Abstract
Pseudomonas aeruginosa causes severe and persistent infections in immune compromised individuals and cystic fibrosis sufferers. The infection is hard to eradicate as P. aeruginosa has developed strong resistance to most conventional antibiotics. The problem is further compounded by the ability of the pathogen to form biofilm matrix, which provides bacterial cells a protected environment withstanding various stresses including antibiotics. Quorum sensing (QS), a cell density-based intercellular communication system, which plays a key role in regulation of the bacterial virulence and biofilm formation, could be a promising target for developing new strategies against P. aeruginosa infection. The QS network of P. aeruginosa is organized in a multi-layered hierarchy consisting of at least four interconnected signaling mechanisms. Evidence is accumulating that the QS regulatory network not only responds to bacterial population changes but also could react to environmental stress cues. This plasticity should be taken into consideration during exploration and development of anti-QS therapeutics.
Keywords: quorum sensing, IQS, PQS, las, rhl, Pseudomonas aeruginosa, virulence, environmental factors
Introduction
Pseudomonas aeruginosa is a ubiquitous, gram-negative bacterium that thrives in diverse habitats and environments. Usually a commensal on the host body, P. aeruginosa is capable of transforming into an opportunistic pathogen when there is a breach of host tissue barriers or a suppressed immune system (Van Delden and Iglewski, 1998). P. aeruginosa is an important nosocomial pathogen, affecting a wide category of patients convalescing in hospitals. They include patients with cystic fibrosis and other lung diseases, traumatized cornea, burns, Gustilo open fractures, long-term intubated patients, the immune-compromised and elderly individuals. The infections caused by P. aeruginosa are usually resistant to treatment by multiple antibiotics and can lead to severe and persistent infections (Bonomo and Szabo, 2006; Chernish and Aaron, 2003; Doshi et al., 2011; Tan, 2008). This translates into further complications and secondary fungal infections, extension of hospital stay, therapeutic failure, and in some cases, premature death of cystic fibrosis patients (Henry et al., 1992; Kosorok et al., 2001; Rabin et al., 2004; Tan, 2008). Because P. aeruginosa grows and survives in various environmental conditions, it makes acquiring an infection extremely easy and outbreaks of extreme drug-resistant strains are common among hospital wards and intensive care units.
It is believed that understanding the regulatory mechanisms with which P. aeruginosa governs virulence gene expression may hold the key to develop alternative therapeutic interventions to control and prevent the bacterial infections (Fig. 1). The recent research progresses show that a bacterial cell-cell communication mechanism, widely known as quorum sensing (QS), plays a key role in modulating the expression of virulence genes in P. aeruginosa. The term quorum sensing was proposed two-decades ago by three renowned microbiologists based on the bacterial population density-dependent regulatory mechanisms found in several microbial organisms, including Vibrio fischeri, Agrobacterium tumefaciens, P. aeruginosa and Erwinia carotovora (Fuqua et al., 1994). Since then, various QS systems have been found in many bacterial pathogens, which are commonly associated with the regulation of virulence gene expression and biofilm formation (Deng et al., 2011; Ng and Bassler, 2009; Pereira et al., 2013; Whitehead et al., 2001). Typically, quorum sensing bacteria produce and release small chemical signals, and at a high population density, the accumulated signals interact with cognate receptors to induce the transcriptional expression of various target genes including those encoding production of virulence factors. While QS becomes a popular concept, it is worthy to note that opinions arose on whether QS is the most-fitted term for mechanistic explanation of the above-mentioned bacterial group behavior. The point of contention stemmed from the fact that autoinducer concentration, the key determinant of “quorum” as defined by QS, was not simply a function of bacterial cell density, but a combined output of many factors such as diffusion rate and spatial distribution, and hence alternative terms such as “diffusion sensing”, “efficiency sensing” and “combinatorial quorum sensing” were proposed (Hense et al., 2007; Redfield, 2002; Cornforth et al., 2014). Whilst interesting, these alternative opinions await further experimental endorsement and by far QS remains as the most rigorously tested mechanism of bacteria cell-cell communication and collective responses.
Figure 1.
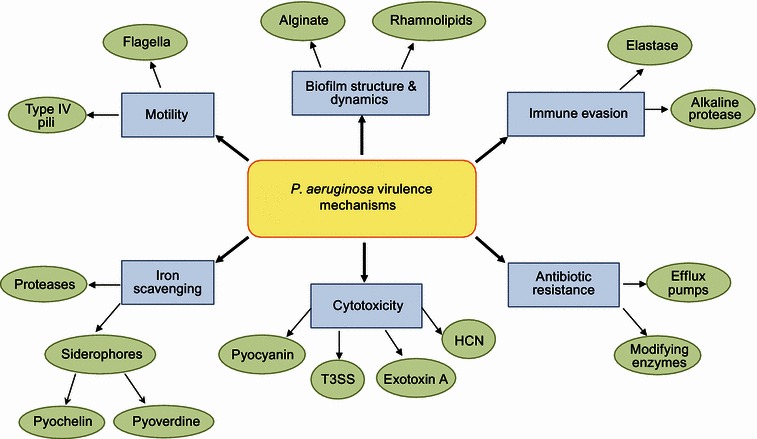
Virulence mechanisms employed during P. aeruginosa infections
Given its importance as a human pathogen, P. aeruginosa has been the subject of intensive investigations and become one of the model organisms in QS research. The research progresses in the last two decades have unveiled a sophisticated hierarchy QS network in this pathogen, which consists of a few sets of connected systems, including las, iqs, pqs and rhl. Particularly, recent findings show that the QS network in P. aeruginosa is highly adaptable and capable of responding to external biostress cues, which provides the pathogen flexibility in the control of virulence gene expression. It would not be surprising that other bacterial pathogens may have also evolved similar flexible QS systems which could respond to changed environmental conditions. This is an important factor to consider in the development of quorum sensing inhibitors (QSIs) as therapeutics, since bacteria routinely encounters adverse environmental conditions when infecting host organisms. This review will provide an overview on the QS systems in P. aeruginosa, focusing on a recently discovered integrated quorum sensing system (IQS), and on the interactions between all the four QS systems and how environmental cues could affect the QS hierarchy.
