Abstract
The data from genome-wide association studies (GWAS) in humans are still predominantly analyzed using single-marker association methods. As an alternative to single-marker analysis (SMA), all or subsets of markers can be tested simultaneously. This approach requires a form of penalized regression (PR) as the number of SNPs is much larger than the sample size. Here we review PR methods in the context of GWAS, extend them to perform penalty parameter and SNP selection by false discovery rate (FDR) control, and assess their performance in comparison with SMA. PR methods were compared with SMA, using realistically simulated GWAS data with a continuous phenotype and real data. Based on these comparisons our analytic FDR criterion may currently be the best approach to SNP selection using PR for GWAS. We found that PR with FDR control provides substantially more power than SMA with genome-wide type-I error control but somewhat less power than SMA with Benjamini–Hochberg FDR control (SMA-BH). PR with FDR-based penalty parameter selection controlled the FDR somewhat conservatively while SMA-BH may not achieve FDR control in all situations. Differences among PR methods seem quite small when the focus is on SNP selection with FDR control. Incorporating linkage disequilibrium into the penalization by adapting penalties developed for covariates measured on graphs can improve power but also generate more false positives or wider regions for follow-up. We recommend the elastic net with a mixing weight for the Lasso penalty near 0.5 as the best method.
Keywords: genome-wide association study, penalized regression, false discovery rate, linkage disequilibrium
THE goal of genome-wide association studies (GWAS) in humans and model organisms is to select a small subset of DNA markers, typically single-nucleotide polymorphisms (SNPs), which are in strong linkage disequilibrium (LD) with functional polymorphisms affecting a biomedical/clinical trait of interest. The selected markers are then replicated in other GWAS, fine-mapped, and further validated. GWAS may be viewed as a large-scale variable selection problem, with several millions of common SNPs, measured directly or imputed, available in current studies in humans.
GWAS practitioners still strongly rely on single-marker analysis (SMA), including linear regression for continuous phenotypes, with control of the genome-wise error rate (GWER) [a special case of the family-wise error rate (FWER)], which accounts for the multiplicity of the entire genome. Assuming that a biomedical trait of interest is affected by multiple polymorphisms with “detectable” effects, SMA fits an incorrect model, and it is sensible to consider alternative methods that test all or subsets of markers simultaneously. This approach requires a form of penalized regression (PR) as the number of SNPs is much larger than the sample size.
A major practical issue with the use of penalized regression is the determination of “optimal” values for the tuning parameter(s) and the lack of an error rate associated with the selection of SNPs. Common approaches for tuning parameter value determination include cross-validation (CV) (e.g., Kohavi 1995) and the use of a model selection criterion such as Akaike’s information criterion (AIC) (Akaike 1974, 1977), the Bayesian information criterion (BIC) (Schwarz 1978), or the extended Bayesian information criterion (EBIC) (Chen and Chen 2008). These approaches work well for prediction but not for the goal of variable selection as most of these criteria lead to an unacceptable number of false positives and do not provide a measure of error associated with the selected SNPs. This distinction is important as the best predictive models may contain covariates that are not important, while the model with all important covariates may not be the best predictive model.
The absence in PR of the control of an appropriate error rate such as the FWER or the false discovery rate (FDR) (Benjamini and Hochberg 1995; Sabatti and Freimer 2003) has been recognized by several authors who have employed different strategies, including data permutation (Ayers and Cordell 2010), stability selection (Meinshausen and Bühlmann 2010) with FDR control (Ahmed et al. 2011), and multistage (data splitting) approaches (Meinshausen et al. 2009; Wasserman and Roeder 2009) to control FWER or FDR. However, these approaches require much more computation and may not be feasible on a genome-wide scale, and/or the provision of an error rate estimate comes at the expense of reduced power.
A second practical issue is that a number of PR methods differing in the penalty function have been proposed, and hence it is unclear whether any and which of these methods should be preferred in the context of variable selection in GWAS. Moreover, some authors (Kim and Xing 2009; Liu et al. 2011) have suggested that penalties should incorporate LD, but it is unclear whether such penalties produce any gain in power and/or a reduction in the false positive findings.
The purpose of this contribution is to review penalized regression methods in the context of GWAS, to extend selected PR methods by incorporating FDR control and to assess their performance in comparison with single-marker (SNP) analysis, and to provide recommendations on the use of these methods in GWAS practice. PR methods are compared with SMA on realistically simulated GWAS, consisting of genotype data on single and multiple chromosomes and a continuous phenotype, and on real data.
Materials and Methods
Data simulation
To simulate GWAS data with realistic patterns of LD, we used the software HAPGEN2 (Su et al. 2011), which produces genotyped individuals by resampling from a set of reference haplotypes. We used the haplotypes of the 60 Caucasians of European origin (CEU) in HapMap2 (International HapMap Consortium 2007). For most simulation scenarios, SNP genotypes were simulated for a single chromosome (chromosome 21). For an additional simulation scenario, SNP genotypes were simulated for three chromosomes (19, 21, and 22). The number of SNPs genotyped across all 22 autosomes in the HapMap2 population is 3,849,034, and the numbers of SNPs on chromosomes 19, 21, and 22 are 56,607, 50,165, and 54,786, respectively. Following the simulation of each SNP data set, SNPs were removed if they had a minor allele frequency (MAF) <0.01 or if their absolute correlation with another SNP exceeded 0.999, reducing the number of SNPs on average to 25,033, 21,519, and 22,199 for chromosomes 19, 21, and 22, respectively.
To determine suitable sample sizes for the data simulation, we performed power calculations, using QUANTO (Gauderman and Morrison 2001). A functional or causal SNP affecting the phenotype is referred to as a quantitative trait locus (QTL). The sample sizes were determined as those needed to detect an isolated QTL by SMA, with a heritability (variance explained by the QTL over total variance) of 0.10, with a power of 50%, and with a P-value-based significance threshold of (5.5 × 10−8 × 3,849,034/50,165) for chromosome 21 and (5.5 × 10−8 × 3,849,034/(56,607 + 50,165 + 54,786)) for the three-chromosome simulation, using the GWER P-value threshold of 5.5 × 10−8 (see below). The required sample sizes were N = 201 and N = 222, respectively.
For the chromosome 21 simulations, two scenarios were considered: (1) two isolated QTL with heritability of 0.10 each and hence a total heritability of 0.20 and (2) eight QTL comprising a group of four QTL with weak pairwise LD (0.01 ≤ r2 ≤ 0.1) and two groups of two QTL each with within-group LD of r2 ≈ 0.5. All QTL had MAF > 0.05. For scenario 2, the four QTL in one group had a heritability of 0.05, and the remaining four QTL had a heritability of 0.04. Taking into account LD among the eight QTL in scenario 2, the total heritability was 0.48 [computed as where is the allelic dose of SNP k in individual i, is the regression coefficient for SNP k, and is the residual variance]. Moreover, for scenario 2 the “effective” heritability of each individual QTL in the context of the single-marker model was ≈ 0.10 due to LD among QTL. Hence, SMA had the same expected power for QTL detection under both scenarios. A total of 200 replicate simulations were performed, in which the QTL positions and effects were kept constant. The phenotype was simulated based on an additive model with Q QTL, where Y denotes the phenotype. Heritability of a single QTL j was computed as Effective heritability (increased due to LD with other QTL) was computed as above but by replacing with
For the multichromosome simulation, chromosome 19 did not harbor any QTL, chromosome 21 harbored the eight QTL of scenario 2, and chromosome 22 harbored two isolated QTL with heritability of 0.10 and MAF > 0.05 as in scenario 1. The total heritability was 0.68.
Single-marker regression
For single-marker regression, variable selection is performed by choosing a cutoff value for the P-values, determined by some method of multiple-testing control, most commonly the GWER (or FWER), which accounts for the multiplicity of the entire genome. The GWER-based P-value threshold is obtained by estimating an “effective number of independent tests” (e.g., International HapMap 2005; Dudbridge and Gusnanto 2008) by permutation or analytic approximation. The International HapMap Consortium permutation-based estimated significance threshold is 5.5 × 10−8 for two-sided tests of SNPs, which we used here. As an alternative to the stringent GWER threshold, we investigate the FDR (Benjamini and Hochberg 1995). For FDR control, we use the Benjamini–Hochberg (BH) procedure (Benjamini and Hochberg 1995) shown to control the FDR under positive-regression dependence (Benjamini and Yekutieli 2001), and this condition is believed to hold for GWAS (Sabatti and Freimer 2003). We also use Benjamini and Yekutieli’s (2001) BY approach, which ensures FDR control under any form of dependence but can be very conservative. Additionally, we consider the local FDR (Efron and Tibshirani 2002; Efron 2003), where the P-values, assumed to represent a mixture of null and alternative hypotheses, are transformed to z-scores that are modeled by a mixture distribution, or
| (1) |
where f0(z) is the density of z under the null hypothesis, f1(z) is the density under the alternative hypothesis, f(z) is the (overall) mixture density of z, and π0 is the proportion of true null hypotheses. Based on (1), the local FDR is
| (2) |
which is also a posterior probability that the null hypothesis is true given z. The FDR is a lower bound on locFDR because it is the expectation of locFDR within a tail area (Efron 2003). Sun and Cai (2007) developed an oracle testing procedure that minimizes the false negative rate (the expected proportion of false negatives among all nonrejections) subject to a constraint on the FDR. This procedure is a thresholding rule based on the locFDR and is implemented with an adaptive procedure that is asymptotically valid and optimal if the estimates of π0, f0, and f in (2) are consistent. We estimated f(z) and π0, using the simple nonparametric approach described in Storey et al. (2005). Despite the correlation structure of the z-values, f0 was fitted well by the N(0, 1) null distribution (see Supporting Information, Figure S1 as a typical representative of the data simulation replicates). We also used the consistent estimators of π0, f0, and f in equation 2 of Jin and Cai (2007). We refer to this procedure with the two sets of estimators for π0, f0, and f as LFDR1 and LFDR2. The proportion of true nulls π0 is expected to be ≤ 1.0 in the context of GWAS.
Penalized regression methods
All penalized regression methods for the multiple-regression model (assuming a centered response variable y and standardized SNP covariates x) can be represented by the estimator
| (3) |
where is a vector of p regression coefficients, yi is the continuous trait measurement on individual i, xik is allelic dose of SNP k in individual i, ei is a residual, N is sample size, P(.) is a penalty function, and is a vector of tuning or penalty parameters with typically one or two components. PR methods differ in the specification of P(.). Variable selection, by setting unimportant coefficients to zero, occurs if the penalty has a singularity at zero. Below we consider only such penalties. Given the extensive literature, we do not review these methods in detail but rather present their penalty functions and discuss some properties relevant to their application to GWAS.
Desirable properties for penalized regression methods have been established by several authors and include sparsity (variable selection is enabled by automatically setting small coefficients to zero), continuity (an estimator that is continuous in data avoids instability in model prediction), asymptotic unbiasedness (bias should be low, in particular for large true coefficients), and the (strong) oracle property (Fan and Li 2001; Fan and Peng 2004; Kim et al. 2008; Zhang 2010). The Lasso (Tibshirani 1996) performs poorly with regard to the unbiasedness property, prompting the development of other penalties to overcome this problem. The adaptive Lasso (Zou 2006), the smoothly clipped absolute deviation (SCAD) (Fan and Li 2001), and the minimax concave penalty (MCP) (Zhang 2010) possess the (strong) oracle property.
