Abstract
Epigenetics studies the emergence of different phenotypes from a single genotype. Although these processes are essential to cellular differentiation and transcriptional memory, they are also widely used in all branches of the tree of life by organisms that require plastic but stable adaptation to their physical and social environment. Because of the inherent flexibility of epigenetic regulation, a variety of biological phenomena can be traced back to evolutionary adaptations of few conserved molecular pathways that converge on chromatin. For these reasons chromatin biology and epigenetic research have a rich history of chasing discoveries in a variety of model organisms, including yeast, flies, plants and humans. Many more fascinating examples of epigenetic plasticity lie outside the realm of model organisms and have so far been only sporadically investigated at a molecular level; however, recent progress on sequencing technology and genome editing tools have begun to blur the lines between model and non-model organisms, opening numerous new avenues for investigation. Here, I review examples of epigenetic phenomena in non-model organisms that have emerged as potential experimental systems, including social insects, fish and flatworms, and are becoming accessible to molecular approaches.
KEY WORDS: Chromatin, Epigenetics, Genomics, Polyphenism
Introduction
When Conrad H. Waddington coined the term ‘epigenetics’ (Waddington, 1942) he could not have dreamed of the controversy and debate that would still surround this word almost 75 years later (Berger et al., 2009; Bird, 2007; Bonasio et al., 2010a; Ptashne, 2007). Waddington wished to establish a new biological discipline that would study the mechanistic action of genes in the context of development (Waddington, 1942); that discipline exists today and it is called ‘developmental genetics’. In the meantime, the meaning of epigenetics has evolved and, without dwelling on the controversies, for the remainder of this review I will take it to mean the inheritance of phenotypic traits that occurs without changes in the DNA sequence (Bonasio et al., 2010a; Riggs et al., 1996).
It is important to note at the outset that epigenetic inheritance can take place during meiosis and affect multiple generations, giving rise to the phenomenon of transgenerational epigenetic inheritance (Heard and Martienssen, 2014), but it also occurs commonly during mitosis (Fig. 1A,B). In the latter case, epigenetic inheritance is responsible for the maintenance of discrete transcriptional states that give rise to different phenotypes originating from the same genotype (Bonasio et al., 2010a). Specifically, the molecular mechanisms that maintain cell identity over multiple rounds of cell division, making sure that lineage-committed cells remain true to their lineage, are epigenetic in nature and are the subject of intense investigation, because they underpin the ability of multicellular organisms to develop from a single fertilized egg and hold great promise for cancer treatment and regenerative medicine (Amabile and Meissner, 2009; Dawson and Kouzarides, 2012). Finally – and most confusingly – the same molecular pathways that are responsible for mitotic and meiotic inheritance of epigenetic states might also participate in the stabilization of transcriptional patterns in post-mitotic cells (Fig. 1C), such as, for example, neurons (Bonasio, 2012). In this case, no biological information is transmitted to a following generation, but the same molecular machinery might be involved. Some molecular biologists prefers to include this in a broader usage of ‘epigenetics’ (Bird, 2007), although it creates a slippery slope, whereby any form of gene regulation is epigenetics, which, I suspect, is the crux of the controversy surrounding this term.
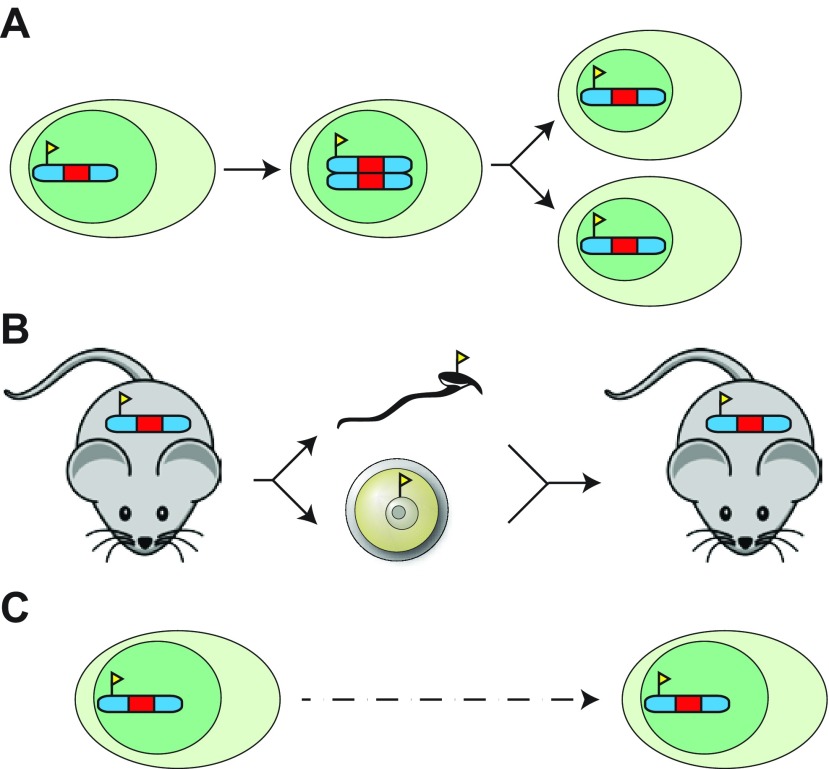
Fig. 1.
Types of epigenetic memory. (A) Mitotic epigenetic inheritance controls the replication of epigenetic marks (yellow flag on chromosome) throughout DNA replication in S phase (middle cell) and throughout cell division in M phase (right cells). Epigenetic marks that can be inherited in this fashion include DNA methylation and, to some extent, certain histone modifications. (B) Meiotic epigenetic inheritance controls transgenerational transmission of information that does not reside in the DNA sequence. Several cases are known in plants and a handful in vertebrates such as mouse. In many cases, although the epigenetic state is visibly transmitted, the nature of the epigenetic signal in the germ cells (yellow flag, middle) remains unknown. (C) In certain cases the same epigenetic signals that transmit information throughout cell division are used to stabilize transcriptional patterns in terminally differentiated, non-dividing cells. Although the inheritance-based definition of epigenetics would exclude such mechanisms, because of the shared molecular features they are nonetheless of interest to epigenetic research.
Although precise terminology is essential to scientific investigation, the energy spent in the attempts to define an abstract concept such as epigenetics might be misdirected, as stated previously (Nanney, 1958). Cells and organisms need to stabilize phenotypic responses to changing environments. Some of these phenotypic states must persist after the initial stimulus has subsided, often for long period of times, and, on occasion, must be re-established after cell division and/or organismal reproduction. Regardless of what we choose to call the underlying molecular mechanisms, they are fundamental to biology and multicellular life could not exist without them.
The variety of biological phenomena in which evolution has taken advantage of the epigenetic machinery, as reviewed below, is a testimony to the fact that our definitions are of little consequence and that by trying to make them narrower and more accurate we might be losing sight of the forest for the trees.
Molecular epigenetics
The reader interested in the molecular details of how epigenetic information is encoded, stored and utilized by the cell should refer to the many reviews that have been published on the topic (Bonasio et al., 2010a; Margueron and Reinberg, 2010). Here, I will simply introduce relevant terminology so that the remainder of this review is comprehensible to a broad range of biologists.
The eukaryotic genome is packaged inside the nucleus into a complex of DNA, RNA and proteins called ‘chromatin’. Several features of chromatin have the potential to encode epigenetic information at the molecular level (Fig. 2). The first epigenetic mark to be discovered was DNA methylation (Holliday, 1987; Holliday and Pugh, 1975). This is a chemical modification that is restricted to cytosine residues in eukaryotes and can alter the binding properties of a DNA sequence, to regulate its function (Bird, 2002). DNA methylation is copied onto newly synthesized strands of DNA by enzymes of the DNMT1 family, explaining its epigenetic nature (Goll et al., 2005).
