Abstract
Transposable elements are a common source of genetic variation that may play a substantial role in contributing to gene expression variation. However, the contribution of transposable elements to expression variation thus far consists of a handful of examples. We used previously published gene expression data from 37 inbred Drosophila melanogaster lines from the Drosophila Genetic Reference Panel to perform a genome-wide assessment of the effects of transposable elements on gene expression. We found thousands of transcripts with transposable element insertions in or near the transcript and that the presence of a transposable element in or near a transcript is significantly associated with reductions in expression. We estimate that within this example population, ∼2.2% of transcripts have a transposable element insertion, which significantly reduces expression in the line containing the transposable element. We also find that transcripts with insertions within 500 bp of the transcript show on average a 0.67 standard deviation decrease in expression level. These large decreases in expression level are most pronounced for transposable element insertions close to transcripts and the effect diminishes for more distant insertions. This work represents the first genome-wide analysis of gene expression variation due to transposable elements and suggests that transposable elements are an important class of mutation underlying expression variation in Drosophila and likely in other systems, given the ubiquity of these mobile elements in eukaryotic genomes.
Keywords: transposable elements, gene expression, rare alleles of large effect, DGRP
IN Drosophila, a substantial fraction of heritable phenotypic variation is thought to be due to rare alleles of large effect in mutation–selection balance at low frequencies in natural populations (Mackay 2010). This “rare alleles of large effect” hypothesis, which states that many common phenotypes are caused by individually rare alleles, may also explain the “missing heritability” of current genome-wide association studies (GWAS) in humans, in which significant associations between disease and common markers explain only a small fraction of heritable variation in complex disease risk (Manolio et al. 2009). In Drosophila, as in other species, studies linking phenotype to genotype have thus far predominantly focused on single nucleotide polymorphisms (SNPs) observed in several lines, though more recently the contributions of other types of variation have been examined, such as copy number variation (Stranger et al. 2007) and other non-SNP complex variation, such as small repeats and insertion/deletion variation (Massouras et al. 2012).
The low frequencies of non-SNP variants in natural populations make them appealing candidates to be rare alleles of large effect. Transposable element (TE) insertions are a particularly appealing class of such variants in Drosophila for several reasons. First, insertions in natural populations are typically at low frequency (Charlesworth and Langley 1989; Cridland et al. 2013). Second, TE insertions as a class have been associated with variation in bristle number in Drosophila melanogaster (Mackay and Langley 1990; Long et al. 2000). Third, the mutation rate due to TE mobilization is high, relative to SNPs (Nuzhdin and Mackay 1994; Viera and Biemont 1997). To date, the contribution of TEs to complex trait variation in Drosophila has been best documented in the context of variation in abdominal bristle number (Mackay and Langley 1990; Long et al. 2000; Macdonald et al. 2005; Gruber et al. 2007), but there has been no systematic effort to include TE insertions in the study of complex traits in this system.
To study the genome-wide impact of TE insertions on a complex trait, we have integrated a set of high-quality TE insertion calls from the Drosophila Genetic Reference Panel (DGRP) (Mackay et al. 2012; Cridland et al. 2013) with data on expression level variation from the same lines (Ayroles et al. 2009). This dataset allows us to directly compare variation in expression level between inbred lines with different TE insertion phenotypes. We expect that if TEs do affect expression level, and that the effect is primarily a decrease in expression, we will find an excess number of TE-containing transcripts at the bottom tail of the distribution of expression measures for a given transcript. We find that a large number of transcripts have nearby transposable element insertions in one or more inbred lines studied. We demonstrate that a significant amount of gene expression variation in D. melanogaster is due to rare transposable element insertions of large effect. We also expect that the location, into which a TE inserts, relative to the structure of the gene, will affect the way in which the TE affects expression. We find that the effects of insertions can vary substantially depending upon the location of the insertion relative to various gene features and that the average effects of transposable element insertions diminish as insertions occur further from the gene start and stop positions. Our finding highlights the importance of incorporating transposable element variation into future work on understanding phenotypic variation in Drosophila and in other species.
Materials and Methods
Sequence data
We acquired Illumina platform paired-end sequencing data for 38 inbred lines (Mackay et al. 2012). Sequence data were downloaded from http://www.hgsc.bcm.tmc.edu/projects/dgrp/ and include lines from both freeze 1 and freeze 2 of the DGRP (Mackay et al. 2012; Supporting Information, Table S1). The data were aligned to the D. melanogaster reference genome version 5 (www.flybase.org) as described in Cridland et al. (2013). Mean sequence coverage for these lines ranged from 5.5× to 174.5× with a mean of 38.2× and a standard deviation of 44.3.
Expression data
We downloaded expression data from ArrayExpress, accession no. E-MEXP-1594, for the same 38 inbred lines for which we had paired-end sequencing data (Ayroles et al. 2009). This gives us expression data paired with genotype data for 38 inbred lines. There are four arrays per line, two each from males and females. The array used is the Affymetrix Drosophila 2.0 array that was designed using version 3 of the D. melanogaster reference (www.flybase.org). We loaded the raw data files into R (R Development Core Team 2008) using the Bioconductor Affy package (Gautier et al. 2004). We used the “rma” function with default settings to perform background correction and normalization. This function calculates the mean expression level for a set of probes that constitute a particular transcript from a gene. We then took the mean value of each probe set expression level for the two arrays from each line/sex combination.
