Abstract
Purpose
Expression of CBL, an ubiquitin ligase, is decreased in 60% of human pancreatic ductal adenocarcinomas (PDACs) and is associated with shorter overall survival. We sought to determine how low CBL directly contributes to clinically more aggressive PDAC.
Experimental Design
Human PDACs were stained for CBL, pEGFR, and EGFR. CBL-low was modeled in PDAC cells (Panc-1, L3.6pl, AsPC-1) via transient transfection (siRNA) or stable knockdown (shRNA). Cell viability and apoptosis were measured by MTT assays and FACS. Immunoblot and a phospho-receptor tyrosine kinase (pRTK) array were used to probe signal transduction. NOD-scid-IL2Rγnull mice were subcutaneously implanted with PDAC or PDACCBL-low cells on opposite flanks and treated with gemcitabine ± erlotinib for ≥4 weeks.
Results
There was an inverse correlation between CBL and pEGFR protein expression in 12 of 15 tumors. CBL knockdown increased PDAC resistance to gemcitabine and 5-FU by upregulating pEGFR (Y1068), pERK, and pAKT. A pRTK array of PDACCBL-low cells revealed additional activated tyrosine kinases but all to a much lower magnitude than EGFR. Increased chemoresistance from low CBL was abrogated by the EGFR inhibitor erlotinib both in vitro and in vivo. Erlotinib + gemcitabine treated PDACCBL-low cells exhibited greater apoptosis by cleaved PARP, Caspase 3 and Annexin V/PI.
Conclusions
Low CBL causes chemoresistance in PDAC via stress-induced EGFR activation that can be effectively abrogated by EGFR inhibition. These results suggest that dysregulation of ubiquitination is a key mechanism of EGFR hyperactivation in PDAC and that low CBL may define PDAC tumors likely to respond to erlotinib treatment.
Keywords: CBL, pancreatic cancer, chemoresistance, EGFR, ubiquitin
Introduction
Over the past decade, the number of people who die annually from pancreatic ductal adenocarcinoma (PDAC) has been slowly increasing in the United States (US). This trend is in strike contrast to the decreasing death rates for other major cancers(1). If the current incidence and mortality continue, PDAC will become the 2nd leading cause of cancer-related deaths in the US by 2020(2). A main contributor to this poor prognosis is PDAC’s notorious unresponsiveness to cytotoxic and targeted therapies. Identification of distinct patient subgroups, by well-defined biomarkers, who are likely to respond to specific therapies has been less successful in PDAC than other solid tumors due to the presence of only a few high-prevalence, unifying molecular changes. (3,4). However, there are two notable examples that reveal this strategy may still work to improve treatment responses. First, three distinct molecular subtypes of PDAC were identified, only one of which was responsive to gemcitabine treatment(5). Second, from the phase III clinical trial comparing gemcitabine plus the EGFR inhibitor erlotinib to gemcitabine alone, subgroup analysis identified those who developed a severe skin rash achieved nearly double the overall survival with combination therapy than those who did not(6). In light of these findings, there has been renewed interest in the identification of biomarkers to identify PDACs likely to respond to these established therapies. The identification and validation of such biomarkers is an unmet clinical need in PDAC(7).
The CBL (named after Casitas B-lineage Lymphoma) family of proteins is comprised of c-CBL, CBL-b, and CBL-c (hereafter collectively referred to as CBL). c-CBL and CBL-b are ubiquitously expressed and function as E3 ubiquitin-protein ligases and multifunctional adaptor proteins involved in regulation of signal transduction, lymphocyte signaling, and the actin cytoskeleton(8,9). CBL-c is a truncated form that is expressed predominantly in epithelial cells; its in vivo functions are poorly understood(10). The E3 ligase domain of CBL mediates the formation of a covalent bond between ubiquitin and protein substrates leading to their trafficking to the endosome for lysosomal degradation. Homozygous deletions or missense mutations in regions affecting the ubiquitin-related function of CBL are commonly found in myeloid neoplasms and are oncogenic. CBL’s ubiquitin targets are commonly tyrosine kinases (TKs), including Epidermal Growth Factor Receptor (EGFR)(8). In a survival-based whole genome multi-dimensional array analysis of 25 human PDACs with high tumor cell content (and low stromal volume), we have reported low CBL transcript expression correlates with shorter disease-free survival in human PDAC(11) and further validated this observation in 42 independent samples. Moreover, low CBL expression was a frequent event, as 60% of resected PDACs had low mRNA levels (defined as below the mean transcript level for all tumors). However, the mechanism by which reduced CBL directly contributes to poor patient survival is not yet known.
EGFR is a cell surface receptor that is activated by binding to extracellular protein ligands which leads to autophosphorylation of the intracellular domain, homo- or hetero-dimerization with other ERBB family members, and downstream signaling predominantly through the MAPK and AKT pathways(12). To maintain physiologic signaling levels, ligand-activated EGFR is internalized, undergoes CBL-mediated ubiquitination, and is targeted for destruction in lysosomes. EGFR has been previously shown to have biologic significance in PDAC. It is upregulated in 60 – 80% of human tumors(13), involved in early tumor initiation(14), and directly correlates with poor prognosis(15). The EGFR inhibitor, erlotinib, is the only molecular targeted therapy shown to improve survival in PDAC in a phase III clinical trial in patients with metastatic disease(6). EGFR dysregulation in PDAC is not due to genome level changes, as it is neither amplified (as seen in breast and gastric cancers) nor mutated (as seen with lung cancer). Rather, recent work in PDAC has confirmed previous findings in other solid tumors that EGFR is (hyper)-activated by autocrine ligand production in response to cytotoxic chemotherapy(16). To our knowledge, CBL has not been previously explored as a possible mechanism of EGFR dysregulation leading to cell autonomous chemoresistance.
The objective of this study was to determine the mechanism whereby low CBL might directly contribute to worse patient survival. We hypothesized that CBL functions as an important negative feedback mechanism to EGFR signaling in PDAC that when suppressed results in enhanced tumor growth and chemoresistance. We found that erlotinib abrogates the chemoresistance afforded by low CBL in vitro and in vivo.
Materials and Methods
Patients and samples
This study was approved by the UCLA Institutional Review Board and the UCLA Office of Animal Research Oversight.
Immunohistochemistry
Archival formalin-fixed, paraffin-embedded human tumor samples were incubated at 60°C for 1 hour, deparaffinized in xylene, and rehydrated with graded alcohol washes. Antigen retrieval was performed by heating in 0.01M sodium citrate buffer at 100°C for 15 minutes (CBL), or in Tris-EDTA buffer at 95°C for 30 minutes (pEGFR, EGFR), followed by quenching of endogenous peroxidase with 3% hydrogen peroxide. After blocking for 1 hour with 5% donkey serum in phosphate-buffer solution (PBS) at room temperature (RT), primary antibody was added to serial sections, pEGFR 1:100, EGFR 1:100 (Cell Signaling Technology, Inc.), and CBL 1:50 (Abcam) and incubated overnight at 4°C. 1:250 biotin-conjugated anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Inc.) was subsequently added and developed using Elite Vectastain ABC kit (Vector Laboratories).
