Abstract
Resveratrol possesses a strong anticancer activity exhibited as the induction of apoptosis through p53 activation. However, the molecular mechanism and direct target(s) of resveratrol-induced p53 activation remain elusive. Here, the Ras-GTPase activating protein SH3 domain binding protein 1 (G3BP1) was identified as a potential target of resveratrol, and in vitro binding assay results using resveratrol (RSVL)-conjugated Sepharose 4B beads confirmed their direct binding. Depletion of G3BP1 significantly diminishes resveratrol-induced p53 expression and apoptosis. We also found that G3BP1 negatively regulates p53 expression by interacting with ubiquitin-specific protease 10 (USP10), a deubiquitinating enzyme of p53. Disruption of the interaction of p53 with USP10 by G3BP1 interference leads to suppression of p53 deubiquitination. Resveratrol, on the other hand, directly binds to G3BP1 and prevents the G3BP1/USP10 interaction, resulting in enhanced USP10-mediated deubiquitination of p53 and consequently increased p53 expression. These findings disclose a novel mechanism of resveratrol-induced p53 activation and resveratrol-induced apoptosis by direct targeting of G3BP1.
Keywords: Resveratrol, G3BP1, p53 and USP10
Introduction
The tumor suppressor p53 plays a central role in the regulation of cell cycle, DNA repair, apoptosis, senescence and angiogenesis (1). The importance of p53 in cancer development is underscored by the observation that the p53 pathway is frequently inactivated by mutations or other defects in more than 50% of all human cancers (1, 2). Therefore, p53 is recognized as a desirable target for cancer prevention and therapy.
Resveratrol (trans-3,5,4’-truhydroxystilbene; RSVL), a natural phytoalexin found in grapes (3) and peanuts (4, 5), is considered as a potential anticancer agent since its inhibitory effects on carcinogenic processes (e.g., initiation, promotion and progression) was first reported in 1997 (6). Resveratrol has been shown to possess an apoptosis-dependent anticancer activity (7–12), and several groups have demonstrated that the p53 activation is implicated in resveratrol-induced apoptosis (13–15). However, the molecular mechanism and direct target(s) of resveratrol-induced p53 expression remain elusive.
Ras-GTPase activating protein SH3 domain binding protein 1 (G3BP1) was first identified as a protein that interacts with the SH3 domain of Ras-GTPase activating protein (Ras-GAP) (16) and is known to be involved in several pathways including Ras signaling (16, 17), c-myc mRNA turnover (18, 19), NF-kappaB signaling (20) and HER2 signaling (21). G3BP1 is also overexpressed in several human cancers including head and neck, breast and colon cancers (21–24). Interestingly, Soncini, et al. (25) discovered the interaction of G3BP1 with ubiquitin-specific protease 10 (USP10), and they concluded that G3BP1 might restrict the deubiquitinating activity of USP10. USP10 is known to directly interact with p53 and specifically deubiquitinate p53, which leads to p53 activation and stabilization (26). However, no one has reported the function of G3BP1 in mediating USP10-regulated deubiquitination of p53.
In this study, we identified G3BP1 as a novel target of resveratrol. We also found that G3BP1 negatively regulates p53 expression by interacting with USP10. G3BP1 binds to USP10 and therefore disrupts the interaction of USP10 with p53, leading to suppression of p53 deubiquitination. Resveratrol, on the other hand, directly binds to G3BP1 and disrupts the G3BP1/USP10 interaction, resulting in enhanced USP10-regulated deubiquitination of p53. Here, we propose a novel mechanism of resveratrol-induced p53 activation that occurs through the direct targeting of G3BP1.
Results
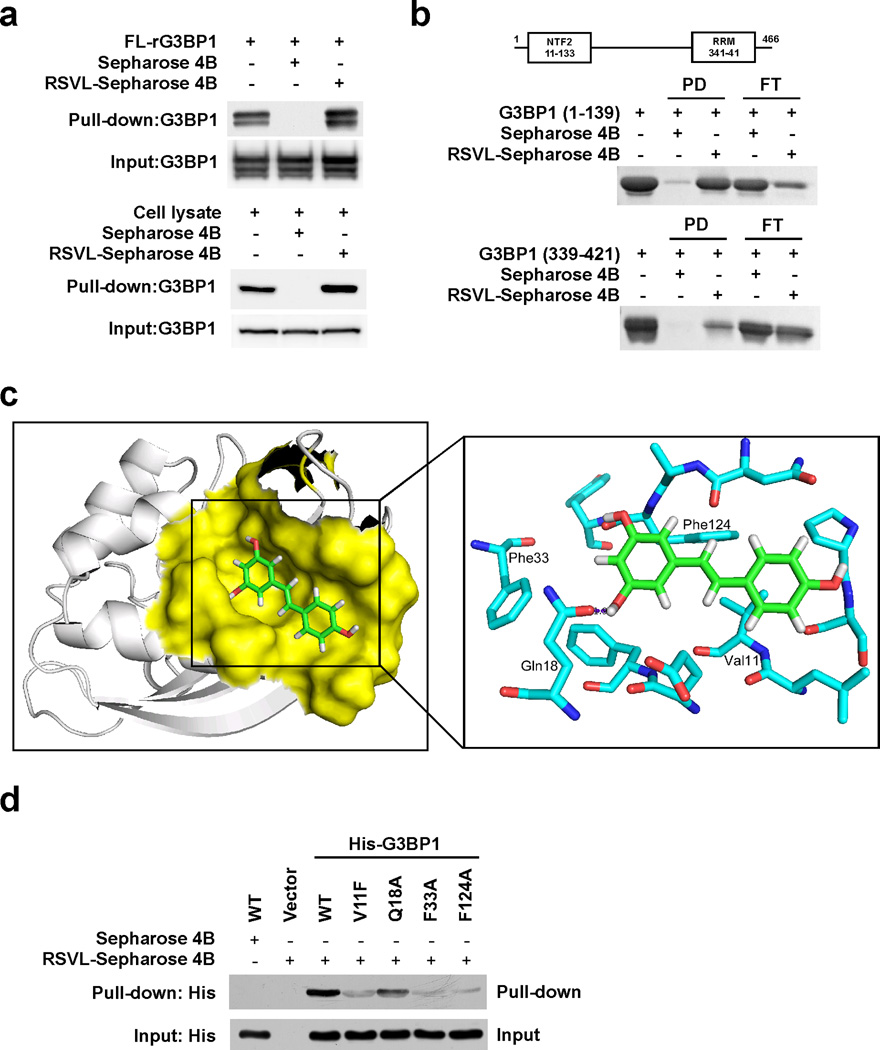
Resveratrol interacts with G3BP1 at the NTF2-like domain
To identify novel molecular targets of resveratrol-induced p53 expression and apoptosis, we first screened binding partners for resveratrol by LC-MS/MS analysis and found G3BP1 as a potential binding protein of resveratrol (Supplemental Table 1). We then performed binding assays using resveratrol-conjugated Sepharose 4B beads, and confirmed that recombinant full-length G3BP1 (rG3BP1) and endogenous G3BP1 bound to resveratrol (Fig. 1a). G3BP1 consists of two domains, the NTF2-like domain at the N-terminus and the eukaryotic RNA recognition motif (RRM) domain at the C-terminal end. To identify which domain is involved in the binding with resveratrol, we purified the recombinant NTF2-like (residues 1–139) and RRM (residues 339–421) domains of G3BP1, and performed in vitro binding assays (Fig. 1b). A strong interaction with resveratrol-conjugated beads was observed for the NTF2-like domain, whereas the RRM domain showed only a weak binding with resveratrol-conjugated beads. We concluded that the NTF2-like domain of G3BP1 is critical for interacting with resveratrol. Because the crystal structure of the NTF2-like domain of G3BP1 is available, we performed ligand docking. According to our generated binding model (Fig. 1c), resveratrol forms hydrophobic interactions with G3BP1 at Val11, Phe33 and Phe124, and the hydroxyl group of resveratrol forms a hydrogen bond with the side chain of Gln18 of G3BP1. To confirm our plausible binding model, we constructed single-point mutants of G3BP1, and transfected them into HEK 293 cells. The result of the binding assay (Fig. 1d) showed that the interaction of G3BP1 with resveratrol was dramatically decreased by a single-point mutation of G3BP1 at Val11, Phe33 or Phe124, indicating that these amino acids are essential for resveratrol binding.
Figure 1. Resveratrol interacts with G3BP1 at the NTF2-like domain.
(a) Resveratrol interacts with G3BP1. Recombinant full-length G3BP1 (FL-rG3BP1; 200 ng) or whole cell lysates from SK-MEL-5 cells (500 µg) were incubated with control or resveratrol-conjugated Sepharose 4B beads, and then the proteins bound to the beads were analyzed by Western blotting. (b) Resveratrol interacts with G3BP1 at the NTF2-like domain. The purified NTF2-like domain (residues 1–139) or RRM domain (residues 339–421) of G3BP1 (400 µg) was incubated with control or resveratrol-conjugated Sepharose 4B beads. The binding proteins were subjected to SDS-PAGE and then stained with Coomassie Blue. Most of the RRM domain protein appeared in the flow-through fraction and washed out during the washing steps. PD: pull-down, FT: flow-through. (c) Computational docking model of resveratrol with the NTF2-like domain of G3BP1. The structure is shown in ribbon diagram with overlapped surface representation in yellow color (insert). Resveratrol molecule is shown as green sticks, and amino acid residues surrounding resveratrol are shown as cyan sticks. Oxygens are colored red and nitrogens are colored blue. (d) Resveratrol interacts with G3BP1 at Val11, Phe33 and Phe124. HEK 293 cells were transfected with the indicated plasmids for 48 h. Whole cell lysates were incubated with control or resveratrol-conjugated Sepharose 4B beads, and then the proteins bound to the beads were analyzed by Western blotting.
