Abstract
CRISPR/Cas9 system of RNA-guided genome editing is revolutionizing genetics research in a wide spectrum of organisms. Even for the laboratory mouse, a model that has thrived under the benefits of embryonic stem (ES) cell knockout capabilities for nearly three decades, CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 technology enables one to manipulate the genome with unprecedented simplicity and speed. It allows generation of null, conditional, precisely mutated, reporter, or tagged alleles in mice. Moreover, it holds promise for other applications beyond genome editing. The crux of this system is the efficient and targeted introduction of DNA breaks that are repaired by any of several pathways in a predictable but not entirely controllable manner. Thus, further optimizations and improvements are being developed. Here, we summarize current applications and provide a practical guide to use the CRISPR/Cas9 system for mouse mutagenesis, based on published reports and our own experiences. We discuss critical points and suggest technical improvements to increase efficiency of RNA-guided genome editing in mouse embryos and address practical problems such as mosaicism in founders, which complicates genotyping and phenotyping. We describe a next-generation sequencing strategy for simultaneous characterization of on- and off-target editing in mice derived from multiple CRISPR experiments. Additionally, we report evidence that elevated frequency of precise, homology-directed editing can be achieved by transient inhibition of the Ligase IV-dependent nonhomologous end-joining pathway in one-celled mouse embryos.
Keywords: CRISPR, genome editing, nonhomologous end joining, mouse knockouts
PHENOTYPIC characterization of mutations is the most accurate and widely used method for elucidating in vivo gene functions and the genetics of diseases. Generation of human disease models is constrained by available genetic tools for a given model system. The laboratory mouse is the most widely used mammalian model due to its powerful genetics, embryonic stem (ES) cell technology, and routine transgenesis and mutagenesis. Traditional gene knockouts produced by gene targeting in ES cells usually produce null mutations; strategies to generate more subtle changes to proteins involve multiple rounds of manipulation in ES cells or forward genetic approaches such as ENU mutagenesis. The discoveries of sequence-specific nucleases have allowed researchers to precisely manipulate embryonic genomes in a wide range of experimental models (including mouse, rat, pig, fish, rabbit, fruit fly, frog, rhesus monkey, etc.), obviating the need for ES cells as an essential intermediate. This new genre of genome editing technologies involves generation of DNA double strand breaks (DSBs) in precise genomic locations by targetable nucleases and exploiting cellular repair machinery to produce mutations. The recently developed clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system is revolutionizing genetics not only in organisms in which gene targeting was not previously possible, but also in the laboratory mouse, where ES cell technology has enabled gene targeting and genome manipulation for nearly three decades.
The “CRISPR” system is a versatile prokaryotic antiviral defense mechanism providing adaptive immunity for a host bacterium against extrachromosomal genetic material (Horvath and Barrangou 2010). This RNA-guided bacterial innate immune system essentially involves three distinct steps: (1) acquisition of foreign DNA, (2) synthesis and maturation of CRISPR RNA (crRNA) followed by formation of RNA-Cas nuclease protein complexes, and (3) target recognition by crRNA and destruction of foreign DNA by Cas nuclease cleavage (Aida et al. 2014; Mashimo 2014; Sander and Joung 2014). Three different types of CRISPR-Cas systems have been described (Makarova et al. 2011). However, due to the simplicity, high efficiency, and multiplexing capability of the type II CRISPR/Cas system, it has been adopted as the genome editing technology of choice. The type II system utilizes a single Cas9 nuclease sufficient to cleave the target DNA specified by crRNA. The ability of targeting any genomic location opened new genome manipulation possibilities. In addition to genome editing, the system was quickly developed as a tool to regulate gene expression. Here we provide an overview of current advancements in this rapidly evolving technique to manipulate the mouse genome.
Mutagenic Capabilities of the CRISPR/Cas9 System
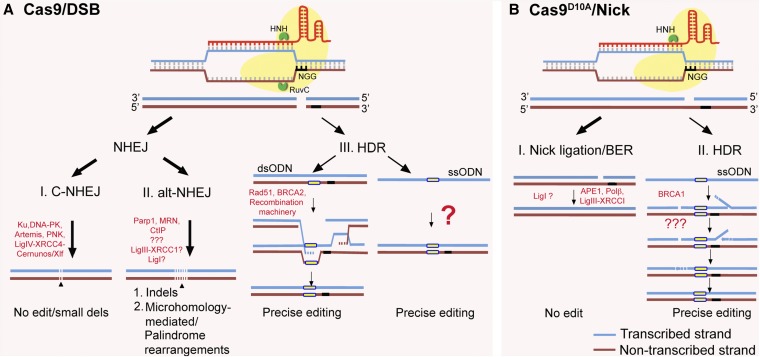
The versatility of CRISPR/Cas9 as a genome editing tool arises from its ability to recognize virtually any sequence in the genome and introduce a controlled break in the DNA. These breaks are repaired by error-prone or high-fidelity cellular mechanisms. The nuclease activity of the CRISPR/Cas9 system is guided by two noncoding RNA elements: (1) crRNA containing 20 bp of unique target sequence (spacer sequence) and (2) tracrRNA (trans-activating crRNA). The crRNA:tracrRNA duplex (also termed guiding RNA or gRNA) directs Cas9 nuclease to target DNA in the genome via complementary base pairing between the spacer on the crRNA and the complementary sequence (called protospacer) on the target DNA. Target specificity of Cas9 protein relies on the presence of specific nucleotides 3′ to the protospacer sequence, termed the protospacer adjacent motif (PAM). The Cas9 RNA-guided endonuclease from Streptococcus pyogenes, spCas9, requires a 5′-NGG–3′ PAM, whereas Cas9 from S. thermophilus (stCas9) and Neisseria meningitidis (nmCas9) require 5′-NNAGAAW-3′ PAM (W = A or T) and 5′-NNNNGATT–3′ PAM motifs, respectively (Hou et al. 2013). These Cas9 variants with different PAM dependencies increase the frequency of targetable loci in genome, however spCas9 (referred to here as Cas9) has been the most broadly used targetable nuclease. Both nuclease domains of Cas9, HNH and RuvC, independently introduce a nick in complementary and noncomplementary strands, respectively, 3 bp upstream of the PAM, thus generating a DSB (Jinek et al. 2012). CRISPR/Cas9-generated DSBs activate cellular DNA damage responses that repair the damage (Figure 1). The nonhomologous end-joining (NHEJ) mechanism is a “quick-fix” DSB repair pathway that ligates the two broken DNA ends. NHEJ repair is divided into two subclasses: (i) Ku- and XRCC4/Ligase IV-dependent, or “canonical” (C-NHEJ); and (ii) Ligase I- or Ligase III-dependent alternative end joining (a-EJ or alt-NHEJ) (Betermier et al. 2014). DSB repair by C-NHEJ is faster and may result in precise (nonmutagenic) end joining or small deletions. However, failure to repair DSB by C-NHEJ may lead to more extensive resection of DNA ends and repair by alt-NHEJ (Figure 1). Alt-NHEJ can yield a variety of mutations including: point mutations, indels ranging from one to hundreds of nucleotides, interchromosomal translocations, pericentric inversions, palindrome-catalyzed deletions, and microhomology-mediated deletions. These often disrupt open reading frames, effectively creating gene knockouts (Choi and Meyerson 2014).
Figure 1.
Schematic showing the proposed cellular repair pathways operating at CRISPR/Cas9-generated DNA breaks (A) or nicks (B). (A) gRNA targeted Cas9 having HNH and RuvC domains induces a DNA break on complementary and noncomplementary strands, respectively. These DSBs may be repaired predominantly by the less error-prone C-NHEJ pathway (I). If C-NHEJ fails, unrepaired DSB sites are recognized by PARP1 thus entering the alt-NHEJ (II) pathway. The Ku-unprotected DNA ends are resected and ultimately ligated by either Ligase III or Ligase I, thus generating longer indels at targeted loci. Alternatively, presence of donor template (ssODN or dsODN) carrying designed mutation (yellow box) may promote homology-directed repair (III) leading to precise editing. Although the exact mechanism of DNA repair using ssODNs is still unknown, CRISPR/Cas9-mediated precise editing with ssODNs is relatively efficient. (B) Cas9 nickase (Cas9D10A), bearing a mutation in RuvC nuclease domain, cleaves the DNA strand complementary to gRNA. The nick is predominantly repaired by the error-free BER pathway or simply undergoes nick ligation (I). In the presence of a ssODN, the nick may also be repaired by BRCA1-dependent HDR (II), generating a precise mutation.
In addition to creating disruptive mutations by error-prone repair, Cas9-generated DSBs can be repaired by high-fidelity homology-directed repair (HDR) mechanisms (Figure 1). HDR uses a homologous template, usually a sister chromatid under natural circumstances, to repair DNA damage if DNA replication has already occurred. Therefore, codelivering the site-specific nuclease with an alternative repair template, such as a plasmid or single-stranded oligodeoxynucleotide (ssODN) bearing locus-specific homology and any intended sequence changes, enables precise mutations at or near the induced DSB. This approach facilitates generation of alleles with precise mutations and allows researchers to mimic human gene variants associated with diseases.
