Abstract
The extent of chromatin compaction is a fundamental driver of nuclear metabolism . Yta7 is a chromatin-associated AAA-ATPase, the human ortholog of which, ANCCA/ATAD2 transcriptionally activates pathways of malignancy in a broad range of cancers. Yta7 directly binds histone H3, and bulk chromatin exhibits increased nucleosomal density in yta7Δ mutants. The suppression of yta7Δ mutant growth and transcriptional phenotypes in budding yeast by decreased dosage of histones H3 and H4 indicates the acute sensitivity of cells to deviations in nucleosome spacing. This study investigated the global changes in chromatin structure upon Yta7 loss or overexpression and determined which of these effects reflected direct Yta7 activity. Metagene analysis of Yta7’s genome-wide localization indicated peak binding of Yta7 just downstream of the transcription start site. Cells lacking Yta7 exhibited increased nucleosome density within genes downstream of the +1 nucleosome, as defined by decreased internucleosomal distance, resulting in progressively 5′-shifted nucleosomes within the gene. In contrast, cells overexpressing Yta7 displayed profound 3′-shifts in nucleosome position and reduced nucleosome density within genes. Importantly, Yta7-bound regions were enriched for nucleosomal shifts, indicating that Yta7 acted locally to modulate nucleosome spacing. The phenotype of cells lacking both Yta7 and Rtt106, the histone H3/H4 chaperone, indicated that Yta7 functions in both Rtt106-dependent and Rtt106-independent ways to modulate nucleosome spacing within genes. This study suggested that Yta7 affected nucleosome density throughout the gene by both blocking Rtt106 from entering the gene, as shown previously at HTA1, and facilitating the loss of nucleosomes from the 5′-end.
Keywords: Yta7, ANCCA, ATAD2, Rtt106, nucleosome positioning
THE size constraints of the nucleus necessitate condensation of eukaryotic DNA into chromatin. The fundamental subunit of chromatin is the nucleosome, 147 bp of DNA wound about the histone octamer (Luger et al. 1997). Each octamer typically contains two copies each of the canonical histones H2A, H2B, H3, and H4. All chromatin-dependent processes—transcription, replication, recombination, and repair—are affected by the position and occupancy of nucleosomes.
The YTA7 gene, conserved from yeast to humans, encodes the only bromodomain-containing AAA-ATPase in Saccharomyces cerevisiae. The human ortholog ANCCA (ATPase Nuclear Coactivator Cancer-Associated), also known as ATAD2, is a coactivator of genes controlled by both the estrogen and the androgen receptors. Increased expression of ANCCA/ATAD2 is associated with breast, prostate, lung, and endometrial cancers with poor prognoses (Zou et al. 2009; Caron et al. 2010; Kalashnikova et al. 2010; Raeder et al. 2013; Zhang et al. 2013). Studies of Yta7 in the budding yeast S. cerevisiae have the potential to shed mechanistic light on the activity of the human oncogenic ortholog. However, yeast studies on the role of Yta7 have been largely limited to analysis of a handful of genes, most notably the canonical histone genes.
In S. cerevisiae there are two copies of each canonical histone gene. H2A and H2B genes are organized into two loci, each with an H2A and H2B gene pair (HTA1-HTB1, HTA2-HTB2). Genes encoding H3 and H4 are similarly organized as two gene pairs (HHT1-HHF1, HHT2-HHF2). In asynchronously growing cells, Yta7 is highly enriched at all four canonical histone gene pairs. Interestingly, Yta7 has been described variously as a repressor of all canonical histone genes (Gradolatto et al. 2008, 2009) and, conversely, as an activator of the primary H2A gene, HTA1 (Fillingham et al. 2009). Other analyses of histone gene transcript levels in cells lacking Yta7 revealed no significant effect other than reduction of HTA1 messenger RNA (mRNA) (Lombardi et al. 2011; Zunder and Rine 2012).
Yta7 is also required for optimal induction of galactose-induced and sporulation-regulated genes. Their induction appears to be direct as Yta7 associates with the GAL gene cluster only upon transcriptional induction. Interestingly, Yta7 exhibits prominent enrichment within the 5′-end of open reading frames (ORFs) upon activation (Lombardi et al. 2011). In accordance with Yta7’s enrichment in ORFs, Yta7 interacts in vivo with Chd1 (Lambert et al. 2009, 2010), a chromatin remodeler that slides nucleosomes (Lusser et al. 2005) and promotes the transition between transcriptional initiation and elongation (Simic et al. 2003; Skene et al. 2014), and with Spt16 (Tackett et al. 2005; Lambert et al. 2009, 2010; Kurat et al. 2011), the largest subunit of the FACT complex, which facilitates transcriptional elongation by destabilizing nucleosomes (Formosa 2008). Furthermore, Yta7 contains two histone-interaction domains: a noncanonical bromodomain and an acidic N-terminal region. Although bromodomains typically bind acetylated lysines, in vitro analyses suggest that Yta7’s bromodomain preferentially interacts with an unacetylated and unmethylated N-terminal tail of histone H3 (Gradolatto et al. 2008, 2009). Direct interactions between Yta7 and other histones have not been reported. Yta7’s putative AAA-ATPase domain is of the NSF/Cdc48/Pex family, which typically form hexameric complexes and participate in unfolding or manipulating proteins (Erzberger and Berger 2006). Indeed, the human ortholog ANCCA/ATAD2 assembles into oligomers, and its ATPase activity is required for its coactivator function (Zou et al. 2007; Revenko et al. 2010). Likewise, mutating the Walker A (K460A) or Walker B (E519Q) motifs of Yta7’s AAA-ATPase domain results in severe loss of function, comparable to the null, indicating that Yta7’s AAA-ATPase is essential for its function (Kurat et al. 2011; Lombardi et al. 2011).
The sensitivity of yta7Δ mutants to changes in the dosage of histones H3 and H4 offers insight into the in vivo importance of Yta7. Specifically, yta7Δ mutants grow better when a copy of a gene encoding either H3 or H4 is deleted (Collins et al. 2007; Costanzo et al. 2010). This growth-defect suppression is not observed upon decreased dosage of the genes encoding H2A or H2B. Many of the yta7Δ mutant’s transcriptional defects are also partially suppressed by decreased dosage of histones H3 and H4 (Lombardi et al. 2011). Consistent with this suppression, we previously found that cells lacking Yta7 appear to exhibit increased levels of chromatin-incorporated histone H3, as determined by co-immunoprecipitation (ChIP) at select loci. Additionally, yta7∆ cells exhibited decreased nucleosome spacing, as assayed by micrococcal nuclease digestion of bulk chromatin. In this study, we mapped precisely those changes in chromatin structure sensitive to Yta7 loss or overexpression and determined which of these effects reflected direct Yta7 activity.