Lasso, elastic net, and adaptive lasso:
The Lasso (Tibshirani 1996) employs the L1 penalty, or
| (4) |
If there are groups of variables with strong pairwise correlations, it has been empirically observed that the Lasso tends to select only one variable from each group. This behavior is in contrast with single-marker regression, which selects all markers in sufficiently strong LD with a QTL.
The elastic net (EN) (Zou and Hastie 2005) combines the Lasso (L1) penalty, enabling it to perform variable selection, with the ridge (L2) penalty, enabling it to deal with multicollinearity, or
| (5) |
In contrast with the Lasso, the EN may be expected to coselect SNPs in strong LD. Here, EN analysis was performed with three different values for λ2: 0.9, 0.5, and 0.3. Lasso and EN are implemented in the R package glmnet (Friedman et al. 2010).
Zou (2006) proposed the adaptive Lasso to obtain an oracle procedure, which has the penalty
| (6) |
where the weights wk are computed from an initial set of solutions for the coefficients. The adaptive Lasso makes sense intuitively by placing smaller weights on the important regressors to reduce the shrinkage applied to their coefficients. Here we used SMA, Lasso with CV, and ridge regression with CV as the initial estimators.
MCP and SCAD:
The MCP and SCAD penalties depend on two tuning parameters; both begin with the same penalization as the Lasso but increasingly reduce the penalization farther away from zero (e.g., Breheny and Huang 2011). Because in preliminary studies we found little difference between the MCP and SCAD results and SCAD is expected to perform somewhere between MCP and Lasso (Breheny and Huang 2011), here we consider only the MCP with penalty function
| (7) |
For the second tuning parameter in MCP (λ2), four values were evaluated (3, 10, 30, and 100), with the largest value approaching the Lasso. MCP is implemented in the R package ncvreg (Breheny and Huang 2011).
Selection of tuning parameter values: Current state:
A major practical issue with the use of penalized regression for GWAS is the need to determine optimal values for the tuning parameter(s). The “usual” approaches include CV and the use of model selection criteria such as AIC, BIC, and EBIC. While CV and AIC are generally useful for prediction rather than model/variable selection, BIC tends to select a true sparse model but does not achieve sufficient sparsity in high-dimensional very sparse settings such as linkage (QTL) mapping and GWAS for which modified BIC criteria have been proposed (e.g., Bogdan et al. 2004). Moreover, the BIC criterion is a function of the degrees of freedom (d.f.) of a model, and the d.f. are not straightforward to obtain in PR (e.g., Ye 1998; Zou et al. 2007; Zhang 2010).
Given the shortcomings of model selection criteria, a superior approach should be obtained by combining PR with the control of an error rate such as FWER or FDR. Ayers and Cordell (2010) used data permutation to determine the value of the tuning parameter. Multistage strategies have also been suggested as an approach to performing error control with PR (Meinshausen et al. 2009; Wasserman and Roeder 2009). Meinshausen et al. randomly split the data in half multiple (B) times. Each time the first half was used for variable screening using Lasso with CV and related methods, and the second half was used to compute adjusted P-values, based on which the FWER and the FDR can be controlled asymptotically. We used Lasso with CV for tuning parameter value selection in the screening step. We expected this method to have reduced power relative to our single-step FDR-controlling methods (see below) due to the data splitting and the use of multiple regression, which reduces the significance of two (highly) correlated variables representing alternative hypotheses. Performing the two steps of screening and variable selection both on the entire data set as in Sun et al. (2010) should improve power; these authors used their Iterative Adaptive Lasso in the screening step and backward selection instead of multiple regression in the second (cleaning) step. However, there is no analytical proof of (asymptotic) FDR (or FWER) control, and we have therefore not further considered a two-step approach performed on the entire data set.
Most recently, Sampson et al. (2013) have proposed a local FDR-based method for selecting the value of the tuning parameter specifically for the adaptive Lasso. The advantage of this approach is that it uses the local FDR based on which optimal oracle procedures were developed for SMA (Sun and Cai 2007; Cai and Sun 2009).
The penalized unified multiple-locus association (PUMA) method (and software) implements some penalized regression methods for GWAS with an efficient algorithm for generalized linear models and with preselection of SNPs based on SMA (Hoffman et al. 2013) to make these methods applicable to large GWAS data sets. It performs heuristic model and SNP selection. PUMA implements MCP with a two-dimensional model search across its two tuning parameters (2D-MCP), and we chose this method here.
A different type of multivariate method for SNP selection in GWAS was proposed by Zuber et al. (2012). For a continuous phenotype, this method computes correlation-adjusted marginal correlations (CAR scores) as where is a vector of marginal correlations between each SNP dose (X) and the phenotype (Y), and is the correlation matrix among the SNPs, which is estimated by a computationally efficient shrinkage method. Note that if there are non-SNP study covariates, the phenotype (Y) must be replaced with the residuals from a linear model containing these covariates. For SNP selection, CAR scores are provided as input to the fdrtools R package that computes P-values, FDR, and local FDR values based on its empirical null modeling (Strimmer 2008). We refer to this method as CAR.
Selection of tuning parameter values: Control of the false discovery rate
Here we propose two approaches for combining penalized regression with FDR control, which can be applied to all penalized regression methods considered here and do not require data splitting or subsampling. The first approach is based on data permutation and the second on an analytic approximation.
FDR control using data permutation:
Let λ denote the single (Lasso) or the first (MCP, EN) tuning parameter. Penalized regression [with coordinate descent algorithms (Friedman et al. 2007)] is efficiently performed by computing the coefficient solutions on a grid of (say, 1000) λ-values, starting from a maximum value λmax at which all solutions are zero and ending at a minimum value λmin that is zero or produces an excessively large model, with the solutions from any previous λ-value serving as a warm start for the next λ. Therefore, the original data set is analyzed on this grid of λ-values, and the number of nonzero coefficients is determined at each λ, denoted by R(λ). Then B permuted data sets (with random reorderings of the continuous vector of phenotypes) are analyzed on the same grid of λ-values, and the number of nonzero coefficients is again determined for each permuted data set b (b = 1, … , B) and λ-value, denoted by F(b, λ). Then for each λ-value, we compute the permutation FDR as the average number of nonzero solutions in the permuted data over the number of nonzero solutions in the original data, We note that here we define the FDR as E(F)/R, where F denotes the number of false positives and R the number of rejections (recall that in PR a null hypothesis is rejected if the corresponding coefficient solution is nonzero). This definition is different from the original definition E(F/R) and thus does not take into account the dependency between F and R, but is easier to work with.
Analytic FDR control:
We now present an approximate, analytic FDR method that was originally proposed in Breheny (2009). This approach can be applied to all penalized regression methods considered here, but we illustrate it for the Lasso and the MCP. We assume that the predictors have been standardized such that and We write the objective function in terms of a single predictor (k), as in Friedman et al. (2007) for the Lasso and in Breheny and Huang (2011) for the MCP [setting λ = λ1 and γ = λ2 in (7)],
| (8) |
where and Differentiating (8) with respect to βk yields the solutions
| (9) |
Based on (9), we consider the probability of a false positive, or We again define the FDR as E(F)/R and approximate the numerator with
| (10) |
In general, FDR control based on (10) is expected to be conservative, similar to the BH FDR, because in (10) we sum over all variables rather than the (unknown) true null variables. However, this should not matter in the context of GWAS where the proportion of null variables is very close to 1.
We rewrite the probability of a false positive as
| (11) |
The distribution of the estimated residuals (r) is unknown and complicated but may be approximated by the normal distribution or even simpler by such that This is an approximation for multiple reasons, including the normality assumption, the mean of zero (correct for least squares where the coefficients are estimated unbiasedly but not for PR), and the independence or zero covariances. Then
| (12) |
Using the “natural” estimator (12) becomes constant for all k and
| (13) |
The FDR estimation (13) is noisy when the model becomes saturated; this is not a problem in practice when FDR evaluation starts from a model with all coefficients set to zero (λmax) and terminates once the desired level of the FDR has been stably exceeded.
When applying the FDR estimation described for the Lasso and the MCP to the EN, Equation 13 needs to be modified to
| (14) |
Penalized regression with fusion-type penalties
It is well known that the Lasso tends to select a single predictor from a group of predictors that have strong pairwise correlations (in the extreme the Lasso does not have a unique solution when two variables are perfectly collinear). For this reason, we prefer the EN (see below), but some authors (Kim and Xing 2009; Liu et al. 2011) have suggested that fused Lasso-type penalties developed for covariates measured on graphs would be more appropriate than the Lasso in such situations. An expectation was that such penalties may increase power and decrease the false positive rate. Fusion-type penalties impose pairwise similarity on the coefficient solutions of highly correlated predictors or encourage sparsity in the differences among the values of the regression coefficients. Pairwise similarity can be imposed on the effects of correlated (SNP) regressors by adding an extra penalty to the existing penalty term, depending on the type of PR method used (here the Lasso). We first consider an added penalty of the form (referred to as LD2lasso)
| (15) |
where Sk is a set of SNPs that are correlated to SNP k, rkm denotes the (Pearson) correlation coefficient among the allelic doses of SNPs k and m, and h(rkm) is a function of rkm that we specified as h(rkm) = |rkm| or h(rkm) = (rkm)2. Set Sk can be limited to adjacent SNPs (Liu et al. 2011) or contain all SNPs whose absolute correlation with SNP k exceeds a certain threshold (here). Taking absolute values of the coefficients accounts for the fact that two correlated SNPs can have similar effect sizes but opposite signs due to the arbitrary coding of the SNP alleles. We note that Li and Li (2008, 2010) used a similar penalty for the general case of (genomics) variables whose dependency structure can be represented as a graph. Their penalty is induced by the Laplace matrix of the graph and differs from (15) by dividing the regression coefficients by the square root of the degree of the corresponding variables, which is justified by the argument that variables with more connections should be allowed to have larger coefficients. This argument does not apply in the GWAS context, and we therefore use (15). Using (3) with the Lasso penalty and the added penalty in (15), the LD2lasso objective function for a single coefficient k can be written as
| (16) |
where
Then one can show that the LD2lasso solution satisfies
| (17) |
For LD2lasso with analytic FDR control, the probability of a false positive analogous to (12) is
| (18) |
where in contrast with (12) the threshold (κ) applied to is now a function of the LD2lasso solutions of other SNPs in strong LD with the current SNP.
An alternative to (15) is a penalty that we refer to as the LD1lasso penalty and is essentially identical to a penalty term in the general and graphical fused Lasso (e.g., Kim and Xing 2009):
| (19) |
Computation of the LD2lasso solution (17) was implemented with a straightforward pathwise coordinate descent algorithm, because its objective function consists of a convex, a continuously differentiable, and a separable term (of convex functions) (Tseng 1988, 2001; Friedman et al. 2007; Chen et al. 2010). This does not apply to the LD1lasso, and hence we focused on the LD2lasso, which seemed to perform better (results not shown).