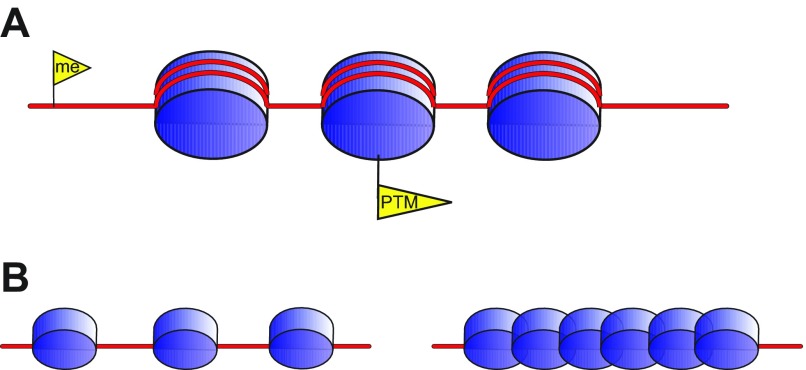
Fig. 2.
Molecular encoding of epigenetic information. Chromatin is composed of DNA (red line) packaged around histone octamers (blue discs) into nucleosomes. (A) In addition to the DNA sequence, biological information can be encoded by chemical changes to the DNA, such as methylation (me) or histone PTMs (yellow flags). (B) Higher-order chromatin structures might also be vehicles of epigenetic information. In this case loose (left) versus dense, compact (right) chromatin correspond to active and repressed chromatin, respectively.
The basic unit of chromatin, the nucleosome, is composed of two copies each of four short polypeptides known as histones, which form a discoid structure around which 147 bp of DNA are wrapped (Luger et al., 1997). In addition to the globular domain, histones have disordered N- and C-terminal tails that protrude from the disc and are available to enzymes that place post-translational modifications (PTMs), such as acetylation, phosphorylation, methylation and others (Kouzarides, 2007; Margueron et al., 2005). These are the so-called ‘histone marks’ on which most chromatin research has been increasingly focused ever since their discovery (Allfrey et al., 1964), and even more so in recent years, after the advent of genome-wide technologies. Histone marks, especially acetylation, can function by directly fine-tuning the biophysical properties of chromatin (Marmorstein, 2001), but by and large they are believed to form binding surfaces for chromatin-binding factors, or histone ‘readers’ (Ruthenburg et al., 2007a), that mediate their downstream functional consequences. PTMs are also found on the globular domain of histones, but, with few exceptions, the biological function of this group of marks is not as well understood (Mersfelder and Parthun, 2006).
A central goal of current research in chromatin biology is to understand how the complex machinery that writes and erases histone marks recognizes its targets in the genome and how a combination of different histone marks are interpreted as separate signals by histone readers (Ruthenburg et al., 2007b). It is becoming increasingly clear that the large amount of noncoding transcription that has been reported in most genomes gives rise to a wide variety of noncoding RNAs (ncRNAs), some of which appear to function as recruitment or regulatory signals that direct these chromatin modifiers to act on the relevant genes (Bonasio and Shiekhattar, in press; Castel and Martienssen, 2013; Rinn and Chang, 2012).
Finally, the higher-order, three-dimensional organization of chromatin, both in the short range, such as enhancer-promoter looping (Kagey et al., 2010) and in the long range, such as topological domains and chromosome territories (Cremer and Cremer, 2001; Dixon et al., 2012; Phillips-Cremins et al., 2013), has also been long speculated to participate in the regulation of gene expression (Fraser and Bickmore, 2007; Misteli, 2007) and, possibly, epigenetic inheritance. Research in this field has been challenging, because of a lack of quantitative approaches; however, things are likely to change thanks to the development of conformation capture approaches (Dekker et al., 2002) and continued progress in the description of the chromatin fiber, such as a recent study that reported the structure of the 30 nm fiber as a double-helix of double helices (Song et al., 2014).
Epigenetics in model organisms
Few fields of molecular biology have seen such a widespread use of different model organisms as epigenetics has. The reason might be that disparate observations of unexpected non-mendelian inheritance of observable traits made in various model organisms all turned out to be due to epigenetic effects (Allis et al., 2007). First came positional effect variegation in Drosophila (Muller, 1941), whereby a gene that is repositioned near repressed heterochromatic regions of the chromosome as a result of a translocation is expressed in a variegated manner, giving rise to mosaicism despite a homogeneous underlying genotype. Genetic screens for enhancers and suppressors of variegation led to the identification of several genes whose product acted on chromatin (Schotta et al., 2003). Another strong link between epigenetic phenomena and histone marks was first established in Drosophila and originated from the study of the genes belonging to the Polycomb group (PcG) and trithorax group (trxG), which were shown decades ago to mediate the epigenetic maintenance of cell identity and embryonic patterning (Ringrose and Paro, 2004). The biochemical characterization of the protein complexes containing the products of these genes revealed that they act on chromatin by either placing or recognizing some of the most intensely studied histone marks, such as the methylation of histone H3 lysine 27 (H3K27me) and H3K4me3, which are linked to PcG and trxG function, respectively.
Much insight into epigenetics has come from the study of plants, particularly maize and Arabidopsis thaliana. The phenomenon of paramutation in maize, first described in 1958 (Brink, 1958) is a classic example of epigenetics, because the effects of a certain allele present in the ancestors are manifest in the progeny, even after the allele has been crossed out for several generations (Chandler, 2007). In fact, it can be argued that plants, lacking the ability to escape detrimental changes in their surrounding environment by physical relocation, are particularly dependent on epigenetic mechanisms to adapt and survive. This might explain why epigenetic phenomena that are only now beginning to be understood in mammals, including transgenerational inheritance and RNA-mediated chromatin changes, have been dissected in much more detail in plants.
The most traditional model organism of all, the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe have also contributed to our understanding of epigenetic phenomena. Although these are monocellular organisms that, at first sight, would seem to have little need for epigenetics, they in fact contain genomic regions that must be kept silent in most circumstances, most notably subtelomeric sequences and the mating type locus (Allis et al., 2007). S. cerevisiae has lost most epigenetic machinery, including DNA methylation and the enzymes involved in several of the histone marks common in plants and metazoans, but still utilizes a complex network of histone acetylation and deacetylation to maintain silencing at heterochromatin regions (Grunstein and Gasser, 2013). However, S. pombe has retained H3K9 methylation, a hallmark of constitutive heterochromatin, and, in fact, meiotic segregation of this histone mark and the corresponding segregation of silencing was one of the first formal demonstrations of the transmission of epigenetic information in cis by a chromatin-based mechanism (Grewal and Klar, 1996).
Finally, epigenetic research has made great strides toward understanding gene regulation and cell identity in mammals. For this, embryonic stem cells from mice and, more recently from humans, as well as induced pluripotent stem cells [pluripotent cells derived from differentiated cells by epigenetic ‘reprogramming’ (Takahashi et al., 2007; Takahashi and Yamanaka, 2006)] have quickly become the cell systems of choice. Several epigenetic phenomena, such as X chromosome inactivation and genomic imprinting (Lee and Bartolomei, 2013), have been studied prevalently in mammals.
Open questions in epigenetics
Despite these fundamental discoveries in epigenetics made in model organisms, many more questions remain unanswered. In several cases the identity of the ‘true’ epigenetic signal remains a matter of controversy, although few would doubt that proximate signals include histone marks, DNA methylation and, at least in some species, ncRNAs of various sorts (Bonasio et al., 2010a). It seems natural that some of these specific, mechanistic questions should be investigated in the same model organisms that have already contributed so much to progress in the field. However, components of these epigenetic pathways are increasingly implicated in the regulation of organism-level phenotypes that cannot be easily modeled in cell culture and are not always available for investigation in suitable model organisms.