Materials and Methods
Yeast strains and media
All yeast strains were derived from W303-1a (listed in Supporting Information, Table S3). One-step integration of knockout cassettes and C-terminal epitope tags constructs was performed as described (Longtine et al. 1998; Goldstein and McCusker 1999; Puig et al. 2001). All gene disruptions were confirmed by 5′- and 3′-junction PCR of both the insertion allele and the wild-type allele. Strains containing integrated pGAL1::YTA7 were constructed by homologous insertion into YTA7’s native locus as described (Longtine et al. 1998; Lombardi et al. 2011). All epitope tagging was confirmed by immunoblotting. Yta7-TAP (Tandem Affinity Purification) provided full Yta7 function (Lombardi et al. 2011).
Micrococcal nuclease digestion
Digests were performed largely as described (Liu et al. 2005) with the following modifications. Cultures of 100 ml of cells at 0.65–0.75 OD600 were cross-linked with 1% formaldehyde for 20 min at room temperature. Cross-linking was quenched by addition of glycine to a final concentration of 300 mM. Cells were washed once in cold water and then resuspended in 4 ml/g spheroplast buffer [100 mM KPO4 (pH 7.5), 1.2 M sorbitol, 0.5 mM CaCl2, 7 mM β-mercaptoethanol] and warmed for 15 min at 37°. Spheroplasting was achieved by adding Zymolyase 100T (0.3 mg/sample) and incubating for 30 min at 37°. Spheroplasts were washed once with spheroplast buffer and then resuspended in 0.75 ml digest buffer [10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.075% NP-40, 5 mM MgCl2, 1 mM CaCl2, 1 mM β-mercaptoethanol]. The amount of micrococcal nuclease (Sigma Aldrich, N3755) was empirically determined for cells grown in each medium to ensure >90% mononucleosomal DNA following digestion. Spheroplasts in digestion buffer were prewarmed for 5 min at 37°, and then 7 units and 4.5 units of micrococcal nuclease were added to YPD and YPGal samples, respectively. Chromatin was digested for 20 min at 37°. Digestion was halted by placing samples on ice and adding EDTA to a final concentration of 10 mM.
Digestion products were obtained by collecting the supernatant after spinning the samples at 13,000 × g. Protein was digested with 800 µg proteinase K (Invitrogen) in 0.5% SDS for 2 hr at 37°. Reversal of cross-linking was performed at 65° overnight. Samples were then RNase-treated, and the DNA was extracted and quantified by Nanodrop and Qubit.
ChIP analysis
For each replicate, 600 ml of culture was grown to OD600 0.6–0.7 and then cross-linked with 1% formaldehyde for 45 min at room temperature. Cross-linking was quenched by addition of glycine to a final concentration of 300 mM. Cells were then harvested and washed twice with TBS. To facilitate lysis and sonication, the cell pellet was split into four aliquots and lysed with 0.5-mm zirconia beads in FA lysis buffer (Aparicio et al. 2005) using the MP Fastprep-24. Chromatin was isolated at 74,000 × g for 36 min and then washed in FA lysis buffer for 1 hr at 4°. Sonication yielded an average sheared DNA size of 300–350 bp.
For the Yta7-TAP immunoprecipitations (IPs) from cells grown in YPD medium, sonicated chromatin from 150 ml of culture was incubated with 30 μl of IgG Sepharose (GE Healthcare) for 1.5 hr at 4°. Resin washing, IP elution, and DNA purification were performed as described (Aparicio et al. 2005) with the four eluates from the original 600-ml culture combined. Yta7-TAP IPs from cells grown in YPGal medium, in which Yta7 was overexpressed or expressed from its native promoter, were performed as above, except the resin volume for both IPs was increased to 120 μl of IgG Sepharose, the level required to bind the majority of overexpressed Yta7. Quantitative PCR was performed on precipitated DNA using an MX3000P qPCR machine (Agilent) and the Dynamo HS SYBR Green qPCR kit (NEB) to ensure enrichment of IP/Input at known Yta7 targets.
Library construction and sequencing
Libraries were constructed using the Illumina Tru-Seq library preparation kit with the following modifications. Upon end repair, samples were purified using a Qiagen MinElute column. Adapters were used at a 50-fold diluted concentration from that provided. For MNase samples, 10 cycles of PCR were used, followed by size selection for 200–300 bp. The 12 samples were pooled for multiplexing using a quantitative PCR (qPCR) Next Generation Sequencing (NGS) Library Quantification Kit (Agilent) and run in a single lane on the Illumina HiSeq 2000 for 100-bp paired-end reads. For ChIP samples, 16 cycles of PCR were used for all amplifications, followed by size selection for 250- to 400-bp fragments. The seven IP samples and seven input samples (Glu: 1 No tag, 2 WT; Gal: 1 No tag, 2 WT, 2 Yta7oe) were multiplexed by qPCR as above and run in a single lane for 100-bp single reads.
MNase-Seq analysis
All read pairs were trimmed to remove adapter contamination. Between 11,234,119 and 22,076,125 reads were mapped to the University of California at Santa Cruz (UCSC) sacCer3 genome with the Burrows–Wheeler Aligner (BWA) v0.7.8 using the linear-time IS algorithm for BWA alignment index construction. Default parameters were used for mapping with BWA. For visualization, all samples were scaled to a total of 10,000,000 reads. The average reads per base pair for each sample were plotted relative to the transcriptional start site annotations used previously (Jiang and Pugh 2009) (Figure 1A and Figure 4, A and B). BAM files were converted to BED format and used as input to nucleosome position from sequencing (NPS) (Zhang et al. 2008b), which was used to identify between 53,290 and 56,541 nucleosome centers for each sample. Parameters for NPS were empirically optimized to maximize the annotation of visible nucleosome patterns in a genome browser. For subsequent analysis, all nucleosomes were assumed to occupy the 147 bp flanking the identified nucleosome center. Each nucleosome position relative to the transcriptional start site (e.g., −1, +1, etc.) was assigned using a model derived from the method of Jiang and Pugh (2009). Then the distribution of the nucleosome centers at each position was plotted and smoothed with a Hanning window (Oppenheim and Schafer 1989) with a bin denominator of 10 bp (compare Figure 1B to Figure S4) for ease of visualization (Figure 1B and Figure 4C). Differences between the means of the nucleosome distributions were compared across samples and assessed for significance with a t-test. Python and Bash code for read handling and all MNase-Seq analysis is included in the published codebase, as is the parameter file for NPS. Genome browser shots (Figure S2, Figure S3, and Figure S9) were made using the UCSC Genome Browser (http://genome.ucsc.edu) (Kent et al. 2002).
