Abstract
In the last two decades, genetic and genomic studies have revealed the astonishing diversity and ubiquity of microorganisms. Emergence and expansion of the human microbiome project has reshaped our thinking about how microbes control host health—not only as pathogens, but also as symbionts. In coral reef environments, scientists have begun to examine the role that microorganisms play in coral life history. Herein, we review the current literature on coral-microbe interactions within the context of their role in evolution, development, and ecology. We ask the following questions, first posed by McFall-Ngai et al. (2013) in their review of animal evolution, with specific attention to how coral-microbial interactions may be affected under future environmental conditions: (1) How do corals and their microbiome affect each other's genomes? (2) How does coral development depend on microbial partners? (3) How is homeostasis maintained between corals and their microbial symbionts? (4) How can ecological approaches deepen our understanding of the multiple levels of coral-microbial interactions? Elucidating the role that microorganisms play in the structure and function of the holobiont is essential for understanding how corals maintain homeostasis and acclimate to changing environmental conditions.
Keywords: coral, holobiont, metamorphosis, biological, symbiosis, pollution and global change, ecosystem, bacterial interactions
Introduction
Multicellular hosts harbor communities of beneficial microbes (Bosch and McFall-Ngai, 2011). Compelling evidence illustrates how microorganisms have facilitated the origin and evolution of animals and are integral parts of all animal life (McFall-Ngai et al., 2013). The coral holobiont is comprised of the coral animal and its associated microorganisms consisting of bacteria, archaea, fungi, viruses, and protists including the dinoflagellate algae Symbiodinium (Rohwer et al., 2002). The coral holobiont is a dynamic system, whose members fluctuate depending on environmental conditions and daily requirements (Shashar et al., 1993; Tanner, 1996; Ainsworth et al., 2011). In their review of animal evolution, McFall-Ngai et al. (2013) proposed that our understanding of microbes' role in the evolution of animal partners could be increased by examining mutual interactions and reciprocal influences during development and genomic evolution. In this review, we follow their framework to evaluate the role coral-microbe associations have played in (1) genome evolution in both host and microbial partners, (2) shaping and driving coral development, (3) modulating holobiont homeostasis, and (4) defining the ecology of interactions in the coral holobiont. We aim to review the current literature on coral-microbe interactions in order to create a consolidated resource of knowledge in an emerging and quickly developing field. As the symbiosis between corals and Symbiodinium represents a long-standing field of investigation (Trench, 1983, 1993; Weber and Medina, 2012) we will focus in this review on our emerging understanding of other microbial components of the holobiont that we collective refer to as the microbiota.
How have corals and microorganisms affected each other's genomes?
Coral microbiome studies have revealed vast bacterial diversity co-existing with different host species (Rohwer et al., 2001; Dinsdale et al., 2008; Sunagawa et al., 2010; Morrow et al., 2012). Microbial associations over evolutionary time scales are likely to contribute to genome differentiation in both the host and its associated microbial partners, a process coined as hologenome evolution (Rosenberg et al., 2007). In light of evidence from multiple systems, we will discuss the different scenarios in which host-microbial interactions shape each other's genomes. These include co-evolutionary patterns, metabolic complementation, genomic modification through either genome reduction/expansion, or genetic exchange through horizontal gene transfer (HGT).
Bacterial genome evolution in the holobiont
Bacterial genome size reflects evolutionary dependency of microbial species engaged in obligate symbiosis with multicellular hosts (Moran et al., 2008). Obligate symbionts exhibit the smallest genome sizes when compared to facultative endosymbionts or free-living bacteria (McCutcheon and Moran, 2012). Data on genome size reduction come from studies on insect endosymbiosis, but similar trends are likely to be observed in other tightly coupled host-microbe associations (Gerardo, 2013). While much of the evolutionary history between corals and their bacterial symbionts is yet to be elucidated, patterns of transmission are likely playing a role in host and symbiont genome evolution. If a bacterial symbiont is both vertically transmitted and an endosymbiont, its genome will be more likely to be reduced in size relative to other bacterial mutualists that have free-living stages. The coral-associated bacterial genomes that have been sequenced to date are from cultured facultative coral associates with a presumed free-living state and contain genomes that are of a similar size as their non-coral associated relatives (Reshef et al., 2008; Mavromatis et al., 2010; Santos et al., 2011; Bondarev et al., 2013; Neave et al., 2014). For example, the recently described Verrucomicrobia species Coraliomargarita akajimensis (3.7 Mb genome) was isolated from water adjacent to the coral Galaxea fascicularis (Mavromatis et al., 2010) is within the size range estimated for other recently sequenced Verrucomicrobia genomes from aquatic habitats (Martinez-Garcia et al., 2012). Other coral-associated genomes from Proteobacteria exceed 5 Mb which is typical for free-living Proteobacteria. These include an alpha-Proteobacterium Pseudovibrio strain FO-BEG1 (5.9 Mb genome) isolated from an enrichment culture of a black band diseased coral (Bondarev et al., 2013), the gamma-Proteobacteria Endozoicomonas montipora LMG24815 (5.6 Mb genome) isolated from Montipora aequituberculata (Yang et al., 2010) and Vibrio coralliilyticus a coral pathogen with a 5.5 Mb genome (Santos et al., 2011; Kimes et al., 2012).
Thanks to major developments in sequencing technology, we are no longer limited to studying the small fraction of coral-associated microbes that can be cultured. Indeed, if corals harbor co-evolved microbial symbionts these may not be readily cultured due to fastidious nutritional requirements and metabolic complementation. Culture-independent approaches such as sequencing of 16S rRNA genes has revealed high proportions of novel diversity associated with corals (Frias-Lopez et al., 2002; Rohwer et al., 2002; de Castro et al., 2010; Sunagawa et al., 2010; Fernando et al., 2014) and the dynamic genomic diversity of the microbiota has been revealed through metagenomics revealing shifts in the proportions of microbial genes for functions such as nutrient cycling and virulence following environmental perturbations (Yokouchi et al., 2006; Dinsdale et al., 2008; Vega Thurber et al., 2009). Despite these advances, identifying specific microbial activities in the holobiont and linking these to individual microbial populations that may share a co-evolutionary history with corals remains a challenge. Genome sequencing of single cells and expanded novel-cultivation-based approaches for isolation-based studies are potential tools to characterize currently uncultured microbial symbionts. Such efforts will improve the taxonomy of coral-associated microbes and will enable us to test hypothesis of coevolution and co-diversification in the coral holobiont.
Emerging evidence for metabolic complementation
In addition to the well-established mutualistic nutritional benefits between corals and Symbiodinium (Muscatine and Porter, 1977; Muscatine, 1990; Shoguchi et al., 2013), other reciprocal relationships of metabolic-complementation could extend beyond the coral-Symbiodinium spectrum for example, to include metabolic interactions between bacteria and Symbiodinium or bacteria and the coral host. Metabolic complementation, where each partner produces gene products necessary for survival of the other, may be reflected in the genome composition of the partners (Gerardo, 2013). In a recent analysis of a tri-partite insect endosymbiosis (i.e., two bacterial endosymbionts and the host), extreme genome reduction was correlated with shared metabolic pathways across three taxa (Husnik et al., 2013). The time of symbiont acquisition however played a key role in driving gene loss in the different bacterial lineages.
The mainly oligotrophic habitat of corals, makes associations driven by metabolic complementation desirable, for instance association between diazotrophic bacteria and M. cavernosa has been shown to improve nitrogen fixation (Lesser et al., 2004). Corals are open systems, in which both obligate endosymbionts and other microbes may be closely interacting with the animal host in a temporally dynamic fashion, increasing the chance for such metabolic cooperation to develop. Indeed, the recently sequenced genome of the coral Acropora digitifera lacked a key enzyme that synthesizes the essential amino acid cysteine from homocysteine or serine (Shinzato et al., 2011; Dunlap et al., 2013). Cysteine thione β synthase (CBS) is missing also from two other Acropora species but it has been reported in multiple corals (Shinzato et al., 2011), suggesting metabolic complementation by microbial symbionts within Acropora (e.g., algal or other).
Evidence that metabolic interaction between holobiont members shaped genome evolution may take the form of genetic systems acquired by the host genome by HGT. Predicted genes for bacterial metabolic pathways i.e., the shikimic acid pathway for biosynthesis of aromatic amino acids, and the glyoxylate pathway for biosynthesis of carbohydrates, are present in the genome of the sea anemone Nematostella due to proposed HGT (Starcevic et al., 2008; Jackson et al., 2011). Similar evidence for potential HGT of the bacterial shikimic acid pathway into the coral A. digitifera has been presented (Shinzato et al., 2011; Shoguchi et al., 2013) pointing to the possible acquisition of the pathway by a common ancestor of the Anthozoa.
The role of HGT is largely unexplored within coral holobiont but new whole genome data from multiple members associated with a single coral host will reveal how prevalent HGT is in coral reef symbioses. Indeed studies from the human gut microbiome have shown that HGT is rampant among gut microbial associates where conditions may parallel those associated with the coral polyp (e.g., high bacterial densities and selective pressures from host) (Smillie et al., 2011). We will soon see an increase in whole genome data from multiple coral hosts, algal symbionts, as well as microbial genomes from these hosts, which will open our horizons to understand the extent of metabolic complementation and rate of HGT across holobiont members.
How do microorganisms influence coral development?
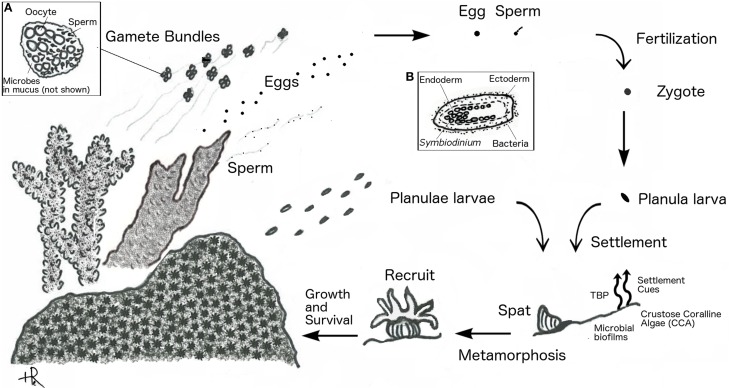
Microbial interactions can play a role at each of the stages in a coral's life cycle and even be crucial in settlement and metamorphosis. Scleractinian corals reproduce asexually through budding and fragmentation, and sexually by gamete spawning or larval brooding. Spawning corals release large quantities of eggs and sperm—either directly or packaged in gamete bundles—into the water column where fertilization and development occurs. Brooding corals, in contrast, release sperm but retain unfertilized eggs inside the gastrovascular cavity of their polyps. The sperm fertilize brooded eggs internally, where they develop before being released as planula larvae. A successful larval life cycle concludes with substrate selection, settlement, and metamorphosis to a polyp (Harrison, 2011). We examine how coral-microbe interactions facilitate the coral life cycle. We define the following transitions: (1) fertilization and spawning – fertilization of gametes and formation of a motile pelagic planula larva (2) settlement and metamorphosis – the selection of appropriate settlement substrate by the free-swimming planula and metamorphosis into polyp (3) coral colony formation – the asexual division of a single polyp into a juvenile colony (4) gamete formation and spawning – sexually mature colonies that produce and release gametes or brood larvae (Figure 1).
Figure 1.
Coral reproductive cycle. Spawning corals (shown as branching) release gamete bundles, eggs, or sperm, and brooding corals (shown as massive) release planulae. Inset (A) shows close up of gamete bundle. Inset (B) shows close up of planula larva.
Gametogenesis and spawning
Gamete formation in corals is not currently known to involve microbial partners, however emerging evidence of vertical and horizontal transmission supports a role for beneficial bacterial populations that associate with coral spawn. The transmission of symbionts in corals has been a subject of careful investigation for several decades and although much more is known about specific modes of transmission for the dinoflagellate algal symbiont Symbiodinium similar mechanisms of transmission may be shared with other microbial symbionts. Algal symbiont transmission is thought to occur in a mainly vertical fashion in brooding corals, with larvae acquiring Symbiodinium from the mother polyp before release (Trench, 1983; Baker, 2003). Vertical transmission has also been observed in several spawning corals that seed their eggs with Symbiodinium (Apprill et al., 2009; Padilla-Gamino et al., 2012). In contrast, externally fertilized larvae generally acquire Symbiodinium from the environment (horizontal transmission) (Trench, 1983; Baker, 2003).
Although less is known about transmission of other non-Symbiodinium microbial symbionts in the holobiont, several studies document both vertical transmission (transfer of symbionts from parent to offspring) and horizontal transmission (uptake of symbionts from the environment) of Bacterial populations. Vertical transmission in Porites astreoides was recently demonstrated. Ectoderm-associated bacteria from the parent colony, mainly Roseobacter spp., were observed in both newly released, and in 4-day old planulae, with densities increasing after settlement (Sharp et al., 2012). In contrast, many coral larvae appear bacteria-free after fertilization and only acquire bacteria as planula or polyps. Apprill et al. (2009) found that 3-day old, but not newly released, planulae of Pocillopora meandrina contained internalized bacterial cells; with Roseobacter spp., as the primary constituents. Horizontal transmission was also documented in the study by Sharp et al. (2010) which followed gametes and planulae of seven broadcast spawners and found no bacterial associations until after the first polyp stage.
Recent studies point to the water column and to the mucus layer surrounding the gametes of spawning corals as media for transfer of symbionts (Symbiodinium and bacterial species) from parent colony to larvae after gamete release (Apprill et al., 2009; Ceh et al., 2012, 2013; Sharp et al., 2012). Both brooding and broadcast spawning corals were observed to release bacteria during spawning, comprised primarily of Roseobacter spp. and Alteromonas spp. (Ceh et al., 2013). The mucus coats of gamete bundles are associated with bacterial populations similar to those found in the parent colony leading to speculation that vertical transmission of these populations may occur either prior to spawning by seeding of the mucus coat with maternally-derived bacterial symbionts, or post-spawning through uptake of microbial associates released by the parent colony into seawater (Apprill et al., 2009; Ceh et al., 2012, 2013; Sharp et al., 2012).
Beneficial activities of the bacterial holobiont members acquired during spawning are unknown and remain an important avenue for ongoing studies. Antimicrobial activity of resident microbes may serve a protective function for coral spawn or newly hatched larvae, although antimicrobial activity associated with extracts from egg bundles themselves was found in only 1 of 11 coral species (Montipora digitata) from the Great Barrier reef (Marquis et al., 2005). As pointed out by Sharp and Ritchie (2012), M. digitata uniquely among the 11 coral species tested, seeds its egg bundles with Symbiodinium, leading the authors to speculate a potentially protective role for Symbiodinium-derived metabolites. Although a low prevalence of solvent-extractable antimicrobial activity was observed in this study, it is possible that other protective compounds such as colonization inhibitors, or antimicrobial peptides (AMP), may contribute to the protection of spawn and larvae against uncontrolled bacterial colonization, as has been suggested by studies in adult corals and in the Cnidarian, Hydra (Jung, 2009; Vidal-Dupiol et al., 2011; Franzenburg et al., 2013; Krediet et al., 2013).
Settlement and metamorphosis
Despite a substantial number of studies investigating the potential role of microorganisms in the induction of larval settlement and metamorphosis, a thorough understanding of all the mechanisms and players involved is still lacking. In corals, metamorphosis and settlement are tightly coupled processes where a coral individual transitions from a motile planula larva to a sedentary polyp. Emerging evidence suggests that bacteria have a fundamental role in moderating metamorphosis and settlement of larvae in the marine environment and suggest that this event may be triggered by diffusible or potentially contact-mediated signals (Dobretsov and Qian, 2004; Hadfield, 2011; Dobretsov et al., 2013; Shikuma et al., 2014).