We downloaded the most recent NetAffx annotation file from www.affymetrix.com (version 32 updated on June 9, 2011), which updates transcript information for the probes on the Drosophila 2.0 array to the D. melanogaster reference version 5.31 and updates coordinates to the version 5 reference sequence. We also downloaded the transcript annotation file from the D. melanogaster reference version 5.31 and identified the transcription start and stop locations for all transcripts.
Because the expression levels of lowly expressed genes are often poorly estimated (Baldi and Long 2001), we restricted our analysis to the top 75% of expression measures for each sex (Table S2).
Removal of regions of identity by descent
Because we are interested specifically in rare alleles in a population, we wanted to remove additional copies of any alleles that may be present due to relatedness between individuals. A TE that is present in multiple individuals due to close relatedness may still be rare with respect to a population, and we would therefore want to include one copy of this TE in the dataset. Previous work has indicated a large amount of identity by descent (IBD) in the DGRP, indicating the presence of closely related individuals within the panel (Cridland et al. 2013). We calculated IBD between the 38 lines by downloading single nucleotide polymorphism data from http://www.hgsc.bcm.tmc.edu/projects/dgrp/) (Mackay et al. 2012). Following Cridland et al. (2013), we performed an all-by-all comparison between lines, examining sliding windows of 1 Mb across the genome with 100 kb steps between windows (Figure S1). Windows that were >95% identical between two lines were marked as IBD and the region was masked in the line with lower coverage. One line was removed entirely because it was >95% IBD over >50% of its genome with another line. For this pair, we removed the line with the lowest coverage. On average, 5.1% of the genome was masked per line, standard deviation 7.3%, suggesting that in general the lines in the DGRP core 40 are not closely related to each other. Masking regions of the genome in related individuals will reduce the sample size for any analysis in that region of the genome, but should not have any other effect with respect to the results of the analysis.
Transposable element calls
Transposable elements were identified in each line following the method described in Cridland et al. (2013), which contains a complete description of how these calls were performed. The pipeline from Cridland et al. (2013), which includes freeze 1, but not freeze 2 data, was run on the new samples from DGRP “freeze 2” (see above). Source code for this pipeline is available at www.molpopgen.org/data.html. The TE calls for those lines are available as Table S1). TEs were identified using the version 5 reference sequence. For each element we detected, we evaluated the same region of the genome in every other line, generating either a “presence,” “absence,” or “no call” information for each TE in all 37 lines, excluding masked regions. Because TEs were called with short-read data, we were able to ascertain the presence or absence of a TE with high confidence; however, identifying the TE family for an individual insertion is much more uncertain with short reads and thus this information was not considered in these analyses. On average, we were able to make a positive presence or absence call in 34.9/37 lines, with a standard deviation of 3.03 calls.
Gene location categories
For each transcript with a TE insertion, we classified the insertion based on the location of the TE relative to the transcript (Table 1). Insertion categories were constructed so as to cover every portion of the span of the transcript. Categories are nonoverlapping with respect to a given transcript, but may overlap with respect to different transcripts; for example, a TE may fall in the 5′ region upstream of one transcript and in the 3′ region downstream of another transcript. Multiple transcripts from the same gene may also be correlated in expression level and thus a nearby TE may affect multiple transcripts from the same gene similarly.
Table 1. TE insertions within and near transcripts.
| Region | No. TE insertions | Expected insertions |
|---|---|---|
| Within exon | 235 | 1096 |
| Introns ≤400 bp | 67 | 130 |
| Within 200 bp of acceptor site | 61 | 90 |
| Within 200 bp of donor site | 63 | 100 |
| Within first intron | 527 | 783 |
| Not within first intron | 754 | 1124 |
| ≤500 bp of TSS | 170 | 247 |
| 501 bp to 2 kb of TSS | 389 | 599 |
| >2 kb of TSS | 1609 | 2126 |
| ≤500 bp of TES | 200 | 234 |
| 501 bp to 2 kb of TES | 333 | 544 |
| >2 kb of TES | 1515 | 1987 |
We cataloged TE insertions in the following genomic regions: (A) exons, intronic regions that are near exon boundaries including (B) introns <400 bp, (C) within 200 bp of a donor site, or (D) within 200 bp of an acceptor site, intronic regions that are more distant from exon boundaries, ≥200 bp from an acceptor or donor site, including (E) the first intron in the gene, as there is evidence that first introns may harbor more cis-regulatory variation than other introns (Marais et al. 2005), (F) not in the first intron in the gene, and regions upstream and downstream of the gene including (G) 0–500 bp 5′ of the transcription start site (TSS), (H) 501 bp to 2 kb 5′ of the TSS, (I) 2001 bp to 10 kb 5′ of the TSS, (J) 0–500 bp 3′ of the 3′ end of the transcript, (K) 501 bp to 2 kb 3′ of the 3′ end of the transcript, and (L) 2001 bp to 10 kb 3′ of the 3′ end of the transcript.