CRISPR/Cas9 as a mouse genome editor
Multiple studies in various human and mouse cell lines showed that the CRISPR/Cas9 system is a powerful genetic tool able to generate various types of mutations (Jinek et al. 2012; Cong et al. 2013; Jinek et al. 2013). Cells transfected with plasmids encoding Cas9 and gRNA can undergo efficient genome editing. The technique was further developed and applied to modify genes in several other model organisms such as Drosophila (Yu et al. 2013), Caenorhabditis elegans (Dickinson et al. 2013; Friedland et al. 2013), and zebrafish (Chang et al. 2013; Hwang et al. 2013; Jao et al. 2013). Simply injecting Cas9 mRNA and gRNA into early embryos resulted in efficient genome editing. However, it was the pioneering work in the mouse that revealed the full potential of the CRISPR/Cas9 system to quickly and efficiently generate genetically modified animals. Shen et al. (2013) reported site-specific cleavage of an endogenous eGFP locus by co-injecting a chimeric gRNA with “humanized” Cas9 mRNA into one-cell stage mouse embryos. This approach produced successful disruption of the endogenous gene, although with a relatively low targeting efficiency (14–20% of newborns). However, even this “low” frequency was remarkable, given that targeting by injection of DNA templates alone occurs rarely if at all in embryos (Brinster et al. 1989). A breakthrough study by Wang et al. (2013) demonstrated the remarkable efficacy of the CRISPR/Cas9 system to target single or multiple genes simultaneously (Table 1). Moreover, this study showed the ability to induce precise genome editing (point mutations) by HDR in mouse embryos. These capabilities opened a world of possibilities for making diverse types of genomic modifications and in a multiplexed manner. For example, in a project to study checkpoints in meiosis (Bolcun-Filas et al. 2014), we were attempting to generate female mice homozygous for mutations in three different genes (Trip13, Trp53, and TAp63), but traditional breeding was problematic and slow. Therefore, we performed simultaneously CRISPR mutagenesis of these genes and obtained control and triply mutant genotypes, the latter of which displayed the expected phenotype of restored fertility (our unpublished data). These phenotypic results were obtained only 6 weeks after embryo injections were performed, in contrast to years it would have taken to generate and breed triple homozygotes from individually targeted ES cells.
Table 1. CRISPR/Cas9-mediated targeting and its efficiency for various mouse/rat genes.
| Reference | Gene(s) | CRISPR targeted/ total born pups (%) | Concentration (Cas9/gRNA/HDR template ng/μl) | Mode of delivery | Nuclease | Type of editing event |
|---|---|---|---|---|---|---|
| Shen et al. (2013)a | Pouf5-IRES-EGFP (knock in) | 1/5 (20) | 20/20 | N/A | NLS-flag-linker-Cas9 mRNA/DSB | NHEJ |
| CAG-EGFP (transgene) | 1/7 (14.2) | 20/20 | N/A | NLS-flag-linker-Cas9 mRNA/DSB | NHEJ | |
| Wang et al. (2013) | Tet1 | 10/12 (83.3) | 100/50 | Cyto | Cas9 RNA/DSB | NHEJ |
| Tet2 | 19/22 (86.4) | 100/50 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Tet3 | 11/15 (73.3) | 100/50 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Tet1+Tet2 (2 gRNAs) | 28/31 (90.3) | 100/50 each | Cyto | Cas9 RNA/DSB | NHEJ | |
| Tet1b | 6/9 (66.6) | 100/50/100g | Cyto | Cas9 RNA/DSB | HDR | |
| Tet2b | 9/15 (60) | 100/50/100g | Cyto | Cas9 RNA/DSB | HDR | |
| Tet1+Tet2b (2 gRNAs) | 1/14 (7.1) | 100/50 each/ 100g each | Cyto | Cas9 RNA/DSB | HDR | |
| Tet1+Tet2 (2 gRNAs) | 6/10 (60) | 100/50 each/ 100g each | Cyto | Cas9 RNA/DSB | HDR | |
| Li et al. (2013)a | Th | 8/9 (88.8) | 25/12.5 | Cyto | Cas9 RNA/DSB | NHEJ |
| Rheb | 3/4 (75) | 25/12.5 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Uhrf2 | 11/12 (91.6) | 25/12.5 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Th | 0/10 (0) | 1 | Pronuc | SP6-Cas9 plasmid/DSB | NHEJ | |
| Th | 1/11 (9) | 2.5 | Pronuc | SP6-Cas9 plasmid/DSB | NHEJ | |
| Mc4rc | 13/15 (86.6) | 25/12.5 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Mc3rc | 1/15 (6.6) | 25/12.5 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Ran et al. (2013a) | Mecp2b (2 gRNAs) | 61/61 (100) | 100/50 each | Cyto | Cas9(D10A) RNA/double nick | NHEJ |
| Mecp2b | 34/37 (91.8) | 100/50 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Yang et al. (2013a)a | Tet1 +Tet2b (2 gRNAs) | 3/15 (20) | 100/50 each/ 200g each | Cyto | Cas9 RNA/DSB | HDR |
| Nanogb | 86/936 (9.1) | 100/50/200h | Cyto | Cas9 RNA/DSB | HDR | |
| Nanog | 7/86 (8.1) | 100/50/200h | Cyto | Cas9 RNA/DSB | HDR | |
| Nanogb | 7/75 (9.3) | 5/2.5/10h | Pronuc | Cas9 RNA/DSB | HDR | |
| Oct4b | 47/254 (18.5) | 100/50/200h | Cyto | Cas9 RNA/DSB | HDR | |
| Oct4 | 3/10 (30) | 100/50/200h | Cyto | Cas9 RNA/DSB | HDR | |
| Oct4b | 13/72 (30) | 5/2.5/10h | Pronuc | Cas9 RNA/DSB | HDR | |
| Sox2 | 12/35 (34.3) | 100/50/200g | Cyto | Cas9 RNA/DSB | HDR | |
| Mecp2 (2 gRNAs) | 16/98 (16)d, 5/98 (5)e | 100/50 each/200g each | Cyto | Cas9 RNA/DSB | HDR | |
| Fujii et al. (2013) | Rosa26 | 7/7 (100) | 10/10 | Cyto | Cas9 RNA/DSB | NHEJ |
| Hprt (2 gRNAs) | 17/18 (94.4) | 10/10 each | Cyto | Cas9 RNA/DSB | NHEJ | |
| Hprt (2 gRNAs) | 5/18 (27.7) (∼10 kb deletion) | 10/10 each | Cyto | Cas9 RNA/DSB | NHEJ | |
| Mashiko et al. (2013) | Cetn1 | 10/17 (58.8) | 5 | Pronuc | Cas9 pX330/DSB | NHEJ |
| Prm1 | 2/3 (66.6) | 5 | Pronuc | Cas9 pX330/DSB | NHEJ | |
| Cetn1 | 5/20 (25.0) | 10/1 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Prm1 | 4/4 (100) | 40/10 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Sung et al. (2014)a | Prkdc | 21/37 (56.7) | 50/250 | Cyto | Cas9 RNA/DSB | NHEJ |
| Foxn1 | 54/58 (93.1) | 10/100 | Cyto | Cas9 RNA/DSB | NHEJ | |
| Foxn1b | 15/17 (88.2) | 200/400i | Pronuc | Cas9 protein/DSB | NHEJ | |
| Foxn1b | 10/14 (71.4) | 200/400i | Cyto | Cas9 protein/DSB | NHEJ | |
| Shen et al. (2014)a | Ar (2 gRNAs) | 5/20 (25) | 20/10 each | Cyto/Pronuc | Cas9 protein/DSB | NHEJ |
| 3/8 (37.5)f | 20/10 each | Cyto/Pronuc | Cas9(D10A) RNA/double nick | NHEJ | ||
| 10/12 (83.3)f | 20/10 each | Cyto/Pronuc | Cas9(H840A) RNA/double nick | NHEJ | ||
| Fujii et al. (2014)a | Rosa26 (4 gRNAs) | 6/6 (100; 83% with ∼1-kb deletion) | 100/10 each | Cyto | Cas9(D10A) RNA/double nick | NHEJ |
| Horii et al. (2014) | Tet1 (Ex4) | 9/9 (100) | 50/20 | Cyto | Cas9 RNA/DSB | NHEJ |
| 5/5 (100) | 50/20 | Pronuc | Cas9 RNA/DSB | NHEJ | ||
| 4/5 (80) | 5 | Pronuc | Cas9 pCAG-hCas9/DSB | NHEJ | ||
| Tet1 (Ex7) | 10/19 (52.6) | 50/20 | Cyto | Cas9 RNA/DSB | NHEJ | |
| 7/15 (46.7) | 50/20 | Pronuc | Cas9 RNA/DSB | NHEJ | ||
| 2/8 (25) | 5 | Pronuc | Cas9 pCAG-hCas9/DSB | NHEJ | ||
| Yen et al. (2014)a | Tyr (2 gRNAs) | 10/12 (83.3) | 5/6.67 each | Cyto | Cas9 RNA/DSB | NHEJ |
| 22/28 (78.5) | 5/6.67 each | Pronuc | Cas9 RNA/DSB | NHEJ |
Boldface numbers represent mice edited for two genes/loci. Cyto/Pronuc, cytoplasmic/male pronuclear.
