Abstract
Genetic interactions can strongly influence the fitness effects of individual mutations, yet the impact of these epistatic interactions on evolutionary dynamics remains poorly understood. Here we investigate the evolutionary role of epistasis over 50,000 generations in a well-studied laboratory evolution experiment in Escherichia coli. The extensive duration of this experiment provides a unique window into the effects of epistasis during long-term adaptation to a constant environment. Guided by analytical results in the weak-mutation limit, we develop a computational framework to assess the compatibility of a given epistatic model with the observed patterns of fitness gain and mutation accumulation through time. We find that a decelerating fitness trajectory alone provides little power to distinguish between competing models, including those that lack any direct epistatic interactions between mutations. However, when combined with the mutation trajectory, these observables place strong constraints on the set of possible models of epistasis, ruling out many existing explanations of the data. Instead, we find that the data are consistent with a “two-epoch” model of adaptation, in which an initial burst of diminishing-returns epistasis is followed by a steady accumulation of mutations under a constant distribution of fitness effects. Our results highlight the need for additional DNA sequencing of these populations, as well as for more sophisticated models of epistasis that are compatible with all of the experimental data.
Keywords: diminishing returns, epistasis, experimental evolution
A central feature of evolutionary adaptation is that the space of potential innovations can vary with the evolutionary history of a population. Examples are common in the microbial world: the ability to import a nutrient may be beneficial only if a mechanism has evolved to utilize it (Quandt et al. 2014), while a previously advantageous drug resistance mutation can be rendered obsolete by the acquisition of a second resistance allele (Weinreich et al. 2006). This capacity for evolutionary feedback is quantified in terms of epistasis, which measures how the effect of a mutation depends on the genetic background in which it arises. In principle, epistasis can lead to widespread historical contingency and can fundamentally alter the dynamics of adaptation (Wright 1932; Gould 1989). But in practice, the long-term evolutionary impact of epistasis remains largely uncharacterized.
Empirical patterns of epistasis are most commonly measured using a direct approach (see de Visser and Krug 2014 for a recent review). Candidate mutations are introduced into a set of genetic backgrounds via crossing or other genetic reconstruction techniques, and the fitnesses of the reconstructed genotypes are measured using competitive fitness assays or related proxies. These data yield a functional relationship between the fitness effect of a mutation and its genetic background, with the traditional pairwise epistasis emerging as a special case when the backgrounds are single mutants. We use the term microscopic epistasis to refer to these measurements, since they track the background dependence of individual mutations. Of course, the precise background dependence of any given mutation is essentially an empirical matter: the fitness effects depend on the biological details of the organism, its environment, and the identities of the mutations themselves. Empirical estimates of these quantities therefore provide valuable insight into the physiological and biophysical properties of the organism (Segrè et al. 2005; St. Onge et al. 2007; Costanzo et al. 2010; Kinney et al. 2010).
With enough reconstructions, one can also obtain information about the larger-scale structure of the fitness landscape. For example, one can assess whether interactions between mutations are generally antagonistic or synergistic (Jasnos and Korona 2007) or estimate the prevalence of sign epistasis (Weinreich et al. 2006) and overall levels of modularity (Segrè et al. 2005). These questions are typically quantified using statistical summaries of the microscopic epistasis (e.g., the distribution of pairwise epistasis values), which are aggregated over a large ensemble of mutations and genetic backgrounds. While the biological interpretation of these statistics is sometimes unclear, they can in principle exhibit regular and generalizable patterns. This makes them potentially amenable to comparison with simple fitness landscape models, such as Fisher’s geometrical model (Fisher 1930), the uncorrelated landscape (Kingman 1978; Kauffman and Levin 1987), and the NK landscape (Kauffman and Weinberger 1989). Yet while these statistics are designed to capture global properties of the fitness landscape, they are still fundamentally microscopic in nature, since they can be estimated only from microscopic (i.e., reconstruction-based) measurements. As such, they suffer from the same throughput limitations as any other reconstruction-based method, and one can focus only on a small subset of possible genotypes.
However, an evolving population typically explores a much larger number of genotypes than are feasible to reconstruct experimentally. The evolutionary dynamics depend on the entire distribution of fitness effects (“the DFE”) and on how this distribution varies among different genetic backgrounds. We denote this background-dependent DFE by the fraction of mutations with fitness effect s in genetic background In contrast to the statistics above, the background dependence of the DFE is a macroscopic form of epistasis, since it includes no information about the background dependence of any individual mutation. Like many macroscopic quantities, there is not a one-to-one correspondence between the underlying microscopic epistasis and its macroscopic manifestation. For example, one can imagine a scenario where epistasis changes the identities of beneficial mutations after every substitution, but in a way that preserves the overall shape of the DFE. In this case, the widespread patterns of microscopic epistasis are masked at the macroscopic level, and the dynamics of adaptation will be indistinguishable from a nonepistatic scenario. At the opposite extreme, the DFE can change even without any microscopic epistasis once selection starts to deplete the finite supply of beneficial mutations. In this case, the dynamics of adaptation will show signatures of macroscopic epistasis even though there are no direct interactions between mutations.
Despite the potential importance of macroscopic epistasis in evolutionary adaptation, it remains less well characterized than its microscopic counterpart. In principle, it is possible to measure the background dependence of the DFE directly, by assaying the fitness of large libraries of random mutants (Silander et al. 2007; MacLean et al. 2010; Miller et al. 2011; Bank et al. 2014). However, such studies suffer from throughput limitations similar to those of the microscopic approach above. These throughput limitations are compounded by the fact that the most important changes in the DFE, from an evolutionary perspective, are often located in difficult-to-sample regions such as the high-fitness tail (Good et al. 2012).
To avoid these issues, a number of studies have focused on the evolutionary outcomes themselves, associating observed differences in the adaptability of different strains with differences in the underlying DFE (Burch and Chao 2000; Silander et al. 2007; Barrick et al. 2010; Woods et al. 2011; Kryazhimskiy et al. 2012, 2014; Perfeito et al. 2014). In principle, this approach offers the greatest sensitivity for detecting relevant differences in the DFE among related genetic backgrounds. However, it does so by transforming the measurement into an inverse problem: the patterns of macroscopic epistasis must ultimately be inferred from the dynamics of a few observable quantities (e.g., changes in fitness over time or across experimental treatments), which depend on the complex population genetics of an evolving microbial population (Lang et al. 2013; Frenkel et al. 2014). Thus, while it is easy to demonstrate the existence of macroscopic epistasis with this approach, it is difficult to associate the observed differences in adaptability with the precise changes in the underlying DFE. This, in turn, has made it hard to distinguish between competing models of epistasis when interpreting the results of these evolution experiments (Frank 2014; Kryazhimskiy et al. 2014).
In this article, we propose a general framework for quantifying patterns of macroscopic epistasis from observed differences in adaptability. We then use this framework to investigate the role of epistasis in a well-studied laboratory evolution experiment in Escherichia coli (Wiser et al. 2013). By analyzing the differences in the dynamics of adaptation through time, we can make inferences about the changes in the DFE that have accumulated over the course of the experiment. These changes constitute the most basic form of epistasis that arises during adaptation to a constant environment. Similar to Kryazhimskiy et al. (2009), we focus on two simple summaries of the dynamics: the competitive fitness and the total number of genetic changes relative to the ancestor. We use a combination of theory and numerical simulations to investigate how well these data are explained by several popular models of macroscopic epistasis, including the recently proposed diminishing-returns model of Wiser et al. (2013). We find that fitness measurements alone have little power to discriminate between different models of epistasis, while the addition of genetic information is sufficient to rule out many existing explanations of the data. Together, these results highlight the need for more sophisticated models of macroscopic epistasis that are compatible with all of the experimental data, as well as additional DNA sequence data to test their predictions.