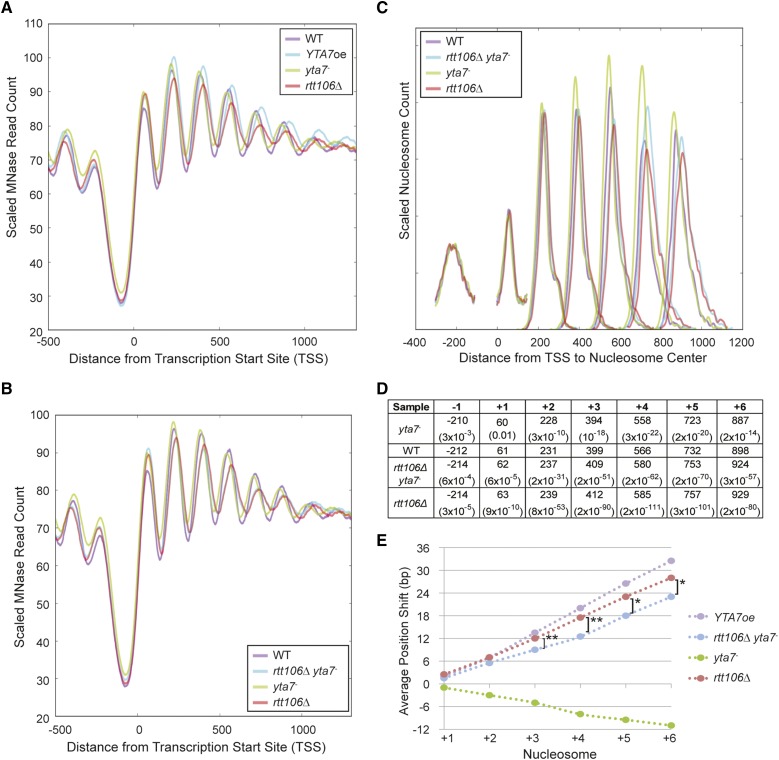
Figure 1.
Yta7 regulated nucleosome spacing within genes. (A) The distribution of MNase-Seq read counts relative to all TSS, or metagene analysis, for wild type (WT), cells not expressing Yta7 (yta7-) and cells overexpressing Yta7 (YTA7oe). yta7- cells exhibited a 5′-shift in nucleosome position, whereas YTA7oe cells exhibited a 3′-shift in nucleosome position relative to wild type. (B) The distribution of nucleosome centers relative to the TSS for called nucleosomes. MNase-seq data were used to identify nucleosome centers, and the distance of each nucleosome center from the TSS was plotted. Distances were smoothed with a Hanning window. The relative nucleosome count was normalized per position. The distribution of unsmoothed densities is shown in Figure S4. (C) Average distance of the indicated nucleosome’s center to the TSS and the associated P-value (two-tailed t-test) compared to wild type. (D) The average shift for the indicated nucleosomes in mutant compared to media-matched wild-type samples (YPD for yta7- and YPGal for YTA7oe). Nucleosome centers were identified as in C but for two biological replicates per strain. Upstream and downstream are indicated by negative and positive, respectively.
Figure 4.
Loss of Rtt106 increased nucleosome spacing. (A) The distribution of MNase-Seq read count relative to the TSS for wild type (WT), cells overexpressing Yta7 (YTA7oe), cells not expressing Yta7 (yta7-), and cells lacking Rtt106 (rtt106Δ). YTA7oe and rtt106Δ cells exhibited a 3′-shift in nucleosome position relative to wild type. (B) The distribution of MNase-Seq read count relative to the TSS for wild type (WT), cells not expressing Rtt106 or Yta7 (rtt106Δ yta7-), cells not expressing Yta7 (yta7-), and cells lacking Rtt106 (rtt106Δ). (C) The distribution of nucleosome centers relative to the TSS for called nucleosomes. MNase-seq data depicted in B were used to identify nucleosome centers, and the distance of each nucleosome center from the TSS was plotted. Distances were smoothed with a Hanning window. The relative nucleosome count was normalized per position (Table S2). (D) Average distance of the indicated nucleosome’s center to the TSS and the associated P-value (two-tailed t-test) compared to wild type. Samples are listed in 5′–3′ order of mean nucleosome position. (E) The average shift for the indicated nucleosomes in mutant compared to media-matched wild-type samples (YPD for yta7- and YPGal for YTA7oe). Nucleosome centers were identified as in D but for two biological replicates per strain. Upstream and downstream are indicated by negative and positive, respectively. Astericks indicate called nucleosome position means significantly distinct between rtt106Δ and rtt106Δ yta7-. *P < 0.001; **P < 10−6 (two-tailed t-test).
ChIP-Seq analysis
Reads were trimmed as for MNase-Seq reads. Between 3,373,008 and 11,418,304 reads were mapped as with the MNase-Seq reads, but with duplicate reads removed to purge presumptive PCR duplicates. Reads with a quality score of <37 were removed. As with MNase-Seq reads, each sample was scaled to 10,000,000 reads for visualization purposes. Yta7-bound regions were identified with MACS v1.4.2 (Zhang et al. 2008a) using the corresponding no-tag ChIP samples to generate the background model, and subpeaks were split using PeakSplitter (Salmon-Divon et al. 2010) with default parameters. For Figure 2A, the scaled read count per base pair was plotted for all loci with a summit occurring within the distances indicated in the individual figure legends. For Figure S7, the UCSC Genome Browser (Kent et al. 2002) was used as above.
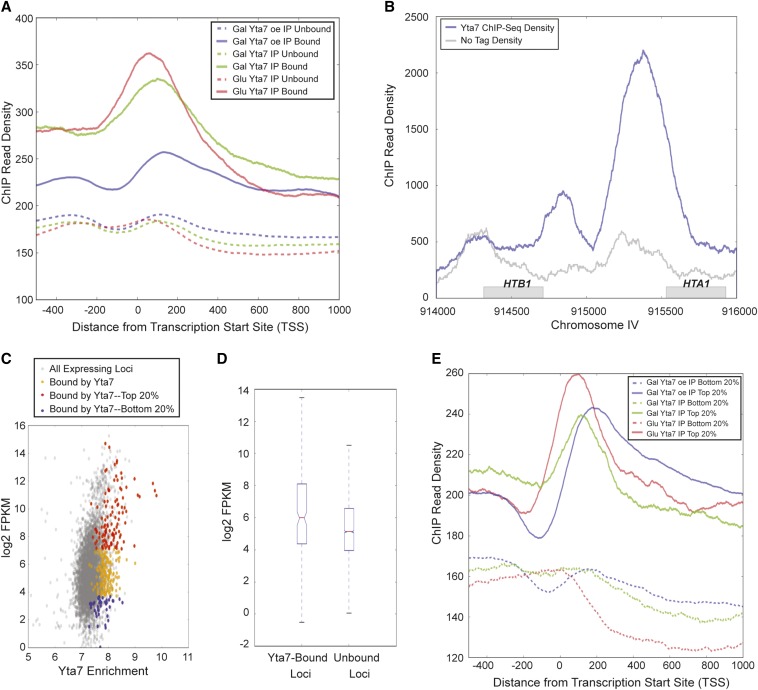
Figure 2.