Cell culture
Human pancreatic cancer cell lines Panc-1 and AsPC-1 cells were obtained in 2005 from the American Type Culture Collection (Rockville, MD). L3.6pl cells were obtained in 2010 from Hong Wu (UCLA). Since receipt, the cells have not been subsequently authenticated. Panc-1 and L3.6pl cells were maintained at low passage in DMEM (Gibco, Life Technologies) +10% fetal bovine serum (FBS) (Gemini Bio-Products) + 1× Penicillin-Streptomycin (PS) (Gibco). AsPC-1 cells were passaged in RPMI (Gibco) + 10% FBS + 1× PS.
siRNA knockdown of CBL
Experimentally validated CBL siRNA was purchased from QIAGEN. Utilizing the RNAi Human/Mouse Starter Kit (QIAGEN) optimization of the fast-forward transfection protocol was performed in each cell type. For each experiment, 3×105 cells in 6-well (western) or 2×104 cells in 96-well (MTT assay) were plated from a single cell suspension, then 5nM CBL or 5nM AllStars Negative Control (neg con) siRNA with HiPerfect Transfection Reagent was added. After overnight incubation, the transfection media was changed to fresh media and transfection efficiency was qualitatively assessed by fluorescent imaging (CBL siRNA-3’ 6-FAM, AllStars Negative Control siRNA-Alexa Fluor 555). Cells were then incubated for 24 hours at optimal growth conditions, prior to start of assays involving serum-deprivation, chemotherapy.
Immunoblots
To directly probe modulation of EGFR activation by epidermal growth factor (EGF) ligand with or without CBL knockdown, Panc-1 cells were first transfected with CBL or neg con siRNA as above, then serum-starved for 4 hours. EGF 10ng/mL (BD Biosciences) was then added to the cells, which were lysed at 5, 20, and 60 minutes to assess the kinetics of EGFR activation/downregulation. Cells were washed with PBS at 4°C, lysed with 2% SDS buffer containing phosphatase inhibitors and mechanical scraping, collected and kept on ice. Data shown is for the 20-minute time point. Lysates were then sonicated for 5 seconds and Bradford assays were performed to measure protein concentrations for each sample. 35µg of protein were then loaded into each well of a 10% acrylamide gel. Samples were then resolved by SDS-PAGE and transferred to a PVDF membrane. Membranes were first blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline + 0.1% Tween20 (TBST), washed ×3 in TBST, then incubated overnight at 4°C in 5% BSA in TBS containing primary antibody at 1:1000 dilution unless otherwise noted: CBL, pAKT 1:500, AKT, pEGFR (Y1068), pEGFR (Y1045), EGFR 1:2000, pERK, ERK (Cell Signaling). β-actin 1:5000 (Sigma) was used to normalize sample loading. After washing, Anti-mouse- 1:10,000 or Anti-rabbit- 1:5,000 HRP-conjugated secondary antibody (Jackson Labs) in 5% milk was incubated for 1 hour at RT, again washed ×3, before addition of Amersham ECL prime (GE Healthcare Life Sciences) and chemiluminescent imaging on a ChemiDoc XRS+ (BioRad).
After siRNA treatment and with cells in the log phase of proliferation, we assessed CBL modulation of EGFR activation in the presence of cytotoxic chemotherapy agents gemcitabine (Gem) (Sagent Pharmaceuticals) and 5-fluorouracil (5-FU) (APP Pharmaceuticals). These were added to Panc-1 and L3.6pl cell lines at their IC50 values (Panc-1: 1uM and 10uM(17,18), L3.6pl: 25nM and 1µM(19,20) respectively) in serum-free media (SFM). After 48 hours in SFM ± chemotherapy, lysates were collected and analyzed via western blot as above. To broaden the scope from EGFR to other RTK pathways, parallel samples were analyzed using the Proteome Profiler™ Human Phospho-RTK Array Kit (R&D Systems Inc). Blots of combination therapy, Gem/5-FU ± erlotinib 10µM (OSI Pharmaceuticals) were also performed.
Apoptosis was assessed via western blot of PARP and Caspase 3 and their cleaved products (Cell Signaling).
MTT Assays
Cells were plated at equal density (2×104) in 96-well plates, treated with siRNA, then Gem/5-FU ± erlotinib were added in SFM at the concentrations above. At 24, 48 and 72 hours from the start of chemotherapy treatment, media was aspirated from each well, and 12mM MTT (Life Technologies) was added in fresh phenol-red free media and incubated for 4 hours at 37°C. Cells were then lysed in 10% SDS-0.01M HCl, and after 12h the quantity of dissolved formazan was measured via automated plate reader at 570nM. All samples and controls were analyzed in triplicate, and each experiment was repeated twice. Shown is data from one representative experiment.
FACS
Utilizing the Annexin V-FITC Apoptosis Detection Kit (BD Biosciences) cells were grown in 6 well plates, transfected with siRNA, then 48 hours in chemotherapy as above. Cells and media were collected, washed, and stained with FITC-Annexin V and PI in binding buffer along with appropriate controls. Total number of Annexin V and PI positive cells were counted using a FACScan flow cytometer (Becton Dickinson) and graphed as percentage of total cell number (FlowJo 9.3.2, Tree Star, Inc.). Error bars are the SEM for 3 independent experiments.
shRNA knockdown of CBL
Lentiviruses were produced by transfecting HEK293T packaging cells in polyethylenimine (Polysciences) with a 3-plasmid system. Briefly, DNA for transfections was prepared by mixing pCMV-Δ8.9, pCMV-VSVG with pLKO.1 plasmid, either as an empty vector or containing one of 6 unique sequences from the TRC library predicted to knockdown CBL (Open Biosystems, Thermoscientific). Lentiviral supernatants were harvested at 24h post-transfection, filtered, and frozen at −80°C for long-term storage.
L3.6pl and Panc-1 cells were transduced with lentivirus in the presence of 8µg/ml polybrene (Sigma, St. Louis, MO). Cells were incubated for 24h, and the media was changed to remove virus particles. Stable knockdown was achieved by 7-day selection in puromycin 1µg/mL. After experimental validation of ≈75% knockdown efficiency by qRT-PCR, the mature antisense sequence 5’-TACCTTTAATTTCACATCGGC-3’ was chosen for in vivo studies.
qRT-PCR
Utilizing the QIAGEN® system, total RNA was extracted from cells (miRNeasy Mini Kit), reverse transcribed (miScript® II Reverse Transcription Kit) and then qRT-PCR was performed (miScript® SYBR Green PCR Kit) with primers for CBL and RPL13A as a housekeeping control. The CBL primer sequences were referenced from the RTPrimerDB(21). RPL13A primers were designed with forward sequence 5’-CATCGTGGCTAAACAGGTACTG-5’ and reverse 5’-GCACGACCTTGAGGGCAGCC-5’. Primers were obtained from Integrated DNA Technologies.