We also restricted our analysis to the euchromatic portions of the genome, defined as (X: 300,000–20,800,000; 2L: 200,000–20,100,000; 2R: 2,300,000–21,000,000; 3L: 100,000–21,900,000; and 3R: 600,000–27,800,000), avoiding centromeric and telomeric regions where high TE density makes individual TE calls more difficult to resolve uniquely (Cridland et al. 2013).
Statistical analysis
For each line containing a TE presence/absence call for a specific transcript/TE pair, we first calculated expression rank for each sex separately. We then compared normalized rank between sexes. For the majority of transcripts there was little difference in the normalized rank between sexes for the line harboring a TE insertion (mean difference in rank 0.01, standard deviation 0.31; see Table S3 for the list of transcripts with large differences between sexes). Therefore, we focus on sex-averaged expression levels for the 9235 transcripts common to the top 75% of expression measures for both males and females.
We next converted sex-averaged expression level into expression rank (from lowest to highest across DGRP inbred lines). To control for variation in sample size across positions, we then normalized ranks by dividing by the sample size (total number of lines without missing or masked data at each site). These normalized ranks provide the basis for our downstream analysis. We hypothesize that if TEs decrease the expression of nearby genes we will see an excess of TE-containing transcripts in the lowest normalized rank categories. Therefore, in addition to the true mapping of expression to lines, we generated 10,000 mappings where line labels were randomly permuted with respect to expression measures. In these permutations, we shuffled expression levels between lines while keeping the identity of the line that contains the TE insertion consistent. These randomly permuted mapping allow us to empirically derive the null distributions for the number of TE-containing transcripts that will be found at any normalized rank. By comparing the expected number of TE-containing transcripts to the observed number of TE-containing transcripts we can identify normalized ranks that have either an excess or a deficit of TE-containing transcripts.
We also calculated a z-score for each transcript/TE pair in a given region for each sex-averaged expression measure, z = (value of the line with the TE − population mean for lines without the TE)/(population standard deviation for lines without the TE). This z-score indicates the number of standard deviations away from the population mean any observed expression level actually is. We expect that TE-containing transcripts will have an excess of very negative z-scores.
Results
Rare transposable element alleles
We identified transposable elements in a set of 37 unrelated inbred lines from the DGRP (Ayroles et al. 2009; Mackay et al. 2012; Cridland et al. 2013). A set of 4376 TE insertions was private to individual lines (Table S1). Private TEs represent 86.6% of all TE insertion events detected in this set of lines, consistent with previous reports characterizing the site frequency spectrum of TEs in Drosophila (Charlesworth and Langley 1989; Cridland et al. 2013). Sequence coverage impacted our de novo transposable element identification rate (Figure S2; Cridland et al. 2013) and therefore the number of private TEs detected in each line varied substantially (mean 118.3, standard deviation 95.3). Since our focus is on understanding how TEs impact gene expression, and the majority of TE insertions are private to a single individual in a population, we limit our analyses to these private insertions.
Transposable elements near transcripts
After limiting our analysis to the 9235 transcripts common to the top 75% of expression measures for both males and females (see Methods), we identified 3889 different transcripts (42.1% of the total number of transcripts) for which at least one DGRP line with a TE was in or within 10 kb of that transcript (Table S2), hereafter TE-associated transcripts. For consistency with a previous reanalysis of the same expression data (Massouras et al. 2012), we considered TEs within 10 kb of a transcript as potential cis-regulators of the gene. Of TE-associated transcripts, 54.2% had a single DGRP line harboring a single nearby TE. TE-associated transcripts with more than one nearby TE could either have multiple DGRP lines each harboring a single nearby TE (1726 transcripts) or a single DGRP line could harbor multiple nearby TEs (388 transcripts). Accounting for this redundancy resulted in a total of 7107 transcript/TE pairs, which make up our dataset for analyzing the effect of TE insertions of expression levels. These transcript/TE pairs correspond to 3358 TE insertions, or 76.7% of private TEs found within 10 kb of transcripts common to the top 75% of expression measures. Of these TEs, 44% were within 10 kb of only one transcript, while 56% of TEs were within 10 kb of multiple transcripts.
Transcript location categories
Because TEs that insert in different locations relative to a gene may have different functional effects, we divided transcripts and their surrounding sequence into mutually exclusive categories that accounted for all sequence within each transcript plus 10 kb to either side of the transcript (Table 1). A priori, we predict that TE insertions into exons are likely to dramatically reduce expression levels. However, insertions into upstream and downstream elements such as promoters and enhancers (Smith and Corces 1991) and near intronic sequences affecting alternative splicing (Varagona et al. 1992) may also affect gene expression levels. Finally, TE insertions may also generate new regulatory elements via their insertion that could result in increased expression levels (Chung et al. 2007).