Yta7 localized to the 5′-end of ORFs. (A) The distribution of Yta7 ChIP-seq reads relative to the TSS for bound and unbound loci. Bound regions were called using MACs peak caller, comparing immunoprecipitated signal between strains containing epitope-tagged Yta7 and untagged strains. Unbound regions are all other loci. (B) Yta7-TAP ChIP-seq read count compared to ChIP-seq of untagged sample (No Tag) across the HTB1-HTA1 locus. (C) A scatterplot between Yta7 ChIP enrichment (Glu Yta7 IP) and expression level of verified genes in YPD (Nagalakshmi et al. 2008). Yta7 ChIP enrichment is log2 of the mean ChIP-seq read density over a gene. (D) Box plot comparison of Yta7-bound and unbound regions as a function of log2 FPKM. (E) As in A, but for genes within the highest and lowest quintiles of expression.
RNA-Seq analysis
Gene expression was assessed using the RNA-Seq reads deposited as SRR002051.fastq in the Sequence Read Archive. These are the reads corresponding to oligo(dT) isolations intended to enrich for mRNA. Reads were mapped and Fragments Per Kilobase of transcript per Million mapped reads (FPKM) was calculated using the suite of Bowtie v2.2.2, Tophat v2.0.11, and Cufflinks v2.2.1. All loci with measurable gene expression (n = 5729) were used in Figure 2A. For Figure S8, the FPKM of Yta7-bound genes was calculated, and unbound genes with optimally similar expression were selected to create the expression-matched gene set. Some genes that were successfully annotated with nucleosome positions contained no measureable expression in the RNA-seq data set and were thus excluded from the analysis. Medians of the compared sets matched to less than one-tenth of one FPKM.
Code and code availability
All Bash and Python code used for read mapping, quality control, feature annotation (including NPS parameters and postprocessing scripts and MACS and PeakSplitter scripts), visualization, and statistical analysis is available at https://github.com/matthewddavis/lombardi. A README file is included to describe the steps required to recreate all figures, although some configuration may be required, depending on the hardware and software environment.
Results
Yta7 regulated nucleosome spacing within genes
Our previous finding indicated that bulk chromatin in cells lacking Yta7 (yta7Δ) exhibited an apparent decreased internucleosomal spacing relative to wild type. Therefore, we sought a high-resolution picture of how this bulk effect mapped to gene architecture across the genome. To this end, we performed micrococcal nuclease digestion followed by high-throughput sequencing (MNase-Seq). To assess both the loss- and gain-of-function phenotypes of YTA7, we utilized a strain with YTA7 under the control of the GAL1 promoter (pGAL1::YTA7-TAP). In this strain, YTA7 was repressed in cells grown in glucose-containing medium or overexpressed in cells grown in galactose-containing medium (designated as yta7- and YTA7oe, respectively). Yta7 protein levels were approximately sevenfold overexpressed in the galactose-grown cells and undetectable in the glucose-grown cells (our unpublished observation). Previous work established that the TAP tag had no discernible impact on Yta7 function (Lombardi et al. 2011).
Mapping the sequence reads from the mononucleosomes produced by MNase digestion with respect to the transcription start site (TSS) clearly demonstrated a 5′-shift in nucleosome position in cells not expressing YTA7 (yta7-) for nucleosomes downstream of the +1 nucleosome, as evident in the metagene analysis (Figure 1A; as heat maps, Figure S1A). Conversely, cells overexpressing YTA7 (YTA7oe) exhibited a profound 3′-shift in position for nucleosomes downstream of the +1 nucleosome (Figure S2). To quantify these shifts, nucleosomes were individually called using NPS software (Zhang et al. 2008b) to determine nucleosome centers (Figure S3). Nucleosomes were then annotated as established (Jiang and Pugh 2009) with respect to position within a given ORF, e.g., +1, and their midpoints were plotted with respect to the TSS (Figure 1B and Table S1). This analysis allowed for statistical evaluation of the changes in nucleosome center position between strains. Both the mutant and the overexpressing strains displayed increasing deviation from wild type for progressively 3′-nucleosomes, with their most salient deviation from wild-type position at the +4 and +5 nucleosomes (Figure 1 B and C). The position of the +1 nucleosome was the least significantly altered within the ORF. This increased deviation moving into the ORF was recapitulated by parallel analysis with other biological replicates (Figure S5). Thus, there appeared to be increased deviation from wild-type position moving farther into the ORF, suggesting the accumulation of an iterative offset with each nucleosome.
To test this possibility, the average shift compared to wild type for each nucleosome was plotted for cells lacking or overexpressing Yta7 (Figure 1D). In YTA7oe cells, the shift in average nucleosome position increased steadily downstream of the +1 nucleosome. The downstream shift of each nucleosome followed a nearly linear trend, indicating that downstream of the +1 nucleosome each subsequent nucleosome had an internucleosomal spacing ∼6 bp longer than in wild type. Thus, nucleosomes were less densely spaced in cells overexpressing Yta7 compared to wild type. In contrast, nucleosomes in cells lacking Yta7 exhibited upstream shifts relative to their wild-type position, reflecting a ∼2-bp decrease in internucleosome spacing. Similar to the shifts in YTA7oe cells, the magnitude of the shifts accumulated progressively farther into the gene, indicating a progressive 5′-shift due to decreased internucleosomal spacing. Thus, the lack and overproduction of Yta7 resulted in opposing progressive nucleosome shifts within ORFs, with cells lacking Yta7 exhibiting the decrease in internucleosomal spacing anticipated from our earlier bulk chromatin analyses.
Yta7 localized to the 5′-end of ORFs
To achieve a high-resolution map of Yta7’s chromatin associations and determine which nucleosomal shifts might be due to direct Yta7 activity, ChIP followed by high-throughput sequencing (ChIP-Seq) was performed. To ensure that the localization data set could be reliably compared to the MNase-Seq data, α-TAP immunoprecipitations were performed on wild-type Yta7-TAP in cells grown in both glucose and galactose media, as well as overexpressed Yta7-TAP (galactose). Yta7-enrichment peaks, or “bound” regions, were called using MACS (Zhang et al. 2008a) relative to DNA immunoprecipitated from cells in which Yta7 was not TAP-tagged. Peak calling identified 568, 560, and 732 Yta7-bound regions for wild-type Yta7 (Glu), wild-type Yta7 (Gal), and overexpressed Yta7 (Gal), respectively (Figure S6 and Figure S7). Interestingly, for all samples, the majority of peak summits fell within 450 bp of a TSS. To determine where Yta7 bound with respect to the TSS, the Yta7 ChIP read counts for bound and unbound regions were plotted relative to all TSS. In cells expressing Yta7 from the native YTA7 promoter, peak Yta7 localization occurred just downstream of the TSS [Figure 2A, glucose (Glu), solid red line, and galactose (Gal), solid green line], coincident with the position of the +1 nucleosome. Upon overexpression, Yta7’s peak localization still occurred shortly inside the ORF (Figure 2A, solid blue line), although the increased concentration of Yta7 appeared to slightly broaden its distribution in the ORF. In general, Yta7 enrichment dropped sharply moving into the ORF, but all strains exhibited moderate enrichment in the promoter (compare unbound to bound sequences from -400 to 0, Figure 2A). As a validation of the peak calling for Yta7, all the known binding sites of Yta7 at the histone genes were recovered (Figure 2B).