Mosaicism reported.
Editing efficiency reported in mice blastocysts.
Editing efficiency reported in rat blastocysts.
Two lox P integrations in one allele.
Two lox P integrations in two alleles.
Data calculated from gel picture.
HDR template: ssODN.
HDR template: dsDNA.
Concentrations in nanomoles/L.
The possibility to direct Cas9 to any genomic locus/loci by providing specific guiding RNA offers a unique tool for geneticists to modify the mouse genome in vivo in many different ways. In addition to null alleles, the CRISPR/Cas9 system can be used to generate conditional floxed alleles (Yang et al. 2013a). It also opens the possibility to engineer larger deletions in one step; simultaneously injecting two gRNAs homologous to loci separated by up to ∼10 kb yielded interstitial deletions via NHEJ-mediated ligation (Fujii et al. 2013) (see Table 1). This new technology also offers scientists a rapid means to overcome a common problem—lack of specific antibodies to an endogenous protein—via epitope tagging. CRISPR/Cas9-stimulated HDR was used to generate mice carrying V5-tagged, GFP, or mCherry fluorescent fusion proteins in the Sox2, Oct4, and Nanog genes, respectively (Yang et al. 2013a).
A longstanding technical challenge has been the generation of mice carrying subtle genomic modifications such as point mutations. Such alterations are useful for functional analysis of transcription factor binding or phosphorylation sites or for testing the impact of single nucleotide polymorphisms (SNPs). ES cell-based targeting technology enables introduction of subtle genomic modifications, but it is a lengthy process involving multiple steps (reviewed by Menke 2013). In most cases, the desired mutation is introduced into the ES cell genome via a “knock-in” gene targeting strategy, followed by removal of selectable marker cassette by Cre/Lox or Flp/FRT recombinases (Hubner et al. 2008). Precise mutations can also be induced in ES cells using oligonucleotides (Aarts et al. 2006; Papaioannou et al. 2012), but low spontaneous efficiency of precise editing prevented widespread use until the ZNF or Cas9-induced DSBs increased the efficiency of targeted editing (Chen et al. 2011; Mali et al. 2013b). DSB-stimulated genome editing by HDR is usually less efficient than NHEJ-mediated mutational outcomes (Table 1), an issue addressed later, but still occurs at substantial levels. As discussed later, efficient oligonucleotide-mediated precise editing stimulated by targeted CRISPR/Cas9 DSBs can be done in a single step by injection into mouse and rat embryos (Wang et al. 2013; Yoshimi et al. 2014).
A major concern and potential limitation of the RNA-guided nuclease system is the possibility of cleavage and deleterious editing at other sites in the genome (off-target sites) in addition to the on-target site (Fu et al. 2013). This could confound phenotypic analyses of CRISPR/Cas9-generated mouse mutants, particularly in founder animals. However, in contrast to evidence for substantial off-target site editing in cell-based systems (Fu et al. 2013), emerging data indicate that CRISPR-induced editing events are highly specific in mouse embryos (Wang et al. 2013; Yang et al. 2013a). The rarity or lack of off-target site editing in embryo injection experiments could be attributed to the transient expression of Cas9 protein from mRNA compared to the extended expression from a plasmid in cell transfection experiments. Moreover, the use of immortalized or cancer cell lines for off-target analyses might misrepresent what occurs in normal cells. Elevated levels of editing at off-target sequences could be due to the aberrant DNA repair mechanisms characteristic of those cell types (Smith et al. 2014; Veres et al. 2014). Whole genome sequencing of CRISPR-targeted human induced pluripotent stem cells (iPSCs) showed a low risk of off-target mutations (Smith et al. 2014; Veres et al. 2014). Altogether, this suggests that careful selection of genomic target sites minimizes the likelihood of off-target mutations in mouse embryos or normal cell types. This issue is discussed further in the section titled Optimal design parameters and pitfall avoidance.
If off-target activity of Cas9 remains a concern, for example in eventual human therapeutic applications, further modification of the CRISPR/Cas9 system may be the answer. One strategy has been to use a nickase version of the Cas9 nuclease (Cas9n), achieved by making an aspartate-to-alanine substitution (D10A) in the RuvC domain or histidine-to-alanine substitution (H840A) in the HNH domain (Jinek et al. 2012; Cong et al. 2013; Ran et al. 2013a). Site-specific nicks are predominantly repaired by high-fidelity base excision repair pathway (BER) or HDR events (Figure 1), but rarely by NHEJ (Cong et al. 2013; Mali et al. 2013a). To increase NHEJ editing efficiency without compromising specificity, a “dual nicking” strategy was applied to efficiently generate DSBs. The D10A Cas9 nickase, directed by a pair of gRNAs targeting opposite strands of target locus efficiently creates a staggered-end DSB that is recombinogenic. This strategy has been successfully used in mouse zygotes to generate indels and knock-in alleles (Ran et al. 2013a; Fujii et al. 2014).
CRISPR/Cas9 as an RNA-guided gene regulator
The ability to direct Cas9 to specific genomic locations has been exploited for applications beyond genome editing. Researchers have modified the CRISPR system into a powerful tool for targeted regulation of gene expression by generating catalytically deficient Cas9 variants in which both the RuvC and HNH nuclease domains have been inactivated (Gilbert et al. 2013; Qi et al. 2013). In the absence of any nuclease activity, the Cas9 protein acts as RNA-guided DNA-binding protein (dCas9) with the ability to activate or silence gene expression depending on the tethered protein. For example, dCas9 fused with artificial transcription factors (ATFs) and supplied with a gRNA was used as a “CRISPR-on” system that acts as a synthetic transcriptional activator (Cheng et al. 2013). On the other hand, the “CRISPRi” system (CRISPR interfering) can be used to downregulate gene expression. gRNA–dCas9 complexes bound to a target gene can physically interfere with the transcriptional machinery, while dCas9 fused with a transcriptional repressor can silence the gene (Gilbert et al. 2013; Qi et al. 2013). The efficacy of CRISPR-on has also been demonstrated in mouse embryos (Cheng et al. 2013). In this case, one-cell stage embryos were co-microinjected with dCas9 fused to three copies of the minimal VP16 transcriptional domain (VP48), vector-borne Nanog-EGFP, and gRNAs targeting the Nanog promoter. The authors demonstrated that multiple gRNAs could bring dCas9VP48 transcriptional activator to the Nanog promoter and efficiently induce EGFP expression in cultured embryos. There have yet to be reports of CRISPRi in mouse embryos that developed to term.
These alternative uses of CRISPRs create many exciting possibilities for manipulating and studying various regulatory networks in vivo. RNA-guided dCas9 fused with a catalytically active protein such as a chromatin-modifying enzyme may be targeted to a specific genomic locus and alter the DNA or chromatin status, thus providing a valuable tool for studying epigenetic regulation or other cellular functions. There is enormous promise for functional studies of genes during development, various diseases, and possibly disease treatment. However, specificity will likely remain a major concern; unlike cleavage specificity, which requires a near perfect match, the binding of Cas9 is less restrained. It has been reported that a single Cas9–gRNA complex species can bind thousands of genomic sites, despite various degrees of mismatch (Wu et al. 2014). Therefore, applications based on catalytically inactive Cas9 need to be vetted for specificity.
If You Want a Mutant Mouse, Should You Use the CRISPR/Cas9 System?
Many straightforward types of RNA-guided genome editing in vivo are remarkably simple and rapid. For this reason, we believe that this will become the preferred method for generating and obtaining mutant and genetically modified mouse models. For most applications, it is much faster than the conventional method using gene targeting in ES cells, which can involve many months of targeting vector construction, selection, and validation of targeted clones and achieving germline transmission from at least one clone. However, for straightforward knockout projects, the days of making constructs and targeting in ES cells are essentially over due to the worldwide International Knockout Mouse Consortium (IKMC). Knockout alleles are already available for most genes (currently nearly 18,000), according to the International Mouse Phenotyping Consortium (IMPC) website https://www.mousephenotype.org, which has the goal of making knockouts in all mouse genes in a defined genetic background and determining the phenotypes of the mutants. Details of the available mouse alleles (currently >4,300), ES cells, and/or targeting constructs (close to 20,000) are available at the IMPC website. Many of the ES cell lines with targeted alleles have the feature of being rendered “conditional ready” (i.e., containing a loxP-flanked exon), so that the function of a gene can be evaluated in particular cell types via CRE-mediated excision (Skarnes et al. 2011). Here, we briefly describe factors that one may consider on the route to obtain a mutant animal.
