual cells toward their differentiation (Fig. 2). This ensures that multiple cell types can be differentiated through division of labor between cell types (Arendt, et al., 2009), such as stimuli acquisition and information conductance and processing in sensory organs (Pan, et al., 2012). Modifying such cell fate determining genes through topologically restricted transcription factors may have enabled differentiation of specific cell types in specific areas, ultimately evolving into cellular assemblies serving a specific function. Understanding cellular diversification requires an understanding of the regulation of cell fate decision making proteins in specific cell types (Lai, et al., 2013, Stergachis, et al., 2013) that interact to develop the cells needed for the function of a specific cellular assembly.
Fig. 2.
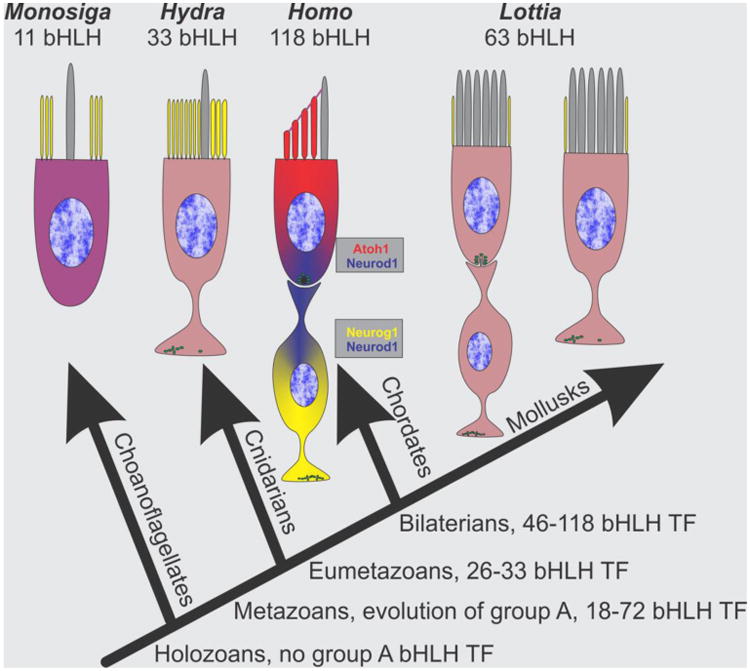
Available data on basic helix-loop-helix transcription factors (bHLH TFs) relevant for the evolution of human hair cells and sensory neurons are shown. Note that Monosiga has 11 bHLH TFs, none of which are orthologous to Metazoans. Eumetazoans have sensory cells with axons and display asymmetric distribution of microvilli (yellow) and kinocilium (gray). In mammals, the three bHLH TFs are partially overlapping to drive neuronal (Neurog1, Neurod1) and hair cell (Atoh1, Neurod1) development. A superficially similar arrangement of sensory cells and sensory neurons is found in some mollusks but which bHLH genes are expressed in these cells is unknown. Given the distribution in protostomes and deuterostomes, mollusk and vertebrates sensory cell evolution without an axon likely indicates functional conservation. Modified after (Pan, et al., 2012).
The importance of the evolution of this cellular decision making network of group A bHLH genes (Fig. 2) in interaction with the Sox genes for eyes, ears and the CNS cannot be underestimated. For example only four bHLH genes and their orthologs (Ascl1, Atoh1/7, Neurog1/2 and Neurod1) are necessary for the development of ∼90% of the neurons in the mammalian brain [cerebellum, cochlear nuclei, large areas of the cortex and sensory cells and neurons of the eye and ear (Bermingham, et al., 2001, Pan, et al., 2009)] and all sensory neurons bringing information into the brain. Indeed, most of the group A bHLH genes are associated with neurosensory development (Chen, et al., 2011, Imayoshi and Kageyama, 2014) and seem to be linked to the evolution of neurons (Fig. 2). It seems that the evolution of the Ascl-like family and Atoh-like family are among the earliest group A bHLH gene that evolved already in sponges out of bHLH genes present in unicellular organisms such as choanoflagellates (Fig. 2). Indeed, recent data suggests that group A bHLH transcription factors, relevant for neuronal development, exist in metazoans that have no trace of a nervous system, raising the issue of function of such neuronally-associated group A bHLH genes in sponges and placozoans (Gyoja, 2014). Understanding the evolution of the bHLH gene network and how it became associated with neurosensory development could provide insight into the cellular decision making that stably transforms metazoan ectoderm into neurosensory cells. In fact, bHLH genes cloned from sponges and injected into developing frog embryo can induce formation of neurons in ectoderm (Richards, et al., 2008) comparable to bHLH genes of vertebrates (Lee, et al., 1995). Recent data (Gyoja, 2014) suggest that the molecular evolution of group A bHLH genes, needed for neuronal differentiation, predate the cellular evolution of neurosensory cells (genes to differentiate neurons evolved before neurons). Alternatively, assuming that ctenophores are the sister group of all metazoans (Martindale, 2013, Martindale and Lee, 2013), the presence of these transcription factors in sponges and placozoans may indicate that regulatory network of these metazoans is either secondarily reduced or a parallel evolution to that found in ctenophores (Ryan, 2014).
Consistent with the data on group A bHLH genes (Gyoja, 2014), most recent work identified true Sox gene members of the high mobility group-box (HMG) group of transcription factors already in early metazoans (Schnitzler, et al., 2014). Experimental work in various bilaterians has shown that members of the SoxB, SoxC, SoxD and SoxF families are essential to specify and maintain neurosensory precursors through the clonal expansion of neurosensory precursor cells (NSP cells) to generate the cellular basis of neurosensory development (Guth and Wegner, 2008). In particular SoxB genes are essential to establish neuroectodermal precursor cell fate (Reiprich and Wegner, 2014), possibly by preparing cells to respond to bHLH genes with neurosensory differentiation (Bylund, et al., 2003). Closer examination shows a regulatory network interaction between Sox genes and bHLH genes such that Sox genes are not only essential to set the stage for bHLH genes to initiate differentiation. In many instances Sox genes counteract bHLH gene actions by maintaining progenitor status and enhance proliferation. In essence, SoxB genes and bHLH genes act like ‘frenemies’: SoxB genes are needed to set the stage for actions of bHLH genes, play some yet not fully determined role in their expression, but ultimately have to be inhibited by bHLH genes to allow neurosensory differentiation (Dabdoub, et al., 2008, Reiprich and Wegner, 2014). Both bHLH genes and Sox genes are essential for neurosensory development and their interactions within each transcription factor family (Imayoshi and Kageyama, 2014, Reiprich and Wegner, 2014) and between these transcription factors is essential for neuronal and neurosensory development of vertebrates (Fig. 3). The transition from Sox to bHLH gene expression may be regulated itself by antagonistic actions such as Gdf11 and other factors (Gokoffski, et al., 2011). Unfortunately, our insights into the evolution of neurosensory systems have not yet gone beyond identifying relevant orthologs of important transcription factors but not when these feedback loops evolved. Specifically, we need to show experimentally how the intricate gene regulatory network of feed-forward and feed-back loops evolved to regulate differentiation of the three principal cell types of the vertebrate nervous system: neurosensory cells, astrocytes and oligodendrocytes (Fig. 3).
Fig. 3. The interactions of Sox and bHLH genes in neurosensory differentiation of a mouse.
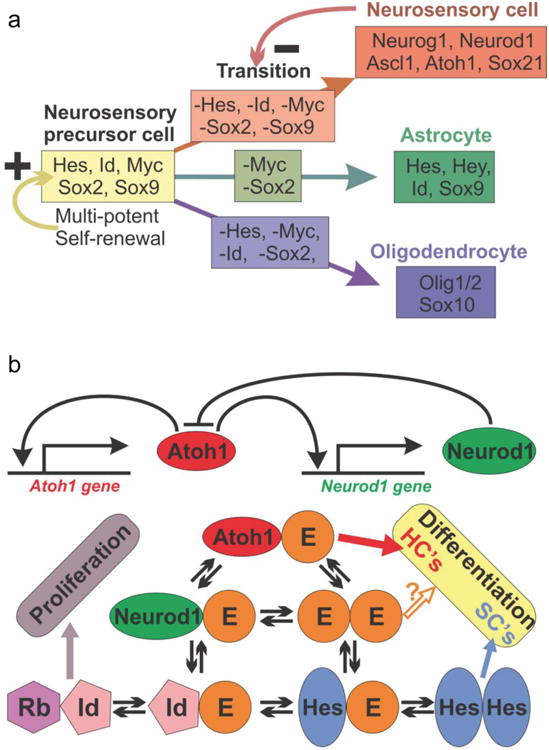
Experimental data in mice have shown a complex interaction of Sox genes and bHLH genes in the progression of neurosensory cell fate commitment and differentiation. a) Sox2 and Sox9 are essential genes in neurosensory precursor cells that ensure self-renewal of precursors but also commitment to the neurosensory lineage. This appears to be in interaction with several bHLH genes that are later found in astrocytes (Hes, Hey, Id). Virtually all of these neurosensory precursor genes are turned off in the neurosensory lineage (arrow with – on transition) and mostly bHLH genes are activated, including Sox21 that antagonizes Sox2. Oligodendrocyte precursors also shut off neurosensory precursor genes but are characterized by a different set of bHLH genes (Olig1/2) and Sox10 and Sox2 for terminal differentiation. bHLH transcription factors can form complex interactions in a given cell that can undergo periodic changes (oscillates) and their signal can undergo context dependent variation between gene expression and suppression. Data in mice and flies suggest that all proneural transcription factors compete for the E-proteins (Tcf3,4,12) to form heterodimers for proper binding. Thus, the level of all proneuronal bHLH TFs (here Atoh1 and Neurod1) and available E-proteins as well as their binding preference will determine how much signaling of heterodimers will occur. Importantly, E-proteins can also interact with Hes/Hey factors and the inhibitors of DNA binding (Ids), limiting availability of E-proteins for proneuronal protein heterodimerization, proportionally to the affinity and concentration of all these interactive partners. In essence, the binding properties and frequency of the binding partners will determine whether a cell is differentiating as a neuron/hair cell, a supporting/glial cell, or is continuing proliferation as a prosensory precursor. HC, hair cell and SC, supporting cell. Modified after (Forrest, et al., 2014, Imayoshi and Kageyama, 2014, Pan, et al., 2012, Reiprich and Wegner, 2014).
Embedded into this emerging concept of molecular evolution of expanding Sox and bHLH gene networks and their cross-regulation to drive neurosensory development is the need of micro RNA for the development of neurons and sensory cells in bilaterians. Specific micro RNAs (miR-124) are evolutionarily conserved and evolved first with bilaterians (Campo-Paysaa, et al., 2011, Peterson, et al., 2009). miR-124 has been shown to be essential for neuronal development through the regulation of chromatin remodeling (Yoo and Crabtree, 2009, Yoo, et al., 2009) and the downregulation of Sox9 (Cheng, et al., 2009). Experimental work in mammals demonstrated that the absence of miR-124 (and other micro RNA species) causes rapid degeneration of developing neurons (Kersigo, et al., 2011, Rosengauer, et al., 2012). Almost as conserved as miR-124 is miR-183 (Peterson, et al., 2009). miR-183 is associated with sensory systems (Pierce, et al., 2008) and is crucial for their development (Soukup, et al., 2009). The regulatory potential of miR's is obvious: expression of just one micro RNA can transform fibroblasts into neurons and such transformation is greatly enhanced by Neurod1 (Yoo, et al., 2009). This indicates that micro RNA evolved to cooperate with certain bHLH gene apparently to facilitate the action of those bHLH genes by suppressing specific Sox genes (Reiprich and Wegner, 2014). More data are needed to understand how many more miR's can play such a role. It is possible that the rapid expansion of bHLH and Sox gene families required additional gene regulation to fine tune the needed detailed cell fate decision. Obviously, the ability of micro RNA to regulate large sets of transcription factor translation could be a prerequisite for the evolution of a more complex neuronal and sensory system of bilaterians (Fritzsch, et al., 2007, Peterson, et al., 2009).