One example is brain function and behavior (Bonasio, 2012). The brain is a complex organ and its ontogeny must access the same epigenetic pathways of gene regulation that contribute to development of all other organs. However, chromatin modifications and possibly other epigenetic signals appear to have dedicated roles in the adult brain. Mutations in several known epigenetic regulators cause neurological phenotypes (Jakovcevski and Akbarian, 2012), and at least in the case of the methyl-cytosine binding protein MeCP2 it was demonstrated that the gene contributes to the phenotype in the adult brain (Luikenhuis et al., 2004), suggesting a functional – rather than developmental – role. The culprit for this connection might be DNA methylation, because its levels respond dynamically to brain activity (Guo et al., 2011), and because its potential for long-term memory of transcriptional states resonates with the need for the long-term stability of neuronal states necessary for higher brain function. But the connections are not restricted to DNA methylation. Increasing experimental evidence suggests that histone acetylation and possibly methylation are required for learning and memory (Day et al., 2013; Gupta et al., 2010), and even the development of long-lasting relationships among individuals (Wang et al., 2013).
Another example is offered by the potential role of epigenetic pathways in regulating longevity. It can hardly be a coincidence that the mammalian enzymes at the center of much research – and controversy (Guarente, 2013) – on the molecular regulation of longevity, the so-called ‘sirtuins’, are deacetylases related to Sir2, one of the few histone modifiers responsible for epigenetic silencing in S. cerevisiae (Houtkooper et al., 2012). Considering that the germline is virtually immortal, it has been proposed that the aging clock is by and large driven by epigenetic deterioration, and that understanding the root causes of this deterioration might allow us to stop or even reverse the clock (Rando and Chang, 2012).
A third and final example is that of transgenerational inheritance. Although examples of this phenomenon in plants abound, it remains unclear whether the few cases characterized in other model organisms are the exception or the rule (Heard and Martienssen, 2014). How widespread is this phenomenon? What molecular features other than the DNA sequence has the required information content and long-term stability to be transmitted into the germline and affect the phenotype of a whole new organism? The recent – and also controversial – observation that some paternal experiences, specifically fear-conditioning in response to an olfactory stimulus, might be transmitted to subsequent generations of mice (Dias and Ressler, 2014) has renewed interest in the epigenetics of brain function and behavior. If such a phenomenon were found to be widespread and its molecular mechanism identified it would cause a paradigm shift in the way we think about epigenetic inheritance, learning and behavior, with ramifications that are currently difficult to fully envision.
Cases of epigenetic regulation of whole-organism phenotypes are common in nature, just not in the conventional model organisms, which were selected for specific characteristics, including homogeneous and stable phenotypes, ease of culture in captivity and fast generation times. As a matter of fact, evidence of major epigenetic effects on visible phenotypes would have probably disqualified organisms from being selected, as evidenced by Linnaeus' horrified reaction when first told about epigenetic variants of the toadflax Linaria vulgaris, which he named Peloria, the Greek word for ‘monster’ (Gustafsson, 1979). The great progress in genomic and genetic tools justifies taking a second look at the variety of life forms that use epigenetic mechanisms to better fit in their environment.
Developmental and phenotypic plasticity
Another way of defining epigenetics is to refer to those processes that allow different phenotypes to arise from a single genotype, known as polyphenisms (West-Eberhard, 2003). Polyphenism is particularly common in insects and brought to an extreme by eusocial insects, as discussed below, but it is also found in other arthropods and, to some extent, in vertebrates. Polyphenisms can originate during development and give rise to distinct, stable phenotypes in the adult individuals (morphs), or it can be plastic, allowing an individual to change its phenotype in a metastable fashion (typical of epigenetic changes) as its environment changes.
Epigenetic metamorphoses of the water flea Daphnia
Daphnia is a genus of freshwater microcrustaceans that has fascinated biologists for more than a century. Its use for epigenetic research has been advocated before (Burggren, 2014; Harris et al., 2012) but a Daphnia genome was sequenced only recently (Colbourne et al., 2011), marking the entry of this organism into the arena of molecular epigenetic research. The key aspects of Daphnia biology that make it valuable to epigeneticists are its ability to reproduce both sexually and asexually and the fact that parthenogenetic individuals, which are genetic clones of the mother, can nonetheless display diverse phenotypes depending on their exposure to certain stimuli during development.
One specific case is the induction of defense structure by exposure to predators (Tollrian and Dodson, 1999). For example, the presence of chemicals released by predators in the water causes the development of a protective cranial extension known as a ‘helmet’ (Fig. 3A). In Daphnia cucullata, these structures are also observed in subsequent generations not directly exposed to the stimulus (Agrawal et al., 1999). However, the fact that the F2 showed smaller effects – which may be an example of the epigenetic ‘washout’ discussed by Burggren in this special issue (Burggren, 2015) – suggests that the modified phenotype might be a consequence of exposure of the F1 germline during embryonic development and not of bona fide transgenerational epigenetic inheritance. Nonetheless, given that similar induced defense mechanisms are also observed in other animals and plants (Agrawal et al., 1999), studying their molecular bases in Daphnia might reveal common paradigms in epigenetic gene regulation.
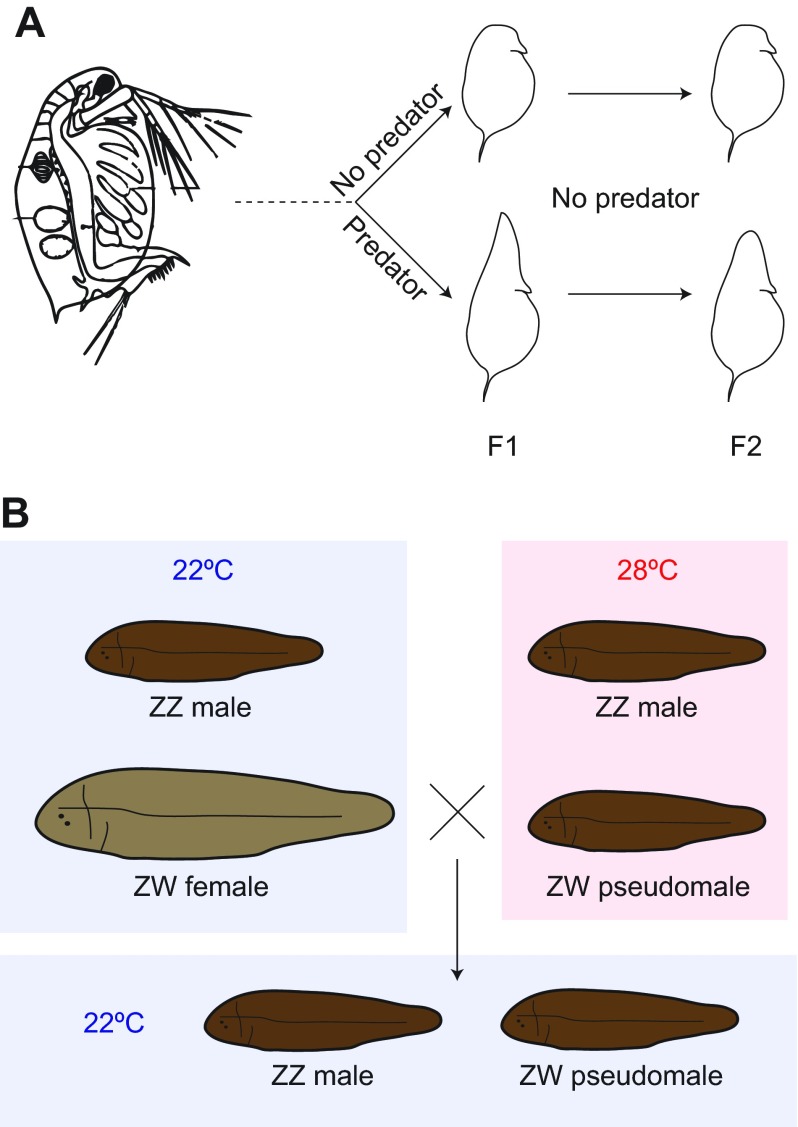
Fig. 3.