To investigate Yta7’s relationship with transcription, several analyses were performed. First, the extent of Yta7 binding was compared with previously published expression data represented as log2 FPKM (Nagalakshmi et al. 2008). This scatterplot clearly indicated that Yta7 bound more genes in the top quintile of expression than in the bottom quintile (Figure 2C). Second, comparison of the distributions of the log2 FPKM values for Yta7-bound and unbound loci illustrated that Yta7-bound loci had a greater mean log2 FPKM than unbound loci (Figure 2D). However, this significant increase in mean log2 FPKM (P < 1.5 * 10−12; two-tailed t-test) belied the broad distribution of transcript abundances. To assess to what extent this broad distribution might be due to false-positive bound regions, the Yta7 ChIP read count was plotted for the top and bottom quintiles of transcript abundance, independent of peak calling (Figure 2E). This analysis clearly indicated a greater immunoprecipitation read count for loci in the top quintile of expression. Taken together, these three analyses indicated a positive correspondence between Yta7 binding and expression level.
Yta7 modulated nucleosome spacing in cis
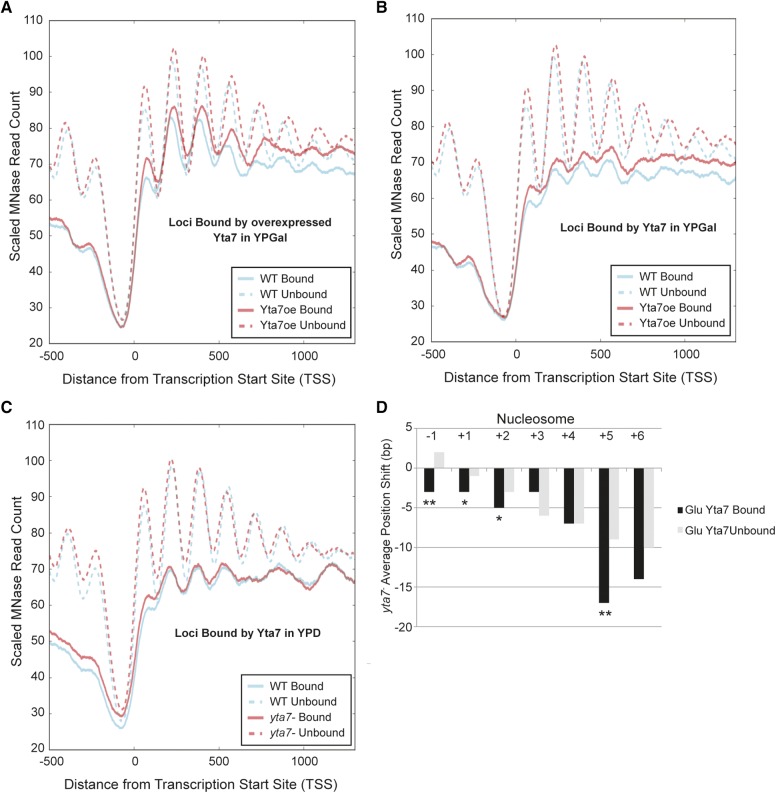
With Yta7 ChIP-seq suggesting that ∼8–11% of genes were stably bound by Yta7, the nucleosomal effects discernible at the genome-wide level could represent an indirect global effect or the signal of Yta7 affecting nucleosome position in cis at its target loci diluted by the many loci to which Yta7 did not bind. To differentiate between these possibilities, we utilized the marked 3′-shift in ORF nucleosome position exhibited in cells overexpressing Yta7 to determine if Yta7-bound regions exhibited more profound shifts than unbound regions. Comparisons of the MNase read count for wild type and YTA7oe at loci bound by overexpressed Yta7 indicated a strong enrichment for 3′ nucleosomal shifts relative to wild type at bound loci vs. unbound loci (Figure 3A). Interestingly, unbound loci retained shifts at the +5 nucleosome and downstream, suggesting the ability of overexpressed Yta7 to also affect nucleosome positioning at sites with which it was not stably associated, as measured by this assay (see Discussion). However, upon Yta7 overexpression, binding of Yta7 strongly predicted the magnitude of 3′ nucleosomal shifts with respect to the TSS. Importantly, this correlation between nucleosomal shifts and Yta7 binding was retained when considering unbound loci with matched transcriptional activity as those bound by Yta7 (Figure S8).
Figure 3.
Yta7 modulated nucleosome spacing in cis. (A) The distribution of MNase-Seq read count relative to the TSS for wild type (WT) and cells overexpressing Yta7 (YTA7oe) for regions bound and unbound by overexpressed Yta7 in YPGal. (B) As A, but at regions bound and unbound by normally expressed Yta7 in YPGal. (C) The distribution of MNase-Seq read count relative to the TSS for wild type and cells not expressing Yta7 (yta7-) for regions bound and unbound by Yta7 in YPD. (D) For regions bound and unbound by Yta7 in YPD, the difference in mean nucleosome position between wild type and yta7- . *P ≤ 0.05; **P ≤ 0.001 (bootstrap simulation, one-tailed).
To address whether Yta7 overexpression might be artificially supporting a relationship between Yta7 binding and nucleosome displacement, the enrichment for 3′-shifts in the YTA7oe strain relative to wild type was determined at loci bound by Yta7 expressed from its native promoter in the same media condition as overexpression (YPGal). This analysis likewise demonstrated a 3′-shift upon Yta7 overexpression greater at Yta7-bound loci compared to unbound loci, indicating that the correlation between nucleosomal shifts and Yta7 binding was retained when considering loci bound by endogenous expression levels of Yta7 (Figure 3B). Cells lacking Yta7 demonstrated an opposing 5′-shift (Figure 1, A and B) of a lesser magnitude than the 3′-shift displayed by the cells overexpressing Yta7. However, even this lower-magnitude shift was discernibly and significantly enriched between bound and unbound loci (Figure 3, C and D). Overall, these data suggested that Yta7 modulated nucleosome spacing downstream of the +1 nucleosome in cis and had a lesser, although detectable, effect on loci not detectably bound by Yta7.