A basic knockout
Creating a mutant allele via CRISPR is simple. A gRNA DNA template for transcription can be prepared and gRNA synthesized the day that synthetic primers are received (see below for using overlap PCR to generate the template). The gRNA plus Cas9 mRNA or protein (both available commercially) are then microinjected into single-celled embryos, and mice are born in 3 weeks, start to finish. This basic method banks on error-prone NHEJ to create a frameshift mutation in a key part of the gene. Before jumping ahead with CRISPRs to produce a mutant allele, one should first consider the availability of preexisting resources. If such a mouse already exists “on the shelf” and is readily available, then this is probably the easiest and cheapest route to obtain your animal. However, if there are importation issues (for example, if the source stock has a pathogen that is not allowed into your institutional animal facility), or if the mutant stock is frozen in a repository, then the CRISPR route may be more expedient and cost effective. However, it is important to realize that a new CRISPR allele may not exactly recapitulate a published allele, leading to potential phenotypic differences. Thus, some characterization (such as protein or mRNA analysis) of a presumably null CRISPR allele is important. Another potential consideration is genetic background, which can dramatically alter phenotype if your new CRISPR allele differs from that used in prior studies. However, if one has reason to study a gene mutation on a particular strain background that differs from that of available alleles, then the CRISPR route becomes attractive, since the RNAs can be microinjected into embryos of any strain. This yields a co-isogenic line (the entire genetic background is derived from the recipient strain, except for the mutated nucleotides), which is superior to generating a congenic line by breeding, in which much “passenger” DNA remains from the originating strain.
If a mouse is not available but ES cells are, then consider the following. In our experience, it can take several months to complete paperwork and obtain targeted and validated ES cells from IKMC repositories. Then, also depending on the source and genotype of parental ES cells, the germline competence of the ES cells can be less than ideal, and two to three independent targeted lines should be acquired to optimize chances of getting germline chimeras. Then, one must factor in the costs of the ES cell microinjections, which are similar to microinjections for CRISPRs. On the other hand, small frameshift deletions in the CRISPR scenario are not necessarily guaranteed to be null. However, from a single CRISPR microinjection, numerous alleles are typically obtained and careful sequence analysis can identify a mutant allele that disrupts or abrogates protein function.
For subtle mutations
Probably the most powerful application of RNA-guided genome editing is the generation of subtle genomic mutations, such as changing crucial amino acids or transcription factor binding sites. As mentioned earlier, traditionally this has involved a multistep targeting strategy in ES cells and allele transmission through chimeras (Menke 2013). CRISPR/Cas9-mediated editing is simpler and faster. Mice bearing single nucleotide changes can be generated within a month and either directly analyzed or bred to establish mutant stocks for subsequent analyses. There is no need for elaborate targeting constructs. Single stranded oligodeoxynucleotides carrying desired mutations can be synthesized and injected into embryos along with gRNA and Cas9, and this can be utilized by the embryo as a homologous recombination repair template. ssODN templates of up to 200 bp in length (centered on the Cas9-induced break) can be synthesized by commercial vendors. Because one typically desires precise changes at specific genomic locations, targeting may be limited by the availability of nearby PAM sites. It is recommended to use the closest PAM and gRNA to the edited sequence to prevent retargeting of the edited allele by Cas9 (discussed in more detail below). However, we have successfully induced a precise mutation 30 bp away from the PAM site used by a gRNA (our unpublished observation).
Epitope tagging or making floxed alleles
CRISPR/Cas9 can also address other problems encountered by mouse researchers such as lack of specific antibodies or the desire to detect a protein of interest in vivo. Small epitope tags (V5, HA, Flag, etc.) can be placed into protein coding sequences by HDR using a ssODN containing an epitope tag and homology arms of ∼40–60 bp (Yang et al. 2013a). Larger tags or fluorescent markers (GST, mCherry, or GFP) require dsDNA templates with homology arms of ∼1–3 kb at either side of DSB sites (Yang et al. 2014). If a conditional allele is desired but one is not available from the IKMC or other repositories, LoxP or FRT sites can be introduced by CRISPR/Cas9 via HDR using ssODN templates (Yang et al. 2013a). To detect simultaneous editing at both sites on the same chromosome (allele), one must design a clever strategy. It is advantageous to introduce unique restriction enzyme sites in addition to LoxPs, as it will help later to discriminate floxed alleles from alleles carrying single LoxP sites in trans.
Larger scale events such as deletions
Previously, generating large deletions (a few kilobases to over a megabase) in the mouse genome was achieved via ES cells engineered to have precisely positioned LoxP sites (produced in two sequential rounds of targeting) followed by Cre-mediated recombination (Ramirez-Solis et al. 1995), or by irradiation-induced deletions of an ES cell line containing a targeted marker (You et al. 1997). Both required substantial up-front work to derive the required ES cell lines. With CRISPR, the most straightforward approach is to induce two DSBs abutting the region to be deleted by microinjection of gRNAs and Cas9 into mouse embryos and screening for events in which the broken ends were joined by NHEJ. While there have been published reports of this strategy working for deletions that are relatively small (<10 kb) (Fujii et al. 2013), the efficiency of megabase-sized deletions is unclear. It may be advisable to attempt to make CRISPR/Cas9-driven deletions in ES cells until such events prove to be efficient in mouse embryos.
As an alternative to stock maintenance or breeding complex genotypes
Since the generation or acquisition of traditional knockout mice can be difficult and is time consuming, investigators commonly maintain these lines for long periods of time and at significant expense. Alternatively, sperm or embryos could be frozen and the stock taken off the shelf. CRISPR technology provides another alternative. Instead of cryopreservation, mutant mice can be regenerated. Generating double and triple mutants usually requires importing mouse mutants from other laboratories or recovery from cryopreservation followed by many months/years of breeding to obtain homozygous mice for all desired mutations. It is even more difficult when dealing with lethal or infertile mutants and requires multiple holding and mating cages. As described earlier, double and triple mutants can be generated within 2 months with CRISPR technology, which can significantly reduce the costs. This also eliminates the problems with strain background differences as mouse lines obtained from different laboratories might be maintained on different mouse backgrounds. CRISPR editing could be theoretically applied to any mouse strain, although certain strains of mice produce better yields and qualities of embryos than other strains.
Optimal Design Parameters and Pitfall Avoidance
It is of course important to consider the main current drawbacks to the CRISPR system—none of which are terribly problematic—in planning your route to a genetically modified mouse. The drawbacks in our perceived order of concern are as follows: (1) for experiments involving homologous recombination, predominant repair by NHEJ compared to HDR; (2) mosaicism in founder animals; and (3) targeting specificity, especially if phenotypic analysis of founders is desired. In the following paragraphs we will discuss the difficulties and options to improve efficiency and specificity of genome editing using the CRISPR/Cas9 system in mice, starting with the latter.
The DNA cleavage specificity of CRISPR/Cas9 is determined by two major factors: the 20-nt guiding sequence and the presence of the PAM at the 3′ end of the gRNA binding site. This 20-nt gRNA pairs with the complementary DNA sequence in the genome (on target) but also drives binding to thousands of other sequences with imperfect matches (off targets) (Hsu et al. 2013; Wu et al. 2014). Few studies have demonstrated the absolute dependency of Cas9 cleavage activity on the correct 8–12 nucleotides proximal to PAM; however, a few mismatches distal to PAM can be tolerated (Hsu et al. 2013; Sternberg et al. 2014).
When designing a guide sequence for genome targeting, the specificity in terms of off-target potential is crucial. For any chosen genomic locus, a specific CRISPR gRNA can be selected using online design tools such as those developed by Feng Zhang’s group at http://tools.genome-engineering.org (Hsu et al. 2013). It predicts the potential off-target binding sites in the genome and calculates quality scores for each gRNA (high scores reflect higher specificity). It also calculates an off-target hit score based on the number, position, and distribution of mismatches to predicted off-target sequences (scores reflect probability of gRNA binding). A target sequence with the least number of off-target sites, especially in exons, should be selected. We examined potential off-target editing in experiments targeting five different genes. gRNAs were selected to have low off-target hit scores (<2.3) and three or more mismatches. Next-generation (next-Gen) sequence analysis (see below) of 56 of the highest-scoring off-target sites revealed no cases of off-target mutagenesis in a total of 90 founder animals (Supporting Information, Table S1).
Nevertheless, certain experiments seeking to mutate specific sites may happen to have very similar sequences elsewhere in the genome, raising the possibility of off-target events. It has been proposed from cell-based studies that use of a truncated gRNA sequence of 17–18 nucleotides (referred as “tru-gRNA”) can decrease nonspecific targeting, and that transfecting synthetic gRNA increases specificity as opposed to plasmid-encoded gRNA, which can direct continuous synthesis (Cho et al. 2013; Fu et al. 2014). In addition, “paired nicks” (as described above) is an alternative to reducing off-target editing in various human cells (Mali et al. 2013a; Cho et al. 2014) and mouse (Ran et al. 2013a). Off-targets having only single nicks will be repaired by nonmutagenic repair pathways, whereas dual nicks recognized as DSBs at target site will result in editing.