Examples of epigenetic plasticity in development. (A) In Daphnia, alternative phenotypes can emerge as a consequence of exposure to predator-derived chemicals (kairomones). In this case, genetically identical F1 (products of parthenogenesis) present differences in the form of a defensive structure called a ‘helmet’. The presence of the helmet can persist in later generations despite the absence of the originating stimulus. (B) In the half-smooth tongue sole Cynoglossus semilaevis, sex is genetically determined by the presence of two Z chromosomes (male) or one Z and one W chromosome (female). However, higher temperatures during the juvenile phase induce conversion of ZW females to ‘pseudomales’. Interestingly, the ZW progeny of these pseudomales will develop into more pseudomales even if the temperature remains low, suggesting an epigenetic transmission of the sex conversion.
In other Daphnia species, exposure to predator-derived chemicals induces the development of a strengthened exoskeleton and small protrusions in the neck region, known as ‘neckteeth’ (Tollrian and Dodson, 1999). Although the transgenerational inheritance of this trait has not been tested, its development can be induced by transient exposure during the first instars and is therefore a case of developmental plasticity controlled by external stimuli. Even if the predator only appears briefly during embryonic development, a signaling cascade results in the activation of an alternative developmental trajectory, which results in a different adult phenotype that is maintained epigenetically for the life of the individual.
A final aspect of Daphnia epigenetics worth mentioning in this context is that many Daphnia species are important test organisms in aquatic toxicology (Martins et al., 2007). The possibility that environmental exposure alters the epigenome and affects not only the individual, but also its descendants is a subject of intensive investigation in vertebrate biology. With its faster generation times and continuous exposure to the aquatic environment, Daphnia might constitute an important model system to address some of these questions. A pioneering study that analyzed changes in DNA methylation upon exposure to polluting agents found no evidence of transgenerational epigenomic damage (Vandegehuchte et al., 2009), although only one locus was considered. Future, broader comparisons of the Daphnia methylomes in clean and polluted waters might reveal new mechanisms by which environmental changes are recorded in the epigenome and affect future generations.
Epigenetic sex reversal in the tongue sole
Among the many types of adult phenotypic plasticity, one of the most egregious is the case of sex reversal, which is surprisingly common throughout the tree of life, affecting several species of insects, fishes, amphibians and reptiles. One particularly interesting example is that of the half-smooth tongue sole Cynoglossus semilaevis, a commercially important flatfish that is extensively cultured in China. The genome of this fish was recently sequenced and revealed that its Z and W sex chromosomes evolved from an ancestral autosome pair starting ~30 million years ago (Chen et al., 2014). Possibly because of the relatively recent origin of the sex chromosomes, this fish species retained an environmental sex-reversal pathway in addition to its genetic sex-determination system based on ZZ and ZW chromosome pairs. Changes in temperature induce juvenile ZW females to develop into fertile ZW ‘pseudomales’. Interestingly, ZW offspring that receive the Z chromosome from the ‘pseudofather’ also develop into pseudomales, and in this case without requiring the temperature change (Fig. 3B). In other words, an environmental stimulus causes changes in the Z chromosome of ZW individuals that are transmitted in an epigenetic manner to the next generation and that result in the development of phenotypic males despite a female karyotype (Chen et al., 2014).
Although the molecular mechanisms responsible for this astounding phenotypic plasticity remain undefined, changes in transcription and DNA methylation at the dmrt1 locus, which has roles in sex determination in chicken and other animals (Matson and Zarkower, 2012; Smith et al., 2009), correlate well with a possible causal role in this environmental sex reversal (Chen et al., 2014). A full survey of the tongue sole's DNA methylome failed to reveal obvious epigenetic culprits for the sex reversal, but only fully developed organisms were analyzed (Shao et al., 2014). A comparison of gene expression and DNA methylation profiles in juveniles before and after the temperature shift that causes sex reversal might provide deeper insight in this process.
In the case of the tongue sole, sex reversal poses yet another epigenetic conundrum, which is how to deal with dosage compensation. In species with different sex chromosomes, genes on the common sex chromosome (Z or X) are present in two copies in the homogametic sex (e.g. XX females in mammals and ZZ males in birds) but only one copy in the heterogametic sex. This difference in genetic dosage is compensated by complex epigenetic processes, such as, for example, X chromosome inactivation in mammals (Augui et al., 2011). The components of the dosage-compensation machinery, as could be expected, are deployed differentially in the two sexes, raising the question of how dosage compensation can be regulated during a phenotypic sex reversal such as the one found in the tongue sole. Initial studies showed that dosage compensation is incomplete between males and females and almost non-existent between males and pseudomales, except for a narrow 2 Mb region, suggesting that these ‘young’ sex chromosomes have yet to evolve a complete compensatory mechanism (Shao et al., 2014). It would be interesting to analyze whether the occurrence of environmental sex reversal is more common in species with ‘young’ sex chromosomes, because the difficulties of reverting the epigenetic regulation of dosage compensation might, in effect, prohibit such processes in other species.
Social interactions and brain epigenetics in cichlid fish
With their wide variety of biology and ethology, fish might also serve as versatile models to study the epigenetics of behavior. One dramatic case of polyphenisms giving rise to alternative social behavior (polyethism) is seen in the African cichlid fish Astatotilapia (Haplochromis) burtoni. Male A. burtoni exist in two, interchangeable, phenotypes: a dominant male that owns territory and is reproductively competent (about 10–20% of the male population) and a non-dominant male that does not reproduce. The differences in reproductive behavior are accompanied by distinct physiology, such as smaller testes in non-dominant males and visible phenotypic traits, such as the bright coloration of dominant males compared with the dull coloration of non-dominants (Fernald, 2012).
Despite the dramatic differences between these two social categories, the distinct phenotypes are not encoded in different DNA sequences within the genome. In fact, when presented with a larger, ‘super-dominant’ fish, formerly dominant males switch to the non-dominant phenotype, with visible changes in coloration appearing as early as a few minutes after exposure. These are accompanied by changes in behavior and physiology, which culminate, about 2 weeks later, in the loss of reproductive competence. This switch is fully reversible: removal of the super-dominant male will cause the converted male to return to its dominant status (Fernald, 2012). At the cellular and molecular level, the transition from non-dominance to dominance is accompanied by the activation of immediate-early genes in the brain (Burmeister et al., 2005), specifically in the so-called ‘social behavior network’ composed of brain nuclei that are involved in social behavior in vertebrates (Maruska et al., 2013b). Interestingly, the same brain network was activated when dominant males were forced to lose their status, but the genes involved were different (Maruska et al., 2013a).
These rapid changes in gene expression must be accompanied by more stable reorganization of the transcriptome so that an individual who just lost (or won) a confrontation does not lose sight of its new place in society. Thus, the metastable switch in social status in A. burtoni is an ideal candidate to understand the role of epigenetic pathways in stabilizing alternative brain states and behavioral repertoires, and with its genome now available (Fernald, 2012), we anticipate that it will provide much needed insight on the role of the social environment in shaping genomic output.
Extreme epigenetics
The examples discussed here are meant to whet the reader's appetite for epigenetic research in non-model organisms. Next, I will present two special cases that stand out as extreme uses of epigenetic regulation of the genome: specifically, the pronounced polyphenisms and polyethisms upon which the societies of ants and bees are constructed, and the outstanding regenerative capacity of planarians.
