Abstract
Several lines of evidence suggest that the circadian clock is constructed of multiple molecular feedback oscillators that function to generate robust rhythms in organisms. However, while core oscillator mechanisms driving specific behaviors are well described in several model systems, the nature of other potential circadian oscillators is not understood. Using genetic approaches in the fungus Neurospora crassa, we uncovered an oscillator mechanism that drives rhythmic spore development in the absence of the well-characterized FRQ/WCC oscillator (FWO) and in constant light, conditions under which the FWO is not functional. While this novel oscillator does not require the FWO for activity, it does require the blue-light photoreceptor CRYPTOCHROME (CRY); thus, we call it the CRY-dependent oscillator (CDO). The CDO was uncovered in a strain carrying a mutation in cog-1 (cry-dependent oscillator gate-1), has a period of ∼1 day in constant light, and is temperature-compensated. In addition, cog-1 cells lacking the circadian blue-light photoreceptor WC-1 respond to blue light, suggesting that alternate light inputs function in cog-1 mutant cells. We show that the blue-light photoreceptors VIVID and CRY compensate for each other and for WC-1 in CRY-dependent oscillator light responses, but that WC-1 is necessary for circadian light entrainment.
Keywords: circadian clock, oscillator, cryptochrome, FRQ-less oscillator, FRQ-WCC oscillator
CIRCADIAN clocks, composed of molecular transcription/translation-based feedback loop (TTFL) oscillators, generate daily rhythms in gene expression, physiology, and behavior in all kingdoms of life. The circadian clock provides a mechanism for organisms to anticipate cyclic changes in the environment to carry out specific tasks at advantageous times of the day. During investigations of clock mechanisms, several studies have revealed evidence for the existence of multiple autonomous oscillators in cells and/or tissues. First, there exist free-running rhythms of different periods in the same organism (Morse et al. 1994; Sai and Johnson 1999; Cambras et al. 2007). Second, residual rhythmicity is found in some strains defective in known oscillator components (Loros and Feldman 1986; Stanewsky et al. 1998; Emery et al. 2000; Collins et al. 2005). Third, some tissue-specific oscillators are constructed differently from core oscillators located in the brains of insects and animals (Stanewsky et al. 1998; Emery et al. 2000; Ivanchenko et al. 2001; Krishnan et al. 2001; Collins et al. 2005). Thus, multiple oscillators may exist both within, and among, cells in organisms with differentiated tissues. Furthermore, recent data revealed circadian rhythms in the oxidation state of highly conserved peroxiredoxin in the absence of transcription and thus a functional TTFL, adding additional levels of complexity to the circadian clock system (Edgar et al. 2012).
The fungus Neurospora crassa is a leading model for studying the circadian clock (Bell-Pedersen 2000; Heintzen and Liu 2007; Lakin-Thomas et al. 2011; Baker et al. 2012) and light signaling (Linden et al. 1997; Bell-Pedersen et al. 2001; Liu 2003; Merrow et al. 2006; Chen and Loros 2009). In N. crassa, the core FRQ/WCC oscillator (FWO) contains the negative elements FREQUENCY (FRQ) and FRQ-interacting RNA helicase (FRH), the blue-light photoreceptor WHITE COLLAR 1 (WC-1), and WHITE COLLAR 2 (WC-2). WC-1 and WC-2 form a complex, called the WCC that functions as a positive element in the oscillator loop. WCC binds the frq promoter and directly activates transcription of the frq gene (Froehlich et al. 2003). As FRQ protein accumulates, it interacts with itself and FRH (Cheng et al. 2001, 2005) and then binds to and promotes the phosphorylation and inactivation of the WCC (Schafmeier et al. 2005; He et al. 2006). Inhibition of WCC activity results in reduced frq transcription and FRQ protein levels. Once FRQ protein levels are sufficiently decreased, FRQ/FRH-directed inhibition of the activity of the WCC is released, and the cycle reactivates the next day. The FWO controls daily rhythms in expression of ∼20% of the genome and overt rhythms in asexual spore development (conidiation) (Vitalini et al. 2006). The conidiation rhythm, typically measured using the race tube assay, has a period in constant dark (DD) conditions of ∼22 h in wild-type strains (Loros and Dunlap 2001).
Several studies revealed that rhythms can persist in the absence of a functional FWO under certain growth conditions and/or in specific genetic backgrounds, providing evidence to suggest the existence of additional oscillators in N. crassa cells (Loros and Feldman 1986; Aronson et al. 1994; Merrow et al. 1999; Ramsdale and Lakin-Thomas 2000; Dragovic et al. 2002; Correa et al. 2003; Granshaw et al. 2003; Christensen et al. 2004; He et al. 2005; de Paula et al. 2006; Brody et al. 2010; Hunt et al. 2012). The term FLO (frq-less-oscillator) was coined to collectively describe these putative circadian and/or noncircadian oscillators (Iwasaki and Dunlap 2000). Indeed, in most cases, rhythms attributed to FLOs were shown to lack one or more of the three canonical clock properties, including the generation of a free-running rhythm of ∼24 hr in the absence of environmental cues, entrainment of the free-running rhythm to 24 hr by environmental cues, and temperature compensation of the clock (Lakin-Thomas 2000; Baker et al. 2012). In entrainment of the clock, the period of the rhythm becomes equal, on average, to an imposed environmental cycle, and a unique stable phase relationship is established between the imposed environmental cycle and the entrained oscillator (Johnson et al. 2003). Synchronization is distinguished from entrainment in that the cycle output occurs in response to the stimulus in a set time frame and does not depend on the length of the imposed environmental cycle. Temperature compensation of the clock means that the rate is relatively independent of temperature within the physiological range, with a temperature coefficient (Q10) near 1. The lack of full circadian properties of the FLOs led to suggestions that the FWO serves as a pacemaker in N. crassa cells, driving rhythms in downstream, so-called slave FLOs (Dunlap and Loros 2005). In this model, the FLOs are intrinsically rhythmic but require the FWO for full circadian properties. Furthermore, the number of FLOs in Neurospora cells is not known. While studies have been undertaken to identify molecular components of the FLOs (Lombardi et al. 2007; Shi et al. 2007; Yoshida et al. 2008; Schneider et al. 2009; Lakin-Thomas et al. 2011; Li et al. 2011; Hunt et al. 2012), little is understood regarding their nature and role in the circadian system.
In an attempt to identify key components of the FLOs, we carried out a genetic screen for mutations that enhance rhythmicity in strains that are deficient in both positive and negative components of the FWO. We identified a mutation called cry-dependent oscillator gate-1 (cog-1) that displayed robust rhythms in conidiation in constant light (LL), independently of the FWO. The oscillator controlling these rhythms fulfills two of the three criteria for a circadian oscillator: it free-runs under constant conditions with a period close to 1 day and is temperature-compensated. However, while cog-1 mutant strains were synchronized by light/dark (LD) cycles independently of the photoreceptor and clock component WC-1, circadian entrainment in LD required WC-1. Finally, we show that the blue-light photoreceptor CRYPTOCHROME (CRY), a core component of the mammalian circadian oscillator (Griffin et al. 1999), is necessary for the novel oscillator activity in Neurospora cells in LL. We therefore call the oscillator the CRY-dependent oscillator (CDO),
Materials and Methods
Strains
The N. crassa strains used in this study are listed in Table 1, their periods under different growth conditions in Table 2, and growth rates of representative strains in Table 3. The vvd knockout (KO) strain was obtained from Christian Heintzen (Heintzen et al. 2001). The Δwc-1::hph strain was obtained from Jay Dunlap (Dartmouth Medical School), and Δwc-1::bar was generated in our lab (Bennett et al. 2013). The cry KO [Fungal Genetics Stock Center (FGSC) 12981] was generated by the N. crassa KO project (Colot et al. 2006) and obtained from the FGSC. All strains used in this study carry the ras-1bd mutation, which clarifies the conidiation rhythm on race tubes (Sargent et al. 1966; Belden et al. 2007). The ras-1bd strain serves as the clock wild-type (WT) control strain. To generate the cog-1 mutation, a strain defective in the positive and negative arm of the FWO (wc-2234W, ras-1bd, Δfrq) was mutagenized by ultraviolet (UV) light according to standard procedures (Davis and De Serres 1970). The construction of double and triple photoreceptor deletion strains is described in Supporting Information, Figure S1, A and B. To complement Δcry, cog-1 cells, a 4.9-kb PCR fragment containing the cry gene was cloned into pCR-BluntBar that contains the bar gene for selection and transformed to Δcry, cog-1 cells using standard Neurospora transformation techniques (Pall and Brunelli 1993). PCR was used to verify the genotypes of double and triple photoreceptor mutant strains from genomic DNA isolated from 7-day-old slant cultures as described (Jin et al. 2007) (Figure S1C). The PCR primers used to verify the KO strains are listed in Table S1.