Combating CRISPR Robustness and NHEJ
A complicating issue with the CRISPR system is actually the robustness of Cas9. The majority of gRNA-targeted Cas9-induced DSBs are eventually channeled to an error prone NHEJ repair pathway (Deriano and Roth 2013; Betermier et al. 2014). Indeed, with proper design and technical competence with microinjections into embryos, many or most founder mice will be homozygous for edited (often null) alleles and potentially can be analyzed phenotypically without further breeding. However, this robustness becomes problematic when the goal is to induce precise mutations into a locus via HDR with an introduced template. If a DSB is first repaired by NHEJ in a manner that precludes subsequent Cas9:gRNA recognition or cutting (for example by mutating the PAM site), then the desired modification is thwarted. If HDR is the first repair event at a given DSB, but the edited locus does not disrupt the PAM site or gRNA base pairing, then the locus can be subject to recutting and mutagenic NHEJ outcomes. Indeed, we have observed numerous cases of multiple events occurring at the same locus in mouse embryos. For example, while trying to introduce point mutations by HDR, we identified alleles in which partial insertion of the donor ssODN sequence was accompanied by adjacent rearrangements (our unpublished observations). To suppress these undesired outcomes, we suggest designing the donor repair template to introduce changes proximal to, or within the PAM site (to non-NGG or NAG) so as to block subsequent rebinding or cutting by the gRNA:Cas9 complex (Hsu et al. 2013). This would also prevent gRNA/Cas9 from recognizing and cutting the repair template itself (see below). However, such alterations will only be possible if the coding sequence is not altered in an unacceptable manner.
Another drawback of Cas9:gRNA complex robustness is that microinjection of Cas9 mRNA and gRNA into single-celled zygotes often causes genetic mosaicism in founder animals. Cas9:gRNA is delivered to zygotes during the period of active DNA replication, thus the editing could be achieved either prior to or after a particular locus has been replicated, with the latter potentially resulting in mosaicism. Additionally, editing may happen after first embryonic division, due to persistence of Cas9:gRNA complexes, also causing mosaicism. We (unpublished results) and others (Yang et al. 2013a; Ma et al. 2014; Yen et al. 2014) have observed mosaic animals carrying three or more alleles. A recent study reported surprisingly high percentage of mosaic mice (up to 80%) generated by CRISPR targeting of the tyrosinase gene (Tyr) (Yen et al. 2014). We have observed a varying frequency of mosaicism, 11–35%, depending on the gene/locus (our unpublished data). We hypothesize that mosaicism is related to the timing of targeted locus replication in the zygote. Targeting early replicating regions would have a higher chance of generating genetic mosaicism as there would be four copies subjected to editing if Cas9 acts upon replicated DNA. The complexity of allelic variations in mice generated by such Cas9:gRNA embryo injections requires detailed genotype analysis (addressed below). We speculate that plasmid-based delivery, as is done in cultured cells, would exacerbate the problem of persistent and uncontrolled expression of Cas9. Alternatively, the amount and thus activity of Cas9 can be regulated by direct delivery of recombinant Cas9 protein (Kim et al. 2014b; Ramakrishna et al. 2014). Direct delivery of recombinant Cas9 protein to human cell lines can reduce potential off-target editing due to the short life span of Cas9 protein within cells (Kim et al. 2014b; Ramakrishna et al. 2014). Sung et al. (2014) reported successful editing by injecting ribonucleoprotein (Cas9 protein:guide RNA) into zebrafish and mouse embryos. Therefore, microinjecting Cas9 protein instead of RNA, possibly in conjunction with ssODN donor templates that introduce PAM site or adjacent sequence changes, may help reduce mosaicism and frequency of undesired mutagenic repair.
Genotyping of Founder Mice
The remarkable efficiency of CRISPR editing in mouse zygotes enables one to conduct experiments in which multiple independent loci are mutated simultaneously. For example, one might have the goal of analyzing double and triple mutant founder mice. However, Cas9-generated DSBs directed at multiple loci or within different parts of the same gene (for example, when LoxP sites are being introduced) can lead to complex allelic outcomes. In such multiplexed approaches, each founder mouse will carry a different combination of mutated alleles, overlaid with the issue of mosaicism at each locus. Therefore, detailed genotyping is necessary before phenotypic analyses.
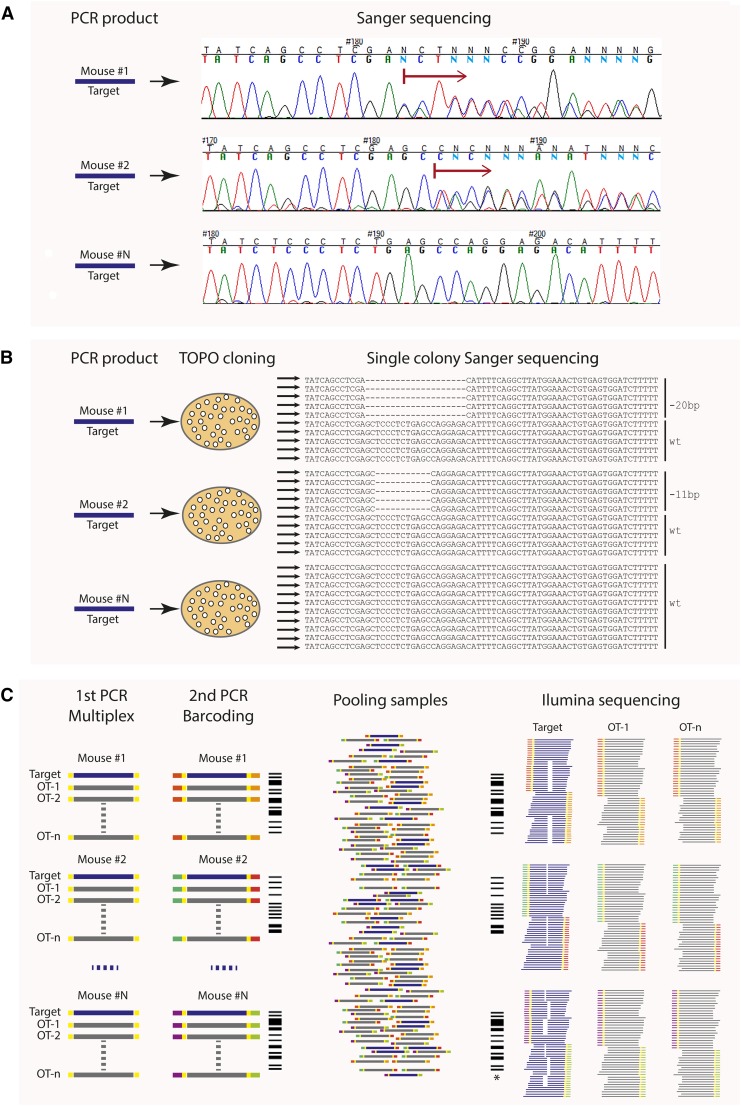
Various genotyping strategies have been described for analysis of CRISPR edited (CRISPRed) mice. The most commonly used assay utilizes SURVEYOR nuclease, which detects and cleaves heteroduplexes formed between wild-type (WT) and indel-containing amplicon strands from CRISPRed mice (Guschin et al. 2010). The loss or gain of a restriction site within the PAM or gRNA recognition sites or discernible size alterations of PCR amplicons following gel electrophoresis are other straightforward alternatives. It has also been proposed to exploit Cas9–gRNA as a sequence-specific “restriction enzyme” for genotyping (Kim et al. 2014a), based on the idea that gRNA-driven editing events can destroy the recognition site, and thus WT amplimers but not edited alleles will be cleavable by Cas9. However, none of these assays can reveal the exact nature of the induced mutation. DNA sequencing is required to reveal the exact lesion, and this information can be used to assess the potential impact on encoded protein. However, direct Sanger sequencing of amplicons from founder mice can be problematic; the presence of two or more different alleles (the latter in the case of mosaics) results in overlapping and asynchronous chromatograms (Figure 2A). One way to overcome this problem is to clone the PCR products into plasmids, followed by Sanger sequencing of a sufficient number of independent clones to identify all alleles in a CRISPR-derived founder (Figure 2B). However, this method is time consuming and may not recover all allelic variants, especially in mosaics.
Figure 2.
Sequencing-based methods to identify CRISPR-edited alleles in founder mice. (A) Sanger sequencing of PCR products around gRNA binding site. PCR amplification from mouse tail biopsy DNA will generate a mixture of two or more (mosaic) amplicons representing allelic variants in the mouse. This can cause overlapping peaks on the chromatogram (red arrow) and difficulty in identifying the mutation(s). (B) Sequencing of plasmid-cloned PCR products. Each clone contains one amplicon/allelic variant present in a mouse. This requires sequencing at least 10 single colonies per targeting event per mouse (e.g., one gene × 20 founder mice × 10 colonies = 200 sequences). In the case of multiplexed editing, proportionately more clones must be sequenced. (C) Next-Gen-based multiplexed sequencing. This method also allows testing for off-target (OT) events and the presence of mosaicism. Target and OT PCR products from one founder mouse are labeled with unique barcode. All PCR products from up to 96 mice (one mouse = one barcode) are pooled together and sequenced. *, mosaic animal.