Results
Fitness and mutation trajectories in the long-term evolution experiment
In the long-term evolution experiment (LTEE) conducted by Lenski and collaborators, 12 populations of E. coli were founded from a single common ancestor (Lenski et al. 1991) and propagated in a constant environment for >60,000 generations (see Wiser et al. 2013 for a recent summary of experimental details). A central observable quantity is the fitness of the evolved populations, which can be measured using competition assays with a marked ancestor. If fi and ff denote the frequencies of the evolved strain at the beginning and end of the competition assay, then the (log-)fitness, X, is given by
| (1) |
where Δt is the duration of the competition in generations. Note that this definition of fitness differs from the traditional measure W reported in previous studies of the LTEE. Although the two measures are correlated, Equation 1 provides a more direct connection to the population genetic theory described in the following sections.
Using the fitness assays reported in Wiser et al. (2013), we calculated the fitness defined by Equation 1 for each population at ∼40 time points during the first 50,000 generations of evolution (see Appendix). We plot the fitness trajectories for the six complete, nonmutator populations in Figure 1A. Measurement error estimated from replicate assays is substantial (SE ∼ 3%, Figure 1C), leaving us with little power to distinguish fluctuations in individual trajectories. Instead, we pool all six populations and focus on the average fitness trajectory (Figure 1B). Bootstrap resampling from the errors in Figure 1C suggests that the measurement error in is smaller (SE ∼ 1%) and more normally distributed (Figure 1D). However, even for the average fitness trajectory, the fluctuations between neighboring time points still fall within experimental uncertainty, so we can obtain robust inferences only from long-term trends in the data.
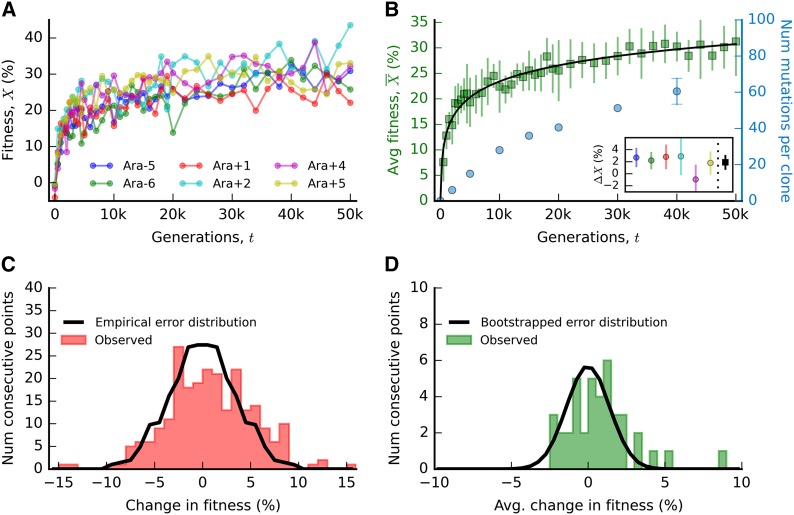
Figure 1.
Fitness and mutation trajectories in the LTEE. (A) Individual fitness trajectories for the six complete, nonmutator populations analyzed by Wiser et al. (2013). Each point is the average of two independent competition assays, with fitness estimated from Equation 1. (B) The average fitness trajectory for the six populations in A (green squares), with ±2 SE confidence intervals. For comparison, the solid line depicts the logarithmic trajectory in Equation 2, with estimated parameters Xc ≈ 4.6 × 10−2 and v0 ≈ 7.7 × 10−4. The blue circles depict the average number of mutations in single clones sampled from the LTEE (see Appendix), with ±2 SE confidence intervals for time points with more than one sampled population. Inset shows independent measurements of the change in fitness, ΔX, between generations 40,000 and 50,000 for the six populations in A (left, circles) and their average (right, square), with ±2 SE confidence intervals. (C) The change in fitness between consecutive time points, pooled across all six populations in A. The black curve shows the empirical distribution of measurement errors, defined as half of the difference between replicate fitness measurements. (D) The average change in fitness between consecutive time points. The black curve shows the bootstrapped distribution of measurement errors, obtained by repeatedly averaging six randomly chosen errors from the empirical distribution in C. The standard deviation of this distribution is
The most striking trend is the pronounced slowdown in the rate of adaptation during the course of the experiment: nearly two-thirds of the total fitness occurred during the first 5000 generations of evolution. This deceleration is inconsistent with a constant DFE, which would predict that the average fitness increases linearly with time. Instead, the slowdown in the rate of adaptation has long been interpreted as a signature of diminishing-returns epistasis, consistent with the approach to a fitness plateau (Lenski and Travisano 1994). Previous work has argued that the shape of the deceleration is best captured by a logarithmic fitness trajectory,
| (2) |
where v0 gives the initial rate of fitness increase and Xc controls the severity of the slowdown (Sibani et al. 1998; Kryazhimskiy et al. 2009; Wiser et al. 2013); the best-fit parameters are shown in Figure 1B. The shape of this trajectory, in combination with more precise measurements of the change in fitness between generations 40,000 and 50,000 (ΔX, see inset in Figure 1B), has been used to argue that fitness is still increasing in the LTEE (Wiser et al. 2013), rather than asymptoting to a fitness peak (Lenski and Travisano 1994).
More recent work has tried to use the shape of the fitness trajectory to make inferences about the underlying model of epistasis in the LTEE (Kryazhimskiy et al. 2009; Wiser et al. 2013; Frank 2014). However, to truly distinguish between different models, we must move beyond the simple curve fitting implied by Equation 2 and postulate a set of concrete population genetic models that can be used to generate predictions for The likelihood of the observed fitness trajectory can then be written in the form
| (3) |
Here, is the probability distribution of the data vector in the underlying model, which depends on some set of parameters θ, and is the distribution of measurement errors, which we assume to be independent and normally distributed with variance for each time point of (Figure 1) and for (Figure 1B, inset). By computing this likelihood, we can assess the fit of a given model using standard statistical techniques (see Appendix). In contrast to the curve-fitting approach of earlier work, this method correctly accounts for inherent stochasticity of the evolutionary process, which can lead to correlated fluctuations in the observed fitness trajectory. Yet in practice, it is often difficult to compute the likelihood in Equation 3 because the model distribution is unknown. This is largely due to the large population size of the LTEE (N ≈ 3 × 107), which makes it difficult to analyze even the simplest population genetic models (Desai and Fisher 2007). To avoid these issues, we use computer simulations of the model to obtain accurate predictions of the fitness trajectory (see Appendix), computing the approximate-likelihood function as
| (4) |
Unfortunately, regardless of the method used for inference, we will demonstrate that there is little power to distinguish between different models of epistasis based on the fitness trajectory alone. As noted by Frank (2014), it is relatively easy to devise an epistatic model that reproduces the observed fitness trajectory in Figure 1B, and we outline several specific examples below. Fortunately, the average fitness trajectory is not the only quantity that has been measured in the LTEE. DNA sequences from a small number of clones are available for several time points in a subset of the lines (Barrick et al. 2009; Wielgoss et al. 2011, 2013). Although these genetic data are more sparse than the fitness measurements, they provide a crucial window into the the molecular changes responsible for the observed patterns of fitness evolution. In Figure 1B, we plot the average number of genetic differences between the ancestor and a set of clones sampled from the nonmutator populations (see Appendix). When viewed as a function of time, this mutational trajectory is the natural genetic analog of the average fitness trajectory Any evolutionary model that purports to explain the long-term trends in must also be consistent with the observed values of As we will see below, this turns out to be much more informative than fitting the fitness trajectory on its own.