In summary, the molecular data on Sox and bHLH transcription factors as well as micro RNAs suggest the evolution of unique sets of genes within these families of transcription and translation regulating factors. What remains unclear is how that complex gene regulatory network evolved to be associated with the transformation of ectoderm to neurosensory precursor cells, to regulate the clonal expansion of such precursor cells and to guide the differentiation into the principal cell types of the neurosensory system of vertebrates. It is possible that the increasing complexity of these interactions through multiple feed-back and feed-forward loops was enabled with a novel regulatory layer, the micro RNAs. More experimental data on the functional interaction of Sox and bHLH genes in diploblastic metazoans is needed to verify how genomic evolution relates to evolution of functional interactions as revealed experimentally in vertebrates and flies. In particular miR's hypothetical function in nervous system evolution of bilaterians requires experimental verification to establish the possibly essential function of these bilaterian specific regulatory elements for neurosensory development and evolution, possibly through misexpression of these miR's in diploblasts.
1.B. Evolving intercellular communication mechanisms through regulated release of synaptic and dense core vesicle
The preceding paragraph highlighted basic aspects of molecular evolution of certain transcription factors and their interactions, including interactions with micro RNA. However, while the essential function of these genes is clear for normal development and differentiation of stem cells, it remains mostly unclear how these embryonic transcription factors regulate the adult neurosensory phenotype. For example, one essential aspect of neurons and sensory cells is the cellular communication via the release of synaptic transmitters to elicit a response in the target cell. The seemingly neurosensory specific aspect of vesicular release evolved out of general vesicular release mechanisms already present in single-celled organisms. In essence, basic molecular aspects needed for synaptic interactions have been identified in the genome of single-celled organisms (Sebé-Pedrós, et al., 2013), sponges and placozoans (Cheng, et al., 2009, Srivastava, et al., 2010), all of which are lacking neurons and synapses. Current data suggest that the basic molecular machinery needed for exocytosis in single-celled organisms was supplemented in neurosensory cells with additional molecules for fast vesicular release necessary for neuronal communication. As with bHLH gene function, such as Myc that evolved perhaps to govern cell-cycle transitions of single cells, such classical synaptic release molecules as SNARE's (soluble NSF-attachment protein receptor) are already found in yeast and therefore before synaptic communication evolved. Thus a major aspect of neuronal communication, vesicle release at presynaptic terminals, evolved out of a general exocytosis process through interactions of SNARE proteins with SM (Sec1/Munc18-like) proteins (Südhof and Rizo, 2011). The differences in interactions of SM proteins and SNARE complexes to catalyze membrane fusion has been modeled in an evolutionary context to define essential features of this process that can be adjusted through additions of other proteins to allow the fast neuronal transmitter release needed at synapses (Xia, et al., 2012). These modifications in neurons consist of chaperons such as Hsc70 and synucleins and their dysfunction can result in neurodegeneration. Synaptotagmins and synaptic “active zone” proteins such as Munc13 and RIMs are central components that adopt the generalized vesicular release mechanism of single-celled organisms such as yeast to the needs of rapid synaptic vesicle release in neurons (Südhof, 2012). Recent work has identified for the first time the quantitative ratios of 60 proteins needed to release and recycle synaptic vesicles (Wilhelm, et al., 2014). It was also shown that many components critical for synaptic functions are present in diploblastic animals that may or may not have neurons (Ryan, et al., 2013, Srivastava, et al., 2010). With this detailed, stoichiometric molecular understanding of the most basic process of neuronal communication at hand, one can now start to sort out the evolution of all partner proteins and their diversification to generate the synaptic proteins needed for vesicular release. Ultimately it needs to be established how transmitter choice and synaptic specializations such as ribbon or non-ribbon synapses of vertebrates and insects (Matkovic, et al., 2013, Weiler, et al., 2014) tie into the emerging network of cell fate decision making transcription factors and their downstream genes.
In summary, evolution of synaptic release proteins indicate a molecular transformation of a general vesicular release mechanism of pre-metazoans into a specialized synaptic vesicle release mechanism in metazoans that was further modified in neurons. This transformation parallels the evolution of the gene network of transcription factors to regulate neurosensory development. Possible causalities between the developmental transcription factor network to guide neuronal development and the molecular network to govern synaptic communication as an essential aspect of the function of adult neurosensory networks need to be established experimentally in the future.
1.C. Evolving a communication system to stabilize connections: Neurotrophin evolution and function to secure lasting neuronal connections
Tightly interwoven with the evolution of the synaptic release mechanism is the release of peptides in small and large dense core vesicles to provide a different time-table of cellular interactions. Among these proteins is a set of specialized proteins, the neurotrophins (von Bartheld and Fritzsch, 2006), that signal through modified receptor tyrosine kinases (Hallbook, et al., 2006) to promote the survival of neurons. These trophic interactions evolved out of tyrosine kinase signaling already known for single-celled organisms (Fairclough, et al., 2013, Sebé-Pedrós, et al., 2013). Neurotrophins and their receptors are essential for the viability of the peripheral nervous system (PNS) and cause extensive loss in vertebrate embryos when they are deleted. Neurotrophins and other neurotrophic factors (Lindahl, et al., 2014, Lindholm and Saarma, 2010) are indispensable for proliferation and survival and are the molecular mediators of ‘programmed cell death’ that is widespread in the CNS of many developing organisms (Oppenheim, 1991) to achieve numerical matching between independently developing sets of neurons. In addition, neurotrophic factors have been implicated to correct aberrant connections through pruning of such connections (Oppenheim, 1991). However, experimentally mis-wired peripheral innervations are not corrected (Mao, et al., 2014) even in systems that depend 100% on neurotrophins for their survival (Fritzsch, et al., 2004). Thus while neurotrophins play a central role in regulating cell death, pruning of aberrant connections seems to be limited and retention of aberrant connections seems to be of little consequences (Taylor, et al., 2012). Moreover, even deletion of two neurotrophins has only a limited effect on the CNS (Silos-Santiago, et al., 1997), questioning the basic assumptions underlying the neurotrophin theory (Dekkers and Barde, 2013). Molecular differences in protostomian and deuterostomian neuron development dependency on neurotrophins may indicate a lineage-specific use of such cell survival regulation factors but also high levels of conservation of some survival factors, including those responsible for the retention of specific subsets of dopaminergic neurons across phyla (Lindstrom, et al., 2013). When those neurotrophic support systems evolved and what was the selective advantage of having a system of survival factors is at the moment unclear.
With this background on the molecular evolution of neurosensory cells in mind, this article will explore aspects of neurosensory evolution of metazoans, with a focus on the evolution of neurosensory cells and organs as well as patterning of the deuterostome brain. This review aims to discuss the molecular basis of locally distinct cellular decision making processes, orchestrated by local patterning events (Srinivasan, et al., 2014) into developing specialized parts of the brain or specific organ systems. This review is organized to reflect the currently accepted systematic relationships among metazoans (Fig. 1), in particular deuterostomes (Bourlat, et al., 2006, Nosenko, et al., 2013, Osigus, et al., 2013, Peterson, et al., 2013, Satoh, 2008, Swalla and Smith, 2008).
2. From single sensory cells and ‘skin-brains’ to sensory organs and a central nervous system: evolving patterning processes and reorganizing them through embryonic transformation
Generating complex sensory organs or a CNS requires that molecular regulations of cellular development happen in the right cells at the right time to achieve the desired outcome in terms of cellular differentiation (Stergachis, et al., 2013) and to coalesce specific cell types into unique aggregates able to perform a distinct function, for example a sensory organ. Such topological regulation and orchestration of intercellular interactions require a multitude of different diffusible signals [Shh, Fgf, Wnt, BMP (Chen and Streit, 2013, Streit, et al., 2000)] that will result in localized expression of regional selector genes [for example, Pax genes in sensory organ development (Fortunato, et al., 2014)] that translate patterning signals into local, cellular action. Modeling the patterning changes that transform a cnidarian-like organism into a bilaterian suggests that skin patterning evolved as a consequence of mesoderm invagination (Meinhardt, 2013) to induce the neural plate and surrounding placodes (Schlosser, et al., 2014). Consistent with this gene-centric perspective proposed here is the fact that the molecular basis of patterning of the developing sensory organs and CNS seemingly evolved in pre-metazoans (Fairclough, et al., 2013, Fortunato, et al., 2014, Sebé-Pedrós, et al., 2013). Even amoebazoa, such as the slime mold (Fig. 1), have sophisticated patterning processes to differentiate the homogenous population of amoeba into a stalk and a spore cell type (Chattwood, et al., 2013), activation of G protein-coupled receptors are essential for directional growth to ensure mating in yeast cells (Martin and Arkowitz, 2014) and gene networks regulate sexual cycles in fungi (Ait Benkhali, et al., 2013).
As outlined in the previous section, it is conceivable that sophisticated molecular networks that enable multicellular interactions of single cells for reproduction purposes became the basis of the cellular interaction in metazoans. However, in metazoans this molecular machinery for cellular communications evolved to pattern the body and its organs, such as the CNS and sensory organs. Obviously, patterning of the body happened already at the level of diploblasts and many molecules found in these patterning processes in bilaterians can be identified in diploblasts that show only 2 (sponges) or 4 (placozoans) distinct cell types. The ideas of a progressive complication of specification of cell types in sponges and placozoans (Osigus, et al., 2013) followed by the evolution of gastrulation have recently been blurred with the claims that ctenophores maybe the sister group of all other metazoans (Ryan, et al., 2013). In fact, it has been claimed that the ‘gastrulation’ of ctenophores is molecularly and structurally distinct from that of other metazoans (Martindale, 2013, Martindale and Lee, 2013). Ctenophores also have an unusual development of the apical sensory organ (Schnitzler, et al., 2014), a gravity sensing system that uses cilia to hold an otoconia mass so that changes in position affect the beating cilia, the organs of motility in these organisms, to change the course of swimming (Tamm, 2014). The nervous system of these animals consists of an assembly of apical neurons and giant fibers that run along the eight strings of combs. Possibly light sensitive cells are molecularly identified, but no response to light has been recorded, indicating that gravity sensing with the single apical sensory organ is the major sensory input for orientation in ctenophores. These unique features as well as a strange double symmetric organization could indicate that ctenophores may indeed be uniquely derived from an unknown metazoan ancestor and have evolved certain features, including a sophisticated multicellular gravity sensing organ and nervous system in parallel to other metazoans that, nevertheless, share many neuronal patterning genes (bHLH, Sox) and molecules related to synaptic function (Ryan, 2014). However, others have argued against this scenario and propose an evolution of metazoan with ctenophores being a highly derived sister group of coelenterates (Nosenko, et al., 2013, Osigus, et al., 2013), arguments we have adopted here (Fig. 1).