Table 1. Strains used in this study.
| Strain | Genotype | Strain no. | Source/reference |
|---|---|---|---|
| WT | matA, ras-1bd | DBP369 | FGSC 1858 |
| WT | mata, ras-1bd | DBP368 | FGSC 1859 |
| cog-1 | matA, ras-1bd, cog-1 | DBP694 | This study |
| cog-1 | mata, ras-1bd, cog-1 | DBP695 | This study |
| ∆frq | mata, ras-1bd, frq10 | DBP287 | Aronson et al. (1994) |
| ∆frq | matA, ras-1bd, frq10 | DBP776 | FGSC 7490 |
| ∆frq, cog-1 | matA, ras-1bd, frq10, cog-1 | DBP831 | This study |
| ∆wc-1 | matA, ras-1bd, ∆wc-1hyg | DBP580 | Lee et al. (2003) |
| ∆wc-1, cog-1 | matA, ras-1bd, ∆wc-1hyg, cog-1 | DBP696 | This study |
| ∆wc-1bar | matA, ras-1bd; ∆wc-1bar | DBP1223 | Bennett et al. (2013) |
| ∆wc-1bar, cog-1 | mata, ras-1bd, ∆wc-1bar, cog-1 | DBP1369 | This study (DBP1223 × DBP695)a |
| ∆vvd | mata, ras-1bd, ∆vvdhyg | DBP693 | Heintzen et al. (2001) |
| ∆vvd | matA, ras-1bd, ∆vvdhyg | DBP1634 | This study (DBP369 × DBP693) |
| ∆vvd, cog-1 | matA, ras-1bd, ∆vvdhyg, cog-1 | DBP1335 | This study (DBP693 × DBP694) |
| ∆vvd, cog-1 | mata, ras-1bd, ∆vvdhyg, cog-1 | DBP1638 | This study (DBP695 × DBP1335) |
| ∆cry | ∆cryhyg, mata, ras-1bd | DBP963 | Laboratory stock |
| ∆cry, cog-1 | ∆cryhyg, matA, ras-1bd, cog-1 | DBP1022 | This study (DBP963 × DBP694) |
| ∆cry, ∆wc-1 | ∆cryhyg, matA, ras-1bd, ∆wc-1bar | DBP1598 | This study (DBP963 × DBP1223) |
| ∆cry, ∆wc-1 | ∆cryhyg, mata, ras-1bd, ∆wc-1bar | DBP1599 | This study (DBP963 × DBP1223) |
| ∆cry, ∆wc-1, cog-1 | ∆cryhyg, matA, ras-1bd, ∆wc-1bar, cog-1 | DBP1645 | This study (DBP1022 × DBP1369) |
| ∆cry, ∆vvd | ∆cryhyg, mata, ras-1bd, ∆vvdhyg | DBP1640 | This study (DBP693 × DBP963) |
| ∆cry, ∆vvd | ∆cryhyg, matA, ras-1bd, ∆vvdhyg | DBP1600 | This study (DBP693 × DBP963) |
| ∆cry, ∆vvd, cog-1 | ∆cryhyg, matA, ras-1bd, ∆vvdhyg, cog-1 | DBP1601 | This study (DBP1022 × DBP1638) |
| ∆vvd, ∆wc-1 | matA, ras-1bd, ∆vvd hyg, ∆wc-1bar | DBP1639 | This study (DBP693 × DBP1223) |
| ∆vvd. ∆wc-1 | mata, ras-1bd, ∆vvdhyg, ∆wc-1bar | DBP1637 | This study (DBP693 × DBP1223) |
| ∆vvd, ∆wc-1, cog-1 | matA, ras-1bd, ∆vvdhyg, ∆wc-1bar, cog-1 | DBP1635 | This study (DBP1335 × DBP1369) |
| ∆vvd, ∆wc-1, cog-1 | mata, ras-1bd, ∆vvdhyg, ∆wc-1bar, cog-1 | DBP1636 | This study (DBP1335 × DBP1369) |
| ∆cry, ∆vvd, ∆wc-1 | mata, ∆cryhyg, ras-1bd, ∆vvdhyg, ∆wc-1bar | DBP1593 | This study (DBP1599 × DBP1634) |
| ∆cry, ∆vvd, ∆wc-1 | matA, ∆cryhyg, ras-1bd, ∆vvdhyg, ∆wc-1bar | DBP1592 | This study (DBP1599 × DBP1634) |
| ∆cry, ∆vvd, ∆wc-1, cog-1 | matA, ∆cryhyg, ras-1bd, ∆vvdhyg, ∆wc-1bar, cog-1 | DBP1597 | This study (DBP1022 × DBP1636) |
| ∆cry, ∆vvd, ∆wc-1, cog-1 | mata, ∆cryhyg, ras-1bd, ∆vvdhyg, ∆wc-1bar, cog-1 | DBP1596 | This study (DBP1022 × DBP1636) |
Derived from the indicated cross (×) between lab strains.
Table 2. Period of strains.
| DD | LL | |||||||
|---|---|---|---|---|---|---|---|---|
| cog-1+ | cog-1- | cog-1+ | cog-1- | |||||
| Genotype | Period (hr) | % Rhythmic | Period (hr) | % Rhythmic | Period (hr) | % Rhythmic | Period (hr) | % Rhythmic |
| WT | 22.3 ± 0.8a n = 60b | 100 | 22.7 ± 0.5, n = 77 | 100 | AR | 0 | 17.1 ± 1.3, n = 194 | 100 |
| Δwc-1 | AR | 0 | 23.9 ± 1.3, n = 77 | 40 | AR | 0 | 25.1 ± 0.8, n = 191 | 100 |
| Δfrq | AR | 0 | 21.7 ± 2.7, n = 34 | 50 | AR | 0 | 23.0 ± 2.0, n = 80 | 75 |
| Δcry | 22.2 ± 0.5, n = 60 | 100 | 22.4 ± 0.5, n = 60 | 100 | AR | 0 | AR | 0 |
| Δvvd | 21.7 ± 0.4, n = 60 | 100 | 23.2 ± 0.9, n = 77 | 100 | AR | 0 | 12.9 ± 1.7, n = 47 | 100 |
| Δvvd, Δwc-1 | AR | 0 | 21.3 ± 1.8, n = 44 | 60 | AR | 0 | 23.0 ± 1.3, n = 62 | 100 |
| Δcry, Δwc-1 | AR | 0 | AR | 0 | AR | 0 | AR | 0 |
| Δcry, Δvvd | 22.2 ± 0.5, n = 60 | 100 | 22.2 ± 0.5, n = 60 | 100 | AR | 0 | AR | 0 |
| Δcry, Δvvd, Δwc-1 | AR | 0 | AR | 0 | AR | 0 | AR | 0 |
| LD 6:6 | LD 9:9 | LD 12:12 | LD 14:14 | |||||
| cog-1+ | cog-1- | cog-1+ | cog-1- | cog-1+ | cog-1- | cog-1+ | cog-1- | |
| Period (h) | Period (h) | Period (h) | Period (h) | Period (h) | Period (h) | Period (h) | Period (h) | |
| WT | 12.1 ± 0.1, n = 172 | 11.9 ± 0.1, n = 179 | 18.1 ± 0.1, n = 90 | 17.8 ± 0.3, n = 96 | 23.9 ± 0.2, n = 84 | 23.9 ± 0.2, n = 84 | 27.3 ± 0.5, n = 68 | 27.6 ± 0.4, n = 74 |
| Δwc-1 | AR | 11.9 ± 0.1, n = 126 | AR | 17.9 ± 0.2, n = 74 | AR | 23.8 ± 0.2, n = 61 | AR | 27.8 ± 0.2, n = 61 |
| Δfrq | ND | ND | ND | ND | ND | ND | ND | ND |
| Δcry | 12.0 ± 0.1, n = 162 | 12.0 ± 0.2, n = 150 | 17.8 ± 0.3, n = 78 | 17.9 ± 0.1, n = 83 | 24.2 ± 0.2, n = 69 | 23.8 ± 0.3, n = 70 | 27.9 ± 0.4, n = 53 | 27.8 ± 0.3, n = 70 |
| Δvvd | 12.0 ± 0.1, n = 168 | 11.9 ± 0.2, n = 190 | 17.8 ± 0.2, n = 84 | 17.8 ± 0.2, n = 108 | 23.6 ± 0.2, n = 72 | 23.5 ± 0.4, n = 99 | 27.8 ± 0.2, n = 62 | 27.8 ± 0.2, n = 72 |
| Δvvd, Δwc-1 | AR | 12.0 ± 0.5, n = 74 | AR | 17.8 ± 0.3, n = 91 | AR | 23.9 ± 0.4, n = 60 | AR | 27.7 ± 0.5, n = 60 |
| Δcry, Δwc-1 | AR | 12.0 ± 0.5, n = 68 | AR | 18.1 ± 0.5, n = 88 | AR | 24.0 ± 0.2, n = 63 | AR | 27.4 ± 1.7, n = 63 |
| Δcry, Δvvd | 12.0 ± 0.5, n = 155 | 12.1 ± 0.1, n = 132 | 17.8 ± 0.2, n = 101 | 17.8 ± 0.2, n = 88 | 23.6 ± 0.2, n = 60 | 23.5 ± 0.2, n = 60 | 27.9 ± 0.2, n = 60 | 28.0 ± 0.2, n = 60 |
| Δcry, Δvvd, Δwc-1 | AR | AR | AR | AR | AR | AR | AR | AR |
AR, arrhythmic; ND, not determined.
Values represent mean ± SD.
n, number of bands per strain used to calculate period.