Because individual TEs can be associated with multiple transcripts, they can fall into multiple location categories, though only one location category with respect to a given transcript. We calculated the expected number of TE insertions that would fall into each location category, assuming that TEs would fall in a given category in numbers proportional to that category’s representation in the genome. For examples, only 6% of TEs are found in exons (Table 1) compared to the 25% of TEs we would expect to find in exonic regions, as 25% of the D. melanogaster genome sequence is protein coding. This observation is consistent with previous observations (Kaminker et al. 2002; Cridland et al. 2013). Other regions that are likely to harbor regulatory variation, such as regions near splice acceptor or donor sites, also had fewer TE insertions than would be expected (Table 1). Regions furthest from the TSSs or transcription end sites (TESs) had the most TE insertions (Table 1), though all regions had fewer than the expected number of TEs, suggesting a general pattern of a reduction in TEs near genes.
Transposable elements as causative mutations
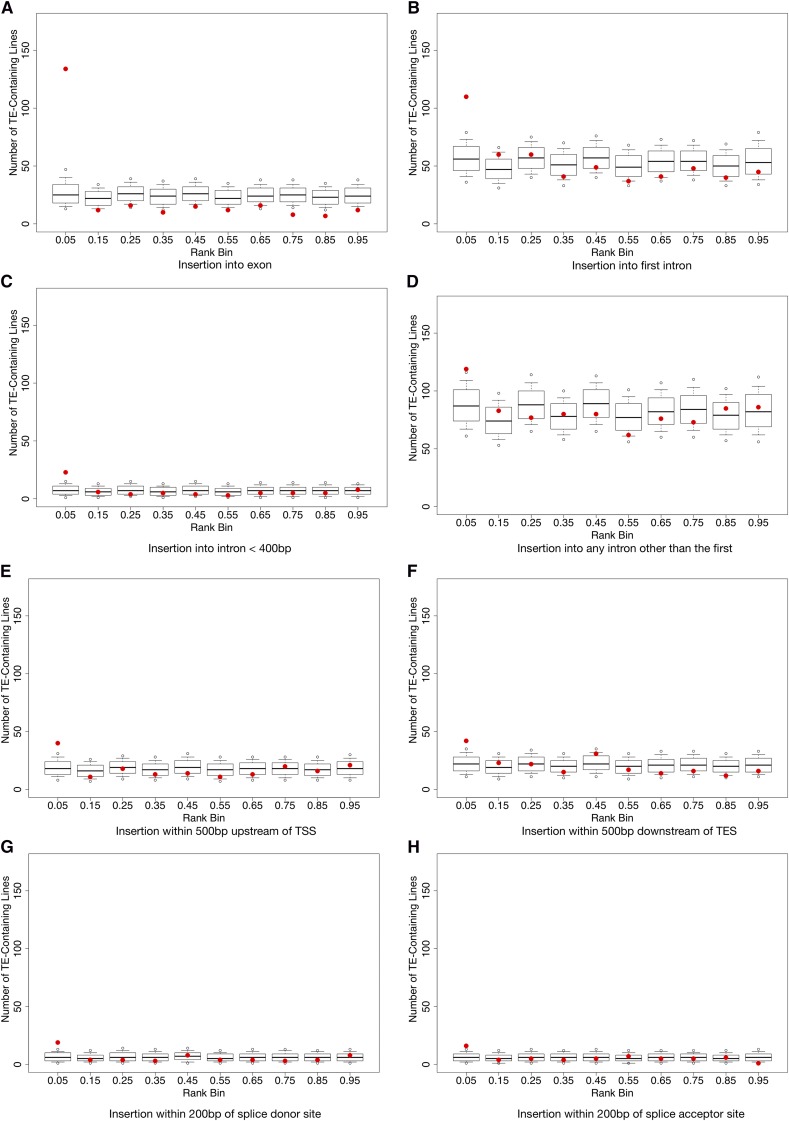
We calculated the normalized rank for each transcript’s expression measure for each line such that a lower normalized rank indicates lower expression of the transcript. We find that the presence of a TE insertion in or near a protein-coding gene is often strongly associated with a reduction in gene expression level as indicated by the excess of TE-containing transcripts being found in the lowest ranked bin of expression levels compared to the null distribution (Figure 1). The strongest effect is seen for TE insertions in exons (Figure 1A) and the second-largest effect was for TE insertions into the first intron of a gene (Figure 1B), which was much more dramatic than insertions into smaller introns (Figure 1C) or insertions into any other intron (Figure 1D). These observations lend credence to the belief that the first introns of Drosophila genes are likely to contain regulatory information (Marais et al. 2005). To our surprise, insertions of TEs in the 500 bp immediately upstream of a predicted TSS (Figure 1E) and insertions in the 500 bp immediately downstream of a predicted polyA site (Figure 1F) had detectable but subtle effects. Finally TE insertions within 200 bp of a splice donor or acceptor site have measurable but modest effects (Figure 1, G and H), suggesting the splicing machinery is fairly tolerant of TE insertions close to these sites.
Figure 1.