Enhanced larval settlement and metamorphosis has been observed in response to crustose coralline algal (CCA) surfaces (Morse et al., 1988) that represent a consortium of a red alga and a complex community of bacteria and archaea. Bacterial isolates from CCA surfaces (Pseudoalteromonas spp.) were found to form biofilms that were sufficient to increase metamorphosis rates of Acropora willisae and A. millepora by up to 50% (Negri et al., 2001). Complex bacterial biofilms formed from shallow reef waters were also sufficient to induce high levels of larval settlement and metamorphosis for A. micophthalma and were superior to biofilms formed in deep-water suggesting a connection between spatial variability of marine biofilm community composition and efficiency of larval settlement (Webster et al., 2004). Finally, antibiotic treatment of larval cultures was shown to be sufficient to inhibit settlement, suggesting that bacterial activity is necessary for settlement induction (Vermeij et al., 2009).
Efforts to uncover the molecular mechanism behind induction of coral metamorphosis by various Pseudoalteromonas strains isolated from CCA surfaces led to isolation of the inducing compound tetrabromopyrrole (TBP) (Tebben et al., 2011). TBP was sufficient for inducing metamorphosis of A. millepora at environmentally relevant concentrations. However, larvae induced by TBP, or by Pseudoalteromonas biofilms, metamorphosed without settlement (Tebben et al., 2011). In contrast, A. millepora larvae exposed to complex CCA both metamorphosed and settled, leading the authors to speculate that TBP production by Pseudoalteromonas strains has a net negative effect on coral recruitment and may have antifouling properties (Tebben et al., 2011). In contrast, a recent study of a diverse set of Caribbean corals documented that TBP extracted from Pseudoalteromonas isolates, as well as produced synthetically, stimulated both metamorphosis and settlement (Sneed et al., 2014). Thus, it is clear that TBP is a diffusible signal produced by coral reef Pseudoalteromonas strains that stimulates coral larval metamorphosis, however TBP may have divergent impacts on net larval recruitment depending on the timing of larval settlement.
Coordination of metamorphosis and settlement may involve detection of multiple types of signals produced by biofilms and CCA including both diffusible signals such as TBP and contact mediated signals located on biofilm surfaces. A contact-mediated mechanism for induction of larval settlement of Hydroides elegans, a polychaete worm, was recently established for a Pseudoalteromonas strain (P. luteoviolacea)—where settlement is triggered by larval contact with bacterial cell surface displays of phage-tail like proteins (Huang et al., 2012; Shikuma et al., 2014). Certain populations of bacteria may also deter coral settlement through diffusible signals, for example larvae were observed to avoid settling on tiles adjacent to benthic cyanobacteria hypothesized to produce toxic secondary metabolites (Kuffner et al., 2006). It is likely that the combination of CCA and biofilms comprised of specific bacterial population(s) produce multiple signals that together provide adequate settlement cues for coordinated metamorphosis and settlement in coral larvae. Such coordinated cues may help larvae from diverse invertebrate taxa identify sites optimal for recruitment, and may represent long standing relationships between larvae and bacterial partners in biofilms on marine surfaces (Hadfield, 2011).
Coral colony formation
Once settlement and metamorphosis is achieved, the coral recruit (i.e., juvenile polyp) must overcome competition with other benthic organisms to grow and form a robust adult colony. Bacteria may mediate the survival of coral recruits in the proximity of adult coral colonies or other coral competitors such as macro- and turf algae (Smith et al., 2006; Marhaver et al., 2013). Enrichment of pathogenic and opportunistic bacteria in the vicinity of adult coral colonies plays a macro-scale role in reef community structure through enhanced mortality of nearby conspecific coral recruits (Marhaver et al., 2013). This effect, first described as the Janzen-Connell hypothesis for diversification of terrestrial forests and may also contribute to the high diversity of invertebrate species observed in coral reefs (Marhaver et al., 2013).
Enrichment of bacteria near primary producers such as benthic algae may also negatively impact the survival of coral recruits. Elevated mortality (up to 100%) of coral nubbins grown with algal competitors and appeared to be mediated by factors able to pass through a fine mesh (Smith et al., 2006). These negative effects were mitigated by the addition of the antibacterial compound ampicillin (Smith et al., 2006) suggesting that bacterial activities, rather than algal toxins, mediated the inhibitory effects of algal proximity on coral recruits. A similar result was obtained by Vermeij et al. (2009), who examined the post-settlement survival of Montipora capitata in the presence and absence of macroalgae and ampicillin where the addition of ampicillin increased recruit survivorship from around 30 to 60% (Vermeij et al., 2009). Exudation of organic carbon by primary producers may enrich for heterotrophic bacteria (Discussed further in Section How can Ecological Approaches Deepen our Understanding of the Multiple Levels of Coral–Microbial Interactions?), including pathogens and opportunists that may impact the survival of recruits. Early survivorship of coral recruits can be increased through gregarious settlement in a species-specific manner (Rivera and Goodbody-Gringley, 2014) and whether coral-to-coral allorecognition or competition during settlement is influenced by coral associated microbial populations is a question worthy of investigation.
Sexual maturation and reproduction
Once settled, recruits grow and reach reproductive maturity. Brooding corals tend to breed year round, releasing smaller quantities of well-developed larvae, while spawning corals release gametes in large, often synchronous, spawning events a few times a year. Timing is generally thought to be mediated by the lunar cycle in most species (Babcock et al., 1986; Tanner, 1996). Over the course of spawning events the composition of bacterial assemblages associated with adult colonies remains relatively stable (Apprill and Rappe, 2011; Ceh et al., 2012) with a notable exception being an increased abundance of several microbial populations hypothesized to play a role during reproduction (Ceh et al., 2012, 2013). During spawning Ceh et al. (2012) observed increases in the proportion of α-Proteobacterial 16S rRNA gene sequences (particularly Roseobacter spp.) in the microbiota of three different coral species: Acropora tenuis, Pocillopora damicornis, and Tubastrea faulkneri, and subsequently documented increases of Roseobacter spp. and Alteromonas spp. in filtered seawater concurrent with release of gametes from A. tenuis, or larvae from P. damicornis (Ceh et al., 2013). The authors hypothesized that release of these microbial populations enriched the bacterioplankton with seed populations for colonization of gametes and larvae. Speculation on potential beneficial functions for these reproductive-associated bacteria include antimicrobial protection and roles in induction of larval settlement (Ceh et al., 2013) however the nature of mutualistic interaction, if any, remains unknown.
Coral spawn that does not proceed through development, decays and represents a source of nutrients and carbon to the water column and benthos that may stimulate growth of microbial populations. During a 4-day mass spawning event in Heron Island on the Great Barrier Reef the abundance of bacteria in the water column nearly doubled and increased 3-fold in the sediments (Patten et al., 2008). The impact of increased microbial activity and abundance stimulated by spawn decay on reef water quality is mediated by the physical characteristics of the reef region i.e., dilution capacity and flushing rate. In shallow bays of the Great Barrier Reef blooms of respiring bacteria, and subsequent hypoxia, in response to decaying coral spawn have been associated with mass mortalities of corals and other benthic invertebrates (Simpson et al., 1992). In contrast, in Kaneohe Bay, Hawaii, a bay with comparatively higher flushing rates, slight changes in bacterial production and microbial community composition in the water column were only observed when spawning coincided with a low tide where the dilution-capacity of the bay was at a minimum (Apprill and Rappe, 2011). Release of nutrients and bacterial populations during coral spawning may impact the early colonization of planulae by bacterial associates although this impact is likely to be influenced by physical process such as flow, mixing, and initial nutrient loads.
How is homeostasis maintained between corals and their microbial symbionts?
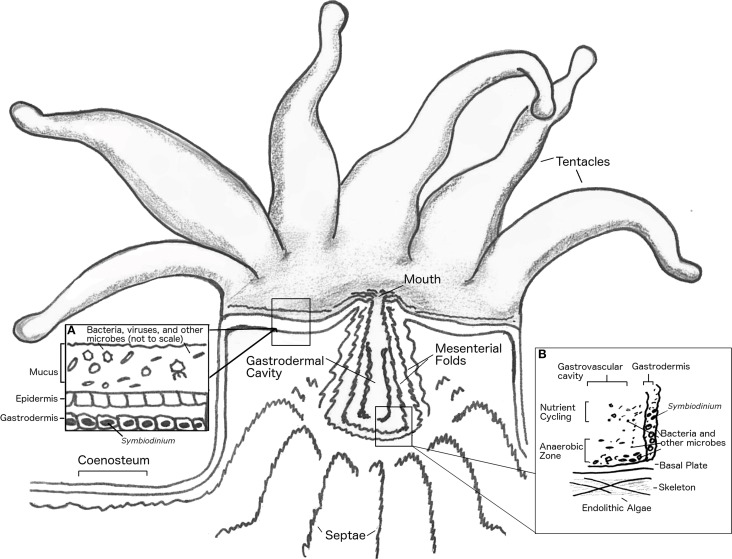
The coral holobiont, from pre-larval to adult stages, is a complex system of diverse organisms that coexist and interact. This complex system is capable of fixing nutrients and building biomass and carbonate structures in nutrient poor waters. When one or more components of the holobiont fail to function as required for system stability, bleaching (Symbiodinium loss) and/or tissue-death may result and lead to progressive death of the entire coral colony. The control and regulation of activities in the holobiont to maintain emergent function is termed homeostasis. In the coral holobiont, like any other animal holobiont, stressors that threaten homeostasis may be biological, chemical, or physical, and an initial stressor may lead to cascading interactions that trigger additional responses. To assess how homeostasis is maintained among components of the coral holobiont this section will focus on how physical and chemical gradients shape the microbiota in different habitats within the coral animal, and consider how perturbation of these gradients influence holobiont function. In particular, we will examine the following habitats: (1) the coral mucus and epidermal surface, (2) intracellular and interstitial spaces, (3) the gastrovascular cavity, and (4) the skeleton (Figure 2).
Figure 2.
Anatomy of coral polyp. Basal body wall not shown. Inset (A), shows close up of mucus layer, epidermis and upper gastrodermis. Inset (B), shows close up of coral gastrovascular cavity.
Coral mucus and epidermal surface
The tissue of the coral organism contains numerous secretory cells that are responsible for production of a continuous mucus layer that is typically a few hundred micrometers thick (Brown and Bythell, 2005; Jatkar et al., 2010; Garren and Azam, 2012). The formation and roles of coral mucus have been reviewed in detail (Brown and Bythell, 2005). Coral mucus serves multiple functions to maintain holobiont homeostasis that may vary in relative importance depending on the coral species. As a permeable barrier between the coral tissue and seawater that can be periodically shed by a mechanism functionally analogous to mucus clearing in other metazoans, the mucus layer is the first line of defense against biofouling, pathogen invasion, and shading by sedimentation (Brown and Bythell, 2005). In addition, the mucus layer may serve as a trap for nutrient-bearing particles (including bacteria) that are periodically ingested by the coral as a mode of suspension feeding (Coles and Strathman, 1973; Muscatine, 1973). Mucus layers are enriched in photosynthate and coral waste products and thus represent a resource-rich habitat that is readily colonized by marine bacteria (Brown and Bythell, 2005). Mucus layers accumulate dense assemblages of bacteria that share similarity with the bacterial composition of seawater (Bourne and Munn, 2005; Lema et al., 2012) and are also structured by interaction with members of the coral holobiont through both top-down process (i.e., removal/selection) (Ritchie, 2006; Franzenburg et al., 2013) and bottom-up processes (i.e., resource availability) (Nakajima et al., 2009) as discussed further in Section How can Ecological Approaches Deepen our Understanding of the Multiple Levels of Coral–Microbial Interactions? Coral mucus layers are complex habitats, defined by chemical, physical and biological gradients (Brown and Bythell, 2005), thus it is not surprising that spatial heterogeneity of mucus-associated bacterial community composition in adult corals can be observed over the scale of a single coral colony (Daniels et al., 2011). Indeed, over a diurnal cycle the mucus of some corals may cycle from being fully-oxygenated due to the photosynthetic metabolism of Symbiodinium to anaerobic at night when respiration rates exceed oxygenesis—this may be enhanced by physical compartmentalization of mucus-bearing structures on the coral colony (Carlton and Richardson, 1995; Kuhl et al., 1995; Brown and Bythell, 2005).
Although the mucus layer is densely colonized by microorganisms, microscopic examination of the coral epidermis beneath the mucus layer reveals a nearly sterile environment (Johnston and Rohwer, 2007). Surface coral cilia have been implicated in deterring the attachment of fouling organisms by generating surface currents on the order of 2 mm per second (Johnston and Rohwer, 2007; Garren et al., 2014). Although coral mucus and ciliary currents are physical deterrents to colonization of coral surfaces, it has been recently demonstrated that the coral pathogen V. coralliilyticus breaches this barrier through rapid bursts of chemotaxis (i.e., directional swimming relative to a gradient) and chemokinesis (i.e., acceleration in response to a chemical concentration) guided by gradients of infochemicals such as DMSP that are produced by the holobiont and may increase during periods of temperature stress (Garren et al., 2014). Exposure to V. coralliilyticus leads to rapid death of coral tissue (Ben-Haim et al., 2003a; Santos et al., 2011). Thus, the dynamic coral mucus layer is a critical, and traversable, barrier that is essential to maintaining a healthy balance of surface-colonizing microorganisms.
Coral cells and interstitial spaces
In corals, the intracellular habitat of endodermal (gastrodermal) cells is dominated by Symbiodinium, which are housed in coral generated membrane vesicles i.e., the “symbiosome” as individuals, diads, or triads (Trench, 1983, 1993). The secondary membrane around the algal endosymbiont protects the alga from coral immune responses and creates a microenvironment within the cytoplasm of low nutrients and high pCO2/low pH conducive to exudation of photosynthate rather than algal biomass synthesis (Trench, 1983, 1993; Venn et al., 2009). To date, studies with detailed ultrastructural imaging of the symbiosome do not report the co-occurrence of bacteria (e.g., Venn et al., 2009; Pernice et al., 2012). Yet obtaining axenic (bacteria-free) cultures of Symbiodinium has proven difficult, suggesting that Symbiodinium associated bacteria may be essential for their sustained growth in culture (Ritchie, 2011). Despite efforts to remove microbial contamination from the culture, analysis of the Symbiodinium genome also revealed the presence of alphaproteobacterial sequences (Shoguchi et al., 2013). This organism matched closely to Parvibaculum lavamentivorans, and to sequences found in seawater samples (Schleheck et al., 2011). Whether Symbiodinium maintains intracellular bacteria is unknown.
The activity of Symbiodinium may shape several intracellular and interstitial physicochemical gradients in the coral holobiont. Reactive oxygen production is associated with the activity of coral phagocytotic immune cells (Mullen et al., 2003) and with oxygenic photosynthesis by Symbiodinium that generates free oxygen radicals in and around coral gastrodermal cells (Kuhl et al., 1995; Banin et al., 2003). Resulting gradients of reactive oxygen within the holobiont may structure microbial populations according to their ability to mitigate oxidative stress through enzymatic activity [e.g., superoxide dismutase (SOD) and catalase]. Virulence of Vibrio shiloi, a pathogen of Symbiodinium in the coral Oculina patagonica required SOD to initiate infection (Banin et al., 2003). While a mutant V. shiloi strain with inactivated SOD could adhere to and penetrate cells of O. patagonica, intracellular survival was impaired, preventing establishment of infection (Banin et al., 2003). These results suggest that high reactive oxygen concentrations proximal to Symbiodinium are a barrier to microbial colonization of the intracellular niche, and may play a role in mediating colonization of microorganisms elsewhere in the holobiont where reactive oxygen is produced.