Yta7 antagonized the H3/H4 chaperone Rtt106
Rtt106 is a chaperone for newly synthesized H3/H4 histones (Li et al. 2008; Clemente-Ruiz et al. 2011; Zunder et al. 2012) and also controls the transcription of histone genes by binding to the regulatory regions of these genes (Fillingham et al. 2009; Ferreira et al. 2011; Zunder and Rine 2012). As the localization of Rtt106 is increased at the canonical histone genes in cells lacking Yta7, it has been posited that Yta7 may inhibit Rtt106 binding to these genes. (Fillingham et al. 2009; Zunder and Rine 2012). Interestingly, Rtt106’s association with histone H3 increases in cells lacking Yta7 (Zunder and Rine 2012), although it is unclear if this increased chaperone–H3 interaction is specific to Rtt106 or is broadly true of H3/H4 chaperones. To probe the functional relationship between Yta7 and Rtt106, MNase-seq was also performed on isogenic rtt106Δ and rtt106Δ yta7- double mutants. rtt106Δ strains exhibited decreased nucleosomal density, as detected by progressive 3′ nucleosomal shifts downstream of the +1 nucleosome (Figure 4A). Plotting the average nucleosome shift in rtt106Δ mutants compared to wild type for ORF nucleosomes +1 through +6 established that the internucleosomal distance is ∼5 bp longer in cells lacking Rtt106 (Figure 4E). Thus, interestingly, cells lacking Rtt106 exhibited decreased nucleosome density despite having increased histone transcript levels (Fillingham et al. 2009; Ferreira et al. 2011; Kurat et al. 2011; Zunder and Rine 2012). This decrease in nucleosome density would be consistent with the loss of H3/H4 deposition expected in a chaperone mutant, with the increased histone gene transcription a likely compensatory response.
Strikingly, the rtt106Δ mutant’s ORF nucleosome distribution tightly paralleled the distribution in cells overexpressing Yta7 (Figure 4, A and E), consistent with Yta7 inhibiting Rtt106 function. If Yta7 functioned solely to inhibit Rtt106, the rtt106Δ and the rtt106Δ yta7- double mutants should theoretically be indistinguishable. Alternatively, if their activities were independent, the phenotype of the rtt106Δ yta7- double mutants should be additive. Given that the yta7- and rtt106Δ single mutants displayed opposite nucleosomal-shift phenotypes (5′ and 3′, respectively), the phenotype of the rtt106Δ yta7- double mutants in this scenario would result in a mild 3′-shift. However, the phenotype of the rtt106Δ yta7- double mutants was a partially rescued version of the rtt106Δ single-mutant phenotype (Figure 4, B, C, and E). When comparing nucleosome positions of mutants to wild type, the shifts exhibited by the rtt106Δ yta7- double mutant were not as 3′ as those of the rtt106Δ single mutant (Figure 4D and Figure S9). To determine if the phenotype of the rtt106Δ yta7- double mutant was statistically distinct from the rtt106Δ single mutant, the nucleosome center distributions of the two strains were compared. The distributions of the +3, +4, +5, and +6 nucleosomes of the rtt106Δ yta7- double mutant were statistically distinct from the rtt106Δ single mutant (P < 10−6, 10−8, 0.0005, and 0.001, respectively, two-tailed t-test; Figure 4E). Thus, the phenotype of the rtt106Δ yta7- double mutant was distinguishable from the rtt106Δ single mutant, but there was a greater requirement for Yta7 in regulating nucleosome position when Rtt106 was present in cells than when Rtt106 was deleted. Therefore, Yta7’s effects on nucleosome density apparently occurred both through inhibition of Rtt106 and through a function(s) independent of Rtt106.
Discussion
Profound 3′-shifts upon Yta7 overexpression
The increase in nucleosome density previously observed in bulk chromatin from cells lacking Yta7 (Lombardi et al. 2011) was recapitulated genome-wide by MNase-Seq methods, allowing these changes to be mapped onto gene architecture. Increasing 5′-shifts of ORF nucleosomes moving downstream of the +1 nucleosome, as occurred in yta7- mutants, also occur in mutants of known chromatin remodelers: isw1, chd1, swi/snf, and ino80 mutants (Yen et al. 2012; van Bakel et al. 2013). Impressively, overexpressing Yta7 resulted in nucleosomal shifts, as reflected by decreased nucleosome density, of a similar magnitude as those obtained by deleting an H3/H4 histone chaperone (Rtt106). As assessed by GAL induction, boundary function, and Rtt106 inhibition, the ATPase function of Yta7 is critical to its in vivo function (Kurat et al. 2011; Lombardi et al. 2011) and is presumably the source of energy behind Yta7’s capacity to affect chromatin remodeling. The progressive 3′ nucleosomal shifts downstream of the +1 nucleosome, opposite the phenotype of yta7- mutants, indicated an increased internucleosomal distance of ∼6 bp, resulting in a cumulative offset of ∼33 bp at the +6 nucleosome.
In principle, it was possible that this observed decrease in nucleosome density in cells overexpressing Yta7 was due to the previously noted ∼30% decrease in total H3 levels in the YTA7oe mutant (Lombardi et al. 2011). However, the current consensus is that a decrease in H3 level alone does not affect nucleosome position in vivo, but rather decreases nucleosome occupancy (Celona et al. 2011; Gossett and Lieb 2012). Moreover, cells lacking Rtt106 exhibited a decrease in nucleosome density similar to cells overexpressing Yta7, yet do not display a significant decrease in H3 levels (Zunder and Rine 2012). Because loci bound by Yta7 demonstrated a marked enrichment in nucleosomal shifts above those observable genome-wide, the simplest view was that Yta7 was capable of perpetrating these shifts by local action in cis. Thus, the decreased nucleosome density observed upon Yta7 overexpression may be due to local decreases in histone H3 levels, and hence nucleosome density, caused by Yta7 activity.
Maintaining a balance between Rtt106 and Yta7
The decreased nucleosome density of the rtt106∆ mutant was consistent with the loss of H3/H4 deposition predicted in a chaperone mutant. Indeed, cryptic transcription, which is associated with decreased nucleosome shielding within ORFs, has been observed in cells lacking Rtt106, as well as other H3/H4 chaperones (Imbeault et al. 2008; Silva et al. 2012). Importantly, this decrease in nucleosome density occurred despite an increase in histone transcript levels (Fillingham et al. 2009; Ferreira et al. 2011; Kurat et al. 2011; Zunder and Rine 2012), indicating a critical distinction between chromatin compaction and histone gene transcription.