The most striking feature of the mutation trajectory in Figure 1 is the sheer number of mutations that have accumulated during the experiment. Although the full data no longer support the constant substitution rate observed in the first 10,000 generations of evolution (Barrick et al. 2009), the number of mutations in the later portion of the experiment is still much higher than one might expect based on the fitness trajectory. Roughly half of all mutations accumulated after the first 10,000 generations, when rate of fitness increase had already slowed substantially. Of course, some unknown fraction of these mutations are likely to be selectively neutral, as these accumulate continuously at the neutral mutation rate Un (Birky and Walsh 1988). There are no a priori estimates of Un, but evidence from the synonymous substitution rate and mutation accumulation lines suggests that a reasonable upper bound is Utot ≈ 7 × 10−4 (Supporting Information, File S1). With this estimate, <30 neutral mutations should have accumulated by generation 40,000, which suggests that most of the ∼ 60 observed mutations in Figure 1 are beneficial. In fact, the substitution rate in the first 10,000 generations is so rapid that many of these beneficial mutations must be segregating in the population at the same time. Given that the typical fitness effect of a fixed mutation is at most ∼10% (Khan et al. 2011), the fixation time of a successful mutation is much longer than the maximum possible waiting time between mutations. As a result, these mutations must compete for fixation within the population—a process known as clonal interference (Gerrish and Lenski 1998; Desai and Fisher 2007). This will prove to be an important factor in the theoretical analysis below.
Macroscopic epistasis from a finite genome
Although a decelerating fitness trajectory is a clear signature of macroscopic epistasis (i.e., a changing DFE), this does not necessarily imply that microscopic epistasis must be at work. The DFE can change even in the absence of epistasis provided that the length of the genome is finite. Given enough time, the population will eventually exhaust the supply of beneficial mutations, and the rate of adaptation will slow substantially. Thus, this nonepistatic scenario offers one of the simplest possible explanations for the decelerating rate of adaptation in the LTEE, provided that it can also quantitatively reproduce the trajectories in Figure 1.
In the simplest version of this model, the beneficial DFE evolves according to the mean-field dynamics,
| (5) |
where Lb is the number of sites and pfix(s) is the fixation probability of a new mutation. Equation 5 accounts for the fact that, once a beneficial mutation fixes, a second mutation at that site is not likely to be beneficial, effectively removing this site from the beneficial portion of the DFE; the overall normalization of ρb(s, t) will therefore decrease as more mutations are driven to fixation. The rate of change of the DFE in Equation 5 is inversely proportional to Lb, and it vanishes in the limit that Lb → ∞ as expected. In a true “finite-sites” model, each of the Lb beneficial mutations corresponds to a single site in the genome, and the ratio Ub/Lb is set by the per-site mutation rate μ. However, Equation 5 also describes the evolution of the DFE in a generalized “running out of mutations” model—for example, there could be Lb genes that are beneficial to knock out or Lb modules to improve (Tenaillon et al. 2012; Kryazhimskiy et al. 2014). In these cases, Lb represents the total number of nonredundant mutations, e.g., the number of genes to knock out, and Ub/Lb is the target size of each module. Note that this model assumes that all modules share the same target size; the variable target size case is treated in more detail in File S1.
Given a solution for the time-dependent DFE in Equation 5, the expected fitness and mutation trajectories are given by
| (6a) |
| (6b) |
| (6c) |
Unfortunately, both Equations 5 and 6 are difficult to solve in general, since the fixation probability also depends on the DFE (Good et al. 2012). Despite this difficulty, we can gain considerable qualitative insight by focusing on the strong-selection, weak-mutation (SSWM) limit, where the fixation probability is given by Haldane’s formula, pfix(s) ≈ 2s (Haldane 1927). In this limit, the evolution of the DFE greatly simplifies, and the distribution of beneficial fitness effects is given by
| (7) |
where ρ0(s) is the DFE in the ancestral background. The average fitness and mutation trajectories can then be obtained by substituting Equation 7 into Equation 6 and evaluating the resulting integral. For example, arguments from extreme value theory suggest that the ancestral DFE may often be exponential (Gillespie 1984; Orr 2002), which leads to an average fitness trajectory of the form
| (8) |
where and However, while this trajectory shares the same qualitative deceleration as the data in Figure 1, it predicts a much sharper deceleration in the adaptation rate than is actually observed (Figure S1). This shows that for a fixed DFE shape, we will not always be able to quantitatively reproduce the observed fitness trajectory with our finite-sites model.
However, the situation changes if we are allowed to arbitrarily tune the shape of the DFE to match to the observed fitness trajectory. In particular, we find that the adaptation rate for the DFE in Equation 7 is proportional to the Laplace transform of s−2ρ0(s), which leads to an inverse relation of the form
| (9) |
In other words, we can reproduce a particular fitness trajectory within our finite-sites model by choosing the ancestral DFE to match the expression above. Note that Equation 9 implicitly assumes that the inverse Laplace transform exists and yields a proper probability distribution. This places certain constraints on the fitness trajectories that we can reproduce with this model, e.g., requiring that is monotonically decreasing. The intuitive reason for this restriction is clear from the definition of the model: exhausting the supply of beneficial mutations can never lead to an increasing adaptation rate, no matter how exotic the ancestral DFE is. Note, however, that deleterious mutations (McCandlish et al. 2014a,b), clonal interference (Desai and Fisher 2007), and the fixation of mutator phenotypes (Wiser et al. 2013) can complicate this picture considerably. The other apparent limitation of this model is that the fitness trajectory must be bounded, since the maximum possible fitness that can be attained is At first glance, this would seem to preclude the logarithmic trajectory in Equation 2, which has no maximum value. However, since experimental trajectories are observed only over a finite time window, 0 ≤ t ≤ tmax, we can always satisfy this restriction in practice by assuming that For example, the logarithmic fitness trajectory in Equation 2 corresponds to an ancestral DFE of the form
| (10) |
where ϵ ≪ Xc/v0tmax is a lower cutoff chosen to maintain normalization (see File S1). The fitting parameters in Equation 2 are given by Xc = Lbσϵ and v0 = 2NUbϵσ2.
A similar argument shows that we can also reproduce a given mutation trajectory (subject to the same technical constraints), provided that the ancestral DFE satisfies
| (11) |
However, while we can fit a broad class of fitness and mutation trajectories by choosing the appropriate ancestral DFE, we do not have complete freedom to fit both quantities at the same time. In the weak mutation limit, the average fitness and mutation trajectories in our finite-sites model are related by
| (12) |
regardless of the choice of ancestral DFE. By choosing ρ0(s) to fit the fitness trajectory, we severely constrain the shape of the mutation trajectory (and vice versa), with only an overall scale NUb/L that can be tuned to fit the data. For example, the logarithmic fitness trajectory in Equation 2 implies a constant substitution rate
| (13) |
where 〈s〉f ≈ σ/log(1/ϵ). This linear increase is inconsistent with the mutation trajectory in Figure 1, which starts to show deviations from linearity after generation 10,000.
However, a potential caveat with this analysis is that the mutation trajectory in Equation 13 (and much of the analysis preceding it) depends on our assumption of the weak-mutation limit, which requires that NUb ≪ 1. This is often not self-consistent: in the LTEE, the weak-mutation analysis typically leads us to infer parameter values that violate the weak-mutation assumptions. For example, in the finite-sites model defined by Equation 10, the fitted values of Xc and v0 in Figure 1 require that NUb ≥ 3, which violates the weak-mutation condition used to derive Equations 10 and 13. Thus, we must turn to our computational framework to rigorously compare this model with the data.