Be this as it may, the evolution of complicated sensory organs dedicated to gravity and light sensing in both ctenophores and cnidarians indicates that sensory organ evolution can be combined with limited development of a nervous system and certainly predates evolution of the sophisticated CNS of bilaterians (Fig. 1). It is possible that the limited motor abilities of diploblastic animals are incompatible with the evolution of a complex nervous system that could integrate sensory stimuli better with a sophisticated motor output provided by mesoderm diversification, requiring only localized ganglion-like concentrations of neurons in an epithelial network (Satterlie, 2011). We like to argue that the ability to generate a refined motor output is a prerequisite to evolve the enhanced computational power of a larger set of interneurons that connect an increasingly sophisticated sensory input to an equally sophisticated motor output (Straka, et al., 2014).
For the remaining article we will built on the idea that the molecular ability to form neurosensory cells of complex sensory organs, such as statocysts and eyes for orientation in space (Fritzsch and Straka, 2014, Lamb, 2013) evolved prior to the bilaterian CNS to process sensory information to guide the limited motor output of animals comparable to ctenophores and cnidarias. Obviously, the evolution of complicated sensory organs is mostly associated with motile diploblastic life forms but they are topographically (opposite the mouth in ctenophores, around the mouth in cnidarians) and structurally so distinct between these two forms of diploblastic animals that they may have independently evolved out of molecular (Gyoja, 2014, Schnitzler, et al., 2014) and cellular (Fritzsch and Straka, 2014) precursors. Experimental gene swapping as conducted in sensory system development of mice and flies (Wang, et al., 2002) is needed to verify the function of homologous neuronal transcription factors in diploblasts and bilaterians to further understand the significance of the obvious molecular and anatomical differences in diploblast and bilaterian sensory system development.
3. ‘Splitting hairs’: molecular transformation of single neurosensory cells to neurons and hair cells of the ear
Evolution of the vertebrate CNS with its enormous numbers of distinct neuronal cell types and extensive local and long range connections (Nieuwenhuys, 2002) is beyond the scope of this brief review. However, concentrating on sensory organ evolution in this paragraph will provide a basic insight into general features of gene network evolutions that sort out just two distinct cell types, hair cells and sensory neurons. Evolution of both sensory neurons and hair cells out of the single neurosensory cells of metazoan ancestors (Fig. 2) is a molecularly reasonably understood case of cellular diversification; it follows the predicted multiplication of bHLH genes (Gyoja, 2014) combined with changes in cell fate determination (Fritzsch, et al., 2010, Pan, et al., 2012). Two genes have been associated with neuron and hair cell development, respectively, through loss of function experiments: Neurog1 and Atoh1 (Fritzsch, et al., 2010). The loss of hair cells in mutants of Neurog1 (Ma, et al., 2000) prompted the idea of a lineage relationship between neuronal and hair cell precursors. This idea of a lineage relationship of sensory neurons and hair cells has been widely accepted among evolutionary biologists (Patthey, et al., 2014) but controversies exist among developmental biologists regarding the details of the lineage relationships among all neurosensory cells of the ear (Fritzsch, et al., 2006, Raft, et al., 2007). Thus, while the severe loss of hair cells as a consequence of neuronal loss (Matei, et al., 2005) has been experimentally verified to be due to lineage relationships for the vestibular neurosensory cells of the ear, the relationship of neurons to hair cells is not yet experimentally clarified for the mammalian cochlea (Raft, et al., 2007).
Recent data have complicated the picture of molecular interactions to sort different cell fate in lineage related cells further (Fig. 3) by showing that several other bHLH genes also play a role in ear development (Kruger, et al., 2006), in part acting redundantly to other bHLH genes. For example, the bHLH gene Neurod1 is expressed in both neurons and hair cells and seems to suppress Atoh1 expression entirely (neurons) or limit its expression (hair cells; Fig. 3). As a consequence of loss of Neurod1, Atoh1 is more profoundly expressed in neurons and converts neurons into hair cells (Jahan, et al., 2010). In contrast to the astonishing fate reversal in neurons to hair cells in the absence of Neurod1, the effect of loss of Neurod1 on hair cells is more subtle and results in alteration of hair cell types, but not in hair cell conversion. In the absence of Neurod1, inner hair cell-like cells that express inner hair cell molecular signature genes such as Fgf8, appear among outer hair cells (Jahan, et al., 2013).
These data suggest a more complex interaction of various bHLH genes (Fig. 3) to define the fate of neurosensory precursors (Forrest, et al., 2014, Jahan, et al., 2013). Most pertinent for this effect of Neurod1 is the undisclosed interaction of multiple bHLH genes, including the Hes and Hey genes that are expressed following Delta/Notch upregulation (Raft and Groves, 2014). Essentially, the level and number of co-expressed bHLH genes will determine the future fate of a given cell (Fig. 3) in conjunction with the level of expression of other genes that maintain proliferative neurosensory precursors such as Sox2 (Dabdoub, et al., 2008). Evolution of Group A bHLH genes in diploblasts and their multiplication in bilaterians (Gyoja, 2014) may have generated the molecular basis to evolve the different cell types found in the vertebrate ear that form a complex network of related transcription factors engaged in a stepwise transformation of proliferating neurosensory precursors into the two distinct neurosensory types of the ear, the neurons and hair cells.
3.A. Evolving a dedicated precursor population to increase a localized ectodermal transformation
While it is possible that the evolutionary origin of the neurosensory cells of the ear predates the evolution of the vertebrate ear out of placodes (Fritzsch and Straka, 2014), the molecular evolution of placodes can now be partially traced in chordates (Schlosser, et al., 2014). Chief among ectodermal patterning genes identifying the otic placode are genes also known experimentally to be essential for proper regulation of bHLH gene expression. For example, Eya1/Six1 affects ear development in a dose-dependent fashion (Zou, et al., 2008) and regulates neurosensory gene expression through binding to the enhancer elements of bHLH genes (Ahmed, et al., 2012a, Ahmed, et al., 2012b). One of the earliest markers of the otic placode, Pax2/8, belong to an ancient network of selector genes (Fortunato, et al., 2014). Pax2/8 are essential for neurosensory development of the ear that is severely disrupted in Pax2/8 double null mutants (Bouchard, et al., 2010). Another factor known for gene expression regulation in placodal precursors, Gata3 (Schlosser, et al., 2014), is essential for ear neurosensory development, in particular of the newly evolved (Fritzsch, et al., 2013) mammalian organ of Corti (Duncan and Fritzsch, 2013). Many other factors have been identified as being early markers of the otic placode and have been tested for their significance through genetic manipulation, but the basic problem is this: multiple genes need to be coordinately expressed in the placodal region to ensure the transformation of ectoderm into neurosensory cells of the ear.
Combined, these data suggest a scenario that progressively transforms the capacity of ectoderm to develop neurosensory cells through the assembly of a network of neurosensory gene regulating transcription factors (Chen and Streit, 2013). This network regulates the localized expression of neurosensory development-mediating Sox and bHLH genes, apparently starting with the Sox2 and Neurog1 as the earliest expressed Sox2 and bHLH gene in the developing mammalian ear (Ma, et al., 2000, Ma, et al., 1998, Mak, et al., 2009, Puligilla, et al., 2010). Several important transcription factors of ear placode development are part of an ancient network predating sensory organ evolution (Bouchard, et al., 2010, Fortunato, et al., 2014) and are associated with choanocytes, out of which hair cells evolved (Fritzsch and Straka, 2014). Hair cells can induce surrounding cells to form vesicles and this ability may have been at the basis of ear formation. For example, hair cells induced in Neurod1 mutants in the developing ganglion form mini-vesicles that organize other cells of the ganglion to develop a continuous epithelium (Jahan, et al., 2010, Jahan, et al., 2013) and hair cells can in vitro organize vesicles around them (Koehler, et al., 2013). In a way, the otic placode can be viewed as an embryonic adaptation that aggregates sensory cell precursors into a single region through the localized Sox and bHLH expression driven by multiple ancient transcription factors (Fortunato, et al., 2014) that in turn are regulated by Fgfs (Chen and Streit, 2013, Fritzsch, et al., 2006). Understanding the evolution of the otic placode to an ear vesicle will require unraveling the molecular basis of the ability of hair cells to induce vesicle formation and its heterochronic shift from hair cells to placodal cells in vertebrates.
3.B. Switching gears: the importance of multiple bHLH genes for smooth transitions of fate
Ectodermal transformation to form either single sensory cells, as in insects, or multiple sensory cells and neurons, as in vertebrates, requires ultimately the expression of Sox and bHLH genes to change the fate of ectodermal cells into neurosensory cells (Imayoshi and Kageyama, 2014, Reiprich and Wegner, 2014). While this general function in particular of bHLH genes has long been established through experimental induction of neurons after bHLH gene mRNA injection into developing Xenopus (Lee, et al., 1995), further analysis has shown a puzzling co-expression of several bHLH genes in the developing ear (Jahan, et al., 2010), not all of which result in loss of a specific cell type in mutants. The expression of these multiple bHLH genes to achieve transformation of ectodermal cells into neurosensory cells follows an increasingly sophisticated patterning process of the ectoderm (Schlosser, et al., 2014, Streit, et al., 2013) that readies these cells to respond with differentiation to the upregulation of bHLH genes as a final step to consolidate this decision making process. Work over the last few years has transformed the simple one gene-one cell type idea generated by early knockout studies that eliminated in Atoh1 null mice all hair cells (Bermingham, et al., 1999) and in Neurog1 null mice all neurons (Ma, et al., 1998) into a more complicated perspective of an interactive gene network (Rue and Garcia-Ojalvo, 2013). In particular, work on Neurod1 mutants suggests a sophisticated cross-regulation of multiple bHLH transcription factors (Jahan, et al., 2010, Jahan, et al., 2013, Ma, et al., 2000) that requires a quantitative assessment of binding to the various enhancer regions through interactions with the ubiquitous E-proteins (Forrest, et al., 2014) as well as maintaining a proliferative precursor status through interactions with the Sox and Id proteins (Fig. 3). This complicated intracellular gene network is apparently accompanied by an equally sophisticated intercellular network of Delta/Notch interactions that replaces the past simple lateral inhibition model (Sprinzak, et al., 2011).
While this complexity of bHLH gene expression has long been noticed, it is now becoming clear that this expression is more than noise generated by stochastic gene expression (Johnston and Desplan, 2014, Stergachis, et al., 2013). More specifically, it appears that the rich co-expression of several bHLH genes allow for coordinated transition of cellular states toward diversification from a single precursor (Fig. 3), as has been described as a general principle of neuronal differentiation through coordinated expression level variation (Imayoshi and Kageyama, 2014, Roybon, et al., 2009). The differential interaction of bHLH genes also results in the differential down-regulation of Sox genes (Bylund, et al., 2003), possibly enhanced through positively regulating miRs that set the stage for normal hair cell differentiation (Kersigo, et al., 2011).