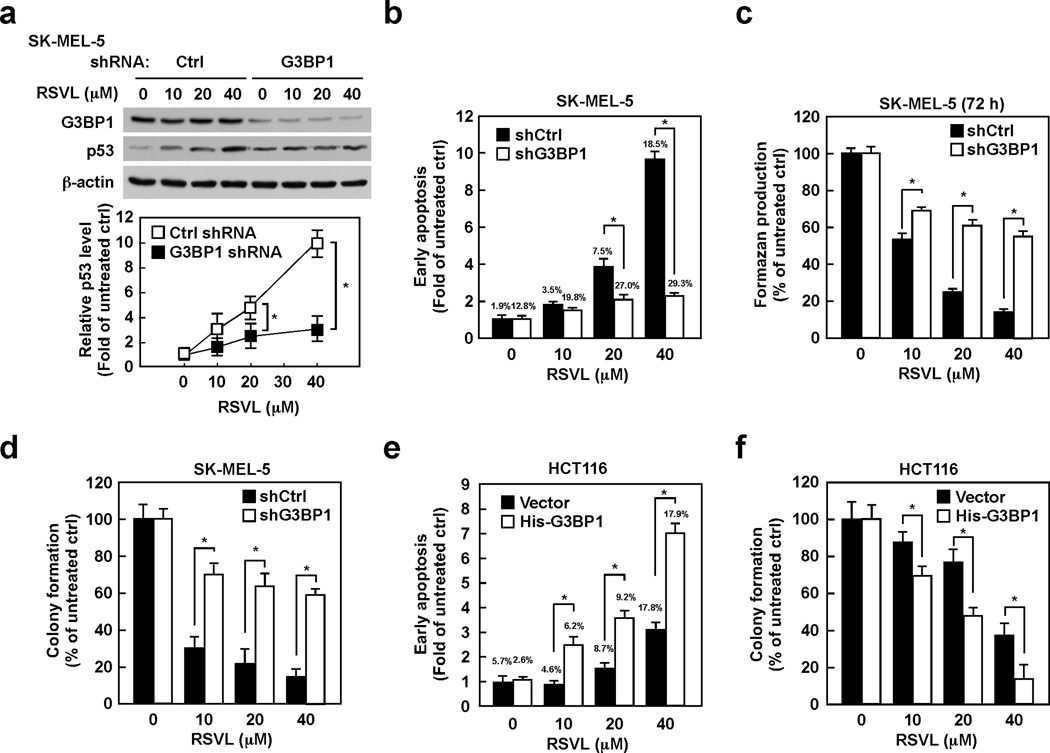
G3BP1 plays an important role in resveratrol-induced p53 expression and apoptosis
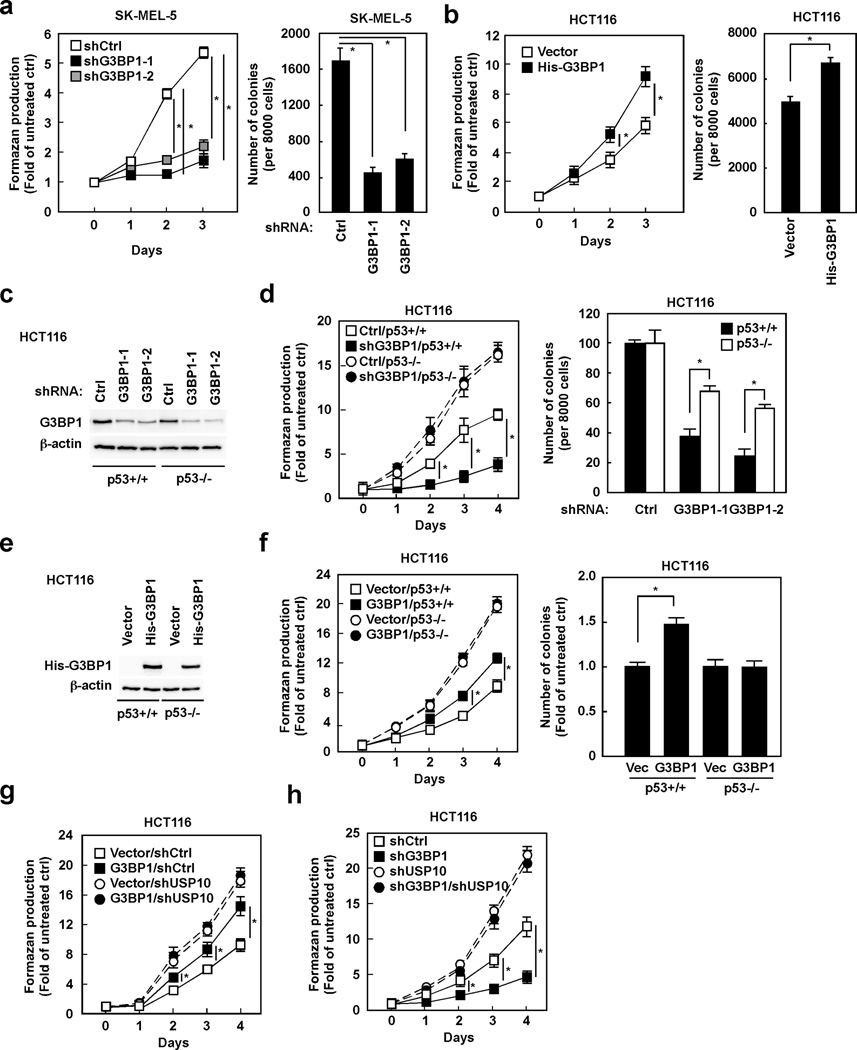
We were interested in revealing the molecular mechanism of resveratrol-induced p53 expression. The expression levels of G3BP1 were therefore determined in various p53 wildtype cancer cell lines, and high expression of G3BP1 was observed in SK-MEL-5 human melanoma skin cells (Supplemental Fig. 1a). Consistent with previous publications (13–15), resveratrol-induced p53 expression and apoptosis were observed in SK-MEL-5 cells (Supplemental Fig. 1b,c), resulting in inhibition of proliferation (as assessed using the MTS assay) and anchorage-independent cell growth (Supplemental Fig. 1d,e). Because G3BP1 was identified as a promising target of resveratrol, we determined whether G3BP1 is implicated in resveratrol-induced p53 expression. Interestingly, resveratrol-induced p53 expression was dramatically diminished by depletion of G3BP1 in SK-MEL-5 cells (Fig. 2a). The same phenomenon was observed in HCT116 human colon cancer cells that have mid-level expression of G3BP1 (Supplemental Fig. 1f). Resveratrol-induced apoptosis was substantially reduced by G3BP1 depletion in both SK-MEL-5 (Fig. 2b) and HCT116 cells (Supplemental Fig. 1g). Moreover, knocking down G3BP1 expression diminished sensitivity to resveratrol’s effect on the proliferation and anchorage-independent growth of SK-MEL-5 cells (Fig. 2c,d). The same phenomenon on anchorage-independent growth was observed in HCT116 cells (Supplemental Fig. 1h). On the other hand, HCT116 cells overexpressing G3BP1 were more sensitive to resveratrol in the induction of apoptosis and inhibition of anchorage-independent growth compared with cells expressing the control vector (Fig. 2e,f). These results strongly indicate that G3BP1 is implicated in resveratrol-induced p53 expression and apoptosis.
Figure 2. G3BP1 plays an important role in resveratrol-induced p53 expression and apoptosis.
(a) G3BP1 is implicated in resveratrol-induced p53 expression. SK-MEL-5 cells expressing the indicated shRNAs were treated with resveratrol (0–40 µM) for 24 h and then whole cell lysates were analyzed by Western blotting. Densitometric analysis of relative protein levels was normalized against β-actin. (b) Resveratrol-induced apoptosis is diminished by depletion of G3BP1. SK-MEL-5 cells expressing the indicated shRNAs were treated with resveratrol (0–40 µM) for 48 h and then cellular apoptosis was determined by flow cytometry. (c) The effect of resveratrol on cell proliferation is diminished by knocking down G3BP1 expression. SK-MEL-5 cells expressing the indicated shRNAs were treated with resveratrol (0–40 µM) for 72 h and then formazan production was determined at the indicated time points by MTS assay. (d) The effect of resveratrol on anchorage-independent growth is diminished in cells expressing G3BP1 shRNA. SK-MEL-5 cells expressing the indicated shRNAs were grown in soft agar with resveratrol (0–40 µM) for 7 days and then the colonies were counted. (e) G3BP1-overexpressing cells are more sensitive to resveratrol-induced apoptosis. HCT116 cells were transfected with the indicated constructs for 48 h and then treated with resveratrol (0–40 µM) for 72 h. Cellular apoptosis was determined by flow cytometry. (f) G3BP1-overexpressing cells are more sensitive to resveratrol’s effect on anchorage-independent cell growth. HCT116 cells transfected with the indicated constructs were grown in soft agar with resveratrol (0–40 µM) for 7 days and then the colonies were counted. All data are represented as means ± S.D. from 3 independent experiments (*p < 0.05).