A second class of physicochemical gradients may be shaped by exudation of organic molecules by intracellular Symbiodinium. In addition to photosynthate, which may represent a source of nutrition for associated bacteria, an increasing diversity Symbiodinium-derived of chemical compounds has been recognized. For instance, Symbiodinium produce microsporine amino acids, which localize to the coral mucus and are hypothesized to protect the holobiont against UV radiation (Banaszak et al., 2000) and to serve as antioxidants (Yakovleva et al., 2004). Symbiodinium also produce DMSP, which is metabolized by a wide variety of microbial taxa (Raina et al., 2009) and as previously discussed, serves as an infochemical for chemotactic pathogens (Garren et al., 2014). How the profile of compounds released by Symbiodinium varies based on clade or sub-clade is not well-known but may influence the composition of the associated microbiota. Coral microbial communities have been shown to differ based on the clade of Symbiodinium present in juveniles of two Acroporid coral species under both stressed and non-stressed conditions (Littman et al., 2009, 2010) supporting the hypothesis that unknown factors associated with different Symbiodinium genotypes influence the coral microbiota composition.
Residence of bacterial populations in intracellular habitats affords close proximity to host- or Symbiodinium-derived ATP and nutrients, and protection from wandering immune cells and predators. In most known cases intracellular occurrence of bacteria in corals has been linked to pathogenesis (e.g., V. shiloi, V. coralliilyticus) (Ben-Haim et al., 1999, 2003a,b; Banin et al., 2003). However, intracellular association of non-pathogenic bacteria has been observed in two Cnidarian classes—the hydrozoans Hydra magnipapillata and Hydra oligactic and the anthozoan coral Montastrea cavernosa (Lesser et al., 2004; Fraune and Bosch, 2007; Chapman et al., 2010) and has been implicated in cultivation-independent surveys of holobiont microbiota composition. The genome sequence of H. magnipapillata revealed a near complete genome for a betaproteobacterial strain (family Comamonadaceae) with some evidence of genome-reduction associated with symbiosis (Chapman et al., 2010). In H. oligactic, stable associations with Rickettsiales were observed in both laboratory and field polyps that were harbored in secondary membranes within epithelial cells suggesting these were endosymbionts (Fraune and Bosch, 2007). In M. cavernosa, a nitrogen-fixing cyanobacterium was found within coral cells (Lesser et al., 2004) and subsequently shown to supplement the nitrogen demand of the host (Lesser et al., 2007). The abundance of Symbiodinium has been positively correlated with the abundance of nitrogen-fixing bacteria (Olson et al., 2009) which may provide a substantial nutritional benefit to all members of the coral holobiont. Association of nitrogen-fixing bacteria with corals have been reported in several other studies (e.g., Chimetto et al., 2008; Lema et al., 2012; Olson and Lesser, 2013). Nitrogen-fixing taxa include various rhizobia closely related to intracellular nitrogen-fixing symbionts of legumes, suggesting rhizobial coral associates may play a similar role (Lema et al., 2012).
Bacterial aggregates associated with coral gastrodermal tissue have been documented in early studies of both reef-building corals and anemones (reviewed in Work and Aeby, 2014). Among 21 coral genera examined, prevalence of cell-associated microbial aggregates (CAMA) was linked to coral genera; notably CAMA were absent in all samples examined from the genus Montipora (Work and Aeby, 2014). Based on histological staining profiles the authors speculated that some of the bacteria resident in CAMA were Rickettsia or Chlamydia—two taxonomic groups containing obligate intracellular species. CAMA are currently not known to be intracellular or extracellular based on current ultrastructural characterization. Widespread association of bacterial aggregates with cells of adult corals, whether intra- or extra-cellular, may point to functional mutualism, and further work to elucidate the species composition and activity associated with these aggregates will shed light on this interesting hypothesis.
Gastrovascular cavity and lumen
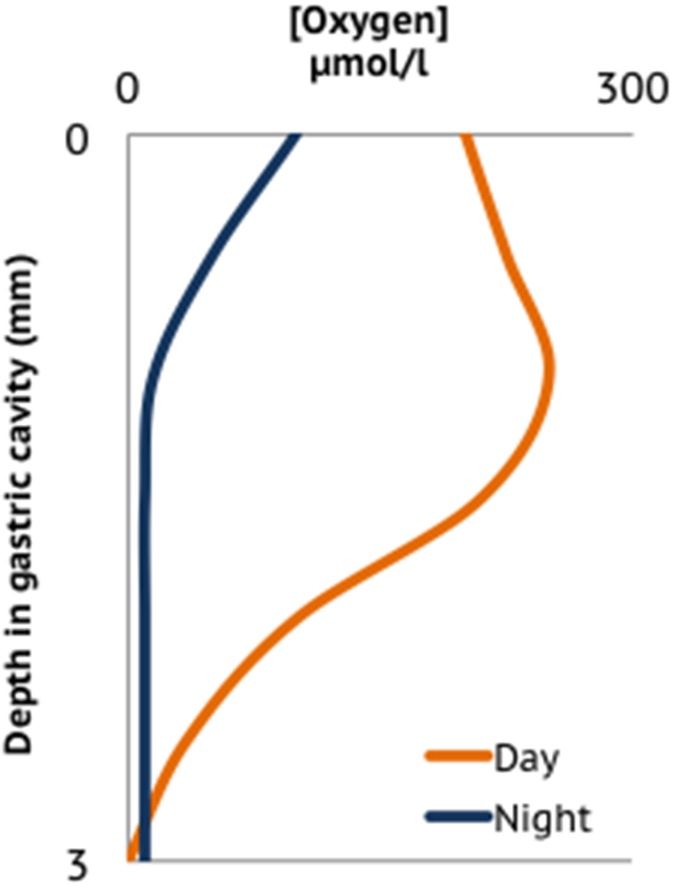
The coral gastric cavity and associated lumen form an enclosed environment with properties distinct from surrounding reef waters and other coral tissues. The gastrovascular cavity is exposed to periodic influxes of both nutrients and a rich assemblage of microbial populations borne in seawater, detritus consumed via suspension feeding, and on coral prey items. Microorganisms in ingested detritus or coral prey may represent transiently associated populations and are hypothesized to contribute to the large diversity of microorganisms observed associated with corals (Fernando et al., 2014). Similar to guts of other organisms, the microorganisms in the coral gastrovascular cavity may provide their host with essential nutrients such as vitamins and amino acids, while participating in food digestion. Using fiber optic microsensors in the only study of its kind to date, Agostini et al. (2012), measured the physical properties at different depths in the gastric cavity of G. fascicularis and observed gradients in dissolved O2, pH, alkalinity, and nutrient concentrations that varied with diel oxygenic photosynthesis of associated Symbiodinium. Elevated levels of vitamin B12, phosphate, and nitrogen species were observed in the gastric cavity, where vitamin B12 was hypothesized to be a nutrient produced by bacteria for coral or Symbiodinium uptake. Dissolved oxygen decreased from the coral mouth to the bottom of the cavity, where consistently low or anoxic conditions prevailed, perhaps creating a permanently anoxic microenvironment (Agostini et al., 2012) (Figure 3) and overall, pH was lower in the cavity compared to overlying seawater. The microbial community in gastric fluids was 100-fold more concentrated than in the surrounding seawater (i.e., >107 cells/ml) and the community composition included ribotypes observed in gut microbiomes of other marine and terrestrial taxa (shrimp, bees, and humans) (Agostini et al., 2012). This data suggests that the coral gastrovascular cavity may be more similar to other animal guts in terms of microbiota structure, and possibly function, than previously expected.
Figure 3.
Oxygen profile in the gastrovascular cavity of a coral polyp adapted from averaged data presented in Agostini et al. (2012).
Coral skeleton
The coral skeleton provides a sheltered environment, offering protection from stressors such as UV light and predation (Shashar et al., 1997). Several studies have examined microbial composition of coral skeletons, finding diverse, and distinct communities of microbial eukaryotes (Shashar et al., 1997), and bacterial communities that contain many of the same bacterial types observed in coral fragments with live tissue (Frias-Lopez et al., 2002; Sweet et al., 2011; Fernando et al., 2014). In a study of bacterial diversity of Mussismilia spp. corals, Fernando et al. (2014) observed that 8.9% of the bacterial 16S rRNA operational taxonomic units (OTUs) from 12 coral individuals were also observed in a bare skeleton sample. Interestingly, skeleton-associated OTUs made up a greater proportion of the bacterial richness for sequence types found in multiple coral samples, up to 77% of the richness of OTUs recovered from at least 11 of the 12 corals (Fernando et al., 2014), suggesting that skeleton-associated microbial populations are a significant source of diversity in the coral holobiont and may contain stable associates. Coral skeletons from three coral species maintained in aquaria revealed bacterial community profiles that did not cluster in a host species-specific manner, in contrast to mucus and tissue samples (Tremblay et al., 2011) raising interesting questions about the extent to which conditions in the coral skeleton are influenced by interactions with the other habitats in the coral holobiont. In 11-day old juvenile polyps of P. astreoides bacterial cells observed by fluorescent in situ hybridization are seen adjacent to tissues surrounding developing coral skeleton (Sharp et al., 2012). The reason for this localization is unclear, though the authors suggest future work should explore whether bacteria may influence deposition of the carbonate skeleton in developing corals. Indeed, the mineral nucleating properties of bacterial cell wall material is well-established and can be both induced in the laboratory, and observed in natural mineral formations (Aloisi et al., 2006).
The activity of the endolithic microbial community is elusive; far fewer studies have focused on skeletal associates than on tissue- and mucus-associated bacteria. Skeleton-associated microbes have been implicated in nitrogen fixation and as a potential source of food and nutrients during bleaching events (Shashar et al., 1994; Le Campion-Alsumard et al., 1995a; Fine and Loya, 2002). Endolithic algae can be observed macroscopically, and blooms are triggered when coral paling or bleaching reduces competition with Symbiodinium for light. Endolithic algae can be recognized as green bands a few millimeters beneath the coral tissue layer (Fine et al., 2006; Carilli et al., 2010). The endolith Ostreobium is distributed among a wide diversity of corals and as a micro-borer can negatively affect the robustness of the coral skeleton (Lukas, 1974; Gutner-Hoch and Fine, 2011). Fungi are also observed in microscopic examination of coral skeletons (Le Campion-Alsumard et al., 1995b). Coral associated fungal assemblages are diverse (Wegley et al., 2007; Amend et al., 2012), can change composition based on environmental conditions (Le Campion-Alsumard et al., 1995b; Bentis et al., 2000; Amend et al., 2012; Yarden, 2014), increase relative to other holobiont members in stressed corals (Vega Thurber et al., 2009; Littman et al., 2011), and in some cases cause coral disease (Bourne et al., 2009). While it is likely that microbial communities in the coral skeleton are distinct from those in tissue-associated habitats due to different physicochemical conditions and localized primary production by endolithic algae, the nature of interactions between endolithic microbiota and the rest of the holobiont is unclear as there are not sufficient studies with both the spatial and taxonomic resolution to distinguish microbial populations associated with live tissue from those associated with healthy coral skeletons.
How can ecological approaches deepen our understanding of the multiple levels of coral-microbial interactions?
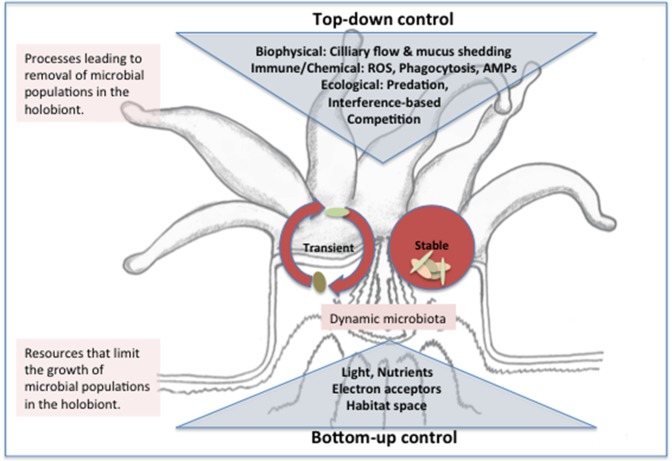
The coral microbiota structure reflects the outcome of structuring in the distinct physicochemical micro-environments within the coral holobiont (Section How is Homeostasis Maintained between Corals and their Microbial Symbionts?) and ecological interactions. In homeostasis, microbial populations are subject to the balance of resource limitation (bottom-up control) and antagonistic interactions that lead to population extinction (top-down control) (Figure 4). Deviations from homeostasis arise when the balance of top-down and bottom-up processes is perturbed, e.g., through alteration of environmental conditions. Cascading responses may in turn lead to growth of pathogens or opportunistic microorganisms, shutting down emergent functions and destroying the scaffold of the coral holobiont with ecosystem-scale consequences.
Figure 4.
Conceptual model of top and bottom down control of the microbiota structure in the coral holobiont. Stable microbes may be introduced to the holobiont through horizontal or vertical transmission and persist in ecological niches within the coral polyp where growth (or immigration) rates balance removal pressures from biophysical processes and immune or ecological interactions. Transient microbes enter the holobiont from environmental sources (e.g., seawater, prey items, or suspension feeding) and removal rates exceed growth/immigration rates such that a dynamic and high diversity microbiota results. Transient and stable populations compete for resources including nutrients, light and space and the outcome of resource-based competition (bottom up control) ultimately determines population growth rate and thus ability to persist when subject to removal. Whether a population is categorized as stable or transient may depend on the timeframe considered. Abbreviations: AMP, antimicrobial peptides; ROS, reactive oxygen species.
Bottom-up control of microbial community structure and activity through resource availability
The microbial communities of several species of corals are hypothesized to harbor stable associates of the coral host, including strains of the nitrogen-fixing cyanobacteria Cyanothece (Lesser et al., 2004, 2007; Olson and Lesser, 2013), alphaproteobacterial strains, such as Roseobacter spp., which are transferred from parent to offspring during reproduction, (Sharp et al., 2010, 2012; Ceh et al., 2013), antimicrobial members persisting in the coral mucus such as Exiguobacterium spp., Photobacterium spp., and Halomonas sp. (Ritchie, 2006; Krediet et al., 2013) and populations such as Endozoicomonas spp. that are found at high proportion across multiple types of corals (Yang et al., 2010; Morrow et al., 2012, 2014; Bayer et al., 2013a,b; Pike et al., 2013; Neave et al., 2014). Resources for maintenance metabolism coupled with favorable physical and chemical conditions are necessary for any organism to persist in a habitat. If the organism is subject to removal (e.g., by predation, or physical dilution) then biomass growth must balance removal rates for persistence. In the coral holobiont, like other ecosystems, competition for resources including light, inorganic nutrients (N, P, and Fe), electron acceptors (O2, SO4), and organic carbon mediates the structure of the populations that persist and coexist. Perturbations that relieve resource limitation have both holobiont and ecosystem scale consequences.
Competition for light and electron acceptors in the holobiont
Light and electron acceptors are critical resources that exert bottom-up control on interactions between coral hosts and bacteria. While the role of light as a resource for Symbiodinium is quite apparent, photosynthetic microbes in the holobiont such as the cyanobacteria and anoxygenic phototrophs also need access to light energy for growth (Lesser et al., 2004; Magnusson et al., 2007). Microbial phototrophs likely compete with Symbiodinium for light—indeed shading by the pigments in the coral polyp can be so severe that epiphytic algae residing in the skeleton beneath the coral tissue of Porites spp. are estimated to receive 1–4% of incident photosynthetically active radiation (PAR) (Shashar and Stambler, 1992) and values as low as 0.1% transmitted PAR have been reported for Favia corals (Shibata and Haxo, 1969).