3.C. Reversing decisions: the molecular basis of stability and flexibility of the cellular decision making process in the ear
Consistent with the insight that cell fate decision making is a process and not a single step is the reversal of such decision at various stages of commitment (Fig. 3). Such decision reversals are particularly interesting to evaluate experimentally the level of fixation of such decision making processes at an individual cellular level. For example, the hypothesis of lineage relationship of neurons, hair cells and supporting cells (Fritzsch, et al., 2006, Ma, et al., 2000) implied that either or both can be converted up to a certain level of differentiation into the other neurosensory cell type of the ear. Such evidence was ultimately provided by showing that simple elimination of Neurod1 can convert differentiating neurons into hair cells (Jahan, et al., 2010). Likewise, the distinction between supporting cells and hair cells can be reversed at even later stages of development through misexpression of Atoh1 in supporting cells (Cox, et al., 2014, Liu, et al., 2014). In contrast, a recent attempt to transform differentiating hair cells into neurons through misexpression of Neurog1 did not result in any transdifferentiation (Jahan, et al., 2012). However, this negative result could relate, among many things, to the inability of Neurog1 to bind to the Atoh1 enhancer to increase expression enough to be meaningful for any differentiation.
Other data on ear development indicate rather dramatic differences in cell fate decision making already prior to bHLH gene expression, likely regulated by Sox genes (Reiprich and Wegner, 2014). On the one extreme are hair cell precursors that are already committed prior to Atoh1 expression, perhaps through Sox2 (Kiernan, et al., 2005), possibly at the time they exit the cell cycle (Kopecky, et al., 2013), to differentiate as hair cell so that they cannot be reverted into a different fate even if powerful neurogenic factors are expressed (Jahan, et al., 2012). On the other hand are neurons and supporting cells that even in a late stage in their decision making process can be transdifferentiated into hair cells (Jahan, et al., 2010, Mizutari, et al., 2013). Simply speaking, all otocyst-derived cells have the potential, possibly as a default state, to differentiate into hair cells if provided with Atoh1, even at a late stage. In contrast, future hair cells are already committed to hair cell fate prior to Atoh1 expression under the guidance of yet to be specified transcription factor(s). This distinction between cell fate commitment - as supported by cell cycle exit - and cell fate execution – as shown by Atoh1 expression - is particularly obvious in the apex of the cochlea. In the apex, a delay of several days exists between cell cycle exit and onset of Atoh1 expression (Fritzsch, et al., 2005, Jahan, et al., 2013, Matei, et al., 2005).
When exactly such fate reversals are irreversible in the ear neurosensory precursors remains to be experimentally evaluated. If properly understood, it has the potential to reverse existing decisions and convert any cell type in the organ of Corti into hair cells, as recently suggested (Mizutari, et al., 2013). It is important for this fate reversal to understand the molecular basis of such decision making to enhance the frequency of the outcome and the stability of the induced transdifferentiation. In this context, level of expression of differentiation-inducing bHLH genes, as mediated by early transcription factors in the otic placode (Ahmed, et al., 2012a, Ahmed, et al., 2012b), ensures quantitative correct expression of interacting transcription factors (Fig. 3) to differentiate the right type of cell at the right place (Jahan, et al., 2013). Translation of this transdifferentiation ability into the clinic will require a more detailed understanding to ensure the reconstitution of a functional organ of Corti, possibly the best strategy to restore some hearing in profoundly deaf people (Zine, et al., 2014).
4. Uncoupling specification from morphogenic transformation: the case of Mauthner cells specification and exo-gastrulae gene expression
Diploblasts have limited aggregation of neurons in combination with an epithelial nerve net (Satterlie, 2011) but may have sophisticated sensory organs rivaling in complexity of those of vertebrates with a much more complex nervous system (Fritzsch and Straka, 2014). It is possible that the bilaterian ancestor had a partially aggregated nervous system like that of Xenoturbella (Raikova, et al., 2000) or acorn worms (Bullock, 1965) with little to no metameric organization. It appears less likely that a metameric organization around a well-developed CNS of ancient metazoans devolved to form the partially centralized epithelial nerve nets found in diploblasts (Matus, et al., 2007b, Satterlie, 2011) and the simple CNS found in some basal bilaterians (Brown, et al., 2008, Fritzsch and Glover, 2006, Harzsch and Muller, 2007, Raikova, et al., 2000). A non-metamerically organized nervous system with no specialized sensory organs seems to be the most parsimonious assumption for diploblasts and, by logical extension, bilaterian ancestors (Bourlat, et al., 2006, Budd, 2001, Satoh, 2008, Swalla and Xavier-Neto, 2008). The different patterns of nervous system of extant deuterostomes are considered here to be independently derived from such bilaterian ancestors, reflecting either independent formation of a CNS (acorn worms, cephalochordates, urochordates, vertebrates) or show a transformation into a pentameric nerve net (echinoderms). If correct, this hypothesis implies that CNS formation comes about by aggregating epidermal nerve cells of diploblasts into a brain, possibly paralleling the condensation of distributed epithelial sensory cells into sensory organs. A byproduct of this hypothesis of condensation of the CNS and sensory organs, would be the de novo appearance of embryonic adaptations (Arendt, et al., 2004, Fritzsch, et al., 2007, Northcutt and Gans, 1983, Sauka-Spengler and Bronner-Fraser, 2008) to lead to brain and sensory organ formation such as neural plate, the placodes and the neural crest (Schlosser, et al., 2014, Steventon, et al., 2014), all of which are embryonic adaptations with poorly-defined cellular and molecular precursors in deuterostomes (Fritzsch and Northcutt, 1993, Sauka-Spengler and Bronner-Fraser, 2008).
Among deuterostomes, only vertebrates achieved not only the aggregation of all neurons into a CNS, but in addition aggregated nearly all peripheral sensory cells into discrete sensory organs and developed a unique set of innervations via neural crest-derived sensory neurons (Fritzsch and Northcutt, 1993, Gans and Northcutt, 1983, Sauka-Spengler and Bronner-Fraser, 2008) and placode-derived sensory neurons (Chen and Streit, 2013, Fritzsch, et al., 2007). The developmental mechanism to achieve this aggregation of sensory cells in vertebrates is the formation of special embryonic tissue, the neural crest and neurogenic placodes (Begbie, et al., 1999, Northcutt and Gans, 1983, Streit, 2007). While much work has concentrated on the specific gene networks that promote embryonic formation of placodes or neural crest (O'Neill, et al., 2012, Ohyama, et al., 2007, Streit, 2007), much less work has been dedicated toward the molecular mechanism of suppression of neuronal fate determination in the remaining ectoderm. It is now clear that BMP upregulation, combined with limited to no expression of Fgf's and FgfR's will maintain a non-neuronal fate in the ectoderm of chordates (Bertrand, et al., 2003, Delaune, et al., 2005, Fritzsch, et al., 2006) but not in hemichordates (Lowe, et al., 2006). That ectoderm default state of chordates is indeed neurogenic has been demonstrated by overexpressing proneuronal bHLH genes in the ectoderm, revealing a transformation into neurons (Lee, et al., 1995, Ma, et al., 1996) that even holds for bHLH genes not associated with neuronal development such as bHLH genes isolated from sponges (Richards, et al., 2008). It is important to realize that this transformation is limited to ectoderm and does not expand to mesoderm, indicating the unique ability of ectoderm to respond to pro-neuronal bHLH genes with differentiation as much as mesoderm can respond to the bHLH gene MyoD.
While neuronal patterning, including the formation of placodes, correlates with the inductive interactions of the underling mesoderm, it needs to be stressed that patterning of the neural plate precedes and can be partially independent of mesoderm induction, possibly reflecting ancient patterning mechanisms predating bilaterians. For example, Hox genes and Krox20 expression develops in exogastrulae in which the ectoderm is not in contact with mesoderm (Doniach, et al., 1992) and patterning of the neural plate in chicken predates gastrulation (Streit, et al., 2000). Even more difficult to reconcile with the ‘traditional’ vertical induction model through endomesoderm involution is the fact that the Mauthner cell, a pair of giant neurons in rhombomere 4 of many aquatic vertebrates, exit the cell cycle at Nieuwkoop and Faber (NF) stage 12, during gastrulation and prior to neurulation in frogs (Lamborghini, 1980). Moreover, explants of the future hindbrain during neurulation may generate Mauthner cells (Stefanelli, 1950), suggesting that cells are specified at the level of the embryonic equivalent of the ancestral adult ‘skin brain’ found in acorn worms Logically, Mauthner cells develop normally when all mitosis is arrested after they have become postmitotic (Harris and Hartenstein, 1991). Giant fibers, such as Mauthner cell fibers, are known for acorn worms where they form a fast response system also known for many other non-vertebrates (Bullock, 1965) and such giant fibers already exist in diploblasts (Satterlie, 2011). Moreover, Mauthner cells are in rhombomere 4 that also gives rise to the facial branchial motoneurons, possibly the most conserved motoneurons of the brainstem (Dufour, et al., 2006, Elliott, et al., 2013).
These developmental experimental data strongly support the notion that ectodermal patterning, including specification of neurons, predates the development and evolution of the deuterostome CNS. It is possible that such giant cells and their fast motor output to induce escape responses were already specified in an epithelial neuronal network of ‘skin brains’ to induce escape through the stimulation by rudimentary sensory organs. Clearly, identifying the molecular basis of this pre-neuronal plate patterning event that may predate mesoderm/ectoderm interaction mediated by gastrulation, could shed light on how much ectodermal patterning existed in animals without gastrulation. After all, Sox2 expression increases prior to gastrulation (Yanai, et al., 2011) and is already highly expressed in neuroectoderm of NF stage 11 embryos (Sasai, et al., 2008). Such early expression in areas of future neuronal development is consistent with the idea developed in mutant mice that Sox2 is an early marker for neuronal fate determination. The alternative, that such data reflect novel developmental reorganizations of amphibians and chickens without any evolutionary significance is possible, would conflict with significant evidence for an hourglass model of vertebrate development and evolution (Akhshabi, et al., 2013, Stergachis, et al., 2013), most likely also relevant for vertebrate hindbrain development and evolution, including the evolution of ectodermal patterning events.