Consider a group of SNPs in strong LD and containing a single SNP that is causal for the phenotype. In this situation, the Lasso (and other penalties not including fusion-type penalties) will tend to select a single or few SNPs, while LD2lasso/LD1lasso would have nonzero solutions for several or all SNPs in this group. Hence for PR without fusion-type penalties, a QTL region for subsequent fine-mapping may be identified as the SNP(s) with nonzero solution plus other SNPs in LD with these SNP(s) above some threshold (e.g., |r| > 0.5). For LD2lasso/LD1lasso, a QTL region may be identified as including all (contiguous) SNPs with nonzero coefficients.
Analyses of multiple chromosomes
The analysis of multiple chromosomes may be regarded as a multigroup analysis, which we first discuss in the context of SMA. For a given biomedical trait of interest, many chromosomes may show no signal, while other chromosomes may show sparse signals or even extended stronger signals. To control the GWER such a grouping structure is irrelevant. However, for FDR control there is no unique way to proceed in the presence of a group structure. As argued by Efron (2008), the usual pooled FDR analysis, where the group structure is simply ignored, can be overly conservative or overly liberal for any particular group. Efron also established that a separate analysis, where FDR control (at the same level) is performed separately for each group and subsequently the results are simply combined, is valid in terms of achieving overall (i.e., across groups) FDR control (at that same level). Efron’s concern about joint FDR analysis of all groups (here chromosomes) within an experiment or study applies directly to SMA with FDR control in GWAS. While this also applies to PR with FDR control, here a separate analysis of each chromosome implies that QTL on other chromosomes are not included in the PR multimarker model, potentially lowering detection power and diminishing the postulated advantage of an all-markers over single-marker analysis. Finally, for most GWAS, one expects that the proportions of true null hypotheses are close to 1 and very similar across groups (chromosomes), so there may be little difference between joint and separate analyses and group-based analyses described below.
SMA:
Besides these two basic strategies of pooled and separate analyses, some specialized methods that take the group structure into account have recently been proposed in the context of univariate analysis (SMA) (Cai and Sun 2009; Hu et al. 2010). Cai and Sun estimated the local FDR within each group and then applied the (approximate oracle) thresholding procedure to the combined set of local FDR values. For SMA of multiple chromosomes, we compared (i) pooled analysis based on the BH method or the local FDR thresholding procedure, (ii) separate analysis based on BH or the local FDR procedure, and (iii) the local FDR-based grouping procedure of Cai and Sun (2009).
Sun and Cai (2009) extended their earlier method to dependent test statistics, using a hidden Markov model (HMM). Wei et al. (2009) combined this method with the group-based method of Cai and Sun (2009) in the context of GWAS and implemented it in the R package PLIS. We tested this method on the single chromosome 21 simulation with eight true QTL, where it produced a very high number of false positives (irrespective of whether the alternative component was fitted with a mixture or a single normal distribution), and hence we did not further pursue the PLIS method.
Penalized regression:
For PR with FDR control, we also compared separate and pooled analyses. For the pooled analysis, the SNPs from all three chromosomes were fitted jointly with the Lasso penalty, and the penalty parameter value was chosen based on controlling the FDR at level 0.05. For the separate analysis, the Lasso analysis was performed separately for each chromosome by omitting the SNPs on the other chromosomes and choosing a separate penalty parameter value to control the FDR at level 0.05 for each chromosome.
Comparison among methods
We define a causal true positive (CTP) as a QTL (defined earlier as a functional or causal SNP affecting the phenotype) that is significant (significant means identified by any of the methods considered). We define a linked true positive (LTP) as a SNP that is significant and has an absolute correlation with a QTL above a certain threshold (T) (note that T is a threshold on the absolute correlation coefficient, not the squared coefficient). We define a CLTP as being either a CTP or an LTP. Methods are compared, across the replicate simulations, based on (1) true positive rate (TPR)1 = number of CTPs/number of QTL, (2) TPR2 = number of QTL detected by at least one CLTP (CTP or LTP)/number of QTL, and (3) threshold-specific empirical tFDR = number of significant SNPs that are not a CLTP at a given T/number of significant SNPs. For criterion 2, we used T = 0.9, 0.7, and 0.5 for an increasingly relaxed definition of TP. For criterion 3, we used T = 0.5, 0.3, and 0.25. The first value (T = 0.5) was chosen because our simulation was designed such that a QTL had a power of 0.5 to be detected and computed using Quanto for a given heritability, sample size, and chromosome-wide P-value threshold, and this power decreased to < 0.01 for a SNP correlated with the QTL at 0.5 (assuming that SNP and QTL have the same allele frequency). The second value (T = 0.3) is just below the threshold of “useful LD” (r = 0.316 or r2 = 0.1) as defined by Kruglyak (1999) and Pritchard and Przeworski (2001) as the value at which the sample size is increased at most 10-fold. The last value (T = 0.25) is close to the threshold used in a previous comparison study (Ayers and Cordell 2010). For completeness, we also provide tFDR values at the remaining thresholds used for TPR2 (0.9, 0.7).
It is not straightforward to evaluate the empirical FDR in GWAS simulations due to LD. We experimented with alternative ways of defining the tFDR, such as partitioning a chromosome into 100-kb windows and computing tFDR (for a given T) as the number of windows in which the significant SNPs are all false positives divided by the number of windows that contain at least one significant SNP. Such an alternative approach did not change any results significantly, so we report tFDR for the first definition.
The results are presented in figures while more detailed results in the form of tables are available in File S1; the tables include standard errors for all average (across replicates) tFDR, TPR1, and TPR2 values that were used for computing P-values for differences in these values between methods or for a one-sided test of average tFDR to exceed level 0.05.
Results and Discussion
Single-chromosome simulation with two or eight QTL: Single-marker analysis
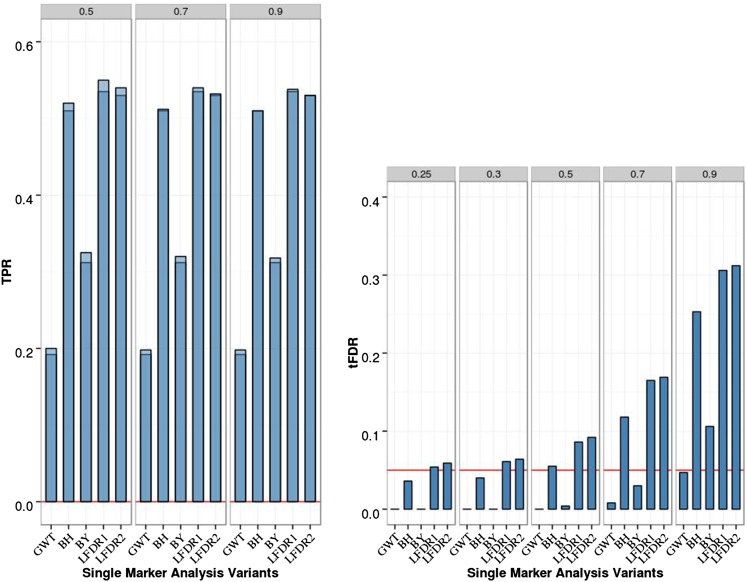
For two isolated QTL, SMA results are summarized in Figure 1 (and Table S1 in File S1). For SMA with GWER control [referred to as genome-wide threshold (GWT) in Figure 1] that controlled the FWER conservatively, the TPR was low (0.2) as expected. For SMA with FDR control, BH and BY controlled the FDR at level 0.05 based on two (three) of the three empirical threshold-specific FDRs (tFDRs), while the local FDR-based approximate oracle procedures LFDR1 and LFDR2 had tFDR values all > 0.05 although not significantly (at nominal P-value of 0.05). The TPR was somewhat higher (0.31) for BY compared with the GWT, while it was substantially higher for BH, LFDR1, and LFDR2 (> 0.5).
Figure 1.
Single-marker analysis (SMA) of chromosome 21 data with two isolated QTL, 200 replicates, and sample size N = 201. Empirical power is shown [defined as true positive rate (TPR)] on the left: Dark blue represents TPR1 as defined in the main text (TPR including only the significant QTL), and light blue represents TPR2 (TPR also including significant SNPs linked to QTL at absolute correlation threshold 0.5, 0.7, or 0.9). Empirical thresholded false discovery rate (tFDR) with absolute correlation thresholds 0.25, 0.3, 0.5, 0.7, and 0.9 is shown on the right: Red horizontal line represents the tFDR value of 0.05. GWT, genome-wide threshold that represents the P-value threshold 5.5 × 10−8; BH, Benjamini–Hochberg; BY, Benjamini–Yekutieli; LFDR1 and LFDR2, local FDR-based thresholding using different estimators of the proportion of true null hypotheses (see main text for details).
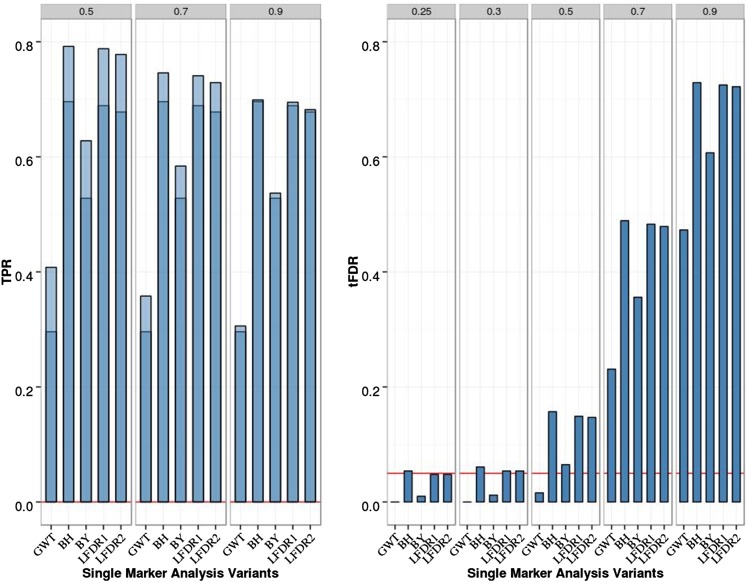
For the eight-QTL scenario (Figure 2 and Table S2 in File S1), TPR was increased for the FDR-based SMA methods relative to the two-QTL case (≥ 0.7 for BH, LFDR1, and LFDR2; ∼0.6 for BY compared with ≤ 0.4 for GWT). All FDR methods controlled the FDR in the sense of producing at least one tFDR value that was not significantly > 0.05 (nominal P-value). The smallest P-value for exceeding level 0.05 by a tFDR value at T = 0.3 was P = 0.034 for SMA-BH.
Figure 2.
Single-marker analysis (SMA) of chromosome 21 data with eight QTL, 200 replicates, and sample size N = 201. Empirical power [defined as true positive rate (TPR)] is on the left: Dark blue represents TPR1 as defined in the main text (TPR including only the significant QTL), and light blue represents TPR2 (TPR also including significant SNPs linked to QTL at absolute correlation threshold 0.5, 0.7, or 0.9). Empirical thresholded false discovery rate (tFDR) with absolute correlation thresholds 0.25, 0.3, 0.5, 0.7, and 0.9 is shown on the right: Red horizontal line represents the tFDR value of 0.05. GWT, genome-wide threshold that represents the P-value threshold 5.5 × 10−8; BH, Benjamini–Hochberg; BY, Benjamini–Yekutieli; LFDR1 and LFDR2, local FDR-based thresholding using different estimators of the proportion of true null hypotheses (see main text for details).
