Abstract
Autophagy is a conserved process involved in lysosomal degradation of protein aggregates and damaged organelles. The role of autophagy in cancer is a topic of intense debate, and the underlying mechanism is still not clear. The hypoxia inducible factor 2α (HIF2α), an oncogenic transcription factor implicated in renal tumorigenesis, is known to be degraded by the ubiquitin-proteasome system (UPS). Here we report that HIF2α is in part constitutively degraded by autophagy. HIF2α interacts with autophagy-lysosome system components. Inhibition of autophagy increases HIF2α, while induction of autophagy decreases HIF2α. The E3 ligase von Hippel Lindau (VHL) and autophagy receptor protein p62 are required for autophagic degradation of HIF2α. There is a compensatory interaction between the UPS and autophagy in HIF2α degradation. Autophagy inactivation redirects HIF2α to proteasomal degradation, while proteasome inhibition induces autophagy and increases the HIF2α-p62 interaction. Importantly, clear cell renal cell carcinoma (ccRCC) is frequently associated with mono-allelic loss and/or mutation of autophagy related gene ATG7, and low expression level of autophagy genes correlates with ccRCC progression. The protein levels of ATG7 and beclin 1 are also reduced in ccRCC tumors. This study indicates that autophagy plays an anticancer role in ccRCC tumorigenesis, and suggests that constitutive autophagic degradation of HIF2α is a novel tumor suppression mechanism.
Keywords: RCC, autophagy, proteasome, HIF2α, VHL, p62
Introduction
Clear cell renal cell carcinoma (ccRCC) is the most common form of kidney cancer. The majority of cases exhibit von Hippel Lindau (VHL) gene deletions or mutations (1, 2). VHL protein forms an E3 ubiquitin ligase complex with elongins B and C to mediate the ubiquitination of hypoxia inducible factors, HIF1α and HIF2α, and the degradation via the ubiquitin-proteasome system (UPS) (1, 3). In ccRCC, HIF1α functions as a tumor suppressor and its expression is often silenced (1, 4–6), while HIF2α is an oncogenic transcription factor driving the expression of target genes involved in angiogenesis, glycolysis and tumor growth (1, 6, 7). Microenvironmental hypoxia and the genetic inactivation of VHL and other proteasome degradation pathway components result in HIF2α accumulation, strongly contributing towards ccRCC development (8–10).
Macroautophagy (referred as autophagy hereafter) is a major intracellular degradation system responsible for the breakdown of long lived proteins, protein aggregates and damaged organelles (11, 12). Autophagic degradation can also be a selective process mediated by receptor protein p62 and substrate protein ubiquitination (13, 14). Proteasome inhibition induces autophagy and polyubiquitinated protein aggregation (15, 16), indicating that autophagy may compensate for proteasome inactivation. However, the compensatory function of proteasome upregulation during autophagy inactivation is not well studied.
The role of autophagy in cancer is context dependent, and it can be either tumor suppressive or tumor promoting (17). The autophagy gene BECN1 is mono-allelically deleted in human breast and ovarian tumors and functions as a haploinsufficient tumor suppressor in mice (18–20), supporting an anticancer role of autophagy. However, it is necessary to validate the genetic alterations of other autophagy genes in copy number, expression level and mutation frequency in different cancers (17). In ccRCC, multiple mutations were shown to exist in the phosphatidyl inositol 3-kinase pathway, as well as in MTOR itself (21), which may inhibit autophagy. Additionally, autophagy inducers reduce growth of ccRCC cells (22). Based on these findings, we reasoned that autophagy might serve as a tumor suppressive process in ccRCC.
In this study, we show that autophagy collaborates with the UPS to degrade HIF2α. ccRCC is frequently associated with mono-allelic loss and/or mutation of the critical autophagy related gene ATG7, and low expression level of autophagy genes correlates with ccRCC progression.
Results
Inhibition of autophagy results in HIF2α accumulation
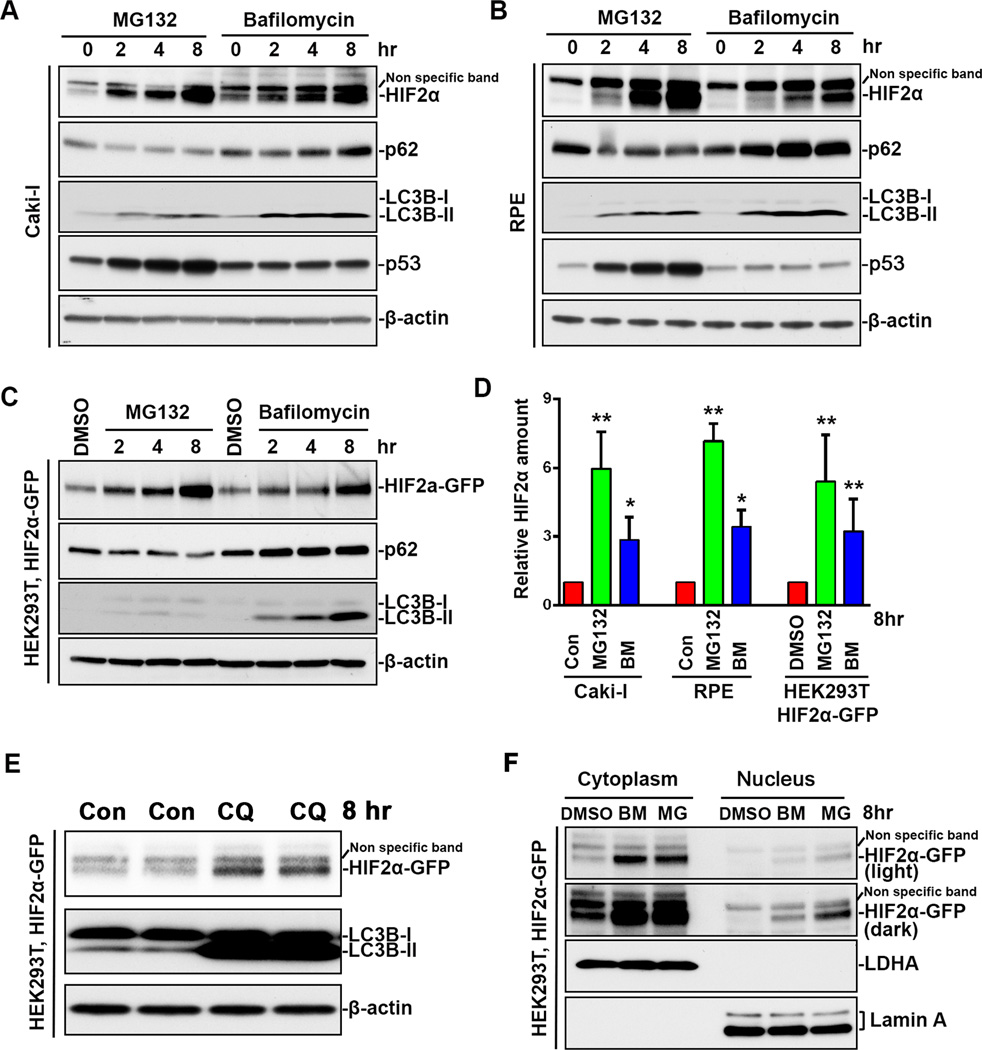
HIF2α is degraded by the proteasome after oxygen-dependent prolyl hydroxylation and VHL-dependent ubiquitination (1, 3). Under normoxia, HIF2α protein levels were very low in the VHL wildtype Caki-I RCC and in the retinal pigment epithelium (RPE) cell lines (Figure 1A, 1B), indicating that HIF2α is constitutively degraded in the presence of intact VHL. When these cell lines were treated with the proteasome inhibitor MG132, there was a gradual accumulation in HIF2α protein levels (Figure 1A, 1B). After 8 hours of treatment, HIF2α levels increased 6–7 fold (Figure 1A, 1B, 1D). These results confirmed that the proteasome was involved in constitutive degradation of HIF2α.
Figure 1.
Both proteasome inhibition and autophagy inhibition induced HIF2α accumulation. (A) Caki-1 cells, (B) RPE cells were treated with 10 µM MG132 or 200 nM bafilomycin A1 for indicated time. (C) HEK293T stable cell line expressing HIF2α-GFP were treated with DMSO, 10 µM MG132 or 200 nM bafilomycin A1 for indicated time. Whole cell lysates were analyzed by immunoblot using antibodies against HIF2α, p62, LC3B, VHL, GFP p53 or β-actin. (D) Quantitation of HIF2α relative amount. HIF2α band intensity was analyzed using ImageJ software. HIF2α levels in non-treated cells or DMSO treated cells were normalized to 1. Data represent mean±S.D., n=3. *, p<0.05, **, p<0.001, compared with control cells. Con, control. MG, MG132. BM, bafilomycin A1. (E) HEK293T stable cell line expressing HIF2α-GFP were treated with 50 µM chloroquine for 8 hrs. CQ, chloroquine. (F) HEK293T stable cell line expressing HIF2α were treated with DMSO, 10 µM MG132 or 200 nM bafilomycin A1 for 8 hrs. Cytoplasmic and nuclear extracts were separated and analyzed by immunoblot using antibodies against GFP, cytoplasm protein LDHA and nuclear membrane protein Lamin A.