The phenotypic similarity between cells without the H3/H4 histone chaperone Rtt106 and cells overexpressing Yta7 was consistent with previous proposals of inhibition of Rtt106 by Yta7. The potential interaction between these two proteins was addressed by double-mutant analysis. Formally, there were three possible relationships between Rtt106 and Yta7 with respect to the control of nucleosome spacing that could explain their opposite mutant phenotypes. In one possibility, as mentioned previously, Yta7 functions only to inhibit Rtt106, which itself promotes closer nucleosome spacing due to its chaperone activity. In this case, the double mutant would exhibit the spacing of the rtt106∆ mutant. A second possibility is that Rtt106 inhibits the activity of Yta7, which promotes larger internucleosome spacing. In that case, the double mutant would be expected to have the closer spacing of the yta7- single mutant. The third possibility is that Rtt106 and Yta7 function independently to control nucleosome spacing. In that case, the double mutant would be expected to have an intermediate spacing between that of either single mutant. Upon testing, the rtt106Δ yta7- double mutant exhibited a phenotype consistent with the first and third possibilities, as it was very similar to the rtt106∆ single mutant yet statistically distinct from it. Thus, rtt106∆ was not entirely epistatic to yta7- as would be predicted in a pure model of Rtt106 inhibition by Yta7. The loss of Yta7 from cells lacking Rtt106 did result in 5′ nucleosomal shifts (relative to the rtt106∆ mutant), but of a lesser magnitude than in cells containing Rtt106. Thus, the phenotype of yta7- cells was likely due to both Rtt106-dependent and Rtt106-independent roles of Yta7.
Action at a distance
Genome-wide analysis indicated that Yta7’s peak enrichment occurred at the 5′-end of ORFs and exhibited a positive correspondence with gene expression level. Yta7’s apparent peak localization at the 5′-ends of ORFs is supported by Yta7’s previously determined in vivo interaction partners: Spt16 (Tackett et al. 2005; Lambert et al. 2009, 2010; Kurat et al. 2011), the largest subunit of the FACT complex, which facilitates elongation by destabilizing nucleosomes (Formosa 2008), and Chd1 (Lambert et al. 2009; Lambert et al. 2010), a nucleosome remodeler that promotes the transition between transcriptional initiation and elongation (Simic et al. 2003; Lusser et al. 2005; Skene et al. 2014). Furthermore, the positive correlation between gene transcript levels and Yta7 localization (Figure 2, C–E) bore out the previous findings of Yta7’s physical interaction with PolII (Tackett et al. 2005; Kurat et al. 2011) and Yta7’s localization to induced genes upon activation (Lombardi et al. 2011). Additionally, the enrichment of nucleosomal shifts in yta7- or YTA7oe mutants at loci determined to be bound by Yta7 indicates that Yta7 localization analysis successfully predicts functional sites.
Recent studies indicate artifactual ChIP enrichment of many diverse proteins, including nuclear-localized GFP, at highly transcribed genes (Park et al. 2013; Teytelman et al. 2013). This artifact is greatly minimized by comparing tagged and untagged IP samples, rather than IP to input (Park et al. 2013). Thus, although not immune to this bias of increased ChIPability at highly transcribed genes, the Yta7 peaks determined were generated using the stricter criteria of Yta7-TAP IP compared to untagged IP. Moreover, artifactual ChIP-seq signals at highly expressed genes are relatively uniform throughout the transcribed region and lack the pronounced 5′ bias shown here (Park et al. 2013; Teytelman et al. 2013).
Given that Yta7 localization predicted nucleosome-shift magnitude, it follows that Yta7 activity at these loci modulated their nucleosome spacing. How Yta7 is utilizing its AAA-ATPase activity at the 5′-end of ORFs to affect these changes throughout the ORF is unclear. Given that double-mutant analysis suggested that some of these shifts were mediated in a Rtt106-dependent manner, Yta7 may be directly inhibiting Rtt106’s ORF association as at the canonical histone genes (Fillingham et al. 2009; Kurat et al. 2011; Zunder and Rine 2012). Specifically, Yta7’s localization at the 5′-end of HTA1 inhibits Rtt106’s association with the ORF (Fillingham et al. 2009). Interestingly, the amount of H3 associated with Rtt106 also increases in cells lacking Yta7 (Zunder and Rine 2012). Therefore, it is possible that Rtt106 interacts more with chromatin in cells lacking Yta7. Specifically, as in the example of HTA1, in Yta7’s absence, Rtt106 may become enriched at loci where Yta7 would normally be positioned. Furthermore, although Rtt106’s role in replication-coupled nucleosome assembly has been well characterized, Rtt106 also interacts with the replication-independent H3/H4 chaperone complex HIRA (Fillingham et al. 2009) and is recruited to highly transcribed ORFs (Imbeault et al. 2008). In fact, Rtt106 is required for new histone deposition at highly transcribed genes (Imbeault et al. 2008). Thus, Yta7’s presence at the 5′-end of ORFs may limit Rtt106’s ability to incorporate H3/H4 upon transcription, resulting in the observed decreased density of nucleosomes upon Yta7 overexpression. However, given that nucleosomal shifts are also observed at ORFs not bound by Yta7, it is clear that either Yta7’s enzymatic activity promotes transient interactions that we lacked the sensitivity to detect or that Yta7 can also inhibit Rtt106 when off of chromatin.
Yta7’s Rtt106-independent modulation of nucleosome spacing throughout the ORF from the 5′-end suggests a link to transcription-coupled retrograde movement of nucleosomes. Specifically, Yta7 occupies a location in the ORF where “old” nucleosomes on transcribed genes eventually end up due to transcription-mediated retrograde nucleosome movement (Weiner et al. 2010; Radman-Livaja et al. 2011). One way that Yta7 could control nucleosome density throughout a gene from its 5′ position would be to promote the loss of these old nucleosomes as they are passed behind RNA polymerase II. Yta7 could promote loss of old nucleosomes directly or by modifying the activity of Spt16 and Chd1. In this model, if Yta7 were absent, nucleosomes would be packed more densely in the ORF or, if Yta7 were overexpressed, lost more efficiently. Taken together, we posit a model in which Yta7 uses its ability to hydrolyze ATP and bind H3 to maintain nucleosome ORF balance by inhibiting Rtt106’s incorporation of H3/H4 and facilitating H3/H4 eviction, possibly as nucleosomes are passed 5′ in the ORF by the retrograde movement coupled to transcription. Thus, direct inhibition of Rtt106 and facilitation of H3/H4 eviction would explain both the Rtt106-dependent and Rtt106-independent aspects of Yta7’s function.
Acknowledgments
For fruitful discussions and support, we thank Mike Botchan, Barbara Meyer, Robert Fisher, and Jay Hollick. We also thank Rachel Zunder, Deborah Thurtle, and Steven A. Baker for technical advice and critiques of this work. This work was supported by National Institutes of Health grants GM31105 and T32 GM 007232.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.168039/-/DC1.
Sequences have been deposited in the National Center for Biotechnology Information Short Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRP044112.