As discussed previously (Section How is Homeostasis Maintained between Corals and their Microbial Symbionts?), oxygenic photosynthesis renders most of the coral interior oxic during the day, however diurnal patterns of anoxia/hypoxia in the polyp gastrovascular cavity (Agostini et al., 2012), coral surface (mucus layer) (Shashar et al., 1993; Brown and Bythell, 2005), skeleton (Kuhl et al., 2008), and in coral tissues compromised by contact with stagnant water or sediment (Kuhl et al., 1995; Weber et al., 2012), enables anaerobic forms of bacterial respiration and fermentation to occur within the holobiont. Bacterial diversity surveys of the coral holobiont have consistently recovered sequences most similar to anaerobic bacteria (Williams et al., 1987; Carlton and Richardson, 1995; Kuhl et al., 1995; Kellogg, 2004; Kline et al., 2006; Closek et al., 2014). While many aerobic heterotrophic bacteria can carry out fermentation in the absence of accessible external electron acceptors, other populations can access the relatively high concentrations of dissolved sulfate in seawater as a terminal electron acceptor for sulfate reduction, producing toxic sulfides as a by-product. Although sulfate-reducing bacteria (SRB) are detected in 16S rRNA gene surveys of presumably healthy corals (Sunagawa et al., 2010; Fernando et al., 2014), toxic sulfides generated by increased growth of SRB can contribute to coral tissue death (Carlton and Richardson, 1995; Bourne et al., 2009, 2011; Richardson et al., 2009). SRB, in particular Desulfovibrio spp., are involved in the progression and transmission of Black Band Disease (Frias-Lopez et al., 2004; Richardson et al., 2009; Sato et al., 2011) and possibly other coral diseases. Thus, resource limitation for oxygen may lead to destabilizing proliferation of bacterial populations that generate sulfide.
Limitation of holobiont microbiota by carbon and nutrients
Corals lacking Symbiodinium obtain nearly all of their macro- and micronutrients from predation and suspension feeding. Symbiotic corals on the other hand supplement a major proportion of their carbon demand by utilization of exuded photosynthate, while still acquiring nutrients from external sources (Trench, 1970, 1983, 1993; Muscatine and Porter, 1977). High relative abundance of microbial cells in the coral gastrovascular cavity likely participate in food digestion in a similar manner as other animal gut microbiota (Agostini et al., 2012). Dense assemblages of microorganisms are also found in coral mucus layers, and mucus has been observed to be a carbon source for bacterial growth (Nakajima et al., 2009). Since photosynthate accumulates within intracellular symbiosomes and is passed directly to the coral host, this source of carbon may not be directly available to the heterotrophic microbiota until it is metabolized and released as waste or mucus. Thus, microorganisms that reside within the coral holobiont may encounter a feast-famine dynamic of carbon and nutrient loading depending on coral feeding behavior and mucus quality.
Macro- and micronutrient availability may also limit microbiota activity in the coral holobiont. Nitrogen limitation is indicated by the success of nitrogen-fixing cyanobacteria in the coral M. cavernosa (Lesser et al., 2004, 2007) and the widespread distribution of the nifH gene in reef building corals—a gene encoding a nitrogenase that mediates the costly metabolic process of nitrogen fixation (Lema et al., 2012; Olson and Lesser, 2013). Additional evidence for nutrient limitation in the holobiont comes from amendment experiments and analysis of nutrient-enrichment gradients where elevated waterborne nutrients are linked to proliferation of endolithic algae in coral skeleton (Fabricius, 2005) and increased density of Symbiodinium in the coral host (Marubini and Davies, 1996; Tanaka et al., 2013) (Section Disruptive Effects of Carbon and Nutrient Enrichment). Upregulated expression of transporters for nutrient uptake have recently been correlated with nutrient limitation in bacterioplanktonic populations (Harke and Gobler, 2013) and future multi-“omics” work targeting the holobiont transporter profiles of microbial populations may shed light on the global dynamics of nutrient limitation across the multiple habitats of the holobiont.
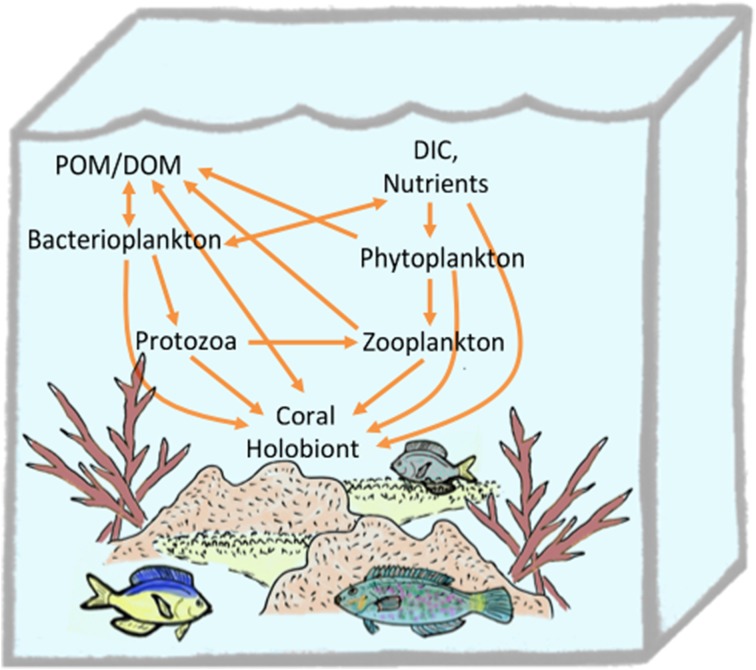
In addition to predation and suspension feeding, corals may obtain nutrients directly. Scleractinian corals are capable of uptaking amino acids (Drew Ferrier, 1991; Al-Moghrabi et al., 1993; Grover et al., 2008; Tremblay et al., 2012), ammonium (Burris, 1983), and dissolved and particulate organic matter from the surrounding seawater (Sorokin, 1973; Tremblay et al., 2012). Indeed, 32P-radiolabeling experiments with bacteria or inorganic phosphate demonstrate that six coral species assimilate phosphate directly from seawater, or as labeled bacterioplankton via suspension feeding, with higher efficiency for the latter (Sorokin, 1973). The reverse was true for carbon uptake where 14C-radiolabelled low molecular weight dissolved organic matter (DOM) was taken up more efficiently that the carbon-equivalent of bacterial cells (Sorokin, 1973). Microbes are well-established as agents that enable the transfer of DOM to higher trophic levels through the microbial loop (Azam et al., 1983; Azam and Cho, 1987). Assimilation of DOM into the coral holobiont may also be converted to animal biomass and mucus that is accessible to higher trophic levels (Wild et al., 2004; Naumann et al., 2010). Recently uptake of DOM and conversion to detritus by the sponge holobiont has been shown to play a major role in recycling nutrients in reef ecosystems via a cycle termed the “sponge loop” (de Goeij et al., 2013). Although photosynthetic activity by Symbiodinium in the coral holobiont places corals as net exporters of DOM into reef ecosystems (Wild et al., 2004), uptake of some forms of DOM by the coral holobiont contributes to efficient recycling of fixed carbon and nutrients on reef ecosystems that allows dense assemblages to thrive in nutrient poor regions (e.g., a “coral loop”) (Figure 5).
Figure 5.
Trophic connections of the coral holobiont in the planktonic food web.
Disruptive effects of carbon and nutrient enrichment
Within the holobiont, and at the ecosystem scale, nutrient-limitation (oligotrophy) appears to be critical for the healthy function of most reefs dominated by coral. Perturbation of the reef environment by exposure to nutrients (N, P, and Fe) or dissolved organic carbon (DOC) has been linked to changes in microbial community structure and adverse physiological consequences for corals (Table 1). Nutrient enrichment is recognized as one of the strongest drivers of coastal habitat degradation and has been associated with increased frequencies of coral disease and bleaching (Vega Thurber et al., 2014). Studies of nutrient enrichment in experiments conducted at the local reef scale (Ward and Harrison, 2000; Koop et al., 2001; Bell et al., 2007; Vega Thurber et al., 2014), under controlled aquarium conditions (Muscatine et al., 1989; Marubini and Davies, 1996; Tanaka et al., 2013) or observations over in situ gradients (Loya et al., 2004) provide insights into the impacts of N and/or P addition on holobiont physiology (reviewed in Fabricius, 2005). Stimulated growth of Symbiodinium accompanies nutrient additions, which can lead to enhanced calcification and rates of skeletal elongation (Muscatine et al., 1989; Marubini and Davies, 1996) but also may increase the risk of bleaching after UV or thermal stress induced production of reactive oxygen by Symbiodinium photosystems (Lesser, 1996; Vega Thurber et al., 2014). Nutrient addition stimulates growth of endolithic algae that act as microborers and elevate rates of skeletal bioerosion, resulting in a more fragile coral skeleton (Koop et al., 2001). Other impacts linked to nutrient enrichment include reduced rates of embryogenesis and reduced overall reproductive success (Ward and Harrison, 2000; Koop et al., 2001; Loya et al., 2004; Bell et al., 2007). Nutrient amendments were associated with increased overall growth rates for Symbiodinium-bearing (but not aposymbiotic) acroporid coral juveniles, promoting recruitment success during competition with benthic cyanobacteria (Tanaka et al., 2013) illustrating complex outcomes depending on the duration of nutrient enrichment and coral life stage.
Table 1.
Effects of physicochemical shifts on the coral holobiont.
| Stressor Direction | Response (and representative references) |
|---|---|
| Organic carbon | • Growth of heterotrophic bacteria and increased abundance (Kline et al., 2006; Nelson et al., 2013) |
| ↑ | • Reduced dissolved oxygen (Simpson et al., 1992) |
| • Enrichment of virulent bacteria or virulence genes (Vega Thurber et al., 2009; Nelson et al., 2013) | |
| Nutrients (N, P or Fe) | • N/P/Fe: Increased algal growth and activity in benthos and plankton and DOM exudation (Bell et al., 2007; Kelly et al., 2012) |
| ↑ | • N/P/Fe: Enrichment of virulent bacteria or virulence genes (Vega Thurber et al., 2009; Kelly et al., 2012) |
| • N/P: Increased proportion of viral sequences (Vega Thurber et al., 2008) | |
| • N/P: Decreased larval production (Ward and Harrison, 2000; Koop et al., 2001; Loya et al., 2004) | |
| • P: increased rates of skeletal extension, contributing to decreased skeletal density (Koop et al., 2001) | |
| • P: increased rates of bioerosion by microborers (Koop et al., 2001; Carreiro-Silva et al., 2012) | |
| Dissolved oxygen | • Survival of dark-incubated Montipora peltiformis in anoxic water for up to 96 h (Mass et al., 2010). |
| ↓ | • Enhanced mortality of Montipora peltiformis in anoxic water with other stressors (low pH, elevated sulfide, contact with organic rich sediment)(Mass et al., 2010). |
| • Nighttime anoxia at tissue/water interface of healthy corals (Shashar et al., 1993), in the gastrovascular cavity (Agostini et al., 2012), and in coral tissue consumed by black band disease (Carlton and Richardson, 1995). | |
| Sedimentation | • Stimulated microbial activity from organic carbon and nutrients leading to reduced pH, elevated sulfide and coral tissue death (Mass et al., 2010) |
| ↑ | • Reduced mass transfer across coral surface (oxygen, nutrients, waste) (Rogers, 1990) |
| • Shading and decreased photosynthesis (Rogers, 1990) | |
| • Increased mucus production and sloughing, along with nitrogen uptake from sediment sources (Mills and Sebens, 2004) | |
| Light/UV | • Elevated rates of oxygenic photosynthesis (Shashar et al., 1993; Mass et al., 2010) |
| ↑ | • Incident light above photoacclimation thresholds, or associated with thermal stress, associated with formation of reactive oxygen and free radicals (Downs et al., 2002), photosystem and host tissue/DNA damage (Lesser and Farrell, 2004) |
| ↓ | • Low light: Reduced coral growth rates for species reliant on photosynthetic Symbiodinium for nutrition (Muscatine, 1973) |
| Temperature | • Symbiodinium loss (bleaching) (Glynn, 1991, 1993; Brown, 1997; Brandt and McManus, 2009) |
| ↑ | • Increased abundance and virulence of pathogens (Ben-Haim et al., 1999, 2003b; Banin et al., 2000; Cervino et al., 2004) |
| • Increased abundance of viral sequences (Vega Thurber et al., 2008, 2009; Littman et al., 2011) | |
| • Decreased larval recruitment to CCA (Webster et al., 2011) | |
| pH/pCO2 | • Experimental pH gradient (pH 7.3 and 8.2): increased % pathogens (Meron et al., 2011) |
| ↓ ↑ | • Natural pH/pCO2 gradient pH 7.3–8.1: no change in % pathogens (Meron et al., 2012) |
| • Natural pCO2 gradient: microbiota shift where proportions of Endozoicomonas spp. decrease while Halomonas spp. increase (Morrow et al., 2014). | |
| • Low pH: Induction of viruses (Vega Thurber et al., 2008) | |
| • Low pH: Decreased larval recruitment to CCA (Webster et al., 2013) |
Addition of DOC, directly or indirectly via nutrient-stimulated primary production, leads to high heterotrophic bacterial abundance and degradation of reef quality (Simpson et al., 1992; Kline et al., 2006; Kelly et al., 2012; McDole et al., 2012). DOC inputs may stimulate heterotrophic microbial activity and growth on reefs in a compound-dependent manner (Nelson et al., 2013). In a recent study comparing carbon exudates of corals and their algal competitors, the composition of coral-derived carbon exudates was shown to match the profile of neutral sugars in the seawater and to exert little influence on microbial community structure after seawater amendment. In contrast, exudates from reef algae were enriched in the labile neutral sugars rhamnose, galactose, fucose, and mannose+xylose (Nelson et al., 2013) and these exudates, when added to seawater, promoted growth of a subset of bacteria enriched in virulence factors (Nelson et al., 2013). Blooms of heterotrophic respiring bacteria after DOC amendment may also have indirect effects on coral physiology, such as by depleting dissolved oxygen concentrations which may render coral tissue anoxic. Whether the negative outcomes of DOC enrichment on reef health are mediated by growth of pathogens that cause disease or by impairment of coral defenses through DOC-triggered stress (e.g., local anoxia) is unclear and indeed may be mediated by both.
Top-down control of coral holobiont structure in a dynamic environment
Top-down forcing of holobiont structure through immune interactions, predation, and interference-based competition allows certain microbes to grow/persist in the holobiont while others are eliminated. Like all animal tissues, corals represent rich stores of nutrients kept out of bounds from potential microbial consumers by tissue barriers and immune functions. The epithelium and immune system of healthy corals are critical lines of defense to prevent indiscriminant microbial digestion of viable tissues and to allow stable coexistence of coral tissue and a microbiota. In addition, emerging research indicates that other components of the holobiont (i.e., the microbiota and perhaps Symbiodinium) contribute to this stable coexistence through a complex chemical ecology of antimicrobial compounds and signals that modulate microbial activity. Environmental stresses that disrupt coordinated activities of the coral holobiont pose a great risk to coral homeostasis by weakening top-down control of microbiota structure and enabling growth of opportunistic microbes that proliferate at the expense of the coral host.
Immune interactions
Emerging knowledge regarding immunity in corals is reviewed in Mullen et al. (2003), Augustin and Bosch (2010), Palmer and Traylor-Knowles (2012). Coral immunity includes inducible components such as migrating phagocytotic cells that destroy invading microorganisms, lectins that promote aggregation of microbial cells for efficient phagocytosis, encapsulation via prophenoloxidase (PPO)-catalyzed melanization to inactivate and contain invading pathogens, and systemic functions such as expression of chitinase and 1,2 beta-glucanase enzymes that weaken microbial cell walls (Mullen et al., 2003; Augustin and Bosch, 2010; Palmer and Traylor-Knowles, 2012). Predicted mechanisms for sensing microorganisms are emerging from coral genome sequence analysis (Shinzato et al., 2012) but have not reached the stage of functional validation. Mechanistic studies for immune defenses in Hydra spp. that are conserved in other metazoans shed light on potential mechanisms in corals (Augustin and Bosch, 2010; Augustin et al., 2010; Hamada et al., 2013). Such mechanisms include sensing of microbial cells in interstitial and extracellular spaces via by cell membrane-bound toll-like receptors (TLR). Microbial invasion into intracellular spaces can be sensed by enzymes detecting foreign nucleic acids (nucleotide binding and oligomerization domain like receptors) (Augustin and Bosch, 2010; Augustin et al., 2010; Hamada et al., 2013). In both cases, sensors direct a signal cascade resulting in enhanced production of antimicrobial peptides and serine protease inhibitors (Augustin et al., 2009; Jung, 2009). In Hydra, species-specific microbial communities and regulation of the microbiome over development is shaped by the innate immune system (e.g., TLR signaling) and maternally provisioned antimicrobial peptides (AMP) (Fraune et al., 2009; Franzenburg et al., 2013).