An alternative is to use deep sequencing of PCR-amplified target loci from founder mice produced from different experiments. The high throughput also permits simultaneous analysis of potential off-target sites. We used this approach (Figure 2C; Materials and Methods) to identify almost all mutated alleles in 90 CRISPRed mice and to reveal mosaicism that was not detected by plasmid clone sequencing or direct sequencing of PCR amplicons. We failed to identify a 203 bp deletion found by PCR sequencing, suggesting that the small size of amplicons (we used 350–450 bp) used in next-Gen sequencing may limit detection of indels. As mentioned earlier, no editing was observed at a combined 56 potential off-target sites. Since the next-Gen sequencing component has such high capacity, economies of scale can be achieved by pooling mice from multiple projects, ideally coordinated by the institutional transgenic facility.
Practical Recommendations, Alternative Methods, and Possible Improvements
Methods of microinjection
Highly efficient genome editing in mouse embryos can be achieved by simple delivery of editing reagents to zygotes. Since Cas9 nuclease can be injected in multiple forms, as either plasmid, mRNA, or protein, it may necessitate a particular delivery method. Plasmids require transcription, therefore pronuclear injection may be the preferred way, but one should consider that integration of the plasmid will occur in a subset of embryos, and this may not be desirable. Cas9 mRNA injection directly to cytoplasm should facilitate translation, while Cas9 protein could be injected directly to the pronucleus—the site of enzymatic activity. It would seem logical that microinjecting into one pronucleus alone could diminish the chances of achieving biallelic mutations. However, this is not the case as we and others have observed, suggesting that ssODNs and gRNA either freely diffuse across the nuclear membranes or the events occur shortly after nuclear breakdown. The pronuclear microinjection of gRNA and Cas9, in a manner essentially identical to what is used for generating transgenic mice, can be easily adapted by most transgenic facilities. Facilities equipped with a Piezo-electric micromanipulator can opt for cytoplasmic injections as reported (Wang et al. 2013; Yang et al. 2013a). Horii et al. (2014) performed an extensive comparison study suggesting that cytoplasmic injection of a gRNA and Cas9 mRNA mixture as the best delivery method. Although the overall editing efficiency in born pups yielded by pronuclear vs. cytoplasmic RNA injection seems to be comparable (Table 1), the latter method generated two- to fourfold more live born pups. Injection of plasmid DNA carrying Cas9 and gRNA to the pronucleus was the least efficient method in terms of survival and targeting efficiency (Mashiko et al. 2013; Horii et al. 2014). Injection into pronuclei seems to be more damaging to embryos than injection of the same volume or concentration of editing reagents to the cytoplasm. It has been shown that cytoplasmic injection of Cas9 mRNA at concentrations up to 200 ng/μl is not toxic to embryos (Wang et al. 2013) and efficient editing was achieved at concentrations as low as 1.5 ng/μl (Ran et al. 2013a). In our hands, injecting Cas9 mRNA at 50–150 ng/μl and gRNA at 50–75 ng/μl first into the pronucleus and also into the cytoplasm as the needle is being withdrawn, yields good survival of embryos and efficient editing by NHEJ in live born pups (our unpublished observations).
While NHEJ-driven editing is highly efficient in mouse embryos (Table 1), there are fewer available data on homology-driven repair from ssODN or dsDNA templates. Two studies reported successful HDR-driven editing by co-injecting (along with gRNA and Cas9 mRNA) ssODNs into the cytoplasm at 100 or 200 ng/μl (Wang et al. 2013; Yang et al. 2013a). In similar experiments, we found that injections involving ssODN as repair templates decreased embryo survival in a dose-dependent manner (our unpublished observations). We believe that the presence of ssODN (at high concentration) in the nucleus may elicit a DNA damage response and result in embryo arrest and death (Nur et al. 2003). This problem might be ameliorated by cytoplasmic injections alone or by decreasing the concentration of ssODN in pronuclear injections (to 10–20 ng/μl). Yang et al. (2013a) also found that dsDNA (circular plasmid) microinjected into the cytoplasm at 200 ng/μl or into the pronucleus at 10 ng/μl yielded good embryo survival and editing efficiency.
Cas9 and gRNA synthesis
The simplicity of CRISPR editing reagents makes it available to many researchers. Cas9 protein, mRNA, and gRNA may be purchased from vendors or prepared in the lab in a few simple steps. Plasmids carrying the Cas9 gene driven by the T7 promoter can be used for in vitro transcription to generate Cas9 mRNA for injection (Yang et al. 2014). Cas9 protein can be produced and purified using affinity purification methods. gRNA can also be transcribed in vitro from plasmids or PCR products. So far, cloning of gRNA seed sequence in the form of complementary annealed oligonucleotides into plasmids containing a chimeric guiding RNA expression cassette appears to be the most used method. These expression vectors are available from Addgene (www.addgene.org/CRISPR/), courtesy of labs that developed these vectors. Plasmids carrying gRNA seed sequences may be microinjected directly to the embryos (Li et al. 2013) or may be used for in vitro transcription to produce the gRNA (Shen et al. 2013; Wang et al. 2013). Due to the need of plasmid-based cloning and sequence verification, this whole process usually takes 2–3 days but is highly efficient. Alternatively, a cloning-free method can be used (Bassett et al. 2013; Fujii et al. 2013; Gagnon et al. 2014). It utilizes two long oligos sharing an overlapping region: (1) a CRISPR-forward primer containing the T7 promoter and 18–20 nt targeting sequence followed by overlapping sequence (boldface type) (5′-GAAATTAATACGACTCACTATAGGN(18-20)GTTTTAGAGCTAGAAATAGC-3′) and (2) a common reverse oligo containing the remaining chimeric gRNA sequence and the complementary overlapping sequence (boldface type) (5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′). These two oligos anneal in the overlap region, and serve as a template for PCR amplification. Overlap PCR will produce a gRNA template for in vitro transcription (for details, see Bassett et al. 2013).
Repair template (ssODN vs. dsDNA)
CRISPR/Cas9 editing can also be used for generating larger modifications (i.e., insertion of reporter genes or antibiotic resistance markers) by providing dsDNA donor repair templates carrying homology arms flanking the site of alteration. We are unaware of any systematic studies to assess the optimal length of homology arms in the donor template used in mouse embryos using this system. However, it has been shown in Drosophila embryos that donor templates with total homology of 2–4 kb were the most efficient in editing induced by zinc finger nucleases (Beumer et al. 2013). When constructing DNA donor templates, the homology arms should be designed in such a way as to prevent gRNA binding and cleavage of repair template by Cas9. Silent changes or naturally existing genetic variations (such as SNPs) can be introduced to PAM or protospacer sequences in homology arms to abolish recognition and cutting by Cas9 (Yang et al. 2014).
For many applications, synthetic ssODNs successfully replace the need for larger gene targeting plasmids and require no experimental effort. They also yield higher editing frequencies than dsDNA repair templates (Chen et al. 2011; Ran et al. 2013b). As shown for DNA nicks, DSB repair using ssDNA and dsDNA templates might involve different repair machineries resulting in different efficiency of editing (Davis and Maizels 2014). Local abundance of repair template may direct the repair toward HDR instead of NHEJ. Cells contain only one endogenous repair template during G1 phase (homologous chromosome) but three after S phase (one sister and two nonsister chromatids). Alternative templates in the form of ssODN or dsDNA injected to the oocyte will outnumber endogenous template/s by thousands or millions. dsDNA templates are usually longer than ssODNs; therefore, the same absolute amount of DNA will carry fewer molecules. Since DNA concentration can negatively affect viability of embryos, injecting less DNA but more molecules, as in the case of ssODNs, might translate to higher embryo survival and editing efficiency. Additionally, linear dsDNA templates might be integrated in the genome, causing potential deleterious effects. One cell-based study showed that optimal editing is achieved when the ssODN template is centered around the Cas9 cut site, and the desired edit site is located within 10 bp (Yang et al. 2013b). The authors also tested various lengths of ssODNs (30–110 nt) and their orientations with respect to the gRNA, finding that 70 nucleotide-long oligonucleotide templates complementary to gRNA enabled the highest editing efficiency. This is interesting, considering that gRNA/Cas9 can bind and cleave the complementary ssODN in vitro (Gasiunas et al. 2012). It remains unclear if the same applies to editing in embryos.
Inhibition of C-NHEJ increases the frequency of HDR events
Currently, there are few reports using CRISPR/Cas9 to generate precise point mutations in the mouse genome. Wang et al. (2013) reported highly efficient HDR-mediated mutagenesis (60–80%) in the Tet1 and Tet2 genes (Table 1). While trying to generate point mutations in gametogenesis genes, we experienced a relatively low frequency (5–29%) of desired HDR events that was locus influenced (see Table 2). We surmise that the frequency of HDR editing events is lower in part because HDR is in competition with NHEJ. DSBs repaired either by C-NHEJ or alt-NHEJ may preclude subsequent HDR, and furthermore, HDR events can be followed by subsequent recleavage and NHEJ, as long as gRNA identity is present.