Communicating editor: A. Gasch
Literature Cited
- Aparicio, O., J. V. Geisberg, E. Sekinger, A. Yang, Z. Moqtaderi et al., 2005 Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. Chapter 21: Unit 21. 23. [DOI] [PubMed] [Google Scholar]
- Caron C., Lestrat C., Marsal S., Escoffier E., Curtet S., et al. , 2010. Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene 29: 5171–5181. [DOI] [PubMed] [Google Scholar]
- Celona B., Weiner A., Di Felice F., Mancuso F. M., Cesarini E., et al. , 2011. Substantial histone reduction modulates genomewide nucleosomal occupancy and global transcriptional output. PLoS Biol. 9: e1001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente-Ruiz M., Gonzalez-Prieto R., Prado F., 2011. Histone H3K56 acetylation, CAF1, and Rtt106 coordinate nucleosome assembly and stability of advancing replication forks. PLoS Genet. 7: e1002376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. R., Miller K. M., Maas N. L., Roguev A., Fillingham J., et al. , 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810. [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger J. P., Berger J. M., 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35: 93–114. [DOI] [PubMed] [Google Scholar]
- Ferreira M. E., Flaherty K., Prochasson P., 2011. The Saccharomyces cerevisiae histone chaperone Rtt106 mediates the cell cycle recruitment of SWI/SNF and RSC to the HIR-dependent histone genes. PLoS ONE 6: e21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J., Kainth P., Lambert J. P., van Bakel H., Tsui K., et al. , 2009. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell 35: 340–351. [DOI] [PubMed] [Google Scholar]
- Formosa T., 2008. FACT and the reorganized nucleosome. Mol. Biosyst. 4: 1085–1093. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Gossett A. J., Lieb J. D., 2012. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet. 8: e1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A., Rogers R. S., Lavender H., Taverna S. D., Allis C. D., et al. , 2008. Saccharomyces cerevisiae Yta7 regulates histone gene expression. Genetics 179: 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradolatto A., Smart S. K., Byrum S., Blair L. P., Rogers R. S., et al. , 2009. A noncanonical bromodomain in the AAA ATPase protein Yta7 directs chromosomal positioning and barrier chromatin activity. Mol. Cell. Biol. 29: 4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault D., Gamar L., Rufiange A., Paquet E., Nourani A., 2008. The Rtt106 histone chaperone is functionally linked to transcription elongation and is involved in the regulation of spurious transcription from cryptic promoters in yeast. J. Biol. Chem. 283: 27350–27354. [DOI] [PubMed] [Google Scholar]
- Jiang C., Pugh B. F., 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 10: R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalashnikova E. V., Revenko A. S., Gemo A. T., Andrews N. P., Tepper C. G., et al. , 2010. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 70: 9402–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C. F., Lambert J. P., van Dyk D., Tsui K., van Bakel H., et al. , 2011. Restriction of histone gene transcription to S phase by phosphorylation of a chromatin boundary protein. Genes Dev. 25: 2489–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. P., Mitchell L., Rudner A., Baetz K., Figeys D., 2009. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol. Cell. Proteomics 8: 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. P., Fillingham J., Siahbazi M., Greenblatt J., Baetz K., et al. , 2010. Defining the budding yeast chromatin-associated interactome. Mol. Syst. Biol. 6: 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhou H., Wurtele H., Davies B., Horazdovsky B., et al. , 2008. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134: 244–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. L., Kaplan T., Kim M., Buratowski S., Schreiber S. L., et al. , 2005. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 3: e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi L. M., Ellahi A., Rine J., 2011. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc. Natl. Acad. Sci. USA 108: E1302–E1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J., 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Lusser A., Urwin D. L., Kadonaga J. T., 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat. Struct. Mol. Biol. 12: 160–166. [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., et al. , 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim A. V., Schafer R. W., 1989. Discrete-Time Signal Processing. Prentice-Hall, Englewood Cliffs, NJ. [Google Scholar]
- Park D., Lee Y., Bhupindersingh G., Iyer V. R., 2013. Widespread misinterpretable ChIP-seq bias in yeast. PLoS ONE 8: e83506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., et al. , 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Radman-Livaja M., Verzijlbergen K. F., Weiner A., van Welsem T., Friedman N., et al. , 2011. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 9: e1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeder M. B., Birkeland E., Trovik J., Krakstad C., Shehata S., et al. , 2013. Integrated genomic analysis of the 8q24 amplification in endometrial cancers identifies ATAD2 as essential to MYC-dependent cancers. PLoS ONE 8: e54873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenko A. S., Kalashnikova E. V., Gemo A. T., Zou J. X., Chen H. W., 2010. Chromatin loading of E2F-MLL complex by cancer-associated coregulator ANCCA via reading a specific histone mark. Mol. Cell. Biol. 30: 5260–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M., Dvinge H., Tammoja K., Bertone P., 2010. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A. C., Xu X., Kim H. S., Fillingham J., Kislinger T., et al. , 2012. The replication-independent histone H3–H4 chaperones HIR, ASF1, and RTT106 co-operate to maintain promoter fidelity. J. Biol. Chem. 287: 1709–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R., Lindstrom D. L., Tran H. G., Roinick K. L., Costa P. J., et al. , 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22: 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene P. J., Hernandez A. E., Groudine M., Henikoff S., 2014. The nucleosomal barrier to promoter escape by RNA polymerase II is overcome by the chromatin remodeler Chd1. eLife 3: e02042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett A. J., Dilworth D. J., Davey M. J., O’Donnell M., Aitchison J. D., et al. , 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169: 35–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman L., Thurtle D. M., Rine J., van Oudenaarden A., 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc. Natl. Acad. Sci. USA 110: 18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H., Tsui K., Gebbia M., Mnaimneh S., Hughes T. R., et al. , 2013. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 9: e1003479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A., Hughes A., Yassour M., Rando O. J., Friedman N., 2010. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 20: 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K., Vinayachandran V., Batta K., Koerber R. T., Pugh B. F., 2012. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., et al. , 2008a Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Shin H., Song J. S., Lei Y., Liu X. S., 2008b Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics 9: 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sun Y., Li Y., Fang Z., Wang R., et al. , 2013. ANCCA protein expression is a novel independent poor prognostic marker in surgically resected lung adenocarcinoma. Ann. Surg. Oncol. 20(Suppl. 3): S577–S582. [DOI] [PubMed] [Google Scholar]
- Zou J. X., Revenko A. S., Li L. B., Gemo A. T., Chen H. W., 2007. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. USA 104: 18067–18072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. X., Guo L., Revenko A. S., Tepper C. G., Gemo A. T., et al. , 2009. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 69: 3339–3346. [DOI] [PubMed] [Google Scholar]
- Zunder R. M., Rine J., 2012. Direct interplay among histones, histone chaperones, and a chromatin boundary protein in the control of histone gene expression. Mol. Cell. Biol. 32: 4337–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder R. M., Antczak A. J., Berger J. M., Rine J., 2012. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc. Natl. Acad. Sci. USA 109: E144–E153. [DOI] [PMC free article] [PubMed] [Google Scholar]