Autophagy is another major intracellular degradation system, but its role in HIF2α degradation is unknown. To this end, we treated Caki-I cells and RPE cells with bafilomycin A1, which blocks autophagic degradation by inhibiting autophagosome-lysosome fusion and acidification (23). As a result of the blockage of autophagic flux, bafilomycin A1 treatment led to accumulation of the autophagy receptor protein p62 and the autophagosome marker protein, lipidated form of LC3 (LC3-II) (23) (Figure 1A, 1B). After 8-hour incubation with bafilomycin A1, HIF2α protein levels increased 3-fold (Figure 1A, 1B, 1D). p53 is another transcription factor that is subjected to polyubiquitination-mediated proteasomal degradation (24). We observed that, in Caki-I and RPE cell lines, p53 was only stabilized by MG132 treatment but not bafilomycin A1 treatment (Figure 1A, 1B). These results indicated that bafilomycin A1 increased HIF2α protein level specifically by inhibiting the autophagy-lysosome system but not by inhibiting or compromising UPS activity. To exclude the possible influence of mRNA transcription and vehicle-treatment on HIF2α protein accumulation, we established a HEK293T stable cell line expressing exogenous HIF2α-GFP and treated cells with DMSO as control. Compared with DMSO, MG132 and bafilomycin A1 induced a gradual accumulation of HIF2α (Figure 1C, 1D). Treatment with chloroquine, another autophagy-lysosome inhibitor, also led to an accumulation of HIF2α (Figure 1E). These results collectively suggest that HIF2α is constitutively degraded not only by the proteasome but also by autophagy under normoxic conditions.
Although HIF2α is known as a transcription factor, we observed that it was mainly localized in the cytoplasm in DMSO-treated HEK293T cells, and it was almost undetectable in nuclear extracts (Figure 1F). These results indicate that, in addition to its role as transcription factor in the nucleus, HIF2α may have an unidentified function in cytoplasm, or is sequestered in the cytoplasm as part of the cellular regulation of HIF-mediated transcription. Interestingly, MG132 and bafilomycin A1 increased HIF2α levels in both the nuclear and the cytoplasmic extracts (Figure 1F). Nuclear membrane protein Lamin A was only found in the nuclear fraction, and the cytoplasmic marker protein lactate dehydrogenase (LDH) was only found in the cytoplasmic fraction, which confirmed that our fractions were pure. These results indicate that, although autophagy degrades proteins in the cytoplasm (11), the accumulated HIF2α in the cytoplasm during autophagy inhibition may also translocate to the nucleus.
Consistent with previous reports that proteasome inhibition activated autophagy (15, 16), MG132 treatment was also observed to induce autophagy in Caki-I, RPE and HEK293T cells, as indicated by the increase in LC3B-II and decrease in p62 (Figure 1A, 1B, 1C). These data suggest that autophagy plays a compensatory role during proteasome inhibition.
VHL is required for HIF2α accumulation during autophagy Inhibition
Proteasome-mediated protein degradation is a selective process, regulated through the high level of specificity of E3 ligases for substrate proteins. Although autophagic degradation was originally described as a non-selective process, it was recently found that protein ubiquitination was also required for degradation of some autophagy substrate proteins (13, 14). Autophagy receptor protein p62 interacts with ubiquitin via its C-terminal UBA domain, and interacts with LC3 via its LIR motif. In this way, p62 recruits and delivers ubiquitinated proteins to the nucleating autophagosome for degradation (13, 14). VHL is the known E3 ligase for HIF2α ubiquitination. Since we observed that HIF2α was subjected to both proteasomal and autophagic degradation (Figure 1), we investigated the requirement of VHL in both degradation processes. To this end, we compared HIF2α stabilization in a VHL-deficient parental 786-O cell line and in a 786-O stable cell line that expresses wild-type VHL.
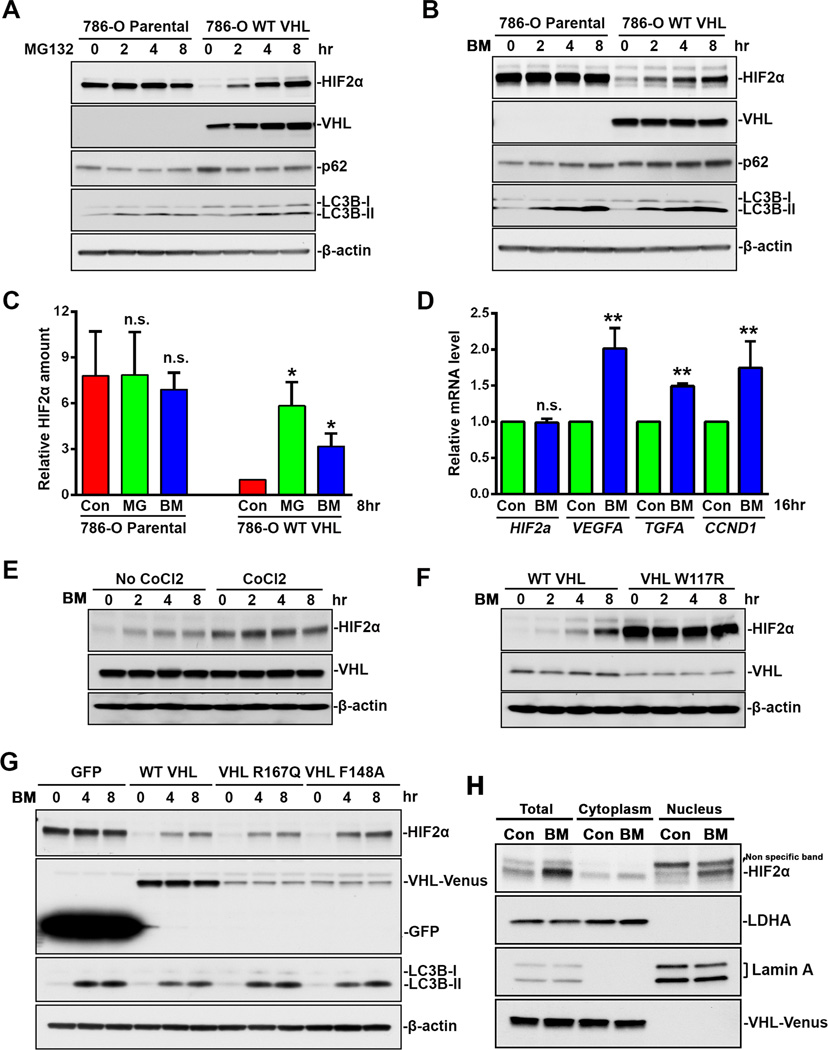
The basal level of HIF2α in 786-O parental cells was relatively high (Figure 2A–2C), and was dramatically reduced with the stable expression of exogenous VHL (Figure 2A–2C). These results indicate that 786-O cells are incapable of degrading HIF2α due to the absence of functional VHL. In 786-O parental cells, neither MG132 nor bafilomycin A1 could significantly increase the already high HIF2α protein levels (Figure 2A–2C). Both agents led to accumulation of HIF2α protein in VHL-expressing 786-O cells (Figure 2A–2C). These results indicate that VHL is required not only for proteasome-mediated HIF2α degradation, but also for autophagy-mediated degradation. In contrast to HIF2α, VHL protein level was only enhanced by MG132 (Figure 2A) but not bafilomycin A1 (Figure 2B), suggesting that VHL was exclusively degraded by the proteasome, and bafilomycin A1-induced HIF2α accumulation was not due to the side effect of proteasomal inhibition.
Figure 2.
VHL-dependent HIF2α accumulation during autophagy inhibition. (A) 786-O parental cells or 786-O stable cell line expressing VHL-wt-Venus were treated with 10 µM MG132 for indicated time. (B) 786-O parental cells or 786-O stable cell line expressing VHL-wt-Venus were treated with 200 nM bafilomycin A1 for indicated time. (C) Quantitation of HIF2α relative amount. HIF2α band intensity was analyzed using ImageJ software. HIF2α levels in control 786-O stable cell line expressing VHL-wt-Venus cells were normalized to 1. Data represent mean±S.D., n=3. **, p<0.001, compared with control cells. (D) HIF2α target gene expression. 786-O stable cell line expressing VHL-wt-Venus were treated with 200 nM bafilomycin A1 for 16 hrs. Total RNA were analyzed by Real-Time PCR using primers specific for HIF2α, VEGFA, TGFA and CCND1. mRNA level in control cells are normalized to 1. Data represent mean±S.D., n=3. **, p<0.001, compared with control cells. (E) 786-O cells were treated with 200 nM bafilomycin A1 for indicated time with or without 8-hr co-treatment with 100 µM CoCl2. (F) 786-O stable cell lines expressing HA-wt-VHL or HA-W117R-VHL were treated with 200 nM bafilomycin A1 for indicated time. (G) 786-O stable cell lines expressing GFP, VHL-wt-Venus, VHL-R167Q-Venus or VHL-F148A-Venus were treated with 200 nM bafilomycin A1 for indicated time. (H) 786-O stable cell lines expressing VHL-wt-Venus were treated with 200 nM bafilomycin A1 for 8 hrs. Cytoplasmic and nuclear extracts were separated. Cell lysates were analyzed by immunoblot using antibodies against HIF2α, p62, LC3B, VHL, GFP, LDHA, LaminA or β-actin. Con, control. MG, MG132. BM, bafilomycin A1.