Normalized rank expression of transposable elements. Observed numbers of TE-containing lines per rank bin vs. 10,000 permutations. Red dots indicate the observed number of TE-containing lines; box plots show permutations. Box plot tails indicate the 2.5% and the 97.5% confidence intervals. Open circles above and below the box plots indicate the 0.5% and the 99.5% confidence interval. (A) TE is in an exon, (B) TE is in a 1st intron, (C) TE is in an intron ≤400bp in length, (D) TE is not in 1st Intron, (E) TE ≤ 500bp from TSS, (F) TE ≤ 500bp from TES, (G) TE ≤ 200bp from a donor site, and (H) TE ≤ 200bp from an acceptor site.
A TE insertion associated with a low expression rank is a strong candidate to be the mutation causing the expression change. Based on this rationale, we consider TE insertions in the lowest 5% of ranked expression values to be putatively causative mutations. Our genome-wide tally of the number of TE causative mutations is the excess number of times the TE-containing DGRP line from a TE-associated transcript was found in the lowest 5% of expression bin over the expected number of times based on our permutations of the data (Table 2). We find 403 TE-associated transcripts where the TE-containing line is in the lowest 5% bin of expression measures over all location categories (Table 2). These 403 TE-associated transcripts are associated with 363 different genes, so this effect is clearly not due to a large number of splice variants at a small number of genes.
Table 2. Causative mutations.
| Category | Observed | Expected | O–E | % functional |
|---|---|---|---|---|
| Within exon | 101 | 7.1 | 93.9 | 93.0 |
| Introns ≤400 bp | 14 | 2.1 | 11.9 | 85.0 |
| Within 200 bp of acceptor site | 5 | 1.8 | 3.2 | 64.0 |
| Within 200 bp of donor site | 12 | 1.8 | 10.2 | 85.0 |
| Within first intron | 38 | 15.3 | 22.7 | 59.7 |
| Not within first intron | 41 | 24.4 | 16.6 | 40.5 |
| ≤500 bp of TSS | 20 | 5.4 | 14.6 | 73.0 |
| 501 bp to 2 kb of TSS | 17 | 11.9 | 5.1 | 30.0 |
| >2 kb of TSS | 55 | 60.4 | −5.4 | −9.8 |
| ≤500 bp of TES | 19 | 6.0 | 13.0 | 68.4 |
| 501 bp to 2 kb of TES | 13 | 10.0 | 3.0 | 23.1 |
| >2 kb of TES | 68 | 56.2 | 11.8 | 17.4 |
| Total | 403 | 202.4 | 200.6 |
% functional = (observed–expected)/observed.
Permutations of the data suggest that there are 200.6 more TE-associated transcripts found in the lowest 5% of expression ranks than are expected by chance alone. Thus, we estimate that 200.6/403, or 49.7%, of the TE-associated transcripts with low expression values are, in fact, causal mutations. Further, considering the 9235 transcripts in the genome that we analyzed, we estimate that 2.2% of transcripts contain a TE with a dramatic effect on expression level, within this example population. We view this as a minimum estimate—given that TE insertions are likely deleterious when inserted near genes (Cridland et al. 2013), increasing the sample size beyond the 37 lines examined here would likely identify more genes with TE insertions affecting gene expression as an increase in sample size would also include an increase in the number of rare alleles to be examined.
Table 2 shows that exons have the highest number of TE insertions estimated to be causative. These insertions into exons make up 46.8% of the total number of causative events inferred genome-wide (Table 2). Insertions into intronic regions near exon boundaries are also highly enriched for causative mutations, with introns overall containing 32.2% of the causative mutations (Table 2). Surprisingly, insertions upstream of the TSS or downstream of the TES do not contribute greatly as expression QTL (eQTL) (∼20% of putative functional events). Although the likelihood of an insertion being functional is clearly a function of its distance from the TSS or TES (Table 2), the absolute number of insertions is inversely proportional to distance from the transcript (Table 1), so more distant insertions contribute similarly to expression variation as close ones. An additional, perhaps surprising, observation is that insertions upstream of the TSS are not that much more likely to be functional than those downstream of the TES.
Effect sizes of transposable element insertions
To estimate the effect size of private TE insertions on gene expression levels, we carried out a parametric analysis. This analysis constructed a z-score for each transcript/TE pair that represents the expression level of the DGRP line harboring the private TE for that transcript relative to the variation in the same expression measure over the TE-free lines. If the general effect of the presence of a TE insertion is to decrease expression levels, we would expect to see a general pattern of negative z-scores for TE-associated transcripts.
Across all location categories we find that the mean z-score for transcripts from lines with TE insertions was −0.23, or roughly a quarter of a genetic standard deviation. This indicates an overall decrease in expression for a transcript harboring a nearby TE (Table 3). A Q-Q plot (Figure S3A) of observed z-score statistics against the same dataset with phenotypes permuted with respect to genotypes shows an increased incidence of very large z-scores in the observed data once z-scores are smaller than −2. There is a roughly twofold enrichment of transcripts (445 observed vs. 227 expected based on permutations) with expression less than −2 in the actual vs. permuted dataset, suggesting on the order of 198 transcript/TE pairs are impacted out of 4886.