Table 3. Growth rates of strains with various oscillator functions.
| DD | LL | |||
|---|---|---|---|---|
| Genotype | Growth rate (cm/day) | Oscillator function | Growth rate (cm/day) | Oscillator function |
| WT | 4.0 ± 0.2a | FWO | 4.0 ± 0.2 | None |
| cog-1 | 3.3 ± 0.3 | FWO, CDO | 3.5 ± 0.2 | CDO |
| Δwc-1 | 4.3 ± 0.4 | None | 4.3 ± 0.3 | None |
| Δwc-1, cog-1 | 3.7 ± 0.2 | CDO | 3.2 ± 0.2 | CDO |
| Δcry | 3.8 ± 0.4 | FWO | 4.5 ± 0.4 | None |
| Δcry, cog-1 | 3.8 ± 0.4 | FWO | 4.8 ± 0.4 | None |
Values are mean ± SD; n ≥ 12.
To determine if the cog-1 mutation is dominant or recessive, a heterokaryon of strain matA, arg-5, ras-1bd and matA, trp-3, ras-1bd, cog-1 was generated. Heterokaryons were selected by growth on minimal media as described (Davis and De Serres 1970). Heterokayotic strains with nuclear ratios of 1:1, as determined by plating on selective media, were examined for rhythmicity on race tubes.
Growth conditions
All vegetative cultures were maintained on 1× Vogel’s, 2% glucose, minimal medium with the appropriate supplements as required (Vogel 1956; Davis and De Serres 1970). Sexual crosses were performed on Westergaard’s crossing agar plates (Westergaard and Mitchell 1947). KO strains containing the hph marker were maintained on Vogel’s minimal medium supplemented with 200 μg/ml hygromycin B (Sigma Aldrich, St. Louis). KO strains containing the bar gene were maintained on nitrate-free Vogel’s medium supplemented with 250 μg/ml glufosinate (Sigma Aldrich). The composition of race tube media was 11.5 ml of 1× Vogel’s, 0.1% glucose, 0.17% arginine, and 1.5% agar. After autoclaving, race tubes were allowed to dry for 7 days. The dried race tubes were inoculated with mycelia or conidia from 7-day-old slants and grown for 1 day at 25° and then transferred to the indicated conditions in Percival growth chambers (Perry, IA). The growth front was marked at the time of transfer. Light was from cool white fluorescent bulbs, unless otherwise indicated. Light intensity was measured with a VWR Scientific Dual Range Light Meter (Radnor, PA) and maintained at 1200 lx in all light experiments, unless otherwise indicated. To examine the effects of different wavelengths of light on the CDO, four different light sources (red: 60 lx; blue: 80 lx; green: 64 lx; white: 72 lx) were used in Percival incubators. Temperature recording in the incubators was accomplished using an EasyLog EL-USB thermometer (Lascar Electronics, Erie, PA). The growth fronts of the race tubes were marked at 24-hr intervals using a red light for cultures in DD and at lights on for cultures in LD cycles. Race tubes were scanned with an EPSON scanner (Long Beach, CA), and growth rates and periods were calculated. n is the number of conidial bands measured on replicate race tubes. The percentage rhythmic was calculated as the number of individual race tubes displaying at least five clear conidial bands for which we could measure period out of the total number of race tubes examined for each strain under a given growth condition.
Western blots
Protein was extracted from ground tissue (Garceau et al. 1997), and 100 µg of total protein was run on 10% SDS PAGE gels and transferred to polyvinylidene difluoride membranes (IPVH00010; Millipore, Billarica, MA). CRY protein was detected by immunoblotting with primary anti-CRY antibody (polyclonal, 1:1000 v/v, a gift from Jay Dunlap) and secondary goat anti-rabbit horseradish peroxide 2-conjugated antibody (diluted 1:20,000 v/v, 170-6515, Bio-Rad, Hercules, CA). Protein bands were visualized using a chemiluminescence kit (SuperSignal West Femto, ThermoFischer Scientific, Waltham, MA) according to the manufacturer’s instructions. The membranes were subsequently stained with amido black [0.1% amido black (Sigma-Aldrich), 10% acetic acid, 25% isopropanol] for protein-level normalization.
Results
CDO functions independently of the FWO
To identify components of N. crassa FLOs, we carried out a screen for mutations that would enhance developmental rhythms on race tubes in strains that were defective in both the positive and negative arms of the FWO. This approach ruled out any possible residual activity from the FWO. The wc-2234W, Δfrq strain was mutagenized by UV light to 50% survival, and a mutant strain, originally identified as Light Mutant 1 and renamed cog-1, that had robust conidiation rhythms in LL was identified (Figure 1 and Table 2). The cog-1 mutant strain was outcrossed multiple times to the clock WT strain ras-1bd to isolate the cog-1 mutation from all other mutations. The cog-1 mutant phenotype segregated with a 1:1 ratio in crosses, suggesting that the rhythmic phenotype is due to a single gene mutation. The resulting ras-1bd, cog-1 strain (called cog-1) was used for all subsequent analyses.
Figure 1.

Identification of the cog-1 mutant strain that uncovers the CDO. A FWO mutant strain (wc-2234W, ∆frq) was mutagenized by UV light, and the resulting strains were assayed in LL (1200 lx) at 25° for rescue of developmental rhythms on race tubes. The cog-1 mutation displayed rhythmic development under these conditions, whereas the parental strain was arrhythmic. The direction of growth is from left to right, and the black marks correspond to 24 hr of growth.
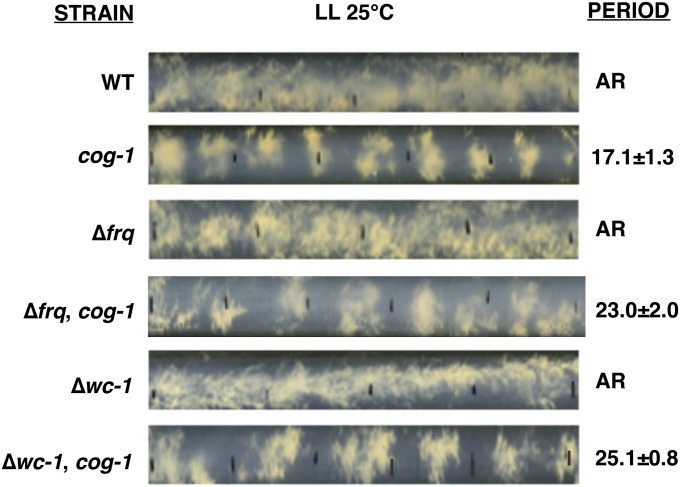
To confirm that the cog-1 mutation rescued developmental rhythms in strains that lack a functional FWO, the cog-1 strain was crossed to frq-null (Δfrq) and wc-1-null (Δwc-1) strains. Progeny that were Δfrq, cog-1 and Δwc-1, cog-1 were isolated and examined, along with control sibling strains, for rhythmicity in LL (Figure 2 and Table 2). Consistent with previous observations, the WT strain was arrhythmic in LL (Crosthwaite et al. 1995; Elvin et al. 2005). Strains that lacked FWO components were also arrhythmic in LL. However, rhythms in conidial development within the circadian range were observed in all strains that contained the cog-1 mutation. In the absence of the FWO components, the period of the cog-1 rhythm was almost 24 hr, whereas cog-1 strains with a functional FWO had a shorter period. These data suggested that the FWO and the CDO genetically interact in LL. While all cog-1 and Δwc-1, cog-1 replicate race-tube cultures displayed rhythms in LL (100%), the number of rhythmic race tubes of Δfrq, cog-1 was reduced to 75% (Table 2).
Figure 2.
The cog-1 mutation restores rhythms in strains that lack a functional FWO in LL. Representative photographs of race tubes of the indicated strains in LL (1200 lx) at 25°. Black lines on the tubes represent 24-hr growth fronts. The period of the rhythmic strains is shown on the right (±SD, n ≥ 60). “AR” indicates that the strain was arrhythmic.
CDO free-runs under constant conditions and is temperature-compensated
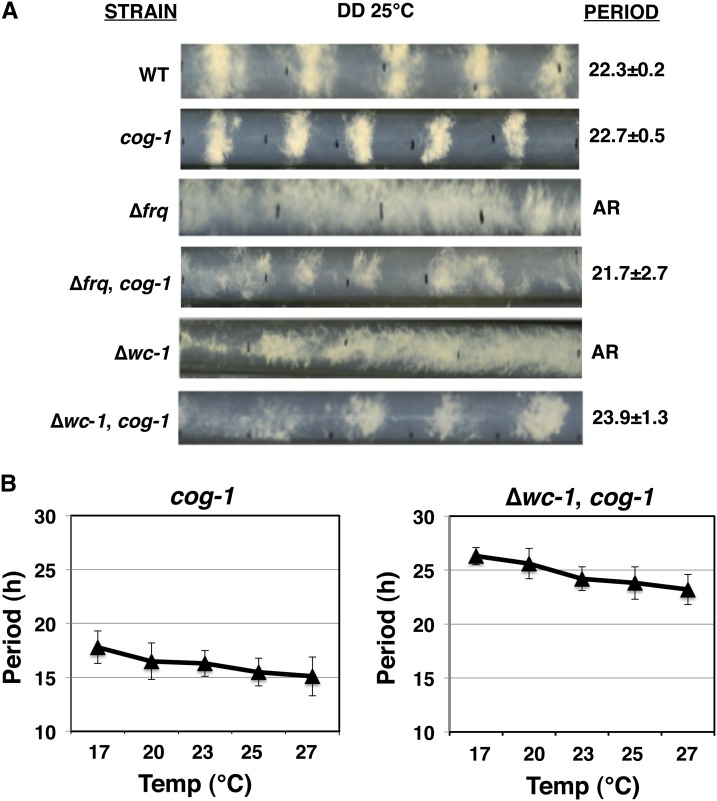
Using heterokaryon analyses, we determined that the cog-1 mutation was recessive (Figure S2), suggesting cog-1 is a loss-of-function mutation that uncovers an autonomous CDO. To test if the CDO has properties of a circadian oscillator, cog-1 strains were further examined for free-running rhythms in DD, light entrainment, and temperature compensation. Consistent with the LL data, and independent of the FWO, strains containing the cog-1 mutation are rhythmic in DD (Figure 3A). However, the robustness of the rhythms (% rhythmic) in cog-1 strains that also lacked components of the FWO was decreased in DD, as compared to LL (Table 2), and robustness of the rhythms in cog-1 strains was independent of whether the clock was initially synchronized in these cells by a light and/or a temperature transition (data not shown). Together, these data suggest that in DD the FWO overrides and/or enhances the CDO rhythms. Consistent with the FWO overriding the CDO in DD, the cog-1 period in DD was similar to the WT strain (22 hr), whereas when only the CDO is expressed (cog-1 in LL), the period is ∼17 hr (Figure 2).