Preclinical trial in NSG mice
After selection in puromycin, two transformed L3.6pl and Panc-1 cell lines, pLKO.1CBL shRNA and pLKO.1neg con shRNA, were expanded by serial passaging. After three passages they were trypsinized, collected and counted. 2×105 pLKO.1neg con shRNA cells were injected into the right flank and 2 ×105 pLKO.1CBL shRNA into the left flank of NOD-scid-IL2Rγnull mice (n=18) and randomly assigned to 1 of 3 groups. By the fourth day post-injection, all mice had developed palpable tumors. At 7 days post-injection, treatment began with Gem 50mg/kg administered by subcutaneous injection twice weekly (n=6). A second treatment group received both Gem + erlotinib 25mg/kg by oral gavage daily (n=6). Control animals received injections and oral gavage of PBS (n=6). Gem only treated animals also received oral gavage PBS. Tumors were measured with digital calipers for length, width, and height every 3–4 days and volume estimated(22). At 22 days in the L3.6pl xenografts several control animals had at least 1 tumor measuring ≥15mm and were euthanized per protocol. At 28 days, L3.6pl Gem treated animals had met this same endpoint and this experiment was concluded. The Panc-1 xenografts were slower growing, and animals were survived out an additional 2 weeks until meeting study endpoints. Necropsies were performed and tumors were analyzed ex vivo for weight, volume, and immunoblotting to verify stable CBL knockdown. In order to validate our findings, the entire experiment was then repeated, with similar trends in tumor growth characteristics and observed group differences. Shown is the data from the first experiment.
Statistical analysis
Statistical analysis was performed with SPSS 20.0.0.1 (IBM, New York, NY). The relationship between categorical variables was examined using Pearson’s chi-square test and difference between means were evaluated by Student t-test or one-way ANOVA as appropriate with significance defined as P<0.05. Image Lab software (BioRad) was used for densitometry analysis of western blots. For ease of comparison, reported values were first normalized to β-actin as a loading control then multiplied by a constant to reach the lowest whole integer. Unless otherwise stated, error bars ± SD.
Results
CBL expression in human PDAC is inversely correlated with EGFR expression and activation
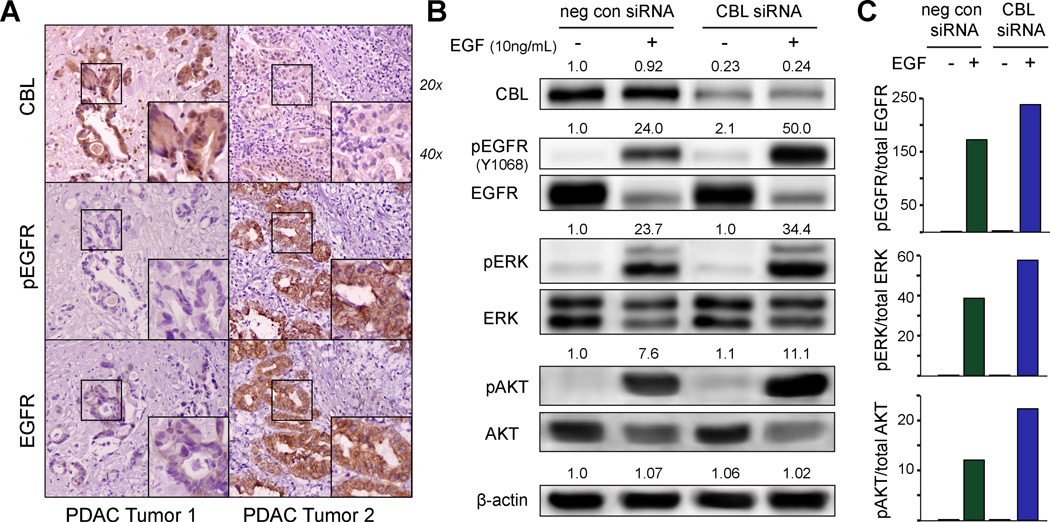
Our previous in silico and in vitro published findings(11) of 67 human PDACs revealed that CBL mRNA expression is decreased in 60% of tumors and significantly correlated with patient survival (i.e. lower CBL was associated with shorter survival). EGFR is one of the most commonly reported CBL targets and is upregulated in at least 60% of human PDAC(13). Therefore, we hypothesized that low CBL leads to higher EGFR expression and activation in human PDAC. Staining of primary human PDAC tissues for CBL, pEGFR, and EGFR revealed an inverse correlation between CBL and pEGFR in 12 of 15 (80%, P=0.01) tumors, and CBL and EGFR expression in 11 of 15 (73%, P=0.03). Fig. 1A shows representative images of strongly positive or negative CBL expression and its inverse correlation with pEGFR and EGFR in tumor cells. Notably, CBL, EGFR, and p-EGFR staining were low in the stroma.
Figure 1.
CBL expression in human pancreatic ductal adenocarcinoma (PDAC) is inversely correlated with EGFR expression and activation. A, immunohistochemistry staining of human PDAC tumors stained for CBL, pEGFR, and EGFR reveals an inverse expression in 12 of 15 tumors (2 representative individual patient samples are shown), suggesting a possible regulatory effect of CBL on EGFR. B, correspondingly, when stimulated by EGF ligand, immunoblots of Panc-1 cells with CBL knockdown show increased phosphorylation of EGFR (Y1068) when compared to the isogenic parental cell line, with increased downstream pERK and pAKT (columns 2 versus 4). Relative densitometry values normalized to β-actin as a loading control are displayed above each phosphoprotein band and, C, plotted as a bar graph.
To further confirm CBL regulation of EGFR expression and activation, Panc-1 PDAC cells with or without siRNA-induced low CBL (hereafter referred to as Panc-1CBL-low or Panc-1 cells, respectively) were cultured in the presence or absence of EGFR ligand, EGF. In the absence of ligand, low CBL conferred only a mild increase in activation of EGFR at its autophosphorylation (Tyrosine 1068) site (Fig. 1B, columns 1 vs. 3). However, in the presence of ligand, low CBL significantly enhanced EGFR activation (Fig. 1B–C, columns 2 vs. 4). Moreover, EGF treatment of Panc-1CBL-low cells also resulted in increased activation of pERK and pAKT (Fig. 1B–C, columns 2 vs. 4); these downstream mediators were not activated in Panc-1CBL-low cells in the absence of exogenous ligand. These results suggest that low CBL increased ligand-dependent EGFR activation, as well as downstream MAPK and AKT signaling, which may lead to improved cell proliferation and survival.