Although targeted studies in corals are few, production of AMPs play profound roles in mediating host-microbe interactions in other invertebrate symbioses (Zasloff, 2002; Ryu et al., 2008). Production of host-derived AMPs is emerging as a trait found among multiple cnidarian taxa. In 2006, aurelin, an AMP from the Scyphozoan Aurelia aurita was described revealing a novel protein structure with no homology to previously identified AMPs (Ovchinnikova et al., 2006). Subsequent work in Hydra revealed Hydramycin (Jung, 2009) followed by identification of multiple species-specific AMPs responsible for intra-generic differences in microbiota structure (Franzenburg et al., 2013). In 2011, damicornin, the first AMP isolated from corals was described (Vidal-Dupiol et al., 2011) and was shown to be constitutively expressed in ectodermal cells of P. damicornis and released in response to surface immune damage. Interestingly, damicornin gene expression was repressed in colonies challenged with V. coralliilyticus (Vidal-Dupiol et al., 2011), suggesting AMP repression is a potential virulence mechanism of the pathogens. Findings that P. damicornis extracts induced in response to mechanical stress have potent activity against the coral pathogen V. coralliilyticus (Geffen and Rosenberg, 2005), imply the presence of yet unknown additional inducible antimicrobial factors. A second coral-derived AMP was reported from the Brazilian octocoral Phyllogorgia dilatata (de Lima et al., 2013). The prevalence, diversity and significance of AMPs in coral development are only beginning to be explored.
Antagonistic interactions: interference based competition, predation and phage infection
While coral gene expression may lead to production of antibiotic compounds and AMPs, other members of the holobiont provide additional top-down control on holobiont structure through interference-based competition, predation and phage infection. Antimicrobial compounds associated with coral mucus and produced by resident bacteria inhibit growth of certain populations of bacteria, including those noted as opportunistic pathogens (Ritchie, 2006; Rypien et al., 2010). Symbiotic bacteria can also act to inhibit growth of other bacterial strains, often invaders that are detrimental to coral health, a relationship described as the coral probiotic hypothesis by Reshef et al. (2006). For instance, certain types of alphaproteobacteria, gammaproteobacteria, bacilli, Pseudoalteromonas sp., and Pseudovibrio sp. were found to significantly inhibit growth of V. coralliilyticus (Rypien et al., 2010; Vizcaino et al., 2010). Alphaproteobacteria and gammaproteobacteria were antagonist against V. shiloi and two members of Pseudoalteromonas spp. were able to inhibit V. shiloi growth (Rypien et al., 2010). In a study employing transposon mutagenesis and assessment of mutant fitness in the Aiptasia model, growth on mucus of the white pox pathogen, Serratia marcescens, required the enzyme N-acetyl-glucosaminidase, the activity of which could be inhibited by native coral microbiota, through a small molecule isolated from a microbial member of the coral mucus, Exiguobacterium sp. (Krediet et al., 2013). This study illustrates that modulation of microbial activity through interference-based competition can impact the fitness of certain microbes in the holobiont and exert control of microbial community structure, in this case through elimination of a coral pathogen.
The role of non-coral predators in the holobiont is not well-constrained although there is evidence that viruses and protozoa contribute to the mortality of coral and Symbiodinium cells. Ciliates have been observed infecting coral tissue leading to tissue death (Antonius and Lipscomb, 2000; Croquer et al., 2006) and preying on intracellular Symbiodinium leading to Brown Band Syndrome in the Great Barrier Reef (Bourne et al., 2008) and White Syndrome in Hawaiian Montipora corals (Work et al., 2012). Whether Ciliates are exclusively associated with coral disease or also interact with the healthy holobiont is not well-established. Other eukaryotic grazers such as Thraustochytrid protists may prey on dense assemblages of bacteria and assemble at the mucus layer (Raghukumar, 1991; Harel et al., 2008). Viruses that infect coral and Symbiodinium have been observed in the holobiont (Wegley et al., 2007; Marhaver et al., 2008; Patten et al., 2008; Correa et al., 2013) and may mediate the health of corals and exacerbate stressful conditions. Herpes-like viral sequences were observed in viral metagenomes from Diploria strigosa (Marhaver et al., 2008) and later analysis of Porites compressa viral metagenomes from stress and control treatments revealed a higher proportion of Herpes-like sequences under stressed conditions (i.e., low pH, elevated temperature, and nutrient enrichment) (Vega Thurber et al., 2008). Genes for Herpes-like viruses were found in other cnidarian genomes suggesting that coral viral infections are common (Vega Thurber et al., 2008).
Bacteriophage sequences have been observed in all coral viral metagenomes (Wegley et al., 2007; Marhaver et al., 2008; Correa et al., 2013) and the diversity of bacteriophage-like sequences associated with corals parallels and perhaps contributes to the diversity of the microbiota (reviewed in Vega Thurber and Correa, 2011). The presence of phage in the mucus layer of corals has been proposed as a mechanism of defense against invading microorganisms (Barr et al., 2013). Binding between phage capsid proteins [immunoglobin (Ig)-like protein domains] and glycoproteins from mucus suggest that bacteriophage are readily incorporated into mucus layers (Barr et al., 2013). The widespread distribution of Ig-like proteins in viral metagenomes from cnidarian (and human) mucus suggests that metazoan mucosal surfaces and phage coevolved to maintain phage adherence. This work presents the fascinating possibility that bacteriophage from the environment are easily incorporated into the coral mucus layer and thus, these may act as an additional defense mechanism to control holobiont microbial community structure through phage infection and lysis (Barr et al., 2013).
Deviations from homeostasis
Variation in physicochemical conditions can have distinct impacts on the coral holobiont community. Among healthy corals the bacterial community composition has been found to vary seasonally (Koren and Rosenberg, 2006) suggesting that shifting ecological drivers (bottom up and top down control) as well as shifting composition of the source assemblages from which immigration occurs leads to a dynamic microbiota that is compatible with a state of homeostasis in the holobiont. In contrast, numerous studies document shifts in coral microbiota following perturbation of physicochemical conditions that may lead to the destabilization and ultimately death of the holobiont (Table 1). Little is known about factors that determine whether the holobiont can maintain homeostasis during, or recover from, physicochemical perturbations. A general trend in coral perturbation studies is that opportunistic pathogens including vibrios and alteromonads increase following exposure to a stressor (Vega Thurber et al., 2009; Littman et al., 2011; Meron et al., 2011) suggesting that activities of virulent microbes may accelerate the impact of a stressor, and contribute to the breakdown of holobiont functioning due to changes in host response or due to pathogenicity (Rosenberg et al., 2007; Bourne et al., 2009).
Conclusions
Scleractinian corals have survived dramatic environmental changes during their evolutionary history including changes in sea level, pH, and temperature—however current anthropogenically-induced changes are occurring at rates unprecedented in the geological record (Glynn, 1991, 1993; Hoegh-Guldberg, 1999; Pandolfi et al., 2003; Hoegh-Guldberg et al., 2007; Mora, 2008). Whether extant corals will survive under current regimes of rapid change will depend upon their ability to adapt to new conditions on anthropogenic timescales. It is likely that coral-associated microbiota will be linked to the capacity of corals to ultimately adapt to new environmental conditions. In plant systems, it has become increasingly clear that crop improvement through microbiome optimization is preferable to plant trait selection (Coleman-Derr and Tringe, 2014). In a similar fashion coral-associated microbes may mediate the resilience of corals to stresses such as resistance to coral-specific pathogens (Reshef et al., 2006; Rosenberg et al., 2007) and perhaps to other stressors.
Our understanding of coral physiology and interaction as a holobiont is now enhanced by the expanded scientific focus on the diverse members of the coral microbiota. Access to whole genome data from the major players in a single holobiont, in particular, organisms that may be vertically transmitted from parent colony to offspring, will enable researchers to develop new, readily testable hypotheses regarding metabolic complementation, genome expansion/reduction, coevolutionary patterns, and rates of HGT across all genomes in a holobiont. Such information will yield insight into the adaptations that have enabled coral holobionts to be so successful in nutrient poor environments.
In order to motivate future work to refine our mechanistic understanding of microorganisms as coral partners through evolution, development, and in ecological interactions within the holobiont we have selected eight questions to highlight based on recent literature (Table 2). Continued research is essential to promote a detailed understanding of the mechanisms shaping the coral holobiont on ecological and evolutionary timescales as pressures emerge for reefs to acclimate and adapt to conditions of global change.
Table 2.
Questions for future research.
| • What are the mechanisms of nutritional and defensive mutualisms between holobiont members? |
| This is a fundamental question to understand the ecology and physiology of the holobiont. The answers may be of biomedical interest as novel antimicrobial compounds or mechanisms that disrupt pathogen colonization or virulence without bactericidal or bacteriostatic activity (e.g., Krediet et al., 2013) may be involved. |
| • What is the diversity of vertically-transmitted microorganisms in coral reproduction? |
| • Do these populations represent obligate or facultative mutualisms? |
| Novel cultivation-based approaches and genome sequencing or single-cell genomics to recover populations that resist cultivation are necessary. Such improvements will enable testing of hypotheses regarding coevolution and codiversification between corals and their microbiota. |
| • What is the role of Symbiodinium in shaping microbiota acquisition in gametes, larvae, and adult coral colonies? |
| • Do algae produce chemical signals that mediate allelopathic recognition or modulate colonization of the microbiota? |
| • How do chemical signals produced by CCA biofilms promote settlement and/or metamorphosis of coral larvae and how is receipt of these signals coordinated to optimize recruitment? |
| • What microbial taxa and activities are associated with coral cell-associated microbial aggregates (CAMAs) and what factors mediate their distribution in the coral polyp? |
| • Do the enclosed microorganisms interact with the coral tissue as mutualists, parasites, or commensals? |
| • Since several studies have documented lower levels of microbial colonization in Montipora spp. compared to other corals (Marquis et al., 2005; Work and Aeby, 2014), to what extent is antimicrobial activity specific to the coral species or their associated Symbiodinium? |
| • What role does the endolithic microbial community play in the homeostasis of the coral holobiont? |
| A potentially transformative avenue for inquiry is exploring the relationship between bacterial activity and skeleton formation in juvenile corals as recently suggested (Sharp et al., 2012). |
| • What are the molecular mechanisms of the host/holobiont stress response that allow proliferation of pathogen-like bacteria (e.g., Vibrio spp., Alteromonas spp.)? |
| Emerging coral holobiont transcriptomics studies will shed new light on how the coral holobiont responds to stresses and exists as a robust and resilient system when in a healthy state. |
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Support for this work was provided to Janelle R. Thompson by the National Research Foundation of Singapore through the Center for Environmental Sensing and Modeling (CENSAM) and by the MIT Civil and Environmental Engineering Department; to Hanny E. Rivera by the MIT Presidential Fellowship; to Collin J. Closek by the National Geographic Society; and to Mónica Medina by NSF projects IOS 1146880 and IOS 0926906. Finally, we would like to thank Dr. Kevin Penn for helpful discussions and for reviewing a draft of the manuscript.
References
- Agostini S., Suzuki Y., Higuchi T., Casareto B. E., Yoshinaga K., Nakano Y., et al. (2012). Biological and chemical characteristics of the coral gastric cavity. Coral Reefs 31, 147–156 10.1007/s00338-011-0831-6 [DOI] [Google Scholar]
- Ainsworth T. D., Wasmund K., Ukani L., Seneca F., Yellowlees D., Miller D., et al. (2011). Defining the tipping point: a complex cellular life/death balance in corals in response to stress. Sci. Rep. 1:160. 10.1038/srep00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Moghrabi S., Allemand D., Jaubert J. (1993). Valine uptake by the sleractinian coral Galaxea fascicularis: characterization and effect of light and nutritional stress. J. Comp. Physiol. B 163, 365–362. [Google Scholar]
- Aloisi G., Gloter A., KrOger M., Wallmann K., Guyot F., Zuddas P. (2006). Nucleation of calcium carbonate on bacterial nanoglobules. Geology 34, 1017–1020 10.1130/G22986A.1 [DOI] [Google Scholar]
- Amend A. S., Barshis D. J., Oliver T. A. (2012). Coral-associated marine fungi form novel lineages and heterogeneous assemblages. ISME J. 6, 1291–1301. 10.1038/ismej.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonius A., Lipscomb D. (2000). First protozoan coral-kliller in the Indo-Pacific. Atoll Res. Bull 481, 1–21 10.5479/si.00775630.481 [DOI] [Google Scholar]
- Apprill A., Marlow H. Q., Martindale M. Q., Rappe M. S. (2009). The onset of microbial associations in the coral Pocillopora meandrina. ISME J. 3, 685–699. 10.1038/ismej.2009.3 [DOI] [PubMed] [Google Scholar]
- Apprill A., Rappe M. S. (2011). Response of the microbial community to coral spawning in lagoon and reef flat environments of Hawaii, USA. Aquat. Microb. Ecol. 62, 251–266 10.3354/ame01471 [DOI] [Google Scholar]
- Augustin R., Bosch T. C. (2010). Cnidarian immunity: a tale of two barriers, in Invertebrate Immunity, ed Soderhall K. (New York, NY: Landes Biosciences and Springer Science; ), 1–16. [DOI] [PubMed] [Google Scholar]
- Augustin R., Fraune S., Bosch T. C. G. (2010). How Hydra senses and destroys microbes. Semin. Immunol. 22, 54–58. 10.1016/j.smim.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Augustin R., Siebert S., Bosch T. C. (2009). Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra's innate immune system. Dev. Comp. Immunol. 33, 830–837. 10.1016/j.dci.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Azam F., Cho B. (1987). Bacterial utilization of organic matter in the sea, in Ecology of Microbial Communities, eds Fletcher M., Gray T. R. G., Jones J. G. (Cambridge, UK: Cambridge University Press; ), 261–281. [Google Scholar]