Figure 3.
The CDO cycles in DD and is temperature-compensated. (A) The cog-1 mutation restores circadian rhythms in development to frq-null and wc-1-null strains in DD. Representative race-tube pictures of the indicated strains are shown from cultures grown in DD at 25° and labeled as in Figure 2. (B) cog-1 rhythms are temperature-compensated. Plots of the period (h) vs. temperature (°C) are shown for the indicated strains (±SD, n ≥ 60).
To determine if the CDO rhythm is temperature compensated, we assayed the developmental rhythm in cog-1 and ∆wc-1, cog-1 strains in LL, conditions under which the FWO is not functional but the CDO rhythm on race tubes is pronounced. While most biochemical reactions are temperature-dependent, with Q10 temperature coefficient values of 2–3, temperature-compensated clocks have Q10 values of between 0.8 and 1.4 (Sweeney and Hastings 1960). The period of the rhythm differed between the cog-1 and ∆wc-1, cog-1 strains as expected; however, the Q10 values measured for both strains between 17° and 27° was 1.2, confirming that the CDO is temperature-compensated (Figure 3B).
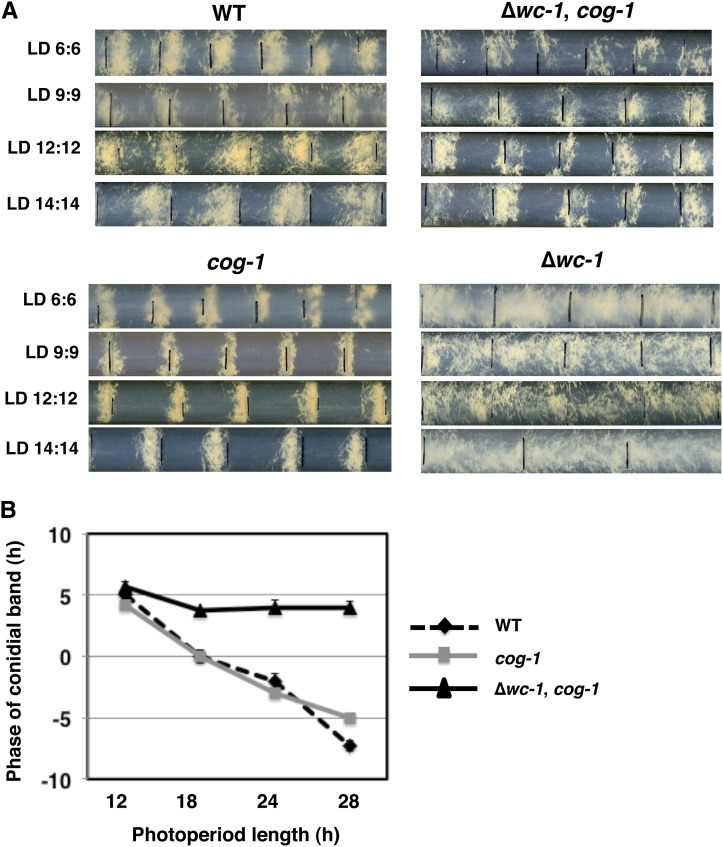
The third defining property of a circadian clock is entrainment. The best way to demonstrate entrainment is to examine environmental cycles with periods that are not equal to 24 hr (Johnson et al. 2003). If the rhythm entrains to environmental cycles it will display a period that equals the length of the environmental cycle and have stable phase angles that differ in different environmental cycle lengths. If instead, the rhythms display periods equal to the cycle length, but have similar phase angles in different cycles, then the rhythms are synchronized (driven), not entrained, by the imposing cycle. Thus, to investigate entrainment of the CDO, we examined strains that display the CDO in different LD cycle lengths (Figure 4A). For WT, cog-1, and ∆wc-1, cog-1 strains, the period of the rhythm matched the imposed LD cycle length. Strains with functional WC-1 displayed conidiation in different phases in LD cycles of different duration, consistent with entrainment. However, the phase angles in ∆wc-1, cog-1 strains stayed relatively constant, with conidiation occurring just after lights on in the different LD cycles, indicative of an LD-driven rhythm (Figure 4B).
Figure 4.
WC-1 is required for light entrainment of the circadian clock. (A) Race-tube cultures were exposed to different LD photoperiods (h). For example, LD 6:6 indicates a 12-hr photoperiod with cycles of 6 hr light and 6 hr dark. Representative race-tube pictures are shown for the indicated strains. Black lines on the race tubes denote when the light was turned on. (B) Plot of the phase of the conidiation band in relation to lights on in strains that showed light responses in each of the photoperiods from A (±SEM, n ≥ 5). A positive number indicates that the conidiation band occurred after lights on, and a negative number indicates that the band occurred prior to lights on. In some cases, the error bar is smaller than the symbol.
To rule out the possibility that the synchronization of ∆wc-1, cog-1 strains in LD cycles was due to changes in temperature, rather than to changes in light, we carefully monitored the temperature of the incubators with a temperature-recording device. The maximum temperature variance recorded was an increase in temperature in the light of 0.5° in each cycle. A 1° temperature change was not sufficient to drive the developmental rhythm in ∆wc-1 cells (Figure S3). Therefore, we can rule out the possibility that the rhythms observed in ∆wc-1, cog-1 strains in LD cycles was due to 0.5° cycles in the incubators. Together, these data demonstrate that WC-1 is required for stable entrainment of the circadian clock to light in N. crassa and that photoreceptors other than WC-1 are capable of responding to light in ∆wc-1, cog-1 cells.