Consistent with the changes in HIF2α protein level, inhibition of autophagy by bafilomycin A1 also increased the expression of HIF2α target genes, including vascular endothelial growth factor A (VEGFA), transforming growth factor A (TGFA) and cyclin D1 (CCND1), but it did not affect the expression of HIF2α itself (Figure 2D). It should be noted that although HIF2α shares some target genes with HIF1α (3), the 786-O cell line is deficient in HIF1α (5), so the increased target gene expression exclusively represents HIF2α transcriptional activity.
The data above show that VHL was required for autophagic degradation of HIF2α. One possibility is that VHL is required for autophagic activity. However, comparable accumulation of LC3B-II was induced by MG132 and bafilomycin A1 in 786-O cells with or without stably expressed VHL (Figure 1D), which indicated that VHL deficiency did not affect autophagosome formation and subsequent fusion with the lysosome. Another possibility is that HIF2α ubiquitination is required for autophagic degradation, and is dependent on the E3 ligase activity of VHL (1, 3), since p62 is known to interact with and deliver ubiquitinated proteins to the autophagosome (13, 14). To test this possibility, we first prevented VHL-mediated HIF2α ubiquitination using hypoxia mimetic CoCl2 (25). As we expected, CoCl2 treatment stabilized HIF2α but prevented additional accumulation induced by bafilomycin A1 (Figure 2E). Second, we assessed HIF2α levels in 786-O cells expressing the HIF-binding incompetent VHL mutant W117R (26). The 786-O cell line stably expressing VHL W117R showed much higher HIF2α protein levels than those expressing wild type VHL (Figure 2F), indicating its inability to degrade HIF2α. No further increase in HIF2α amount was observed in cells expressing VHL W117R mutant after bafilomycin A1 treatment (Figure 2F). On the other hand, the expression of partially HIF ubiquitination competent VHL R167Q or VHL F148A isoforms (27) in 786-O cells reduced the basal level of HIF2α, and enabled 786-O cells to accumulate HIF2α in response to bafilomycin A1 treatment (Figure 2G). These results showed that pharmacologic or genetic inhibition of E3 ligase activity of VHL blocked bafilomycin A1-induced HIF2α accumulation, and suggested that VHL E3 ligase activity was required for autophagy-mediated HIF2α degradation.
Furthermore, endogenous HIF2α was mainly found in the cytoplasmic fraction of 786-O stable cell line expressing VHL. Bafilomycin A1 treatment increased HIF2α more obviously in the nucleus than in the cytoplasm, while the E3 ligase VHL was only found in the cytoplasm, indicating that the cytoplasm is the location of cellular regulation of HIF, and the accumulated HIF2α during autophagy inactivation translocates to the nucleus. Compared with the results we obtained with HIF2α-GFP (Figure 1F), we suspect that the GFP tag might delay cytoplasmic-nuclear translocation.
Induction of autophagy promotes VHL-dependent HIF2α degradation
The results above show that the inhibition of the autophagy system led to the accumulation of HIF2α. We further studied the changes of HIF2α during autophagy induction. mTOR is a negative regulator of autophagy. mTOR phosphorylates Ulk1 at Ser 757, and prevents Ulk1 activation and autophagy induction (28). mTOR inhibition by starvation or via rapamycin has been reported to induce autophagy (11).
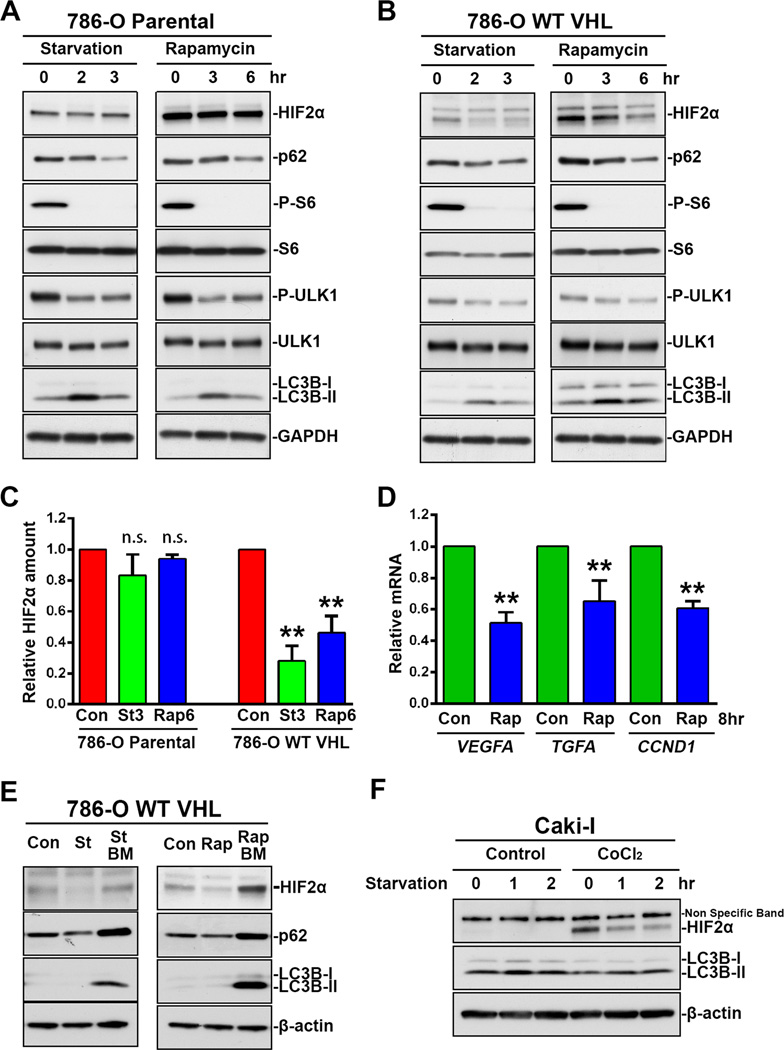
When both 786-O parental cells and 786-O cells stably expressing VHL were exposed to starvation or treated with rapamycin, mTOR activity was inhibited as indicated by the decrease in ribosomal protein S6 phosphorylation and Ulk1 phosphorylation at Ser 757, and autophagy was induced as indicated by the decrease in p62 protein level and the transient increase of LC3B-II (Figure 3A, 3B). However, an obvious decrease in HIF2α protein level was only observed in 786-O cells expressing VHL (Figure 3B, 3C) but not in 786-O parental cells (Figure 3A, 3C), indicating that VHL was required for autophagic degradation of HIF2α during autophagy induction. Consistent with the changes in HIF2α protein level, rapamycin treatment also decreased the expression of HIF2α target genes, including VEGFA, TGFA and CCND1 (Figure 3D). Furthermore, starvation- or rapamycin-induced HIF2α decrease was blocked by the presence of bafilomycin A1 (Figure 3E), confirming that HIF2α degradation was mediated by the autophagy-lysosome system. Unlike HIF2α, the degradation of p62 by autophagy was not significantly influenced by the expression of VHL (Figure 3A, 3B). As an autophagy receptor protein, p62 functions to interact with more than one autophagy substrates, and VHL inactivation is not expected to attenuate p62 degradation significantly. Interestingly, replacement of old DMEM with pre-warmed fresh DMEM decreased HIF2α protein (Starvation 0, Figure 3A, 3B), indicating that HIF2α was unstable, especially in response to environmental stimuli.
Figure 3.
VHL-dependent HIF2α decrease during autophagy induction. (A) 786-O parental cells and (B) 786-O stable cell lines expressing VHL-wt-Venus were cultured in DMEM (0) or EBSS (starvation) for 2 or 3 hrs, or treated with 100 nM rapamycin for 3 or 6 hrs. (C) Quantitation of HIF2α relative amount. HIF2α band intensity was analyzed using ImageJ software. HIF2α levels in control cells were normalized to 1. Data represent mean±S.D., n=3. **, p<0.001, compared with control cells. Con, control. St3, starvation 3hr. Rap6, rapamycin 6 hr. (D) HIF2α target gene expression. 786-O stable cell line expressing VHL-wt-Venus were treated with 100 nM rapamycin for 8 hrs. Total RNA were analyzed by Real-Time PCR using primers specific for VEGFA, TGFA and CCND1. mRNA level in control cells are normalized to 1. Data represent mean±S.D., n=3. **, p<0.001, compared with control cells. Con, control. Rap, rapamycin. (E) 786-O stable cell lines expressing VHL-wt-Venus were treated with 100 nM rapamycin or cultured in EBSS (starvation) in the presence or absence of 200 nM bafilomycin A1. Con, control. St, starvation. Rap, rapamycin. BM, bafilomycin A1. (F) Caki-I cells with or without 6-hr pretreatment with 100 µM CoCl2 were washed with PBS 3 times, and followed by continuous culture in DMEM or EBSS (starvation) for 1 or 2 hrs. Cell lysates were analyzed by immunoblot using antibodies against HIF2α, p62, S6, phosphor-S6, LC3B or β-actin.