Polyphenism and polyethism in eusocial insects
As one of the most diverse groups of animals on the planet, insects are also specialists in exhibiting multiple alternative phenotypes within a single species and, therefore, from a single set of genomic information. The types and specific cases of polyphenism are too many to list here and I refer the reader to the several excellent reviews on the topic (Evans and Wheeler, 2001; Simpson et al., 2011). I will point out that among the most impressive – and economically important – examples of non-eusocial insect polyphenism and polyethism are the phase transitions displayed by locusts such as Locusta migratoria and Schistocerca gregaria, as reviewed elsewhere in this issue (Ernst et al., 2015).
The availability of such a variety of polyphenic blueprints in the insect world might explain why the large majority of eusocial animals belong to this group. Indeed, colonies of eusocial insects (ants, termites, some bees) depend heavily on the existence of pronounced polyphenisms among their members (Bonasio, 2014), as they are founded on the principle of division of labor (Hölldobler and Wilson, 1990). These insect colonies are composed of different categories of adult individuals (castes) with different morphologies and physiologies and, most importantly, carrying out different tasks within the colonies, which require distinct sets of behaviors. Typically, most individuals are functionally sterile and occupy themselves with colony maintenance (foraging, defense, construction, cleaning), with reproductive rights restricted to one or few well-protected individuals: queens in the case of haplodiploid hymenoptera and royal couples (kings and queens) in the case of termites (Hölldobler and Wilson, 1990). The differences between queens and workers are not limited to reproductive biology. For example, queens live longer; the longest documented lifespan of a queen ant is 28.5 years, compared with the worker's lifespan of just a few months (Jemielity et al., 2005; Keller, 1998; Keller and Genoud, 1997).
Because of these pronounced differences between related individuals and because, at least in most species, castes are not pre-assigned to embryos via genetic mechanisms (Fig. 4A), the world of social insects has attracted the attention of epigenetics research (Bonasio, 2012). A boost to this interest was provided by the initial observation that the genome of the eusocial honeybee Apis mellifera, unlike that of Drosophila, encoded orthologs for all classes of DNA methyltransferases (Honeybee Genome Sequencing Consortium, 2006), which was followed by similar observations in ants (Bonasio et al., 2010b) and termites (Terrapon et al., 2014). Differences in DNA methylation between castes have been observed in bees and ants and correlated with caste-specific gene expression (Bonasio et al., 2012; Lyko et al., 2010). At least in the case of honeybees, DNMT activity was reported to be causally linked to caste determination in that inhibition of DNMTs resulted in spontaneous development of queens from otherwise worker-destined larvae (Kucharski et al., 2008). The downstream mechanisms for this effect remain in large part mysterious, although the regulation of alternative splicing by differential DNA methylation is a promising candidate (Foret et al., 2012; Li-Byarlay et al., 2013). Interestingly, similar cases of splicing regulation by DNA methylation have been reported in human cells (Shukla et al., 2011), suggesting that these regulatory mechanisms might be more widespread than previously thought.
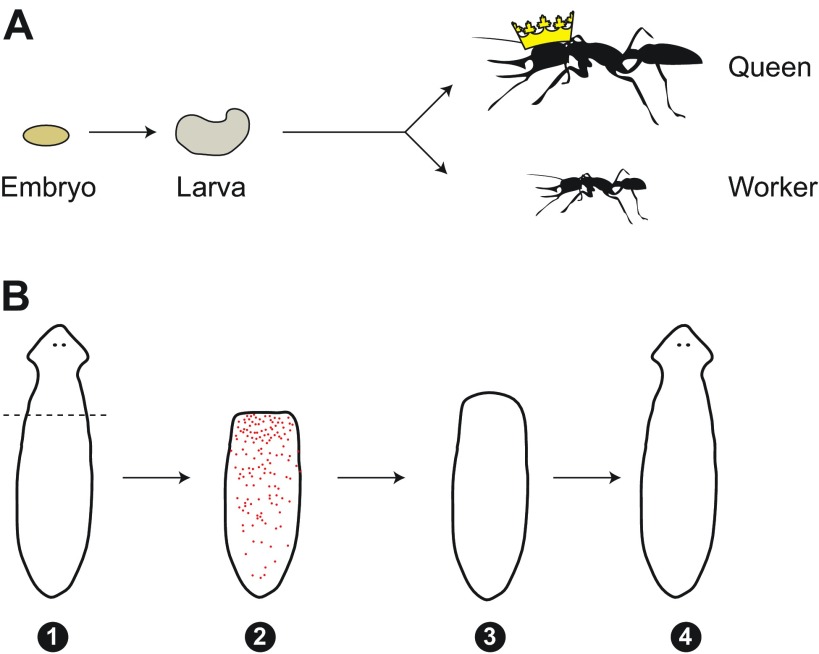
Fig. 4.
Extreme epigenetics. (A) Eusocial insects, such as ants, encode different developmental destinies in the same genome. In most species embryos (left) and larvae (middle) are genetically indistinguishable, but they give rise to entirely different adults (right); specifically, reproductives (queens) and non-reproductives (workers). These differ not only in size and morphology, but also in physiology and behavior. (B) Adult planarians can regenerate all body tissues and structures after amputation (1). Pluripotent adult stem cells known as neoblasts (red dots) migrate to the wound site (2), create a regenerating structure called blastema (3), which eventually restores all organs of the adult animal, including the nervous system (4).
Research on epigenetic pathways other than DNA methylation has been lagging behind, with only two studies published on histone post-translational modifications: one reporting their presence in chromatin from Apis mellifera (Dickman et al., 2013) and one unveiling a potential role for the acetyl-transferase CBP in shaping caste-specific patterns of H3K27 acetylation in different worker castes of the carpenter ant Camponotus floridanus (Simola et al., 2013). Moreover, we still lack a complete annotation of ncRNAs in eusocial insects, which will probably provide additional pieces of this epigenetic puzzle. The availability of several ant and bee genomes, with many more in the pipeline, as well as the first report of successful transgenesis in Apis (Schulte et al., 2014), has opened the doors to molecular research on one of biology's longstanding fascinations: the sophisticated, organized societies of eusocial insects.
Regeneration in flatworms
Another case of extreme epigenetics is the astounding regenerating ability of freshwater planarians (Reddien and Sánchez Alvarado, 2004) (Fig. 4B). These flatworms (phylum Platyhelminthes) have the capacity to regenerate large regions of their bodies at the adult stage, including missing organs and complex structures, starting from minuscule fragments. Planarians have been under the scrutiny of biologists since the original observation of their regenerative capacities in the 18th century and were of particular interest to Thomas Hunt Morgan, who pioneered their systematic study (Morgan, 1898). Although several other metazoans have the ability to regenerate, planarians offer several advantages that have propelled them to the forefront of research on the molecular mechanisms of regeneration (Sánchez Alvarado, 2006).
Planarian regeneration relies on the function of proliferating neoblasts, a population of adult stem cells that populate the wound after dissection and form a structure known as a blastema (Reddien and Sánchez Alvarado, 2004), which is a necessary prerequisite for the regeneration of more complex structures. The ease of gene knockdown by RNA interference (RNAi) in these organisms allowed for forward genetic screens that have identified several genes required for neoblast function and regeneration (Reddien et al., 2005). For example, knockdown of the planarian version of the gene encoding β-catenin results in disrupted anterior–posterior patterning during regeneration and causes dissected animals to grow heads in place of tails at their posterior end (Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2007).