Quorum Sensing Systems in Pseudomonas aeruginosa
History of quorum sensing
The concept of quorum sensing in P. aeruginosa was an extension of the studies based on the prototype luxI-luxR system in Vibrio fischeri, in which luxI encodes the biosynthesis of an acylhomoserine lactone (AHL) signal N-(3-oxohexanoyl)-L-homoserine lactone (OHHL), and luxR encodes an AHL-dependent transcription factor (Eberhard, 1972; Nealson et al., 1970; Stewart and Williams, 1992; Williams et al., 1992). With significant homology to the LuxR protein, LasR in P. aeruginosa was initially identified to be a key regulator in the expression of lasB gene encoding for a metalloprotease elastase (Cook, 1992; Gambello and Iglewski, 1991). Subsequently, LasR was also shown to be required for the transcription of aprA, lasA and toxA, and thus it was thought to be a global regulator of the virulence genes in P. aeruginosa (Gambello and Iglewski, 1991; Gambello et al., 1993; Passador et al., 1993; Toder et al., 1991). LasI, the LuxI equivalent in P. aeruginosa, was proposed to synthesize AHL signals with autoinducing and elastase-regulating properties (Jones et al., 1993). One year later, the actual chemical structure of this Pseudomonas autoinducer (PAI) was characterized as N-(3-oxododecanoyl)-homoserine lactone (OdDHL) (Pearson et al., 1994). PAI is structurally related to the autoinducers discovered in other gram-negative bacteria species (Cao and Meighen, 1993; Eberhard et al., 1981; Zhang et al., 1993).
Shortly after, a second autoinducer, factor 2, was discovered in P. aeruginosa (Pearson et al., 1995). This discovery was made following a puzzling observation that an unusually high concentration of OdDHL was required to activate the lasB promoter (Pearson et al., 1995), suggesting that another factor in PAO1 may be required for lasB activation. The P. aeruginosa factor 2 was structurally identified to be N-butyrylhomoserine lactone (BHL) (Pearson et al., 1995). BHL was not shown to interact with LasR protein directly to activate lasB gene expression, nor does it directly regulate the latter (Pearson et al., 1995), triggering another hunt for its cognate receptor. Within the same year, RhlR, a regulatory protein encoded by the rhamnolipid synthase gene cluster rhlABR, was identified to be the cognate receptor of BHL (Ochsner and Reiser, 1995). The rhlI gene, which encodes the biosynthesis of BHL and sharing significant sequence homologies to luxI and lasI, was found at the downstream of the rhlABR cluster. Expression of RhlI could restore the production of several exoproducts such as elastase, pyocyanin, hemolysin and rhamnolipids, and both RhlI and RhlR are required for the full activation of the rhlABR and lasB promoters (Brint and Ohman, 1995; Ochsner and Reiser, 1995).
The las and rhl quorum sensing systems
These key discoveries in P. aeruginosa QS systems inspired further researches on their functions, regulons and the molecular mechanisms with which the las and rhl circuits activate the expression of QS-responsive genes. The results showed that upon binding with the respective autoinducers OdDHL and BHL, the receptor proteins LasR and RhlR get activated and form complexes. The LasR-OdDHL and RhlR-BHL complexes bind to the conserved las-rhl boxes residing in the promoters of target genes, thereby activating their transcriptional expression (Schuster and Greenberg, 2007; Whiteley and Greenberg, 2001; Whiteley et al., 1999). Transcriptomic studies based on lasI and rhlI mutants revealed that the regulons are on a continuum, with some genes that respond dramatically well to OdDHL (e.g. lasA), some with BHL specificities (e.g. rhlAB), and some equally well to both signals (Schuster and Greenberg, 2006; Schuster et al., 2003). These genes constitute nearly 10% of P. aeruginosa genome, and therefore accounts for a majority of the physiological processes and virulence phenotypes (Schuster and Greenberg, 2006). Some of these key virulence genes are listed for the convenience of discussion (Table 1).
Table 1.
Examples of quorum sensing (QS) regulated virulence factors and their effects to the human host
| QS regulated gene | Protein or virulence factor | Effects to host during infections | Benefits to P. aeruginosa | References |
|---|---|---|---|---|
| lasB | Elastase | Degradation of elastin, collagen, and other matrix proteins | Extracellular iron acquisition from host proteins | Wolz et al. (1994); Yanagihara et al. (2003) |
| lasA | Protease | Disruption of epithelial barrier | Staphylolytic activity, host immune evasion and enhanced colonization | Kessler et al. (1993); Park et al. (2000) |
| toxA | Exotoxin A | Cell death | Establishment of infection; enhanced colonization | Daddaoua et al. (2012); McEwan et al. (2012) |
| aprA | Alkaline protease | Degradation of host complement system and cytokines | Immune evasion and persistent colonization | Laarman et al. (2012) |
| rhlAB | Rhamnosyl-transferases (rhamnolipids) | Necrosis of host macrophage and polymorphonuclear lymphocytes | Immune evasion; biofilm development | Jensen et al. (2006); Lequette and Greenberg (2005) |
| lecA | Lectin (galactophilic lectin) | Paralysis of airway cilia | Establishment of infection; enhanced colonization | Adam et al. (1997) |
| hcnABC | Hydrogen cyanide | Cellular respiration arrest; Poorer lung function | Enhanced colonization | Ryall et al. (2008); Solomonson (1981) |
| phzABCDEFG, phzM | Pyocyanin | Oxidative effects dampen host cellular respiration and causes oxidative stress; Paralysis of airway cilia; Delayed inflammatory response to P. aeruginosa infections through neutrophil damage | Establishment of infection; enhanced colonization; immune evasion | Denning et al. (1998); Jackowski et al. (1991); Lau et al. (2004) |
LasR also induces the expression of RsaL, a transcriptional repressor of lasI. Binding of RsaL to the bidirectional rsaL-lasI promoter inhibits the expression of both genes, which generates a negative feedback loop that counteracts the positive signal feedback loop mentioned earlier, thereby balancing the levels of OdDHL (Rampioni et al., 2007). Whilst LasR/OdDHL and RsaL do not compete for the same binding site on the lasI promoter region, the repression by RsaL is stronger than the activation by LasR (Rampioni et al., 2007). RsaL also inhibits the expression of some QS target genes such as biosynthetic genes of pyocyanin and cyanide (Rampioni et al., 2007). A range of positive and negative regulatory proteins were subsequently identified and they control the las and rhl systems in a variety of ways. Noteworthy are the regulatory effects of QscR and VqsR, which are homologues of LuxR. QscR forms heterodimers with LasR/OdDHL and RhlR/BHL and prevents their binding with the promoter DNA of downstream responsive genes, therein dampening the las and rhl QS signalling effects (Ledgham et al., 2003a). QscR also binds to OdDHL and utilize it for activating its own regulon (Chugani et al., 2001; Fuqua, 2006; Schuster and Greenberg, 2006). VqsR is a positive regulator of the las QS system and is itself regulated by the LasR/OdDHL complex (Li et al., 2007). More recently, an anti-activator QslA was identified, which binds to LasR via protein-protein interaction and prevents the interaction of the latter with promoter DNA of the las responsive genes. The inhibitory effect of QslA on LasR is irrespective of OdDHL concentrations. By disrupting the ability of LasR to trigger the expression of downstream genes and cause a QS response, QslA controls the overall QS activation threshold (Seet and Zhang, 2011). There are quite a few other super-regulators of the AHL-based QS systems which are summarized in the table below (Table 2). In addition, quorum quenching enzymes, which degrade AHL signals, the AHL-acylases PvdQ and QuiP, are also involved in balancing the level of AHL signals in P. aeruginosa (Huang et al., 2006; Sio et al., 2006).