Single-chromosome simulation with two isolated QTL: PR with FDR control
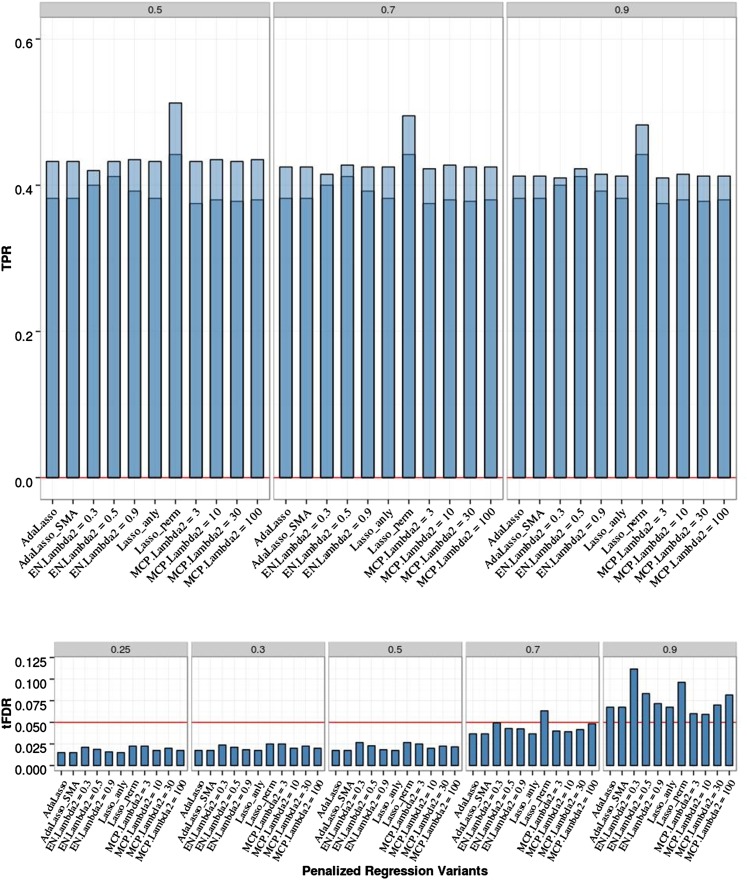
We verified the expectation that Lasso with standard tuning parameter value selection based on CV produces an unacceptably high empirical FDR (> 0.5; results not shown). The results for PR with FDR control for this scenario are presented in Figure 3 and Table S3 and Table S4 in File S1. Differences in empirical tFDR and TPR among all PR methods with analytic FDR control were small; there was a very slight advantage for the elastic net with the mixing parameter set to 0.5 (EN50); Lasso and MCP with different values for the second tuning parameter (from λ2 = 3 to λ2 = 100) produced almost identical results. All PR methods were able to control the FDR by producing tFDR values < 0.05 for all T ≤ 0.5. In terms of power and using the EN50 to represent PR, in comparison with SMA-BH, the average TPR1 and TPR2 values of EN50 were lower with P-values for the differences ranging from 0.004 to 0.013. Comparing the EN50 with SMA-BY, the average TPR1and TPR2 values of EN50 were higher with all P-values ∼0.002. The Lasso with permutation FDR control had slightly higher tFDR values compared with the Lasso with analytic FDR control; TPR1 and TPR2 of the permutation Lasso were similar to those of SMA-BH and higher than those of the Lasso with analytic FDR control (smallest P-value for the difference = 0.01). The results for Lasso with permutation FDR were stable across different sets of permutations (100–400 permutations).
Figure 3.
Penalized regression (PR) analyses of chromosome 21 data with two isolated QTL, 200 replicates, and sample size N = 201. Empirical power [defined as true positive rate (TPR)] is shown on the top: Dark blue represents TPR1 as defined in the main text (TPR including only the significant QTL), and light blue represents TPR2 (TPR also including significant SNPs linked to QTL at absolute correlation threshold 0.5, 0.7, or 0.9). Empirical thresholded false discovery rate (tFDR) with absolute correlation thresholds 0.25, 0.3, 0.5, 0.7, and 0.9 is shown on the bottom: Red horizontal line represents the tFDR value of 0.05.The following penalized regression methods (in the order shown) are represented: AdaLasso, adaptive Lasso where all three initial estimators (SMA, Lasso with CV, and ridge regression with CV) produced the same result; EN_Lambda2 = x, elastic net with Lasso portion of λ2 = x (x = 0.3, 0.5, 0.9); Lasso_anly, Lasso with analytical FDR control; Lasso_perm, Lasso with permutation FDR control; MCP_lambda2 = x, minimax concave penalty with λ2 = x (x = 3, 10, 30, 100).
Single-chromosome simulation with eight QTL: PR with FDR control
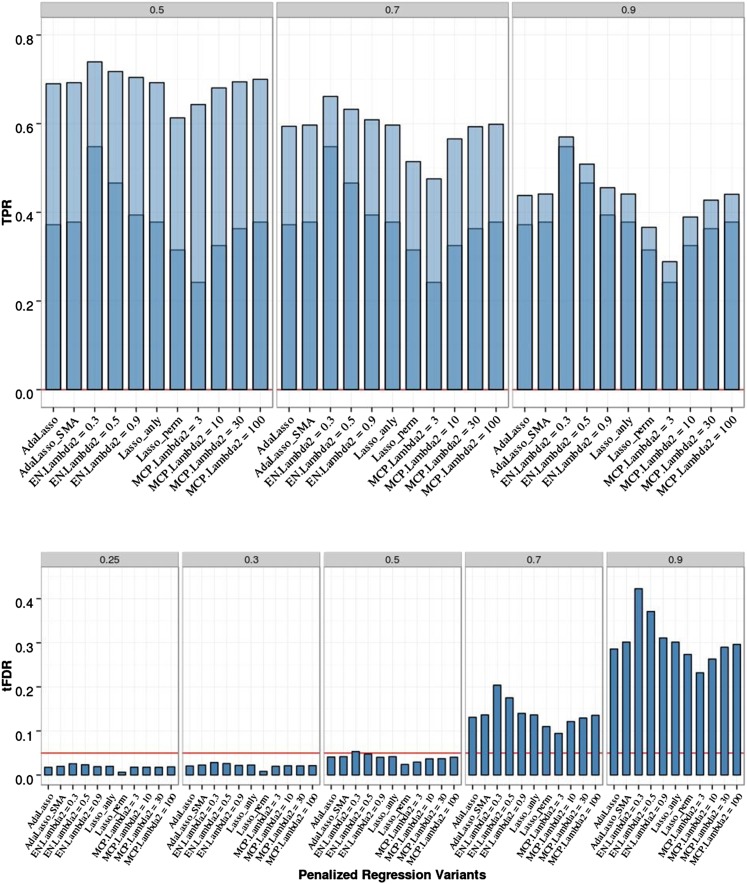
The results for this scenario are presented in Figure 4 and Table S5 and Table S6 in File S1. All PR methods were able to control the FDR by producing tFDR values below (or not significantly above) 0.05 for all T ≤ 0.5. Most of the differences in empirical tFDR and TPR among different PR methods with analytic FDR control remained small but some were more pronounced for the eight-QTL data relative to the two-QTL data: EN50 had a higher TRP1 and TRP2 than the Lasso, with a P-value of 2 × 10−5 for the difference in TPR1 and P = 0.001 for TPR2 at T = 0.9; MCP with low values for the second tuning parameter (λ2 = 3, 10) had lower TPR1 (P-values 1.3 × 10−14 and 0.003, respectively) and TPR2 (P-values from 2 × 10−15 to 0.03 and from 0.003 to 0.33, respectively, with the upper end of each range representing TPR2 at T = 0.5) than the Lasso. Power was expectedly higher relative to the two-QTL scenario (for EN50, TPR2 at T = 0.5 increased from 0.43 to 0.73). Comparing the EN50 with SMA-BH, the average TPR1 and TPR2 values of EN50 were lower with P-values for the differences ranging from 5 × 10−22 to 0.001. Comparing the EN50 with SMA-BY, EN50 had a significantly higher value for TPR2 at T = 0.5 (P-value 0.0005) while EN50 had a lower value for TPR1 (P-value 0.005). In general, differences among PR methods were largest for TPR1 and TPR2 at T = 0.9 and were diminished for TPR2 with T = 0.5. Comparing Lasso with permutation vs. analytic FDR estimation, the permutation Lasso had lower TPR1 and TPR2 with P-values for the differences ≤ 0.0006. It is surprising that the permutation method, which was the most powerful approach in the two-QTL scenario, is so conservative in the eight-QTL scenario. This appears to be due to the fact that permuting the phenotypes, while justified under the global null hypothesis of no association between any SNPs and the phenotypes, is conservative when evaluating the significance of a QTL in the presence of substantial signal from other QTL. By permuting the phenotype, which consists of both fixed signal and random error, the contribution of the random errors is overestimated. This does not affect the validity of the permutation approach in terms of properly controlling the FDR but does diminish power in cases like the eight-QTL scenario.
Figure 4.
Penalized regression (PR) analyses of chromosome 21 data with eight QTL, 200 replicates, and sample size N = 201. Empirical power [defined as true positive rate (TPR)] is shown on the top: Dark blue represents TPR1 as defined in the main text (TPR including only the significant QTL), and light blue represents TPR2 (TPR also including significant SNPs linked to QTL at absolute correlation threshold 0.5, 0.7, or 0.9). Empirical thresholded false discovery rate (tFDR) with absolute correlation thresholds 0.25, 0.3, 0.5, 0.7, and 0.9 is shown on the bottom: Red horizontal line represents the tFDR value of 0.05.The following penalized regression methods (in the order shown) are represented: AdaLasso, adaptive Lasso where all three initial estimators (SMA, Lasso with CV, and ridge regression with CV) produced the same result; EN_Lambda2 = x, elastic net with Lasso portion of λ2 = x (x = 0.3, 0.5, 0.9); Lasso_anly, Lasso with analytical FDR control; Lasso_perm, Lasso with permutation FDR control; MCP_lambda2 = x, minimax concave penalty with λ2 = x (x = 3, 10, 30, 100).
The eight-QTL simulation included two groups of two QTL each with within-group r2 ≈ 0.5 (absolute correlation among SNP doses of 0.71). Because of the Lasso’s tendency to select only a single SNP per group and the EN’s ability to deal with colinearity and potential coselection of correlated SNPs (SNPs in LD), we took a closer look at the behavior of Lasso and EN in this situation. Averaged over both groups and the 200 replicate simulations and including LTPs (T = 0.5), the Lasso selected zero SNPs in 22%, one SNP in 43%, two SNPs in 26%, and more than two SNPs in 9% of cases. By comparison, the EN selected zero SNPs in 19%, one SNP in 37%, two SNPs in 31%, and more than two SNPs in 13% of cases. Expectedly the EN selected two and more than two SNPs more frequently but the difference was not substantial.