Low CBL mediates chemoresistance through enhanced EGFR autoactivation
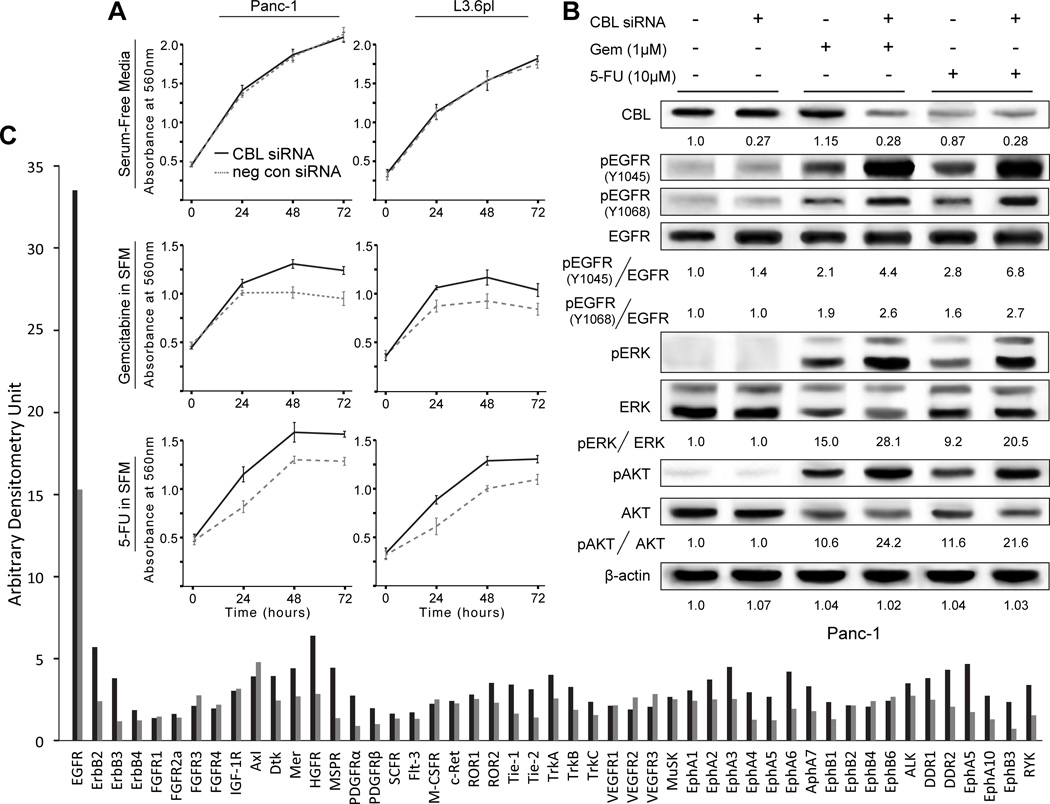
Considering that low CBL increased EGFR-dependent AKT and MAPK signaling, we next sought to determine the functional significance of low CBL on PDAC cells in culture. CBL siRNA treatment of Panc-1, L3.6pl and AsPC-1 PDAC cell lines did not yield increased viability (Fig. 2A and Fig. S1), soft agar growth, or invasion either alone or in the presence of exogenous EGF ligand (data not shown). Given that all of the patients in the previous survival analysis received adjuvant chemotherapy following surgical resection(11), we next tested the functional consequences of low CBL in the presence of chemotherapy. Panc-1CBL-low and L3.6plCBL-low had significantly greater viability than isogenic cells with intact CBL expression when treated with gemcitabine or 5-FU, two chemotherapeutic drugs commonly used for PDAC (Fig. 2A). Phosphoproteomic comparison of gemcitabine and 5-FU treated Panc-1CBL-low and Panc-1 cells in serum-free media indicated that resistance to these two drugs afforded by low CBL is associated with autoactivation of EGFR, and MAPK and AKT signaling (Fig. 2B). The same was confirmed in L3.6plCBL-low low versus L3.6pl cells (Fig. S2). Chemotherapy-treated PDACCBL-low cells have increased EGFR phosphorylation at both autoactivation (Y1068) and CBL binding (Y1045) sites when compared to PDAC cells, further suggesting that low CBL may be responsible for differences in activation levels. This finding differs from Fig. 1B, as in the presence of chemotherapy, exogenous EGF ligand was not needed to yield enhanced EGFR activation and downstream signaling.
Figure 2.
Low CBL mediates enhanced chemoresistance through EGFR autoactivation. A, MTT assay reveals that cell viability is enhanced with CBL knockdown in Panc-1 and L3.6pl cells treated with gemcitabine (Gem) or 5-fluorouracil (5-FU) at 24, 48 and 72 hours (P<0.05). Error bars ± SD. B, when treated with chemotherapy in serum-free conditions, immunoblot analysis of Panc-1 cells reveals that EGFR is autoactivated at it canonical autophosphorylation site (Y1068) with corresponding activation of downstream mediators ERK and AKT (columns 3 and 5 vs. 1). CBL knockdown further enhances autoactivation of EGFR at Y1068 as well as the docking site for CBL binding Y1045, (columns 3 vs. 4 and 5 vs. 6). Relative densitometry values using β-actin as a loading control are reported below each band. C, a human phosphoRTK immunoblot array of L3.6pl cells in gemcitabine (25nM) reveals that pEGFR is highly expressed and increased >2-fold with CBL knockdown.
CBL also downregulates many additional, non-EGFR, TKs implicated in pancreatic tumorigenesis such as SRC(11), IGF-1R(23), PDGFR(24), c-MET(25), and RON(26). The specificity of low CBL for EGFR activation was evaluated using a phospho-TK screen (Fig. 2C and Fig. S3). Gemcitabine treated L3.6plCBL-low cells had >2× activation of EGFR than L3.6pl cells. While at a much lower magnitude (membrane exposure for 5 minutes), Erbb2, 3, and 4; HGFR; and PDGFR all also had significantly greater activation in cells with low CBL. These findings suggest that low CBL expression associated with shorter survival in human PDAC may be due to chemoresistance resulting from enhanced autoactivation of EGFR or other RTKs.
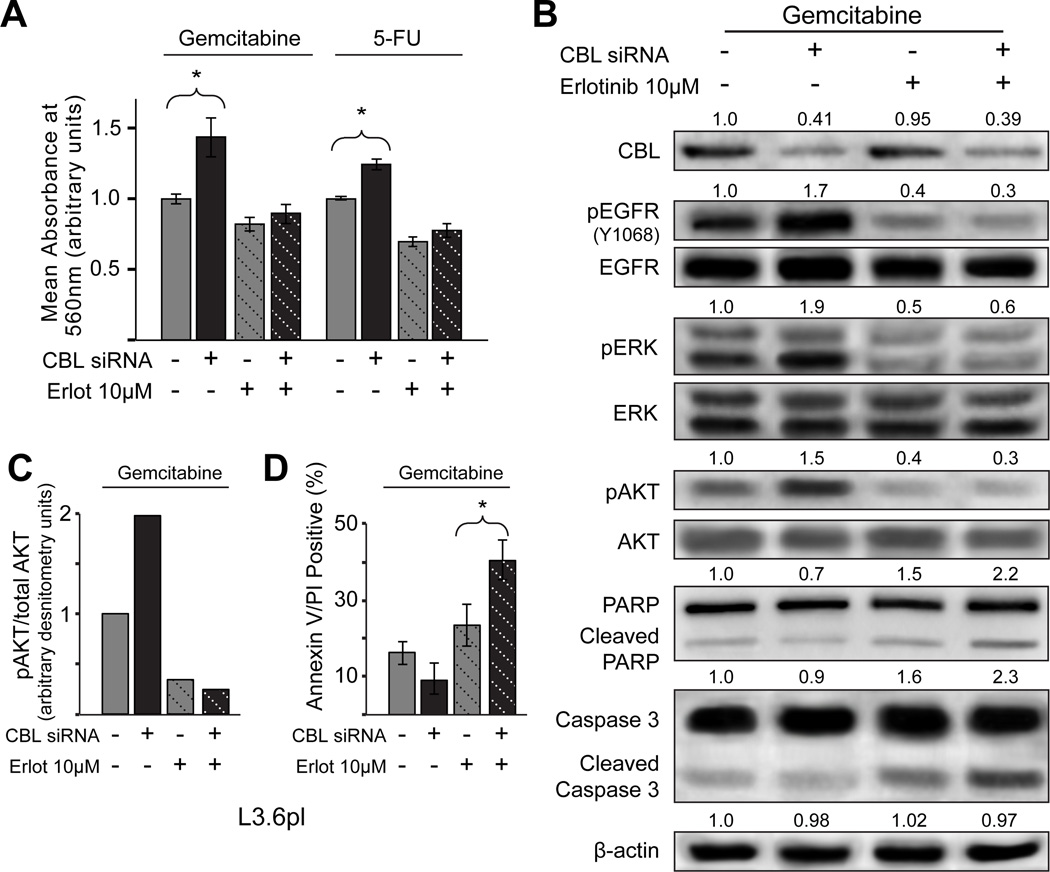
Erlotinib targets the CBL mediated chemoresistance mechanism
To further determine if EGFR autoactivation was the principal mechanism responsible for chemoresistance in PDACs with low CBL, EGFR activation was blocked using the EGFR inhibitor erlotinib in vitro. Fig. 3A reveals that the addition of erlotinib abrogates the viability advantage of L3.6plCBL-low cells as compared to L3.6pl cells in the presence of gemcitabine or 5-FU. Biochemically, erlotinib effectively inhibits EGFR autoactivation and the downstream mediators pAKT and pERK (Fig. 3B–C). Finally, erlotinib also increased gemcitabine-induced apoptosis in L3.6plCBL-low cells as evidenced by cleaved PARP and Caspase 3 immunoblots (Fig. 3B) and Annexin V/PI flow cytometry (Fig. 3D). These results suggest that erlotinib abrogates the chemoresistance afforded by low CBL, implicating EGFR as a key functional target of CBL in human PDAC.