- Azam F., Fenchel T., Field J. G., Gray J. S., Meyerreil L. A., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263 10.3354/meps010257 [DOI] [Google Scholar]
- Babcock R. C., Bull G. D., Harrison P. L., Heyward A. J., Oliver J. K., Wallace C. C., et al. (1986). Synchronous spawnings of 105 scleractinian coral species on the Great Barrier Reef. Mar. Biol. 90, 379–394 10.1007/BF00428562 [DOI] [Google Scholar]
- Baker A. C. (2003). Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 34, 661–689 10.1146/annurev.ecolsys.34.011802.132417 [DOI] [Google Scholar]
- Banaszak A., LaJeunesse T. C., Trench R. K. (2000). The synthesis of mycosporine-like amino acids (MAAs) by cultured, symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 249, 219–233. 10.1016/S0022-0981(00)00192-1 [DOI] [PubMed] [Google Scholar]
- Banin E., Israely T., Kushmaro A., Loya Y., Orr E., Rosenberg E. (2000). Penetration of the coral-bleaching bacterium Vibrio shiloi into Oculina patagonica. Appl. Environ. Microbiol. 66, 3031–3036. 10.1128/AEM.66.7.3031-3036.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin E., Vassilakos D., Orr E., Martinez R. J., Rosenberg E. (2003). Superoxide dismutase is a virulence factor produced by the coral bleaching pathogen Vibrio shiloi. Curr. Microbiol. 46, 418–422. 10.1007/s00284-002-3912-5 [DOI] [PubMed] [Google Scholar]
- Barr J. J., Auro R., Furlan M., Whiteson K. L., Erb M. L., Pogliano J., et al. (2013). Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 10771–10776. 10.1073/pnas.1305923110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T., Arif C., Ferrier-Pages C., Zoccola D., Aranda M., Voolstra C. R. (2013a). Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar. Ecol. Prog. Ser. 479, 75–84 10.3354/meps10197 [DOI] [Google Scholar]
- Bayer T., Neave M. J., Alsheikh-Hussain A., Aranda M., Yum L. K., et al. (2013b). The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl. Environ. Microbiol. 79, 4759–4762. 10.1128/AEM.00695-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P. R., Lapointe B. E., Elmetri I. (2007). Reevaluation of ENCORE: support for the eutrophication threshold model for coral reefs. Ambio 36, 416–424. 10.1579/0044-7447(2007)36[416:ROESFT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y., Banim E., Kushmaro A., Loya Y., Rosenberg E. (1999). Inhibition of photosynthesis and bleaching of zooxanthellae by coral pathogen Vibrio shiloi. Environ. Microbiol. 1, 223–229. 10.1046/j.1462-2920.1999.00027.x [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y., Thompson F. L., Thompson C. C., Cnockaert M. C., Hoste B., Swings J., et al. (2003a). Vibrio coralliilyticus sp nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int. J. Syst. Evol. Microbiol. 53, 309–315. 10.1099/ijs.0.02402-0 [DOI] [PubMed] [Google Scholar]
- Ben-Haim Y., Zicherman-Keren M., Rosenberg E. (2003b). Temperature-regulated bleaching and lysis of the coral Pocillopora damicornis by the novel pathogen Vibrio coralliilyticus. Appl. Environ. Microbiol. 69, 4236–4242. 10.1128/AEM.69.7.4236-4242.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentis C. J., Kaufman L., Golubic S. (2000). Endolithic fungi in reef-building corals (Order: Scleractinia) are common, cosmopolitan, and potentially pathogenic. Biol. Bull. 198, 254–260. 10.2307/1542528 [DOI] [PubMed] [Google Scholar]
- Bondarev V., Richter M., Romano S., Piel J., Schwedt A., Schulz-Vogt H. N. (2013). The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ. Microbiol. 15, 2095–2113. 10.1111/1462-2920.12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch T. C., McFall-Ngai M. J. (2011). Metaorganisms as the new frontier. Zoology (Jena). 114, 185–190. 10.1016/j.zool.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. G., Boyett H. V., Henderson M. E., Muirhead A., Willis B. L. (2008). Identification of a ciliate (Oligohymenophorea: Scuticociliatia) associated with brown band disease on corals of the Great Barrier Reef. Appl. Environ. Microbiol. 74, 883–888. 10.1128/AEM.01124-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W., Harvell C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17, 554–562. 10.1016/j.tim.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Bourne D. G., Muirhead A., Sato Y. (2011). Changes in sulfate-reducing bacterial populations during the onset of black band disease. Isme J. 5, 559–564. 10.1038/ismej.2010.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D. G., Munn C. B. (2005). Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ. Microbiol. 7, 1162–1174. 10.1111/j.1462-2920.2005.00793.x [DOI] [PubMed] [Google Scholar]
- Brandt M., McManus J. (2009). Disease incidence is related to bleaching extent in reef-building corals. Ecology 90, 2859–2867. 10.1890/08-0445.1 [DOI] [PubMed] [Google Scholar]
- Brown B. E. (1997). Coral bleaching: causes and consequences. Coral Reefs 16, S129–S138 10.1007/s003380050249 [DOI] [Google Scholar]
- Brown B. E., Bythell J. C. (2005). Perspectives on mucus secretion in reef corals. Mar. Ecol. Prog. Ser. 296, 291–309 10.3354/meps296291 [DOI] [Google Scholar]
- Burris R. H. (1983). Uptake and assimilation of 15NH4+ by a variety of corals. Mar. Biol. 75, 151–155 10.1007/BF00405997 [DOI] [Google Scholar]
- Carilli J. E., Godfrey J., Norris R. D., Sandin S. A., Smith J. E. (2010). Periodic endolithic algal blooms in Montastraea faveolata corals may represent periods of low-level stress. Bull. Mar. Sci. 86, 709–718. [Google Scholar]
- Carlton R. G., Richardson L. L. (1995). Oxygen and sulfide dynamics in a horizontally migrating cyanobacterial mat: black band disease of corals. Microb. Ecol. 18, 155–162 10.1111/j.1574-6941.1995.tb00173.x [DOI] [Google Scholar]
- Carreiro-Silva M., Kiene W. E., Golubic S., McClanahan T. R. (2012). Phosphorus and nitrogen effects on microbial euendolithic communities and their bioerosion rates. Mar. Pollut. Bull. 64, 602–613. 10.1016/j.marpolbul.2011.12.013 [DOI] [PubMed] [Google Scholar]
- Ceh J., Raina J. B., Soo R. M., van Keulen M., Bourne D. G. (2012). Coral-bacterial communities before and after a coral mass spawning event on ningaloo reef. PLoS ONE 7:e36920. 10.1371/journal.pone.0036920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceh J., van Keulen M., Bourne D. G. (2013). Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness. Microb. Ecol. 65, 227–231. 10.1007/s00248-012-0105-z [DOI] [PubMed] [Google Scholar]
- Cervino J. M., Hayes R. L., Polson S. W., Polson S. C., Goreau T. J., Martinez R. J., et al. (2004). Relationship of Vibrio species infection and elevated temperatures to yellow blotch/band disease in Caribbean corals. Appl. Environ. Microbiol. 70, 6855–6864. 10.1128/AEM.70.11.6855-6864.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J. A., Kirkness E. F., Simakov O., Hampson S. E., Mitros T., Weinmaier T., et al. (2010). The dynamic genome of Hydra. Nature 464, 592–596. 10.1038/nature08830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimetto L. A., Brocchi M., Thompson C. C., Martins R. C., Ramos H. R., Thompson F. L. (2008). Vibrios dominate as culturable nitrogen-fixing bacteria of the Brazilian coral Mussismilia hispida. Syst. Appl. Microbiol. 31, 312–319. 10.1016/j.syapm.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Closek C. J., Sunagawa S., DeSalvo M. K., Piceno Y. M., DeSantis T. Z., Brodie E. L., et al. (2014). Coral transcriptome and bacterial community profiles reveal distinct Yellow Band Disease states in Orbicella faveolata. ISME J. 8, 2411–2422. 10.1038/ismej.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D., Tringe S. G. (2014). Building the crops of tomorrow: advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 5:283. 10.3389/fmicb.2014.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles S. L., Strathman R. (1973). Observations on coral mucus “floc” and their potential trophic significance. Limnol. Oceanogr. 18, 673–678 10.4319/lo.1973.18.4.0673 [DOI] [Google Scholar]
- Correa A. M., Welsh R. M., Vega Thurber R. L. (2013). Unique nucleocytoplasmic dsDNA and +ssRNA viruses are associated with the dinoflagellate endosymbionts of corals. ISME J. 7, 13–27. 10.1038/ismej.2012.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croquer A., Bastidas C., Lipscomp D., Rodrıguez-Martınez R., Jordan-Dahlgren E., Guzman H. (2006). First report of folliculinid ciliates affecting Caribbean scleractinian corals. Coral Reefs 25, 187–191 10.1007/s00338-005-0068-3 [DOI] [Google Scholar]
- Daniels C., Zeifman A., Heym K., Ritchie K. B., Watson C. A., Berzins I., et al. (2011). Spatial heterogeneity of bacterial communities in the mucus of Montastraea annularis. Mar. Ecol. Prog. Ser. 426, 29–40 10.3354/meps09024 [DOI] [Google Scholar]
- de Castro A. P., Araujo S. D., Jr., Reis A. M., Moura R. L., Francini-Filho R. B., Pappas G., Jr., et al. (2010). Bacterial community associated with healthy and diseased reef coral Mussismilia hispida from eastern Brazil. Microb. Ecol. 59, 658–667. 10.1007/s00248-010-9646-1 [DOI] [PubMed] [Google Scholar]
- de Goeij J. M., van Oevelen D., Vermeij M. J., Osinga R., Middelburg J. J., de Goeij A. F., et al. (2013). Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science 342, 108–110. 10.1126/science.1241981 [DOI] [PubMed] [Google Scholar]
- de Lima L. A., Migliolo L., Barreiro e Castro C., Pires Dde O., Lopez-Abarrategui C., Goncalves E. F., et al. (2013). Identification of a novel antimicrobial peptide from Brazilian coast coral Phyllogorgia dilatata. Protein Pept. Lett. 20, 1153–1158. 10.2174/0929866511320100010 [DOI] [PubMed] [Google Scholar]
- Dinsdale E. A., Pantos O., Smriga S., Edwards R. A., Angly F., Wegley L., et al. (2008). Microbial ecology of four coral atolls in the Northern Line Islands. PLoS ONE 3:e1584. 10.1371/journal.pone.0001584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobretsov S., Abed R. M. M., Teplitski M. (2013). Mini-review: Inhibition of biofouling by marine microorganisms. Biofouling 29, 423–441. 10.1080/08927014.2013.776042 [DOI] [PubMed] [Google Scholar]
- Dobretsov S., Qian P.-Y. (2004). The role of epibotic bacteria from the surface of the soft coral Dendronephthya sp. in the inhibition of larval settlement. J. Exp. Mar. Biol. Ecol. 299, 35–50 10.1016/j.jembe.2003.08.011 [DOI] [Google Scholar]
- Downs C. A., Fauth J. E., Halas J. C., Dustan P., Bemiss J., Woodley C. M. (2002). Oxidative stress and seasonal coral bleaching. Free Radic. Biol. Med. 33, 533–543. 10.1016/S0891-5849(02)00907-3 [DOI] [PubMed] [Google Scholar]
- Drew Ferrier M. (1991). Net uptake of dissolved free amino acids by four scleractinian corals. Coral Reefs 10, 183–187 10.1007/BF00336772 [DOI] [Google Scholar]
- Dunlap W. C., Starcevic A., Baranasic D., Diminic J., Zucko J., Gacesa R., et al. (2013). KEGG orthology-based annotation of the predicted proteome of Acropora digitifera: ZoophyteBase – an open access and searchable database of a coral genome. BMC Genomics 14:509. 10.1186/1471-2164-14-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius K. E. (2005). Effects of terrestrial runoff on the ecology of corals and coral reefs: review and synthesis. Mar. Pollut. Bull. 50, 125–146. 10.1016/j.marpolbul.2004.11.028 [DOI] [PubMed] [Google Scholar]
- Fernando S. C., Wang J., Sparling K., Garcia G., Francini-Filho R. B., de Moura R. L., et al. (2014). Microbiota of the major South Atlantic reef building coral Mussismilia. Microb. Ecol. [Epub ahead of print]. 10.1007/s00248-014-0474-6 [DOI] [PubMed] [Google Scholar]
- Fine M., Loya Y. (2002). Endolithic algae: an alternative source of photoassimilates during coral bleaching. Proc. Biol. Sci. 269, 1205–1210. 10.1098/rspb.2002.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine M., Roff G., Ainsworth T. D., Hoegh-Guldberg O. (2006). Phototrophic microendoliths bloom during coral “white syndrome.” Coral Reefs 25, 577–581 10.1007/s00338-006-0143-4 [DOI] [Google Scholar]
- Franzenburg S., Walter J., Kunzel S., Wang J., Baines J. F., Bosch T. C., et al. (2013). Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. U.S.A. 110, E3730–E3738. 10.1073/pnas.1304960110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S., Abe Y., Bosch T. C. (2009). Disturbing epithelial homeostasis in the metazoan Hydra leads to drastic changes in associated microbiota. Environ. Microbiol. 11, 2361–2369. 10.1111/j.1462-2920.2009.01963.x [DOI] [PubMed] [Google Scholar]
- Fraune S., Bosch T. C. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151. 10.1073/pnas.0703375104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J., Klaus J. S., Bonheyo G. T., Fouke B. W. (2004). Bacterial community associated with black band disease in corals. Appl. Environ. Microbiol. 70, 5955–5962. 10.1128/AEM.70.10.5955-5962.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J., Zerkle A. L., Bonheyo G. T., Fouke B. W. (2002). Partitioning of bacterial communities between seawater and healthy, black band diseased, and dead coral surfaces. Appl. Environ. Microbiol. 68, 2214–2228. 10.1128/AEM.68.5.2214-2228.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M., Azam F. (2012). Corals shed bacteria as a potential mechanism of resilience to organic matter enrichment. Isme J. 6, 1159–1165. 10.1038/ismej.2011.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M., Son K., Raina J. B., Rusconi R., Menolascina F., Shapiro O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. Isme J. 8, 999–1007. 10.1038/ismej.2013.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geffen Y., Rosenberg E. (2005). Stress-induced rapid release of antibacterials by scleractinian corals. Mar. Biol. 146, 931–935 10.1007/s00227-004-1505-5 [DOI] [Google Scholar]
- Gerardo N. M. (2013). The give and take of host-microbe symbioses. Cell Host Microbe 14, 1–3. 10.1016/j.chom.2013.07.001 [DOI] [PubMed] [Google Scholar]
- Glynn P. W. (1991). Coral reef bleaching in the 1980s and possible connections with global warming. Trends Ecol. Evol. (Amst.) 6, 175–179. 10.1016/0169-5347(91)90208-F [DOI] [PubMed] [Google Scholar]
- Glynn P. W. (1993). Coral reef bleaching: ecological perspectives. Coral Reefs 12, 1–17. 10.1007/BF0030377925070864 [DOI] [Google Scholar]
- Grover R., Maguer J. F., Allemand D., Ferrier-Pages C. (2008). Uptake of dissolved free amino acids by the scleractinian coral Stylophora pistillata. J. Exp. Biol. 211, 860–865. 10.1242/jeb.012807 [DOI] [PubMed] [Google Scholar]
- Gutner-Hoch E., Fine M. (2011). Genotypic diversity and distribution of Ostreobium quekettii within scleractinian corals. Coral Reefs 30, 643–650 10.1007/s00338-011-0750-6 [DOI] [Google Scholar]
- Hadfield M. G. (2011). Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann. Rev. Mar. Sci. 3, 453–470. 10.1146/annurev-marine-120709-142753 [DOI] [PubMed] [Google Scholar]
- Hamada M., Shoguchi E., Shinzato C., Kawashima T., Miller D. J., Satoh N. (2013). The complex NOD-like receptor repertoire of the coral Acropora digitifera includes novel domain combinations. Mol. Biol. Evol. 30, 167–176. 10.1093/molbev/mss213 [DOI] [PubMed] [Google Scholar]
- Harel M., Ben-Dov E., Rasoulouniriana D., Siboni N., Kramarsky-Winter E., Loya Y., et al. (2008). A new Thraustochytrid, strain Fng1, isolated from the surface mucus of the hermatypic coral Fungia granulosa. FEMS Microbiol. Ecol. 64, 378–387. 10.1111/j.1574-6941.2008.00464.x [DOI] [PubMed] [Google Scholar]