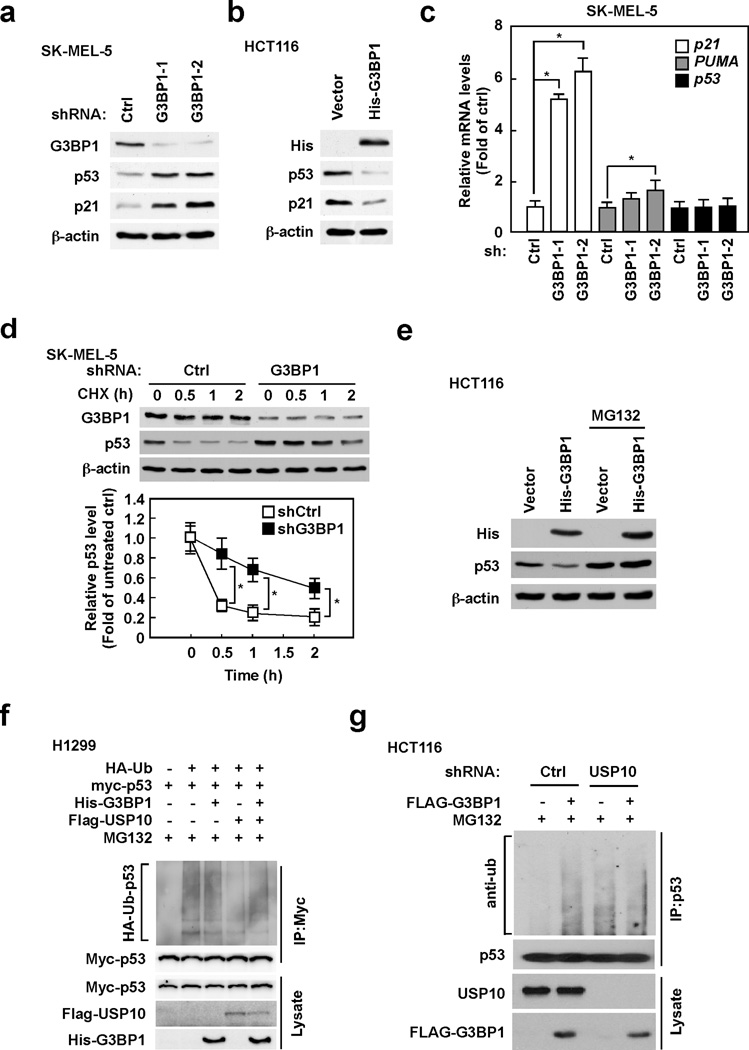
G3BP1 negatively regulates p53 expression by inhibiting USP10-mediated deubiquitination of p53
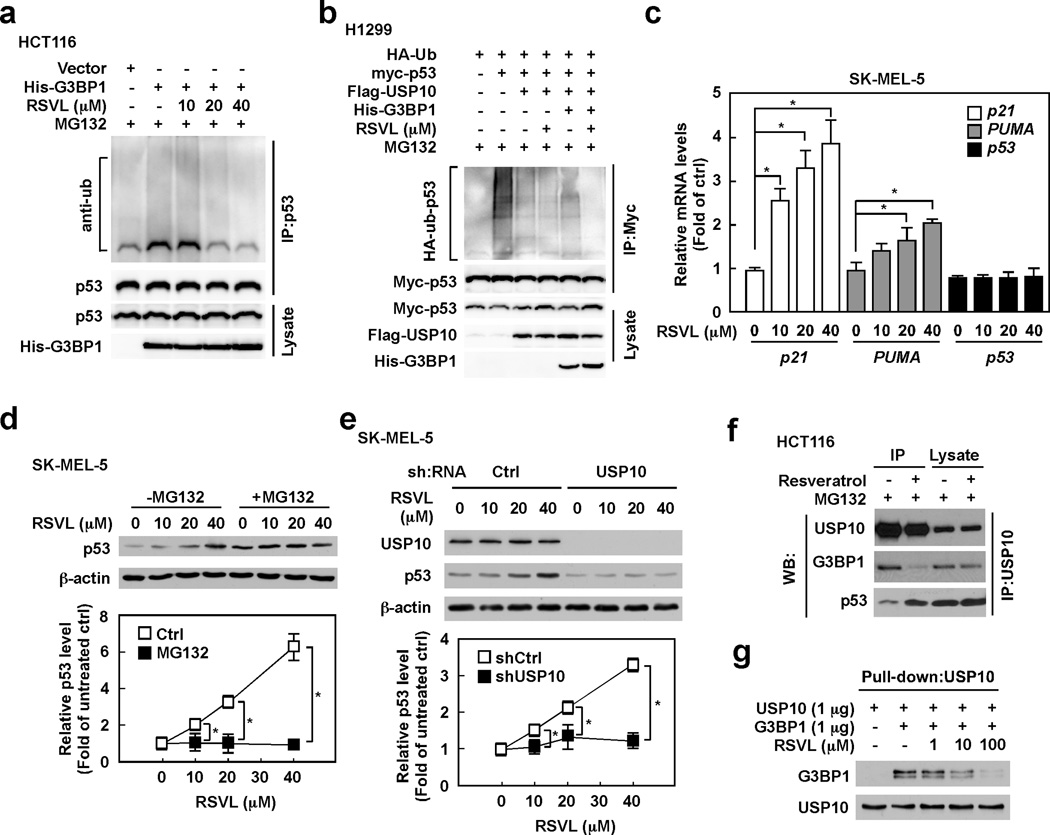
Our results indicate that resveratrol-induced p53 expression is affected by G3BP1 manipulation. We then examined the role of G3BP1 in regulating p53 expression. The depletion of G3BP1 obviously increased the protein levels of p53 and p21, a target of p53, in SK-MEL-5 (Fig. 3a) and HCT116 cells (Supplemental Fig. 2a). HCT116 cells overexpressing G3BP1, on the other hand, exhibited decreased expression levels of p53 and p21 compared with cells expressing a control vector (Fig. 3b). Because another G3BP family protein, G3BP2, was reported to interact with p53 and regulate p53 protein level (27), we examined the effect of G3BP2 in regulating p53 expression. Less effect on p53 protein levels was observed in HCT116 cells overexpressing G3BP2 compared with G3BP1 overexpressing cells (Supplemental Fig. 2b). We therefore believe that G3BP1 plays a more important role in regulating p53 compared to G3BP2. The mRNA levels of p53 and its target genes, p21 and the p53 up-regulated modulator of apoptosis (PUMA), were then determined in G3BP1-manipulated cells. Depletion of G3BP1 dramatically enhanced p21 mRNA level and slightly increased PUMA mRNA levels in SK-MEL-5 cells (Fig. 3c), indicating that G3BP1-mediated p53 is active as a transcription factor. On the other hand, p53 mRNA levels were not changed in G3BP1-depleted SK-MEL-5 cells (Fig. 3c). Furthermore, knocking down G3BP1 expression markedly increased p53 stability (Fig. 3d), and the reduction of p53 levels by G3BP1 overexpression was reversed by treatment with the proteasome inhibitor MG132 (Fig. 3e) in SK-MEL-5 cells, indicating that G3BP1 regulates p53 expression through the proteasome degradation pathway.
Figure 3. G3BP1 negatively regulates p53 expression by inhibiting USP10-mediated deubiquitination of p53.
(a) Depletion of G3BP1 increases the levels of the p53 and p21 proteins. SK-MEL-5 cells were infected with the indicated shRNAs for 48 h and then whole cell lysates were analyzed by Western blotting. (b) Overexpression of G3BP1 reduces the protein levels of p53 and p21. HCT116 cells were transfected with the indicated constructs for 48 h and then whole cell lysates were analyzed by Western blotting. (c) Knock-down of G3BP1 enhances mRNA levels of p21 and PUMA, but has no effect on p53 mRNA levels. SK-MEL-5 cells were infected with the indicated shRNAs for 48 h and then mRNA levels were analyzed by quantitative PCR (qPCR). Relative mRNA levels were normalized against GAPDH. Data are represented as means ± S.D. from 3 independent experiments and statistical significance was analyzed after applying the Bonferroni correction (level of significance after correction is 0.025, 0.05/2). (d) G3BP1 negatively regulates p53 stability. SK-MEL-5 cells expressing the indicated shRNAs were treated with 20 µg/ml cycloheximide (CHX) and harvested at the indicated time points. Whole cells lysates were analyzed by Western blotting. Densitometric analysis of relative protein levels was normalized against β-actin and data are represented as means ± S.D. from 3 independent experiments (*p < 0.05). (e) G3BP1 negatively regulates p53 expression in a proteasome-dependent manner. HCT116 cells were transfected with the indicated constructs for 48 h and then incubated with or without MG132 (10 µM) for an additional 4 h. Whole cell lysates were analyzed by Western blotting. (f) G3BP1 suppresses USP10-mediated deubiquitination of p53. H1299 cells were transfected with the indicated constructs for 48 h and then incubated with MG132 (10 µM) for an additional 4 h. Whole cell lysates were co-immunoprecipitated with a Myc-tagged antibody followed by Western blotting with anti-HA or anti-Myc. (g) G3BP1 regulates ubiquitination of endogenous p53 through USP10. HCT116 cells expressing control or USP10 shRNAs were transfected with the indicated constructs for 48 h and then incubated with MG132 (10 µM) for an additional 4 h. Whole cell lysates were co-immunoprecipitated with a p53 antibody followed by Western blotting with anti-Ub.
Interestingly, using the yeast two-hybrid system, G3BP1 was identified as an USP10 interacting protein (25), and USP10 has been reported as a specific deubiquitinating enzyme of p53 (26). We therefore hypothesized that G3BP1 might affect USP10-mediated deubiquitination of p53. To test this hypothesis, we conducted an in vivo ubiquitination assay using H1299 cells (Fig. 3f). The results showed that G3BP1 had no effect on ubiquitination of p53 (compare lanes 2 and 3). Because USP10 is a p53 deubiquitinating enzyme, p53 ubiquitination was decreased by transfection of USP10 (compare lanes 3 and 4), whereas deubiquitination of p53 by USP10 was inhibited by co-transfection of USP10 and G3BP1 (compare lanes 4 and 5). G3BP1 also inhibited USP10-mediated deubiquitination of endogenous p53 in HCT116 cells (Fig. 3g). Overexpression of G3BP1 enhanced p53 ubiquitination in cells expressing control shRNA (compare lanes 1 and 2). Depletion of USP10 enhanced p53 ubiquitination (compare lanes 1 and 3), whereas enhancement of p53 ubiquitination by G3BP1 was not observed in USP10 depleted cells (compare lanes 3 and 4). Mouse double minute 2 homolog (Mdm2), E3 ubiquitin-protein ligase, is also known as an important regulator of p53. We therefore determined the consequence of Mdm2 and G3BP1-mediated p53 regulation. Depletion of G3BP1 has no significant effect on either Mdm2 protein or mRNA levels (Supplemental Fig. 2c,d). Moreover, the interaction of p53 with Mdm2 was not affected by the manipulation of G3BP1 (Supplemental Fig. 2e), and no interaction of G3BP1 with Mdm2 was observed in SK-MEL-5 cells (Supplemental Fig. 2f), indicating that G3BP1-regulated p53 expression is not dependent on Mdm2. These results suggest that G3BP1 negatively regulates p53 expression by inhibiting USP10-mediated deubiquitination of p53.