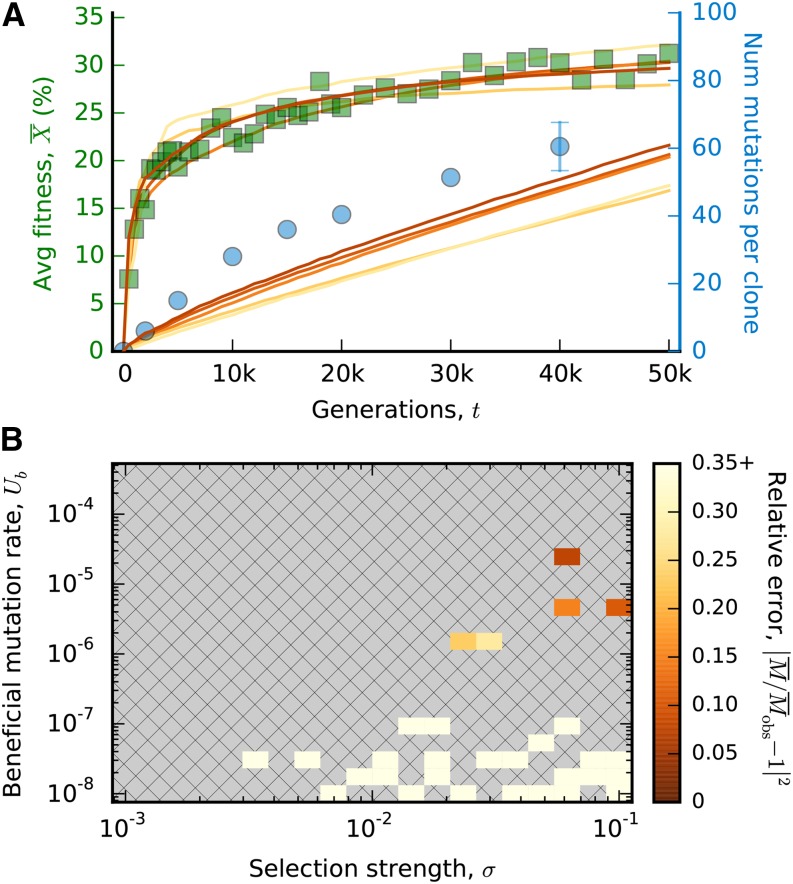
To do so, we performed a grid search over combinations of Ub, Lb, and σ for the ancestral DFE defined by Equation 10 (File S2). The posterior predictive P-value for the fitness trajectory is P ≈ 0.9 (χ2-test, see Appendix), which shows that the finite-sites model can still reproduce the observed fitness trajectory in the presence of clonal interference. Figure 2 shows the average fitness and mutation trajectories for all parameters with P > 0.05. The mutation trajectories also include a best-fit rate of neutral mutations (0 ≤ Un ≤ Utot − Ub), which is fitted to minimize the mean square error from the observed mutation trajectory. Even with this correction, the mutation trajectories remain inconsistent with the data, which allows us to reject the simple finite-sites model in Equation 5.
Figure 2.
Fitting a finite sites model to the LTEE. (A) Simulated fitness and mutation trajectories for the ancestral DFE in Equation 10 with ϵ = 3 × 10−4 (solid lines). We have included all simulated combinations of U, σ, and Lb with P > 0.05 and Each line is colored according to the relative error of the mutation trajectory, after fitting the best-fit neutral mutation rate 0 < Un < Utot by least squares. For comparison, we have also included the observed fitness and mutation trajectories from Figure 1B. (B) The relative error of the mutation trajectory for all simulated parameter combinations. For each combination of U and σ, we plot the minimum error across all simulated values of Lb, and we have included only parameter combinations for which P > 0.05.
Fitness trajectories on an uncorrelated fitness landscape
We next consider an alternative model of macroscopic epistasis—the uncorrelated fitness landscape—which represents the opposite limit of the additive finite-genome models above (Kingman 1978; Gillespie 1984; Kauffman and Levin 1987; Orr 2002). In this model, the fitness of every genotype is drawn independently from the same distribution f(X). In our notation, this implies that the DFE is given by
| (14) |
where denotes the fitness of genotype This uncorrelated landscape contains extensive microscopic epistasis, with the standard deviation of the pairwise epistasis ϵij = sij − si − sj on the same order as si. The fitness effect of a given mutation is therefore barely heritable. However, much of this idiosyncratic microscopic epistasis averages out at the level of the DFE, which depends on the genetic background only through the fitness
The dynamics of adaptation become particularly simple when f(X) is exponentially distributed, since the beneficial portion of the DFE remains exponential (with the same mean) regardless of the fitness. Instead, epistasis primarily acts to reduce the beneficial mutation rate via Ub(X) = Ube−X/σ, where σ is the average fitness effect in the ancestral background. In the weak-mutation limit, this diminishing mutation rate leads to the same logarithmic fitness trajectory as Equation 2, with Xc = σ and v0 = 4NUbσ2 (Kryazhimskiy et al. 2009). Thus, the fitnesses in Figure 1 can also be reproduced in this model of extreme epistasis, in addition to the purely additive model in Equation 10. However, the corresponding mutation trajectory,
| (15) |
contains no free parameters. This form of implies a beneficial substitution rate of essentially zero after t ∼ Xc/v0 generations, which is clearly inconsistent with the data, both on a curve-fitting level (Figure S2) and in simulation (Figure S3). Thus, while the fitness trajectory is consistent with an uncorrelated landscape, this model is again unable to reproduce the observed mutation trajectory.
Global fitness-mediated epistasis
The general patterns of macroscopic epistasis in the uncorrelated landscape can also be realized in other models that have much less microscopic epistasis. For example, a key simplifying assumption of the uncorrelated landscape is that the effective beneficial mutation rate depends only on the fitness of the genetic background and not on its specific genotype. This leads us to consider a broader class of models of the form
| (16) |
where the shape of the DFE is similarly mediated by fitness. This form of epistasis has been implicated in recent genetic reconstruction studies (Chou et al. 2011; Khan et al. 2011; Kryazhimskiy et al. 2014), and it has been hypothesized to describe the patterns of epistasis in the LTEE as well (Kryazhimskiy et al. 2009; Wiser et al. 2013). Most of these studies have focused on an even simpler class of models of the form
| (17) |
where the fitness dependence of the DFE is given by a simple change of scale. We assume by convention that f(0) = 1, so that ρ0(s) represents the ancestral DFE. In the weak mutation limit, the fitness trajectories for Equation 17 must satisfy the implicit relation
| (18) |
where We can then invert this equation to solve for f(X) as a function of the fitness trajectory:
| (19) |
Thus, like the finite-sites model above, we can reproduce a given fitness trajectory with the rescaled DFE in Equation 17 by choosing the correct form for f(X). Note, however, that Equation 19 implicitly assumes that the right-hand side exists and is a real-valued function, which is satisfied for all This is a less restrictive condition than we found for the finite-sites model in Equation 9, which reflects the fact that fitness-mediated epistasis can generate accelerating as well as decelerating fitness trajectories with the appropriate choice of f(X).
We can realize this model microscopically by assuming that fitness effects of individual mutations obey the same scaling relation,
| (20) |
which allows us to make predictions for microscopic quantities like the fitness effects of reconstructed strains. However, there is not complete freedom to choose f(X) in this microscopic model, since the combined effects of a sequence of mutations must commute with each other. The only rescaling that satisfies this commutative property is the linear relation f(X) = 1 − X/Xc, where Xc represents the global fitness maximum. In this case, the fitness effect of each mutation is scaled by the fractional distance to the peak, similar to the “stick-breaking” model of Nagel et al. (2012). In the weak-mutation limit, this model reproduces the hyperbolic fitness trajectory
| (21) |
which has been used to fit the LTEE fitness data in previous studies (Lenski and Travisano 1994). However, as shown by Wiser et al. (2013), Equation 21 provides a relatively poor fit to the observed fitness trajectory (Figure S1), even after accounting for clonal interference (posterior predictive P < 10−3). This allows us to rule out all microscopic models of the form in Equation 20.
For other choices of f(X), Equation 17 will hold only in a statistical sense, with a more complicated pattern of microscopic epistasis than predicted by Equation 20 (File S1). Wiser et al. (2013) have shown that the logarithmic fitness trajectory in Equation 2 can be recovered by setting
| (22) |
However, like the additive and uncorrelated models above, the mutation trajectory in this case is strongly constrained by In the weak mutation limit, the mutation trajectories for Equation 17 must satisfy
| (23) |
with only an overall scale that can be tuned to fit the data. Note that the shape of ρ0(s) remains largely unconstrained by this choice: two beneficial DFEs with the same 〈s〉f can reproduce the same pair of fitness and mutation trajectories, although the required value of Ub will be different.