Single-chromosome simulation with two/eight QTL: Fusion-type penalties
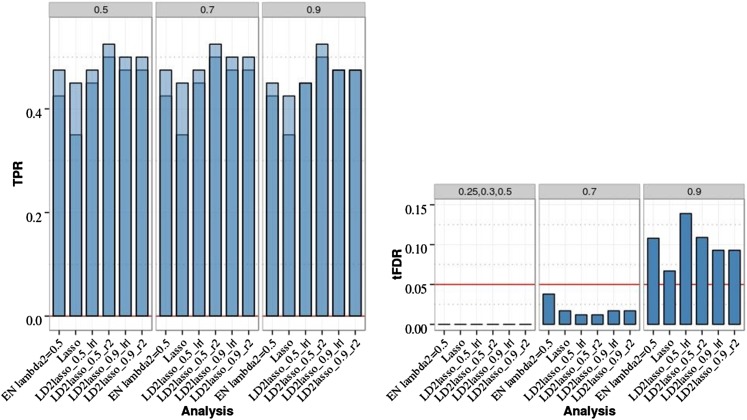
Here we focused on the LD2lasso with φ (the relative weight given to the Lasso penalty) fixed at two different values (0.9, 0.5), with the two LD functions h(r) = |r| and h(r) = r2, and with h(r) = 0 if |r| < 0.85 [for this threshold, the median number of SNP “neighbors” was 2 with interquartile range (IQR) 0–5 and range 0–39] or |r| < 0.50 (median number of SNP neighbors = 14 with IQR 6–25 and range 0–102). Because we did not optimize the implementation of the LD2lasso for computational speed, we compared the LD2lasso with the Lasso and EN50 based on only 20 data replicates that were randomly selected from the 200 replicates. For the chromosome 21 data with two isolated QTL, the LD2lasso achieved higher power than Lasso and EN50 while maintaining FDR control (Figure 5 and Table S7 in File S1). The largest difference was the increase in TPR1 from 0.35 for the Lasso to 0.5 for the best LD2lasso analysis with φ = 0.5 and h(r) = r2 (P-value < 0.05), while all other differences in TPR1 and TPR2 were smaller.
Figure 5.
Empirical power [true positive rate (TPR)] and thresholded false discovery rate (tFDR) for LD2lasso vs. Lasso and elastic net (EN) penalized regression (PR) analyses of chromosome 21 data with two isolated QTL, 20 randomly selected replicates, and sample size N = 201. Dark blue represents TPR1 as defined in the main text (TPR including only the significant QTL), and light blue represents TPR2 (TPR also including significant SNPs linked to QTL at absolute correlation threshold 0.5, 0.7, or 0.9). The empirical tFDR was computed at five absolute correlation thresholds between causal and linked SNPs (T = 0.25, 0.3, 0.5, 0.7, 0.9). The red horizontal line represents the tFDR value of 0.05. EN_Lambda2 = 0.5 denotes the elastic net with Lasso portion of λ2 = 0.5; LD2lasso_0.9_r2 (LD2lasso_0.9_|r|) represents the LD2lasso with weight on the Lasso penalty equal to φ = 0.9 and LD function h(r) = r2 (h(r) = |r|). The LD function was set to zero when the absolute correlation between two SNPs was |r| < 0.85.
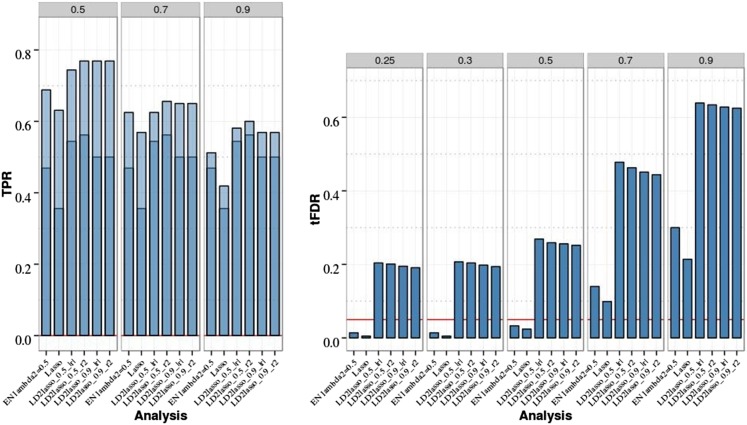
For the chromosome 21 data with eight QTL (Figure 6 and Table S8 in File S1), however, all LD2lasso variants were unable to control the FDR with average tFDR values at the lowest threshold of T = 0.25 near 0.2 (P-value for the differences to FDR level 0.05 < 4 × 10−4). We recall that the eight-QTL data contained a group of four QTL with weak pairwise LD (0.01 ≤ r2 ≤ 0.1); the high tFDR values (0.2–0.26) for LD2lasso were mostly due to SNPs in LD with the group of four causal SNPs at small absolute correlation values. In terms of TPR, the difference in TPR1 between the best LD2lasso analysis with φ = 0.5, h(r) = r2, and |r| < 0.85 (|r| < 0.50) and the Lasso had a P-value of 5.4 × 10−5 (1.7 × 10−5), while the difference in TPR1 between the best LD2lasso and the EN50 had a larger P-value of 0.10 (0.008). TPR2 values were also higher, but most P-values for the differences in TPR2 between the same methods were > 0.05 due to smaller differences (less than half) and the small number of replicates.
Figure 6.
Empirical power [true positive rate (TPR)] and thresholded false discovery rate (tFDR) for LD2lasso vs. Lasso and elastic net (EN) penalized regression (PR) analyses of chromosome 21 data with eight QTL, 20 randomly selected replicates, and sample size N = 201. Dark blue represents TPR1 as defined in the main text (TPR including only the significant QTL), and light blue represents TPR2 (TPR also including significant SNPs linked to QTL at absolute correlation threshold 0.5, 0.7, or 0.9). The empirical tFDR was computed at five absolute correlation thresholds between causal and linked SNPs (T = 0.25, 0.3, 0.5, 0.7, 0.9). The red horizontal line represents the tFDR value of 0.05. EN_Lambda2 = 0.5 denotes the elastic net with Lasso portion of λ2 = 0.5; LD2lasso_0.9_r2 (LD2lasso_0.9_|r|) represents the LD2lasso with weight on the Lasso penalty equal to φ = 0.9 and LD function h(r) = r2 (h(r) = |r|). The LD function was set to zero when the absolute correlation between two SNPs was |r| < 0.85.
Single-chromosome simulation with eight QTL: PR vs. SMA and CAR
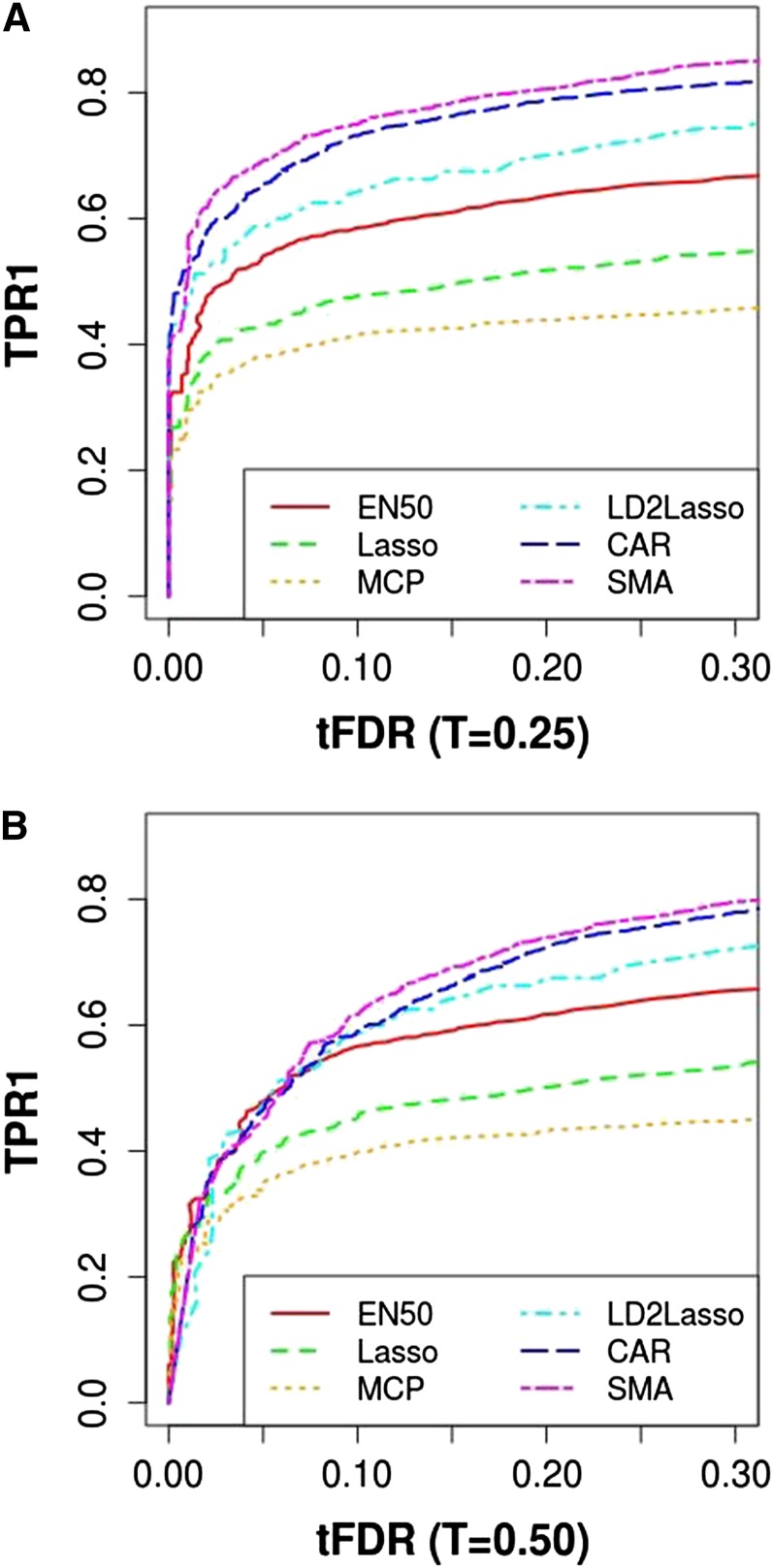
Because all previous comparisons focused on differences among methods in empirical TPR and tFDR for a single FDR cutoff of 0.05, here we present TPR1 vs. tFDR at T = 0.25 and T = 0.50 (Figure 7) and TPR2 (T = 0.50) vs. tFDR at T = 0.25 and T = 0.50 (Figure 8). Figure 7 and Figure 8 were generated by varying the P-value cutoff for SMA and CAR and varying the penalty parameter for PR. In terms of power to detect causal SNPs (TPR1) and when using the most relaxed definition of tFDR (with T = 0.25), SMA dominated all other methods with CAR being second followed by LD2lassso and EN50, with Lasso and MCP (λ2 = 10) performing worst (Figure 7). When using a more stringent definition of false positives with tFDR at T = 0.50, SMA/CAR dominated in terms of TPR1 only for tFDR values > 0.05.
Figure 7.