Table 3. Mean z-scores for transcripts with TEs.
| Category | Mean z-score | N |
|---|---|---|
| Within exon | −3.44 | 249 |
| Introns ≤400 bp | −1.03 | 72 |
| Within 200 bp of acceptor site | −0.90 | 64 |
| Within 200 bp of donor site | −0.67 | 64 |
| Within first intron | −0.37 | 545 |
| Not within first intron | −0.11 | 852 |
| ≤500 bp of TSS | −0.43 | 186 |
| 501 bp to 2 kb of TSS | −0.01 | 418 |
| >2 kb of TSS | −0.05 | 2121 |
| ≤500 bp of TES | −0.52 | 213 |
| 501 bp to 2 kb of TES | −0.04 | 347 |
| >2 kb of TES | −0.02 | 1976 |
Mean z-scores are calculated from the transcript/TE pairs for all transcripts with an insertion in each location category.
Ignoring the four location categories involving TE insertions >500 bp up/down stream of the TSS/TES, constructing a similar Q-Q plot of observed vs. permutation-based z-scores (Figure S3B) showed an increased incidence of large z-scores once z-scores are smaller than −1. There was a roughly twofold enrichment of transcripts with z-score less than −1 (649 observed vs. 385 permuted), and 264 more transcript/TE pairs in this category than we expected based on permutations. Since only 2245 transcript/TE pairs have a TE in an intron, exon, or within 500 bp of TSS/TES, these data suggest TEs inserted in these regions are very likely to have an effect. Across these location categories we find that the mean z-score for transcripts from lines with TE insertions was −0.67, or about two-thirds of a genetic standard deviation. It is noteworthy that an effect size of one genetic standard deviation would be considered large by the standards of the genetics of complex traits (Cantor et al. 2010).
All location categories within 500 bp of a gene showed a negative mean z-score for transcript/TE pairs (Table 3). Transposable elements inserted into exons had the largest effect on gene expression; z-scores for transcript/TE pairs where the TE is in an exonic region had a mean of −3.44, indicating that transcript expression was reduced by an average of more than three standard deviations from the mean due to the presence of a TE. Clearly, this effect size is much larger than those typically identified as eQTL (Cantor et al. 2010). In general, insertions in intronic regions showed larger effect sizes than insertions upstream of the TSS or downstream of the TES. Of location categories involving intronic sequence, insertions falling into first introns show a larger effect size than insertions into other large introns that are at least 200 bp away from an acceptor or donor site. Location categories closest to transcripts also showed lower mean z-scores than regions further away from transcripts.
The effect of a TE insertion on the expression of nearby genes can be many standard deviations from the mean. The most extreme example of this was the gene nessy, where the average expression level of the line with the TE was 40 standard deviations lower than TE free lines. This is likely an effectively null mutation at this gene segregating in nature. Comparing the average standard deviation in mean expression per DGRP line for TE-associated transcripts to transcripts with no TEs in or within 10 kb indicates that TE-associated transcripts have a larger mean standard deviation, 0.33 vs. 0.28, (t-test; P = 1.0e-22). Since the DGRP line with the TE insertion is not used in calculating the standard deviation this suggests that transcripts with TEs may be those that are more tolerant of variation in expression level.
Transposable elements vs. SNPs and other complex variants
Massouras et al. (2012) have reanalyzed the same expression data used here (Ayroles et al. 2009) and attempted to associate complex variants (primarily insertion/deletion events and small duplications rather than TEs) with variation in gene expression levels. They identified 17,501 cis-eQTL associated with 2033 genes (at a 10% false discovery rate), with 499 of these genes having eQTL with similar effect in each sex. Though due to the way statistical testing was carried out in this earlier work it is difficult to estimate how many independent cis-eQTL were discovered for any given gene. We compared our set of genes where the TE-containing transcript was in the lowest 5% of expression measures to the Massouras et al. (2012) set of genes identified as having a cis-eQTL in both sexes to identify genes common to both sets.
We find that 145 of the 499 genes with previously reported cis-eQTL common to both sexes have TEs within 10 kb of the gene and of these 22 genes, a TE in the DGRP line with the lowest level of expression. These numbers suggest that 4.4% of genes they identify as having a non-TE cis-eQTL also have a TE that is categorized as a causative mutation. While it is unclear how the TE and non-TE mutations are contributing to expression variation, and how they may interact, this observation highlights the importance of incorporating TE information into work on expression and phenotypic analyses. Given that our analysis has focused on insertions of large effect, this is a conservative estimate of the number of TEs that may contribute to expression differences. There are also important differences between these studies. First, we only looked at sex-averaged expression, which means that we are likely excluding a number of insertions that affect expression in a sex-specific manner. Second, this study specifically focused on rare variants, while the previous analysis of this dataset was restricted to variants found in at least three lines.