Furthermore, we studied the changes in HIF2α in VHL positive Caki-I cells. The basal level of HIF2α in Caki-I cells was too low to detect the further decrease under starvation conditions, but the accumulated HIF2α by CoCl2 pretreatment was also depleted quickly when the cells were followed by continuous culture in starvation medium EBSS (Figure 3F). These results further confirm that HIF2α is in part subjected to autophagic degradation, and E3 ligase VHL is required for this process.
HIF2α interacts with the autophagy-lysosome system components
So far, we have shown that HIF2α is in part constitutively degraded by autophagy under normoxic conditions. Inhibition of autophagy increased HIF2α levels while induction of autophagy decreased HIF2α levels. To further confirm these findings, we examined the interaction between HIF2α and autophagy-lysosome system components.
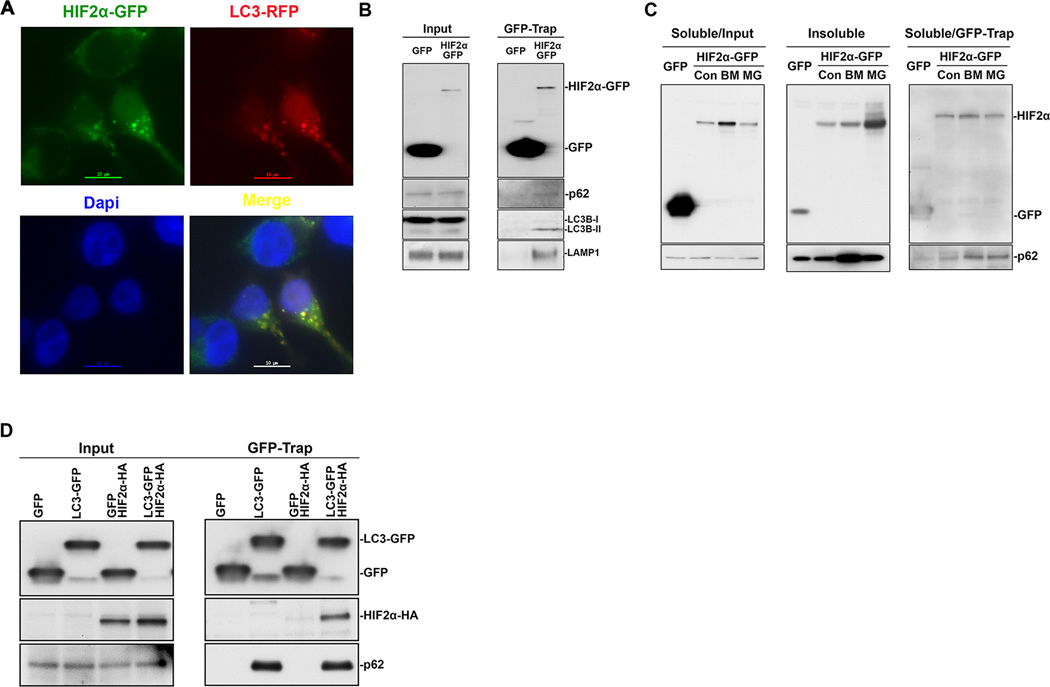
In HEK293T cells, stably expressed HIF2α-GFP was mainly localized to the cytoplasm (Figure 4A), which is consistent with previous fractionation results (Figure 1D). Importantly, some cells contain HIF2α-GFP aggregates (Figure 4A), which are supposed to be degraded by autophagy. We then transiently transfected this cell line with LC3A-RFP, and found that HIF2α-GFP aggregates co-localized with autophagosomes, as indicated by the LC3A-RFP punctate cytoplasmic structures (Figure 4A).
Figure 4.
HIF2α interacted with autophagy and lysosome components. (A) Colocalization between HIF2α-GFP and LC3A-RFP. HEK293T stable cell line expressing HIF2α-GFP were transiently transfected with LC3A-RFP. Representative images with HIF2α-GFP aggregates and autophagosomes were shown. Scale bar, 10um. (B) HEK293T cells were transiently transfected with GFP or HIF2α-GFP. (C) HEK293T cells were transiently transfected with GFP or HIF2α-GFP. After 48 hr, cells were further treated with 200 nM bafilomycin A1 (BM) or 10 µM MG132 (MG) for 6 hr. (D) HEK293T cells were transiently transfected with GFP or LC3A-GFP with or without HIF2α-HA. Cells were lysed in GFB buffer containing 1% Triton X-100. The soluble portion was subjected to immunoprecipation (GFP-Trap) using Chromotek-GFP-Trap beads. The insoluble pellets were dissolved in 1% SDS (Insoluble). The soluble portion (Input) or immunoprecipitated proteins (GFP-Trap) were subjected to immunoblot assay using antibodies against HA, GFP, HIF2α, p62 or LAMP1. Con, control. St3, starvation 3hr. Rap6, rapamycin 6 hr.
Next, we transiently expressed GFP or HIF2α-GFP in HEK293T cells, and immunoprecipitated GFP proteins using a GFP-binding antibody derived from Vicuna pacos. Although the expression and immunoprecipitation of GFP were much higher than those of HIF2α-GFP, endogenous autophagy-lysosome components, including p62, LC3 and lysosome-associated membrane protein 1 (LAMP1), were co-immunoprecipitated with HIF2α-GFP but not with GFP (Figure 4B).
Autophagy inhibition by bafilomycin A1 increased the HIF2α protein level in both Triton X-100 soluble and insoluble fractions, indicating part of HIF2α formed aggregates (Figure 4C). Proteasome inhibition by MG132 also induced a dramatic accumulation of HIF2α, with most of the protein found in the Triton X-100 insoluble fraction (Figure 4C). Although there was no obvious change in p62 in the Triton X-100 soluble fraction, more p62 was co-immunoprecipitated with HIF2α-GFP in the presence of bafilomycin A1 or MG132 (Figure 4C), indicating that proteasome inhibition enhanced the interaction between HIF2α and p62 (Fig. 4C). Next, we transiently expressed GFP or LC3A-GFP with or without HIF2α-HA in HEK293T cells, and observed that HIF2α-HA and endogenous p62 were co-immunoprecipitated with LC3-GFP but not GFP (Figure 4D). These results collectively confirm that HIF2α interacts with autophagy-lysosome system components, and the interaction is enhanced during proteasome inhibition.