Plot of true positive rate TPR1 vs. thresholded false discovery rate (tFDR): TPR1 vs. tFDR (T = 0.25) and TPR1 vs. tFDR (T = 0.50). TPR1 and tFDR are defined in the main text. MCP was run with λ2 = 10. CAR represents correlation-adjusted marginal correlations.
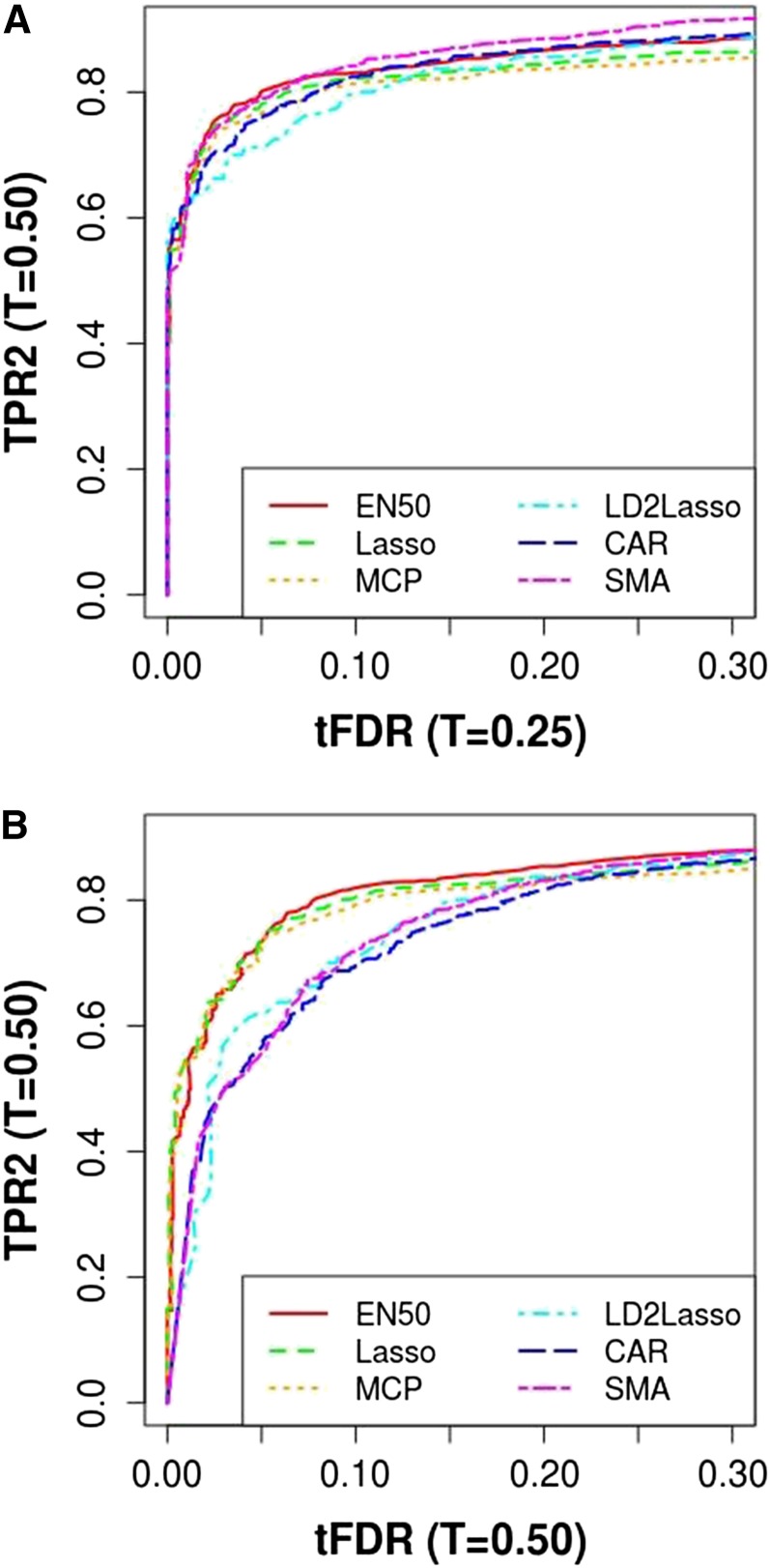
Figure 8.
Plot of true positive rate TPR2 vs. thresholded false discovery rate (tFDR): TPR2 (T = 0.50) vs. tFDR (T = 0.25) and TPR2 (T = 0.50) vs. tFDR (T = 0.50). TPR2 and tFDR are defined in the main text. MCP was run with λ2 = 10. CAR represents correlation-adjusted marginal correlations.
In terms of power to detect QTL with causal or linked SNPs (TPR2 at T = 0.5) (Figure 8) and when using the most relaxed definition of tFDR (with T = 0.25), EN50 and SMA (the latter for higher tFDR (T = 0.25) values) slightly dominated the other methods, with the LD2lasso now performing worst for the most relevant range of tFDR (T = 0.25) from 0.01 to 0.10. When replacing tFDR (T = 0.25) with tFDR (T = 0.50), EN50 performed best (slightly better than Lasso and MCP), with SMA, CAR, and LD2lasso now separated as the worst performers.
Single-chromosome simulation with two/eight QTL: Previous approaches
The multisplit analysis of Meinshausen et al. (2009) was evaluated based on a random subset of 20 replicates of the chromosome 21 two-QTL data and on another random subset of 20 replicates of the chromosome 21 eight-QTL data (Table S9 in File S1). It controlled the FDR quite conservatively (all tFDR values equal to zero) and had low power relative to the single-step PR methods with FDR control, as expected. Power (TPR) was < 0.18 for the two-QTL data and < 0.21 for the eight-QTL data. P-values for the differences in TPR between the multisplit analysis and the Lasso ranged from 0.01 to 0.04 for the two-QTL data and from 2.4 × 10−7 to 4 × 10−4 for the eight-QTL data.
The adaptive Lasso with local FDR control of Sampson et al. (2013) was evaluated using all 200 replicates of the chromosome 21 two-QTL and eight-QTL data (Table S10 in File S1). R code for executing this method was generously provided to us by the first author (H. Yi). For our data, the local FDR adaptive Lasso controlled the FDR at the local FDR threshold of 0.1 (tFDR values < 0.05), but its power (TPR) was much lower than that of the Lasso for both the two-QTL and the eight-QTL data (P-values for the differences in TPR to the Lasso ranged from 1 × 10−12 to 2 × 10−13 and from 2.4 × 10−67 to 2.5 × 10−83, respectively). When raising the local FDR threshold to 0.5, the FDR was not controlled for the two-QTL data (tFDR values highly significantly > 0.05) but was controlled for the eight-QTL data (tFDR values < 0.05) where the TPR values, however, remained significantly below the values of the Lasso.
PUMA was run on the eight-QTL data with 200 replicates, choosing the 2D-MCP method, omitting the SNP preselection step and otherwise using default parameter values. The results are in Table S11 in File S1 for four different P-value thresholds for SNP selection ranging from 1 × 10−7 to 0.05, and these results need to be compared with our MCP with analytic FDR control [and four fixed values of the second tuning parameter λ2 in (9)] in Table S6. The authors (Hoffman et al. 2013) recommended a P-value threshold of 1 × 10−7 for 2D-MCP, which controlled the FDR in terms of the tFDR values in Table S11; the next lower threshold of 1 × 10−6 had tFDR values >0.06 whose one-sided test for exceeding level 0.05 had P-values from 0.055 to 0.11. Overall, TPR1 and TPR2 values for 2D-MCP were significantly below those for MCP with analytic FDR control (P-values for differences from 9 × 10−5 to 8 × 10−29 for 2D-MCP SNP selection threshold 1 × 10−7 and from 0.032 to 2 × 10−22 for 2D-MCP SNP selection threshold 1 × 10−6). To explain these differences, we looked at the values for λ2 for the models chosen in PUMA by AIC over the 200 data sets, which surprisingly had a median of 3.46, with almost all values well below 10. For our own MCP analysis with FDR control we had chosen four values for the second tuning parameter (3, 10, 30, and 100), of which the lowest value performed the worst. We then replaced the heuristic AIC model selection followed by SNP selection of PUMA with our analytic FDR tuning parameter selection, which significantly improved TPR1 and TPR2 (last column of Table S11). The analytic FDR criterion selected models with larger values for λ2 with a median of 33.6 (as well as larger values for λ1, median 0.166 vs. 0.310) This improvement still underperformed the results in Table S6 because PUMA uses a (internally determined) grid of values for λ1 and λ2 in 2D-MCP, which was too sparse for optimal performance of the analytic FDR approach.
Analyses of multiple chromosomes
Here we analyzed the simulated data including chromosomes 19 (no QTL), 21 (eight QTL), and 22 (two QTL) with 200 replicates. For SMA, the average tFDR values for pooled and separate BH analyses and for the local FDR grouping procedure were all > 0.05 for T = 0.25 and 0.3 but most were insignificantly higher (largest P-value of 0.001 for separate BH). Differences in TPR between SMA-based pooled and separate analyses and the grouping procedure of Cai and Sun (2009) were small and insignificant, although there was a trend for increasing TPR from pooled analysis to separate analysis to the grouping procedure (see Table S12 in File S1).
For penalized regression using Lasso PR with analytic FDR control, pooled and separate analyses controlled the FDR in terms of their average tFDR values, and differences in TPR between the pooled and the separate Lasso were small and insignificant (Table S13 in File S1). We therefore compared the separate and pooled analyses for individual chromosomes. From Table S13, we can see that for chromosome 21 (with eight QTL), TPR1 and TPR2 decreased somewhat from separate to pooled analysis with P-values for the differences ranging from 0.02 to 0.05. For chromosome 22 (with only two QTL), in contrast, TPR1 and TPR2 increased significantly as expected from separate to pooled analysis with all P-values < 4 × 10−10.
Analysis of real data
We applied PR and SMA methods to data from the Health ABC GWAS of interleukin 6 soluble receptor (IL-6 SR). The analysis included 786 Health ABC Caucasians who had both GWAS and serum IL-6 SR measurements (a continuous phenotype, approximately normally distributed) available (see File S2 for details). The genotype data included 750,424 SNPs (after standard edits) from chromosomes 1, 3, 4, 6, and 19. Covariates included in the analysis models were age, gender, site of data collection, and one principal component score obtained using Eigenstrat. We analyzed these data by SMA with GWER threshold (SMA-GWT), by SMA with BH FDR control (SMA-BH), and with the elastic net with 50% Lasso penalty (EN50) and analytic FDR control. SMA-GWT is commonly used in practice, and SMA-BH performed best among all SMA methods with FDR control on the simulated data. The elastic net was chosen as the single PR method because it performed best on the simulated data among all PR methods combined with our novel method for FDR control and because differences among PR methods were generally small. For SMA-BH and EN50 (EN50-sep), chromosomes were analyzed separately, and for EN50, a joint analysis of all chromosomes was also performed (EN50-joint).