Table 2.
Super-regulators of QS in P. aeruginosa
| Regulator | Mechanism of action | References |
|---|---|---|
| AlgR2 | Negative transcriptional regulator of lasR and rhlR | Ledgham et al. (2003a); Westblade et al. (2004) |
| DksA | Negative transcriptional regulator of rhlI | Branny et al. (2001); Jude et al. (2003); van Delden et al. (2001) |
| GacA/GacS | Positive transcriptional regulator of lasR and rhlR | Parkins et al. (2001); Reimmann et al. (1997) |
| MvaT | Negative transcriptional regulator (global regulation) | Diggle et al. (2002) |
| QscR | Negative regulator (anti-activator) of LasR protein | Chugani et al. (2001); Ledgham et al. (2003b) |
| QslA | Negative regulator (anti-activator) of LasR and PqsR proteins | Seet and Zhang (2011) |
| QteE | Negative post-translational regulator of LasR and RhlR | Siehnel et al. (2010) |
| RpoN | Negative transcriptional regulator of lasRI and rhlRI | Heurlier et al. (2003); Thompson et al. (2003) |
| RpoS | Negative transcriptional regulator of rhlI | Latifi et al. (1996); Schuster et al. (2004); Whiteley et al. (2000) |
| RsaL | Negative transcriptional regulator of lasI | Bertani and Venturi (2004); de Kievit et al. (1999) |
| RsmA | Negative transcriptional regulator of lasI | Pessi et al. (2001) |
| Vfr | Positive transcriptional regulator of lasR and rhlR | Albus et al. (1997) |
| VpsR | Positive transcriptional regulator of lasI | Juhas et al. (2004) |
Quinolone-based intercellular signaling
The third QS signal, PQS, was purified and characterized in 1999 by Pesci and co-workers when they observed that spent culture media from wild type PAO1 causes a dramatic induction of lasB expression in a lasR mutant of P. aeruginosa, which could not be mimicked by OdDHL or BHL (Pesci et al., 1999). PQS is structurally identified as 2-heptyl-3-hydroxy-4-quinolone, and it is chemically unique from the AHL signals of the las and rhl systems. Originally studied as an antibacterial molecule (Cornforth and James, 1956; Lightbown and Jackson, 1956), this is the first instance that a 4-quinolone compound was reported as a signalling molecule in bacteria. The PQS synthesis cluster has been identified to consist of pqsABCD, phnAB and pqsH (Gallagher et al., 2002). Shortly after the identification of PQS signal, the receptor PqsR (then known as MvfR) has been implicated in the regulation of PQS production (Cao et al., 2001). PqsA is an anthranilate-coenzyme A ligase (Coleman et al., 2008; Gallagher et al., 2002), which activates anthranilate to form anthraniloyl-coenzyme A, initiating the first step of the PQS biosynthesis. A pqsA mutant does not produce any akyl-quinolones (AQs) (Deziel et al., 2004). PqsB, PqsC and PqsD are probable 3-oxoacyl-(acyl carrier protein) synthases and they mediate the conversion of anthranilate into 2-heptyl-4-quinolone (HHQ) by incorporation of β-ketodecanoic acid (Deziel et al., 2004; Gallagher et al., 2002). HHQ is the precursor of PQS and can be intercellularly transmitted between P. aeruginosa cells. HHQ is converted into PQS by the action of PqsH, a putative flavin-dependent monooxygenase that purportedly hydroxylates HHQ at the 3-position (Deziel et al., 2004; Dubern and Diggle, 2008; Gallagher et al., 2002; Schertzer et al., 2009). The transcription of pqsH is controlled by LasR, implying that the PQS system is controlled by the las system (Schertzer et al., 2009). PqsL is also predicted to be a monooxygenase and is most likely to be involved in the synthesis of the AQ N-oxides, (e.g. 4-hydroxy-2-heptylquinoline-N-oxide, HQNO) (Lépine et al., 2004). Disruption in PqsL caused an overproduction of PQS (D’Argenio et al., 2002), probably owing to a blocked AQ N-oxide pathway which leads to an accumulation of HHQ (Deziel et al., 2004; Lépine et al., 2004). In certain strains of P. aeruginosa, accumulation of PQS and HHQ leads to autolysis and cell death (D’Argenio et al., 2002; D’Argenio et al., 2007; Whitchurch et al., 2005). The role of PqsE remains largely unknown, which is a probable metallo-β-lactamase. Mutation of pqsE does not affect PQS biosynthesis (Gallagher et al., 2002), but the mutants failed to respond to PQS (Diggle et al., 2003; Farrow et al., 2008; Gallagher et al., 2002), and did not express the PQS-controlled phenotypes such as pyocyanin and PA-IL lectin production. In contrast, overexpression of PqsE alone led to enhanced pyocyanin and rhamnolipid production, which is otherwise dependent on the PQS signaling system (Farrow et al., 2008). These puzzling phenomena need to be further investigated for elucidating the role of PqsE in the bacterial physiology and virulence.
PqsR is a LysR-type transcriptional regulator that binds to the promoter region of pqsABCDE operon and directly controls the expression of the operon (Cao et al., 2001; Gallagher et al., 2002). The expression of pqsR is in turn controlled by LasR/OdDHL (Camilli and Bassler, 2006). PqsR is the cognate receptor of PQS and also its co-inducer, as the activity of PqsR in inducing the expression of pqsABCDE is dramatically increased when PQS is bound by the receptor (Wade et al., 2005; Xiao et al., 2006b). HHQ was also found to be able to bind to and induce the expression of PqsR, though it does so with ~ 100-fold less potency than PQS (Wade et al., 2005; Xiao et al., 2006a). Mutation of pqsR resulted in non-production of any AQs and pyocyanin (Cao et al., 2001; Gallagher et al., 2002; Schertzer et al., 2009; von Bodman et al., 2008), indicating that PqsR is essential for executing PQS signal transduction.