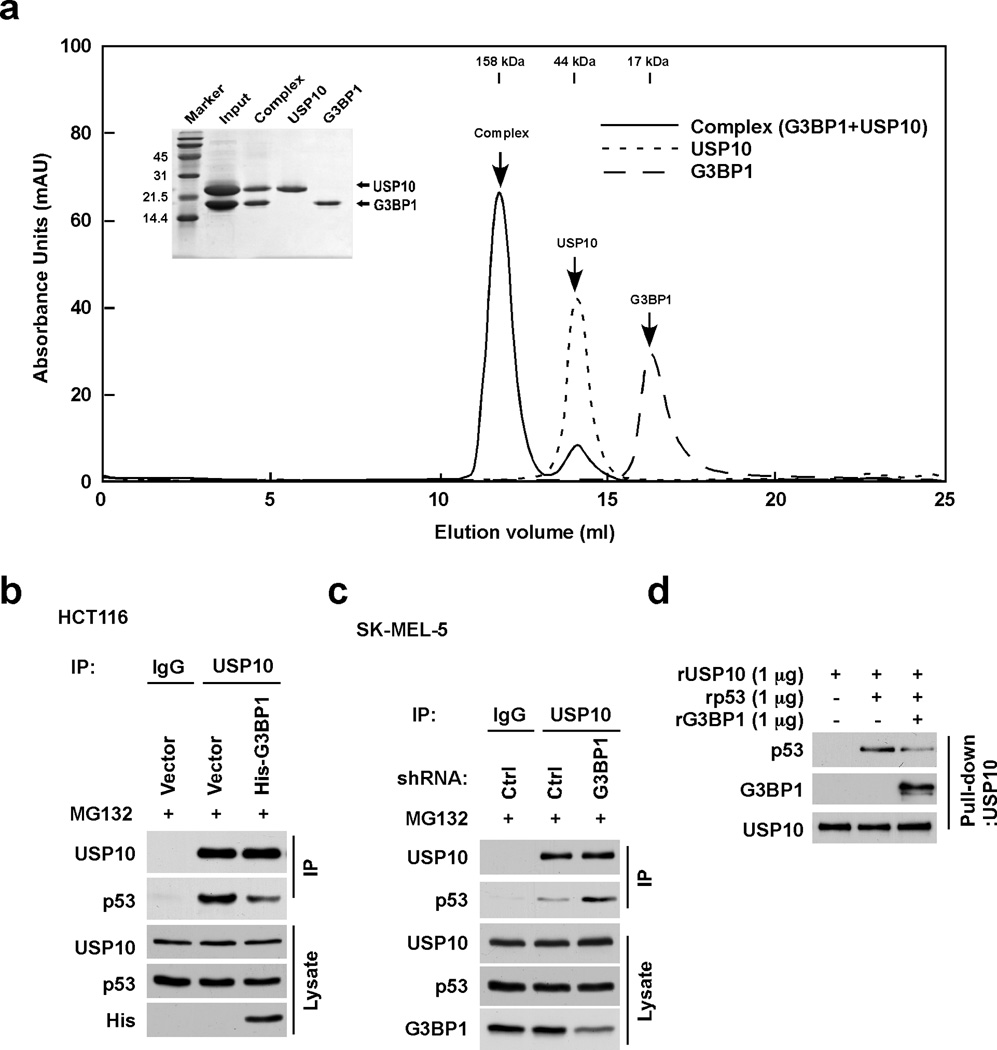
G3BP1 disrupts the interaction of USP10 with p53
Using truncated mutants of both G3BP1 and USP10 transfected into HCT116 cells, we observed that the N-terminal portion of G3BP1 (residues 1–146) interacts with the N-terminus of USP10 (residues 1–100). These data are not shown here because they are consistent with data presented in two recent publications. The groups reported that the NTF2-like domain of G3BP1 (residues 1–138) is required for the interaction with USP10 (28) and that the N-terminal region of USP10 (residues 1–76) is critical for interacting with G3BP1 (29). However, for the first time we show an in vitro interaction of the purified NTF2-like domain of G3BP1 (residues 1–139) and the N-terminal region of USP10 (residues 1–150). Both proteins were eluted on a gel-filtration column as a single peak representing a complex (Fig. 4a). In contrast, the RRM domain of G3BP1 and USP10 were eluted as two separate peaks representing single proteins (data not shown). According to a previous report (26), the N-terminal region of USP10 (residues 1–100) interacts with p53 to deubiquitinate p53. We conjectured that G3BP1 disrupts the interaction of USP10 with p53 by binding to USP10, and consequently suppresses USP10-mediated deubiquitination of p53. The results indicated that overexpression of G3BP1 decreased the USP10/p53 interaction as shown by co-IP assays using USP10 antibodies in HCT116 cells (Fig. 4b). Consistently, down-regulation of G3BP1 increased the USP10/p53 interaction in SK-MEL-5 cells (Fig. 4c). The same phenomenon was observed using p53 antibodies in a co-IP assay (Supplemental Fig. 3a). In addition, an in vitro competition assay showed that the in vitro interaction between USP10 and p53 was clearly decreased in the presence of G3BP1 (Fig. 4d). These results suggest that G3BP1 interrupts the interaction of USP10 with p53 by its own interaction with USP10.
Figure 4. G3BP1 disrupts the interaction of USP10 with p53.
(a) G3BP1 (residues 1–139) interacts with USP10 (residues 1–150) in vitro. Three separate FPLC runs, one run for each protein and the third run with the two proteins combined were merged into one graph, with the SDS-PAGE results inserted to confirm that the first high molecular weight peak is a G3BP1/USP10 complex. The results were plotted with dashed (G3BP1), dotted (USP10), and solid (G3BP1/USP10 complex) lines. G3BP1 eluted from the gel-filtration column as a monomer (MW 18 kDa) and USP10 (MW ~22 kDa) eluted as a dimer with an apparent MW of 44 kDa. The elution volumes for gel-filtration standards are shown at the top. (b) Overexpression of G3BP1 decreases the interaction of USP10 with p53. HCT116 cells were transfected with the indicated constructs for 48 h and then treated with MG132 (10 µM) for 4 h. Whole cell lysates were co-immunoprecipitated with anti-USP10 followed by Western blotting with anti-USP10 or anti-p53. (c) Depletion of G3BP1 enhances the interaction of USP10 with p53. SK-MEL-5 cells were infected with lentivirus encoding the indicated shRNAs for 48 h and then treated with MG132 (10 µM) for 4 h. Whole cell lysates were co-immunoprecipitated with anti-USP10 followed by Western blotting with anti-USP10 or anti-p53. (d) G3BP1 disrupts the interaction between USP10 and p53 in vitro. Recombinant G3BP1, USP10 and p53 were pulled down with anti-USP10 followed by Western blotting with anti-p53, anti-G3BP1 or anti-USP10.
G3BP1 regulates cancer cell proliferation through p53 and USP10
Our results showed that G3BP1 negatively regulates p53 expression by inhibiting USP10-mediated p53 deubiquitination. Because p53 plays a critical role in inducing apoptosis, resulting in inhibition of proliferation in several cancer cell lines, we examined the role of G3BP1 in various cancer cells. Proliferation (as assessed using the MTS assay) and anchorage-independent growth were dramatically reduced by depletion of G3BP1 in SK-MEL-5 (Fig. 5a) and HCT116 cells (Supplemental Fig. 4a). An enhancement of proliferation and anchorage-independent growth were observed in G3BP1 overexpressing HCT116 cells (Fig. 5b). To demonstrate whether G3BP1-regulated cancer cell proliferation is p53 dependent, G3BP1 was manipulated in HCT116 expressing wildtype p53 (p53+/+) and p53 deficient (p53−/−) cells. Depletion of G3BP1 inhibited the proliferation and anchorage-independent growth of HCT116 p53+/+ cells, whereas the effects were significantly reduced in HCT116 p53−/− cells (Fig. 5c,d). The inhibition of cell proliferation by G3BP1 depletion was also diminished by double knock-down of G3BP1 and p53 in SK-MEL-5 cells (Supplemental Fig. 4b). Moreover, overexpression of G3BP1 enhanced proliferation and anchorage-independent growth of HCT116 p53+/+ cells, whereas these effects were not observed in HCT116 p53−/− cells (Fig. 5e,f). These results strongly indicated that G3BP1 modulates cancer cell proliferation in a p53-dependent manner. We then determined the consequence of USP10 in G3BP1-regulated cancer cell proliferation (Fig. 5g). Overexpression of G3BP1 enhanced proliferation of HCT116 cells expressing control shRNA. Consistent with a previous report (26), cells expressing USP10 shRNA increased cell proliferation. However, enhancement of cell proliferation by G3BP1 was not observed in cells expressing USP10 shRNA. Furthermore, the double knock-down of G3BP1 and USP10 rescued the inhibition of proliferation induced by G3BP1 depletion in HCT116 (Fig. 5h) and SK-MEL-5 cells (Supplemental Fig. 4c). These results suggest that G3BP1-regulated cancer cell proliferation is dependent on USP10 and p53.
Figure 5. G3BP1 regulates cancer cell proliferation through p53 and USP10.