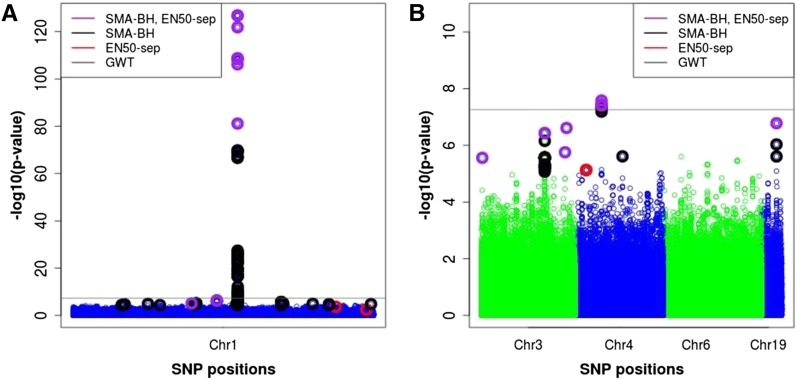
Chromosome 1 represents a special situation in that it contains a region with an extremely strong QTL signal. The very stringent SMA-GWT threshold identified 74 SNPs for this chromosome, many more than the 15 EN50-sep SNPs (see Table 1). Of these 74 SNPs, 61 were in LD above the absolute correlation threshold of 0.25 with EN50-sep SNPs, and the remaining 13 SNPs had largest absolute correlations with EN50-sep SNPs between 0.20 and 0.24 and therefore likely still overlap with EN50-sep SNPs (given the very strong signal) rather than represent independent signals. The EN50-sep selected a small subset of the peak area SNPs that had the smallest SMA P-values (Figure 9). While the SMA-GWT SNPs were all located in the area of the major peak, several EN50-sep SNPs were located in regions clearly separated from the peak, which mostly overlapped with SNPs detected by SMA-BH except for one EN50-sep SNP (see Figure 9). Expectedly, SMA-BH identified the largest number of SNPs (161), of which 101 were in LD above the absolute correlation threshold of 0.25 with EN50-sep SNPs. The remaining 60 SMA-BH SNPs had largest absolute correlations with EN50-sep SNPs between 0.07 and 0.24, indicating together with Figure 9 that SMA-BH selected some SNPs physically well separated from and likely not in LD with any EN50 SNPs. To check on the interpretation of the SNP–SNP correlation values, we simulated independent SNPs for N = 786 representing 11 different MAF values between 0.01 and 0.5 (assuming Hardy–Weinberg equilibrium) and computed all pairwise absolute correlations over 100,000 replications, yielding 75th, 95th, and 99th percentiles just below 0.025, 0.07, and 0.13, respectively, with a maximum value of 0.31. Based on our simulation studies (in particular the eight-QTL simulation), SMA-BH may have higher power than EN50-sep (by ∼10%) but also may not completely control the FDR, so the 60 SMA-BH SNPs with very small correlations to EN50-sep SNPs may represent some additional true signals and some false positives. We also reran EN50-sep on chromosome 1 by excluding all peak area SNPs but including a single peak area SNP (with the smallest P-value) in the model without shrinkage, but this analysis did not identify any additional SNPs (such as any of the 60 SMA-BH SNPs) compared to the previous EN50-sep analysis.
Table 1. Analysis of the real data with single-marker analysis, using the genome-wide threshold P-value < 5.5 × 10-8 (SMA-GWT) or Benjamini–Hochberg FDR control (SMA-BH) applied separately to the data on each chromosome and elastic net penalized regression with analytic FDR control applied separately to each chromosome (EN50s) or jointly across all five chromosomes (EN50j).
| Analysis Method | Significant SNPs | Chromosome | ||||
|---|---|---|---|---|---|---|
| 1 | 3 | 4 | 6 | 19 | ||
| SMA-GWT | No. sig | 74 | 0 | 8 | 0 | 0 |
| ∩EN50s | 9/17/42/61 | 0/0/0/0 | 6/8/8/8 | 0/0/0/0 | 0/0/0/0 | |
| SMA-BH | No. sig | 161 | 30 | 10 | 0 | 3 |
| ∩EN50s | 13/23/49/101 | 5/30/30/30 | 6/9/9/9 | 0/0/0/0 | 1/1/3/3 | |
| EN50j | No. sig | 14 | 3 | 3 | 0 | 0 |
| ∩EN50s | 14/14/14/14 | 2/2/2/2 | 2/2/2/2 | 0/0/0/0 | 0/0/0/0 | |
| EN50s | No. sig | 15 | 5 | 8 | 0 | 1 |
| ∩SMA-GWT | 9/9/9/9 | 0/0/0/0 | 6/6/6/6 | 0/0/0/0 | 0/0/0/0 | |
| ∩SMA-BH | 13/13/13/14 | 5/5/5/5 | 6/6/6/6 | 0/0/0/0 | 1/1/1/1 | |
No. sig denotes the number of significant SNPs, and ∩ denotes overlap with. The four numbers for overlap (e.g., of SMA-GWT with EN50s in row 3) represent the number of significant SMA-GWT SNPs that are identical to a significant EN50s SNP / correlated ≥ 0.8 with a significant EN50s SNP / correlated ≥ 0.5 with a significant EN50s SNP / correlated ≥ 0.25 with a significant EN50s SNP.
Figure 9.
Plot of –log10(P-value) for SNPs on chromosomes 1 (left), 3, 4, 6, and 19 (right) for the real data. The purple, black, and red circles represent significant SNPs selected by both SMA with BH FDR control (SMA-BH) and EN50 (elastic net with 50% weight on L1), by SMA-BH only, and by EN50 only, respectively. EN50-sep denotes EN50 applied to each chromosome separately. The gray line is the genome-wide threshold of 5.5 × 10−8, above which SNPs were selected by SMA-GWT. The y-axis represents raw P-values from SMA.
On chromosome 4, SMA-GWT, SMA-BH, and EN50-sep identified similar numbers of SNPs, 8 for SMA-GWT (all correlated with EN50 SNPs), 10 for SMA-BH (all but 1 correlated with EN50 SNPs), and 8 for EN50 (6 of which were correlated with SMA-GWT or SMA-BH SNPs).
On chromosomes 3 and 19, no QTL were identified with the stringent SMA-GWT, while SMA-BH and EN50-sep identified the same QTL (Table 1 and Figure 9). For chromosome 3, the 30 SMA-BH SNPs were all in LD (absolute correlation threshold of 0.25) with the 5 EN50-sep SNPs, and for chromosome 19, the 3 SMA-BH SNPs were all in LD with the 1 EN50-sep SNP. No SNPs were identified by any method on chromosome 6. Overall, these results show that SMA-BH and EN50-sep identify very similar candidate regions in a genome scan; that EN50 (and other PR methods) will select a subset of SNPs in a cluster of correlated SNPs, here those with the smallest SMA P-values; and that for follow-up (fine-mapping) studies, all EN50 (or other PR) identified SNPs plus a set of SNPs correlated with the identified SNPs (above a certain threshold) should be considered.
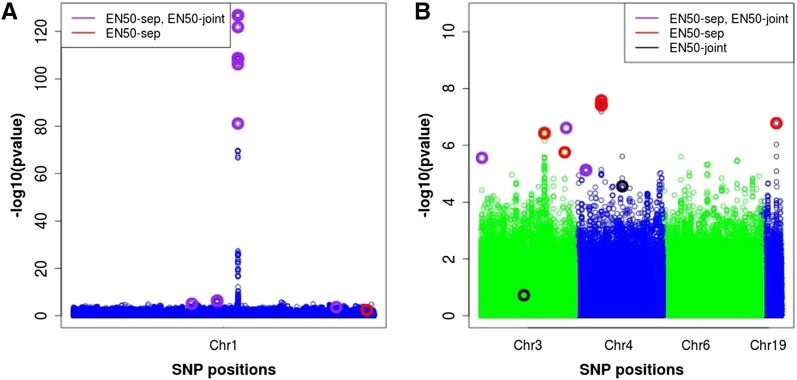
Finally, the joint analysis of all chromosomes by EN50 with FDR control (EN50-joint), compared to the separated analyses (EN50-sep), identified fewer SNPs (20 SNPs for EN50-joint compared with 29 SNPs for EN50-sep). EN50-joint identified one SNP on chromosome 3 and another on chromosome 4 that were not in LD with EN50-sep SNPs. The EN50-joint SNP on chromosome 3 is likely a false positive due to its −log10(P-value) being much smaller than those of all other identified (by any method) SNPs (Figure 10). The EN50-joint SNP on chromosome 4, however, was in LD (absolute correlation > 0.9) with a SMA-BH SNP.
Figure 10.
Plot of −log10(P-value) for SNPs on chromosomes 1 (left), 3, 4, 6, and 19 (right) for the real data. EN50-sep (EN50-joint) denotes the elastic net with 50% weight on L1 applied to each chromosome separately (to all chromosomes jointly). The purple, red, and black circles represent significant SNPs selected by both EN50-sep and EN50-joint, by EN50-sep only, and by EN50-joint only, respectively. The y-axis represents raw P-values from SMA.
Concluding remarks
The goals of this study were to provide a review and an independent comparison of penalized regression methods with single-marker analysis for variable (SNP) selection in GWAS and to implement and evaluate penalty/tuning parameter value selection by FDR control. A recent application of PR to GWAS (Waldmann et al. 2013, p. 1) arrived at this conclusion: “Hence, we can conclude that it is important to analyze GWAS data with both the lasso and the elastic net and an alternative tuning criterion to minimum MSE is needed for variable selection.” Here we provided such an alternative criterion.
We provided both a simple, approximate analytic method and a permutation-based method for FDR control in PR. The analytic method performed consistently well although being somewhat conservative. The permutation method expectedly had higher power (was less conservative) than the analytic method in the two-QTL scenario but was more conservative than the analytic method in the eight-QTL scenario. This initially unexpected behavior of the permutation method appears to be due to the fact that permuting the phenotypes means permuting some signal along with the random error and thus overestimates the amount of random error in the data. This causes the permutation method to become increasingly conservative with increasing numbers and effect sizes of causal SNPs (QTL). Because our simulated data had small sample sizes (∼200) and contained QTL jointly explaining 20 or 48% of the phenotypic variance (for the two- or eight-QTL data, respectively), this problem of the permutation method is more pronounced in our simulation than in real GWAS data characterized by very large sample sizes (thousands to tens of thousands) and small SNP effects (explaining ≤ 1% of the overall variation). However, the problem may manifest itself also in real data, when there are larger numbers of causal SNPs or causal SNPs with larger effect sizes as in our Health ABC GWAS. This is unfortunate because the main advantage of multimarker over single-marker approaches is that they are able to use the signal from other markers in determining significance. It may be possible to overcome this limitation by estimating the residuals using the fitted model and then permuting the residuals instead of the phenotype; we are currently investigating the performance of this approach.
Based on our comparisons with previous strategies for SNP selection in PR, it appears that our analytic FDR criterion is currently the best approach to SNP selection when using PR for GWAS, and this approach should be appealing to practitioners who desire a measure of error, in particular in terms of FDR, associated with the selected SNPs. Finally we note that our simple analytic FDR method can also be applied to penalized logistic regression.
When the focus is on variable selection (here the identification of a causal SNP by itself or a linked SNP) and not on estimation or prediction, our results indicate that among all PR methods investigated here, a version of the elastic net (EN50) performs better than the other PR methods in most situations, although differences were often small. Based on our analyses of simulated and real data, the EN50 should identify QTL regions that are very similar to those identified by SMA combined with BH FDR control. In contrast with SMA-BH, EN50 identifies a QTL region with a single SNP or few SNPs, and hence subsequent fine-mapping should include the EN50 SNPs plus additional SNPs in LD with the EN50 SNPs above a certain threshold. Expectedly, EN50 and SMA-BH have substantially more power than SMA with the genome-wide type-I error threshold.