Autophagy collaborates with the proteasome to degrade HIF2α
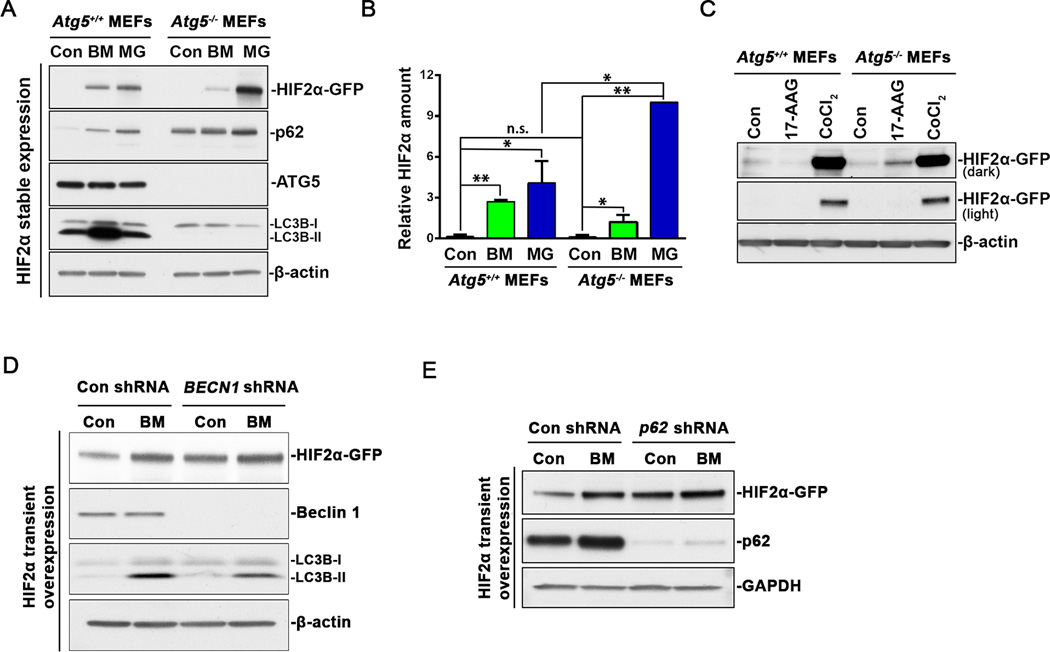
To further investigate the role of autophagy in HIF2α degradation, we studied the turnover of HIF2α in Atg5 knockout mouse embryonic fibroblasts (MEFs). The endogenous murine HIF2α level was either too low to detect even in the presence of MG132 or CoCl2 or it was not recognized by the antibody (NB100-122, Novus Biologicals) (Data not shown). We then established MEF stable cell lines expressing human HIF2α-GFP. In Atg5+/+ MEFs, the basal level of the stably expressed exogenous HIF2α was still almost undetectable, but treatment with bafilomycin A1 or MG132 induced an accumulation of HIF2α, confirming that both autophagy and the proteasome were involved in HIF2α degradation (Figure 5A, 5B). In Atg5−/− MEFs, LC3B-II was undetectable even in the presence of bafilomycin A1, and the baseline of p62 level was much higher than that in Atg5+/+ MEFs (Figure 5A, 5B) (29), indicating inactivation of autophagy by Atg5 knockout. Surprisingly, no obvious increase was observed in stably expressed HIF2α in Atg5−/− MEFs (Figure 5A, 5B). These observations suggest that autophagy inactivation might redirect HIF2α to the proteasome for efficient degradation. As we expected, MG132 treatment increased HIF2α to a much greater extent in Atg5−/− MEFs than in Atg5+/+ MEFs (Figure 5A, 5B), indicating the proteasome was responsible for more HIF2α degradation in Atg5−/− MEFs to compensate for autophagic inactivation. When both proteasomal and autophagic degradation were blocked by CoCl2, the accumulation of HIF2α was comparable in Atg5+/+ and Atg5−/− MEFs (Figure 5C), confirming the expression of HIF2α was not affected by Atg5 knockout. Interestingly, bafilomycin A1 treatment still slightly increased HIF2α protein level in Atg5−/− MEFs, possibly through inhibition of Atg5-independent alternative autophagy (30). Current data indicate that autophagy degrades protein aggregates and the proteasome is responsible for degrading soluble proteins (15). Since autophagy inhibition redirected HIF2α to proteasomal degradation (Figure 5A, 5B), we hypothesized that shuttling by heat shock proteins (HSPs) was probably required for such a redirection process. Treatment with HSP90 inhibitor 17-AAG induced an increase in HIF2α protein level in Atg5−/− MEFs but not in Atg5+/+ MEFs (Figure 5C), implying that HSPs were probably involved in the disaggregation of HIF2α and redirection to proteasomal degradation.
Figure 5.
Atg5 knockout favors proteasomal degradation of HIF2α. (A) Atg5+/+ and Atg5−/− MEFs stably expressing HIF2α-GFP were treated with 10 µM MG132 or 200 nM bafilomycin A1 for 8 hr. (B) Quantitation of HIF2α relative amount. HIF2α band intensity was analyzed using ImageJ software. HIF2α levels in MG132-treated Atg5−/− MEFs were normalized to 10. Con, control. Data represent mean±S.D., n=3. **, p<0.001, compared with same treatment in Atg5+/+ MEFs. Con, control. BM, bafilomycin A1. MG, MG132. (C) Atg5+/+ and Atg5−/− MEFs stably expressing HIF2α-GFP were treated with 2 µM 17-AAG or 100 µM CoCl2 for 8 hr. (D) HEK293T cells stably expressing control shRNA or BECN1 shRNA were transiently transfected with HIF2α-GFP. After 48 hr, cells were further treated with 200 nM bafilomycin A1 (BM) for 6 hr. Con, control. BM, bafilomycin A1. (E) HEK293T cells stably expressing control shRNA or p62 shRNA were transiently transfected with HIF2α-GFP. After 48 hr, cells were further treated with 200 nM bafilomycin A1 (BM) for 6 hr. Con, control. BM, bafilomycin A1. Con, control. BM, bafilomycin A1. MG, MG132. Whole cell lysates were analyzed by immunoblot using antibodies against GFP, p62, ATG5, LC3B, β-actin or GAPDH.
The data described above show that an Atg5 deficiency did not increase stably expressed HIF2α, and this was probably due to the compensatory function of the proteasome. Since protein transient overexpression has been shown to overload the proteasome and thus block compensatory regulation (31), we then studied the turnover of transiently overexpressed HIF2α in BECN1 or p62 knockdown HEK293T cells. Compared with control shRNA, the expression of BECN1 shRNA dramatically reduced beclin1 protein levels and bafilomycin A1-induded LC3B-II accumulation, indicating an efficient knockdown and impairment of autophagy activity (Figure 5D). Importantly, BECN1 knockdown led to an increase in the baseline of transiently overexpressed HIF2α (Figure 5D). The inhibition of autophagic flux by bafilomycin A1 significantly increased HIF2α protein level in control knockdown cells but not in BECN1 knockdown cells (Figure 5D). These data demonstrate that HIF2α turnover is truly autophagy dependent. Similarly, p62 knockdown also increased the basal level of overexpressed HIF2α (Figure 5E), which confirmed the importance of p62 as an autophagy receptor in HIF2a degradation.
The autophagy pathway is altered in human ccRCC samples
It has been reported that oncogenic protein HIF2α level is higher in more advanced ccRCC (9, 32). Our data show that HIF2α was in part degraded by autophagy, and we reasoned that autophagy pathway was probably genetically altered in ccRCC. To explore this possibility, we systematically examined gene expression levels, gene copy number alterations, and mutation rates of autophagy related genes in ccRCCs from the Cancer Genome Atlas (TCGA) database.
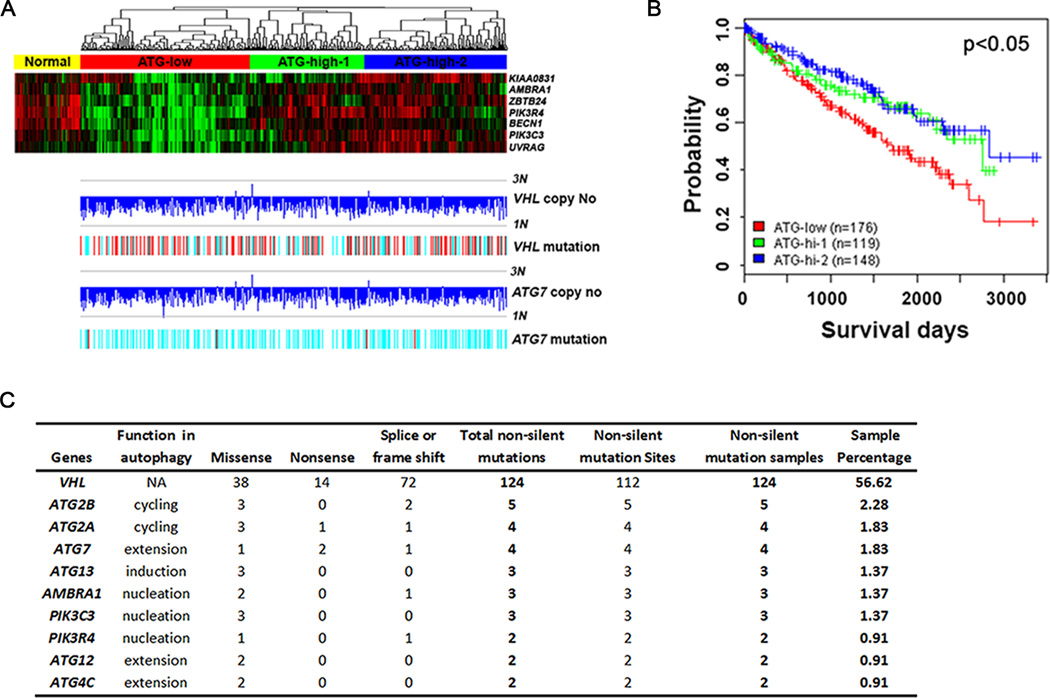
First, we found that the expression of autophagy genes functioning in the nucleation step was coordinated, and varied in ccRCC samples. The class III phosphoinositide 3 kinase (PI3K) complex, which consists of VPS34, VPS15 and beclin 1, is important for autophagosome nucleation. Beclin 1 binding proteins, such as ATG14L, UVRAG, AMBRA1 and BIF1, also positively regulate autophagosome nucleation and maturation (33–35). Using K means grouping of a composite gene expression score for these 7 genes encoding these proteins, we divided ccRCC samples into 3 groups, the autophagy low expressing (ATG-low, 176 tumors), the autophagy high expressing subgroup 1 (ATG-high-1, 119 samples) and the autophagy high expressing subgroup 2 (ATG-high-2, 148 samples) (Figure 6A). The expression level of these genes in normal kidney tissues was higher than that in the ATG-low ccRCC group, while relatively close to that in the ATG-high groups (Figure 6A). These results imply that ccRCCs with low autophagy gene expression might be related to more aggressive tumors. Kaplan-Meier analysis showed that the ccRCCs with low autophagy gene expression had poorer survival, and ccRCCs with high autophagy gene expression had longer survival. There was no obvious difference between two high autophagy gene expression subgroups (Figure 6B).