CRY is essential for CDO rhythms
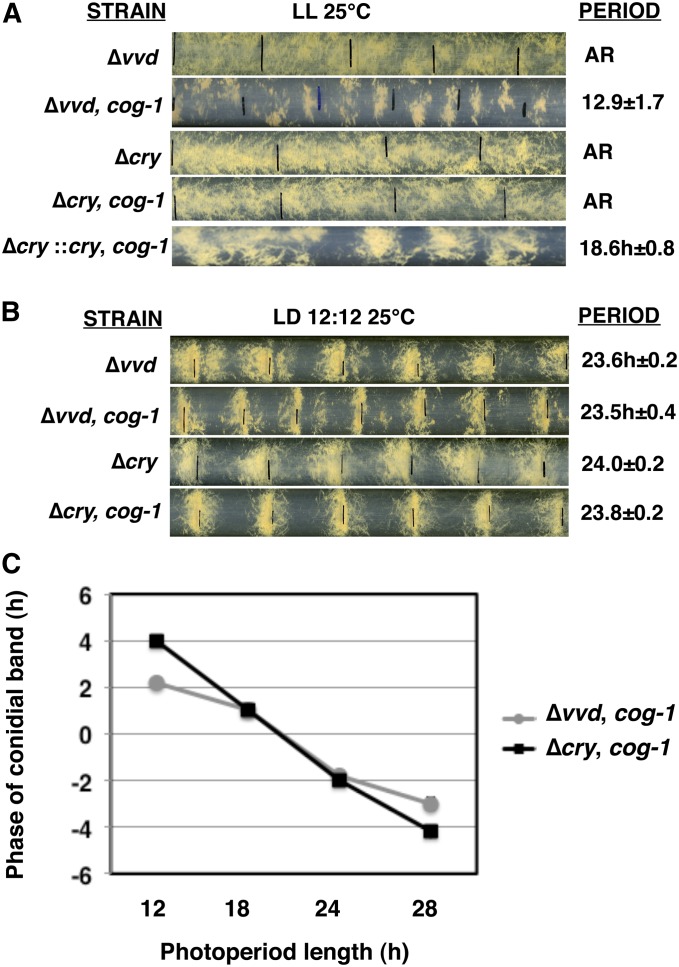
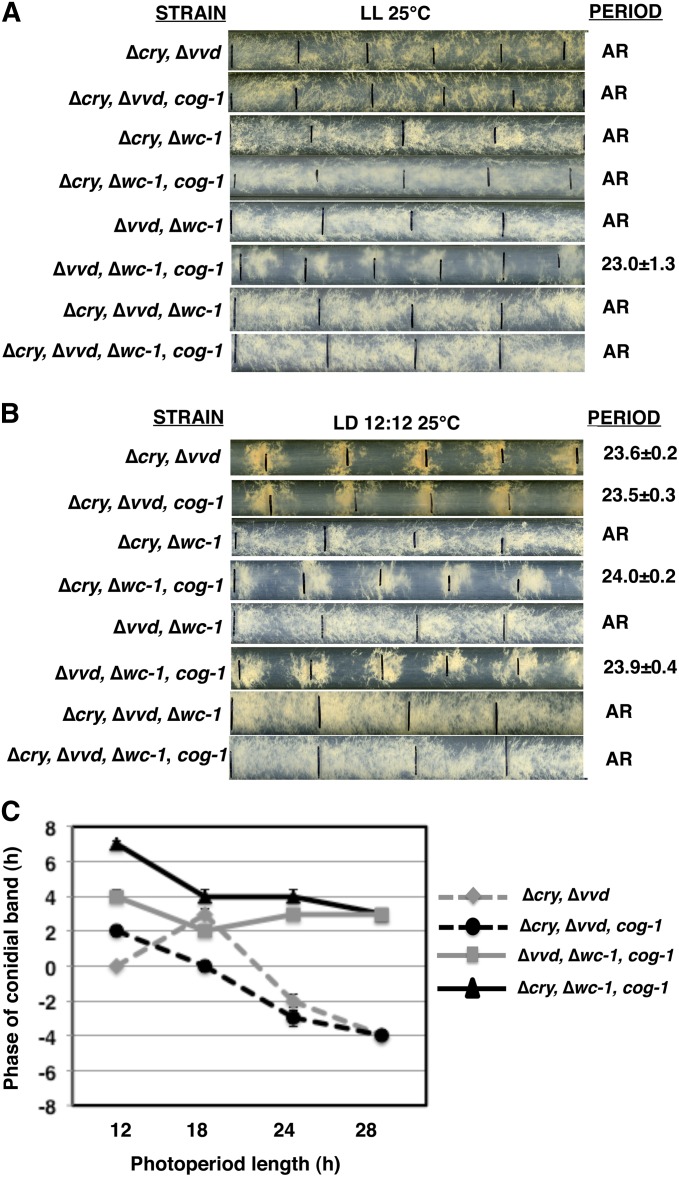
The N. crassa clock is known to be responsive to only blue light (Sargent and Briggs 1967). Consistent with this, the response of the CDO to light is restricted to the blue-light range (Figure S4). Therefore, to identify the photoreceptors responsible for light responses in the ∆wc-1, cog-1 strain, we crossed the cog-1 mutation to strains carrying deletions of the other known blue-light photoreceptors in N. crassa, VVD (Zoltowski et al. 2007) and CRY (Froehlich et al. 2010) to generate ∆vvd, cog-1, and ∆cry, cog-1 strains. These strains, and the control siblings, were assayed for rhythms in LL and for entrainment in LD cycles at 25° (Figure 5). In the ∆vvd, cog-1 strain in LL, rhythms in development were observed (Figure 5A); however, the period of the rhythm was reduced, and more variable, as compared to cog-1 (compare Figure 5A to Figure 2). The ∆vvd, cog-1 strains also entrained to LD cycles (Figure 5, B and C). Surprisingly, in ∆cry, cog-1 strains grown in LL, the developmental rhythms were abolished. The rhythm was rescued by ectopic transformation of the cry gene into the ∆cry, cog-1 strain (∆cry::cry, cog-1) (Figure 5A). In addition, the ∆cry, cog-1 strain entrained normally to LD cycles (Figure 5, B and C). Entrainment of ∆vvd, cog-1 and ∆cry, cog-1 strains to LD cycles is consistent with the presence of a functional FWO in these cells. Together, these data demonstrated that CRY is necessary for function of the CDO in LL and that both CRY and VVD are dispensable for light responses in cog-1 cells. However, the WC-1 photoreceptor likely compensated for the loss of individual VVD or CRY photoreceptors in these strains.
Figure 5.
The blue-light photoreceptor CRY, but not VVD, is necessary for CDO function, and other photoreceptors can compensate for rhythms in LD cycles. Representative race-tube photographs of the indicated photoreceptor mutant strains are shown in LL at 25° (A) and in LD 12:12 cycles at 25° (B) and labeled as in Figure 2. (C) Plot of the phase of the conidiation bands in relation to lights on for the indicated strains (±SEM, n ≥ 5).
To determine if light responses in the ∆vvd, cog-1 and ∆cry, cog-1 strains are due to the presence of WC-1, we generated double photoreceptor mutants in the cog-1 mutant background and examined rhythms in LL and responses to light in LD cycles at 25°. Consistent with a requirement for CRY in CDO function, all of the cog-1 double photoreceptor mutants that harbor cry deletions were arrhythmic in LL (Figure 6A and Table 2). Of the double photoreceptor mutants, only the ∆vvd, ∆wc-1, cog-1 strain displayed rhythms in LL. The period of the rhythm of ∆vvd, ∆wc-1, cog-1 cells was similar to ∆wc-1, cog-1 (Figure 2), but was significantly longer than ∆vvd, cog-1 (Figure 5A and Table 2), suggesting that while VVD may modify the period of the CDO, VVD is not necessary for CDO rhythms. In LD cycles, light responses were observed in all of the double photoreceptor mutant strains that harbored the cog-1 mutation (Figure 6B), and, consistent with our earlier results, entrainment to light required WC-1, but not VVD and CRY (Figure 6C).
Figure 6.
CRY and VVD are not required for light entrainment of the circadian clock. (A) Race-tube cultures of the indicated photoreceptor mutant strains were grown in LL and (B) were exposed to different LD photoperiods (h) and labeled as in Figure 2. (C) The phases of the conidiation band in relation to lights on were plotted (±SEM, n ≥ 5) as in Figure 4B.
We next generated strains that lacked all three blue-light photoreceptors, with and without the cog-1 mutation, and assayed the developmental rhythm in LL and LD cycles at 25° (Figure 6, A and B; Table 2). Independent of the cog-1 mutations, the triple photoreceptor deletion mutant strains were arrhythmic under both lighting conditions. Arrhythmicity in the triple photoreceptor mutant cells may be due to a lack of synchronization of the clock by light in the culture or to loss of function of the oscillators. To distinguish these possibilities, we used a temperature shift from 30° to 25° to synchronize the cells (Gooch et al. 1994; Liu et al. 1998). In support of loss of clock function in the triple photoreceptor mutant, the strain was arrhythmic following a temperature shift (Figure S5). Together, these data indicated that any of the three blue-light photoreceptors are able to substitute for each other to drive rhythms in LD cycles in strains containing the cog-1 mutation; however, in all cases, WC-1 was required for normal circadian entrainment to light.
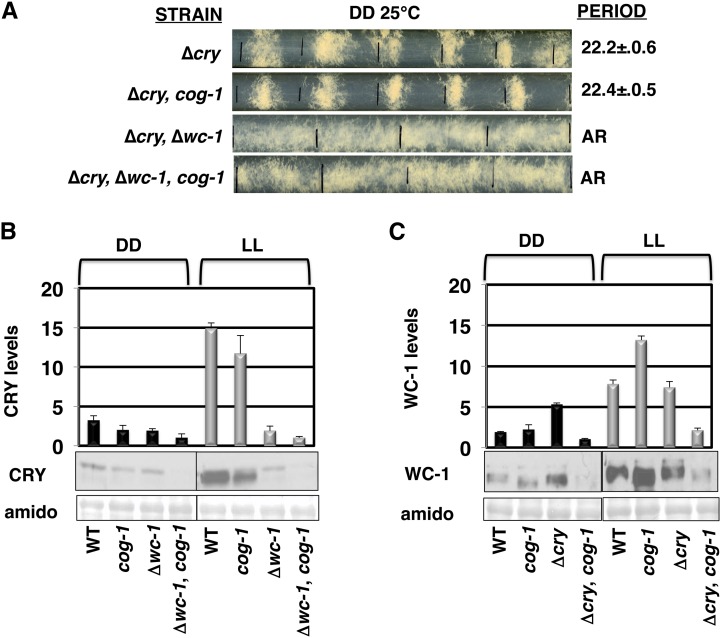
To further examine the requirement for CRY in the function of the CDO, we assayed the photoreceptor mutant strains in DD at 25°. In strains where the FWO was functional, rhythms with periods close to WT were observed independently of cry or cog-1 (Figure 7A and Table 2). In wc-1 deletion strains, development is arrhythmic in DD as expected, but the cog-1 mutation rescued rhythmicity (Figure 3). Consistent with a central role for CRY in the CDO, rescue of rhythmicity in ∆wc-1 cells by cog-1 depended upon CRY, as evidenced by arrhythmic development in the ∆cry, ∆wc-1, cog-1 strain (Figure 7A). Together, these data suggest that CRY is necessary for the function of the CDO in LL (Figure 5A) and in DD (Figure 7A).
Figure 7.