Incorporating fusion-type penalties developed for covariates measured on graphs may improve power but can also generate more false positives or rather wide QTL regions for follow-up, in situations when there are multiple, moderately correlated causal SNPs located on the same chromosome and false positives are defined as any significant SNPs correlated with a causal SNP below an absolute value of 0.25.
When applying PR to large genome-wide data sets for joint (pooled) analysis of all chromosomes, prescreening SNPs based on SMA P-values is still necessary due to the high memory requirements for holding the entire SNP genotype matrix in memory (see Hoffman et al. 2013 for more details). We note that if prescreening is performed, then for tuning parameter value selection by our analytic FDR method the number of markers (p) in (13) should be set to the total number of markers (prior to preselection) to maintain control of the FDR (which should produce the same result as what would be obtained without preselection). It would, however, be prudent to perform PR with FDR control both on the entire set of SNPs from all chromosomes (pooled analysis) and separately on each chromosome (separate analysis). Our results from the simulated and real data representing several chromosomes indicate that the pooled analysis does not necessarily provide better power for all chromosomes.
We note that currently there is substantial interest in extending GWAS of single phenotypes to high-dimensional phenotypes (e.g., Marttinen et al. 2012). While high-dimensional phenotypes allow aspects of modeling that are not feasible with a single phenotype, the results of the current study still provide useful information for the design of analysis methods for GWAS of high-dimensional phenotypes.
Finally, code used for data simulation and code for EN50 analysis with analytic FDR control are provided in File S3. An example simulated data set with eight QTL on chromosome 21 is available at the Dryad Digital Repository (http://doi.org/10.5061/dryad.hc445).
Acknowledgments
This work was supported by the National Institute of General Medical Sciences grant R01HG005254 (to I.H.), by the National Institute on Aging grant 1R01AG032098-01A1 and the National Heart, Lung, and Blood Institute grant R01HL101250 (to Y.L.), and by a Virginia Tech Genetics, Bioinformatics, and Computational Biology Ph.D. program fellowship (to H.Y.). The Health ABC study was supported by the National Institute on Aging contracts N01AG62101, N01AG62103, and N01AG62106. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract HHSN268200782096C.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.167817/-/DC1.
Communicating editor: F. Zou
Literature Cited
- Ahmed, I., A.-L. Hartikainen, M.-R. Järvelin, and S. Richardson, 2011 False discovery rate estimation for stability selection: application to genome-wide association studies, pp. 1–20 in Statistical Applications in Genetics and Molecular Biology. [DOI] [PubMed] [Google Scholar]
- Akaike H., 1974. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19: 716–723. [Google Scholar]
- Akaike, H., 1977 On entropy maximization principle, pp. 27–41 in Application of Statistics, edited by P. R. Krishnaiah. North-Holland, Amsterdam, The Netherlands.
- Ayers K. L., Cordell H. J., 2010. SNP selection in genome‐wide and candidate gene studies via penalized logistic regression. Genet. Epidemiol. 34: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Benjamini Y., Yekutieli D., 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Bogdan M., Ghosh J. K., Doerge R., 2004. Modifying the Schwarz Bayesian information criterion to locate multiple interacting quantitative trait loci. Genetics 167: 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breheny P., Huang J., 2011. Coordinate descent algorithms for nonconvex penalized regression, with applications to biological feature selection. Ann. Appl. Stat. 5: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breheny, P. J., 2009 Regularized methods for high-dimensional and bi-level variable selection. Ph.D. Thesis, University of Iowa, Iowa City, IA.
- Cai T. T., Sun W., 2009. Simultaneous testing of grouped hypotheses: finding needles in multiple haystacks. J. Am. Stat. Assoc. 104: 1467–1481. [Google Scholar]
- Chen J., Chen Z., 2008. Extended Bayesian information criteria for model selection with large model spaces. Biometrika 95: 759–771. [Google Scholar]
- Chen, X., S. Kim, Q. Lin, J. G. Carbonell, and E. P. Xing, 2010 Graph-structured multi-task regression and an efficient optimization method for general fused Lasso. arXiv:1005.3579.
- Dudbridge F., Gusnanto A., 2008. Estimation of significance thresholds for genomewide association scans. Genet. Epidemiol. 32: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B., 2003. Robbins, empirical Bayes, and microarrays. Annals of Statistics 31: 366–378. [Google Scholar]
- Efron B., 2008. Simultaneous inference: When should hypothesis testing problems be combined? Ann. Appl. Stat. 2: 197–223. [Google Scholar]
- Efron B., Tibshirani R., 2002. Empirical Bayes methods and false discovery rates for microarrays. Genet. Epidemiol. 23: 70–86. [DOI] [PubMed] [Google Scholar]
- Fan J., Li R., 2001. Variable selection via nonconcave penalized likelihood and its oracle properties. J. Am. Stat. Assoc. 96: 1348–1360. [Google Scholar]
- Fan J., Peng H., 2004. Nonconcave penalized likelihood with a diverging number of parameters. Ann. Stat. 32: 928–961. [Google Scholar]
- Friedman J., Hastie T., Höfling H., Tibshirani R., 2007. Pathwise coordinate optimization. Ann. Appl. Stat. 1: 302–332. [Google Scholar]
- Friedman J., Hastie T., Tibshirani R., 2010. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33: 1. [PMC free article] [PubMed] [Google Scholar]
- Gauderman, W. J., and J. Morrison, 2001 QUANTO documentation. Technical report no. 157. Department of Preventive Medicine, University of Southern California, Los Angeles.
- Hoffman G. E., Logsdon B. A., Mezey J. G., 2013. PUMA: a unified framework for penalized multiple regression analysis of GWAS data. PLoS Comput. Biol. 9(6): e1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. X., Zhao H., Zhou H. H., 2010. False discovery rate control with groups. J. Am. Stat. Assoc. 105: 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium , 2005. A haplotype map of the human genome. Nature 437: 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Cai T. T., 2007. Estimating the null and the proportion of nonnull effects in large-scale multiple comparisons. J. Am. Stat. Assoc. 102: 495–506. [Google Scholar]
- Kim S., Xing E. P., 2009. Statistical estimation of correlated genome associations to a quantitative trait network. PLoS Genet. 5: e1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Choi H., Oh H.-S., 2008. Smoothly clipped absolute deviation on high dimensions. J. Am. Stat. Assoc. 103: 1665–1673. [Google Scholar]
- Kohavi, R., 1995 A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI’95 Proceedings of the 14th International Joint Conference on Artificial intelligence. Morgan Kaufmann Publishers, San Francisco, Vol. 2, pp. 1137–1145. [Google Scholar]
- Kruglyak L., 1999. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 22: 139–144. [DOI] [PubMed] [Google Scholar]
- Li C., Li H., 2008. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 24: 1175–1182. [DOI] [PubMed] [Google Scholar]
- Li C., Li H., 2010. Variable selection and regression analysis for graph-structured covariates with an application to genomics. Ann. Appl. Stat. 4: 1498–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., K. Wang, S. Ma, and J. Huang, 2011 Accounting for linkage disequilibrium in genome-wide association studies: a penalized regression method. Technical report. Department of Statistics and Actuarial Science, University of Iowa, Iowa City, IA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marttinen P., Gillberg J., Havulinna A., Corander J., Kaski S., 2012. Genome-wide association studies with high-dimensional phenotypes. Stat. Appl. Genet. Mol. Biol. 12: 413–431. [DOI] [PubMed] [Google Scholar]
- Meinshausen N., Bühlmann P., 2010. Stability selection. J. R. Stat. Soc. Ser. B Stat. Methodol. 72: 417–473. [Google Scholar]
- Meinshausen N., Meier L., Bühlmann P., 2009. P-values for high-dimensional regression. J. Am. Stat. Assoc. 104: 1671–1681. [Google Scholar]
- Pritchard J. K., Przeworski M., 2001. Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatti C., Freimer N., 2003. False discovery rate in linkage and association genome screens for complex disorders. Genetics 164: 829–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. N., Chatterjee N., Carroll R. J., Müller S., 2013. Controlling the local false discovery rate in the adaptive Lasso. Biostatistics 14: 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G., 1978. Estimating the dimension of a model. Ann. Stat. 6: 461–464. [Google Scholar]
- Storey J. D., Akey J. M., Kruglyak L., 2005. Multiple locus linkage analysis of genomewide expression in yeast. PLoS Biol. 3: e267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K., 2008. A unified approach to false discovery rate estimation. BMC Bioinformatics 9: 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z., Marchini J., Donnelly P., 2011. HAPGEN2: simulation of multiple disease SNPs. Bioinformatics 27: 2304–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Cai T. T., 2007. Oracle and adaptive compound decision rules for false discovery rate control. J. Am. Stat. Assoc. 102: 901–912. [Google Scholar]
- Sun W., Cai T. T., 2009. Large‐scale multiple testing under dependence. J. R. Stat. Soc. Ser. B Stat. Methodol. 71: 393–424. [Google Scholar]
- Sun W., Ibrahim J. G., Zou F., 2010. Genomewide multiple-loci mapping in experimental crosses by iterative adaptive penalized regression. Genetics 185: 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R., 1996. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. B 58: 267–288. [Google Scholar]
- Tseng, P., 1988 Coordinate Ascent for Maximizing Nondifferentiable Concave Functions. Massachusetts Institute of Technology, Laboratory for Information and Decision Systems, Cambridge, MA. [Google Scholar]
- Tseng P., 2001. Convergence of a block coordinate descent method for nondifferentiable minimization. J. Optim. Theory Appl. 109: 475–494. [Google Scholar]
- Waldmann P., Mészáros G., Gredler B., Fuerst C., Sölkner J., 2013. Evaluation of the lasso and the elastic net in genome-wide association studies. Front. Genet. 4: 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman L., Roeder K., 2009. High dimensional variable selection. Ann. Stat. 37: 2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., Sun W., Wang K., Hakonarson H., 2009. Multiple testing in genome-wide association studies via hidden Markov models. Bioinformatics 25: 2802–2808. [DOI] [PubMed] [Google Scholar]
- Ye J., 1998. On measuring and correcting the effects of data mining and model selection. J. Am. Stat. Assoc. 93: 120–131. [Google Scholar]
- Zhang C.-H., 2010. Nearly unbiased variable selection under minimax concave penalty. Ann. Stat. 38: 894–942. [Google Scholar]
- Zou H., 2006. The adaptive lasso and its oracle properties. J. Am. Stat. Assoc. 101: 1418–1429. [Google Scholar]
- Zou H., Hastie T., 2005. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B Stat. Methodol. 67: 301–320. [Google Scholar]
- Zou H., Hastie T., Tibshirani R., 2007. On the “degrees of freedom” of the lasso. Ann. Stat. 35: 2173–2192. [Google Scholar]
- Zuber V., Silva A. P. D., Strimmer K., 2012. A novel algorithm for simultaneous SNP selection in high-dimensional genome-wide association studies. BMC Bioinformatics 13: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]