5. Concentrating and involuting the nervous system: molecular parallelisms to placodes
While the previous paragraph explored the possible importance of ectodermal patterning events prior to gastrulation, most metazoans gastrulate and use this process of vertical interaction of the involuting endo- mesoderm to pattern the ectoderm. Patterning the body and the nervous system (Paulin, 2014) resulted in regional concentrations of neurons in the nerve net out of patterning the ectoderm of diploblastic ancestors (Meinhardt, 2004, Meinhardt, 2013). These ‘skin brains’ have ectoderm that consists of many neurons between simple skin cells (Fritzsch and Glover, 2006, Pani, et al., 2012) with molecular patterning gene expression broadly comparable to the vertebrate brain (Fig. 4). This seems to indicate that the vertebrate brain is essentially a transformed epidermis of the diploblastic ancestor, as predicted in mathematical models (Meinhardt, 2013). Simply speaking, molecular evolution of cell type specifying mechanisms evolved in single-celled organisms. However, such mechanisms were through further evolution of cell-cell interactions integrated into regional specific cell type development. In addition, general topological information derived from patterns set up by the diffusion of morphogens was apparently translated into local neurosensory cell type specification. Importantly, the molecular basis of specific neurosensory cell types (Fritzsch and Straka, 2013) and the molecular basis allowing the local specialization evolved prior to the evolution of brains in skin-brains. Morphogenetic and inductive events may have led to the formation of CNS in bilaterians through the transformation of an already patterned epidermal nerve cell network into a network with localized specialization, possibly multiple times (Meinhardt, 2004, Northcutt, 2012). A ‘deep molecular homology’ has been recognized in the development of various organs (Shubin, et al., 2009). In the CNS this may come about through an ancestral patterning network that evolved prior to sensory organ and CNS evolution. Topological information later evolved to guide convergent evolution (Stern, 2013) of rather different brains and sensory organs in different phyla that may nevertheless share multiple conserved genes guiding cellular differentiation. A case in point is the different use of hedgehog/sonic hedgehog in flies and vertebrates: whereas Hh of flies is needed to define segment boundaries via short range signaling (Stern, 2013), Shh is used in vertebrates to specify dorso-ventral patterning of the CNS via long range signaling (Echelard, et al., 1993, Ingham and McMahon, 2001) instead of adding to the formation of somites (Dias, et al., 2014, Newman, 2014), with somites treated by many as indicative of arthropod ‘segments’ despite the fact that in many vertebrates, these tissue blocks are at different locations on the left and the right side of the body (Fritzsch and Northcutt, 1993).
Fig. 4.
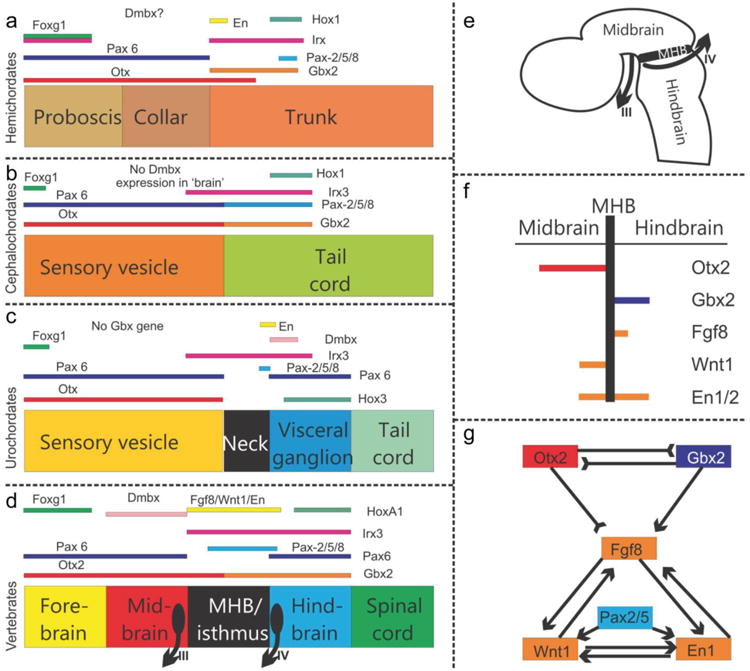
The evolution of gene expression at the midbrain–hindbrain boundary (MHB) is shown for deuterostomes. The MHB of vertebrates shows abutting Otx2 and Gbx2 expression (D-G). This stabilizes the expression of Fgf8 (G), which in turn stabilizes the expression of Wnt1 and engrailed (En1). Mutation of Otx2, Gbx2, Fgf8, or Wnt1 eliminates the MHB. Pax2/5/8 are also expressed at the MHB, whereas the expression of Dmbx occurs immediately rostral to the MHB in the midbrain to later expand into the hindbrain and spinal cord (D). Note the partial overlap of Pax2/5/8 with the caudal expression of Otx2 and the rostral expression of Gbx2 (D). Hemichordates (A) have overlapping expression of Gbx, Otx, Irx and En in the rostral trunk. Pax6 abuts Gbx2 whereas Pax2/5/8 overlaps with the caudal expression of Gbx2. Outgroup data suggest that coelenterates have a Dmbx ortholog, thus raising the possibility that hemichordates (A) also have a Dmbx gene. Cephalochordates (B) have no Dmbx expression in the ‘brain’. Otx abuts with Gbx, like in vertebrates. However, Gbx overlaps with Pax2/5/8 and most of Irx3. Urochordates (C) have no Gbx gene but have a Pax2/5/8 and Pax6 configuration comparable to vertebrates. Dmbx overlaps with the caudal end of the Irx3 expression whereas Dmbx expression is rostral to Irx3 in vertebrates. Together these data show that certain gene expression domains are topographically conserved (Foxg1, Hox, Otx), whereas others show varying degrees of overlap. It is conceivable that the evolution of nested expression domains of transcription factors is causally related to the evolution of specific neuronal features such as the evolution of oculomotor and trochlear motoneurons (D,E) around the MHB. Experimental work has demonstrated that the development of these motor centers depends on the formation of the MHB. Adopted from (Beccari, et al., 2013, Fritzsch and Glover, 2006, Pani, et al., 2012)
Consistent with this theoretical consideration outlined in the previous paragraph are data (Lowe, 2008, Lowe, et al., 2006, Lowe, et al., 2003, Pani, et al., 2012) demonstrating that many patterning genes, previously considered to be associated with CNS neuromeres of craniates, are expressed in the skin nerve plexus of acorn worms and play a distinct role in developmental patterning (Fig. 4). The basic organization of repetitive neuronal elements as well as the evolution of neuromeric boundaries can be molecularly dissected as discrete steps in deuterostome and chordate evolution that progresses from a more basic, non-neuromeric organization (Lowe, et al., 2003) to a fully developed craniate organization with clear association with compartmental boundaries (Beccari, et al., 2013, Fritzsch and Glover, 2006, Murakami, et al., 2005). This is most obvious in the hindbrain organization of the lamprey, vertebrates that have a different organization of neural elements with respect to rhombomere boundaries (Fritzsch, 1998, Murakami, et al., 2004, Murakami, et al., 2005). These overall similarities in expression domains and neuronal organization among bilaterians have long been understood and used to indicate a possible homology at the molecular and structural level (Arendt, 2005, Arendt, et al., 2008, Reichert, 2009, Reichert and Simeone, 1999). More recent data indicate that the limited molecular toolbox may have biased our understanding and may have falsely identified molecular homology among anatomically analogous organs (Lowe, et al., 2006, Newman and Bhat, 2009). Clearly, basic protostomes and many basic bilaterians have no metamerically organized ‘nerves’ radiating out in a specific pattern from epithelial nerve plexus (Bullock, 1965, Fritzsch and Glover, 2006, Harzsch and Muller, 2007). Indeed, the asymmetric but metameric organization of nerves in cephalochordates and basic chordates (Fritzsch and Northcutt, 1993) may reflect a peripheral nerve reorganization as much as the molecular basis of boundary formation in the CNS reflect a step-wise evolution of neuromeric boundaries and their underlying molecular basis (Murakami, et al., 2005). Rhombomeres of vertebrates, by some considered to be equivalent to arthropod segments because of their comparable Hox-code, may reflect different anatomic solutions to compartment formation while maintaining ectodermal pre- patterning and associated genes in an independently evolved CNS. It should be stressed that through most of vertebrate evolution, the majority of neurons of the CNS were concentrated in the spinal cord and this proportion only changed in a few vertebrate radiations with dramatic increase in brain size, notably the mammals (Nieuwenhuys, et al., 1998)
Consistent with this interpretation are the molecularly well-known early steps in CNS formation that reflect the transformation of ectoderm into neuroectoderm. A key player across bilaterians is the Dpp/BMP pathway (Reichert, 2009). However, in addition to downregulation of BMP, chordates require the action of Fgfs (Bertrand, et al., 2003, Fritzsch, et al., 2006, Fritzsch and Glover, 2006) to induce neuroectoderm. Fgf's play no role in neuronal specification in arthropods (Urbach and Technau, 2004) and hemichordates (Lowe, et al., 2006). It remains unclear if this dependence on or absence of Fgfs is linked to the apparent dorso-ventral patterning difference in nervous system induction that has been so long a source of a general body plan reversal ideas (Arendt, 2005, Arendt, et al., 2008, Lowe, et al., 2006) but has been questioned by other lines of evidence (Brown, et al., 2008, Satoh, 2008). In the absence of understanding the full complement of all genes involved in these processes and their interactions, it appears most parsimonious to assume that the known differences are likely to indicate a non-homologous origin of a CNS based on partially homologous transcription factor actions (Newman and Bhat, 2009) out of an epidermal nerve plexus, as initially proposed for the acorn worm (Lowe, et al., 2006, Lowe, et al., 2003). Other known similarities such as the use of different and similar sets of bHLH genes, the ubiquitous use of the Delta-Notch system and certain molecular aspects of proliferation regulation (Arendt, 2005, Reichert, 2009, Urbach and Technau, 2004) could reflect their similar functions in the common diploblast ancestor (Magie, et al., 2007, Martindale, et al., 2004, Matus, et al., 2007a, Matus, et al., 2007b, Mazza, et al., 2007, Putnam, et al., 2007, Seipel, et al., 2004, Yamada, et al., 2007). What needs to be clarified now is how the aggregation of a nerve net of the diploblast ancestor, most likely also present in the common bilaterian ancestor (Fig. 1), has been tied into the emerging and different general embryonic patterning mechanisms (Meinhardt, 2004, Newman and Bhat, 2009, Salazar-Ciudad, et al., 2003) to elicit a local upregulation of specific neural inducers to form a dorsal or ventral CNS in deuterostomes and protostomes, respectively.
6. Conclusion
Understanding the evolution of the generalized developmental cell fate specification through topographically restricted gene expression cascades to initiate and regulate cellular differentiation of neurosensory cells out of ectoderm is a key step in sensory organ or CNS evolution. Cellular fate switching evolved as an essential step in early metazoan evolution to transform an assembly of identical cells into a progressively different set of up to over 200 identifiable cell types found in mammals. To transform the cellular evolution into organ differentiation, molecular mechanisms evolved that regulate proliferation of precursor populations in integration with cell fate commitment. Within this process of establishing cellular diversity, Sox and bHLH genes played a major role to consolidate cell fate commitment into cellular differentiation, possibly as an extension of the temporal aggregation of single-celled organisms for vegetative and sexual reproduction. Sox and bHLH genes are essential regulators of neuronal and neurosensory induction and differentiation in the ectoderm and are embedded in an increasingly complex decision making process that ensures temporal and intensity specific expression of interacting intracellular and intercellular networks of transcription factors to drive topologic cell type and subtype specific differentiation. Understanding the nodes of such networks will allow inducing transdifferentiation in vivo, once gene expression regulation through manipulation of enhancers is understood.
Acknowledgments
This work was supported by NIH (P30 DC 010362, R03 DC013655), NASA Base Program and the OVPR, University of Iowa. We thank Drs. S. Reiprich and U. Ernsberger as well as two unknown reviewers for helpful comments and suggestions to improve this paper.