Figure 6.
Genetic alteration of autophagy genes in ccRCC. (A) Defining ccRCCs into autophagy gene expression subgroups (ATG-low, ATG-high-1 and ATG-high-2) using seven genes involved in autophagy nucleation. Top, gene expression heatmap of 7-gene signature. Samples were ordered by the 7-gene composite score and separated into low and high expression groups by Kmeans. Bottom, corresponding gene copy number changes and mutation of ATG7 and VHL were plotted based on SNP6 data. Copy number levels of 1N and 3N were marked. (B) Kaplan Meier analysis of ccRCC patient survival stratified by autophagy gene expression levels as defined in Fig. 6A. (C) Mutations of autophagy related genes. Data were organized from TCGA level 2 mutation data.
Second, we checked the variation in gene copy number of autophagy genes. ATG7 is one of the key proteins involved in the ubiquitin-like conjugation system that regulates autophagosome extension. Notably, ATG7 is mapped on the p arm of chromosome 3 (3p), where VHL is located. Since most ccRCC tissues are known to harbor loss of 3p, we reasoned that ccRCC was associated with allelic loss of ATG7 at the same time. As we expected, more than 80% of ccRCC tissues lost one copy of VHL and ATG7 (Fig. 6A).
Third, we examined the mutation frequency of autophagy related genes. We found that 56% ccRCC tissues harbored a VHL mutation (Fig. 6A, C). Importantly, 4 out of 219 tumor samples were found to have an ATG7 mutation, and 2 of these were nonsense mutations (Fig. 6C). Such nonsense mutations in ATG7 were also found in lung cancer samples from the TCGA database (data not shown). Furthermore, a list of other autophagy genes involved in different steps also harbored non-silent mutations (Fig. 6C), while such mutations were not found in normal kidney tissues. These data collectively indicate that ccRCC is a heterogeneous tumor, and that autophagy related genes are selectively inactivated in ccRCC at different levels, including copy number loss, decreased mRNA expression and direct mutations.
Importantly, VHL mono-allelic loss and mutations are evenly distributed in three subgroups (Figure 6A). These findings indicate that VHL copy number loss or mutation is a truncal event in the genomic evolution of RCC, and is more important in tumor initiation than in progression. In contrast, it is the variation of autophagy gene expression level that determines patient survival (Figure 6A). Low expression of autophagy genes can be an alternative explanation for HIF2α deregulation in tumors where VHL is not lost or VHL mutants are competent in HIF2α ubiquitination. However, due to the unavailability of HIF2α protein level in TCGA database and the complexity of the regulatory mechanisms of HIF2α and its target genes, it is challenging to generate straightforward evidence to further confirm this correlation.
Autophagy related proteins are reduced in ccRCC samples
The results obtained with the TCGA dataset reveal the mutation and copy number loss of ATG7, and the reduced expression of autophagy genes involved in autophagosome nucleation. To study the changes at a protein level, tissue microarrays that contains triplicated normal kidney samples and ccRCC tumor samples were stained with hematoxylin and eosin (H&E) or immunohistochemical stained with an ATG7 antibody or a beclin1 antibody. Normal kidney tissue showed structures containing proximal tubules and distal tubules, while ccRCC tumors did not contain these structures (Figure 7A). ccRCC samples also showed clear cytoplasm because the intracytoplasmic glycogen and lipids are dissolved during histologic processing (36). Compared with normal kidney tissues, the overall percentage of ATG7 or beclin1 positive cells were significantly reduced in ccRCC (Figure 7A, 7B). These results indicate that the expression of some key autophagy proteins, such as ATG7 and beclin 1, are also reduced at a protein level in ccRCC tissues.
Figure 7.
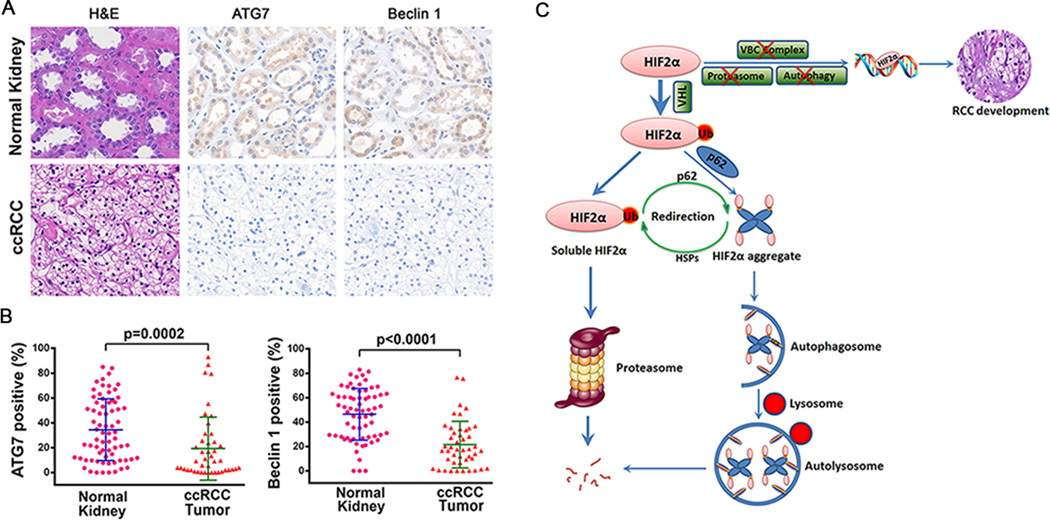
Decreased protein expression of ATG7 and Beclin 1 in ccRCC. Tissue microarrays containing normal kidney and ccRCC sections were subjected to H&E staining or Immunohistochemistry staining using ATG7 antibody or beclin 1 antibody. (A) Representative images were shown. (B) Percentage of ATG7 or beclin 1 positive cells in ccRCC tumors or normal kidneys. Mann-Whitney test was used for statistical analysis. (C) Working model. HIF2α is ubiquitinated by its E3 ligase VHL. Around 2/3 of ubiquitinated HIF2α are soluble and can be degraded by proteasome system. Around 1/3 of ubiquitinated HIF2α interacts with UBA domain of p62 and forms aggregates. HIF2α aggregates are recruited to autophagosome via p62-LC3 interaction and are degraded after fusion with the lysosome. There is a compensatory interaction between the proteasome and autophagy in HIF2α degradation. On the one hand, autophagy inactivation redirects HIF2α to proteasomal degradation, and heat shock proteins are probably involved in refolding of HIF2α aggregates. On the other hand, proteasome inhibition induces autophagy and promotes the HIF2α-p62 interaction and aggregation. When the components involved in VHL-elonginB-C (VBC) complex, proteasome degradation pathway and autophagy degradation pathway are genetically deleted, mutated, or their expression is decreased at the same time, HIF2α cannot be degraded by the proteasome or autophagy. Part of the accumulated HIF2α will translocate to nucleus, drive the expression of its target genes and initiate RCC development.
Discussion
Although it has been widely accepted that there is a relationship between autophagy and cancer, the role of autophagy in tumorigenesis is still a topic of intense debate (17). It is possible that the function of autophagy is context-specific, and it varies depending on tumor type, grade, stage and depth (17). The mono-allelic loss and low expression of BECN1 in breast and ovarian cancers support that autophagy functions as a tumor suppressor in some cases (18). Further comprehensive analyses of other autophagy genes and proteins in different cancers is required (17). In our current study, we examined the genetic alteration of the autophagy pathway in ccRCC, and revealed striking changes in autophagy genes, including ATG7 mono-allelic loss, reduced gene expression and somatic mutations. Importantly, the reduction of autophagy gene expression is associated with shorter patient survival. Taken together with the mono-allelic loss of BECN1, such a broad down-regulation of autophagy related genes provides further support for the tumor suppressive role of autophagy.
Several anti-tumor mechanisms of autophagy have been reported, such as damaged organelle elimination, p62 degradation, genome stabilization, NRF2 and NFkB inactivation, and T lymphocyte attraction (17, 37–39). As a cellular degradation process, it is also likely that autophagy directly degrades oncogenic proteins to suppress tumorigenesis. Here we found that HIF2α, an oncogenic transcription factor that drives RCC tumor initiation and metastasis, was constitutively subjected to autophagic degradation. In support of this conclusion, we found that HIF2α colocalized and interacted with autophagy-lysosome system components. Autophagy inhibition by bafilomycin A1 increased HIF2α, while autophagy induction by starvation or rapamycin decreased HIF2α. These data support the presence of an autophagy-dependent mechanism for suppressing proteins implicated in oncogenesis. It has been widely accepted that HIF2α plays an oncogenic role as a transcription factor. Consistent with this concept, here we also observed that the modulation of autophagy activity by bafilomycin A1 or rapamycin not only affected HIF2α protein level, but also influenced HIF2α target gene expression. Moreover, HIF2α is mainly localized in the cytoplasm under steady state conditions, indicating that HIF2α might also play an oncogenic role in the cytoplasm, and the possible underlying mechanism is currently under investigation. Furthermore, it should be noted that HIF2α is only one of a number potentially oncogenic proteins that is subjected to autophagic degradation, and large-scale screening is necessary to identify additional autophagy substrates that are implicated in renal tumorigenesis. Additionally, it has been recently reported that HIF1α is degraded by chaperone-mediated autophagy (CMA) by not macroautophagy (40). Further investigation will be required to explore the function of CMA in HIF2α degradation and renal cell carcinoma development.