The importance of pqs signaling system in the bacterial infection has been illustrated by a range of studies. Null mutation of the pqs system resulted in reduced biofilm formation and decreased production of virulence factors such as pyocyanin, elastase, PA-IL lectin and rhamnolipids (Cao et al., 2001; Diggle et al., 2003; Rahme et al., 2000; Rahme et al., 1997). PQS is also required for full virulence towards plants (Cao et al., 2001), nematodes (Gallagher et al., 2002) and mice (Cao et al., 2001; Lau et al., 2004). In burn-wound mouse models, the killing abilities of pqsA are attenuated compared to the wild type parental strain (Déziel et al., 2005; Xiao et al., 2006b). Intriguingly, the pqsH mutant did not result in a decrease in virulence in burn-wound mouse model (Xiao et al., 2006b), but displayed a reduced killing on nematodes (Gallagher et al., 2002), hence the importance of PQS in regulation of virulence remains debatable. PQS, its precursor HHQ, and the derivative HQNO (4-hydroxy-2-heptylquinoline-N-oxide), are often found in the sputum, bronchoalveolar fluid and mucopurulent fluid of cystic fibrosis sufferers (Collier et al., 2002). Taken together, this could suggest that the precursors of PQS may play an equally important role as PQS in virulence and infections.
An integrated QS system
Recently, a fourth inter-cellular communication signal has been discovered to be capable of integrating environmental stress cues with the quorum sensing network (Lee et al., 2013). Named as IQS, it belongs to a new class of quorum sensing signal molecules and was structurally established to be 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde. The genes that are involved in IQS synthesis are a non-ribosomal peptide synthase gene cluster ambBCDE. When disrupted, it caused a decrease in the production of PQS and BHL signals, as well as the virulence factors such as pyocyanin, rhamnolipids and elastase. Upon addition of 10 nmol/L IQS to the mutants, these phenotypes could be restored fully, indicating that IQS is a potent inter-cellular communication signal compared with its counterparts (Fig. 2). Further, IQS has been shown to contribute to the full virulence of P. aeruginosa in four different animal host models (mouse, zebrafish, fruitfly and nematode), highlighting the important roles of this new QS system in modulation of bacterial pathogenesis. Importantly, under phosphate depletion stress conditions, IQS was demonstrated to be able to partially take over the functions of the central las system (Lee et al., 2013), providing critical clues in understanding the puzzling phenomenon that the clinical isolates of P. aeruginosa frequently harbour mutated lasI or lasR genes (Ciofu et al., 2010; D’Argenio et al., 2007; Hoffman et al., 2009; Smith et al., 2006).
Figure 2.
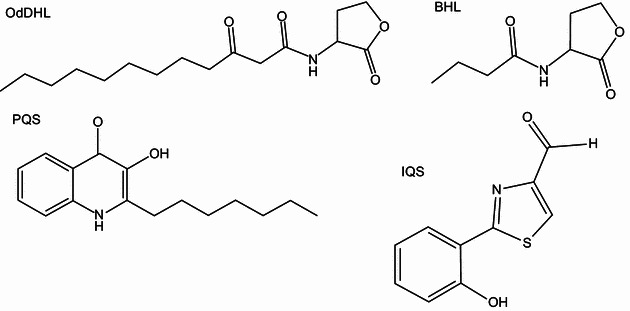
Structures ofP. aeruginosaquorum sensing (QS) signals. Clockwise from left, N-(3-oxododecanoyl)-homoserine lactone (OdDHL); N-butyrylhomoserine lactone (BHL); 2-heptyl-3-hydroxy-4-quinolone (Pseudomonas Quinolone Signal, PQS); 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde (Integrated Quorum Sensing Signal, IQS)
Interconnection between the four QS systems
The QS circuits in P. aeruginosa are organized in a hierarchical manner. At the top of the signalling hierarchy is the las system. When activated by OdDHL, LasR-OdDHL complex multimerizes and activates the transcription of rhlR, rhlI, lasI (hence a positive feedback loop), and other virulence genes that are part of its regulon (Kiratisin et al., 2002; Latifi et al., 1996; Pesci et al., 1997). The RhlR-BHL complex also dimerizes and similarly activates the expression of its own regulon and rhlI, forming the second positive feedback loop (Ventre et al., 2003; Winson et al., 1995). LasR-OdDHL also positively regulates PqsR, the transcriptional regulator of the HHQ/PQS biosynthesis operon pqsABCD, as well as the expression of pqsH, the gene encoding the final converting enzyme of PQS from HHQ (Deziel et al., 2004; Gallagher et al., 2002; Xiao et al., 2006a). PQS, in turn, was found to be able to enhance the transcription of rhlI, thus influencing BHL production and the overall expression of the rhl QS system, thus indirectly modulating the rhl-dependent phenotypes (McKnight et al., 2000; Pesci et al., 1999). Interestingly, pqsR and pqsABCDE expression is inhibited by RhlR/BHL (Cao et al., 2001), suggesting that the ratio of the concentrations between OdDHL and BHL play a decisive role in the dominance of the pqs signaling system (Cao et al., 2001).
With las governing the expression of both pqs and rhl systems, it was often described as being at the top of the QS hierarchy. The rhl system on the other hand, is under the control of both las and pqs, yet many QS-dependent virulence factors are predominantly activated by RhlR-BHL (Latifi et al., 1995; Schuster and Greenberg, 2007; Schuster et al., 2004; Whiteley et al., 1999; Winzer et al., 2000), thus the rhl system functions like a workhorse for the QS command. Since LasR-OdDHL controls the onset and activation of both the pqs and rhl QS circuits, these systems therefore represent a step-wise activation cascade that will be triggered by attainment of a “quorum” in P. aeruginosa cultures. The recently identified IQS was also found to be tightly controlled by LasRI under rich medium conditions. Disruption of either lasR or lasI completely abrogates the expression of ambBCDE and the production of IQS (Lee et al., 2013) (Fig. 3).
Figure 3.