For the logarithmic fitness trajectory in Equation 2, Wiser et al. (2013) have shown that the corresponding mutation trajectory grows as a square root of time:
| (24) |
At first glance, Equation 24 appears to give a decent fit to the observed mutation trajectory (Figure S2), although it systematically overestimates the curvature. However, the best-fit scale 〈s〉f ∼ 5% lies in the clonal interference regime, so we must again turn to our computational framework to rigorously compare this model with the data.
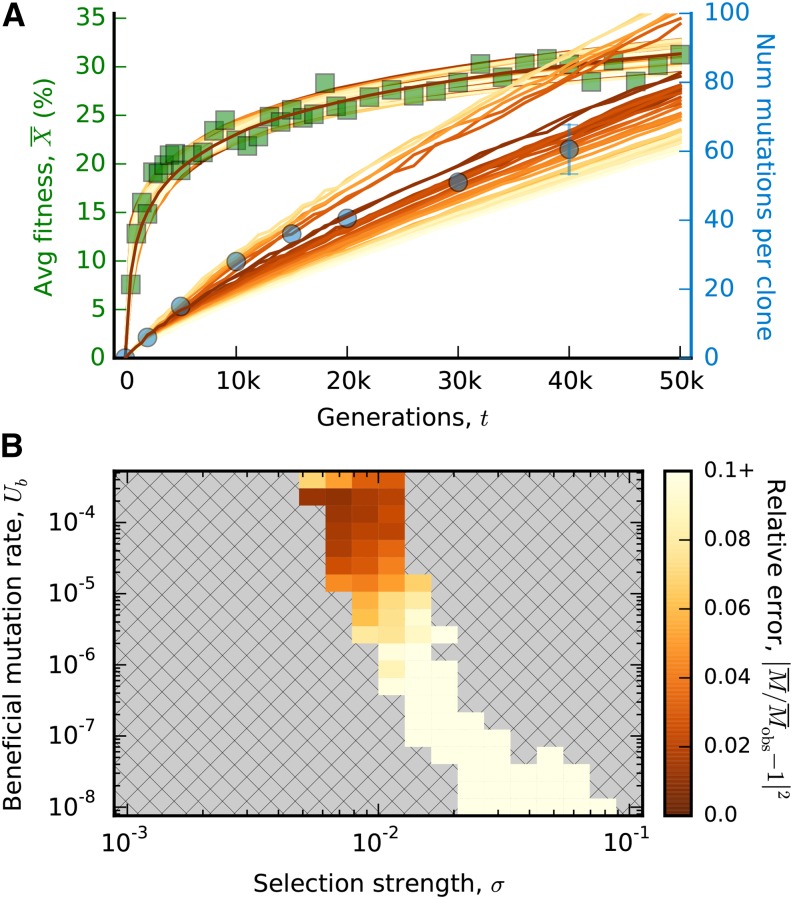
To do so, we performed a grid search over combinations of U, σ, and Xc for an exponential ancestral DFE, ρ0(s) ∝ exp(−s/σ), which was the specific generative model proposed by Wiser et al. (2013). The posterior predictive P-value for the fitness trajectory is P ≈ 0.9, which shows that the global diminishing-returns model can still reproduce the observed fitness trajectory even when NUb > 1. Figure 3A shows the average fitness and mutation trajectories for all parameters with P > 0.05. As expected, there is a large “ridge” of parameter values that reproduce the observed fitness trajectory, but the vast majority of these parameter combinations are inconsistent with the observed mutation trajectory. Those parameters with the best estimates of still display some small systematic errors, underestimating the number of mutations in the first part of the experiment and overestimating them later (Figure 3B). The numerical values of these parameters are also wildly unrealistic, since they predict that the mutation rate to fitness effects >1% is more than a quarter of the genomic point mutation rate. In light of this information, we conclude that the mutation data are inconsistent with the particular global diminishing-returns model proposed by Wiser et al. (2013).
Figure 3.
Fitting a fitness-mediated epistasis model to the LTEE data. (A) An analogous version of Fig. 2A constructed for the global diminishing returns model in Eq. (22) with an exponential ancestral DFE. We have included all simulated combinations of U, σ, and Xc with p > 0.05 and < 0.1. (B) The analogous version of Fig. 2B. Note the change in scale for the relative error in the mutation trajectory.
However, our findings in the SSWM limit above suggest that the precise value of the estimated beneficial mutation rate can dramatically vary with the shape of the ancestral DFE, while the predictions of Equations 19 and 23 are insensitive to this choice. Similar insensitivity to the DFE has been noted in the clonal interference regime as well (Hegreness et al. 2006; Desai and Fisher 2007; Fogle et al. 2008; Good et al. 2012; Fisher 2013). In accordance with this intuition, the inferred parameters become more realistic if we truncate the exponential distribution at smax = 4σ, although the systematic errors in the mutation trajectory remain (Figure S4). Compared to the finite-sites model and the uncorrelated landscape above (as well as the original Wiser et al. 2013 model), this modified version of the global diminishing-returns model is the only one that can plausibly reproduce all of the observed data.
Evidence for two evolutionary epochs
While the discrepancies in the mutation trajectory are too small to reject the fitness-mediated epistasis model outright, these systematic errors still suggest that the model defined by Equation 22 may be missing a key feature of the experiment. This argues for a degree of caution in interpreting the parameters inferred in Figure 3, particularly far into the future where the errors in the fitness and mutation trajectories start to grow larger. Of course, we could continue to postulate more elaborate models of epistasis to account for the mutation trajectory, and with enough additional parameters this approach is likely to be successful. For example, the fitness and mutation trajectories can both be reproduced by generalized finite-sites models where we can tune the individual target sizes (File S1) or by fitness-mediated epistasis models where the overall mutation rate also depends on the fitness. But without a biological basis for choosing among the space of possible models, these additional assumptions are likely to overfit the mutation trajectory and lead to incorrect predictions for other observables (e.g., genetic diversity or variation among lines). Instead, we focus on an alternative class of models that are simpler in some ways, although more complex in others.
Revisiting the original data in Figure 1, we note that we were initially led to consider models of macroscopic epistasis, rather than a constant DFE, because of the differences between the initial part of the experiment (e.g., before generation 10,000) and the later part of the experiment (e.g., after generation 10,000). The differences in the rate of adaptation are so striking that we can clearly rule out a constant DFE even without a rigorous statistical comparison. In contrast, the deceleration in the fitness trajectory in the final 40,000 generations of evolution is much less pronounced. This leads us to consider a simple statistical question: Does the later portion of the experiment actually contain evidence of macroscopic epistasis, or are we simply extrapolating from the strong deceleration in the early part of the fitness trajectory?
We can frame this statistical question as a model of epistasis with two evolutionary epochs: an initial “poorly adapted” phase in the first 10,000 generations followed by a more “well-adapted” phase for the remaining 40,000 generations. We do not attempt to model the initial phase, but instead simply assume that the population is subject to some complicated and unspecified model of epistasis that generates the observed data with probability one. This could account for the fact that the large-effect mutations available at the beginning of the experiment might depend on specific details of the ancestral strain or other experimental details. After this initial phase of adaptation is complete, the population enters a second phase of evolution with negligible macroscopic epistasis. In other words, rather than try to fit a single model of a changing DFE to the whole experiment, we neglect the first 10,000 generations and instead try to fit an evolutionary model with a constant DFE to the last 40,000 generations of evolution.