The proteasome and autophagy system were initially thought to work in parallel, and recent investigations suggested that both degradation systems are functionally linked (15, 16, 41, 42), but the collaboration in degrading an individual substrate that is subjected to both proteasomal and autophagic degradation is not well studied. HIF2α is known to be degraded by the proteasome in a VHL dependent fashion. Here we demonstrate that autophagy also mediated HIF2α degradation in a VHL dependent fashion. Since p62 is a self-oligomeric and stress response protein which binds ubiquitinated proteins via its UBA domain (13, 14, 43), we assumed that once HIF2α is ubiquitinated by VHL, the soluble VHL-elongin C-elongin B-HIF2α fraction would be degraded by the proteasome, while other complexes would be recognized by p62 to form aggregates and be degraded by autophagy. It is possible that the HIF2α overexpression, accumulation or aggregation under stress conditions might redirect cells to initiate or favor autophagic degradation mediated by p62, which is supported by the enhanced interaction between HIF2α and p62 during proteasome inhibition. Importantly, autophagic inactivation by knockout of Atg5 did not led to the accumulation of stably expressed HIF2α at a basal level, which was due to the compensatory function of the proteasome. This study revealed a new collaborative interaction between both degradation systems in handling a co-substrate. With the presence of such a compensatory interaction, chronic autophagy suppression may not lead to accumulation of some autophagy substrates, and an acute genetic inhibition was probably required to transiently increase basal level of these substrates before cells initiate a proteasome-mediated compensatory mechanism.
In ccRCC, frequent mutations of genes encoding VHL, TCEB1 and proteasome pathway components had been reported (8–10), and here we report genetic alterations in the autophagy pathway, including allelic loss, somatic mutations and reduced gene expression. Since our results show that HIF2α can be degraded cooperatively by the proteasome and autophagy in a VHL dependent fashion, it appears that ccRCC cells have evolved to keep HIF2α from being degraded by genetically disrupting the ubiquitination effector proteins and degradation pathways at the same time.
In summary, our data reveal that the oncogenic transcription factor HIF2α is a novel target of autophagy, and there is a compensatory relationship between the proteasome and autophagy in HIF2α degradation. Autophagy plays a tumor suppressive role in ccRCC tumorigenesis probably via constitutive degradation of HIF2α. This study might open a new therapeutic window for ccRCC management by down-regulating HIF2α levels through the simultaneous modulation of autophagy and the proteasome.
Materials and Methods
Antibodies and Reagents
LC3B antibody was prepared by the Eissa laboratory as previously described (43). p62 antibody was from American Research Products, Inc (03-GP62-C) or Santa Cruz Biotechnology, Inc (sc-28359). HIF2α antibody (NB100-122) was from Novus Biologicals. VHL antibody (2738), S6 antibody (2117), P-S6 antibody (2211), beclin 1 antibody (3495), LDHA antibody (3558), Lamin A antibody (2032), ATG5 antibody (8540) and ATG7 antibody (8558) were from Cell Signaling Technology. ATG7 antibody (LS-B330) for immunohistochemistry was from LifeSpan Bioscences. Beclin 1 antibody (B6186) for immunohistochemistry, β-actin antibody (A1978) and rapamycin (R0395) were from Sigma. HA antibody (PRB-101P) was from Covance. LAMP1 antibody was form BD Biosciences Pharmingen (555798). GFP antibody (sc-8334) and p53 antibody (sc-126) were from Santa Cruz Biotechnology. Chromotek-GFP-Trap Resin Agarose (ACT-CM-GFA0050) was from Allele Biotechnology. Bafilomycin A1 (B-1080) was from LC Laboratories. MG132 (474790) was from Calbiochem.
Cell Culture, Transfection and Establishment of Stable Cell Lines
786-O cells, HEK293T cells, RPE cells and Caki-1 Cells were grown in DMEM containing 10% fetal bovine serum. 786-O stable cell lines expressing Venus, VHL-wt-Venus, VHL-R167Q-Venus and VHL-F148A-Venus were infected with retroviral and selected in medium containing 1 mg/ml G418, and 786-O stable cell lines expressing HA-wt-VHL or HA-W117A-VHL were infected with retroviral and selected in medium containing 2 µg/ml puromycin (44). Stable cell lines expressing control shRNA (Sigma, SHC002V Mission Non-Traget ShRNA), p62 shRNA (Sigma, TRCN0000007235), BECN1 shRNA (Thermo Scientific, V3LHS_349509), or HIF2α-GFP were infected with lentiviral particles and selected in medium containing 2 µg/ml puromycin. Lentiviral vectors expressing HIF2α-GFP were produced by Viral Vector Production Core Laboratory at the Baylor College of Medicine.
Cell Fractionation, Cell Lysis, Immunoprecipitation and Immunoblot Analysis
Cell fractionation was carried out using NE-PER nuclear and cytoplasmic extraction reagent (Thermo Scientific, 78835). Cells were lysed in RIPA butter (50 mM Tric-Cl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, 0.1% SDS) for immunoblot analysis, and lysed in GFB buffer (50 mM Bis-tris-propane, pH 7.7, 150 mM NaCl, 10% glycerol, 1% Triton X-100) for GFP-trap using Chromotek-GFP-Trap beads.
RNA Isolation and Real-Time PCR
Total RNA isolation and Real-Time PCR were carried out as previously described (43), GAPDH was used as a control. Primer sequences for RT-PCR: HIF2α, 5´-TTGATGTGGA AACGGATGAA-3´ and 5´-GGAACCTGCTCTTGCTGTTC-3´; VEGFA, 5´-CCCACTGAGGAGTCCAACAT-3´ and 5´-TTTCTTGCGCTTTCGTTTTT-3´; TGFA, 5´-TGTGTCTGCCATTCTGGGTA-3´ and 5´-GACCTGGCAGCAGTGTATCA-3´; CCND1, 5´-CCCTCGGTGTCCTACTTCAA-3´ and 5´-AGGAAGCGGTCCAGGTAGTT-3´; GAPDH, 5´-CAATGACCCCTTCATTGACC-3´ and 5´-TTGATTTTGGAGGGATCTCG-3´.
Tissue Microarray, Image Acquiring and Image Analysis
Human subject protocol (2007-0511) was approved by Institutional Research Board at M.D. Anderson Cancer Center. Tissue microarrays (TMA) were generated and immunohistochemically stained as previously described (45). The slides were scanned with the Vectra image scanning system (Caliper Life Sciences). The percentage of ATG7 or beclin 1 cytoplasmic positive cells in whole tissue sections were analyzed using the Vectra Inform software (Caliper Life Sciences). Mann-Whitney test was used for statistical analysis.
Genetic alteration analysis using TCGA dataset
Level 3 RNA-Seq data, level 3 SNP array data, level 2 somatic mutation data, and clinical data for renal clear cell carcinomas were downloaded from the Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). Clustering analyses were done using the Cluster and TreeView software (available at http://rana.lbl.gov/EisenSoftware.htm). K-means grouping, Cox proportional hazard regression, and Kaplan-Meier log rank test were done using the R software (http://www.r-project.org).
ACKNOWLEDGEMENTS
We acknowledge the TCGA Research network. This work was supported by the Nanomedicine Roadmap Initiative Grant, the Renee Kaye Cure Fur Cancer Grant and the MD Anderson Cancer Center Kidney Cancer Research Program.
Footnotes
Conflict of interest:
The authors declare no conflict of interest.