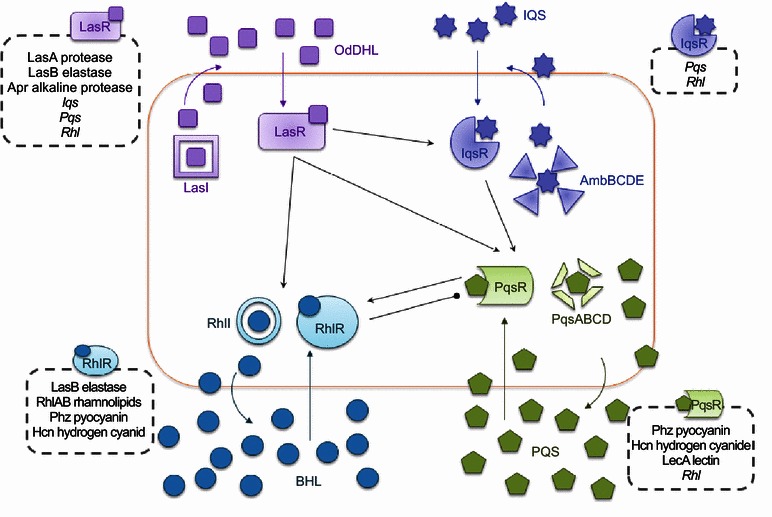
Schematic representation of the four QS signaling networks inP. aeruginosaand their respective regulons. Arrows indicate a stimulatory effect. Perpendicular lines indicate an inhibitory effect
However, exceptions do occur. The lasR mutants were found to have a delayed production of PQS, instead of having an abolished PQS system as previously thought, and PQS could also overcome the dependency on LasR in activating the expression of rhl QS system and production of downstream virulence factors (Diggle et al., 2003). It was subsequently discovered that this could be due to the effects of RhlR, as the lasR and rhlR double mutant had barely any detectable PQS, but when rhlR was overexpressed, the production of PQS, as well as virulence factors such as LasB elastase and LasA protease, are restored (Dekimpe and Deziel, 2009). RhlR was also shown to upregulate the expression of lasI, the most-specific LasR-regulated gene, and OdDHL production was consequently increased (Dekimpe and Deziel, 2009). This indicates that compensation by the rhl QS system could override this hierarchy and maintain the expression of QS-dependent virulence factors in spite of a non-functional central las system. Similarly, the dominance of las on IQS signal production was reversed when P. aeruginosa was subjected to phosphate depletion stress, and the iqs system could up-regulate the expression of pqs and rhl systems and the production of QS-dependent virulence factors in the lasI or lasR mutant (Lee et al., 2013). Low phosphate levels also elevate IQS production in wild type P. aeruginosa (Lee et al., 2013). These findings highlight the importance of environmental factors in modulating the bacterial QS systems and the plasticity of the QS networks in accommodation and exploitation of environmental changes for the benefit of bacterial pathogens. The next section is dedicated to discussion of such examples in details with the aim to shed light on understanding the complicated and sophisticated QS regulatory mechanisms in P. aeruginosa.
Environmental triggers and the QS responses
Evidence is accumulating that environmental stress conditions could exert substantial influence on the QS systems of P. aeruginosa. Starvation, phosphate and iron depletion are known to promote the expression and activity of RhlR in the absence of lasR (Jensen et al., 2006; Van Delden et al., 1998). More recently, it was found that phosphate depletion could induce IQS production even in the absence of functional las system (Lee et al., 2013). This discovery is clinically significant as substantial amount of P. aeruginosa chronic infection isolates bear a loss-of-function las system (Cabrol et al., 2003; Denervaud et al., 2004; Hamood et al., 1996; Schaber et al., 2004; Smith et al., 2006). The roles and the molecular mechanisms with which various environmental cues and host immune factors modulate the QS systems of P. aeruginosa will be discussed separately in the following sections.
Phosphate-depletion stress
Phosphate is essential for all living cells owing to its key roles in signal transduction reactions such as phospho-relay, and as an essential component of the energy molecule ATP, nucleotides, phospholipids and other important biomolecules. Foreseeably, bacterial pathogens may encounter strong competition for free phosphates from host cells during the process of pathogen-host interaction. Therefore, the ability to withstand phosphate starvation and the response mechanisms of harnessing phosphate from external sources is critical for P. aeruginosa survival and establishment of infections. As a result, phosphate-depletion stress has been shown to have far-reaching effects on QS signalling profiles, gene expression, physiology and virulence of bacterial pathogens (Chugani and Greenberg, 2007; Frisk et al., 2004; Jensen et al., 2006; Lee et al., 2013; Zaborin et al., 2009).
When facing with phosphate limitation, P. aeruginosa exhibits increased swarming motility and cytotoxicity towards the human bronchial epithelial cell line 16HBE14o- (Bains et al., 2012), attesting to the strong responses phosphate deprivation could elicit from the pathogen. Additionally, phosphate depletion stress was shown to prompt the up-regulation of iron chelator pyoverdine biosynthesis, which in turn, could result in the inactivation of the phosphate acquisition pathway. When the pyoverdine signalling pathway was interrupted, pyochelin biosynthesis was in turn increased as compensation (Zaborin et al., 2009). This resulted in high amounts of ferric ions to be acquired. Coupled with the dramatic increase in PQS production (part of the phosphate starvation response), the lethal PQS-Fe(III) red coloured complex was formed. When ingested, the red-spotted P. aeruginosa caused rapid mortality in C. elegans, a phenomenon known as “red death” (Zaborin et al., 2009). Such signalling cross-talk demonstrates the interconnectivity between the phosphate and iron acquisition systems in P. aeruginosa, the investment in resources the bacteria makes to maintain their homeostasis, and the deleterious effects on the host when the fine balance is tipped.
The lack of phosphate also dramatically activates the expression of pqsR and the PqsR-regulated pqsABCDE and phnAB genes. Along with the enhanced pqs system, the expression of QS-associated virulence genes responsible for the synthesis of rhamnolipids, phenazines, cyanide, exotoxin A and LasA protease are similarly induced (Bains et al., 2012; Zaborin et al., 2009). This was thought to lead to the acute mortality rate of the host organism Caenorhabditis elegans after being infected by P. aeruginosa that were grown in phosphate starvation medium (Zaborin et al., 2009). These observations correlate and could well be explained by our current knowledge on IQS. With depletion in phosphate, expression of iqs system is induced (Lee et al., 2013), which in turn triggers an up-regulation of the downstream pqs and rhl QS systems, and eventually, an observed boost in QS-associated virulence factors production and killing rates.
It is crucial to note that the two-component sensor-response regulator system PhoBR plays an indispensable role in detection and signal transduction of phosphate stress cues (Anba et al., 1990; Filloux et al., 1988; Hsieh and Wanner, 2010), as disruption of phoB completely abolished the virulence of P. aeruginosa towards C. elegans (Zaborin et al., 2009), and dramatically diminished its swarming motility and cytotoxicity (Bains et al., 2012). PhoB (and the pho regulon) was also shown to participate in the inhibition of biofilm formation, c-di-GMP signal degradation and repression of the type III secretion systems (Haddad et al., 2009), all of which could significantly affect the clinical outcome during P. aeruginosa infections (Abe et al., 2005; Costerton, 2001; Hauser et al., 2002; Hueck, 1998; Roy-Burman et al., 2001). The phoB mutant grows poorly in low phosphate medium and failed to produce the QS-dependent virulence factor pyocyanin (Lee and Zhang, unpublished data). Remarkably, PhoBR is indispensable for coordinating the las-independent, phosphate-dependent IQS signalling activation, wherein the “IQS phenotype” would be abolished in a phoB mutant (Lee et al., 2013). The PhoBR-IQS loop could also explain the observations by Jensen and co-workers, who reported that low phosphate prompted an enhancement of the rhl QS system even when las was functionally absent and this is coordinated by PhoB (Jensen et al., 2006).