Literature
- Ahmed M, Wong EY, Sun J, Xu J, Wang F, Xu PX. Eya1-Six1 interaction is sufficient to induce hair cell fate in the cochlea by activating Atoh1 expression in cooperation with Sox2. Dev Cell. 2012a;22:377–390. doi: 10.1016/j.devcel.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012b;139:1965–1977. doi: 10.1242/dev.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait Benkhali J, Coppin E, Brun S, Peraza-Reyes L, Martin T, Dixelius C, Lazar N, van Tilbeurgh H, Debuchy R. A network of HMG-box transcription factors regulates sexual cycle in the fungus Podospora anserina. PLoS genetics. 2013;9:e1003642. doi: 10.1371/journal.pgen.1003642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhshabi S, Sarda S, Dovrolis C, Yi S. An explanatory evo-devo model for the developmental hourglass. arXiv preprint arXiv. 2013:13094722. doi: 10.12688/f1000research.4583.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B, Colleran C, Leger-Silvestre I, Berger AB, Dez C, Normand C, Perez-Fernandez J, McStay B, Gadal O. Structure-function analysis of Hmo1 unveils an ancestral organization of HMG-Box factors involved in ribosomal DNA transcription from yeast to human. Nucleic acids research. 2013;41:10135–10149. doi: 10.1093/nar/gkt770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D. Genes and homology in nervous system evolution: comparing gene functions, expression patterns, and cell type molecular fingerprints. Theory Biosci. 2005;124:185–197. doi: 10.1007/BF02814483. [DOI] [PubMed] [Google Scholar]
- Arendt D, Denes AS, Jekely G, Tessmar-Raible K. The evolution of nervous system centralization. Philos Trans R Soc Lond B Biol Sci. 2008;363:1523–1528. doi: 10.1098/rstb.2007.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Hausen H, Purschke G. The ‘division of labour’ model of eye evolution. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2009;364:2809–2817. doi: 10.1098/rstb.2009.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Tessmar-Raible K, Snyman H, Dorresteijn AW, Wittbrodt J. Ciliary photoreceptors with a vertebrate-type opsin in an invertebrate brain. Science. 2004;306:869–871. doi: 10.1126/science.1099955. [DOI] [PubMed] [Google Scholar]
- Beccari L, Marco-Ferreres R, Bovolenta P. The logic of gene regulatory networks in early vertebrate forebrain patterning. Mechanisms of development. 2013;130:95–111. doi: 10.1016/j.mod.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Begbie J, Brunet JF, Rubenstein JL, Graham A. Induction of the epibranchial placodes. Development. 1999;126:895–902. doi: 10.1242/dev.126.5.895. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115:615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- Bouchard M, de Caprona D, Busslinger M, Xu P, Fritzsch B. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol. 2010;10:89. doi: 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, Heyland A, Moroz LL, Copley RR, Telford MJ. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Brown FD, Prendergast A, Swalla BJ. Man is but a worm: chordate origins. Genesis. 2008;46:605–613. doi: 10.1002/dvg.20471. [DOI] [PubMed] [Google Scholar]
- Budd GE. Why are arthropods segmented? Evolution & development. 2001;3:332–342. doi: 10.1046/j.1525-142x.2001.01041.x. [DOI] [PubMed] [Google Scholar]
- Bullock T. The nervous system of hemichordates. In: Bullock T, Horridge G, editors. Structure and function in the nervous systems of invertebrates. WH Freeman Comp. Ltd; San Francisco: 1965. pp. 1567–1157t. [Google Scholar]
- Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nature neuroscience. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Campo-Paysaa F, Semon M, Cameron RA, Peterson KJ, Schubert M. microRNA complements in deuterostomes: origin and evolution of microRNAs. Evolution & development. 2011;13:15–27. doi: 10.1111/j.1525-142X.2010.00452.x. [DOI] [PubMed] [Google Scholar]
- Chattwood A, Nagayama K, Bolourani P, Harkin L, Kamjoo M, Weeks G, Thompson CR. Developmental lineage priming in Dictyostelium by heterogeneous Ras activation. eLife. 2013;2 doi: 10.7554/eLife.01067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Dai F, Balakrishnan-Renuka A, Leese F, Schempp W, Schaller F, Hoffmann MM, Morosan-Puopolo G, Yusuf F, Bisschoff IJ, Chankiewitz V, Xue J, Chen J, Ying K, Brand-Saberi B. Diversification and molecular evolution of ATOH8, a gene encoding a bHLH transcription factor. PloS one. 2011;6:e23005. doi: 10.1371/journal.pone.0023005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Streit A. Induction of the inner ear: stepwise specification of otic fate from multipotent progenitors. Hear Res. 2013;297:3–12. doi: 10.1016/j.heares.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature neuroscience. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox BC, Chai R, Lenoir A, Liu Z, Zhang L, Nguyen DH, Chalasani K, Steigelman KA, Fang J, Cheng AG, Zuo J. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development. 2014;141:816–829. doi: 10.1242/dev.103036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, Pevny LH, Kelley MW. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. The regulatory genome: gene regulatory networks in development and evolution. Academic Press; 2010. [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Developmental biology. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS. Early evolution of metazoan transcription factors. Current opinion in genetics & development. 2009;19:591–599. doi: 10.1016/j.gde.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Dekkers MP, Barde YA. Developmental biology. Programmed cell death in neuronal development. Science. 2013;340:39–41. doi: 10.1126/science.1236152. [DOI] [PubMed] [Google Scholar]
- Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- Dias AS, de Almeida I, Belmonte JM, Glazier JA, Stern CD. Somites without a clock. Science. 2014;343:791–795. doi: 10.1126/science.1247575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniach T, Phillips CR, Gerhart JC. Planar induction of anteroposterior pattern in the developing central nervous system of Xenopus laevis. Science. 1992;257:542–545. doi: 10.1126/science.1636091. [DOI] [PubMed] [Google Scholar]
- Dufour HD, Chettouh Z, Deyts C, de Rosa R, Goridis C, Joly JS, Brunet JF. Precraniate origin of cranial motoneurons. Proceedings of the National Academy of Sciences. 2006;103:8727–8732. doi: 10.1073/pnas.0600805103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Fritzsch B. Continued expression of GATA3 is necessary for cochlear neurosensory development. PloS one. 2013;8:e62046. doi: 10.1371/journal.pone.0062046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, McMahon JA, McMahon AP. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- Elliott KL, Houston DW, Fritzsch B. Transplantation of Xenopus laevis tissues to determine the ability of motor neurons to acquire a novel target. PLoS One. 2013;8:e55541. doi: 10.1371/journal.pone.0055541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, Manning G, Lang BF, Haas B, Nusbaum C, King N. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest MP, Hill MJ, Quantock AJ, Martin-Rendon E, Blake DJ. The emerging roles of TCF4 in disease and development. Trends in molecular medicine. 2014 doi: 10.1016/j.molmed.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Fortunato SA, Leininger S, Adamska M. Evolution of the Pax-Six-Eya-Dach network: the calcisponge case study. EvoDevo. 2014;5:23. doi: 10.1186/2041-9139-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B. Of mice and genes: evolution of vertebrate brain development. Brain Behav Evol. 1998;52:207–217. doi: 10.1159/000006564. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Beisel KW, Pauley S, Soukup G. Molecular evolution of the vertebrate mechanosensory cell and ear. Int J Dev Biol. 2007;51:663–678. doi: 10.1387/ijdb.072367bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10-year-old hypothesis. Cell Mol Life Sci. 2010;67:3089–3099. doi: 10.1007/s00018-010-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Glover JC. Evolution of the deuterostome central nervous system: an intercalation of developmental patterning processes with cellular specification processes. In: Kaas JH, editor. Evolution of the Nervous System. Vol. 2. Academic Press; Oxford: 2006. pp. 1–24. [Google Scholar]
- Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, Beisel KW, Wang VY. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Northcutt RG. Cranial and spinal nerve organization in amphioxus and lampreys: evidence for an ancestral craniate pattern. Acta Anat (Basel) 1993;148:96–109. doi: 10.1159/000147529. [DOI] [PubMed] [Google Scholar]
- Fritzsch B, Pan N, Jahan I, Duncan JS, Kopecky BJ, Elliott KL, Kersigo J, Yang T. Evolution and development of the tetrapod auditory system: an organ of Corti-centric perspective. Evol Dev. 2013;15:63–79. doi: 10.1111/ede.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Straka H. Evolution of vertebrate mechanosensory hair cells and inner ears: toward identifying stimuli that select mutation driven altered morphologies. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology. 2013 doi: 10.1007/s00359-013-0865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Straka H. Evolution of vertebrate mechanosensory hair cells and inner ears: toward identifying stimuli that select mutation driven altered morphologies. Journal of comparative physiology A, Neuroethology, sensory, neural, and behavioral physiology. 2014;200:5–18. doi: 10.1007/s00359-013-0865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Tessarollo L, Coppola E, Reichardt LF. Neurotrophins in the ear: their roles in sensory neuron survival and fiber guidance. Prog Brain Res. 2004;146:265–278. doi: 10.1016/S0079-6123(03)46017-2. [DOI] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. Neural Crest and the Origin of Vertebrates: A New Head. Science. 1983;220:268–273. doi: 10.1126/science.220.4594.268. [DOI] [PubMed] [Google Scholar]
- Gokoffski KK, Wu HH, Beites CL, Kim J, Kim EJ, Matzuk MM, Johnson JE, Lander AD, Calof AL. Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development. 2011;138:4131–4142. doi: 10.1242/dev.065870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan R, Shen N, Dror I, Zhou T, Horton J, Rohs R, Bulyk ML. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell reports. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth SI, Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyoja F. A genome-wide survey of bHLH transcription factors in the Placozoan Trichoplax adhaerens reveals the ancient repertoire of this gene family in metazoan. Gene. 2014;542:29–37. doi: 10.1016/j.gene.2014.03.024. [DOI] [PubMed] [Google Scholar]