CRY, but not VVD or WC-1, is required for CDO-generated rhythms in DD. (A) Representative race tubes of the indicated strains labeled as in Figure 2. (B and C) Representative Western blots of CRY protein (B) and WC-1 protein (C) in the indicated strains harvested following 24 hr of growth in DD or LL and plotted above (±SEM, n = 3). The amido-stained membrane is shown below and was used to normalize protein loading.
The cry promoter is bound by the WCC after a light pulse (Smith et al. 2010), providing a direct link between the FWO and the CDO. To examine this link further, we assayed the levels of CRY protein in WT and ∆wc-1 strains with or without the cog-1 mutation in DD and LL (Figure 7B). In cells grown for 24 h in LL, CRY levels were increased relative to the DD samples in WT, and cog-1 cells, but not in ∆wc-1, and ∆wc-1, cog-1 cells, consistent with previous data demonstrating a role for WC-1 in CRY light responses (Froehlich et al. 2010). Low levels of CRY protein were also observed in ∆wc-1 and ∆wc-1, cog-1 cells grown in LL and harvested at different times of the day (Figure S6). Developmental rhythms persist in cog-1 and ∆wc-1, cog-1 cells in LL, but the levels of CRY protein varied widely between these strains, with very low levels in ∆wc-1, cog-1 cells and high levels in cog-1 cells (Figure 7B and Figure S6). Thus, the levels of CRY protein do not appear to correlate with rhythmicity of the strains in LL. We also examined WC-1 steady-state levels in the same cells. WC-1 levels were increased in cells grown in LL for 24 hr as compared to the 24-hr DD samples in WT, cog-1, and ∆cry cells and were slightly elevated in ∆cry, cog-1 cells (Figure 7C). Similar results were observed in cells grown for 12, 15, and 18 hr in DD and LL (Figure S7). Consistent with previous data, we observed that WC-1 levels were elevated in ∆cry cells as compared to WT cells in DD (Olmedo et al. 2010). Surprisingly, this increase in WC-1 levels was not observed in ∆cry, cog-1 cells in DD (Figure 7 and Figure S6), suggesting the possibility that COG-1 functions as a CRY-dependent activator of the WCC. However, similar to CRY protein levels, no correlations between the levels of WC-1 protein and rhythmicity were observed.
CDO activity reduces growth rate
To determine if activity of the CDO has an affect on Neurospora cells, we compared the growth rates of representative strains with variations in FWO and CDO function in DD and LL (Table 3). Under both conditions, CDO activity reduced the growth rate as compared to WT cells. Cells with a cry deletion had a slightly slower growth rate in DD. In cells that lack both FWO and CDO activity in LL and DD, the growth rate was increased as compared to WT cells, suggesting that, while the clock provides an overall adaptive advantage to organisms in natural LD cycles, its activity negatively affects growth of the fungus in DD or LL, similar to results obtained in cyanobacteria (Ma et al. 2013).
Discussion
To investigate the organization of the N. crassa circadian system, we identified the cog-1 mutation that restores rhythms to strains that lack a functional FWO in DD and LL. While the oscillator that drives this rhythm free-runs with a circadian period under constant conditions, and is temperature-compensated in LL, development in strains expressing the CDO, but lacking WC-1, were driven by light, rather than entrained by LD cycles. Thus, the CDO, similar to other reported FLOs (Dunlap and Loros 2004), lacks full circadian properties. In addition, these data revealed that blue-light photoreceptors in addition to WC-1 are capable of perceiving light information to ultimately drive development in the fungus. Surprisingly, any of the three blue-light photoreceptors—VVD, CRY, or WC-1—can compensate for each other in cog-1 mutant light responses. These data support the notion that the photoreceptors function in multi-protein complexes, providing opportunities for crosstalk in response to light (Bayram et al. 2010)
The cog-1 mutation is recessive (Figure S2), suggesting that cog-1 is most likely a loss of function mutation. While we have not yet identified the defective gene product in the cog-1 mutant strain, these data indicate that the gene specified by the cog-1 mutation encodes a protein that either directly or indirectly reduces the activity of the CDO when it is present, rather than as a component of the CDO that is necessary for its function. There are several possible reasons for why the CDO might be repressed via cog-1. For example, the CDO may be required only under certain growth conditions, and shutting off the CDO when those conditions are not met may provide a growth advantage to the organism. Consistent with this hypothesis, we observed that strains with a functional CDO had a reduced growth rate (Table 3). Alternatively, the CDO may be an ancient oscillator that was shut down to allow a more efficient oscillator, the FWO, which fully entrains to environmental conditions, to take over and provide a mechanism for anticipation of environmental cycles.
Our data are consistent with a role for CRY in the function of the CDO, as CRY deletion strains are arrhythmic under conditions in which the CDO is normally expressed (Figure 5, Figure 6, and Figure 7). In plants and insects, CRY is necessary for light entrainment of the circadian clock (Emery et al. 1998, 2000; Stanewsky et al. 1998), and in animals, CRY 1 and CRY 2 function as negative components of the core circadian oscillator (Reppert and Weaver 2002). Some insects, such as the monarch butterfly, have both Drosophila and mammalian versions of CRY, supporting an ancestral-like clock mechanism that involves both light sensing and transcriptional repressor roles for CRY (Zhu et al. 2008). CRY in N. crassa is a member of the CRY-DASH family of proteins, and while it can bind chromophores, it does not appear to have photolyase activity typical of CRY-DASH proteins (Froehlich et al. 2010). Both cry messenger RNA (mRNA) and protein are induced by light, and light induction requires WC-1, consistent with direct binding of the WCC to the cry promoter (Smith et al. 2010). Furthermore, cry mRNA accumulates with a low-amplitude circadian rhythm, peaking in the nighttime (Froehlich et al. 2010). The cry deletion strains show a small decrease in amplitude of a few light-induced genes and a slight phase delay in LD entrainment in otherwise WT strains (Chen et al. 2009). Given the central role of CRY in the insect and animal clockworks, the lack of pronounced circadian or light phenotypes in CRY deletion mutants in Neurospora has, until now, been surprising (Chen and Loros 2009; Froehlich et al. 2010; Olmedo et al. 2010). While our data strongly support a central role for CRY in the CDO, we were surprised to find that steady-state levels of CRY protein do not correlate with the rhythmicity of the strains. For example, the levels of CRY protein are low in Δwc-1, cog-1 and Δwc-1 cells in LL, but only the Δwc-1, cog-1 displays rhythms in development (Figure 2, Figure 7, and Figure S6). The data also pointed to the possibility that cog-1 functions both as a CRY-dependent activator of WC-1 (as evidenced by the low levels of WC-1 protein in Δcry, cog-1 cells as compared to Δcry cells in LL) and as a negative regulator of CDO activity. Taken together, CDO rhythms appear in cells that lack a functional FWO, have low levels (or activity) of CRY protein, and carry the cog-1 mutation. Furthermore, in cog-1 strains in LL, the period is 17 hr and the levels of CRY are high, whereas in Δwc-1, cog-1, cells the period is 25 hr and the levels of CRY are low. These data suggest that low levels (or activity) of CRY correlate with a longer CDO period. These data, together with our results showing no detectable consequences in rhythms in DD in strains completely lacking CRY (and thus a functional CDO), support the idea that the CDO functions downstream of the FWO and is not necessary for developmental rhythms.
Similar to circadian oscillators, we predict that the CDO functions as an autonomous molecular feedback loop, although this still needs to be determined. Because the CDO does not have full circadian properties, requiring WC-1 for LD entrainment, we surmise that it lies downstream of the FWO and may function as a slave oscillator to enhance rhythmic outputs, such as the development rhythm. This observation is reminiscent of previous studies demonstrating that WCC plays a central role in generating FLO-like oscillatory behavior in development in FWO-deficient strains in temperature cycles (Hunt et al. 2012). In LL and naturally occurring LD cycles, the CDO may take on a more prominent role to maintain rhythms during long periods of light when the FWO would normally break down. This idea is congruent with previous suggestions characterizing the role of VVD in LD entrainment (Elvin et al. 2005), in which the FWO oscillator is predicted to fully function in the night, whereas a slave oscillator, such as the CDO, may function during periods of light to complete the downstream rhythmic events. Our observation that the CDO is less robust in DD (Figure 3) also fits with this hypothesis. Additional evidence points to an interaction between the FWO and the CDO, including differences in the period of the rhythm in the cog-1 strains FWO-deficient vs. FWO-sufficient strains grown in LL (Figure 2), and the demonstration that the cry promoter is a direct target of the WCC (Smith et al. 2010). However, because the phase of the rhythm in cry (nighttime peak) does not match the morning activity of the WCC (Froehlich et al. 2010), it is possible that other components of the CDO control cry rhythmicity.