Assuming a constant DFE implies that the fitness and mutation trajectories after generation 10,000 are given by
| (25) |
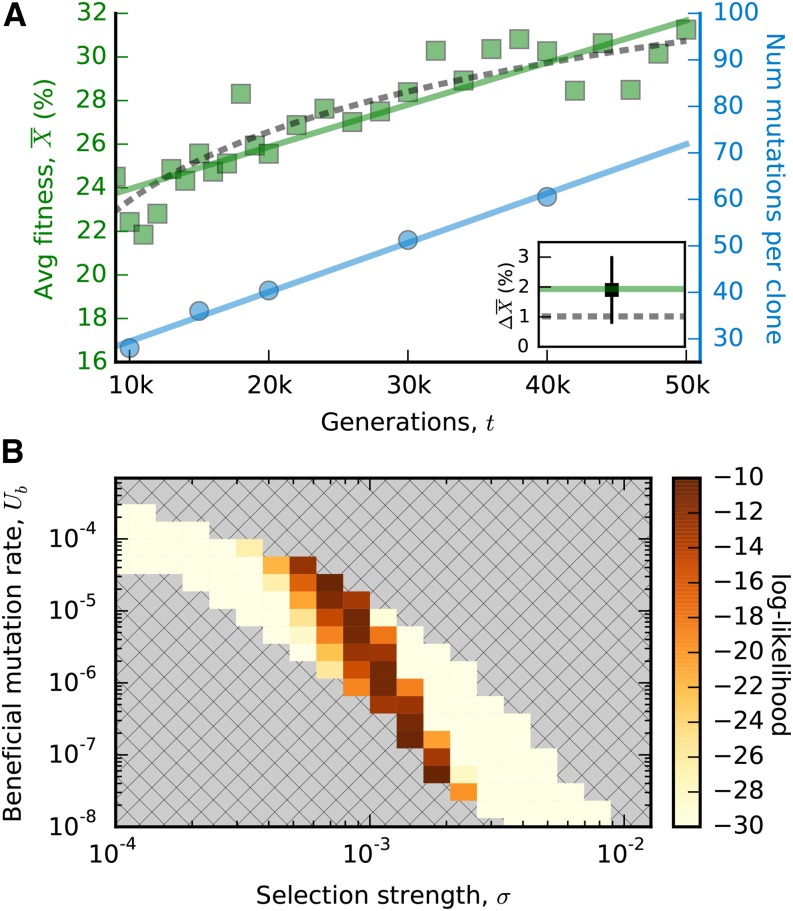
This fitness trajectory has the same number of nominal parameters as the logarithmic curve in Equation 2, although it is important to remember that Equation 2 carries an implicit functional degree of freedom that was used to obtain the logarithmic trajectory in the first place. Figure 4 shows that even on a purely curve-fitting level, the nonepistatic fitness trajectory is only marginally less accurate than its epistatic counterpart. We infer an adaptation rate of v0 ≈ 0.2% per 1000 generations and a substitution rate of R0 ≈ 1.1 per 1000 generations. Although this adaptation rate appears to overestimate the fitness gain after generation 40,000, the more precise fitness assays performed between generations 40,000 and 50,000 corroborate the 2% increase (Figure 4, inset). The fitted values of v0 and R0 can be used to infer the typical fitness effect of a fixed mutation and a corresponding effective mutation rate based on the relations
| (26) |
derived in previous theoretical work (Desai and Fisher 2007). For the values of v0 and R0 above, we find a typical fixed fitness effect of order seff ∼ 2 × 10−3 and an effective mutation rate of order Ueff ∼ 2 × 10−6.
Figure 4.
Fitting a nonepistatic model to the last 40,000 generations of evolution. (A) The average fitness and mutation trajectories from Figure 1B, along with the predictions of the nonepistatic curve in Equation (25) (solid lines). The estimated parameters are v0 ≈ 1.9 × 10−6 and R0 ≈ 1.1 ×10−3. For comparison, the best-fit logarithmic trajectory from Equation 2 is shown by the dashed line. Inset shows predictions for the average change in fitness between generations 40,000 and 50,000 (lines) compared to the independent measurement from the inset in Figure 1B (square). (B) The likelihood of the fitness trajectory for a constant, exponential DFE truncated at smax ≈ 4σ, with a y intercept fitted using maximum likelihood. We have included only parameters whose substitution rates are consistent with the observed mutation trajectory
However, this discussion has so far been based purely on curve fitting and not on a specific generative model of the dynamics. Using our computational framework, we can evaluate the fit of the two-epoch model more rigorously. To do so, we performed a grid search over combinations of U, σ, and Xc for a truncated exponential distribution (smax = 4σ). Recall that there is little power to infer the shape of the DFE in this model; we chose the truncated exponential distribution because its parameters can be directly compared to the best-fit diminishing-returns model in Equation 22. We find that the best nonepistatic models are statistically consistent with the observed fitness trajectory (P ≈ 0.9; χ2-test) and provide only a marginally worse fit than the diminishing-returns models above (Δ log Λ < −3). Moreover, this difference in likelihood vanishes completely if we restrict our attention to the last 35,000 generations of the experiment rather than the last 40,000. Together, these results suggest that there is limited evidence for macroscopic epistasis in the later portion of the LTEE based on the currently available data.
Discussion
Genetic reconstructions provide numerous examples of interactions between the fitness effects of individual mutations. The existence of these interactions is hardly surprising, given the physiological and developmental complexity of most organisms. However, the evolutionary implications of these interactions remain controversial. In this study, we used longitudinal data from a long-term evolution experiment in E. coli to investigate the evolutionary influence of epistasis in a simple empirical setting. We focused on two basic questions: (1) How do naturally occurring patterns of epistasis alter the rate of fitness increase and the accumulation of new mutations in a constant environment? And (2) what are the simplest models of epistasis that are consistent with the observed data?
The first of these questions is largely descriptive and has been the focus of previous work in this experimental system and many others (Lenski and Travisano 1994; Wichman et al. 1999; Silander et al. 2007; Barrick et al. 2009; Kryazhimskiy et al. 2012, 2014; Wiser et al. 2013; Perfeito et al. 2014). The latter question, in contrast, demands a quantitative approach and must account for the fact that the underlying model of epistasis can be observed only through the filter of population genetic stochasticity and measurement error. In this study, we developed a computational framework to account for these confounding factors, which allows us to quantify the consistency of a predicted fitness trajectory using well-established statistical tools. Combined with analytical results in the weak-mutation limit, we used this framework to investigate the compatibility of several popular models of epistasis.
We found that the shape of a decelerating fitness trajectory on its own provides little power to distinguish between different models of epistasis, including finite-sites models that lack any direct interactions between mutations. This suggests that the underlying “symmetry group” or universality class for this observable may be quite large (Frank 2014), which could potentially explain why previous studies have been so successful at fitting the LTEE fitness trajectory with simple epistatic models (Sibani et al. 1998; Wiser et al. 2013; Frank 2014). However, this symmetry is broken as soon as we include information about the number of mutations that have accumulated, and the combination of fitness and genetic data places much stronger constraints on the set of possible models. Of the simple two- and three-parameter models considered here, we found that a variant of the global diminishing-returns model proposed by Wiser et al. (2013) provides the best fit to the observed data, although certain systematic errors still remain. These systematic errors, combined with the weak diminishing-returns signal in the first five mutations in the Ara−1 population (File S1), suggest a degree of caution in interpreting the support for this model.
Instead, we find that the data are equally well explained by a two-epoch model of adaptation, in which an initial burst of macroscopic epistasis is followed by a steady accumulation of mutations under a constant DFE. Although this model offers no insight into the initial (presumably idiosyncratic) phase of adaptation, it provides a more parsimonious explanation for the dynamics in the latter phase of the experiment. Moreover, given that this second phase accounts for three-quarters of the present duration of the LTEE and more than half of the accumulated mutations, it could be argued that it provides a better description of the “typical” dynamics of adaptation in a constant environment than the initial, epistatic phase of the adaptive walk. Under this hypothesis, the widespread diminishing-returns epistasis observed in other experimental systems may simply be a reflection of their comparatively brief duration.