Effect of TEs as a class of variation
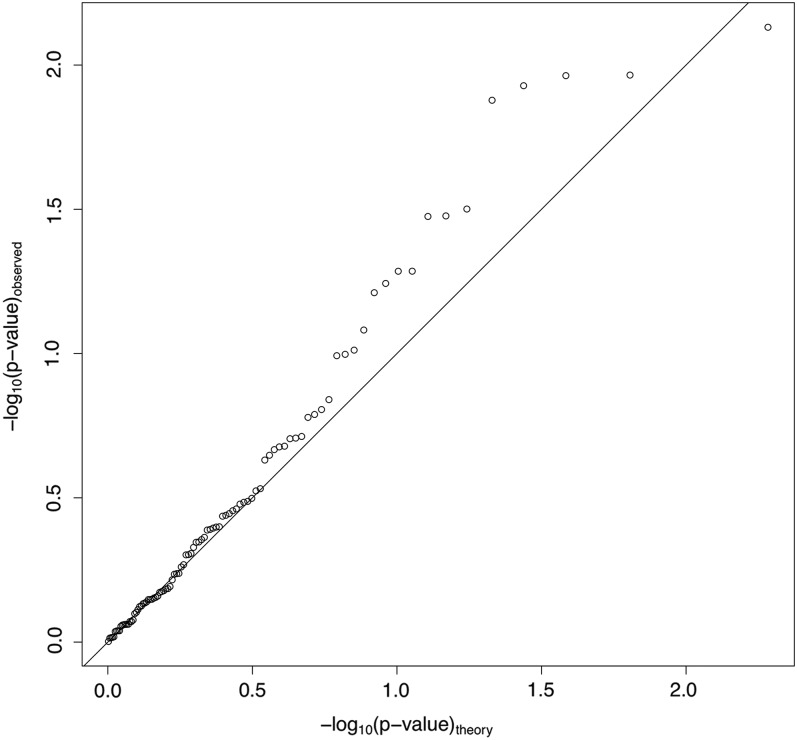
Several previous studies that found associations between TEs and variation have done so by examining TEs as a class of variation, where all TEs are expected to contribute to phenotypic variation in the same way (Mackay and Langley 1990; Long et al. 2000; Macdonald et al. 2005; Gruber et al. 2007). We examined transcripts where four or more DGRP lines had a TE insertion in a particular location category, though each TE insertion was only present in a single DGRP line. We find 96 such transcripts having four or more DGRP lines with a TE in a given location category. For each such transcript we performed a t-test for a difference in gene expression between TE-harboring DGRP lines vs. TE-free DGRP lines. We expect that lines containing TEs will have lower expression levels than lines that do not contain TEs. We then plotted the cumulative distribution of these P-values against their expectation, based on a single permutation, in Figure 2 (on a log-log scale). If TEs impact gene expression, we would expect to see more t-tests with significant P-values in the observed data when compared to the permuted data. The excess of significant transcripts (i.e., points beyond P < 0.05) over that expected based on theory indicates that TEs as a class are likely to impact gene expression. For seven of the eight cases where the t-test was significant, the TE-harboring lines had lower expression than the TE-free lines, consistent with TEs generally partially abolishing gene expression.
Figure 2.
Transposable elements as a class of variation. Probability–probability plot of observed and expected P-values from t-tests of all cases where four or more lines show an independent TE insertion in the same location category for the same transcript.
Discussion
We find that rare allele of large effect (RALE) transposable element insertions contribute substantially to gene expression variation between individuals. We find that TE-associated transcripts are found at the bottom tail of expression measures within our example population for a transcript substantially more often than would be expected. We further observe that within our example population, ∼2.2% of transcripts show expression variation that is due to the presence of a private TE and that many more transcript/TE pairs show a reduction in expression of at least one standard deviation. Although these effects are modest relative to TEs that move a DGRP line into the lowest 5% with respect to gene expression, they are still quite large by the standards of the field of the genetics of complex traits (Cantor et al. 2010). Furthermore, while individual insertions are private to a single DGRP line, as a class of variation these insertions are common and frequently insert into regions that may produce changes in expression. We find similar results when TEs are considered as a class of variation as has been done previously on smaller scales (Mackay and Langley 1990; Long et al. 2000; Macdonald et al. 2005; Gruber et al. 2007).
In general we believe that our results are a conservative estimate of the effects of transposable element insertions on gene expression. First, our estimation of causative mutations only quantifies the numbers of TEs with effect sizes large enough to place a TE-containing line in the smallest 5% quantile with respect to gene expression, that is, a TE with a very large effect size. Second, our analysis is restricted to TEs that are private to a single DGRP line. While this choice reflects the population dynamics of most TE insertions, that they are private to a given individual (Charlesworth and Langley 1989; Cridland et al. 2013), there are still a substantial number of TE insertions that segregate at higher frequencies. These insertions are likely to contribute further to expression variation.
We also identified differences in the number of likely functional insertions and the average effect size of an insertion depending upon the location category of the TE insertion. Our a priori expectations that TE insertions into exonic regions and TE insertions into potential cis-regulatory sequence in intronic regions would reduce expression levels were supported by the data. Given the standard model of how transcription occurs, we also predicted a priori that insertions immediately upsteam of the TSS would have a large effect; however, the effects we observed were much smaller than the effects in exonic regions. We are led to conclude that regions immediate upstream of the TSS are more robust to TE insertions than we initially hypothesized.