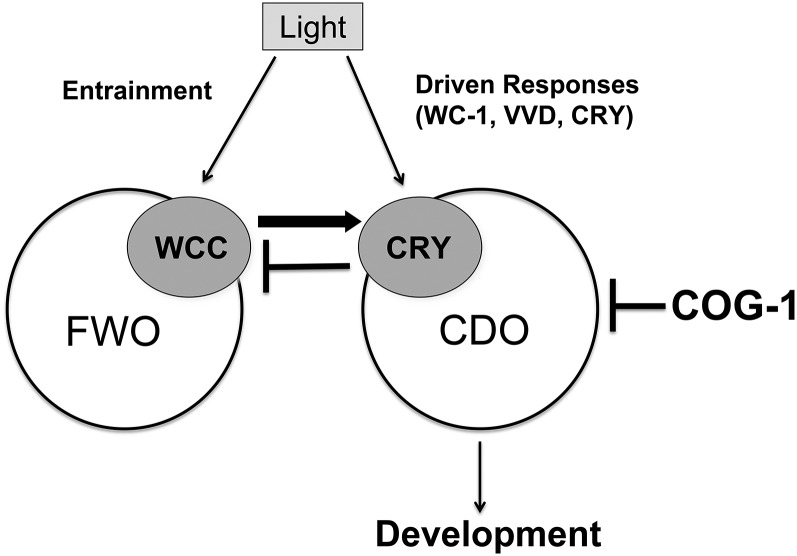
We propose a simple model to provide a framework for future tests of the complexity of the clock (Figure 8). In cog-1-deficient strains, and independent of the FWO, CRY is active and the CDO feedback loop oscillates. How cog-1 represses the CDO under conditions in which the FWO is not operative (such as in mutants of the FWO or in LL) is not known. Light signals may directly affect CRY activity to synchronize the CDO rhythm in LD cycles. In LL, some mechanism would be expected to exist to desensitize CRY activity during chronic light treatment. It has been shown that VVD functions in N. crassa to desensitize photo-transduction pathways during chronic light treatment and plays a role in establishing the phase of the clock in LD cycles (Elvin et al. 2005). Thus, VVD is a good candidate for desensitizing CRY to chronic light to promote CDO function. Consistent with a role for VVD in LL, the period of the CDO rhythm is significantly reduced in Δvvd, cog-1 strains (Figure 5). This model leads to several testable predictions: (i) that the activity of CRY would be increased in cog-1 mutant strains and reduced in WC-1 deletions in LL; (ii) that the activity of CRY would cycle in LL and LD in the absence of WC-1; (iii) that Δvvd will double the cycling of the CDO due to increased CRY activation in LL, which would be dependent on WC-1; (iv) that mutations in cry, or artificially increasing the levels of CRY in Δwc-1, cog-1 strains, would alter the period of the CDO rhythm; and (v) identification of CRY-interacting proteins would uncover additional components of the CDO. In any case, the clock system is likely to be even more complex than depicted here. For example, under specialized growth conditions, developmental rhythms with periods ranging between 6 and 21 hr were observed in vvd mutant alleles in LL that are dependent on WC-1, but not on FRQ (Schneider et al. 2009). In addition, rhythms in the expression of the ccg-16 gene in N. crassa are controlled by a FLO that requires WC-1, but not FRQ, for activity. This FLO, called the WC-FLO, that appears to be both temperature-compensated and entrained to environmental cycles independently of WCC and FRQ (de Paula et al. 2006, 2007).
Figure 8.
Model of the circadian clock composed of the FWO and CDO. See the text for a description of the model.
In summary, this work provides new insights into the complexity of the oscillator system in N. crassa and light responses. The identification of CRY as an CDO component, and the discovery of the cog-1 mutation that uncovered the CDO, will undoubtedly aid in testing models of the CDO, its connections to the environment, and to the FWO. As CRY is considered to be an ancient photoreceptor that has different activities in diverse organisms, including DNA repair, light perception, and running of the circadian clock (Somers et al. 1998; Stanewsky et al. 1998; Reppert and Weaver 2002; Daiyasu et al. 2004; Froehlich et al. 2010), our finding of the CRY-dependent CDO may provide key insights into the evolution of the clock.
Acknowledgments
We thank the Neurospora Program Project (P01GM68087), the Fungal Genetics Stock Center, Jay Dunlap, and Christian Heintzen for strains; Jay Dunlap for CRY antibodies; Kyung Suk Seo and Louis Morgan for the initial identification of cog-1; and Salem Hamiani and Xiuyun Tian for assistance with data collection. We also thank Teresa Lamb for insightful comments on the manuscript and advice throughout the course of this study. This work was supported by grant P01 NS39546 from the National Institutes of Health (to D.B.-P.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169441/-/DC1.
Communicating editor: E. U. Selker
Literature Cited
- Aronson B. D., Johnson K. A., Dunlap J. C., 1994. Circadian clock locus frequency: protein encoded by a single open reading frame defines period length and temperature compensation. Proc. Natl. Acad. Sci. USA 91: 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. L., Loros J. J., Dunlap J. C., 2012. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev. 36: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayram O., Braus G. H., Fischer R., Rodriguez-Romero J., 2010. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet. Biol. 47: 900–908. [DOI] [PubMed] [Google Scholar]
- Belden W. J., Larrondo L. F., Froehlich A. C., Shi M., Chen C. H., et al. , 2007. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 21: 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., 2000. Understanding circadian rhythmicity in Neurospora crassa: from behavior to genes and back again. Fungal Genet. Biol. 29: 1–18. [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D., Crosthwaite S. K., Lakin-Thomas P. L., Merrow M., Okland M., 2001. The Neurospora circadian clock: Simple or complex? Philos. Trans. R. Soc. Lond. B Biol. Sci. 356: 1697–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L. D., Beremand P., Thomas T. L., Bell-Pedersen D., 2013. Circadian activation of the mitogen-activated protein kinase MAK-1 facilitates rhythms in clock-controlled genes in Neurospora crassa. Eukaryot. Cell 12: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody S., Oelhafen K., Schneider K., Perrino S., Goetz A., et al. , 2010. Circadian rhythms in Neurospora crassa: downstream effectors. Fungal Genet. Biol. 47: 159–168. [DOI] [PubMed] [Google Scholar]
- Cambras T., Weller J. R., Angles-Pujoras M., Lee M. L., Christopher A., et al. , 2007. Circadian desynchronization of core body temperature and sleep stages in the rat. Proc. Natl. Acad. Sci. USA 104: 7634–7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Loros J. J., 2009. Neurospora sees the light: light signaling components in a model system. Commun. Integr. Biol. 2: 448–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H., Ringelberg C. S., Gross R. H., Dunlap J. C., Loros J. J., 2009. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. EMBO J. 28: 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang Y., Heintzen C., Liu Y., 2001. Coiled-coil domain-mediated FRQ-FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J. 20: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., He Q., Wang L., Liu Y., 2005. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev. 19: 234–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M. K., Falkeid G., Loros J. J., Dunlap J. C., Lillo C., et al. , 2004. A nitrate-induced frq-less oscillator in Neurospora crassa. J. Biol. Rhythms 19: 280–286. [DOI] [PubMed] [Google Scholar]
- Collins B. H., Dissel S., Gaten E., Rosato E., Kyriacou C. P., 2005. Disruption of Cryptochrome partially restores circadian rhythmicity to the arrhythmic period mutant of Drosophila. Proc. Natl. Acad. Sci. USA 102: 19021–19026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H. V., Park G., Turner G. E., Ringelberg C., Crew C. M., et al. , 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. USA 103: 10352–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa A., Lewis Z. A., Greene A. V., March I. J., Gomer R. H., et al. , 2003. Multiple oscillators regulate circadian gene expression in Neurospora. Proc. Natl. Acad. Sci. USA 100: 13597–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite S. K., Loros J. J., Dunlap J. C., 1995. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell 81: 1003–1012. [DOI] [PubMed] [Google Scholar]
- Daiyasu H., Ishikawa T., Kuma K., Iwai S., Todo T., et al. , 2004. Identification of cryptochrome DASH from vertebrates. Genes Cells 9: 479–495. [DOI] [PubMed] [Google Scholar]
- Davis R. H., de Serres F. J., 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17: 79–143. [Google Scholar]
- de Paula R. M., Lewis Z. A., Greene A. V., Seo K. S., Morgan L. W., et al. , 2006. Two circadian timing circuits in Neurospora crassa cells share components and regulate distinct rhythmic processes. J. Biol. Rhythms 21: 159–168. [DOI] [PubMed] [Google Scholar]
- de Paula R. M., Vitalini M. W., Gomer R. H., Bell-Pedersen D., 2007. Complexity of the Neurospora crassa circadian clock system: multiple loops and oscillators. Cold Spring Harb. Symp. Quant. Biol. 72: 345–351. [DOI] [PubMed] [Google Scholar]
- Dragovic Z., Tan Y., Gorl M., Roenneberg T., Merrow M., 2002. Light reception and circadian behavior in “blind” and “clock-less” mutants of Neurospora crassa. EMBO J. 21: 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J. C., Loros J. J., 2004. The Neurospora circadian system. J. Biol. Rhythms 19: 414–424. [DOI] [PubMed] [Google Scholar]
- Dunlap J. C., Loros J. J., 2005. Analysis of circadian rhythms in Neurospora: overview of assays and genetic and molecular biological manipulation. Methods Enzymol. 393: 3–22. [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Green E. W., Zhao Y., van Ooijen G., Olmedo M., et al. , 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin M., Loros J. J., Dunlap J. C., Heintzen C., 2005. The PAS/LOV protein VIVID supports a rapidly dampened daytime oscillator that facilitates entrainment of the Neurospora circadian clock. Genes Dev. 19: 2593–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., So W. V., Kaneko M., Hall J. C., Rosbash M., 1998. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 95: 669–679. [DOI] [PubMed] [Google Scholar]