- Harke M. J., Gobler C. J. (2013). Global transcriptional responses of the toxic cyanobacterium, Microcystis aeruginosa, to nitrogen stress, phosphorus stress, and growth on organic matter. PLoS ONE 8:e69834. 10.1371/journal.pone.0069834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison P. L. (2011). Coral reefs: an ecosystem in transition, in Sexual Reproduction of Scleractinian Corals, eds Dubinsky Z., Stambler N. (New York, NY: Springer Science and Business Media; ), 59–85. [Google Scholar]
- Hoegh-Guldberg O. (1999). Climate change, coral bleaching and the future of the world's coral reefs. Mar. Freshwater Res. 50, 839–866 10.1071/MF99078 [DOI] [Google Scholar]
- Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742. 10.1126/science.1152509 [DOI] [PubMed] [Google Scholar]
- Huang Y., Callahan S., MHadfield G. (2012). Recruitment in the sea: bacterial genes required for inducing larval settlement in a polychaete worm. Sci. Rep. 2, 228. 10.1038/srep00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnik F., Nikoh N., Koga R., Ross L., Duncan R. P., Fujie M., et al. (2013). Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell 153, 1567–1578. 10.1016/j.cell.2013.05.040 [DOI] [PubMed] [Google Scholar]
- Jackson D. J., Macis L., Reitner J., Wörheide G. (2011). A horizontal gene transfer supported the evolution of an early metazoan biomineralization strategy. BMC Evol. Biol. 11:238. 10.1186/1471-2148-11-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatkar A., Brown B., Bythell J., Guppy R., Morris N., Pearson J. (2010). Measuring mucus thickness in reef corals using a technique devised for vertebrate applications. Mar. Biol. 2, 261–267 10.1007/s00227-009-1313-z [DOI] [Google Scholar]
- Johnston I. S., Rohwer F. (2007). Microbial landscapes on the outer tissue surfaces of the reef-building coral Porites compressa. Coral Reefs 26, 375–383 10.1007/s00338-007-0208-z [DOI] [Google Scholar]
- Jung S. (2009). Hydramacin-1: structure and antibacterial activity of a protein from the basal metazoan Hydra. J. Biol. Chem. 284, 1896. 10.1074/jbc.M804713200 [DOI] [PubMed] [Google Scholar]
- Kellogg C. A. (2004). Tropical Archaea: diversity associated with the surface microlayer of corals. Mar. Ecol. Prog. Ser. 273, 81–88 10.3354/meps273081 [DOI] [Google Scholar]
- Kelly L. W., Barott K. L., Dinsdale E., Friedlander A. M., Nosrat B., Obura D., et al. (2012). Black reefs: iron-induced phase shifts on coral reefs. ISME J. 6, 638–649. 10.1038/ismej.2011.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimes N. E., Grim C. J., Johnson W. R., Hasan N. A., Tall B. D., Kothary M. H., et al. (2012). Temperature regulation of virulence factors in the pathogen Vibrio coralliilyticus. ISME J. 6, 835–846. 10.1038/ismej.2011.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline D. I., Kuntz N. M., Breitbart M., Knowlton N., Rohwer F. (2006). Role of elavated organic carbon levels and microbial activity in coral mortality. Mar. Ecol. Prog. Ser. 314, 119–125 10.3354/meps314119 [DOI] [Google Scholar]
- Koop K., Booth D., Broadbent A., Brodie J., Bucher D., Capone D., et al. (2001). ENCORE: the effect of nutrient enrichment on coral reefs. Synthesis of results and conclusions. Mar. Pollut. Bull. 42, 91–120. 10.1016/S0025-326X(00)00181-8 [DOI] [PubMed] [Google Scholar]
- Koren O., Rosenberg E. (2006). Bacteria associated with mucus and tissues of the coral Oculina patagonica in summer and winter. Appl. Environ. Microbiol. 72, 5254–5259. 10.1128/AEM.00554-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krediet C. J., Ritchie K. B., Alagely A., Teplitski M. (2013). Members of native coral microbiota inhibit glycosidases and thwart colonization of coral mucus by an opportunistic pathogen. ISME J. 7, 980–990. 10.1038/ismej.2012.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner I., Walters L., Becerro M., Paul V., Ritson-Williams R., Beach K. (2006). Inhibition of coral recruitment by macroalgae and cyanobacteria. Mar. Ecol. Prog. Ser. 323, 107–117 10.3354/meps323107 [DOI] [Google Scholar]
- Kuhl M., Cohen Y., Dalsgaard T., Jorgense B. B., Revsbech N. P. (1995). Microenvironment and photosynthesis of zooxanthellae in scleractinian corals studied with microsensors for O2, pH, and light. Mar. Ecol. Prog. Ser. 117, 159–172 10.3354/meps117159 [DOI] [Google Scholar]
- Kuhl M., Holst G., Larkum A. W., Ralph P. J. (2008). Imaging of oxygen dynamics within the endolithic algal community of the massive coral Porites lobata. J. Phycol 44, 541–550 10.1111/j.1529-8817.2008.00506.x [DOI] [PubMed] [Google Scholar]
- Le Campion-Alsumard T., Golubic S., Hutchings P. (1995a). Microbial endoliths in skeletons of live and dead corals: Porites lobata (Moorea, French Polynesia). Mar. Ecol. Prog. Ser. Oldendorf 117, 149–157 10.3354/meps117149 [DOI] [Google Scholar]
- Le Campion-Alsumard T., Golubic S., Priess K. (1995b). Fungi in corals: symbiosis or disease? Interaction between polyps and fungi causes pearl-like skeleton biomineralization. Mar. Ecol. Prog. Ser. 117, 137–147 10.3354/meps117137 [DOI] [Google Scholar]
- Lema K. A., Willis B. L., Bourne D. G. (2012). Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl. Environ. Microbiol. 78, 3136–3144. 10.1128/AEM.07800-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser M. P. (1996). Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in symbiotic dinoflagellates. Limnol. Oceanogr. 41, 271–283 10.4319/lo.1996.41.2.0271 [DOI] [Google Scholar]
- Lesser M. P., Falcón L. I., Rodríguez-Román A., Enríquez S., Hoegh-Guldberg O., Iglesias-Prieto R. (2007). Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian coral Montastraea cavernosa. Mar. Ecol. Prog. Ser. 346, 143–152 10.3354/meps07008 [DOI] [Google Scholar]
- Lesser M. P., Farrell J. H. (2004). Exposure to solar radiation increases damage to both host tissues and algal symbionts of corals during thermal stress. Coral Reefs 23, 367–377 10.1007/s00338-004-0392-z [DOI] [Google Scholar]
- Lesser M. P., Mazel C. H., Gorbunov M. Y., Falkowski P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. 10.1126/science.1099128 [DOI] [PubMed] [Google Scholar]
- Littman R., Willis B. L., Bourne D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3, 651–660. 10.1111/j.1758-2229.2010.00234.x [DOI] [PubMed] [Google Scholar]
- Littman R. A., Bourne D. G., Willis B. L. (2010). Responses of coral-associated bacterial communities to heat stress differ with Symbiodinium type on the same coral host. Mol. Ecol. 19, 1978–1990. 10.1111/j.1365-294X.2010.04620.x [DOI] [PubMed] [Google Scholar]
- Littman R. A., Willis B. L., Bourne D. G. (2009). Bacterial communities of juvenile corals infected with different Symbiodinium (dinoflagellate) clades. Mar. Ecol. Prog. Ser. 389, 45–59 10.3354/meps08180 [DOI] [Google Scholar]
- Loya Y., Lubinevsky H., Rosenfeld M., Kramarsky-Winter E. (2004). Nutrient enrichment caused by in situ fish farms at Eilat, Red Sea is detrimental to coral reproduction. Mar. Pollut. Bull. 49, 344–353. 10.1016/j.marpolbul.2004.06.011 [DOI] [PubMed] [Google Scholar]
- Lukas K. J. (1974). Two species of Chlorophyte genus ostreobium from skeletons of Atlantic and Caribbean reef corals. J. Phycol. 10, 331–335. [Google Scholar]
- Magnusson S., Fine M., Kuhl M. (2007). Light microclimate of endolithic phototrophs in the scleractinian corals Montipora monasteriata and Porites cylindrica. Mar. Ecol. Prog. Ser. 332, 119–128 10.3354/meps332119 [DOI] [Google Scholar]
- Marhaver K. L., Edwards R. A., Rohwer F. (2008). Viral communities associated with healthy and bleaching corals. Environ. Microbiol. 10, 2277–2286. 10.1111/j.1462-2920.2008.01652.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhaver K. L., Vermeij M. J., Rohwer F., Sandin S. A. (2013). Janzen-Connell effects in a broadcast-spawning Caribbean coral: distance-dependent survival of larvae and settlers. Ecology 94, 146–160. 10.1890/12-0985.1 [DOI] [PubMed] [Google Scholar]
- Marquis C. P., Baird A. H., de Nys R., Holmström C., Koziumi N. (2005). An evaluation of the antimicrobial properties of the eggs of 11 species of scleractinian corals. Coral Reefs 24, 248–253 10.1007/s00338-005-0473-7 [DOI] [Google Scholar]
- Martinez-Garcia M., Brazel D. M., Swan B. K., Arnosti C., Chain P. S., Reitenga K. G., et al. (2012). Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of Verrucomicrobia. PLoS ONE 7:e35314. 10.1371/journal.pone.0035314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marubini F., Davies P. S. (1996). Nitrate increases zooxanthellae population density and reduces skeletogenesis in corals Mar. Biol. 127, 319–328 10.1007/BF00942117 [DOI] [Google Scholar]
- Mass T., Genin A., Shavit, U, Grinstein M., Tchernov D. (2010). Flow enhances photosynthesis in marine benthic autotrophs by increasing the efflux of oxygen from the organism to the water. Proc. Natl. Acad. Sci. U.S.A. 107, 2527–2531. 10.1073/pnas.0912348107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K., Abt B., Brambilla E., Lapidus A., Copeland A., Deshpande S. (2010). Complete genome sequence of Coraliomargarita akajimensis type strain (04OKA010-24). Stand. Genomic Sci. 2, 290–299. 10.4056/sigs.952166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon J. P., Moran N. A. (2012). Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26. 10.1038/nrmicro2670 [DOI] [PubMed] [Google Scholar]
- McDole T., Nulton J., Barott K. L., Felts B., Hand C., Hatay M., et al. (2012). Assessing Coral Reefs on a Pacific-Wide Scale Using the Microbialization Score. PLoS ONE 7:e43233. 10.1371/journal.pone.0043233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M., Hadfield M. G., Bosch T. C., Carey H. V., Domazet-Loso T., Douglas A. E., et al. (2013). Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 110, 3229–3236. 10.1073/pnas.1218525110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D., Atias E., Iasur Kruh L., Elifantz H., Minz D., Fine M., et al. (2011). The impact of reduced pH on the microbial community of the coral Acropora eurystoma. ISME J. 5, 51–60. 10.1038/ismej.2010.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meron D., Rodolfo-Metalpa R., Cunning R., Baker A. C., Fine M., Banin E. (2012). Changes in coral microbial communities in response to a natural pH gradient. ISME J. 6, 1775–1785. 10.1038/ismej.2012.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills M., Sebens K. (2004). Ingestion and assimilation of nitrogen from benthic sediments by three species of coral. Mar. Biol. 145, 1097–1106 10.1007/s00227-004-1398-3 [DOI] [Google Scholar]
- Mora C. (2008). A clear human footprint in the coral reefs of the Caribbean. Proc. R. Soc. B Biol. Sci. 275, 767–773. 10.1098/rspb.2007.1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran N. A., McCutcheon J. P., Nakabachi A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- Morrow K. M., Bourne D. G., Humphrey C., Botte E. S., Laffy P., Zaneveld J., et al. (2014). Natural volcanic CO2 seeps reveal future trajectories for host–microbial associations in corals and sponges. ISME J. [Epub ahead of print]. 10.1038/ismej.2014.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow K. M., Moss A. G., Chadwick N. E., Liles M. R. (2012). Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl. Environ. Microbiol. 78, 6438–6449. 10.1128/AEM.01162-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D., Hooker N., Morse A., Jensen R. (1988). Control of larval metamorphosis and recruitment in sympatric agariciid corals. J. Exp. Mar. Biol. Ecol. 116, 193–217 10.1016/0022-0981(88)90027-5 [DOI] [Google Scholar]
- Mullen K. M., Peters E. C., Harvell C. D. (2003). Coral resistance to disease, in Coral Health and Disease, eds Rosenberg E., Loya Y. (Berlin: Springer; ), 377–399. [Google Scholar]
- Muscatine L. (1973). Nutrition of corals, in Biology and Geology of Coral Reefs, eds Jones O. A., Endean R. (New York, NY: Academic Press; ), 77–115 10.1016/B978-0-12-395526-5.50012-2 [DOI] [Google Scholar]
- Muscatine L. (1990). The role of symbiotic algae in carbon and energy flux in reef corals, in Ecosystems of the World: Coral Reefs, ed Dubinsky Z. (Amsterdam: Elsevier; ), 75–88. [Google Scholar]
- Muscatine L., Falkowski P. G., Dubinsky Z., Cook P. A., McCloskey L. R. (1989). The effect of external nutrient resources on the population dynamics of zooxanthellae in a reef coral. Proc. R. Soc. Lond. B 236, 311–324 10.1098/rspb.1989.0025 [DOI] [Google Scholar]
- Muscatine L., Porter J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460 10.2307/1297526 [DOI] [Google Scholar]
- Nakajima R., Yoshida T., Azman B. A. R., Zaleha K., Othman B. H. R., Toda T. (2009). In situ release of coral mucus by Acropora and its influence on the heterotrophic bacteria. Aquat. Ecol. 43, 815–823 10.1007/s10452-008-9210-y [DOI] [Google Scholar]
- Naumann M., Mayr C., Struck U., Wild C. (2010). Coral mucus stable isotope composition and labeling: experimental evidence for mucus uptake by epizoic acoelomorph worms. Mar. Biol. 157, 2521 10.1007/s00227-010-1516-3 [DOI] [Google Scholar]
- Neave M., Michell C., Apprill A., Voolstra C. R. (2014). Whole-Genome Sequences of three symbiotic endozoicomonas bacteria. Genome Announc. 2, e00802–e00814. 10.1128/genomeA.00802-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri A. P., Webster N. S., Hill R. T., Heyward A. J. (2001). Metamorphosis of broadcast spawning corals in response to bacteria isolated from crustose algae. Mar. Ecol. Prog. Ser. 223, 121–131 10.3354/meps223121 [DOI] [Google Scholar]
- Nelson C. E., Goldberg S. J., Wegley Kelly L., Haas A. F., Smith J. E., Rohwer F., et al. (2013). Coral and macroalgal exudates vary in neutral sugar composition and differentially enrich reef bacterioplankton lineages. ISME J. 7, 962–979. 10.1038/ismej.2012.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson N. D., Ainsworth T. D., Gates R. D., Takabayashi M. (2009). Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J. Exp. Mar. Biol. Ecol. 371, 140–146 10.1016/j.jembe.2009.01.012 [DOI] [Google Scholar]
- Olson N. D., Lesser M. P. (2013). Diazotrophic diversity in the Caribbean coral, Montastraea cavernosa. Arch. Microbiol. 195, 853–859. 10.1007/s00203-013-0937-z [DOI] [PubMed] [Google Scholar]
- Ovchinnikova T. V., Balandin S. V., Aleshina G. M., Tagaev A. A., Leonova Y. F., Krasnodembsky E. D., et al. (2006). Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 348, 514–523. 10.1016/j.bbrc.2006.07.078 [DOI] [PubMed] [Google Scholar]
- Padilla-Gamino J. L., Pochon X., Bird C., Concepcion G. T., Gates R. D. (2012). From parent to gamete: vertical transmission of Symbiodinium (Dinophyceae) ITS2 sequence assemblages in the reef building coral Montipora capitata. PLoS ONE 7:e38440. 10.1371/journal.pone.0038440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C. V., Traylor-Knowles N. (2012). Towards an integrated network of coral immune mechanisms. Proc. Biol. Sci. 279, 4106–4114. 10.1098/rspb.2012.1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi J. M., Bradbury R. H., Sala E., Hughes T. P., Bjorndal K. A., Cooke R. G., et al. (2003). Global trajectories of the long-term decline of coral reef ecosystems. Science 301, 955–958. 10.1126/science.1085706 [DOI] [PubMed] [Google Scholar]
- Patten N. L., Harrison P. L., Mitchell J. G. (2008). Prevalence of virus-like particles within a staghorn scleractinian coral (Acropora muricata) from the Great Barrier Reef. Coral Reefs 27, 569–580 10.1007/s00338-008-0356-9 [DOI] [Google Scholar]