REFERENCES
- 1.Jonasch E, Futreal PA, Davis IJ, Bailey ST, Kim WY, Brugarolas J, et al. State of the science: an update on renal cell carcinoma. Molecular cancer research : MCR. 2012 Jul;10(7):859–880. doi: 10.1158/1541-7786.MCR-12-0117. PubMed PMID: 22638109. Pubmed Central PMCID:3399969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008 Aug 1;14(15):4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. PubMed PMID:18676741. Pubmed Central PMCID: 2629664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews Cancer. 2012 Jan;12(1):9–22. doi: 10.1038/nrc3183. PubMed PMID: 22169972. Pubmed Central PMCID:3401912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, et al. Genetic and functional studies implicate HIF1alpha as a 14q kidney cancer suppressor gene. Cancer discovery. 2011 Aug;1(3):222–235. doi: 10.1158/2159-8290.CD-11-0098. PubMed PMID: 22037472. Pubmed Central PMCID: 3202343. Epub 2011/11/01. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shinojima T, Oya M, Takayanagi A, Mizuno R, Shimizu N, Murai M. Renal cancer cells lacking hypoxia inducible factor (HIF)-1alpha expression maintain vascular endothelial growth factor expression through HIF-2alpha. Carcinogenesis. 2007 Mar;28(3):529–536. doi: 10.1093/carcin/bgl143. PubMed PMID: 16920734. [DOI] [PubMed] [Google Scholar]
- 6.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer cell. 2007 Apr;11(4):335–347. doi: 10.1016/j.ccr.2007.02.006. PubMed PMID: 17418410. Pubmed Central PMCID: 3145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanharanta S, Shu W, Brenet F, Hakimi AA, Heguy A, Viale A, et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nature medicine. 2013 Jan;19(1):50–56. doi: 10.1038/nm.3029. PubMed PMID: 23223005. Pubmed Central PMCID: 3540187. Epub 2012/12/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nature genetics. 2012 Jan;44(1):17–19. doi: 10.1038/ng.1014. PubMed PMID: 22138691. [DOI] [PubMed] [Google Scholar]
- 9.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature genetics. 2013 Aug;45(8):860–867. doi: 10.1038/ng.2699. PubMed PMID: 23797736. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013 Jul 4;499(7456):43–49. doi: 10.1038/nature12222. PubMed PMID: 23792563. Pubmed Central PMCID: 3771322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. PubMed PMID: 19653858. Pubmed Central PMCID: 2831538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007 Jul;27(1):135–144. doi: 10.1016/j.immuni.2007.05.022. PubMed PMID: 17658277. Pubmed Central PMCID: 2680670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Molecular cell. 2009 May 15;34(3):259–269. doi: 10.1016/j.molcel.2009.04.026. PubMed PMID: 19450525. [DOI] [PubMed] [Google Scholar]
- 14.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009 Jun 12;137(6):1001–1004. doi: 10.1016/j.cell.2009.05.023. PubMed PMID: 19524504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. The American journal of pathology. 2007 Aug;171(2):513–524. doi: 10.2353/ajpath.2007.070188. PubMed PMID: 17620365. Pubmed Central PMCID: 1934546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007 Jun 14;447(7146):859–863. doi: 10.1038/nature05853. PubMed PMID: 17568747. [DOI] [PubMed] [Google Scholar]
- 17.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012 Jun;12(6):401–410. doi: 10.1038/nrc3262. PubMed PMID: 22534666. Epub 2012/04/27. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999 Dec 9;402(6762):672–676. doi: 10.1038/45257. PubMed PMID: 10604474. [DOI] [PubMed] [Google Scholar]
- 19.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2003 Dec 9;100(25):15077–15082. doi: 10.1073/pnas.2436255100. PubMed PMID: 14657337. Pubmed Central PMCID: 299911. Epub 2003/12/06. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of clinical investigation. 2003 Dec;112(12):1809–1820. doi: 10.1172/JCI20039. PubMed PMID: 14638851. Pubmed Central PMCID: 297002. Epub 2003/11/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012 Mar 8;366(10):883–892. doi: 10.1056/NEJMoa1113205. PubMed PMID: 22397650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turcotte S, Chan DA, Sutphin PD, Hay MP, Denny WA, Giaccia AJ. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer cell. 2008 Jul 8;14(1):90–102. doi: 10.1016/j.ccr.2008.06.004. PubMed PMID: 18598947. Pubmed Central PMCID: 2819422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010 Feb 5;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. PubMed PMID: 20144757. Pubmed Central PMCID: 2852113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends in molecular medicine. 2010 Nov;16(11):528–536. doi: 10.1016/j.molmed.2010.09.002. PubMed PMID: 20932800. Pubmed Central PMCID: 2978905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Y, Hilliard G, Ferguson T, Millhorn DE. Cobalt inhibits the interaction between hypoxia-inducible factor-alpha and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-alpha. The Journal of biological chemistry. 2003 May 2;278(18):15911–15916. doi: 10.1074/jbc.M300463200. PubMed PMID: 12606543. [DOI] [PubMed] [Google Scholar]
- 26.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nature cell biology. 2000 Jul;2(7):423–7. doi: 10.1038/35017054. PubMed PMID: 10878807. [DOI] [PubMed] [Google Scholar]
- 27.Hacker KE, Lee CM, Rathmell WK. VHL type 2B mutations retain VBC complex form and function. PloS one. 2008;3(11):e3801. doi: 10.1371/journal.pone.0003801. PubMed PMID: 19030229. Pubmed Central PMCID: 2583047. Epub 2008/11/26. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011 Feb;13(2):132–141. doi: 10.1038/ncb2152. PubMed PMID: 21258367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, et al. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proceedings of the National Academy of Sciences of the United States of America. 2013 Aug 6;110(32):E2950–E2957. doi: 10.1073/pnas.1307736110. PubMed PMID: 23878245. Pubmed Central PMCID: 3740898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009 Oct 1;461(7264):654–658. doi: 10.1038/nature08455. PubMed PMID: 19794493. [DOI] [PubMed] [Google Scholar]
- 31.Urushitani M, Kurisu J, Tsukita K, Takahashi R. Proteasomal inhibition by misfolded mutant superoxide dismutase 1 induces selective motor neuron death in familial amyotrophic lateral sclerosis. Journal of neurochemistry. 2002 Dec;83(5):1030–1042. doi: 10.1046/j.1471-4159.2002.01211.x. PubMed PMID: 12437574. [DOI] [PubMed] [Google Scholar]
- 32.Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, et al. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer cell. 2002 Jun;1(5):459–468. doi: 10.1016/s1535-6108(02)00071-5. PubMed PMID: 12124175. [DOI] [PubMed] [Google Scholar]
- 33.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nature reviews Molecular cell biology. 2010 May;11(5):329–341. doi: 10.1038/nrm2882. PubMed PMID: 20379207. Epub 2010/04/10. eng. [DOI] [PubMed] [Google Scholar]
- 34.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature cell biology. 2009 Apr;11(4):385–396. doi: 10.1038/ncb1846. PubMed PMID: 19270696. Epub 2009/03/10. eng. [DOI] [PubMed] [Google Scholar]
- 35.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annual review of pathology. 2013 Jan 24;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. PubMed PMID: 23072311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prasad SR, Humphrey PA, Catena JR, Narra VR, Srigley JR, Cortez AD, et al. Common and uncommon histologic subtypes of renal cell carcinoma: imaging spectrum with pathologic correlation. Radiographics : a review publication of the Radiological Society of North America, Inc. 2006 Nov-Dec;26(6):1795–1806. doi: 10.1148/rg.266065010. discussion 806–10. PubMed PMID: 17102051. [DOI] [PubMed] [Google Scholar]
- 37.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer cell. 2008 Apr;13(4):343–354. doi: 10.1016/j.ccr.2008.02.001. PubMed PMID; 18394557. [DOI] [PubMed] [Google Scholar]
- 38.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009 Jun 12;137(6):1062–1075. doi: 10.1016/j.cell.2009.03.048. PubMed PMID: 19524509. Pubmed Central PMCID: 2802318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011 Dec 16;334(6062):1573–1577. doi: 10.1126/science.1208347. PubMed PMID: 22174255. [DOI] [PubMed] [Google Scholar]
- 40.Hubbi ME, Hu H, Kshitiz, Ahmed I, Levchenko A, Semenza GL. Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. The Journal of biological chemistry. 2013 Apr 12;288(15):10703–10714. doi: 10.1074/jbc.M112.414771. PubMed PMID: 23457305. Pubmed Central PMCID: 3624450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Molecular cell. 2009 Feb 27;33(4):517–527. doi: 10.1016/j.molcel.2009.01.021. PubMed PMID: 19250912. Pubmed Central PMCID: 2669153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007 Dec 14;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. PubMed PMID: 18083104. [DOI] [PubMed] [Google Scholar]
- 43.Liu XD, Ko S, Xu Y, Fattah EA, Xiang Q, Jagannath C, et al. Transient aggregation of ubiquitinated proteins is a cytosolic unfolded protein response to inflammation and endoplasmic reticulum stress. The Journal of biological chemistry. 2012 Jun 1;287(23):19687–19698. doi: 10.1074/jbc.M112.350934. PubMed PMID: 22518844. Pubmed Central PMCID: 3366003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Z, German P, Bai S, Feng Z, Gao M, Si W, et al. Agents that stabilize mutated von Hippel-Lindau (VHL) protein: results of a high-throughput screen to identify compounds that modulate VHL proteostasis. Journal of biomolecular screening. 2012 Jun;17(5):572–580. doi: 10.1177/1087057112436557. PubMed PMID: 22357874. Epub 2012/02/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J, Jonasch E, Alexander A, Short JD, Cai S, Wen S, et al. Cytoplasmic sequestration of p27 via AKT phosphorylation in renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009 Jan 1;15(1):81–90. doi: 10.1158/1078-0432.CCR-08-0170. PubMed PMID: 19118035. Pubmed Central PMCID: 3030253. Epub 2009/01/02. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]