In light of this speculation, it is worth commenting on the population genetic parameters estimated in the second, slower phase of the LTEE. When faced with a constant environment, it is natural to expect that a population will eventually enter a slower phase of adaptation once the most obvious beneficial mutations are exhausted. However, this steady state is usually assumed to have a negligible beneficial mutation rate and correspondingly simple evolutionary dynamics. In contrast, the scaled mutation rates that we estimate for the LTEE are surprisingly large (NU ∼ 10–100) and are comparable to rapidly evolving laboratory yeast populations near the beginning of their adaptive walk (Frenkel et al. 2014). Although the fitness effects of these mutations (s ∼ 0.1%) fall below the resolution limit of most fitness assays, the effective population size is large enough that the scaled selection strengths are quite large from a population genetic standpoint (Ns ≳ 104). Interestingly, these scaled beneficial mutation rates and selection strengths are sufficiently large that deleterious mutations are expected to have a negligible influence on the rate of adaptation (Good and Desai 2014). [We also note that extrapolating these estimates to the mutator lines would suggest that the declining mutation rate observed in Ara-1 (Wielgoss et al. 2013) may involve selection for more than just a reduced deleterious load.]
Together, these estimates suggest that even the “slow” phase of the LTEE is characterized by rapid adaptation, in which multiple beneficial mutations compete for fixation at the same time. These dynamics are illustrated by the simulated mutation trajectories in Figure S6, which display an even greater amount of hitchhiking and clonal interference than similar trajectories measured in a recent evolution experiment in yeast (Lang et al. 2013). However, since the individual fitness effects are an order of magnitude smaller, the competition between beneficial mutations occurs over a much longer timescale than is normally observed in experimental evolution. For example, it is not uncommon to find a beneficial mutation that persists for thousands of generations at intermediate frequencies before it accumulates enough additional mutations to sweep to fixation. These transiently stable polymorphisms would suggest adaptive radiation or frequency-dependent selection on a more traditional experimental timescale (e.g., <2000 generations), but they arise here as a natural consequence of the population genetic process.
Of course, the preceding discussion should be treated with a degree of caution, since our present estimates are based on a limited number of clone sequences and relatively noisy fitness measurements. It is possible that additional data would indicate a departure from the constant adaptation rate in the second phase of the experiment or reverse some of the systematic errors of the global diminishing-returns model. Moreover, even perfectly resolved fitness and mutation trajectories will likely be consistent with more than one evolutionary model, just as a perfectly resolved fitness trajectory is consistent with multiple mutation trajectories. Our estimates should therefore be viewed as merely consistent with the available data, rather than strongly supported by them. Nevertheless, our results demonstrate that an easy-to-measure genetic observable such as the mutation trajectory can greatly restrict the set of models that are consistent with a measured fitness trajectory. Additional information about the genetic diversity within the population (e.g., measured from pairwise heterozygosity among clones) will likely provide even more power to distinguish between competing hypotheses. The computational framework developed here provides a powerful and flexible method for incorporating this genetic information as it becomes available.
Acknowledgments
We thank Sergey Kryazhimskiy, Elizabeth Jerison, and Daniel Rice for useful discussions and Joshua Plotkin, David McCandlish, and two reviewers for helpful comments on the manuscript. This work was supported in part by a National Science Foundation (NSF) Graduate Research Fellowship, the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, the Harvard Milton Fund, grant PHY 1313638 from the NSF, and grant GM104239 from the National Institutes of Health. Simulations in this article were performed on the Odyssey cluster supported by the Research Computing Group at Harvard University.
Appendix
Fitness Trajectories
The finesses of the LTEE strains were calculated from Equation 1, using the raw competition assays reported by Wiser et al. (2013). The quantity f/(1 − f) was estimated from the ratio of red and white colonies when plated on arabinose media, and the duration of each competition was Δt = log2(100) ≈ 6.6 generations. The fitness gains between generations 40,000 and 50,000 were calculated in a similar manner, but with a longer competition time of Δt = 3 log2(100) ≈ 19.9 generations. Subsequent analysis of the fitness trajectory was restricted to the six nonmutator populations with complete fitness measurements: Ara−5, Ara−6, Ara+1, Ara+2, Ara+4, and Ara+5 (Wiser et al. 2013). These populations were chosen because they have complete fitness trajectories that can be reasonably expected to evolve under the same population genetic model. Notably, this subset excludes both the citrate-metabolizing population (Ara−3) studied by Blount et al. (2008) and the crossfeeding population (Ara−2) studied by Rozen and Lenski (2000).
Fitness Effects of Individual Mutations
The fitness effects of the five mutations in Figure S5 were calculated from the raw competition assays reported by Khan et al. (2011). The fitness effect s was defined as the difference between the fitnesses of the mutant and background genotypes, which were estimated from the competition assays using the same procedure as above.
The Mutation Trajectory
The total number of genetic changes was estimated from the DNA sequences of clones analyzed by Barrick et al. (2009) and Wielgoss et al. (2011). The finalized mutation calls for the clones in Barrick et al. (2009) were obtained from supplementary tables 1 and 2 of that work, and the mutation calls for the clones in Wielgoss et al. (2011) were obtained from the supplementary data files available at http://barricklab.org/twiki/pub/Lab/SupplementLongTermMutationRates/long-term_mutation_rates.zip. To obtain a more densely sampled mutation trajectory, we included clones from populations that were excluded from the fitness trajectory analysis above. This includes seven clones sampled from the Ara−1 population prior to the spread of the mutator phenotype and two clones sampled from the Ara−3 population prior to the spread of the citrate-metabolizing phenotype. The complete list of included clones is given in Table S1.
Population Genetic Simulations
Simulated fitness and mutation trajectories were obtained from a forward-time algorithm designed to mimic the serial transfer protocol of the LTEE. Between each transfer, lineages are assumed to expand clonally for log2(100) ≈ 6.64 generations at a deterministic exponential growth rate r = r0 + X, where X is the fitness relative to the ancestor. At the transfer step, the population is diluted 100-fold (with Poisson sampling noise) to Nb = 5 × 106 individuals. Mutations accumulate at a constant rate U during the growth phase, but we assumed that they do not significantly influence the fitness of the individual until the next transfer cycle. Thus, mutation was approximated by assuming that each individual has a probability 6.64 ⋅ U of gaining a mutation at the end of a transfer step, with additive fitness effects drawn from the genotype-specific DFE, For the finite-sites model, a discrete ancestral DFE was initialized by drawing L fitness effects from the continuous distribution ρ0(s), with the same realization shared across replicate lines. A copy of our implementation is available at https://github.com/benjaminhgood/ltee_inference.
Likelihood Estimation
The likelihood of each parameter combination was estimated from simulations, using Equation 4. To speed computation, we simulated 18 replicate populations and generated n = 10,000 different 6-population averages by bootstrap resampling. The scaled likelihood Λ, which differs from by a constant factor, was defined as
| (A1) |
where the measurement uncertainties and were estimated from Figure 1.
Statistical Tests
The consistency of each parameter combination was assessed using a χ2 goodness-of-fit test. We simulated 18 replicate populations to estimate and and we generated n = 10,000 different 6-population averages by bootstrap resampling these replicates and adding unbiased Gaussian measurement noise with and . The P-value is then approximated by
| (A2) |
where θ(x) is the Heaviside step function and is the mean square error,
| (A3) |
The posterior predictive P-value for the entire model is then defined by
| (A4) |
where p(θ) is the prior distribution of parameter values.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.172460/-/DC1.