Figure 3.
Erlotinib targets the CBL mediated chemoresistance mechanism. A, when erlotinib (Erlot), a targeted EGFR inhibitor, was added in combination with Gem or 5-FU to cells with CBL knockdown, cell viability was significantly reduced (column 2 vs. 4 and 6 vs. 8, P<0.05), and comparable to the isogenic parental cell line (column 3 vs. 4 and 7 vs. 8). Error bars ± SD. B, utilizing immunoblot to investigate the underlying signaling mechanisms involved we observed that erlotinib inhibition of pEGFR Y1068 decreases pAKT signaling (columns 2 vs. 4) and enhances apoptosis as evidenced by increased cleaved PARP and caspase 3. Relative densitometry units normalized to β-actin are reported above each band. C, densitometry analysis comparing the ratio of pAKT/total AKT again highlights the inhibition of pAKT signaling by erlotinib even with CBL knockdown. D, likewise, a greater percentage of apoptotic cells were observed by Annexin V/PI FACS when CBL knockdown cells were treated with dual therapy Gem + erlotinib. Error bars ± SD.
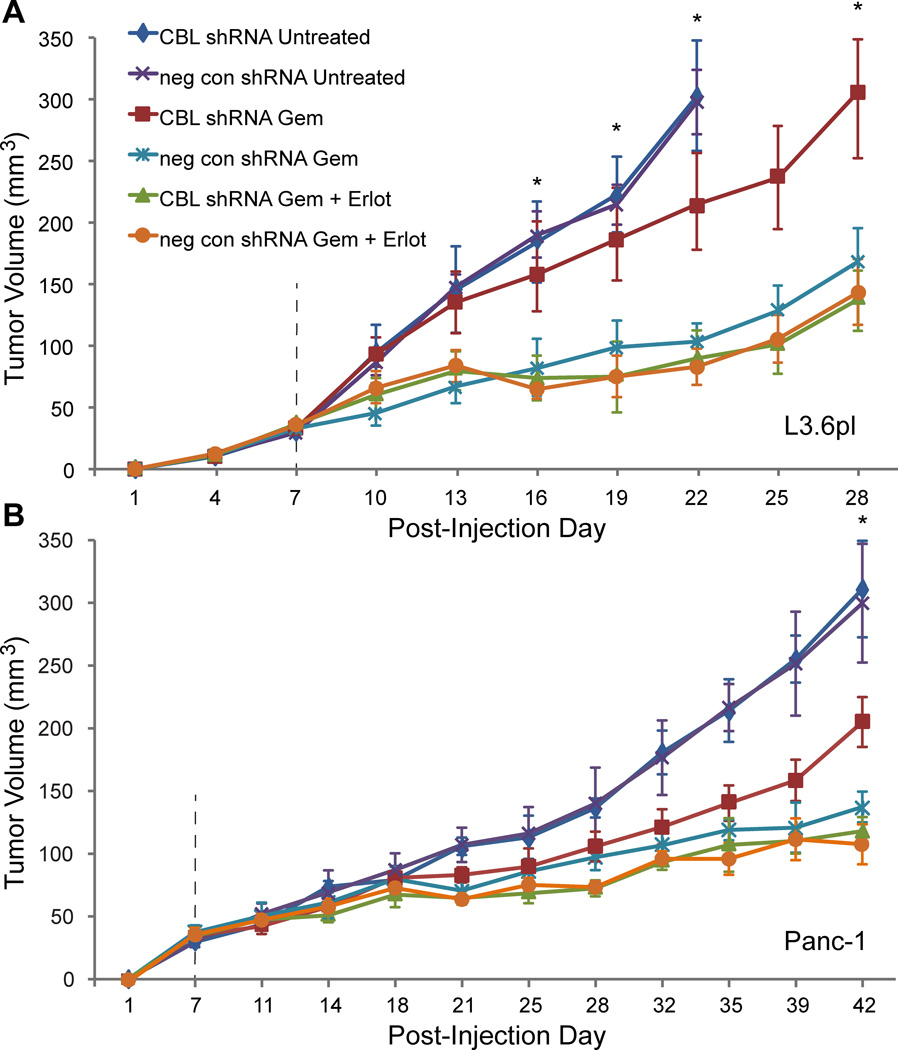
EGFR inhibition abrogates chemoresistance mediated by low CBL in vivo
Expanding on the cell culture results, we next tested the synergy between erlotinib and gemcitabine on growth of PDACCBL-low tumors in vivo. Lenti-viral mediated transduction of L3.6pl and Panc-1 PDAC cells with CBL shRNA yielded low CBL expression (Fig. S4). These cell lines treated with empty vector (PDACneg con shRNA) or CBL shRNA (PDACCBL shRNA) were subcutaneously implanted into the opposite flanks of NOD-scid-IL2Rγnull mice and left untreated, or treated with gemcitabine +/− erlotinib. As with the cell culture findings, CBL-low did not result in larger tumors than the negative controls in untreated mice (Fig. 4A–B). However, CBL-low did confer chemoresistance, as PDACCBL shRNA tumors were approximately twice the size as PDACneg con shRNA tumors in mice treated with gemcitabine. Strikingly, when CBL-low tumors were treated with combination erlotinib and gemcitabine, chemosensitivity to gemcitabine was completely restored as there was no significant difference between PDACCBL shRNA and PDACneg con shRNA tumor growth (Fig. 4A–B and Fig. S5). Analysis of tumor explants via western blots confirmed that low CBL expression was maintained (Fig. S4). In summary, the combined treatment of gemcitabine and erlotinib abrogates the chemoresistance in isogenic PDAC cell lines engineered with low CBL in vivo.