- Hallbook F, Wilson K, Thorndyke M, Olinski RP. Formation and evolution of the chordate neurotrophin and Trk receptor genes. Brain Behav Evol. 2006;68:133–144. doi: 10.1159/000094083. [DOI] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Muller CH. A new look at the ventral nerve centre of Sagitta: implications for the phylogenetic position of Chaetognatha (arrow worms) and the evolution of the bilaterian nervous system. Front Zool. 2007;4:14. doi: 10.1186/1742-9994-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. Coordinated scaling of cortical and cerebellar numbers of neurons. Frontiers in neuroanatomy. 2010;4 doi: 10.3389/fnana.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. The remarkable, yet not extraordinary, human brain as a scaled-up primate brain and its associated cost. Proceedings of the National Academy of Sciences. 2012;109:10661–10668. doi: 10.1073/pnas.1201895109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. The Plant cell. 2012;24:4483–4497. doi: 10.1105/tpc.112.105023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Kageyama R. bHLH factors in self-renewal, multipotency, and fate choice of neural progenitor cells. Neuron. 2014;82:9–23. doi: 10.1016/j.neuron.2014.03.018. [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & development. 2001;15:3059–3087. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Calisto LE, Morris KA, Kopecky B, Duncan JS, Beisel KW, Fritzsch B. Expression of Neurog1 instead of Atoh1 can partially rescue organ of Corti cell survival. PloS one. 2012;7:e30853. doi: 10.1371/journal.pone.0030853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PloS one. 2010;5:e11661. doi: 10.1371/journal.pone.0011661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan I, Pan N, Kersigo J, Fritzsch B. Beyond generalized hair cells: molecular cues for hair cell types. Hear Res. 2013;297:30–41. doi: 10.1016/j.heares.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Desplan C. Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science. 2014;343:661–665. doi: 10.1126/science.1243039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersigo J, D'Angelo A, Gray BD, Soukup GA, Fritzsch B. The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre-mediated microRNA loss. Genesis. 2011;49:326–341. doi: 10.1002/dvg.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–325. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky BJ, Jahan I, Fritzsch B. Correct timing of proliferation and differentiation is necessary for normal inner ear development and auditory hair cell viability. Developmental Dynamics. 2013;242:132–147. doi: 10.1002/dvdy.23910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Schmid T, Kruger S, Bober E, Braun T. Functional redundancy of NSCL-1 and NeuroD during development of the petrosal and vestibulocochlear ganglia. Eur J Neurosci. 2006;24:1581–1590. doi: 10.1111/j.1460-9568.2006.05051.x. [DOI] [PubMed] [Google Scholar]
- Lai H, Meredith D, Johnson E. bHLH Factors in Neurogenesis and Neuronal Subtype Specification. Patterning and Cell Type Specification in the Developing CNS and PNS: Comprehensive Developmental Neuroscience. 2013;1:333. [Google Scholar]
- Lamb TD. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog Retin Eye Res. 2013 doi: 10.1016/j.preteyeres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Lamborghini JE. Rohon‐beard cells and other large neurons in Xenopus embryos originate during gastrulation. Journal of Comparative Neurology. 1980;189:323–333. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Levin TC, Greaney AJ, Wetzel L, King N, Sánchez Alvarado A. The rosetteless gene controls development in the choanoflagellate S. rosetta. eLife. 2014;3 doi: 10.7554/eLife.04070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Danilova T, Palm E, Lindholm P, Voikar V, Hakonen E, Ustinov J, Andressoo JO, Harvey BK, Otonkoski T, Rossi J, Saarma M. MANF is indispensable for the proliferation and survival of pancreatic beta cells. Cell reports. 2014;7:366–375. doi: 10.1016/j.celrep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm P, Saarma M. Novel CDNF/MANF family of neurotrophic factors. Dev Neurobiol. 2010;70:360–371. doi: 10.1002/dneu.20760. [DOI] [PubMed] [Google Scholar]
- Lindstrom R, Lindholm P, Kallijarvi J, Yu LY, Piepponen TP, Arumae U, Saarma M, Heino TI. Characterization of the structural and functional determinants of MANF/CDNF in Drosophila in vivo model. PloS one. 2013;8:e73928. doi: 10.1371/journal.pone.0073928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Fang J, Dearman J, Zhang L, Zuo J. In vivo generation of immature inner hair cells in neonatal mouse cochleae by ectopic atoh1 expression. PloS one. 2014;9:e89377. doi: 10.1371/journal.pone.0089377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ. Molecular genetic insights into deuterostome evolution from the direct-developing hemichordate Saccoglossus kowalevskii. Philos Trans R Soc Lond B Biol Sci. 2008;363:1569–1578. doi: 10.1098/rstb.2007.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Terasaki M, Wu M, Freeman RM, Jr, Runft L, Kwan K, Haigo S, Aronowicz J, Lander E, Gruber C, Smith M, Kirschner M, Gerhart J. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 2006;4:e291. doi: 10.1371/journal.pbio.0040291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe CJ, Wu M, Salic A, Evans L, Lander E, Stange-Thomann N, Gruber CE, Gerhart J, Kirschner M. Anteroposterior patterning in hemichordates and the origins of the chordate nervous system. Cell. 2003;113:853–865. doi: 10.1016/s0092-8674(03)00469-0. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Magie CR, Daly M, Martindale MQ. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Developmental biology. 2007;305:483–497. doi: 10.1016/j.ydbio.2007.02.044. [DOI] [PubMed] [Google Scholar]
- Mak AC, Szeto IY, Fritzsch B, Cheah KS. Differential and overlapping expression pattern of SOX2 and SOX9 in inner ear development. Gene expression patterns : GEP. 2009;9:444–453. doi: 10.1016/j.gep.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Reiprich S, Wegner M, Fritzsch B. Targeted deletion of Sox10 by Wnt1-cre defects neuronal migration and projection in the mouse inner ear. PloS one. 2014;9:e94580. doi: 10.1371/journal.pone.0094580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Arkowitz RA. Cell polarization in budding and fission yeasts. FEMS microbiology reviews. 2014 doi: 10.1111/1574-6976.12055. [DOI] [PubMed] [Google Scholar]
- Martindale MQ. Evolution of development: the details are in the entrails. Current Biology. 2013;23:R25–R28. doi: 10.1016/j.cub.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale MQ, Lee PN. The development of form: Causes and consequences of developmental reprogramming associated with rapid body plan evolution in the bilaterian radiation. Biological Theory. 2013;8:253–264. [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, Morris K, Feng F, Jones K, Lee J, Fritzsch B. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovic T, Siebert M, Knoche E, Depner H, Mertel S, Owald D, Schmidt M, Thomas U, Sickmann A, Kamin D. The Bruchpilot cytomatrix determines the size of the readily releasable pool of synaptic vesicles. The Journal of cell biology. 2013;202:667–683. doi: 10.1083/jcb.201301072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Daly M, Martindale MQ. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evolution & development. 2007a;9:25–38. doi: 10.1111/j.1525-142X.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. FGF signaling in gastrulation and neural development in Nematostella vectensis, an anthozoan cnidarian. Dev Genes Evol. 2007b;217:137–148. doi: 10.1007/s00427-006-0122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza ME, Pang K, Martindale MQ, Finnerty JR. Genomic organization, gene structure, and developmental expression of three clustered otx genes in the sea anemone Nematostella vectensis. J Exp Zoolog B Mol Dev Evol. 2007 doi: 10.1002/jez.b.21158. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Different strategies for midline formation in bilaterians. Nat Rev Neurosci. 2004;5:502–510. doi: 10.1038/nrn1410. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Pattern Formation in Morphogenesis. Springer; 2013. From Hydra to Vertebrates: Models for the Transition from Radial-to Bilateral-Symmetric Body Plans; pp. 207–224. [Google Scholar]
- Mizutari K, Fujioka M, Hosoya M, Bramhall N, Okano HJ, Okano H, Edge AS. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron. 2013;77:58–69. doi: 10.1016/j.neuron.2012.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Pasqualetti M, Takio Y, Hirano S, Rijli FM, Kuratani S. Segmental development of reticulospinal and branchiomotor neurons in lamprey: insights into the evolution of the vertebrate hindbrain. Development. 2004;131:983–995. doi: 10.1242/dev.00986. [DOI] [PubMed] [Google Scholar]
- Murakami Y, Uchida K, Rijli FM, Kuratani S. Evolution of the brain developmental plan: Insights from agnathans. Developmental biology. 2005;280:249–259. doi: 10.1016/j.ydbio.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Jahan I, Bonde G, Sun X, Hildebrand MS, Engelhardt JF, Smith RJ, Cornell RA, Fritzsch B, Bánfi B. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS genetics. 2012;8:e1002966. doi: 10.1371/journal.pgen.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neriec N, Desplan C. Different ways to make neurons: parallel evolution in the SoxB family. Genome Biol. 2014;15:116. doi: 10.1186/gb4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SA. Form and function remixed: developmental physiology in the evolution of vertebrate body plans. J Physiol. 2014;592:2403–2412. doi: 10.1113/jphysiol.2014.271437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SA, Bhat R. Dynamical patterning modules: a “pattern language” for development and evolution of multicellular form. Int J Dev Biol. 2009;53:693–705. doi: 10.1387/ijdb.072481sn. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R. Deuterostome brains: synopsis and commentary. Brain Res Bull. 2002;57:257–270. doi: 10.1016/s0361-9230(01)00668-2. [DOI] [PubMed] [Google Scholar]
- Nieuwenhuys R, Donkelaar HJ, Nicholson C, Smeets W, Wicht H. The central nervous system of vertebrates. Springer; Berlin etc: 1998. [Google Scholar]
- Northcutt RG. Evolution of centralized nervous systems: two schools of evolutionary thought. Proceedings of the National Academy of Sciences. 2012;109:10626–10633. doi: 10.1073/pnas.1201889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcutt RG, Gans C. The genesis of neural crest and epidermal placodes: a reinterpretation of vertebrate origins. Q Rev Biol. 1983;58:1–28. doi: 10.1086/413055. [DOI] [PubMed] [Google Scholar]
- Nosenko T, Schreiber F, Adamska M, Adamski M, Eitel M, Hammel J, Maldonado M, Muller WE, Nickel M, Schierwater B, Vacelet J, Wiens M, Worheide G. Deep metazoan phylogeny: when different genes tell different stories. Molecular phylogenetics and evolution. 2013;67:223–233. doi: 10.1016/j.ympev.2013.01.010. [DOI] [PubMed] [Google Scholar]
- O'Neill P, Mak SS, Fritzsch B, Ladher RK, Baker CV. The amniote paratympanic organ develops from a previously undiscovered sensory placode. Nature communications. 2012;3:1041. doi: 10.1038/ncomms2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- Oppenheim R. Cell death during development of the nervous system. Annual review of neuroscience. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- Osigus HJ, Eitel M, Schierwater B. Chasing the urmetazoon: striking a blow for quality data? Molecular phylogenetics and evolution. 2013;66:551–557. doi: 10.1016/j.ympev.2012.05.028. [DOI] [PubMed] [Google Scholar]
- Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 2009;337:407–428. doi: 10.1007/s00441-009-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan N, Kopecky B, Jahan I, Fritzsch B. Understanding the evolution and development of neurosensory transcription factors of the ear to enhance therapeutic translation. Cell Tissue Res. 2012;349:415–432. doi: 10.1007/s00441-012-1454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani AM, Mullarkey EE, Aronowicz J, Assimacopoulos S, Grove EA, Lowe CJ. Ancient deuterostome origins of vertebrate brain signalling centres. Nature. 2012;483:289–294. doi: 10.1038/nature10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patthey C, Schlosser G, Shimeld SM. The evolutionary history of vertebrate cranial placodes - I: Cell type evolution. Developmental biology. 2014 doi: 10.1016/j.ydbio.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Paulin MG. Springer Handbook of Bio/Neuroinformatics. Springer; 2014. Pattern Formation and Animal Morphogenesis; pp. 73–92. [Google Scholar]