Given that single neoblasts can regenerate all the structure in the adult animal (Wagner et al., 2011), they are pluripotent adult stem cells and, as such, epigenetic pathways must control their ability to regenerate. Indeed, pioneering studies have reported requirements for the chromatin remodeler CHD4 (Scimone et al., 2010), various histone methyltransferase (Hubert et al., 2013) and the H3K9 methylation reader heterochromatin protein 1 (Zeng et al., 2013). Despite the fact that planarians do not seem to contain measurable amounts of methylated cytosines, the methyl-CpG binding protein MBD2/3 is also required for regeneration (Jaber-Hijazi et al., 2013). The latter observation is interesting in light of the fact that there is no consensus on whether planarians methylate their genomes or even on the classes of DNA methyltransferases that they express (Jaber-Hijazi et al., 2013; Regev et al., 1998; Zeng et al., 2013). The orthologs of MBD2/3 and CHD4 are found in the same NuRD complex in vertebrates (Zhang et al., 1999); therefore, it is possible that the effect of MBD2/3 on regeneration is mediated via the NuRD complex and is independent of DNA methylation.
These are only the first steps in dissecting the epigenetic regulation of neoblast pluripotency and differentiation. As it would be expected, histone marks and the machinery that is responsible for their deposition are strongly conserved in planarians (Robb and Sánchez Alvarado, 2014) and characterizing their functions in the context of regeneration might help us understand their role in vertebrate adult stem cells.
Emerging technologies to study non-model organisms
A few years ago the endeavor of establishing a new model organism for molecular research would have been daunting to the point that few researchers would have undertaken it. To seriously consider an organism as a model for molecular biology a few basic requirements must be met. (1) Its genes and regulatory regions should be known. Ideally the entire genome, at least in draft form, should be available and well annotated, so that coding and noncoding genes can be cloned, characterized and their activity quantified. (2) There must be a way to interfere with gene function, ideally by gene targeting or transgenesis or at least by means of transient manipulations. (3) It must be possible to raise the organism in the laboratory, perform artificial crosses and propagate genetically modified lines indefinitely.
Until recently, only a few species met these three requirements and only through the tireless labor of countless scientists over decades. Times have changed. Massive parallel sequencing (also known as next-generation sequencing) has brought about a revolution in the ease and speed of nucleic acid sequencing that could not have been imagined just a decade ago. It cost $3 billion and years of dedicated work by hundreds of researchers to generate a rough draft of the human genome in 2000 (Lander et al., 2001; Venter et al., 2001). As of 2014, the newest instruments can sequence a human genome (to 30 times coverage) in 3 days for about $1000 (Hayden, 2014). Projects are on the way to sequence tens, hundreds, even thousands of genomes from various organism groups, including helmints, birds and insects (i5K Consortium, 2013). In fact, as more and more genomes are published, the rate-limiting steps of de novo genome sequencing, assembly and annotation, become increasingly fast, because they can lean on the annotated genomes of species that are closer in evolutionary (and sequence) distance.
Great strides in RNA research have provided solutions to the second issue, interfering with gene function. Since the late 1990s we have known that small RNA molecules (endogenous or artificial) have the ability to interfere with gene function by degrading or inhibiting the translation of mRNAs with stretches of complementary sequence (Hannon, 2002). This phenomenon, known as RNA interference (RNAi), has become an invaluable tool to manipulate gene levels in cell culture and intact organisms. However, in most organisms RNAi is only transient and in many non-model organisms it requires delivery of double-stranded or small-interfering RNAs by individual injection, which is clearly not desirable for routine experimentation.
To overcome the limitations of RNAi, a true experimental organism should be amenable to genetic manipulation. This has been the real obstacle in establishing new model systems, because in most cases mutagenesis and forward genetics are made impossible by long generation times or difficulty in handling the necessarily large number of individuals involved and reverse genetics, namely gene targeting, was only available in a handful of model organisms or cell types in which homologous recombination happens at an unusually frequent rate (e.g. yeast and mouse embryonic stem cells). These considerations have been swept away by the advent of CRISPR (clustered regularly interspaced short palindromic repeat)-mediated genome editing (Cong et al., 2013). The CRISPR/Cas system is a molecular defense pathway that evolved to protect prokaryotes from foreign genetic material. Small guide RNAs recruit nucleases to complementary RNA or DNA sequences, which are then cut and degraded (Wiedenheft et al., 2012). The utility of such a pathway for molecular biology was grasped by multiple groups and a particular flavor of CRISPR, which uses a nuclease that produces double-stranded breaks in genomic DNA, was adapted to introduce these cuts anywhere in a genome of interest (Cong et al., 2013; Jinek et al., 2012). Because double-stranded breaks are universally resolved either by non-homologous end-joining (which causes small deletions and loss-of-function mutations) or, in the presence of a suitable donor, by homologous recombination, the CRISPR/Cas system allows for the high-efficiency introduction of any type of mutation in a genome of choice.
Although CRISPR-mediated genome editing still requires microinjection of guide RNAs and nucleases into embryos at a suitable stage, it dramatically lowers the barrier to the adoption of new organisms as genetic models and offers the power of genetic approaches in virtually any organism that can be reared in the laboratory.
Closing remarks
In this review, I describe a few examples of non-model organisms on which molecular investigation has already begun. They by no means constitute a complete list and are only meant to illustrate the incredible variety of biological phenomena that could be under epigenetic control and lie outside the realm of conventional model organisms. It is beyond doubt that epigenetic forces shape phenotypes at the cellular and organismal level, before, during and after development. As an epigenetics researcher, I believe in the important contributions of epigenetics to the diversity and sophistication of life on Earth. Nonetheless, I offer a word of caution toward attempts at extending its realm of action to evolutionary times.
At times, the existence of phenotypic plasticity and transgenerational inheritance have been interpreted as supporting evolutionary processes that do not rely on random variation and natural selection (Danchin et al., 2011; Jablonka and Lamb, 2008). I do not believe this is an accurate representation of our understanding of molecular epigenetics (Dickins and Rahman, 2012) and it is dangerously prone to creationist distortions (see, for example, http://creation.com/epigenetics-and-darwin). The examples of polyphenisms and epigenetic plasticity described above are compatible with the notion that multiple phenotypes can be encoded in the same genome, with the appropriate phenotypes activated in response to a stimulus and stabilized by epigenetic mechanisms. Some phenotypes can indeed be maintained through sexual reproduction, resulting in their epigenetic inheritance, on occasion over several generations. In some cases transgenerational epigenetic effects might even confer inheritance of acquired traits via a communication route between the soma and the germline – Darwin's gemmulae (Darwin, 1868) – for which ncRNAs are prime candidates (Heard and Martienssen, 2014; Rechavi et al., 2014; Rechavi et al., 2011). However, a characteristic of epigenetic states is that they are meta-stable (Bonasio et al., 2010a). It is difficult to envision molecular mechanisms by which epigenetic signals could cause an acquired phenotype to be fixed over evolutionary times for thousands of generations. In the absence of such molecular evidence and in deference to Ockham's razor, I continue to support the modern synthesis, which has been greatly successful in explaining inheritance and evolution to date.
In conclusion, many non-model organisms appear to have adapted epigenetic pathways in unique ways to develop alternative phenotypes and behaviors that make them better suited for their environment. The availability of cheap sequencing technologies and efficient genome editing tools will allow us to probe for the first time these phenomena at the molecular level.
FOOTNOTES
Competing interests
The author declares no competing financial interests.
Funding
This work was supported by the National Institutes of Health with grant DP2MH107055 to R.B. Deposited in PMC for release after 12 months.