(a) Down-regulation of G3BP1 decreases proliferation and anchorage-independent growth. SK-MEL-5 cells expressing the indicated shRNAs were plated and then formazan production was determined at the indicated time points by MTS assay. These cells were also grown in soft agar for 7 days and then the colonies were counted. Data are shown as means ± S.D. from 3 independent experiments and statistical significance was analyzed after applying the Bonferroni correction (level of significance after correction is 0.025, 0.05/2). (b) Overexpression of G3BP1 enhances proliferation and anchorage-independent growth of HCT116 cells. Cells transfected with the indicated constructs were plated and then formazan production was determined at the indicated time points by MTS assay. These cells were also grown in soft agar for 7 days and then the colonies were counted. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (c) G3BP1 is depleted in wildtype (p53+/+) and deficient (p53−/−) HCT116 cells by shRNAs. Both cell types were infected with the indicated shRNAs for 48 h and then whole cell lysates were analyzed by Western blotting. (d) The inhibitory effect on proliferation and anchorage-independent growth by G3BP1 depletion was diminished in HCT116 p53−/− cells compared with p53+/+ cells. Cells expressing the indicated shRNAs were plated and then formazan production was determined at the indicated time points by MTS assay. These cells were also grown in soft agar for 7 days and then the colonies were counted. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (e) G3BP1 is overexpressed in HCT116 p53+/+ and p53−/− cells. Both cell types were transfected with the indicated constructs for 48 h and then whole cell lysates were analyzed by Western blotting. (f) G3BP1 enhances proliferation and anchorage-independent growth through p53. HCT116 p53+/+ or p53−/− cells transfected with the indicated constructs were plated and formazan production was determined at the indicated time points by MTS assay. These cells were also grown in soft agar for 7 days and then the colonies were counted. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (g) G3BP1 enhances cancer cell growth through USP10. HCT116 cells expressing control or USP10 shRNAs were transfected with the indicated constructs for 48 h. Formazan production was determined at the indicated time points by MTS assay. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (h) Double knock-down of G3BP1 and USP10 rescues the inhibition of proliferation caused by G3BP1 depletion. HCT116 cells were infected with the indicated shRNAs for 48 h. Formazan production was determined at the indicated time points by MTS assay. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05).
Resveratrol enhances USP10-mediated deubiquitination of p53 by disrupting the G3BP1/USP10 interaction
Our results indicate that G3BP1 regulates p53 expression by inhibiting USP10-mediated deubiquitination of p53. We questioned whether resveratrol mediates G3BP1-induced p53 ubiquitination. Indeed, G3BP1 overexpression dramatically induced p53 ubiquitination, and resveratrol dose-dependent treatment diminished p53 ubiquitination in HCT116 cells (Fig. 6a). We also conducted an in vivo ubiquitination assay in H1299 cells (Fig. 6b). The results showed that the inhibitory effect of resveratrol on p53 ubiquitination was not observed in cells without G3BP1 (compare lanes 3 and 4), but resveratrol strongly inhibited p53 ubiquitination in cells co-transfected with G3BP1 and USP10 (compare lanes 5 and 6), indicating that resveratrol suppresses G3BP1-induced p53 ubiquitination through USP10. The mRNA levels of p53 and its target genes were then determined (Fig. 6c). Resveratrol significantly enhanced p21 and PUMA mRNA levels, whereas p53 mRNA levels were not affected by resveratrol treatment. Additionally, resveratrol-induced p53 expression was not observed in cells treated with MG132 (Fig. 6d) or in cells expressing USP10 shRNA (Fig. 6e). To examine a role of Mdm2 in resveratrol-induced p53 expression, the Mdm2 levels and the interaction of Mdm2 with p53 were analyzed. Mdm2 mRNA levels were slightly increased by resveratrol treatment but no significant differences were observed (Supplemental Fig. 5a). Protein levels of Mdm2 were not affected by resveratrol treatment in SK-MEL-5 or HCT116 cells (Supplemental Fig. 5b). Furthermore, the interaction of Mdm2 and p53 was not changed by resveratrol treatment (Supplemental Fig. 5c), indicating that Mdm2 is not involved in resveratrol-mediated p53 expression. Our in vitro pull-down assay data indicated that resveratrol interacted with G3BP1 at the NTF2-like domain, which is the same region that interacts with USP10. We therefore hypothesized that resveratrol interferes with the G3BP1/USP10 interaction. To test this hypothesis, we conducted co-IP assays in the presence or absence of resveratrol. As expected, the interaction between G3BP1 and USP10 decreased in HCT116 cells treated with resveratrol (Fig. 6f). Interestingly, the USP10/p53 interaction was increased by treatment with resveratrol. An in vitro competition assay also showed that resveratrol inhibited the G3BP1/USP10 interaction in a dose-dependent manner (Fig. 6g). These results strongly indicate that resveratrol enhances USP10-mediated deubiquitination of p53 by disrupting the G3BP1/USP10 interaction.
Figure 6. Resveratrol enhances USP10-mediated deubiquitination of p53 by disrupting the G3BP1/USP10 interaction.
(a) Resveratrol suppresses G3BP1-induced p53 ubiquitination. HCT116 cells transfected with the indicated constructs were treated with resveratrol for 48 h and then incubated with MG132 (10 µM) for 4 h. Whole cell lysates were co-immunoprecipitated with a p53 antibody followed by Western blotting with anti-Ub or anti-p53. (b) Resveratrol suppresses G3BP1-mediated p53 ubiquitination through USP10. H1299 cells were transfected with the indicated constructs for 48 h and then treated with resveratrol for 48 h. After additional incubation with MG132 (10 µM) for 4 h, whole cell lysates were co-immunoprecipitated with a Myc-tagged antibody followed by Western blotting with anti-HA or anti-Myc. (c) Resveratrol enhances mRNA levels of p21 and PUMA, but has no effect on p53 mRNA levels. SK-MEL-5 cells were treated with resveratrol (0–40 µM) for 12 h and then mRNA levels were analyzed by qPCR. Relative mRNA levels were normalized against GAPDH. Data are shown as means ± S.D. from 3 independent experiments and statistical significance was analyzed after applying the Bonferroni correction (level of significance after correction is 0.017, 0.05/3). (d) Resveratrol induces p53 through proteasome degradation. SK-MEL-5 cells were treated with resveratrol for 24 h and then incubated with or without MG132 (10 µM) for an additional 4 h. Whole cell lysates were analyzed by Western blotting. Densitometric analysis of relative protein levels was normalized against β-actin and data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (e) USP10 is implicated in resveratrol-induced p53 expression. SK-MEL-5 cells expressing the indicated shRNAs were treated with resveratrol (0–40 µM) for 24 h and then whole cell lysates were analyzed by Western blotting. Densitometric analysis of relative protein levels was normalized against β-actin and data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (f) Resveratrol inhibits the interaction of G3BP1 with USP10 and enhances the USP10/p53 interaction. HCT116 cells were treated with resveratrol for 24 h and then treated with MG132 (10 µM) for an additional 4 h. Whole cell lysates were co-immunoprecipitated with anti-USP10 followed by Western blotting with anti-USP10, anti-G3BP1 or anti-p53. (g) Resveratrol inhibits the G3BP1/USP10 interaction in vitro. Recombinant G3BP1 and USP10 were incubated with the indicated concentration of resveratrol and then pulled down with anti-USP10 followed by Western blotting with anti-USP10 or anti-G3BP1.
The p53-dependent effect of resveratrol mainly relies on G3BP1
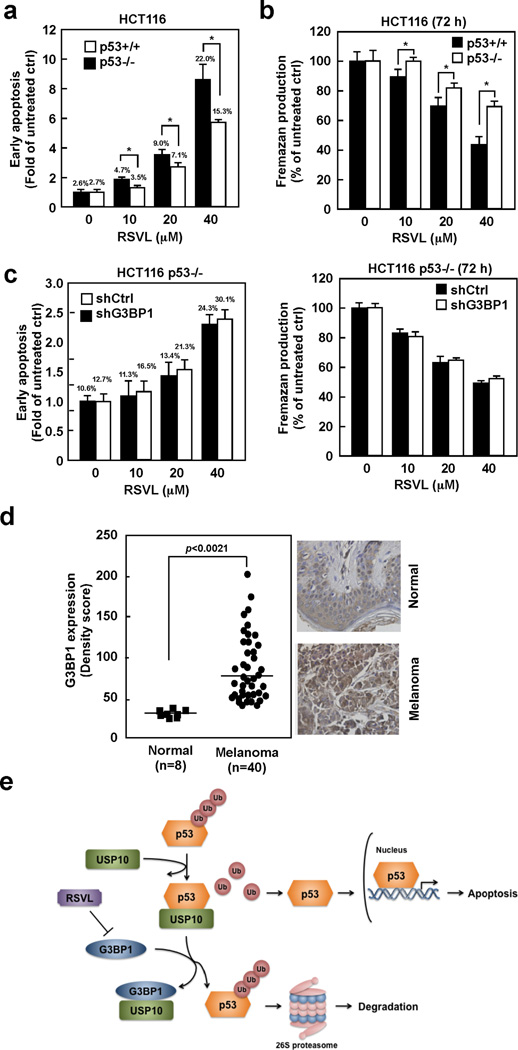
Because resveratrol has been reported to induce p53-dependent as well as p53-independent apoptosis in a certain types of human cancer cells (30), we compared the effect of resveratrol in HCT116 p53+/+ and p53−/− cells. The effects of resveratrol in inducing apoptosis and inhibiting cell proliferation were significantly reduced in HCT116 p53−/− cells compared with HCT 116 p53+/+ cells (Fig. 7a,b). A similar phenomenon of inhibition of proliferation was observed in SK-MEL-5 cells by depletion of p53 (Supplemental Fig. 6a). Although the effect of resveratrol is diminished in p53 depleted or deficient cells, resveratrol still induced apoptosis and inhibited cell proliferation, indicating resveratrol exerts both p53-dependnt and -independent effects. To examine the consequence of G3BP1 in p53-dependent effect of resveratrol, G3BP1 was depleted in HCT116 p53−/− or RPMI7951 (a p53-null human skin malignant melanoma) cells and then cells were treated with resveratrol. Interestingly, the effect of resveratrol on either inducing apoptosis or inhibiting proliferation was not affected by depletion of G3BP1 in HCT116 p53−/− (Fig. 7c) and RPMI7951 (Supplemental Fig. 6b) cells. We therefore concluded that the p53-dependent effect of resveratrol mainly relies on G3BP1. Furthermore, human skin melanoma tissue array analysis showed that the protein levels of G3BP1 were significantly higher in skin melanoma tissue compared with normal skin tissue (Fig. 7d), which is consistent with a role of G3BP1 in promoting cancer cell proliferation. Overall, our study demonstrates that resveratrol directly targets G3BP1, which in turn prevents the G3BP1/USP10 interaction and consequently increases USP10-regulated deubiquitination of p53 (Fig. 7e).
Figure 7. The p53-dependent effect of resveratrol relies mainly on G3BP1.
(a) The effect of resveratrol was diminished in HCT116 p53−/− cells compared with p53+/+ cells. Both cell types were treated with resveratrol (0–40 µM) for 72 h and then cellular apoptosis was determined by flow cytometry. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (b) HCT116 p53−/− cells are less sensitive to resveratrol’s effect on proliferation. Both cell types were treated with resveratrol for 72 h and then formazan production was determined by MTS assay. Data are shown as means ± S.D. from 3 independent experiments (*p < 0.05). (c) The p53-dependent effect of resveratrol mainly relies on G3BP1. HCT116 p53−/− cells expressing the indicated shRNAs were treated with resveratrol for 72 h and then early apoptosis and formazan production were determined by flow cytometry or MTS assay, respectively. Data are shown as means ± S.D. from 3 independent experiments and no significant difference was observed compared with shCtrl cells (*p < 0.05). (d) G3BP1 is overexpressed in human melanoma skin tissue. G3BP1 levels were analyzed by immunohistochemistry using 40 cases of human melanoma skin tissue and 8 cases of normal skin tissue, and then the density score from each sample was determined. Representative cases are shown (right panels). (e) A working model of G3BP1-mediated p53 regulation by resveratrol.
Discussion
G3BP was first identified in 1996 as a protein that binds to the SH3 domain of RasGAP (16). G3BP1 has been reported to regulate several pathways involved in cancer biology including Ras signaling (16, 17). G3BP1 is also reportedly overexpressed in several human cancers (21–24). Furthermore, a novel peptide, GAP161, was shown to block the expression of G3BP1 and induce apoptosis in HCT116 cells (31). Therefore G3BP1 is recognized as a potential target for cancer prevention and therapy. However, a recent report indicated that in conditions where G3BP1 and RasGAP bind to known partners, no interaction between G3BP1 and RasGAP was observed (32). The authors also demonstrated that TAT-RasGAP317–326 does not modulate any of the known G3BP1 functions including modulation of c-myc mRNA levels and sensitization of cancer cells, which raises the question of G3BP1 function. Thus, the molecular mechanisms explaining G3BP1’s role in cancer development are not fully understood.
G3BP1 binds to USP10 and inhibits the deubiquitination activity of USP10 (25). Because we discovered that USP10 is a specific deubiquitinating enzyme of p53 (26), we anticipated that G3BP1 would regulate p53 expression through USP10. As was shown in vitro and in cells, the NTF2-like domain of G3BP1 interacts with the N-terminal portion of USP10. The purified recombinant proteins eluted from a gel filtration column as a complex. When G3BP1 was combined with USP10 at a 2:1 ratio, no single peaks corresponding to individual proteins were observed. We propose that the resulting high molecular weight complex was comprised of 2 molecules of USP10 and 4 molecules of G3BP1 because USP10 was being eluted as a dimer. In this study, we found that G3BP1 enhances the p53 ubiquitination by disrupting the interaction of USP10 with p53, leading to destabilization of p53. Moreover, G3BP1 regulates cancer cell proliferation through p53 and USP10. Thus, we have elucidated a novel mechanism explaining the role of G3BP1 in regulating wildtype p53 cancer development.
The current study also describes G3BP1 as a novel target of resveratrol. Resveratrol is known as one of the most promising anticancer agents isolated from natural products. Many reports have shown that resveratrol possesses an apoptosis-dependent anticancer activity mediated through p53 activation (7–12). However, no report has elucidated the mechanism of resveratrol-induced p53 activation. We found that the resveratrol-induced p53 expression was completely diminished in cells expressing G3BP1 shRNA, and the induction of apoptosis and suppression of anchorage-independent cell growth by resveratrol were obviously decreased by down-regulation of G3BP1. As a mechanism of resveratrol-induced p53 expression, we revealed that resveratrol directly binds to G3BP1 and disrupts the G3BP1/USP10 interaction, releasing USP10 which in turn results in enhanced USP10-mediated deubiquitination of p53. Our results suggest that resveratrol induces p53 expression and apoptosis by directly targeting G3BP1.
Early apoptotic cells were increased from 1.9% to 12.8% by G3BP1 depletion in SK-MEL-5 cells (Fig. 2b). Furthermore, in G3BP1-silenced cells, the p53 levels were enhanced to the levels of p53 in control cells stimulated with 20 µM resveratrol. However, resveratrol treatment (40 µM) could still induce additional apoptosis in G3BP1-depleted cells from 12.8% to 29.3%. As shown in Fig. 7a, resveratrol-induced apoptosis was significantly diminished in HCT116 p53−/− cells compared with p53+/+ cells. However, it was not completely blocked, indicating that resveratrol induced not only p53-dependent apoptosis but also p53-independent apoptosis. Interestingly, we found the effects of resveratrol were not affected by G3BP1 depletion in p53 deficient cells (Fig. 7c, Supplemental Fig. 6b). Although resveratrol could induce p53-dependent and -independent apoptosis, the p53-dependent effect of resveratrol mainly relies on G3BP1. Moreover, depletion of G3BP1 had no effect on proliferation of HCT116 p53−/− (Fig. 5d) or SK-MEL-5 cells expressing p53 shRNA (Supplemental Fig. 4b). However, knocking down G3BP1 expression slightly increased apoptosis from 10.6% to 12.7% (Fig. 7c) and inhibited anchorage-independent growth (Fig. 5d) in HCT116 p53−/− cells, suggesting that G3BP1 might have p53-independent effect. Because G3BP1 has been reported to regulate several pathways involved in cancer development including Ras signaling, NF-kappaB signaling and HER2 signaling (16, 17), those pathways could be implicated in p53-independent effect of G3BP1.
Several groups, including our group, have identified molecular targets of resveratrol such as leukotriene A4 hydrolase and cyclooxygenase-2 (33, 34). Because resveratrol has multiple targets, we also determined the binding of resveratrol with G3BP1 related proteins, including G3BP2, the cytoplasmic poly (A) binding protein (PABP) and NTF2. Results (Supplemental Fig. 7) indicated that resveratrol also binds to G3BP2 and PABP, but not to NTF2, suggesting that G3BP2 and PABP also could be targets of resveratrol-induced p53 expression. Our results showed that overexpression of G3BP1 dramatically decreased p53 levels, whereas overexpression of G3BP2 had less effect on p53 expression levels compared with G3BP1. PABP reportedly regulates p53 translocation to the mitochondria by enhancing acetylation and phosphorylation of p53, but did not affect p53 protein expression (35). We therefore conclude that G3BP1 plays a more important role in p53 expression compared to G3BP2 or PABP. However, the effect of resveratrol on G3BP2 and PABP in cancer development would be interesting to study.
Melanoma is categorized as the most aggressive form of skin cancer, and the number of cases worldwide has doubled in the past 20 years (36). The incidence of melanoma has been increasing at a rate of 6–7% annually and accounts for 75% of deaths from skin cancer. In the United States, the incidence and mortality of melanoma continues to rise faster than that of any other cancer (37). Our results showed that G3BP1 is highly expressed in a human melanoma cell line, and higher expression of G3BP1 is observed in human skin melanoma tissue compared with normal skin tissue. Interestingly, a low frequency (0–10%) of p53 mutation is observed in human melanoma (38–40). This information allows us to conclude that resveratrol might exert highly beneficial preventive or therapeutics effects against melanoma. Overall, our study showed that resveratrol induces p53-dependent apoptosis by directly targeting G3BP1 in p53 wildtype cancers.
Materials and Methods
Cell culture, plasmids, antibodies, and reagents
SK-MEL-5 (p53 wildtype) and RPMI7951 (p53-null) cells were cultured in Eagle’s Minimum Essential Medium (MEM)/10% fetal bovine serum (FBS) with antibiotics containing non-essential amino acids and sodium pyrobate. HCT116 p53+/+ and p53−/− cells were cultured in McCoy's 5a Medium/10% FBS with antibiotics. H1299 (p53-null) cells were cultured in RPMI-1640/10% FBS with antibiotics. HEK 293 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM)/10% FBS. All cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA) and cytogenetically tested and authenticated before the cells were frozen. Each vial of frozen cells was thawed and maintained for a maximum of 8 weeks.
His-G3BP1 was cloned into the BamHI/XbaI sites of the pcDNA4/HisMax vector (Invitrogen, Carlsbad, CA). The Myc-p53, HA-Ub and Flag-HA-USP10 (Addgene plasmid 22543, provided by Dr. W. Harper) (41) plasmids were purchased from Addgene (Cambridge, MA). All G3BP1 plasmids with point mutations were generated by site-directed mutagenesis (Stratagene, La Jolla, CA). Recombinant His-G3BP1, Flag-USP10 or GST-p53 was purchased from Novus Biologicals (Littleton, CO), BPS Bioscience (San Diego, CA) or Millipore (Billerica, MA), respectively.
Anti-G3BP1 (07–1801) and anti-Mdm2 (ab-4) were obtained from Millipore. Anti-p53 (DO-1), anti-G3BP1 (H-10), anti-USP10 (P-18), anti-His-probe (H-3) and anti-β-actin were purchased from Santa Cruz (Dallas, TX). Anti-Myc-tag (9B11), and anti-USP10 were from Cell Signaling (Danvers, MA). Anti-HisG-HRP was obtained from Invitrogen and anti-Flag M2-HRP was from Sigma (St Louis, MO). Anti-HA-HRP (16B12) was purchased from Covance (Princeton, NJ).
RNA interference
The lentiviral expression vectors (PLKO.1-shG3BP1 and PLKO.1-shUSP10) and packaging vectors (pMD2.0G and psPAX) were purchased from OpenBioSystems (Huntsville, AL). G3BP1 shRNA#1: AAACCCAGGGCTGCCTTGGAAAAG. G3BP1 shRNA#2: AAACCCAGGGCTGCCTTGGAAAAG. USP10 shRNA: AAACCCAGGGCTGCCTTGGAAAAG. p53 shRNA: CGGCGCACAGAGGAAGAGAAT.
Lentivirus shRNAs were constructed using the protocol shown on the OpenBioSystems website.
Purification of recombinant G3BP1 and USP10
The NTF2-like (residues 1–139) and RRM (residues 339–421) domains of hG3BP (NP-005745) were cloned into the pET-28a vector (Novagen, Madison, WI, USA). The NTF2-like domain of G3BP1 was expressed in Codon Plus (DE3) RIPL E.coli (Stratagene) and then harvested after 2–2.5 h growth at 37°C following induction with 1 mM isopropyl-1-thio-β-galactopyranoside (IPTG). The RRM domain of G3BP1 was expressed in BL21 (DE3) E.coli (Novagen). The cells were induced with 0.2 mM IPTG and cultured at 15°C overnight. The N-terminal portion of USP10 (residues 1–150) was cloned into the pET-28a vector, expressed in BL21 (DE3) E.coli after 2 h of growth at 37°C following induction with 0.5 mM IPTG. All proteins were purified on HisPur Ni-NTA resin (Thermo Scientific, Waltham, MA, USA) using standard procedures. The lysis and washing buffers were comprised of 30 mM imidazole, 500 mM NaCl, 50 mM NaH2PO4 (pH 8.0), and 10% glycerol. The proteins were eluted with 200–250 mM imidazole, 150 mM NaCl, and 20 mM Tris-HCl (pH 8.0). The second step of purification was performed on an FPLC system using a size exclusion column HiLoad™ 16/60 Superdex™ S200 (GE Healthcare, Pittsburgh, PA) equilibrated with 150 mM NaCl and 20 mM Tris-HCl (pH 8.0).
Resveratrol binding assay
Resveratrol-conjugated Sepharose 4B beads were prepared as described previously (33). Recombinant full-length G3BP1 or a whole cell lysate was incubated with resveratrol-conjugated or control beads at 4°C overnight in NP-40 lysis buffer [50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 0.5% NP-40 and protease inhibitor]. After washing 5 times with NP-40 lysis buffer, the proteins bound to the beads were analyzed by Western blotting. Purified recombinant NTF2-like or RRM domain of G3BP1 (400 µg) was incubated with beads for 30 min at room temperature. The resin was washed 5 times with buffer [150 mM NaCl and 20 mM Tris-HCl (pH 8.0)], and then subjected to SDS-PAGE.
Western blotting
Cells were harvested and disrupted with NP-40 lysis buffer. Whole cell lysates were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking, the membrane was incubated with a specific primary antibody, and then protein bands were visualized with the ECL system after hybridization with a horseradish peroxidase-conjugated secondary antibody.
Computer modeling
The crystal structure of the NTF2-like domain of G3BP1 was obtained from the RCSB Protein Data Bank, PDB entry 3Q90. The subunit labeled chain A was extracted and used in docking. Hydrogens were added consistent with pH 7.0 and all water molecules were removed using the Protein Preparation Wizard in Maestro v9.2. Then the structure was energy-minimized. The chemical structure of resveratrol was prepared using LigPrep v2.5 and then assigned AMSOL partial atomic charge. The program Glide v5.7 was used for ligand docking. Flexible Docking was performed with extra precision (XP) mode. The number of poses per ligand was set to 10 in post-docking minimization and the best 5 poses were output. The other parameters were kept as default.
Anchorage-independent cell growth assay
Cells (8×103 per well) were suspended in basal medium eagle (BME) containing 10% FBS and 0.33% agar and plated on solidified BME containing 10% FBS and 0.5% agar. After incubation for 7 days, the colonies were counted under a microscope using the Image-Pro Plus Software (v.4) program (Media Cybernetics, Rockville, MD).
Apoptosis assay
Cells (1×105 per well) were seeded in 6-cm dishes and then apoptosis was analyzed by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) using the Annexin V-FITC apoptosis detection kit (MBL International Corp., Woburn, MA). Cells stained by Annexin V but not by propidium iodide were determined as early apoptotic cells.
Cell proliferation assay
Cells (2×103 per well) were seeded in 96-well plates and then formazan production was determined using the CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay (Promega, Madison, WI) according to the manufacturer’s instructions. This assay is composed of tetrazolium compound (3-(4,5--dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS) and an electron-coupling reagent (phenazine methosulfate; PMS). MTS is converted into a formazan by dehydrogenase found in metabolically active cells, and the quantity of formazan product is directly proportional to the number of living cells.
Co-immunoprecipitation Assay
Whole cell lysates were pre-cleared with protein A/G agarose beads in NP-40 lysis buffer for 30 min at 4°C and then incubated with 2 µg antibody at 4°C overnight. After incubation with protein A/G agarose beads for 2 h at 4°C, the immunocomplexes were analyzed by Western blotting.
In vivo ubiquitination assay
H1299 cells were transfected with HA-Ub, Myc-p53, Flag-USP10 and/or His-G3BP1 for 48 h and then incubated with 10 µM MG132 for 4 h. Whole cell lysates were co-immunoprecipitated with anti-Myc, and then analyzed by Western blotting.
In vitro binding assay using size exclusion chromatography
The His-tagged proteins, the NTF2-like domain of G3BP1 (residues 1–139) and the N-terminal portion of USP10 (residues 1–150) were purified by a two-step procedure as described above. They both were mixed at a 1:1 molar ratio (~13 nmol) and incubated for 1 h at room temperature. The mixture was loaded onto the Superdex™ 200 10/300 GL column (25 ml) equilibrated with 20 mM Tris-HCl (pH 8.0) containing 150 mM NaCl. A small peak of free USP10 always appeared under these conditions. When G3BP1 was mixed with USP10 at a 2:1 ratio, no free proteins were eluted (data not shown). Similar experiments were performed with the RRM domain and the N-terminal portion of USP10.
In vitro pull-down assay
Recombinant proteins (1 µg each) were incubated with 2 µg antibody in NP-40 lysis buffer at 4°C overnight. After incubation with protein A/G agarose beads for 2 h at 4°C, proteins bound to the beads were analyzed by Western blotting.
Quantitative PCR (qPCR)
Cells (1×105 per well) were seeded in 6-well plates and then total RNA was purified with Trizol (Invitrogen) according to the manufacturer’s instructions. After synthesizing cDNA using the Superscript II Reverse transcription kit (Invitrogen), the quantitative PCR reaction was performed using 7500 Real-Time PCR Systems (Applied Biosystems) with the following primers.
p53: sense 5’-ATGGAGGAGCCGCAGTCAG-3’; antisense 5’-AGAAGCCCAGACGGAAACCG-3’. Mdm2: sense 5’-CAGCTTCGGAACAAGAGACC-3’; antisense 5’-GTCCGATGATTCCTGCTGAT-3’. p21: sense 5'-TCACTGTCTTGTACCCTTGTGC-3’; antisense 5’-GGCGTTTGGAGTGGTAGAAA-3’. PUMA: 5’-GACCTCAACGCACAGTACGA-3’; antisense5’-GAGATTGTACAGGACCCTCCA-3’. GAPDH: sense 5’-AGCCACATCGCTCAGACAC-3’; antisense 5’-GCCCAATACGACCAAATCC-3’.
In vitro competition assay
rG3BP1 (1 µg) was incubated with resveratrol (0, 1, 10 or 100 µM) in NP-40 lysis buffer at 4°C overnight and then incubated with rUSP10 (1 µg) and anti-USP10 (2 µg) for 2 h at 4°C. After incubation with protein A/G agarose beads for an additional 2 h at 4°C, the proteins bound to the beads were analyzed by Western blotting.
Tissue array
A human skin melanoma tissue array (U.S. Biomax, Rockville, MD) was de-paraffinized in xylene and rehydrated in serial concentrations of alcohol. After boiling in 10 mM sodium citrate buffer (pH 6.0) for 12 min, the tissues were incubated with 5% H2O2 for 10 min and then blocked with 50% normal goat serum for 1 h followed by incubation with anti-G3BP1 (1:100) at 4°C overnight. After incubation with a secondary antibody for 1 h, G3BP1 expression was visualized with 3,3'-diaminobenzidine. Images were captured under a microscope and analyzed using the ImageJ software program (v1.37v, National Institutes of Health).
Statistical analysis
All quantitative data are expressed as means ± S.D. The Student’s t-test with Bonferroni correction was used for statistical analysis. The criteria for statistical significance are shown in each figure.
Supplementary Material
Acknowledgements
We thank Dr. Yibin Deng (The Hormel Institute, University of Minnesota) for kindly providing the expression vectors of shRNA p53.
Financial support: This work was supported by AgStar Minnesota (ZD), The Hormel Foundation (ZD), and National Institutes of Health grants CA111536 (ZD), CA172457 (ZD), CA166011 (ZD), R37 CA081064 (ZD), CA148940 (ZL), CA108961 (ZL), the National Basic Research Program of China (973 Program, Grant No. 2013CB530700), National Natural Science Foundation of China (81102011, 81222029 and 31270806) and Kendall-Mayo Fellowship in Biochemistry, the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81221001), Shanghai Science and Technology Committee Modernization of Traditional Chinese Medicine special (11DZ1973801).
Footnotes
Potential conflicts of interest: No potential conflicts of interest
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10(4):431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- 3.Celotti E, Ferrarini R, Zironi R, Conte LS. Resveratrol content of some wines obtained from dried Valpolicella grapes: Recioto and Amarone. J Chromatogr A. 1996;730(1–2):47–52. doi: 10.1016/0021-9673(95)00962-0. [DOI] [PubMed] [Google Scholar]
- 4.Chen RS, Wu PL, Chiou RY. Peanut roots as a source of resveratrol. J Agric Food Chem. 2002;50(6):1665–1667. doi: 10.1021/jf011134e. [DOI] [PubMed] [Google Scholar]
- 5.Sobolev VS, Cole RJ. trans-resveratrol content in commercial peanuts and peanut products. J Agric Food Chem. 1999;47(4):1435–1439. doi: 10.1021/jf9809885. [DOI] [PubMed] [Google Scholar]
- 6.Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science. 1997;275(5297):218–220. doi: 10.1126/science.275.5297.218. [DOI] [PubMed] [Google Scholar]
- 7.Delmas D, Rebe C, Lacour S, Filomenko R, Athias A, Gambert P, et al. Resveratrol-induced apoptosis is associated with Fas redistribution in the rafts and the formation of a death-inducing signaling complex in colon cancer cells. J Biol Chem. 2003;278(42):41482–41490. doi: 10.1074/jbc.M304896200. [DOI] [PubMed] [Google Scholar]
- 8.Estrov Z, Shishodia S, Faderl S, Harris D, Van Q, Kantarjian HM, et al. Resveratrol blocks interleukin-1beta-induced activation of the nuclear transcription factor NF-kappaB, inhibits proliferation, causes S-phase arrest, and induces apoptosis of acute myeloid leukemia cells. Blood. 2003;102(3):987–995. doi: 10.1182/blood-2002-11-3550. [DOI] [PubMed] [Google Scholar]
- 9.Fulda S, Debatin KM. Sensitization for tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by the chemopreventive agent resveratrol. Cancer Res. 2004;64(1):337–346. doi: 10.1158/0008-5472.can-03-1656. [DOI] [PubMed] [Google Scholar]
- 10.Gill C, Walsh SE, Morrissey C, Fitzpatrick JM, Watson RW. Resveratrol sensitizes androgen independent prostate cancer cells to death-receptor mediated apoptosis through multiple mechanisms. Prostate. 2007;67(15):1641–1653. doi: 10.1002/pros.20653. [DOI] [PubMed] [Google Scholar]
- 11.Niles RM, McFarland M, Weimer MB, Redkar A, Fu YM, Meadows GG. Resveratrol is a potent inducer of apoptosis in human melanoma cells. Cancer Lett. 2003;190(2):157–163. doi: 10.1016/s0304-3835(02)00676-6. [DOI] [PubMed] [Google Scholar]
- 12.Pervaiz S. Resveratrol: from grapevines to mammalian biology. Faseb J. 2003;17(14):1975–1985. doi: 10.1096/fj.03-0168rev. [DOI] [PubMed] [Google Scholar]
- 13.She QB, Bode AM, Ma WY, Chen NY, Dong Z. Resveratrol-induced activation of p53 and apoptosis is mediated by extracellular-signal-regulated protein kinases and p38 kinase. Cancer Res. 2001;61(4):1604–1610. [PubMed] [Google Scholar]
- 14.Hsieh TC, Juan G, Darzynkiewicz Z, Wu JM. Resveratrol increases nitric oxide synthase, induces accumulation of p53 and p21(WAF1/CIP1), and suppresses cultured bovine pulmonary artery endothelial cell proliferation by perturbing progression through S and G2. Cancer Res. 1999;59(11):2596–2601. [PubMed] [Google Scholar]
- 15.Joe AK, Liu H, Suzui M, Vural ME, Xiao D, Weinstein IB. Resveratrol induces growth inhibition, S-phase arrest, apoptosis, and changes in biomarker expression in several human cancer cell lines. Clin Cancer Res. 2002;8(3):893–903. [PubMed] [Google Scholar]
- 16.Parker F, Maurier F, Delumeau I, Duchesne M, Faucher D, Debussche L, et al. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol Cell Biol. 1996;16(6):2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pazman C, Mayes CA, Fanto M, Haynes SR, Mlodzik M. Rasputin, the Drosophila homologue of the RasGAP SH3 binding protein, functions in ras- and Rho-mediated signaling. Development. 2000;127(8):1715–1725. doi: 10.1242/dev.127.8.1715. [DOI] [PubMed] [Google Scholar]
- 18.Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, et al. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21(22):7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, et al. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18(7):3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prigent M, Barlat I, Langen H, Dargemont C. IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J Biol Chem. 2000;275(46):36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- 21.Barnes CJ, Li F, Mandal M, Yang Z, Sahin AA, Kumar R. Heregulin induces expression, ATPase activity, and nuclear localization of G3BP, a Ras signaling component, in human breast tumors. Cancer Res. 2002;62(5):1251–1255. [PubMed] [Google Scholar]
- 22.Liu Y, Zheng J, Fang W, You J, Wang J, Cui X, et al. Identification of metastasis associated gene G3BP by differential display in human cancer cell sublines with different metastatic potentials G3BP as highly expressed in non-metastatic. Chin Med J (Engl) 2001;114(1):35–38. [PubMed] [Google Scholar]
- 23.French J, Stirling R, Walsh M, Kennedy HD. The expression of Ras-GTPase activating protein SH3 domain-binding proteins, G3BPs, in human breast cancers. Histochem J. 2002;34(5):223–231. doi: 10.1023/a:1021737413055. [DOI] [PubMed] [Google Scholar]
- 24.Gautier-Bert K, Murol B, Jarrousse AS, Ballut L, Badaoui S, Petit F, et al. Substrate affinity and substrate specificity of proteasomes with RNase activity. Mol Biol Rep. 2003;30(1):1–7. doi: 10.1023/a:1022261925117. [DOI] [PubMed] [Google Scholar]
- 25.Soncini C, Berdo I, Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 2001;20(29):3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- 26.Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140(3):384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MM, Wiederschain D, Kennedy D, Hansen E, Yuan ZM. Modulation of p53 and MDM2 activity by novel interaction with Ras-GAP binding proteins (G3BP) Oncogene. 2007;26(29):4209–4215. doi: 10.1038/sj.onc.1210212. [DOI] [PubMed] [Google Scholar]
- 28.Matsuki H, Takahashi M, Higuchi M, Makokha GN, Oie M, Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 2013;18(2):135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M, et al. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol Cell Biol. 2013;33(4):815–829. doi: 10.1128/MCB.00763-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chow SE, Wang JS, Chuang SF, Chang YL, Chu WK, Chen WS, et al. Resveratrol-induced p53-independent apoptosis of human nasopharyngeal carcinoma cells is correlated with the downregulation of DeltaNp63. Cancer Gene Ther. 2010;17(12):872–882. doi: 10.1038/cgt.2010.44. [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Zhang S, He H, Zhao W, Chen J, Shao RG. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci. 2012 doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annibaldi A, Dousse A, Martin S, Tazi J, Widmann C. Revisiting G3BP1 as a RasGAP binding protein: sensitization of tumor cells to chemotherapy by the RasGAP 317–326 sequence does not involve G3BP1. PLoS One. 2011;6(12):e29024. doi: 10.1371/journal.pone.0029024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oi N, Jeong CH, Nadas J, Cho YY, Pugliese A, Bode AM, et al. Resveratrol, a red wine polyphenol, suppresses pancreatic cancer by inhibiting leukotriene A(4)hydrolase. Cancer Res. 2010;70(23):9755–9764. doi: 10.1158/0008-5472.CAN-10-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zykova TA, Zhu F, Zhai X, Ma WY, Ermakova SP, Lee KW, et al. Resveratrol directly targets COX-2 to inhibit carcinogenesis. Mol Carcinog. 2008;47(10):797–805. doi: 10.1002/mc.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thangima Zannat M, Bhattacharjee RB, Bag J. In the absence of cellular poly (A) binding protein, the glycolytic enzyme GAPDH translocated to the cell nucleus and activated the GAPDH mediated apoptotic pathway by enhancing acetylation and serine 46 phosphorylation of p53. Biochem Biophys Res Commun. 2011;409(2):171–176. doi: 10.1016/j.bbrc.2011.04.094. [DOI] [PubMed] [Google Scholar]
- 36.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 37.Lev DC, Ruiz M, Mills L, McGary EC, Price JE, Bar-Eli M. Dacarbazine causes transcriptional up-regulation of interleukin 8 and vascular endothelial growth factor in melanoma cells: a possible escape mechanism from chemotherapy. Mol Cancer Ther. 2003;2(8):753–763. [PubMed] [Google Scholar]
- 38.Castresana JS, Rubio MP, Vazquez JJ, Idoate M, Sober AJ, Seizinger BR, et al. Lack of allelic deletion and point mutation as mechanisms of p53 activation in human malignant melanoma. Int J Cancer. 1993;55(4):562–565. doi: 10.1002/ijc.2910550407. [DOI] [PubMed] [Google Scholar]
- 39.Albino AP, Vidal MJ, McNutt NS, Shea CR, Prieto VG, Nanus DM, et al. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 1994;4(1):35–45. doi: 10.1097/00008390-199402000-00006. [DOI] [PubMed] [Google Scholar]
- 40.Hartmann A, Blaszyk H, Cunningham JS, McGovern RM, Schroeder JS, Helander SD, et al. Overexpression and mutations of p53 in metastatic malignant melanomas. Int J Cancer. 1996;67(3):313–317. doi: 10.1002/(SICI)1097-0215(19960729)67:3<313::AID-IJC1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.