- Pernice M., Meibom A., Van Den Heuvel A., Kopp C., Domart-Coulon I., Hoegh-Guldberg O., et al. (2012). A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 6, 1314–1324. 10.1038/ismej.2011.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike R. E., Haltli B., Kerr R. G. (2013). Description of Endozoicomonas euniceicola sp nov and Endozoicomonas gorgoniicola sp nov., bacteria isolated from the octocorals Eunicea fusca and Plexaura sp., and an emended description of the genus Endozoicomonas. Int. J. Syst. Evol. Microbiol. 63, 4294–4302. 10.1099/ijs.0.051490-0 [DOI] [PubMed] [Google Scholar]
- Raghukumar S. (1991). Occurence of thraustochytrid fungi in corals and coral mucus. Indian J. Mar. Sci. 20, 176–181. [Google Scholar]
- Raina J. B., Tapiolas D., Willis B. L., Bourne D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. 10.1128/AEM.02567-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshef L., Koren O., Loya Y., Zilber-Rosenberg I., Rosenberg E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073. 10.1111/j.1462-2920.2006.01148.x [DOI] [PubMed] [Google Scholar]
- Reshef L., Ron E., Rosenberg E. (2008). Genome analysis of the coral bleaching pathogen Vibrio shiloi. Arch. Microbiol. 190, 185–194. 10.1007/s00203-008-0388-0 [DOI] [PubMed] [Google Scholar]
- Richardson L. L., Miller A. W., Broderick E., Kaczmarsky L., Gantar M., Stanic D., et al. (2009). Sulfide, microcystin, and the etiology of black band disease. Dis. Aquat. Org. 87, 79–90. 10.3354/dao02083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Prog. Ser. 322, 1–14 10.3354/meps322001 [DOI] [Google Scholar]
- Ritchie K. B. (2011). Bacterial Symbionts of Corals and Symbiodinium. Heidelberg: Springer Berlin. [Google Scholar]
- Rivera H. E., Goodbody-Gringley G. (2014). Aggregation and cnidae development as early defensive strategies in Favia fragum and Porites astreoides. Coral Reefs. 33, 1079–1084 10.1007/s00338-014-1180-z [DOI] [Google Scholar]
- Rogers C. S. (1990). Responses of coral reefs and reef organisms to sedimentation. Mar. Ecol. Prog. Ser. 62, 185–202. 10.3354/meps06218515737355 [DOI] [Google Scholar]
- Rohwer F., Breitbart M., Jara J., Azam F., Knowlton N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20, 85–91 10.1007/s003380100138 [DOI] [Google Scholar]
- Rohwer F., Seguritan V., Azam F., Knowlton N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10 10.3354/meps243001 [DOI] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Rypien K. L., Ward J. R., Azam F. (2010). Antagonistic interactions among coral-associated bacteria. Environ. Microbiol. 12, 28–39. 10.1111/j.1462-2920.2009.02027.x [DOI] [PubMed] [Google Scholar]
- Ryu J. H., Kim S. H., Lee H. Y., Bai J. Y., Nam Y. D., Bae J. W., et al. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. 10.1126/science.1149357 [DOI] [PubMed] [Google Scholar]
- Santos E. O., Alves N., Jr., Dias G. M., Mazotto A. M., Vermelho A., Vora G. J., et al. (2011). Genomic and proteomic analyses of the coral pathogen Vibrio coralliilyticus reveal a diverse virulence repertoire. ISME J. 5, 1471–1483. 10.1038/ismej.2011.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Willis B. L., Bourne D. G. (2011). Successional changes in bacterial communities during the development of black band disease on the reef coral, Montipora hispida. ISME J. 4, 203–214. 10.1038/ismej.2009.103 [DOI] [PubMed] [Google Scholar]
- Schleheck D., Weiss M., Pitluck S., Bruce D., Land M. L., Han S., et al. (2011). Complete genome sequence of Parvibaculum lavamentivorans type strain (DS-1(T)). Stand. Genomic Sci. 5, 298–310. 10.4056/sigs.2215005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp K. H., Distel D., Paul V. J. (2012). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6, 790–801. 10.1038/ismej.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp K. H., Ritchie K. B. (2012). Multi-partner interaction in corals in the face of climate change. Biol. Bull. 223, 66–77. [DOI] [PubMed] [Google Scholar]
- Sharp K. H., Ritchie K. B., Schupp P. J., Ritson-Williams R., Paul V. J. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species. PLoS ONE 5:e10898. 10.1371/journal.pone.0010898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shashar N., Banaszak A. T., Lesser M. P., Amrami D. (1997). Coral endolithic algae: life in a protected environment. Pac. Sci. 51, 167–173. [Google Scholar]
- Shashar N., Cohen N., Loya Y. (1993). Extreme diel fluctuations of oxygen in diffusive boundary layers surrounding stony corals. Biol. Bull. Mar. Biol. Lab. Woods Hole 185, 455–461 10.2307/1542485 [DOI] [PubMed] [Google Scholar]
- Shashar N., Feldstein T., Cohen Y., Loya Y. (1994). Nitrogen fixation (acetylene reduction) on a coral reef. Coral Reefs 13, 171–174 10.1007/BF00301195 [DOI] [Google Scholar]
- Shashar N., Stambler N. (1992). Endolithic algae within corals – life in an extreme environment. J. Exp. Mar. Biol. Ecol. 163, 277–286 10.1016/0022-0981(92)90055-F [DOI] [Google Scholar]
- Shibata K., Haxo F. (1969). Light transmission and spectral distribution through epi- and endozoic algal layers in the brain coral (Favia). Biol. Bull. Mar. Biol. Lab. Woods Hole 136, 461–468 10.2307/1539688 [DOI] [Google Scholar]
- Shikuma N. J., Pilhofer M., Weiss G. L., Hadfield M. G., Jensen G. J., Newman D. K. (2014). Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343, 529–533. 10.1126/science.1246794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C., Hamada M., Shoguchi E., Kawashima T., Satoh N. (2012). The repertoire of chemical defense genes in the coral Acropora digitifera genome. Zool. Sci. 29, 510–517. 10.2108/zsj.29.510 [DOI] [PubMed] [Google Scholar]
- Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., et al. (2011). Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323. 10.1038/nature10249 [DOI] [PubMed] [Google Scholar]
- Shoguchi E., Shinzato C., Kawashima T., Gyoja F., Mungpakdee S., Koyanagi R., et al. (2013). Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408. 10.1016/j.cub.2013.05.062 [DOI] [PubMed] [Google Scholar]
- Simpson C. J., Cary J. L., Masini R. J. (1992). Destruction of corals and other reef animals by coral spawn slicks on Ningaloo Reef, Western Australia. Coral Reefs 12, 185–191 10.1007/BF00334478 [DOI] [Google Scholar]
- Smillie C. S., Smith M. B., Friedman J., Cordero O. X., David L. A., Alm E. J. (2011). Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244. 10.1038/nature10571 [DOI] [PubMed] [Google Scholar]
- Smith J. E., Shaw M., Edwards R. A., Obura D., Pantos O., Sala E., et al. (2006). Indirect effects of algae on coral: algae-mediated, microbe-induced coral mortality. Ecol. Lett. 9, 835–845. 10.1111/j.1461-0248.2006.00937.x [DOI] [PubMed] [Google Scholar]
- Sneed J. M., Sharp K. H., Ritchie K. B., Paul V. J. (2014). The chemical cue tetrabromopyrrole from a biofilm bacterium induces settlement of multiple Caribbean corals. Proc. Biol. Sci. 281. 10.1098/rspb.2013.3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin Y. I. (1973). On the feeding of some scleractinian coral with bacteria and dissolved organic matter. Limnol. Oceanogr. 18, 380–385. 10.4319/lo.1973.18.3.038019046402 [DOI] [Google Scholar]
- Starcevic A., Akthar S., Dunlap W. C., Shick J. M., Hranueli D., Cullum J., et al. (2008). Enzymes of the shikimic acid pathway encoded in the genome of a basal metazoan, Nematostella vectensis, have microbial origins. Proc. Natl. Acad. Sci. U.S.A. 105, 2533–2537. 10.1073/pnas.0707388105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Woodley C. M., Medina M. N. (2010). Threatened corals provide underexplored microbial habitats. PLoS ONE 5:e9554. 10.1371/journal.pone.0009554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet M. J., Croìquer A., Bythell J. C. (2011). Bacterial assemblages differ between compartments within the coral holobiont. Coral Reefs 30, 39–52 10.1007/s00338-010-0695-1 [DOI] [Google Scholar]
- Tanaka Y., Iguchi A., Inoue M., Mori C., Sakai K., Suzuki A., et al. (2013). Microscopic observation of symbiotic and aposymbiotic juvenile corals in nutrient-enriched seawater. Mar. Pollut. Bull. 68, 93–98. 10.1016/j.marpolbul.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Tanner J. E. (1996). Seasonality and lunar periodicity in the reproduction of Pocilloporid corals. Coral Reefs 15, 59–66 10.1007/BF01626077 [DOI] [Google Scholar]
- Tebben J., Tapiolas D. M., Motti C. A., Abrego D., Negri A. P., Blackall L. L., et al. (2011). Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS ONE 6:e19082. 10.1371/journal.pone.0019082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay P., Naumann M. S., Sikorski S., Grover R., Ferrier-Pagès C. (2012). Experimental assessment of organic carbon fluxes in the scleractinian coral Stylophora pistillata during a thermal and photo stress event. Mar. Ecol. Prog. Ser. 453, 63–77 10.3354/meps09640 [DOI] [Google Scholar]
- Tremblay P., Weinbauer M. G., Rottier C., Guérardel Y., Nozais C., Ferrier-Pagès C. (2011). Mucus composition and bacterial communities associated with the tissue and skeleton of three scleractinian corals maintained under culture conditions. J. Mar. Biolog. Assoc. U.K. 91, 649–657 10.1017/S002531541000130X [DOI] [Google Scholar]
- Trench R. K. (1970). Synthesis of a mucous cuticle by a zoanthid. Nature 227, 1155–1156. 10.1038/2271155b0 [DOI] [PubMed] [Google Scholar]
- Trench R. K. (1983). Dinoflagellates in non-parasitic symbioses, in The Dinoflagellates, ed Taylor F. J. R. (London: Blackwell Publications; ), 530–570. [Google Scholar]
- Trench R. K. (1993). Microalgal-invertebrate symbioses – a review. Endocytobiosis Cell Res. 9, 135–175. [Google Scholar]
- Vega Thurber R., Correa A. (2011). Viruses of reef-building scleractinian corals. J. Exp. Mar. Biol. Ecol. 408, 102–113 10.1016/j.jembe.2011.07.030 [DOI] [Google Scholar]
- Vega Thurber R. L., Barott K. L., Hall D., Liu H., Rodriguez-Mueller B., Desnues C., et al. (2008). Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc. Natl. Acad. Sci. U.S.A. 105, 18413–18418. 10.1073/pnas.0808985105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber R. L., Burkepile D. E., Fuchs C., Shantz A. A., McMinds R., Zaneveld J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob. Chang. Biol. 20, 544–554. 10.1111/gcb.12450 [DOI] [PubMed] [Google Scholar]
- Vega Thurber R., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R. A., Angly F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11, 2148–2163. 10.1111/j.1462-2920.2009.01935.x [DOI] [PubMed] [Google Scholar]
- Venn A. A., Tambutte E., Lotto S., Zoccola D., Allemand D., Tambutte S. (2009). Imaging intracellular pH in a reef coral and symbiotic anemone. Proc. Natl. Acad. Sci. U.S.A. 106, 16574–16579. 10.1073/pnas.0902894106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeij M. J., Smith J. E., Smith C. M., Vega Thurber R., Sandin S. A. (2009). Survival and settlement success of coral planulae: independent and synergistic effects of macroalgae and microbes. Oecologia 159, 325–336. 10.1007/s00442-008-1223-7 [DOI] [PubMed] [Google Scholar]
- Vidal-Dupiol J., Ladriere O., Destoumieux-Garzon D., Sautiere P. E., Meistertzheim A. L., Tambutte E., et al. (2011). Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 286, 22688–22698. 10.1074/jbc.M110.216358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino M. I., Johnson W. R., Kimes N. E., Williams K., Torralba M., Nelson K. E., et al. (2010). Antimicrobial resistance of the coral pathogen Vibrio coralliilyticus and Caribbean sister phylotypes isolated from a diseased octocoral. Microb. Ecol. 59, 646–657. 10.1007/s00248-010-9644-3 [DOI] [PubMed] [Google Scholar]
- Ward S., Harrison P. (2000). Changes in gametogenesis and fecundity of acroporid corals that were exposed to elevated nitrogen and phosphorus during the ENCORE experiment. J. Exp. Mar. Biol. Ecol. 246, 179–221. 10.1016/S0022-0981(99)00182-3 [DOI] [PubMed] [Google Scholar]
- Weber M., de Beer D., Lott C., Polerecky L., Kohls K., Abed R. M., et al. (2012). Mechanisms of damage to corals exposed to sedimentation. Proc. Natl. Acad. Sci. U.S.A. 109, E1558–1567. 10.1073/pnas.1100715109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Medina M. (2012). The role of microalgal symbionts (Symbiodinium) in holobiont physiology, in Genomic Insights into the Biology of Algae, ed Piganeau G. (London: Elsevier; ), 119–140 10.1016/B978-0-12-391499-6.00004-9 [DOI] [Google Scholar]
- Webster N. S., Smith L. D., Heyward A. J., Watts J. E. M., Webb R. I., Blackall L. L., et al. (2004). Metamorphosis of a Scleractinian coral in response to microbial biofilms. Appl. Environ. Microbiol. 70, 1213–1221. 10.1128/AEM.70.2.1213-1221.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. S., Soo R., Cobb R., Negri A. P. (2011). Elevated seawater temperature causes a microbial shift on crustose coralline algae with implications for the recruitment of coral larvae. ISME J. 5, 759–770. 10.1038/ismej.2010.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. S., Uthicke S., Botte E. S., Flores F., Negri A. P. (2013). Ocean acidification reduces induction of coral settlement by crustose coralline algae. Glob. Chang. Biol. 19, 303–315. 10.1111/gcb.12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegley L., Edwards R., Rodriguez-Brito B., Liu H., Rohwer F. (2007). Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 9, 2707–2719. 10.1111/j.1462-2920.2007.01383.x [DOI] [PubMed] [Google Scholar]
- Wild C., Huettel M., Klueter A., Kremb S. G., Rasheed M. Y., Jorgensen B. B. (2004). Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature 428, 66–70. 10.1038/nature02344 [DOI] [PubMed] [Google Scholar]
- Williams W. M., Viner A. B., Broughton W. J. (1987). Nitrogen fixagtion (acetylene reduction) associated with the living coral Acropora variabilis. Mar. Biol. 94, 531–535 10.1007/BF00431399 [DOI] [Google Scholar]
- Work T., Aeby G. (2014). Microbial aggregates within tissues infect a diversity of corals throughout the Indo-Pacific. Mar. Ecol. Prog. Ser. 500, 1–9 10.3354/meps10698 [DOI] [Google Scholar]
- Work T. M., Russell R., Aeby G. S. (2012). Tissue loss (white syndrome) in the coral Montipora capitata is a dynamic disease with multiple host responses and potential causes. Proc. Biol. Sci. 279, 4334–4341. 10.1098/rspb.2012.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovleva I., Bhagooli R., Takemura A., Hidaka M. (2004). Differential susceptibility to oxidative stress of two scleractinian corals: antioxidant functioning of mycosporine-glycine. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 139, 721–730. 10.1016/j.cbpc.2004.08.016 [DOI] [PubMed] [Google Scholar]
- Yang C. S., Chen M. H., Arun A. B., Chen C. A., Wang J. T., Chen W. M. (2010). Endozoicomonas montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int. J. Syst. Evol. Microbiol. 60, 1158–1162. 10.1099/ijs.0.014357-0 [DOI] [PubMed] [Google Scholar]
- Yarden O. (2014). Fungal association with sessile marine invertebrates. Front. Microbiol. 5:228. 10.3389/fmicb.2014.00228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokouchi H., Fukuoka Y., Mukoyama D., Calugay R., Takeyama H., Matsunaga T. (2006). Whole-metagenome amplification of a microbial community associated with scleractinian coral by multiple displacement amplification using phi29 polymerase. Environ. Microbiol. 8, 1155–1163. 10.1111/j.1462-2920.2006.01005.x [DOI] [PubMed] [Google Scholar]
- Zasloff M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. 10.1038/415389a [DOI] [PubMed] [Google Scholar]