Overall, this work shows that TEs make important and measurable contributions to gene expression variation. Specifically, a measurable fraction of all transcripts have TE insertions private to a single strain that have an effect on gene expression that would be quite large by the standards of the eQTL community. Furthermore, the presence of TEs of large effect are largely ignored in eQTL and QTL studies and many observed phenotypic differences between individuals may be due, at least in part, to TE insertions. It is also possible that many eQTL are due to TEs linked to common SNP variants that were identified. That being said, it is difficult to estimate the total fraction of previously published eQTLs due either to TE insertions or that are affected by them. Thus our results suggest that TEs should not be ignored in studies attempting to dissect the genetics basis of phenotypic variation, and that experiments should be designed to quantify the extent to which TE insertions can explain previous SNP/phenotype associations. While our work was carried out in D. melanogaster, transposable elements are a common feature of eukaryotic genomes and studies of expression variation in Drosophila and in other species should be aware of the potential effects of TE insertions and incorporate them into their analysis. If rare alleles of large effect turn out to make major contributions to standing variation in human complex disease phenotypes, one wonders what fraction of that effect is due to transposable element insertions in or near genes.
Acknowledgments
We thank Trudy Mackay for early data access. We also thank Dave Begun and members of his lab for helpful comments.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.170837/-/DC1.
Available freely online through the author-supported open access option.
Communicating editor: E. A. Stone
Literature Cited
- Ayroles J. F., Carbone M. A., Stone E. A., Jordan K. W., Lyman R. F., et al. , 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat. Genet. 41: 299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldi P., Long A. D., 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinform. 17: 509–519. [DOI] [PubMed] [Google Scholar]
- Cantor R. M., Lange K., Sinsheimer J. S., 2010. Prioritizing GWAS results: a review of statistical methods and recommendations for their application. Am. J. Hum. Genet. 86: 6–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., 1989. The population genetics of Drosophila transposable elements. Annu. Rev. Genet. 23: 251–287. [DOI] [PubMed] [Google Scholar]
- Chung H., Bogwitz M. R., McCart C., Andrianopoulos A., R. H. Ffrench-Constant, et al, 2007. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics 175: 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland J. M., Macdonald S. J., Long A. D., Thornton K. R., 2013. Abundance and distribution of transposable elements in two Drosophila QTL mapping resources. Mol. Biol. Evol. 30: 2311–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M., Irizarry R. A., 2004. Affy–analysis of Affmetrix GeneChip data at the probe level. Bioinform. 20: 307–315. [DOI] [PubMed] [Google Scholar]
- Gruber J. D., Genissel A., Macdonald S. J., Long A. D., 2007. How repeatable are associations between polymorphisms in achaete-scute and bristle number variation in Drosophila. Genetics 175: 1987–1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker J. S., Bergman C. M., Kronmiller B., Carlson J., Svirskas R., et al. , 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long A. D., Lyman R. F., Morgan A. H., Langley C. H., Mackay T. F. C., 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with bristle number in Drosophila melanogaster. Genetics 154: 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald S. J., Pastinen T., Long A. D., 2005. The effect of polymorphisms in the Enhancer of split gene complex on bristle number variation in a large wild-caught cohort of Drosophila melanogaster. Genetics 171: 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., 2010. Mutations and quantitative genetic variation: lessons from Drosophila. Phil. Trans. R. Soc. B. 365: 1229–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F. C., Langley C. H., 1990. Molecular and phenotypic variation in the achaete-scute region of Drosophila melanogaster. Nature 348: 64–66. [DOI] [PubMed] [Google Scholar]
- Mackay T. F. C., Richards S., Stone E. A., Barbadilla A., Ayroles J. F., et al. , 2012. The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais G., Nouvellet P., Keightley P. D., Charlesworth B., 2005. Intron size and exon evolution in Drosophila. Genetics 170: 481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massouras A., Waszak S. M., Albarca-Aguilera M., Hens K., Holcombe W., 2012. Genomic variation and its impact on gene expression in Drosophila melanogaster. PLoS Genet. 8: e1003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuzhdin S., MacKay T. F., 1994. Direct determination of retrotransposon transposition rates in Drosophila melanogaster. Genet. Res. 63: 139–144. [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2008 R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna. http://www.R-project.org.
- Smith P. A., Corces V. G., 1991. Drosophila transposable elements: mechanisms of mutagenesis and interactions with the host genome. Adv. Genet. 29: 229–300. [DOI] [PubMed] [Google Scholar]
- Stranger B. E., Forrest M. S., Dunning M., Ingle C. E., Beazley C. et al, 2007. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315: 848–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona M. K., Purugganan M., Wessler S. R., 1992. Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viera C., Biemont C., 1997. Transposition rate of the 412 retrotransposable element is independent of copy number in natural populations of Drosophila simulans. Mol. Biol. Evol. 14: 185–188. [DOI] [PubMed] [Google Scholar]