Table 2. CRISPR/Cas9-mediated editing under different conditions.
| Injection condition | Concentration (Cas9/gRNA/ssODN) ng/μl | Two-cell stage embryos/live zygotes immediately after injection (%) | Transferred two-cell embryos (recipients) | Total edited animals/newborns (%) | HDR-mediated repair (%a) | HDR:NHEJb |
|---|---|---|---|---|---|---|
| Tex15 − SCR7 | 0/50/100 | 49/78 (62.8) | — | — | — | |
| Tex15 + SCR7 | 0/50/100 | 29/33 (87.8) | — | — | — | |
| Tex15 − SCR7 | 50/50/100 | 58/74 (74.3) | 58 (3) | 11/17 (64.7) | 2 /17 (5.8) | 1:10 |
| Tex15 + SCR7 | 50/50/100 | 35/40 (87.5) | 35 (2) | 16/16 (100) | 9/16 (56.2) | 1:2.5 |
| Cdk2c | 50/50/100 | 69/86 (80.2) | 69 (3) | 20/27 (74.1) | 10/27 (44.4) | 1:1.5 |
Tex15 + SCR7: microinjection of Tex15 gRNA and Cas9 followed by embryo culture up to two-cell stage in presence of SCR7.
Repair percentage of live born pups carrying at least one HDR event.
Repair event as total number of alleles including all different kinds of alleles in mosaics.
Cdk2, ssODN with additional silent mutations in seed sequence.
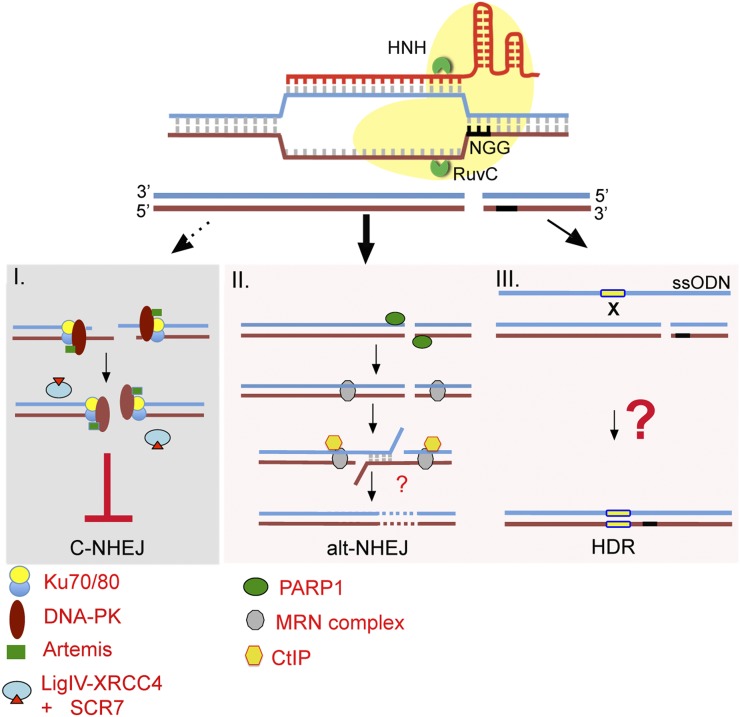
Having encountered this complication, we sought to increase the efficiency of HDR events by suppressing C-NHEJ, which may possibly increase the overall editing efficiency by entering into the error-prone alt-NHEJ pathway. One potential method toward this end came from studies of genome editing in Drosophila embryos mutated for DNA Ligase IV, a component of the C-NHEJ pathway. Co-injection of zinc finger nuclease (ZFN) and circular donor DNA (carrying several kilobases of homology) to lig4−/− mutant embryos led to a dramatic increase in HDR targeting (70%) compared to WT embryos (0%) (Beumer et al. 2008). Inhibition of C-NHEJ, which simply ligates broken blunt ends or introduces smaller deletions (Betermier et al. 2014), induces resection and as a result promotes alt-NHEJ and HDR. Indeed, an increase in overall editing efficiency in the Tyr gene of rats was reported by coexpressing the Exo1 exonuclease with engineered TALENS. Exonuclease-driven end resection shifted the repair toward more mutagenic alt-NHEJ (Beumer et al. 2008). Based on these two reports, we hypothesized that temporal inhibition of the C-NHEJ pathway in mouse embryos could similarly increase total mutagenic editing events by promoting the alt-NHEJ and HDR pathways (Figure 3). Instead of NHEJ-deficient embryos, we used a recently developed Ligase IV inhibitor SCR7 (Srivastava et al. 2012). SCR7 has been shown to directly bind to the DNA binding domain of Ligase IV and thus interfere with the progression of the C-NHEJ events.
Figure 3.
Transient inhibition of C-NHEJ with the Ligase IV inhibitor (SCR7) increases editing efficiency. In the presence of SCR7, DSBs will be predominantly repaired by the highly error-prone alt-NHEJ (II) pathway, generating indels, or HDR-mediated precise editing (yellow box) (III). Thickness of the arrows represents relative interplay of individual pathways involved in the repair of targeted DSB in presence of SCR7.
To determine if SCR7 could be used for suppressing C-NHEJ-mediated events in mouse embryos, we first tested the impact of treating embryos with this drug (5–100 μM). We did not see any adverse effect on embryo development to the two-cell stage. Interestingly, embryos microinjected with gRNA and ssODN followed by overnight culture in 50 μM SCR7 showed a relatively better survival up to the two-cell stage. However, prolonged culture in the presence of SCR7 negatively affected progression of cultured embryos to blastocyst stage. Next, to test if treatment of embryos with SCR7 would increase the HDR:NHEJ ratio of CRISPR editing, we designed a gRNA and ssODN HDR template for targeting a CG-to-TA change in the Tex15 gene. Indeed, SCR7 increased the efficiency of HDR events up to ∼10-fold in resulting pups. SCR7 treatment led to an increased HDR:NHEJ event ratio, from 1:10 to 1:2.5 (Table 2). Interestingly, all animals born from these microinjected embryos cultured in SCR7 were edited. This suggests that the transient suppression of C-NHEJ by SCR7 skewed DSB repair to the alt-NHEJ and HDR pathways, and that in the presence of excessive amounts of donor template (ssODN), homology-driven repair can occur at a higher frequency.
Conclusions: Faster, Better, Cheaper
Powerful genetic tools and physiological similarities to humans have made the laboratory mouse the leading model for study of human gene functions and diseases. However, functional testing in mice of human genetic variants, such as SNPs implicated by genome-wide association studies, has been technically challenging. The CRISPR/Cas9 system has opened a plethora of possibilities for precise genome editing. Now nearly any change mimicking human coding variants can be introduced to the mouse genome. Already much progress has been made to increase targeting specificity and the simplicity of making gene-edited mice. Still, improvements that increase precise editing efficiency, lower mosaicism, and enable more complicated genetic alterations in an efficient manner will increase the power of the system for diverse uses. Developing a better understanding of the repair mechanisms involved in repair of CRISPR/Cas9-induced DNA breaks is one crucial step toward maximizing the system. Additional applications beyond genome editing, such as sequence-specific gene regulation, has huge potential but has yet to be successfully implemented in the mouse. Probably the most anticipated potential of RNA-guided genome editing lies in therapeutic applications. A few studies using cell lines and mice have shown that CRISPR-driven editing can correct disease causing mutations and reverse the phenotypes (Wu et al. 2013; Yoshimi et al. 2014), but the biggest obstacle for therapeutic use of CRISPRs in humans would be the delivery of editing reagents for efficient allele correction in vivo or in stem cells that can be reintroduced into people. For we mouse geneticists, the CRISPR/Cas9 system equips our genetic toolbox with entirely new capabilities and enables us to conduct mouse research faster, better, and cheaper.
Materials and Methods
Creating genome-edited mice
gRNA seed sequences and potential off-target sites were predicted using CRISPR Design Tool at http://crispr.mit.edu/. sgRNAs were produced by cloning annealed complementary oligos into pX330-U6-chimeric_BB-CBh-hSpCas9 at the BbsI site (Cong et al. 2013), generating PCR products containing a T7 promoter sequence, then performing in vitro transcription (MEGAshortscript T7 Transcription kit (Life Technologies). Products were purified using the MEGAclear kit (Life Technologies, cat. no. AM1908). For a detailed protocol, see also Ran et al. (2013b). Microinjections into the pronucleus + cytoplasm of FVB/NJ X B6(Cg)-Tyrc-2J/J embryos were done using standard methods. For experiments involving SCR7, injected embryos were cultured in KSOM media overnight in the presence of 50 μM SCR7 (XcessBio, cat. no. M60082).
Next-Gen sequencing of barcoded multiplexed PCRs for gRNA target and off-target sites
Amplicon design: genomic regions of 350-450 bp around predicted gRNA binding sites (on target and off target; Table S1) were identified and gene specific primers including adapter sequence were designed using the BatchPrimer3 online tool. Next, the first Multiplex PCR was performed as follows: Amplicons were amplified by multiplexing PCR (≤20 amplicons together) using QIAGEN Multiplex PCR Kit (cat no. 206143) and 300 ng of genomic DNA template from each individual mouse (best in 96-well format). The 1st round PCR products were diluted 1:3 and 1ml of individual PCR was used for “barcoding” PCR using unique Illumina MID-p5/p7 index primers combinations. We used CloneID 1x PCR mix (Lucigen, cat no. 30059). Barcoded PCR products (2 μl) from each individual mouse were pooled together and were purified using the Agencourt AMPure XP PCR Purification system (cat no. A63880). The purified multi-amplicon mixture was sequenced on an Illumina MiSeq instrument and a 600 bp v3 kit (PE 2 × 300 bp). Reads sorted by barcodes were analyzed using the Geneious software package.