Iron and PQS signaling system
Unlike phosphate, the modulatory effect of iron starvation on P. aeruginosa QS networks appears to be less direct. A deficiency in iron does lead to notable increases in the expression of genes involved in iron acquisition (ferric uptake siderophores, pyochelin and pyoverdine; ferrous iron transporters like haem and feo), exoenzymes that could cleave iron-bound host proteins (alkaline protease, lasB elastase) and other redox enzymes and toxins (exotoxin A) (Ochsner et al., 2002). Further, the iron depletion stress response was found to lead to an inhibition of oxygen transfer from the atmosphere to liquid P. aeruginosa cultures, thus protecting bacteria cells from oxidative stress. Production of the virulence factor LasB elastase is also significantly increased in these iron depletion cultures (Kim et al., 2003). Although some of the upregulated virulence factors, like alkaline protease and elastase, are known to be regulated by the QS systems of P. aeruginosa (see Table 1), a direct link between iron deprivation and up-regulation of central QS genes such as lasI, lasR, rhlI or rhlR has yet to be found. In a report by Diggle and co-workers, the PQS molecules were found to function as an iron trap when secreted into the extracellular milieu of P. aeruginosa (Diggle et al., 2007). This was hypothesized to serve the purpose of storing up free ferric ions which could subsequently be internalized into the cells by the siderophores, in order to safeguard against a sudden dip in iron concentration. Iron starvation could also trigger a Fur-dependent de-repression of the small regulatory RNAs prrF1 and prrF2 expression. PrrF1 and PrrF2 bind to and inhibit the expression of antABC genes which encode for the anthranilate degradation enzymes AntABC. Since anthranilate is the precursor of PQS biosynthesis, inhibition of its degradation could lead to accumulation of anthranilate, which consequently elevates the concentration of HHQ and PQS in the bacteria cells. This in turn might boost the PQS-PqsR signaling pathway. PqsR was also found to inhibit antABC expression, albeit in a PrrF1,2-independent manner (Oglesby et al., 2008). Taken together, the above findings seem to suggest that iron depletion stress may modulate bacterial virulence through the pqs system, which awaits further investigations.
ANR and oxygen deprivation
Low oxygen tension is a key factor affecting cyanide biosynthesis (cyanogenesis) in P. aeruginosa (Castric, 1994; Castric, 1983). The final product, hydrogen cyanide (HCN), is a highly potent extracellular virulence factor and contributes to high mortality rates during infection of host organisms (Ryall et al., 2008; Solomonson, 1981). Additionally, increase in P. aeruginosa cell density was also shown to remarkably elevate expression of hcnABC, the synthase genes for HCN, and reaches its optimum levels during the transit from exponential to stationary growth phase of the bacteria (Castric et al., 1979). This may suggest a cooperative link between oxygen deprivation and QS in the regulatory mechanism of cyanogenesis, which was subsequently demonstrated through characterization of ANR, a transcriptional regulator associated with bacterial anaerobic growth.
ANR, which is converted into its active form when oxygen tension is low, is a key regulator controlling the expression of arginine deiminase and nitrate reductase. ANR belongs to the FNR (fumarate and nitrate reductase regulator) family of transcriptional regulators and is the main transcriptional regulator that acts in parallel with the QS systems for the expression of hydrogen cyanide biosynthesis genes (Pessi and Haas, 2000). ANR, together with LasR-OdDHL or RhlR-BHL, bind to the promoter region of the hcnABC cluster, exhibiting a synergistic effect brought upon by oxygen limitation stress. Further, the PRODORIC promoter analysis programme predicted the FNR/ANR binding consensus sequences in up to 25% of the predicted QS-controlled promoters, implying that ANR might be an important co-regulator of the QS-dependent virulence genes in anaerobic environments (Schuster and Greenberg, 2006).
Starvation stress
When exposed to unfavourable environments and nutrient starvation, P. aeruginosa must rapidly cope and elicit a prompt response to modify their metabolic profiles for survival. This process is termed as the stringent response and brings about diverse effects ranging from inhibition of growth processes to cell division arrest (Joseleau-Petit et al., 1999; Svitil et al., 1993) and more importantly, a premature activation of the P. aeruginosa QS systems that is independent of cell-density (van Delden et al., 2001). The QS signals BHL and N-hexanoyl-homoserine lactone (HHL) are prematurely produced and PQS synthesis inhibited (Baysse et al., 2005). The spike in BHL QS signal is likely to result in the concomitant increase in production of downstream virulence factors elastase and rhamnolipids (Schafhauser et al., 2014).
The QS-based response is mediated by the stringent response protein RelA. In face of amino acid shortage, uncharged tRNA triggers the activity of the ribosome-associated RelA, which in turn synthesizes ppGpp (nucleotide guanosine 3’,5’-bisdiphosphate), an intracellular signal that enables the bacteria cell to self-perceive their inability in synthesis of proteins (Gentry and Cashel, 1996). When overexpressed, RelA leads to early transcriptional expression of the lasR and rhlR genes, as well as production of QS signals OdDHL and BHL (van Delden et al., 2001), hence leading to the overproduction of the aforementioned QS-dependent virulence factors. Furthermore, RelA and ppGpp was also shown to coordinate the stress response associated with alterations in membrane phospholipid composition and loss of membrane fluidity. When the phospholipid biosynthesis protein LptA was deleted, an increase in relA expression and ppGpp production was observed, which resulted in a premature activation of BHL and HHL QS signals biosynthesis (Baysse et al., 2005).
In a recent study, Schafhauser and co-workers observed that the synthesis of the starvation signal ppGpp negatively regulates the biosynthesis of HHQ and PQS signals, and is required for full expression of both the las and rhl QS systems (Schafhauser et al., 2014). In the relA and spoT double mutant that is unable to synthesize ppGpp, both the las and rhl QS systems are down-regulated, and the production of QS-dependent virulence factors rhamnolipid and elastase are reduced (Schafhauser et al., 2014). Whilst it has been previously reported that ppGpp increases the expression of LasR and RhlR and the resultant downstream factors (Baysse et al., 2005; van Delden et al., 2001), repression on the pqs system by ppGpp is somewhat unexpected. More experiments are required to investigate on the significance of this selective dampening of the pqs system.