Communicating editor: L. M. Wahl
Literature Cited
- Bank C., Hietpas R. T., Wong A., Bolon D. N., Jensen J. D., 2014. A Bayesian MCMC approach to assess the complete distribution of fitness effects of new mutations: uncovering the potential for adaptive walks in challenging environments. Genetics 196: 841–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., et al. , 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247. [DOI] [PubMed] [Google Scholar]
- Barrick J. E., Kauth M. R., Strelioff C. C., Lenski R. E., 2010. Escherichia coli rpob mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27: 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birky C. W., Jr, Walsh J. B., 1988. Effects of linkage on rates of molecular evolution. Proc. Natl. Acad. Sci. USA 85: 6414–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount Z. D., Borland C. Z., Lenski R. E., 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. USA 105: 7899–7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch C. L., Chao L., 2000. Evolvability of an rna virus is determined by its mutational neighbourhood. Nature 406: 625–628. [DOI] [PubMed] [Google Scholar]
- Chou H.-H., Chiu H.-C., Delaney N. F., Segrè D., Marx C. J., 2011. Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., E. D. Spearet al. , 2010. The genetic landscape of a cell. Science 327: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Visser J. A. G., Krug J., 2014. Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15: 480–490. [DOI] [PubMed] [Google Scholar]
- Desai M. M., Fisher D. S., 2007. Beneficial mutation selection balance and the effect of genetic linkage on positive selection. Genetics 176: 1759–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D. S., 2013. Asexual evolution waves: fluctuations and universality. J. Stat. Mech. 2013: P01011. [Google Scholar]
- Fisher R. A., 1930. The distribution of gene ratios for rare mutations. Proc. R. Soc. Edinb. 50: 204–219. [Google Scholar]
- Fogle C. A., Nagle J. L., Desai M. M., 2008. Clonal interference, multiple mutations and adaptation in large asexual populations. Genetics 180: 2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S. A., 2014. Generative models vs. underlying symmetries to explain biological pattern. J. Evol. Biol. 27: 1172–1178. [DOI] [PubMed] [Google Scholar]
- Frenkel E. M., Good B. H., Desai M. M., 2014. The fates of mutant lineages and the distribution of fitness effects of beneficial mutations in laboratory budding yeast populations. Genetics 196: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish P., Lenski R., 1998. The fate of competing beneficial mutations in an asexual population. Genetica 127: 127–144. [PubMed] [Google Scholar]
- Gillespie J., 1984. Molecular evolution over the mutational landscape. Evolution 38: 1116–1129. [DOI] [PubMed] [Google Scholar]
- Good B. H., Desai M. M., 2014. Deleterious passengers in adapting populations. Genetics 198: 1183–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good B. H., Rouzine I. M., Balick D. J., Hallatschek O., Desai M. M., 2012. Distribution of fixed beneficial mutations and the rate of adaptation in asexual populations. Proc. Natl. Acad. Sci. USA 109: 4950–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., 1989. Wonderful Life: The Burgess Shale and the Nature of History. W. W. Norton, New York. [Google Scholar]
- Haldane J., 1927. The mathematical theory of natural and artificial selection, part v: selection and mutation. Proc. Camb. Philos. Soc. 23: 828–844. [Google Scholar]
- Hegreness M., Shoresh N., Hartl D., Kishony R., 2006. An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617. [DOI] [PubMed] [Google Scholar]
- Jasnos L., Korona R., 2007. Epistatic buffering of fitness loss in yeast double deletion strains. Nat. Genet. 39: 550–554. [DOI] [PubMed] [Google Scholar]
- Kauffman S., Levin S., 1987. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128: 11–45. [DOI] [PubMed] [Google Scholar]
- Kauffman S., Weinberger E. D., 1989. The nk model of rugged fitness landscape and its application to maturation of the immune response. J. Theor. Biol. 141: 211–245. [DOI] [PubMed] [Google Scholar]
- Khan A. I., Din D. M., Schneider D., Lenski R. E., Cooper T. F., 2011. Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C., 1978. A simple model for the balance between selection and mutation. J. Appl. Probab. 15: 112. [Google Scholar]
- Kinney J. B., Murugana A., Callan C. G., Jr, Cox E. C., 2010. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc. Natl. Acad. Sci. USA 107: 9158–9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S., Tkaik G., Plotkin J. B., 2009. The dynamics of adaptation on correlated fitness landscapes. Proc. Natl. Acad. Sci. USA 106: 18638–18643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryazhimskiy S. K., Rice D. P., Desai M. M., 2012. Population subdivision and adaptation in asexual populations of Saccharomyces cerevisiae. Evolution 66: 1931–1941. [DOI] [PubMed] [Google Scholar]
- Kryazhimskiy S. K., Rice D. P., Jerison E. R., Desai M. M., 2014. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science 344: 1519–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G. I., Rice D. P., Hickman M. J., Sodergren E., Weinstock G. M., et al. , 2013. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature 500: 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Travisano M., 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc. Natl. Acad. Sci. USA 91: 6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C., 1991. Long-term experimental evolution in Escherichia coli. i. adaptation and divergence during 2,000 generations. Am. Nat. 138: 1315–1341. [Google Scholar]
- MacLean R. C., Perron G. G., Gardner A., 2010. Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186: 1345–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCandlish, D., C. Epstein, and J. B. Plotkin, 2014a The inevitability of unconditionally deleterious substitutions during adaptation. Evolution 68: 1351–1365. [DOI] [PubMed]
- McCandlish, D. M., J. Otwinowski, and J. B. Plotkin, 2014b On the role of epistasis in adaptation. arXiv:1410.2508.
- Miller C. R., Joyce P., Wichman H. A., 2011. Mutational effects and population dynamics during viral adaptation challenge current models. Genetics 187: 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel A. C., Joyce P., Wichman H. A., Miller C. R., 2012. Stickbreaking: a novel fitness landscape model that harbors epistasis and is consistent with commonly observed patterns of adaptive evolution. Genetics 190: 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H., 2002. The population genetics of adaptation: the adaptation of dna sequences. Evolution 56: 1317–1330. [DOI] [PubMed] [Google Scholar]
- Perfeito L., Sousa A., Bataillon T., Gordo I., 2014. Rates of fitness decline and rebound suggest pervasive epistasis. Evolution 68: 150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt E. M., D. E. Deatherage, A. D. Ellington, G. Georgiou, and J. E. Barrick, 2014. Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc. Natl. Acad. Sci. USA 111: 2217–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen D. E., Lenski R. E., 2000. Long-term experimental evolution in Escherichia coli. viii. dynamics of a balanced polymorphism. Am. Nat. 155: 24–35. [DOI] [PubMed] [Google Scholar]
- Segrè D., Deluna A., Church G., Kishony R., 2005. Modular epistasis in yeast metabolism. Nat. Genet. 37: 77–83. [DOI] [PubMed] [Google Scholar]
- Sibani P., Brandt M., Alstrøm P., 1998. Evolution and extinction dynamics in rugged fitness landscapes. Int. J. Mod. Phys. 12: 361–391. [Google Scholar]
- Silander O. K., Tenaillon O., Chao L., 2007. Understanding the evolutionary fate of finite populations: the dynamics of mutational effects. PLoS Biol. 5: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Onge R. P., Mani R., Oh J., Proctor M., E. Funget al. , 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 39: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Rodrguez-Verdugo A., Gaut R. L., McDonald P., Bennett A. F., et al. , 2012. The molecular diversity of adaptive convergence. Science 335: 457–461. [DOI] [PubMed] [Google Scholar]
- Weinreich D. M., Delaney N. F., DePristo M. A., Hartl D. L., 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312: 111–114. [DOI] [PubMed] [Google Scholar]
- Wichman H. A., Badgett M. R., Scott L. A., Boulianne C. M., Bull J. J., 1999. Different trajectories of parallel evolution during viral adaptation. Science 285: 422–424. [DOI] [PubMed] [Google Scholar]
- Wielgoss, S., J. E. Barrick, O. Tenaillon, S. Cruveiller, B. Chane-Woon-Ming et al., 2011 Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli G3 (Bethesda) 1: 183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgoss S., Barrick J. E., Tenaillon O., Wiser M. J., Dittmar J., et al. , 2013. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. USA 110: 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiser M. J., Ribeck N., Lenski R. E., 2013. Long-term dynamics of adaptation in asexual populations. Science 342: 1364–1367. [DOI] [PubMed] [Google Scholar]
- Woods R. J., Barrick J. E., Cooper T. F., Shrestha U., Kauth M. R., et al. , 2011. Second-order selection for evolvability in a large Escherichia coli population. Science 331: 1433–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1932 The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proceedings of the VI International Congress of Genetics, Genetics Society of America, Ithaca, New York, pp. 356–366. [Google Scholar]