Primers for first Multiplex PCR:
FP: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGN(18-20)-3′ [adapter sequence (gene specific sequence)]
RP: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGN(18-20)-3′ [adapter sequence (gene specific sequence)].
Acknowledgments
This work was supported by NIH grant GM45415 to JCS and contract CO29155 from the NY State Stem Cell Program.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169771/-/DC1.
Available freely online through the author-supported open access option.
Communicating editor: O. Hobert
Literature Cited
- Aarts M., Dekker M., de Vries S., van der Wal A., te Riele H., 2006. Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res. 34: e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida T., Imahashi R., Tanaka K., 2014. Translating human genetics into mouse: the impact of ultra-rapid in vivo genome editing. Dev. Growth Differ. 56: 34–45. [DOI] [PubMed] [Google Scholar]
- Bassett A. R., Tibbit C., Ponting C. P., Liu J. L., 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betermier M., Bertrand P., Lopez B. S., 2014. Is non-homologous end-joining really an inherently error-prone process? PLoS Genet. 10: e1004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer K. J., Trautman J. K., Bozas A., Liu J. L., Rutter J., et al. , 2008. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 105: 19821–19826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer, K. J., J. K. Trautman, K. Mukherjee, and D. Carroll, 2013 Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 (Bethesda) pii: g3.112.005439v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E., Rinaldi V. D., White M. E., Schimenti J. C., 2014. Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science 343: 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R. L., Braun R. E., Lo D., Avarbock M. R., Oram F., et al. , 1989. Targeted correction of a major histocompatibility class II E alpha gene by DNA microinjected into mouse eggs. Proc. Natl. Acad. Sci. USA 86: 7087–7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N., Sun C., Gao L., Zhu D., Xu X., et al. , 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Pruett-Miller S. M., Huang Y., Gjoka M., Duda K., et al. , 2011. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8: 753–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. W., Wang H., Yang H., Shi L., Katz Y., et al. , 2013. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23: 1163–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim J. M., Kim J. S., 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31: 230–232. [DOI] [PubMed] [Google Scholar]
- Cho S. W., Kim S., Kim Y., Kweon J., Kim H. S., et al. , 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P. S., Meyerson M., 2014. Targeted genomic rearrangements using CRISPR/Cas technology. Nat. Commun. 5: 3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., et al. , 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L., Maizels N., 2014. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. USA 111: E924–E932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriano L., Roth D. B., 2013. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47: 433–455. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J. A., Khayter C., Maeder M. L., Reyon D., et al. , 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31: 822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Sander J. D., Reyon D., Cascio V. M., Joung J. K., 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W., Kawasaki K., Sugiura K., Naito K., 2013. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 41: e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W., Onuma A., Sugiura K., Naito K., 2014. Efficient generation of genome-modified mice via offset-nicking by CRISPR/Cas system. Biochem. Biophys. Res. Commun. 445: 791–794. [DOI] [PubMed] [Google Scholar]
- Gagnon J. A., Valen E., Thyme S. B., Huang P., Ahkmetova L., et al. , 2014. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 9: e98186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G., Barrangou R., Horvath P., Siksnys V., 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A., Larson M. H., Morsut L., Liu Z., Brar G. A., et al. , 2013. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin D. Y., Waite A. J., Katibah G. E., Miller J. C., Holmes M. C., et al. , 2010. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 649: 247–256. [DOI] [PubMed] [Google Scholar]
- Horii T., Arai Y., Yamazaki M., Morita S., Kimura M., et al. , 2014. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep 4: 4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P., Barrangou R., 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170. [DOI] [PubMed] [Google Scholar]
- Hou Z., Zhang Y., Propson N. E., Howden S. E., Chu L. F., et al. , 2013. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 110: 15644–15649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D., Scott D. A., Weinstein J. A., Ran F. A., Konermann S., et al. , 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31: 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A., Barrett T., Flavell R. A., Davis R. J., 2008. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol. Cell 30: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y., Fu Y., Reyon D., Maeder M. L., Tsai S. Q., et al. , 2013. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31: 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L. E., Wente S. R., Chen W., 2013. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 110: 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., et al. , 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., et al. , 2013. RNA-programmed genome editing in human cells. eLife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. M., Kim D., Kim S., Kim J. S., 2014a Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat. Commun. 5: 3157. [DOI] [PubMed] [Google Scholar]
- Kim S., Kim D., Cho S. W., Kim J., Kim J. S., 2014b Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Qiu Z., Shao Y., Chen Y., Guan Y., et al. , 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 31: 681–683. [DOI] [PubMed] [Google Scholar]
- Ma Y., Shen B., Zhang X., Lu Y., Chen W., et al. , 2014. Heritable multiplex genetic engineering in rats using CRISPR/Cas9. PLoS ONE 9: e89413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K. S., Haft D. H., Barrangou R., Brouns S. J., Charpentier E., et al. , 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P. B., Esvelt K. M., Moosburner M., et al. , 2013a CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K. M., Aach J., Guell M., et al. , 2013b RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D., Fujihara Y., Satouh Y., Miyata H., Isotani A., et al. , 2013. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3: 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashimo T., 2014. Gene targeting technologies in rats: zinc finger nucleases, transcription activator-like effector nucleases, and clustered regularly interspaced short palindromic repeats. Dev. Growth Differ. 56: 46–52. [DOI] [PubMed] [Google Scholar]
- Menke D. B., 2013. Engineering subtle targeted mutations into the mouse genome. Genesis 51: 605–618. [DOI] [PubMed] [Google Scholar]
- Nur E. K. A., Li T. K., Zhang A., Qi H., Hars E. S., et al. , 2003. Single-stranded DNA induces ataxia telangiectasia mutant (ATM)/p53-dependent DNA damage and apoptotic signals. J. Biol. Chem. 278: 12475–12481. [DOI] [PubMed] [Google Scholar]
- Papaioannou I., Simons J. P., Owen J. S., 2012. Oligonucleotide-directed gene-editing technology: mechanisms and future prospects. Expert Opin. Biol. Ther. 12: 329–342. [DOI] [PubMed] [Google Scholar]
- Qi L. S., Larson M. H., Gilbert L. A., Doudna J. A., Weissman J. S., et al. , 2013. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S., Kwaku Dad A. B., Beloor J., Gopalappa R., Lee S. K., et al. , 2014. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 24: 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R., Liu P., Bradley A., 1995. Chromosome engineering in mice. Nature 378: 720–724. [DOI] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Lin C. Y., Gootenberg J. S., Konermann S., et al. , 2013a Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154: 1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013b Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J. D., Joung J. K., 2014. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32: 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhang J., Wu H., Wang J., Ma K., et al. , 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 23: 720–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Zhang W., Zhang J., Zhou J., Wang J., et al. , 2014. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11: 399–402. [DOI] [PubMed] [Google Scholar]
- Skarnes W. C., Rosen B., West A. P., Koutsourakis M., Bushell W., et al. , 2011. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474: 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., et al. , 2014. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15: 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Nambiar M., Sharma S., Karki S. S., Goldsmith G., et al. , 2012. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 151: 1474–1487. [DOI] [PubMed] [Google Scholar]
- Sternberg S. H., Redding S., Jinek M., Greene E. C., Doudna J. A., 2014. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507: 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung Y. H., Kim J. M., Kim H. T., Lee J., Jeon J., et al. , 2014. Highly efficient gene knockout in mice and zebrafish with RNA-guided endonucleases. Genome Res. 24: 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A., Gosis B. S., Ding Q., Collins R., Ragavendran A., et al. , 2014. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang H., Shivalila C. S., Dawlaty M. M., Cheng A. W., et al. , 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Scott D. A., Kriz A. J., Chiu A. C., Hsu P. D., et al. , 2014. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Liang D., Wang Y., Bai M., Tang W., et al. , 2013. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13: 659–662. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang H., Shivalila C. S., Cheng A. W., Shi L., et al. , 2013a One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Wang H., Jaenisch R., 2014. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 9: 1956–1968. [DOI] [PubMed] [Google Scholar]
- Yang L., Guell M., Byrne S., Yang J. L., De Los Angeles A., et al. , 2013b Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 41: 9049–9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen S. T., Zhang M., Deng J. M., Usman S. J., Smith C. N., et al. , 2014. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev. Biol. 393: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K., Kaneko T., Voigt B., Mashimo T., 2014. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat. Commun. 5: 4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Y., Bergstrom R., Klemm M., Lederman B., Nelson H., et al. , 1997. Chromosomal deletion complexes in mice by radiation of embryonic stem cells. Nat. Genet. 15: 285–288. [DOI] [PubMed] [Google Scholar]
- Yu Z., Ren M., Wang Z., Zhang B., Rong Y. S., et al. , 2013. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics 195: 289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]