Response to host factors
It has been traditionally thought that opportunistic pathogens such as Pseudomonas aeruginosa invade hosts with a weakened immune system or attenuated epithelial barrier in a passive manner, until an important observation was made by Wu and colleagues that P. aeruginosa major outer-membrane protein OprF is able to recognize and bind to human T cell-based cytokine interferon gamma (IFN-γ). This in turn activates the rhl QS system and substantially enhances the expression of lecA and production of its encoded virulence protein, galactophilic lectin. Pyocyanin, an additional QS-regulated virulence factor, was also found to be up-regulated in the presence of IFN-γ (Wu et al., 2005). Although IFN-γ was the only cytokine found to activate the rhl QS system and it is not known whether and if yes, how the upstream las and pqs networks are affected, this work presents a direct evidence of the interactions between host-derived immune factors and bacterial membrane proteins, which consequently leads to QS-based responses. In another example, dynorphin, an endogenous κ-receptor agonist, was found to penetrate the bacterial membrane and potently induce the expression of pqsR and pqsABCDE, and lead to increased biosynthesis of PQS, HHQ and the related derivative HQNO. The growth advantage against probiotic gut microorganisms Lactobacillus spp. and virulence towards C. elegans is also remarkably enhanced when P. aeruginosa is exposed to dynorphin (Zaborina et al., 2007). This finding is of particular significance to P. aeruginosa caused gut infections as dynorphin is usually in high concentrations in the intestinal mucosa and epithelial cells, attesting to the remarkable mechanisms utilized by the bacteria to enhance virulence by integrating host opioids into its existing QS circuitry.
Further, human hormones, particularly the C-type natriuretic peptide (CNP) that is produced by endothelial cells and used for maintaining body fluid homeostasis and blood pressure control, was demonstrated to have positive effects on P. aeruginosa virulence. Through activation of the P. aeruginosa membrane natriuretic peptides sensor, CNP induces a rise in intracellular cAMP concentration and lead to the activation of the global virulence activator Vfr, which either alone or together with another regulator PtxR, enhances the synthesis of QS signals OdDHL and BHL, and inhibits the production of PQS. Vfr also drives the increased expression of virulence factors hydrogen cyanide and lipopolysaccharide, thereby elevating the mortality rate in C. elegans infected with CNP-treated P. aeruginosa (Blier et al., 2011).
Most recently, the human host defence peptide LL-37, the only cathelicidin class of cationic antimicrobial peptides synthesized by phagocytes, epithelial cells and keratinocytes, was revealed to exert a positive effect on P. aeruginosa QS and virulence profiles. When stimulated by exogenous LL-37 at physiological concentrations, P. aeruginosa exhibits heightened production of virulence factors pyocyanin, hydrogen cyanide, elastase and rhamnolipids. The PQS signal level is also elevated. LL-37 was also found to decrease the susceptibility of the bacteria to gentamicin and ciprofloxacin antibiotics. These phenotypes were suggested to be mediated by the quinolone response protein and virulence regulator PqsE (Strempel et al., 2013).
Summary and perspectives
Pseudomonas aeruginosa is one of the most notorious opportunistic human pathogens as it employs a variety of virulence factors and mechanisms during infection (Fig. 1). The type of virulence pathways activated is often dependent on the environment conditions and stresses the bacteria encounter. Extensive research over the past two decades has documented numerous instances of environmental cues including the biostresses of host origin, which could dramatically influence the virulence phenotypes of P. aeruginosa. The findings from recent research progresses suggest that these effects could largely be through modulation of the bacterial QS network, which comprises at least four QS signaling mechanisms including las, iqs, pqs and rhl. In particular, the most recently identified IQS highlights how a bacterial QS system could integrate environmental cues with bacterial quorum information. These four systems interact closely with one another giving rise to an intricately linked intercellular communication network. Such a complicated and multi-component QS network may enable P. aeruginosa to accommodate various environmental cues and biostresses (Fig. 4).
Figure 4.
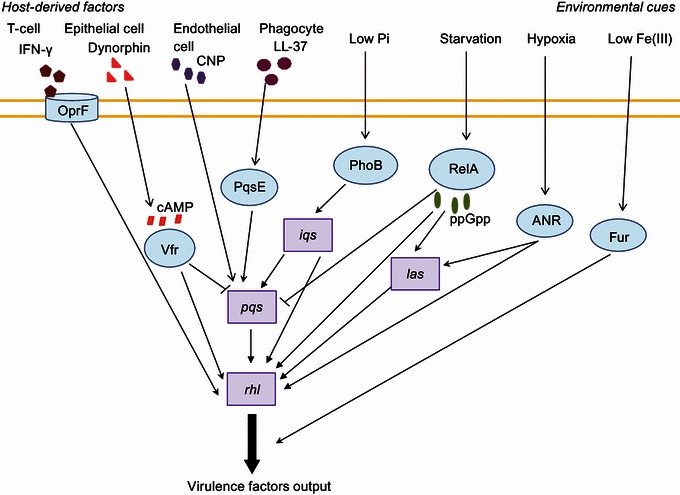
Schematic representation of how environmental conditions and host factors influence theP. aeruginosaQS signaling hierarchy. For simplicity, the QS systems are represented as a whole unit, namely, las, iqs, pqs and rhl
Previous efforts in the design of anti-QS therapeutics were focused primarily on inhibition of the las system (Borlee et al., 2010; Mattmann and Blackwell, 2010). However, in light of the recent discovery that IQS could replace the functions of las in conditions that closely mimics host infection (Lee et al., 2013), coupled with the high mutation frequencies of lasR typical of P. aeruginosa clinical isolates (Ciofu et al., 2010; D’Argenio et al., 2007; Hoffman et al., 2009; Smith et al., 2006), it becomes clear that the ongoing strategies targeting the las system is insufficient, and that the prevalence of IQS system in clinical isolates should be evaluated to ensure development of potent anti-QS therapeutics. Furthermore, we should also keep in mind that there are many unknowns that require further investigations for clear understanding of how the bacterial QS network could act on various environmental cues in regulation of bacterial virulence and biofilm formation. For example, it is not clear how IQS could regulate the downstream pqs and rhl signaling systems and what is the impact of iqs system on the virulence of clinical isolates. Similarly, much remains to be done in understanding whether and if yes, how environmental cues could modulate the las, pqs and rhl systems. Recognition of how the external stressors change the way the QS network is connected may generate tremendous impact on the perspective from which therapeutic interventions could be developed, especially those environmental cues almost always encountered by P. aeruginosa during infections of the host. For instance, successful establishment of an infection and colonization of the cystic fibrosis lung chambers would require P. aeruginosa strains to sense, withstand and respond to deprivation of iron, phosphate, and attacks by lung macrophage-derived factors (Campodonico et al., 2008; Konings et al., 2013; Krieg et al., 1988). Then, as the pathogen transits into a long-term, chronic infection mode, the stresses of living within a biofilm matrix may include oxygen deprivation and nutrient limitation (Jackson et al., 2013; Sauer et al., 2004). Investigation along this line will further advance our understanding of the complicated and sophisticated QS regulatory mechanisms and may continue to generate unexpected interesting findings.