References
- i5K Consortium (2013). The i5K Initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J. Hered. 104, 595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A. A., Laforsch C., Tollrian R. (1999). Transgenerational induction of defences in animals and plants. Nature 401, 60-63. [Google Scholar]
- Allfrey V. G., Faulkner R., Mirsky A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51, 786-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C. D., Jenuwein T., Reinberg D. (2007). Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Amabile G., Meissner A. (2009). Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol. Med. 15, 59-68. [DOI] [PubMed] [Google Scholar]
- Augui S., Nora E. P., Heard E. (2011). Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 12, 429-442. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Kouzarides T., Shiekhattar R., Shilatifard A. (2009). An operational definition of epigenetics. Genes Dev. 23, 781-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6-21. [DOI] [PubMed] [Google Scholar]
- Bird A. (2007). Perceptions of epigenetics. Nature 447, 396-398. [DOI] [PubMed] [Google Scholar]
- Bonasio R. (2012). Emerging topics in epigenetics: ants, brains, and noncoding RNAs. Ann. N. Y. Acad. Sci. 1260, 14-23. [DOI] [PubMed] [Google Scholar]
- Bonasio R. (2014). The role of chromatin and epigenetics in the polyphenisms of ant castes. Brief. Funct. Genomics 13, 235-245. [DOI] [PubMed] [Google Scholar]
- Bonasio R., Shiekhattar R. (2014). Regulation of transcription by long noncoding RNAs. Annu. Rev. Genet. 48, 433-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R., Tu S., Reinberg D. (2010a). Molecular signals of epigenetic states. Science 330, 612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R., Zhang G., Ye C., Mutti N. S., Fang X., Qin N., Donahue G., Yang P., Li Q., Li C., et al. (2010b). Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329, 1068-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R., Li Q., Lian J., Mutti N. S., Jin L., Zhao H., Zhang P., Wen P., Xiang H., Ding Y., et al. (2012). Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 22, 1755-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R. A. (1958). Paramutation at the R locus in maize. Cold Spring Harb. Symp. Quant. Biol. 23, 379-391. [DOI] [PubMed] [Google Scholar]
- Burggren W. W. (2014). Epigenetics as a source of variation in comparative animal physiology – or – Lamarck is lookin’ pretty good these days. [DOI] [PubMed]
- Burggren W. W. (2015). Dynamics of epigenetic phenomena: intergenerational and intragenerational phenotype ‘washout’. J. Exp. Biol. 218, 80-87. [DOI] [PubMed] [Google Scholar]
- Burmeister S. S., Jarvis E. D., Fernald R. D. (2005). Rapid behavioral and genomic responses to social opportunity. PLoS Biol. 3, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S. E., Martienssen R. A. (2013). RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14, 100-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler V. L. (2007). Paramutation: from maize to mice. Cell 128, 641-645. [DOI] [PubMed] [Google Scholar]
- Chen S., Zhang G., Shao C., Huang Q., Liu G., Zhang P., Song W., An N., Chalopin D., Volff J. N., et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253-260. [DOI] [PubMed] [Google Scholar]
- Colbourne J. K., Pfrender M. E., Gilbert D., Thomas W. K., Tucker A., Oakley T. H., Tokishita S., Aerts A., Arnold G. J., Basu M. K., et al. (2011). The ecoresponsive genome of Daphnia pulex. Science 331, 555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C. (2001). Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292-301. [DOI] [PubMed] [Google Scholar]
- Danchin É., Charmantier A., Champagne F. A., Mesoudi A., Pujol B., Blanchet S. (2011). Beyond DNA: integrating inclusive inheritance into an extended theory of evolution. Nat. Rev. Genet. 12, 475-486. [DOI] [PubMed] [Google Scholar]
- Darwin C. (1868). The Variation of Animals and Plants Under Domestication. London: John Murray. [Google Scholar]
- Dawson M. A., Kouzarides T. (2012). Cancer epigenetics: from mechanism to therapy. Cell 150, 12-27. [DOI] [PubMed] [Google Scholar]
- Day J. J., Childs D., Guzman-Karlsson M. C., Kibe M., Moulden J., Song E., Tahir A., Sweatt J. D. (2013). DNA methylation regulates associative reward learning. Nat. Neurosci. 16, 1445-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J., Rippe K., Dekker M., Kleckner N. (2002). Capturing chromosome conformation. Science 295, 1306-1311. [DOI] [PubMed] [Google Scholar]
- Dias B. G., Ressler K. J. (2014). Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins T. E., Rahman Q. (2012). The extended evolutionary synthesis and the role of soft inheritance in evolution. Proc. Biol. Sci. 279, 2913-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman M. J., Kucharski R., Maleszka R., Hurd P. J. (2013). Extensive histone post-translational modification in honey bees. Insect Biochem. Mol. Biol. 43, 125-137. [DOI] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst U. R., Van Hiel M. B., Depuydt G., Boerjan B., De Loof A., Schoofs L. (2015). Epigenetics and locust life phase transitions. J. Exp. Biol. 218, 88-99. [DOI] [PubMed] [Google Scholar]
- Evans J. D., Wheeler D. E. (2001). Gene expression and the evolution of insect polyphenisms. BioEssays 23, 62-68. [DOI] [PubMed] [Google Scholar]
- Fernald R. D. (2012). Social control of the brain. Annu. Rev. Neurosci. 35, 133-151. [DOI] [PubMed] [Google Scholar]
- Foret S., Kucharski R., Pellegrini M., Feng S., Jacobsen S. E., Robinson G. E., Maleszka R. (2012). DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl. Acad. Sci. USA 109, 4968-4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P., Bickmore W. (2007). Nuclear organization of the genome and the potential for gene regulation. Nature 447, 413-417. [DOI] [PubMed] [Google Scholar]
- Goll M. G., Bestor T. H. (2005). Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481-514. [DOI] [PubMed] [Google Scholar]
- Grewal S. I., Klar A. J. (1996). Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86, 95-101. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Gasser S. M. (2013). Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb. Perspect. Biol. 5, a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2013). Calorie restriction and sirtuins revisited. Genes Dev. 27, 2072-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U., Ma D. K., Mo H., Ball M. P., Jang M. H., Bonaguidi M. A., Balazer J. A., Eaves H. L., Xie B., Ford E., et al. (2011). Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S., Kim S. Y., Artis S., Molfese D. L., Schumacher A., Sweatt J. D., Paylor R. E., Lubin F. D. (2010). Histone methylation regulates memory formation. J. Neurosci. 30, 3589-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K., Rink J., Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science. 319, 323-327.PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson A. (1979). Linnaeus' Peloria: the history of a monster. Theor. Appl. Genet. 54, 241-248. [DOI] [PubMed] [Google Scholar]
- Hannon G. J. (2002). RNA interference. Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- Harris K. D., Bartlett N. J., Lloyd V. K. (2012). Daphnia as an emerging epigenetic model organism. Genet. Res. Int. 2012, 147892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden E. (2014). Is the $1,000 genome for real. NATNEWS doi:10.1038/nature.2014.14530. [Google Scholar]
- Heard E., Martienssen R. A. (2014). Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölldobler B., Wilson E. O. (1990). The Ants. Cambridge, MA: Harvard University Press. [Google Scholar]
- Holliday R. (1987). The inheritance of epigenetic defects. Science 238, 163-170. [DOI] [PubMed] [Google Scholar]
- Holliday R., Pugh J. E. (1975). DNA modification mechanisms and gene activity during development. Science 187, 226-232. [PubMed] [Google Scholar]
- Honeybee Genome Sequencing Consortium (2006). Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R. H., Pirinen E., Auwerx J. (2012). Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 13, 225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert A., Henderson J. M., Ross K. G., Cowles M. W., Torres J., Zayas R. M. (2013). Epigenetic regulation of planarian stem cells by the SET1/MLL family of histone methyltransferases. Epigenetics 8, 79-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J. L., Saló E., Adell T. (2008). Silencing of Smed-βcatenin1 generates radial-like hypercephalized planarians. Development 135, 1215-1221. [DOI] [PubMed] [Google Scholar]
- Jaber-Hijazi F., Lo P. J., Mihaylova Y., Foster J. M., Benner J. S., Tejada Romero B., Chen C., Malla S., Solana J., Ruzov A., et al. (2013). Planarian MBD2/3 is required for adult stem cell pluripotency independently of DNA methylation. Dev. Biol. 384, 141-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E., Lamb M. J. (2008). Soft inheritance: challenging the modern synthesis. Genet. Mol. Biol. 31, 389-395. [Google Scholar]