Figure 4.
EGFR inhibition abrogates chemoresistance mediated by low CBL in vivo. PDACCBL shRNA versus PDACneg con shRNA cells were injected into opposite flanks of 2 sets of 18 NOD-scid-IL2Rγnull mice. At 1 week, animals in each set were divided into 3 treatment groups: untreated (n=6), gemcitabine (Gem) (n=6), and Gem + erlotinib (Erlot) (n=6). Tumor volume (V) was then approximated by digital caliper measurement every 3–4 days utilizing the formula V = ½ × length × width × height. The untreated group had unabated growth and met critical endpoints for tumor size by post-injection day 22 in the, A, L3.6pl tumors and by day 42 in the, B, Panc1 tumors with no difference in growth rates for CBL shRNA versus neg con shRNA tumors. All tumors in Gem treated mice showed some responsiveness to treatment, but the CBL shRNA tumors grew to nearly double the size of the neg con shRNA tumors, meeting a critical endpoint for euthanasia by day 28 in the L3.6pl group, at which point this experiment was ended. The Panc-1 experiment was carried out an additional 2 weeks until the untreated group had reached this same endpoint. Animals treated with combination therapy Gem + Erlot had the lowest tumor growth of all groups, and strikingly for these groups, there was no difference in size between CBL shRNA and neg con shRNA tumors. *P<0.05 for CBL shRNA Gem versus other chemotherapy treated groups (ANOVA). Error bars ± SEM.
Discussion
The relative incidence of cancer-related deaths due to PDAC, as compared to other malignancies, is increasing in the US(27). This disturbing trend can be partly attributed to PDAC’s resistance to treatment and particularly aggressive tumor biology(3,4). To gain a better understanding of the rapid clinical progression, recent studies have focused not only on determining the genomic and transcriptomic makeup of this tumor but on identifying the specific changes that are associated with patient survival(5,11,28,29). Our group published a survival-based array analysis of human PDACs which identified and validated that low CBL was directly correlated with prognosis (lower CBL with shorter survival). However, the mechanism underlying CBL’s prognostic impact was not determined. Therefore, the purpose of the current study was to identify how low CBL may lead to shorter survival in PDAC patients.
Using primary human PDAC tissue samples and isogenic cell lines engineered with low CBL, in culture and in vivo, we found that in human PDAC, low CBL increased EGFR activation and yielded resistance to gemcitabine and 5-FU, the two most frequently used chemotherapies in PDAC. Low CBL induced chemoresistance is effectively abrogated by the EGFR inhibitor, erlotinib.
Tumor cell intrinsic autoactivation of EGFR after treatment with cytotoxic chemotherapy has been previously shown to occur in many solid malignancies. Recently, in the context of PDAC using transgenic mice, Miyabayashi et al.(16) determined that EGFR activation was due to gemcitabine-induced cellular stress which led to activation of MAPK, leading to release of EGFR ligands. These findings are further supported by previous work revealing that the ligand releasing enzyme, ADAM17/TACE, is expressed on the surface of 100% of human PDAC tissues and cell lines(30). Likewise, colon cancer cells treated with 5-FU in culture increase ADAM17 expression and release of EGFR-activating ligands: TGFα, amphiregulin, and heregulin(31). Interestingly, EGFR autoactivation may not be a generalized response to all cytotoxic chemotherapies, as only a subset of cancer cells treated with oxaliplatin developed EGFR activation(32). Our results reveal that EGFR is activated with both gemcitabine and 5-FU, the two most commonly used agents in PDAC.
CBL deletions and missense mutations are prevalent in myeloid malignancies(33). Functional studies of specific mutations reveal that CBL has both oncogenic and tumor suppressor functions. The E3 ligase domain regulates RTK internalization through ubiquitination; mutations in this region are generally oncogenic. In contrast, the ring finger domain stabilizes the FAK (RTK) – SRC – actin cytoskeleton promoting cellular motility and AKT signaling, therefore mutations in this region are generally suppressive(8). Furthermore, oncogenic mutations are homozygous in all cases of CML; loss of the wild-type suppressive allele is required for transformation in cell culture and in vivo(34).
The net consequence of specific CBL mutations or loss of CBL expression is less defined in solid malignancies than for CML. This uncertainty is further fueled by the paradoxical finding that CBL mutations outside of the E3 ligase region yielded increased cellular viability and motility in lung cancer cells in culture(35). However, the functional significance of interrupting CBL-dependent processes on solid tumor progression has been implicated, as the addition of a proteasome inhibitor decreased gemcitabine-induced cell death in head and neck cancer cells(36). In the context of PDAC development, E3 ligases, of which CBL is a member, contain oncogenic mutations in a small subset of pancreatic cystic neoplasms(37). Our previous results identified that low CBL expression is common in human PDAC, as 60% of human tumors had low levels on qRT-PCR(11). Taken together, our previous study(11) and current results reveal that low CBL is oncogenic in PDAC. We found that patients with lower CBL expression have shorter overall survival. Loss of the RTK down-regulating E3 ligase function had greater functional significance than loss of adaptor function, as chemoresistance afforded by low CBL was abrogated by EGFR inhibition with erlotinib.
The mechanism underlying low CBL expression in PDAC has also not been identified. Our previous study(11) revealed that CBL mRNA expression was both decreased in 60% of patients and prognostically significant. Genomic deletions of the CBL locus were not present in the in silico arrayed tumors. Previous PDAC sequencing studies have not identified frequent CBL mutations(38). Taken together, these findings implicate an epigenetic or post-transcriptional process as one potential mechanism regulating CBL expression. microRNA 125a-3p was suggested (but not validated) in our previous study, as it was (i) predicted by sequence to bind CBL, (ii) anti-correlated with CBL expression across tumors, and (iii) prognostically significant. However, CBL protein expression was not examined in these 67 human PDACs, leaving open the possibility that regulation may also be occurring on the protein-level.
Importantly, our current study revealed an inverse correlation with CBL expression and EGFR activation (pEGFR) in human PDACs. There are numerous proteins that have been implicated in PDAC tumorigenesis and previously shown to modulate CBL expression: SRC(11), c-MET(25), and PTEN(39). SRC activation, which was also a prognostically significant finding in our previous study, induces CBL destruction and enables EGFR to evade destruction(40). c-MET activation can also yield low CBL(41). PTEN loss destabilizes CBL-EGFR-ubiquitin ligase complexes, yielding increased EGFR activity in cancer cell lines(42). Taken together, these candidate molecular and protein-level mechanisms of low CBL, previously implicated in PDAC tumorigenesis, further suggest that CBL may be an important node involved in PDAC progression. Future studies should be designed to identify the mechanisms involved in loss of CBL function in PDAC.