Acknowledgements
This work was funded by the Biomedical Research Council, Agency for Science, Technology and Research (A*STAR), Singapore, and by the National Natural Science Foundation of China (Grant No. 31330002). We apologize to the scientists who made contributions to the field, but their works have not been cited due to space limitations.
Compliance with Ethics Guidelines
Jasmine Lee and Lian-Hui Zhang declare that they have no conflict of interest and this article does not contain any studies with human or animal subjects performed by the any of the authors.
Abbreviations
- AHL
acylhomoserine lactone
- CNP
C-type natriuretic peptide
- HCN
hydrogen cyanide
- IFN-γ
interferon gamma
- IQS
integrated quorum sensing system
- QS
quorum sensing
- QSIs
quorum sensing inhibitors
References
- Abe A, Matsuzawa T, Kuwae A. Type-III effectors: sophisticated bacterial virulence factors. C R Biol. 2005;328:413–428. doi: 10.1016/j.crvi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Adam EC, Mitchell BS, Schumacher DU, Grant G, Schumacher U. Pseudomonas aeruginosa II lectin stops human ciliary beating: therapeutic implications of fucose. Am J Respir Crit Care Med. 1997;155:2102–2104. doi: 10.1164/ajrccm.155.6.9196121. [DOI] [PubMed] [Google Scholar]
- Albus AM, Pesci EC, Runyen-Janecky LJ, West SE, Iglewski BH. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anba J, Bidaud M, Vasil ML, Lazdunski A. Nucleotide sequence of the Pseudomonas aeruginosa phoB gene, the regulatory gene for the phosphate regulon. J Bacteriol. 1990;172:4685–4689. doi: 10.1128/jb.172.8.4685-4689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains M, Fernandez L, Hancock RE. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol. 2012;78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baysse C, Cullinane M, Dénervaud V, Burrowes E, Dow JM, Morrissey JP, Tam L, Trevors JT, O’Gara F. Modulation of quorum sensing in Pseudomonas aeruginosa through alteration of membrane properties. Microbiology. 2005;151:2529–2542. doi: 10.1099/mic.0.28185-0. [DOI] [PubMed] [Google Scholar]
- Bertani I, Venturi V. Regulation of the N-acyl homoserine lactone-dependent quorum-sensing system in rhizosphere Pseudomonas putida WCS358 and cross-talk with the stationary-phase RpoS sigma factor and the global regulator GacA. Appl Environ Microbiol. 2004;70:5493–5502. doi: 10.1128/AEM.70.9.5493-5502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier A-S, Veron W, Bazire A, Gerault E, Taupin L, Vieillard J, Rehel K, Dufour A, Le Derf F, Orange N. C-type natriuretic peptide modulates quorum sensing molecule and toxin production in Pseudomonas aeruginosa. Microbiology. 2011;157:1929–1944. doi: 10.1099/mic.0.046755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo RA, Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clinical Infectious Diseases. 2006;43(Suppl 2):S49–S56. doi: 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- Borlee BR, Geske GD, Blackwell HE, Handelsman J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl Environ Microbiol. 2010;76:8255–8258. doi: 10.1128/AEM.00499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branny P, Pearson JP, Pesci EC, Kohler T, Iglewski BH, Van Delden C. Inhibition of quorum sensing by a Pseudomonas aeruginosa dksA homologue. J Bacteriol. 2001;183:1531–1539. doi: 10.1128/JB.183.5.1531-1539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brint JM, Ohman DE. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. Journal of Bacteriology. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrol S, Olliver A, Pier GB, Andremont A, Ruimy R. Transcription of quorum-sensing system genes in clinical and environmental isolates of Pseudomonas aeruginosa. J Bacteriol. 2003;185:7222–7230. doi: 10.1128/JB.185.24.7222-7230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campodonico VL, Gadjeva M, Paradis-Bleau C, Uluer A, Pier GB. Airway epithelial control of Pseudomonas aeruginosa infection in cystic fibrosis. Trends Mol Med. 2008;14:120–133. doi: 10.1016/j.molmed.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao JG, Meighen EA. Biosynthesis and stereochemistry of the autoinducer controlling luminescence in Vibrio harveyi. J Bacteriol. 1993;175:3856–3862. doi: 10.1128/jb.175.12.3856-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Krishnan G, Goumnerov B, Tsongalis J, Tompkins R, Rahme LG. A quorum sensing-associated virulence gene of Pseudomonas aeruginosa encodes a LysR-like transcription regulator with a unique self-regulatory mechanism. Proc Natl Acad Sci USA. 2001;98:14613–14618. doi: 10.1073/pnas.251465298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric PA. Hydrogen cyanide production by Pseudomonas aeruginosa at reduced oxygen levels. Can J Microbiol. 1983;29:1344–1349. doi: 10.1139/m83-209. [DOI] [PubMed] [Google Scholar]
- Castric P. Influence of oxygen on the Pseudomonas aeruginosa hydrogen cyanide synthase. Curr Microbiol. 1994;29:19–21. [Google Scholar]
- Castric PA, Ebert RF, Castric KF. The relationship between growth phase and cyanogenesis in Pseudomonas aeruginosa. Curr Microbiol. 1979;2:287–292. [Google Scholar]
- Chernish RN, Aaron SD. Approach to resistant gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr Opin Pulm Med. 2003;9:509–515. doi: 10.1097/00063198-200311000-00011. [DOI] [PubMed] [Google Scholar]
- Chugani S, Greenberg E. The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microbial pathogenesis. 2007;42:29–35. doi: 10.1016/j.micpath.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O, Mandsberg LF, Bjarnsholt T, Wassermann T, Høiby N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology. 2010;156:1108–1119. doi: 10.1099/mic.0.033993-0. [DOI] [PubMed] [Google Scholar]
- Coleman JP, Hudson LL, McKnight SL, Farrow JM, Calfee MW, Lindsey CA, Pesci EC. Pseudomonas aeruginosa PqsA is an anthranilate-coenzyme A ligase. J Bacteriol. 2008;190:1247–1255. doi: 10.1128/JB.01140-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier DN, