Abstract
Although mutations in cardiac myosin binding protein-C (cMyBP-C) cause heart disease, its role in muscle contraction is not well understood. A mechanism remains elusive partly because the protein can have multiple effects, such as dual biphasic activation and inhibition observed in actin motility assays. Here we develop a mathematical model for the interaction of cMyBP-C with the contractile proteins actin and myosin and the regulatory protein tropomyosin. We use this model to show that a drag-activation-competition mechanism accurately describes actin motility measurements, while models lacking either drag or competition do not. These results suggest that complex effects can arise simply from cMyBP-C binding to actin.
Main Text
Myosin binding protein-C (MyBP-C) is a regulatory protein in striated muscle (1). Its function is not well understood, but in cardiac muscle it modulates contraction and relaxation rates and contributes to contractile reserve in response to inotropic stimuli (2). Mutations in MYBPC3, the gene encoding the cardiac isoform, cMyBP-C, cause cardiomyopathies; altered function of cMyBP-C due to posttranslational modification is prevalent in heart failure (3). Understanding cMyBP-C’s role in muscle contraction is therefore important in both health and disease. Here, by developing a mathematical model and fitting published in vitro measurements that serve as a simplified model of the more complex in vivo system, we provide evidence cMyBP-C acts via a drag-activation-competition mechanism.
cMyBP-C interacts with proteins that generate (4,5) and regulate (6,7) muscle force. Muscle contraction is powered by cyclic interactions of myosin with actin, proteins that occur, respectively, in thick and thin filaments. Myosin motors turn chemical energy in ATP into mechanical work by binding to actin and sliding the thin filament relative to the thick filament. In vertebrate striated muscle, this process is regulated by thin-filament associated proteins, troponin and tropomyosin. Tropomyosin filaments wrap around actin, sterically blocking myosin from binding to actin. The block is partly removed when calcium binds to troponin, which changes conformation and moves tropomyosin from the blocked toward the open position (Fig. 1 A).
Figure 1.
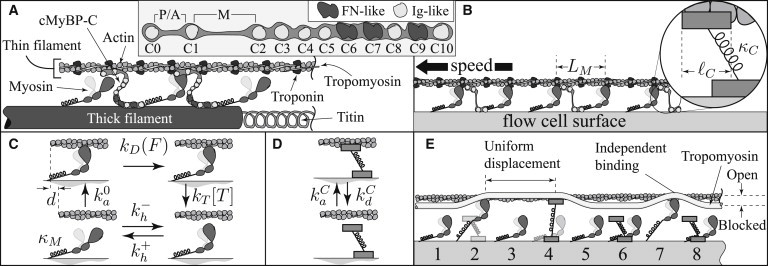
Diagrams of cMyBP-C in sarcomeres, motility assays, and in silico. (A) Cartoon of the proteins in muscle contraction, including cMyBP-C shown extending from the thick filament (myosin) to interact with actin in the thin filament. (Inset) Modular domain structure of cMyBP-C. (B) Cartoon of a motility assay with a regulated thin filament (actin with troponin and tropomyosin) in the presence of an N-terminal fragment of cMyBP-C (C0C3). The average spacing of myosin molecules is LM. (Inset) Model assumptions that cMyBP-C occludes a region of length ℓC on actin and is anchored to the flow cell surface via a linear spring of stiffness κC. (C and D) Myosin’s and cMyBP-C’s interactions with actin, respectively. (E) The complete model, including actin, myosin, tropomyosin, and cMyBP-C N-terminal fragments. Myosin competes with cMyBP-C to bind to actin. Once bound, either cMyBP-C or myosin displaces tropomyosin toward the open position. If nearby molecules bind, they activate intervening molecules (i.e., molecules 2 and 4 activate molecule 3); if distant molecules bind, they do so independently (i.e., molecules 4 and 7 are independent).
Parts of cMyBP-C interact specifically with myosin, actin, and tropomyosin (4,5,7–10). cMyBP-C includes a series of immunoglobulin-like domains and fibronectin type-3-like domains numbered C0–C10 starting from the N-terminus (Fig. 1 A) (11), along with a Pro-Ala-rich sequence and a MyBP-C specific motif (M), which are likely to be at least partially disordered (12). The C-terminal domain of cMyBP-C associates with thick filaments (4,8), while the N-terminal domain interacts with the myosin motor (9), actin (5,6,10,13,14), and tropomyosin (6,7). N-terminal domains, including the M-motif, an ∼100 amino-acid linker sequence between domains C1 and C2, have attracted particular attention because the M-motif contains regulatory sites (11,14) and N-terminal fragments reproduce the action of whole cMyBP-C in many assays.
One such assay is actin motility, where fluorescently-labeled actin filaments, with or without troponin and tropomyosin, are observed moving over a myosin-coated surface. Actin is propelled by the ATP-dependent force generation of the myosin heads (Fig. 1 B). Either in the absence of regulatory proteins or in their presence at high calcium, actin moves smoothly. Increasing amounts of exogenous cMyBP-C uniformly decrease actin speed (6,14). Conversely, in the presence of regulatory proteins at low calcium, when actin is normally stationary because tropomyosin blocks myosin binding, increasing cMyBP-C has a biphasic effect, activating actin motility at low concentrations but decreasing actin speed at higher concentrations (6).
The complex activating and inhibitory effects of cMyBP-C are challenging to explain with a single mechanism. E.g., while activation might be explained by the ability of cMyBP-C N-terminal domains to bind to actin and shift tropomyosin toward the open position (7), inhibition could result either from competition of cMyBP-C with myosin heads for binding to actin (5) or by transient links between cMyBP-C and the flow cell surface that create a viscous drag slowing motility (14). Previously we developed mathematical models for actin motility without regulatory proteins (15) or with them at high and low calcium (16). Here we adapt these models to help distinguish the mechanism(s) by which cMyBP-C affects actin motility (see the Supporting Material).
This model describes three molecular interactions. The first, myosin’s ATP-dependent interaction with actin, is modeled with a four-state scheme (Fig. 1 C). Myosin with ADP and phosphate (Pi) in its active site binds to actin, releases Pi and changes conformation, sliding actin forward a distance d. These steps, not necessarily in that order, result in a transition from an un-/weakly-bound state to a strongly bound state with rate constant ka0. Once strongly bound, myosin releases ADP with force-dependent rate constant , where kBT is the Boltzmann’s constant times temperature, and λ has the units of distance. Myosin then detaches from actin with rate constant kT[T], where [T] is the concentration of ATP. Finally, myosin hydrolyzes ATP and reverses its conformational change in a reversible process with forward rate k+h and reverse rate k−h. This model fits many in vitro measurements, including motility of actin filaments without regulatory proteins, with parameters, k0a = 40 s−1, k0D = 350 s−1, kT = 2 μM−1 s−1, k+h = 100 s−1, k−h = 10 s−1, d = 10 nm, λ = 1.86 nm, and myosin stiffness κM = 0.3 pN/nm (15).
The second molecular interaction in the model is cMyBP-C’s with actin. We assume that cMyBP-C is anchored to the surface of the flow cell, binds specifically to actin, and when bound, prohibits myosin binding over a region of length ℓC. Conversely, if a myosin molecule is bound in that region, cMyBP-C cannot bind to actin. Once bound, cMyBP-C acts as a linear spring of stiffness, κC. Force-independent attachment and detachment occur at rates kaC and kdC, respectively (Fig. 1 D). Although these assumptions seem to presuppose that myosin and cMyBP-C compete for actin binding sites and that cMyBP-C is attached to the surface, competition is abolished if ℓC = 0 and attachment is abolished if κC = 0.