While oncologists have become skeptical about the efficacy of EGFR inhibition in PDAC, we believe that it may still be an effective treatment approach. A close analysis of the phase III trial identifies that while the overall improvement in survival was minimal for all patients (6.37 vs. 5.95 months, P=0.03), the subgroup who developed grade 2 skin rashes (compared to those who did not develop skin changes) derived amongst the best clinical benefit of all combination treatments that have been reported for this disease (median survival 10.5 vs. 5.3 months, P<0.001)(6,43,44). These observations shed hope on the efficacy of this drug for PDAC.
Our findings that erlotinib abrogates chemotherapy resistance associated with low CBL suggest that CBL may be a sensitive pretreatment negative predictive biomarker for erlotinib response. However, the specificity of CBL for erlotinib response may be limited by the diverse CBL target proteins which include other TKs and non-TK targets(8). This concern is further fueled by our phospho-TK array findings which identified enhanced activation of proteins in CBL-low cells in addition to EGFR that have been previously implicated In PDAC progression. Future studies are needed to identify (i) if erlotinib resistant PDACs with low CBL exist and, if so (ii) therapies that abrogate their chemoresistance. Regardless, our findings are the first to suggest that disruptions of the post-TK endosomal/lysosomal destruction process may play a role in the chemoresistance associated with this devastating disease.
Conclusions
In conclusion, our findings reveal that low CBL increases chemotherapy stress-induced EGFR activation in human PDAC, resulting in treatment resistance. Dual treatment with gemcitabine or 5-FU and erlotinib abrogated the chemoresistance of isogenic PDAC cells engineered with low CBL in cell culture and in vivo. These results are clinically significant for PDAC, as they (i) identify the importance of the lysosomal destruction process as a mechanism for EGFR dysregulation and (ii) begin to define low CBL expression as a predictive biomarker of response to the previously FDA approved EGFR inhibitor, erlotinib.
Supplementary Material
Statement of Translational Relevance.
Molecular targeted therapies given alone or in combination with cytotoxic therapy have had limited success in prolonging survival for patients with pancreatic cancer. It is still thought that there are discrete subgroups of patients who may clearly benefit, but without an understanding of the underlying chemoresistance mechanisms, their prospective identification has been hampered. In our previous integrative molecular array analysis, we found that downregulation of the ubiquitin ligase CBL occurred in the majority (60%) of tumors and correlated with decreased patient survival. In this study we identify that this may be due to low CBL enhancing chemoresistance through EGFR activation. In a preclinical in vivo PDAC model this chemoresistance is overcome by EGFR inhibition. These results suggest that patients with lower CBL expression may benefit from an EGFR inhibitor as a part of multi-drug therapy.
Acknowledgments
The authors thank Dr. Sanjeet Patel, UCLA, for HEK293T cells and lentiviral plasmids, and Dr. William Isacoff, UCLA, for providing erlotinib and 5-fluorouracil.
Grant Support
This study was supported by the California Institute of Regenerative Medicine TG2-01169 (B.E. Kadera, T.R. Donahue), Gerald S. Levey Surgical Research Award (B.E. Kadera), Concern Foundation for Cancer Research (T.R. Donahue), Hirshberg Foundation for Cancer Research (T.R. Donahue), CURE: Pilot and Feasibility Study NIH/NIDDKP30 DK41301 (T.R. Donahue), STOP Cancer Foundation (T.R. Donahue), Association for Academic Surgery (T.R. Donahue). NIH 1R01CA187678-01
Footnotes
Conflicts of Interest: None
Author’s Contributions
Conception and design: BEK, TRD
Development of methodology: BEK, TRD, PAT, DWD
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): BEK, NW, LL, AHN, PAT
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): BEK, LL, PAT
Writing, review, and/or revision of the manuscript: BEK, TRD, DWD, PAT, NW, LL
Study supervision: TRD
References
- 1.Howlander N, Noone A-M, Krapcho M, Newman N, Aminou R, Waldron W, et al. SEER Cancer Statistics Review. 2012. May, pp. 1–27. [Google Scholar]
- 2.Martisian LM, Aizenberg R, Rosenzweig A. The Alarming Rise of Pancreatic Cancer Deaths in the United States: Why We Need to Stem the Tide Today. Pancreatic Cancer Action Network [Internet] 2012:1–12. Available from: http://www.pancan.org/section_research/reports/pdf/incidence_report_2012.pdf.
- 3.Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biankin AV, Waddell N, Kassahn KS, Gingras M-C, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nature Medicine. Nature Publishing Group. 2011:1–5. doi: 10.1038/nm.2344. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MJ, Goldstein D, Hamm J, Figer A, Hecht JR, Gallinger S, et al. Erlotinib Plus Gemcitabine Compared With Gemcitabine Alone in Patients With Advanced Pancreatic Cancer: A Phase III Trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 7.Philip PA, Mooney M, Jaffe D, Eckhardt G, Moore M, Meropol N, et al. Consensus report of the national cancer institute clinical trials planning meeting on pancreas cancer treatment. J Clin Oncol. 2009;33:5660–5669. doi: 10.1200/JCO.2009.21.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt MHH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 9.Sanjay A, Horne WC, Baron R. The Cbl family: ubiquitin ligases regulating signaling by tyrosine kinases. Sci STKE. 2001;2001 doi: 10.1126/stke.2001.110.pe40. pe40. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths EK, Sanchez O, Mill P, Krawczyk C, Hojilla CV, Rubin E, et al. Cbl-3-deficient mice exhibit normal epithelial development. Molecular and Cellular Biology. 2003;23:7708–7718. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donahue TR, Tran LM, Hill R, Li Y, Kovochich A, Calvopina JHH, et al. Integrative survival-based molecular profiling of human pancreatic cancer. Clin Cancer Res. 2012;18:1352–1363. doi: 10.1158/1078-0432.CCR-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gullick WJ, Downward J, Parker PJ, Whittle N, Kris R, Schlessinger J, et al. The Structure and Function of the Epidermal Growth Factor Receptor Studied by Using Antisynthetic Peptide Antibodies. Proc R Soc Lond B. The Royal Society. 1985;226:127–134. doi: 10.1098/rspb.1985.0087. [DOI] [PubMed] [Google Scholar]
- 13.Ghaneh P, Kawesha A, Evans JD, Neoptolemos JP. Molecular prognostic markers in pancreatic cancer. J Hep Bil Pancr Surg. Springer-Verlag. 2002;9:1–11. doi: 10.1007/s005340200000. [DOI] [PubMed] [Google Scholar]
- 14.Ardito CM, Grüner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, et al. EGF Receptor Is Required for KRAS-Induced Pancreatic Tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamanaka Y, Friess H, Kobrin M, Büchler MW, Beger H, Korc M. Coexpression of epidermal growth factor receptor and ligands in human pancreatic cancer is associated with enhanced tumor aggressiveness. Anticancer research. 1993;13:565. [PubMed] [Google Scholar]