- Jakovcevski M., Akbarian S. (2012). Epigenetic mechanisms in neurological disease. Nat. Med. 18, 1194-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemielity S., Chapuisat M., Parker J. D., Keller L. (2005). Long live the queen: studying aging in social insects. Age (Dordr.) 27, 241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., et al. (2010). Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller L. (1998). Queen lifespan and colony characteristics in ants and termites. Insectes Soc. 45, 235-246. [Google Scholar]
- Keller L., Genoud M. (1997). Extraordinary lifespans in ants: a test of evolutionary theories of ageing. Nature 389, 958-960. [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693-705. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., Maleszka R. (2008). Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827-1830. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- Lee J. T., Bartolomei M. S. (2013). X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308-1323. [DOI] [PubMed] [Google Scholar]
- Li-Byarlay H., Li Y., Stroud H., Feng S., Newman T. C., Kaneda M., Hou K. K., Worley K. C., Elsik C. G., Wickline S. A., et al. (2013). RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. USA 110, 12750-12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251-260. [DOI] [PubMed] [Google Scholar]
- Luikenhuis S., Giacometti E., Beard C. F., Jaenisch R. (2004). Expression of MeCP2 in postmitotic neurons rescues Rett syndrome in mice. Proc. Natl. Acad. Sci. USA 101, 6033-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F., Foret S., Kucharski R., Wolf S., Falckenhayn C., Maleszka R. (2010). The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Reinberg D. (2010). Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., Trojer P., Reinberg D. (2005). The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15, 163-176. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. (2001). Protein modules that manipulate histone tails for chromatin regulation. Nat. Rev. Mol. Cell Biol. 2, 422-432. [DOI] [PubMed] [Google Scholar]
- Martins J., Oliva Teles L., Vasconcelos V. (2007). Assays with Daphnia magna and Danio rerio as alert systems in aquatic toxicology. Environ. Int. 33, 414-425. [DOI] [PubMed] [Google Scholar]
- Maruska K. P., Becker L., Neboori A., Fernald R. D. (2013a). Social descent with territory loss causes rapid behavioral, endocrine and transcriptional changes in the brain. J. Exp. Biol. 216, 3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruska K. P., Zhang A., Neboori A., Fernald R. D. (2013b). Social opportunity causes rapid transcriptional changes in the social behaviour network of the brain in an African cichlid fish. J. Neuroendocrinol. 25, 145-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson C. K., Zarkower D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersfelder E. L., Parthun M. R. (2006). The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 34, 2653-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. (2007). Beyond the sequence: cellular organization of genome function. Cell 128, 787-800. [DOI] [PubMed] [Google Scholar]
- Morgan T. H. (1898). Experimental studies of the regeneration of Planaria maculata. Dev. Genes Evol. 7, 364-397. [Google Scholar]
- Muller H. (1941). Induced mutations in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 9, 151-167. [Google Scholar]
- Nanney D. L. (1958). Epigenetic control systems. Proc. Natl. Acad. Sci. USA 44, 712-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Reddien P. (2007). Smed-β-catenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327-330. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins J. E., Sauria M. E., Sanyal A., Gerasimova T. I., Lajoie B. R., Bell J. S., Ong C. T., Hookway T. A., Guo C., Sun Y., et al. (2013). Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. (2007). On the use of the word ‘epigenetic’. Curr. Biol. 17, R233-R236. [DOI] [PubMed] [Google Scholar]
- Rando T. A., Chang H. Y. (2012). Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell 148, 46-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O., Minevich G., Hobert O. (2011). Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell 147, 1248-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi O., Houri-Ze'evi L., Anava S., Goh W. S., Kerk S. Y., Hannon G. J., Hobert O. (2014). Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158, 277-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757. [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Lamb M. J., Jablonka E. (1998). The role of DNA methylation in invertebrates: developmental regulation or genome defense? Mol. Biol. Evol. 15, 880-891. [Google Scholar]
- Riggs A., Martienssen R., Russo V. (1996). Introduction. In Epigenetic Mechanisms of Gene Regulation, Vol. 32 (ed. Russo V., Martienssen R., Riggs A.), pp. 1-4 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Ringrose L., Paro R. (2004). Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38, 413-443. [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Chang H. Y. (2012). Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb S. M., Sánchez Alvarado A. (2014). Histone modifications and regeneration in the planarian Schmidtea mediterranea. Curr. Top. Dev. Biol. 108, 71-93. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A. J., Allis C. D., Wysocka J. (2007a). Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 25, 15-30. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007b). Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A. (2006). Planarian regeneration: its end is its beginning. Cell 124, 241-245. [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Dorn R., Reuter G. (2003). Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 14, 67-75. [DOI] [PubMed] [Google Scholar]
- Schulte C., Theilenberg E., Müller-Borg M., Gempe T., Beye M. (2014). Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. USA 111, 9003-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Meisel J., Reddien P. W. (2010). The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development 137, 1231-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C., Li Q., Chen S., Zhang P., Lian J., Hu Q., Sun B., Jin L., Liu S., Wang Z., et al. (2014). Epigenetic modification and inheritance in sexual reversal of fish. Genome Res. 24, 604-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. (2011). CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola D. F., Ye C., Mutti N. S., Dolezal K., Bonasio R., Liebig J., Reinberg D., Berger S. L. (2013). A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 23, 486-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson S. J., Sword G. A., Lo N. (2011). Polyphenism in insects. Curr. Biol. 21, R738-R749. [DOI] [PubMed] [Google Scholar]
- Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., Doran T. J., Sinclair A. H. (2009). The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461, 267-271. [DOI] [PubMed] [Google Scholar]
- Song F., Chen P., Sun D., Wang M., Dong L., Liang D., Xu R. M., Zhu P., Li G. (2014). Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376-380. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861-872. [DOI] [PubMed] [Google Scholar]
- Terrapon N., Li C., Robertson H. M., Ji L., Meng X., Booth W., Chen Z., Childers C. P., Glastad K. M., Gokhale K., et al. (2014). Molecular traces of alternative social organization in a termite genome. Nat. Commun. 5, 3636. [DOI] [PubMed] [Google Scholar]
- Tollrian R., Dodson S. I. (1999). Inducible defenses in Cladocera. In The Ecology and Evolution of Inducible Defenses (ed. Tollrian R., Harvell C. D.), pp. 177-202 Princeton, NJ: Princeton University Press. [Google Scholar]
- Vandegehuchte M. B., Kyndt T., Vanholme B., Haegeman A., Gheysen G., Janssen C. R. (2009). Occurrence of DNA methylation in Daphnia magna and influence of multigeneration Cd exposure. Environ. Int. 35, 700-706. [DOI] [PubMed] [Google Scholar]
- Venter J. C., Adams M. D., Myers E. W., Li P. W., Mural R. J., Sutton G. G., Smith H. O., Yandell M., Evans C. A., Holt R. A., et al. (2001). The sequence of the human genome. Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- Waddington C. (1942). The epigenotype. Endeavor 1, 18-20. [Google Scholar]
- Wagner D. E., Wang I. E., Reddien P. W. (2011). Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332, 811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Duclot F., Liu Y., Wang Z., Kabbaj M. (2013). Histone deacetylase inhibitors facilitate partner preference formation in female prairie voles. Nat. Neurosci. 16, 919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard M. J. (2003). Developmental Plasticity and Evolution. Oxford: Oxford University Press. [Google Scholar]
- Wiedenheft B., Sternberg S. H., Doudna J. A. (2012). RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331-338. [DOI] [PubMed] [Google Scholar]
- Zeng A., Li Y. Q., Wang C., Han X. S., Li G., Wang J. Y., Li D. S., Qin Y. W., Shi Y., Brewer G., et al. (2013). Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J. Cell Biol. 201, 409-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ng H. H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. (1999). Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13, 1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]