The third molecular interaction in the model is cMyBP-C and myosin with regulatory proteins (Fig. 1 E). Assumed negligible at high calcium (pCa 5) (16), these interactions are important at low calcium (pCa 9) where tropomyosin sterically inhibits the binding of both myosin and cMyBP-C to actin (7,17). At low calcium, if either myosin or cMyBP-C binds to actin, tropomyosin is locally displaced (18), favoring the binding of nearby molecules (7,17). Assuming that 1) two nearby bound molecules uniformly displace tropomyosin; 2) two distant bound molecules are independent; and 3) the transition between these regimes is abrupt, it follows that two parameters define this local coupling: , the maximum number of molecules activated by a pair of bound molecules; and ε, the actin binding rate at low calcium relative to the rate at high calcium. This model describes the motility of regulated thin filaments (actin with troponin and tropomyosin) at low calcium with = 11 and ε = 0.003 (16).
We used this model to fit measurements of actin motility with skeletal muscle myosin from two groups of researchers, one group using regulated thin filaments and variable concentrations of the N-terminal cMyBP-C fragments C0C2 and C1C2 (6), and the other using unregulated actin filaments and variable concentrations of C1C2, C0C2, C0C3, and C0C1f (the latter includes part of the M-motif) (14). The two data sets are nearly identical after rescaling for differences in cMyBP-C concentration (Fig. 2). Rescaling consisted of dividing concentrations of C0C2 and C1C2 of Razumova et al. (6) by 10 and 14, respectively. Our qualitative results are independent of rescaling (see the Supporting Material).
Figure 2.
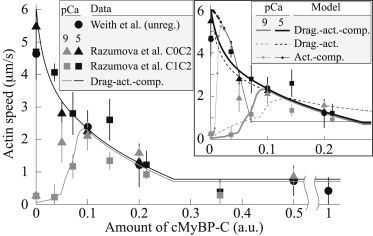
A drag-activation-competition model for MyBP-C is consistent with measurements of actin motility in the absence of regulatory proteins (14) or in their presence at high (pCa 5) and low (pCa 9) calcium (6). (Inset) Models lacking either drag or activation do not fit the data.
Data were fit by the drag-activation-competition model (Fig. 2; theory and data not significantly different, p > 0.05, χ2 test). We varied the model’s three unknown parameters to optimize the fit, PC = kCa/(kCa + kCd), which is cMyBP-C’s affinity for actin, b = κCk0D/κMkCd, which determines the importance of drag, and a = ℓC/LM, which determines the importance of competition. Best-fit parameters are PC = 0.83 ± 0.09, b = 0.13 ± 0.05, and a = 0.9 ± 0.2.
The model requires both competition and drag to fit the data. Fits without competition (a = 0) or without drag (b = 0) are significantly different from the data (p < 0.005, p < 0.001, respectively, χ2 test); fits with competition and drag are significantly better than fits without competition or without drag (p < 0.005, p < 0.001, respectively, F-test). Fitting measurements of competition between myosin and MyBP-C in solution (5) provides independent estimates of PC = 0.9 ± 0.1 and a = 0.8 ± 0.1, consistent with the motility fits, further supporting the view that competition is important in motility (see details in the Supporting Material).
Optimizing the fit of a mathematical model to data allows rigorous testing of putative mechanisms. Because it interacts both with contractile and regulatory proteins, developing a model for cMyBP-C is challenging, inasmuch as few models for regulation have the computational efficiency necessary for optimization. Here, we developed a model and showed that a drag-activation-competition mechanism for cMyBP-C is consistent with actin motility data, whereas models lacking drag or competition are not (Fig. 2, inset). These complex effects can arise simply by cMyBP-C binding to the actin filament: cMyBP-C binding to actin displaces tropomyosin at low calcium, leading to filament activation, whereas increasing cMyBP-C concentration renders binding sites unavailable to myosin, leading to competition. Transient links formed by cMyBP-C binding to actin filaments and to the flow cell surface create a viscous drag that further slows motility. Future use of this model may help distinguish mechanistic effects of cMyBP-C mutations that affect actin binding.
Acknowledgments
This work was supported in part by National Science Foundation grant No. DMS-1413185 and a Hellman Fellowship to S.W.
Supporting Material
Supporting Citations
References (19–21) appear in the Supporting Material.
References
- 1.Offer G., Moos C., Starr R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J. Mol. Biol. 1973;74:653–676. doi: 10.1016/0022-2836(73)90055-7. [DOI] [PubMed] [Google Scholar]
- 2.Winegrad S. Cardiac myosin binding protein C. Circ. Res. 1999;84:1117–1126. doi: 10.1161/01.res.84.10.1117. [DOI] [PubMed] [Google Scholar]
- 3.Xu Q., Dewey S., Gomes A.V. Malignant and benign mutations in familial cardiomyopathies: insights into mutations linked to complex cardiovascular phenotypes. J. Mol. Cell. Cardiol. 2010;48:899–909. doi: 10.1016/j.yjmcc.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 4.Moos C., Offer G., Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J. Mol. Biol. 1975;97:1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- 5.Moos C., Mason C.M., Dubin J.H. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J. Mol. Biol. 1978;124:571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 6.Razumova M.V., Shaffer J.F., Harris S.P. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: evidence for long-lived cross-bridges. J. Biol. Chem. 2006;281:35846–35854. doi: 10.1074/jbc.M606949200. [DOI] [PubMed] [Google Scholar]
- 7.Mun J.Y., Previs M.J., Craig R. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. USA. 2014;111:2170–2175. doi: 10.1073/pnas.1316001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okagaki T., Weber F.E., Reinach F.C. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruen M., Gautel M. Mutations in β-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 10.Shaffer J.F., Kensler R.W., Harris S.P. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J. Biol. Chem. 2009;284:12318–12327. doi: 10.1074/jbc.M808850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barefield D., Sadayappan S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J. Mol. Cell. Cardiol. 2010;48:866–875. doi: 10.1016/j.yjmcc.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karsai A., Kellermayer M.S.Z., Harris S.P. Mechanical unfolding of cardiac myosin binding protein-C by atomic force microscopy. Biophys. J. 2011;101:1968–1977. doi: 10.1016/j.bpj.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Dijk S.J., Bezold K.L., Harris S.P. Earning stripes: myosin binding protein-C interactions with actin. Pflugers Arch. 2014;466:445–450. doi: 10.1007/s00424-013-1432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weith A., Sadayappan S., Warshaw D.M. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J. Mol. Cell. Cardiol. 2012;52:219–227. doi: 10.1016/j.yjmcc.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walcott S., Warshaw D.M., Debold E.P. Mechanical coupling between myosin molecules causes differences between ensemble and single-molecule measurements. Biophys. J. 2012;103:501–510. doi: 10.1016/j.bpj.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walcott S. A differential equation model for tropomyosin-induced myosin cooperativity describes myosin-myosin interactions at low calcium. Cell. Mol. Bioeng. 2013;6:13–25. [Google Scholar]
- 17.Craig R., Lehman W. Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J. Mol. Biol. 2001;311:1027–1036. doi: 10.1006/jmbi.2001.4897. [DOI] [PubMed] [Google Scholar]
- 18.Kad N.M., Kim S., Baker J.E. Single-myosin crossbridge interactions with actin filaments regulated by troponin-tropomyosin. Proc. Natl. Acad. Sci. USA. 2005;102:16990–16995. doi: 10.1073/pnas.0506326102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith D.A., Maytum R., Geeves M.A. Cooperative regulation of myosin-actin interactions by a continuous flexible chain I: actin-tropomyosin systems. Biophys. J. 2003;84:3155–3167. doi: 10.1016/S0006-3495(03)70040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uyeda T.Q.P., Kron S.J., Spudich J.A. Myosin step size. Estimation from slow sliding movement of actin over low densities of heavy meromyosin. J. Mol. Biol. 1990;214:699–710. doi: 10.1016/0022-2836(90)90287-V. [DOI] [PubMed] [Google Scholar]
- 21.Harris D.E., Warshaw D.M. Smooth and skeletal muscle myosin both exhibit low duty cycles at zero load in vitro. J. Biol. Chem. 1993;268:14764–14768. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