- 16.Miyabayashi K, Ijichi H, Mohri D, Tada M, Yamamoto K, Asaoka Y, et al. Erlotinib prolongs survival in pancreatic cancer by blocking gemcitabine-induced MAPK signals. Cancer Research. 2013;73:2221–2234. doi: 10.1158/0008-5472.CAN-12-1453. [DOI] [PubMed] [Google Scholar]
- 17.Hong SP, Wen J, Bang S, Park S, Song SY. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int J Cancer. 2009;125:2323–2331. doi: 10.1002/ijc.24573. [DOI] [PubMed] [Google Scholar]
- 18.Piacentini P, Donadelli M, Costanzo C, Moore PS, Palmieri M, Scarpa A. Trichostatin A enhances the response of chemotherapeutic agents in inhibiting pancreatic cancer cell proliferation. Virchows Arch. 2006;448:797–804. doi: 10.1007/s00428-006-0173-x. [DOI] [PubMed] [Google Scholar]
- 19.Bhutia YD, Hung SW, Patel B, Lovin D, Govindarajan R. CNT1 expression influences proliferation and chemosensitivity in drug-resistant pancreatic cancer cells. Cancer Research. 2011;71:1825–1835. doi: 10.1158/0008-5472.CAN-10-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyu M-A, Kurzrock R, Rosenblum MG. The immunocytokine scFv23/TNF targeting HER-2/neu induces synergistic cytotoxic effects with 5-fluorouracil in TNF-resistant pancreatic cancer cell lines. Biochem Pharmacol. 2008;75:836–846. doi: 10.1016/j.bcp.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Lefever S, Vandesompele J, Speleman F, Pattyn F. RTPrimerDB: the portal for real-time PCR primers and probes. Nucleic Acids Res. 2009;37:D942–D945. doi: 10.1093/nar/gkn777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomayko MM, Reynolds CP. Determination of Subcutaneous Tumor Size in Athymic (Nude) Mice. Cancer Chemother Pharmacol. 1989;24:148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 23.Jaquish DV, Yu PT, Shields DJ, French RP, Maruyama KP, Niessen S, et al. IGF1-R signals through the RON receptor to mediate pancreatic cancer cell migration. Carcinogenesis. 2011;32:1151–1156. doi: 10.1093/carcin/bgr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang RF, Yokoi K, Bucana CD, Tsan R, Killion JJ, Evans DB, et al. Inhibition of platelet-derived growth factor receptor phosphorylation by STI571 (Gleevec) reduces growth and metastasis of human pancreatic carcinoma in an orthotopic nude mouse model. Clin Cancer Res. 2003;9:6534–6544. [PubMed] [Google Scholar]
- 25.Li C, Wu J-J, Hynes M, Dosch J, Sarkar B, Welling TH, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218.e5–2227.e5. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 26.Logan-Collins J, Thomas RM, Yu P, Jaquish D, Mose E, French R, et al. Silencing of RON receptor signaling promotes apoptosis and gemcitabine sensitivity in pancreatic cancers. Cancer Research. 2010;70:1130–1140. doi: 10.1158/0008-5472.CAN-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.American Cancer Society. Cancer facts & figures 2013. Atlanta, GA: 2013. http://www.cancer.org/research/cancerfactsfigures/cancerfactsfigures/cancer-facts-figures-2013. [Google Scholar]
- 28.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackford A, Serrano OK, Wolfgang CL, Parmigiani G, Jones S, Zhang X, et al. SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res. 2009;15:4674–4679. doi: 10.1158/1078-0432.CCR-09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ringel J, Jesnowski R, Moniaux N, Lüttges J, Ringel J, Choudhury A, et al. Aberrant expression of a disintegrin and metalloproteinase 17/tumor necrosis factor-alpha converting enzyme increases the malignant potential in human pancreatic ductal adenocarcinoma. Cancer Research. 2006;66:9045–9053. doi: 10.1158/0008-5472.CAN-05-3287. [DOI] [PubMed] [Google Scholar]
- 31.Kyula JN, Van Schaeybroeck S, Doherty J, Fenning CS, Longley DB, Johnston PG. Chemotherapy-induced activation of ADAM-17: a novel mechanism of drug resistance in colorectal cancer. Clin Cancer Res. 2010;16:3378–3389. doi: 10.1158/1078-0432.CCR-10-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Schaeybroeck S, Karaiskou-McCaul A, Kelly D, Longley D, Galligan L, Van Cutsem E, et al. Epidermal growth factor receptor activity determines response of colorectal cancer cells to gefitinib alone and in combination with chemotherapy. Clin Cancer Res. 2005;11:7480–7489. doi: 10.1158/1078-0432.CCR-05-0328. [DOI] [PubMed] [Google Scholar]
- 33.Kales SC, Ryan PE, Nau MM, Lipkowitz S. Cbl and human myeloid neoplasms: the Cbl oncogene comes of age. Cancer Research. 2010;70:4789–4794. doi: 10.1158/0008-5472.CAN-10-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanada M, Suzuki T, Shih LY, Otsu M, Kato M. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009 doi: 10.1038/nature08240. [DOI] [PubMed] [Google Scholar]
- 35.Tan Y-HC, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K, et al. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS ONE. 2010;5:e8972. doi: 10.1371/journal.pone.0008972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng FY, Varambally S, Tomlins SA, Chun PY, Lopez CA, Li X, et al. Role of epidermal growth factor receptor degradation in gemcitabine-mediated cytotoxicity. Oncogene. 2007;26:3431–3439. doi: 10.1038/sj.onc.1210129. [DOI] [PubMed] [Google Scholar]
- 37.Wu J, Jiao Y, Dal Molin M, Maitra A, de Wilde RF, Wood LD, et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proceedings of the National Academy of Sciences. 2011;108:21188–21193. doi: 10.1073/pnas.1118046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottenhof NA, de Wilde RF, Maitra A, Hruban RH, Offerhaus GJA. Molecular characteristics of pancreatic ductal adenocarcinoma. Patholog Res Int. 2011;2011:620601. doi: 10.4061/2011/620601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill R, Calvopina JHH, Kim C, Wang Y, Dawson DW, Donahue TR, et al. PTEN loss accelerates KrasG12D-induced pancreatic cancer development. Cancer Research. 2010;70:7114–7124. doi: 10.1158/0008-5472.CAN-10-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bao J, Gur G, Yarden Y. Src promotes destruction of c-Cbl: Implications for oncogenic synergy between Src and growth factor receptors. P Natl Acad Sci Usa. 2003;100:2438–2443. doi: 10.1073/pnas.0437945100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai AZ, Durrant M, Zuo D, Ratcliffe CDH, Park M. Met kinase-dependent loss of the E3 ligase Cbl in gastric cancer. J Biol Chem. 2012;287:8048–8059. doi: 10.1074/jbc.M112.339820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vivanco I, Rohle D, Versele M, Iwanami A, Kuga D, Oldrini B, et al. The phosphatase and tensin homolog regulates epidermal growth factor receptor (EGFR) inhibitor response by targeting EGFR for degradation. Proceedings of the National Academy of Sciences. 2010;107:6459–6464. doi: 10.1073/pnas.0911188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoff Von DD, Ramanathan RK, Borad MJ, Laheru DA, Smith LS, Wood TE, et al. Gemcitabine plus nab-paclitaxel is an active regimen in patients with advanced pancreatic cancer: a phase I/II trial. J Clin Oncol. 2011;29:4548–4554. doi: 10.1200/JCO.2011.36.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. New Engl J Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.