- Peterson KJ, Dietrich MR, McPeek MA. MicroRNAs and metazoan macroevolution: insights into canalization, complexity, and the Cambrian explosion. Bioessays. 2009;31:736–747. doi: 10.1002/bies.200900033. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Su YH, Arnone MI, Swalla B, King BL. MicroRNAs support the monophyly of enteropneust hemichordates. Journal of experimental zoology Part B, Molecular and developmental evolution. 2013;320:368–374. doi: 10.1002/jez.b.22510. [DOI] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evolution & development. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L, Harrison M, Paxinos G, Watson C. A developmental ontology for the mammalian brain based on the prosomeric model. Trends in neurosciences. 2013;36:570–578. doi: 10.1016/j.tins.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:714–722. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV, Jurka J, Genikhovich G, Grigoriev IV, Lucas SM, Steele RE, Finnerty JR, Technau U, Martindale MQ, Rokhsar DS. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Raft S, Groves AK. Segregating neural and mechanosensory fates in the developing ear: patterning, signaling, and transcriptional control. Cell Tissue Res. 2014 doi: 10.1007/s00441-014-1917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, Johnson JE, Segil N, Groves AK. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- Raikova OI, Reuter M, Jondelius U, Gustafsson MKS. An immunocytochemical and utrastructural study of the nervous and muscular system of Xenoturbella westbladi (Bilateria inc. sed.) Zoomorphology. 2000;120:107–118. [Google Scholar]
- Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O'Hanlon D, Lin ZY, Chen GI, Easton LE, Ule J, Gingras AC. A Global Regulatory Mechanism for Activating an Exon Network Required for Neurogenesis. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H. Evolutionary conservation of mechanisms for neural regionalization, proliferation and interconnection in brain development. Biol Lett. 2009;5:112–116. doi: 10.1098/rsbl.2008.0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H, Simeone A. Conserved usage of gap and homeotic genes in patterning the CNS. Current opinion in neurobiology. 1999;9:589–595. doi: 10.1016/S0959-4388(99)00002-1. [DOI] [PubMed] [Google Scholar]
- Reiprich S, Wegner M. From CNS stem cells to neurons and glia: Sox for everyone. Cell Tissue Res. 2014 doi: 10.1007/s00441-014-1909-6. [DOI] [PubMed] [Google Scholar]
- Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–1161. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- Rosengauer E, Hartwich H, Hartmann AM, Rudnicki A, Satheesh SV, Avraham KB, Nothwang HG. Egr2:: cre mediated conditional ablation of dicer disrupts histogenesis of mammalian central auditory nuclei. PloS one. 2012;7:e49503. doi: 10.1371/journal.pone.0049503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roybon L, Hjalt T, Stott S, Guillemot F, Li JY, Brundin P. Neurogenin2 directs granule neuroblast production and amplification while NeuroD1 specifies neuronal fate during hippocampal neurogenesis. PloS one. 2009;4:e4779. doi: 10.1371/journal.pone.0004779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rue P, Garcia-Ojalvo J. Modeling gene expression in time and space. Annual review of biophysics. 2013;42:605–627. doi: 10.1146/annurev-biophys-083012-130335. [DOI] [PubMed] [Google Scholar]
- Ryan JF. Did the ctenophore nervous system evolve independently? Zoology. 2014;117:225–226. doi: 10.1016/j.zool.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Smith SA. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar-Ciudad I, Jernvall J, Newman SA. Mechanisms of pattern formation in development and evolution. Development. 2003;130:2027–2037. doi: 10.1242/dev.00425. [DOI] [PubMed] [Google Scholar]
- Sasai N, Yakura R, Kamiya D, Nakazawa Y, Sasai Y. Ectodermal factor restricts mesoderm differentiation by inhibiting p53. Cell. 2008;133:878–890. doi: 10.1016/j.cell.2008.03.035. [DOI] [PubMed] [Google Scholar]
- Satoh N. An aboral-dorsalization hypothesis for chordate origin. Genesis. 2008;46:614–622. doi: 10.1002/dvg.20416. [DOI] [PubMed] [Google Scholar]
- Satterlie RA. Do jellyfish have central nervous systems? The Journal of experimental biology. 2011;214:1215–1223. doi: 10.1242/jeb.043687. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. Evolution of the neural crest viewed from a gene regulatory perspective. Genesis. 2008;46:673–682. doi: 10.1002/dvg.20436. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Patthey C, Shimeld SM. The evolutionary history of vertebrate cranial placodes II. Evolution of ectodermal patterning. Developmental biology. 2014 doi: 10.1016/j.ydbio.2014.01.019. [DOI] [PubMed] [Google Scholar]
- Schnitzler CE, Simmons DK, Pang K, Martindale MQ, Baxevanis AD. Expression of multiple Sox genes through embryonic development in the ctenophore Mnemiopsis leidyi is spatially restricted to zones of cell proliferation. EvoDevo. 2014;5:15. doi: 10.1186/2041-9139-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Irimia M, Del Campo J, Parra-Acero H, Russ C, Nusbaum C, Blencowe BJ, Ruiz-Trillo I. Regulated aggregative multicellularity in a close unicellular relative of metazoa. Elife. 2013;2:e01287. doi: 10.7554/eLife.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipel K, Yanze N, Schmid V. Developmental and evolutionary aspects of the basic helix-loop-helix transcription factors Atonal-like 1 and Achaete-scute homolog 2 in the jellyfish. Developmental biology. 2004;269:331–345. doi: 10.1016/j.ydbio.2004.01.035. [DOI] [PubMed] [Google Scholar]
- Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- Silos-Santiago I, Fagan AM, Garber M, Fritzsch B, Barbacid M. Severe sensory deficits but normal CNS development in newborn mice lacking TrkB and TrkC tyrosine protein kinase receptors. The European journal of neuroscience. 1997;9:2045–2056. doi: 10.1111/j.1460-9568.1997.tb01372.x. [DOI] [PubMed] [Google Scholar]
- Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC evolutionary biology. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Developmental biology. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, LeBon L, Garcia-Ojalvo J, Elowitz MB. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLoS computational biology. 2011;7:e1002069. doi: 10.1371/journal.pcbi.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Hu JS, Currle DS, Fung ES, Hayes WB, Lander AD, Monuki ES. A BMP-FGF Morphogen Toggle Switch Drives the Ultrasensitive Expression of Multiple Genes in the Developing Forebrain. PLoS computational biology. 2014;10:e1003463. doi: 10.1371/journal.pcbi.1003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanelli A. Studies on the development of Mauthner's cell. Genetic Neurology. 1950:161–165. [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, Haugen E, Heimfeld S, Murry CE, Akey JM, Stamatoyannopoulos JA. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DL. The genetic causes of convergent evolution. Nature reviews Genetics. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- Steventon B, Mayor R, Streit A. Neural crest and placode interaction during the development of the cranial sensory system. Developmental biology. 2014 doi: 10.1016/j.ydbio.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straka H, Fritzsch B, Glover JC. Connecting ears to eye muscles: evolution of a ‘simple’ reflex arc. Brain Behav Evol. 2014;83:162–175. doi: 10.1159/000357833. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406:74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- Streit A, Tambalo M, Chen J, Grocott T, Anwar M, Sosinsky A, Stern CD. Experimental approaches for gene regulatory network construction: the chick as a model system. Genesis. 2013;51:296–310. doi: 10.1002/dvg.22359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter GF. Principles of brain evolution. Sinauer Associates; 2005. [Google Scholar]
- Südhof TC. The presynaptic active zone. Neuron. 2012;75:11–25. doi: 10.1016/j.neuron.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC, Rizo J. Synaptic vesicle exocytosis. Cold Spring Harbor perspectives in biology. 2011;3:a005637. doi: 10.1101/cshperspect.a005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sánchez-Pons N. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nature communications. 2013;4 doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalla BJ, Smith AB. Deciphering deuterostome phylogeny: molecular, morphological and palaeontological perspectives. Philos Trans R Soc Lond B Biol Sci. 2008;363:1557–1568. doi: 10.1098/rstb.2007.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swalla BJ, Xavier-Neto J. Chordate origins and evolution. Genesis. 2008;46:575–579. doi: 10.1002/dvg.20477. [DOI] [PubMed] [Google Scholar]
- Tamm SL. Cilia and the life of ctenophores. Invertebrate Biology 2014 [Google Scholar]
- Taylor AR, Gifondorwa DJ, Robinson MB, Strupe JL, Prevette D, Johnson JE, Hempstead B, Oppenheim RW, Milligan CE. Motoneuron programmed cell death in response to proBDNF. Dev Neurobiol. 2012;72:699–712. doi: 10.1002/dneu.20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Neuroblast formation and patterning during early brain development in Drosophila. Bioessays. 2004;26:739–751. doi: 10.1002/bies.20062. [DOI] [PubMed] [Google Scholar]
- von Bartheld CS, Fritzsch B. Comparative analysis of neurotrophin receptors and ligands in vertebrate neurons: tools for evolutionary stability or changes in neural circuits? Brain Behav Evol. 2006;68:157–172. doi: 10.1159/000094085. [DOI] [PubMed] [Google Scholar]
- Wagner A. The molecular origins of evolutionary innovations. Trends in genetics : TIG. 2011;27:397–410. doi: 10.1016/j.tig.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Wang VY, Hassan BA, Bellen HJ, Zoghbi HY. Drosophila atonal fully rescues the phenotype of Math1 null mice: new functions evolve in new cellular contexts. Curr Biol. 2002;12:1611–1616. doi: 10.1016/s0960-9822(02)01144-2. [DOI] [PubMed] [Google Scholar]
- Weiler S, Krinner S, Wong AB, Moser T, Pangrsic T. ATP Hydrolysis Is Critically Required for Function of CaV1.3 Channels in Cochlear Inner Hair Cells via Fueling Ca2+ Clearance. J Neurosci. 2014;34:6843–6848. doi: 10.1523/JNEUROSCI.4990-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BG, Mandad S, Truckenbrodt S, Krohnert K, Schafer C, Rammner B, Koo SJ, Classen GA, Krauss M, Haucke V, Urlaub H, Rizzoli SO. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- Wolber LE, Steves CJ, Tsai PC, Deloukas P, Spector TD, Bell JT, Williams FM. Epigenome-Wide DNA Methylation in Hearing Ability: New Mechanisms for an Old Problem. PloS one. 2014;9:e105729. doi: 10.1371/journal.pone.0105729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Wang YJ, Liang Y, Niu QK, Tan XY, Chu LC, Chen LQ, Zhang XQ, Ye D. The ARID-HMG DNA-binding protein AtHMGB15 is required for pollen tube growth in Arabidopsis thaliana. The Plant journal : for cell and molecular biology. 2014;79:741–756. doi: 10.1111/tpj.12582. [DOI] [PubMed] [Google Scholar]
- Xia T, Tong J, Rathore SS, Gu X, Dickerson JA. Network motif comparison rationalizes Sec1/Munc18-SNARE regulation mechanism in exocytosis. BMC systems biology. 2012;6:19. doi: 10.1186/1752-0509-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A, Pang K, Martindale MQ, Tochinai S. Surprisingly complex T-box gene complement in diploblastic metazoans. Evolution & development. 2007;9:220–230. doi: 10.1111/j.1525-142X.2007.00154.x. [DOI] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–496. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Current opinion in neurobiology. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zine A, Löwenheim H, Fritzsch B. Adult Stem Cells. Springer; New York: 2014. Toward Translating Molecular Ear Development to Generate Hair Cells from Stem Cells; pp. 111–161. [Google Scholar]
- Zou D, Erickson C, Kim EH, Jin D, Fritzsch B, Xu PX. Eya1 gene dosage critically affects the development of sensory epithelia in the mammalian inner ear. Human molecular genetics. 2008;17:3340–3356. doi: 10.1093/hmg/ddn229. [DOI] [PMC free article] [PubMed] [Google Scholar]