- Emery P., Stanewsky R., Hall J. C., Rosbash M., 2000. A unique circadian-rhythm photoreceptor. Nature 404: 456–457. [DOI] [PubMed] [Google Scholar]
- Froehlich A. C., Loros J. J., Dunlap J. C., 2003. Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc. Natl. Acad. Sci. USA 100: 5914–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich A. C., Chen C. H., Belden W. J., Madeti C., Roenneberg T., et al. , 2010. Genetic and molecular characterization of a cryptochrome from the filamentous fungus Neurospora crassa. Eukaryot. Cell 9: 738–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau N. Y., Liu Y., Loros J. J., Dunlap J. C., 1997. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469–476. [DOI] [PubMed] [Google Scholar]
- Gooch V. D., Wehseler R. A., Gross C. G., 1994. Temperature effects on the resetting of the phase of the Neurospora circadian rhythm. J. Biol. Rhythms 9: 83–94. [DOI] [PubMed] [Google Scholar]
- Granshaw T., Tsukamoto M., Brody S., 2003. Circadian rhythms in Neurospora crassa: farnesol or geraniol allow expression of rhythmicity in the otherwise arrhythmic strains frq10, wc-1, and wc-2. J. Biol. Rhythms 18: 287–296. [DOI] [PubMed] [Google Scholar]
- Griffin E. A., Jr, Staknis D., Weitz C. J., 1999. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286: 768–771. [DOI] [PubMed] [Google Scholar]
- He Q., Cheng P., Liu Y., 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev. 19: 1518–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Cha J., Lee H. C., Yang Y., Liu Y., 2006. CKI and CKII mediate the FREQUENCY-dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev. 20: 2552–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzen C., Liu Y., 2007. The Neurospora crassa circadian clock. Adv. Genet. 58: 25–66. [DOI] [PubMed] [Google Scholar]
- Heintzen C., Loros J. J., Dunlap J. C., 2001. The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating, and regulates clock resetting. Cell 104: 453–464. [DOI] [PubMed] [Google Scholar]
- Hunt S., Elvin M., Heintzen C., 2012. Temperature-sensitive and circadian oscillators of Neurospora crassa share components. Genetics 191: 119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanchenko M., Stanewsky R., Giebultowicz J. M., 2001. Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks. J. Biol. Rhythms 16: 205–215. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Dunlap J. C., 2000. Microbial circadian oscillatory systems in Neurospora and Synechococcus: models for cellular clocks. Curr. Opin. Microbiol. 3: 189–196. [DOI] [PubMed] [Google Scholar]
- Jin Y., Allan S., Baber L., Bhattarai E. K., Lamb T. M., et al. , 2007. Rapid genetic mapping in Neurospora crassa. Fungal Genet. Biol. 44: 455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. H., Elliott J. A., Foster R., 2003. Entrainment of circadian programs. Chronobiol. Int. 20: 741–774. [DOI] [PubMed] [Google Scholar]
- Krishnan B., Levine J. D., Lynch M. K., Dowse H. B., Funes P., et al. , 2001. A new role for cryptochrome in a Drosophila circadian oscillator. Nature 411: 313–317. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P. L., 2000. Circadian rhythms: new functions for old clock genes. Trends Genet. 16: 135–142. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P. L., Bell-Pedersen D., Brody S., 2011. The genetics of circadian rhythms in Neurospora. Adv. Genet. 74: 55–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Dunlap J. C., Loros J. J., 2003. Roles for WHITE COLLAR-1 in circadian and general photoperception in Neurospora crassa. Genetics 163: 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Motavaze K., Kafes E., Suntharalingam S., Lakin-Thomas P., 2011. A new mutation affecting FRQ-less rhythms in the circadian system of Neurospora crassa. PLoS Genet. 7: e1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden H., Ballario P., Macino G., 1997. Blue light regulation in Neurospora crassa. Fungal Genet. Biol. 22: 141–150. [DOI] [PubMed] [Google Scholar]
- Liu Y., 2003. Molecular mechanisms of entrainment in the Neurospora circadian clock. J. Biol. Rhythms 18: 195–205. [DOI] [PubMed] [Google Scholar]
- Liu Y., Merrow M., Loros J. J., Dunlap J. C., 1998. How temperature changes reset a circadian oscillator. Science 281: 825–829. [DOI] [PubMed] [Google Scholar]
- Lombardi L., Schneider K., Tsukamoto M., Brody S., 2007. Circadian rhythms in Neurospora crassa: clock mutant effects in the absence of a frq-based oscillator. Genetics 175: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros J. J., Dunlap J. C., 2001. Genetic and molecular analysis of circadian rhythms in Neurospora. Annu. Rev. Physiol. 63: 757–794. [DOI] [PubMed] [Google Scholar]
- Loros J. J., Feldman J. F., 1986. Loss of temperature compensation of circadian period length in the frq-9 mutant of Neurospora crassa. J. Biol. Rhythms 1: 187–198. [DOI] [PubMed] [Google Scholar]
- Ma P., Woelfle M. A., Johnson C. H., 2013. An evolutionary fitness enhancement conferred by the circadian system in cyanobacteria. Chaos Solitons Fractals 50: 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M., Brunner M., Roenneberg T., 1999. Assignment of circadian function for the Neurospora clock gene frequency. Nature 399: 584–586. [DOI] [PubMed] [Google Scholar]
- Merrow M., Boesl C., Ricken J., Messerschmitt M., Goedel M., et al. , 2006. Entrainment of the Neurospora circadian clock. Chronobiol. Int. 23: 71–80. [DOI] [PubMed] [Google Scholar]
- Morse D., Hastings J. W., Roenneberg T., 1994. Different phase responses of the two circadian oscillators in Gonyaulax. J. Biol. Rhythms 9: 263–274. [DOI] [PubMed] [Google Scholar]
- Olmedo M., Ruger-Herreros C., Luque E. M., Corrochano L. M., 2010. A complex photoreceptor system mediates the regulation by light of the conidiation genes con-10 and con-6 in Neurospora crassa. Fungal Genet. Biol. 47: 352–363. [DOI] [PubMed] [Google Scholar]
- Pall M. L., Brunelli J. P., 1993. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet. Newsl. 40: 59–62. [Google Scholar]
- Ramsdale M., Lakin-Thomas P. L., 2000. sn-1,2-diacylglycerol levels in the fungus Neurospora crassa display circadian rhythmicity. J. Biol. Chem. 275: 27541–27550. [DOI] [PubMed] [Google Scholar]
- Reppert S. M., Weaver D. R., 2002. Coordination of circadian timing in mammals. Nature 418: 935–941. [DOI] [PubMed] [Google Scholar]
- Sai J., Johnson C. H., 1999. Different circadian oscillators control Ca(2+) fluxes and lhcb gene expression. Proc. Natl. Acad. Sci. USA 96: 11659–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M. L., Briggs W. R., 1967. The effects of light on a circadian rhythm of conidiation in Neurospora. Plant Physiol. 42: 1504–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent M. L., Briggs W. R., Woodward D. O., 1966. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiol. 41: 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafmeier T., Haase A., Kaldi K., Scholz J., Fuchs M., et al. , 2005. Transcriptional feedback of Neurospora circadian clock gene by phosphorylation-dependent inactivation of its transcription factor. Cell 122: 235–246. [DOI] [PubMed] [Google Scholar]
- Schneider K., Perrino S., Oelhafen K., Li S., Zatsepin A., et al. , 2009. Rhythmic conidiation in constant light in vivid mutants of Neurospora crassa. Genetics 181: 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Larrondo L. F., Loros J. J., Dunlap J. C., 2007. A developmental cycle masks output from the circadian oscillator under conditions of choline deficiency in Neurospora. Proc. Natl. Acad. Sci. USA 104: 20102–20107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M., Sancar G., Dekhang R., Sullivan C. M., Li S., et al. , 2010. Transcription factors in light and circadian clock signaling networks revealed by genomewide mapping of direct targets for Neurospora White Collar Complex. Eukaryot. Cell 9: 1549–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers D. E., Devlin P. F., Kay S. A., 1998. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490. [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., et al. , 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Sweeney B. M., Hastings J. W., 1960. Effects of temperature upon diurnal rhythms. Cold Spring Harb. Symp. Quant. Biol. 25: 87–104. [DOI] [PubMed] [Google Scholar]
- Vitalini M. W., de Paula R. M., Park W. D., Bell-Pedersen D., 2006. The rhythms of life: circadian output pathways in Neurospora. J. Biol. Rhythms 21: 432–444. [DOI] [PubMed] [Google Scholar]
- Vogel H. J., 1956. A convenient growth medium for Neurospora (medium N). Microb. Genet. Bull 13: 2–43. [Google Scholar]
- Westergaard M., Mitchell H. K., 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573–577. [Google Scholar]
- Yoshida Y., Maeda T., Lee B., Hasunuma K., 2008. Conidiation rhythm and light entrainment in superoxide dismutase mutant in Neurospora crassa. Mol. Genet. Genomics 279: 193–202. [DOI] [PubMed] [Google Scholar]
- Zhu H., Sauman I., Yuan Q., Casselman A., Emery-Le M., et al. , 2008. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 6: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowski B. D., Schwerdtfeger C., Widom J., Loros J. J., Bilwes A. M., et al. , 2007. Conformational switching in the fungal light sensor VIVID. Science 316: 1054–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]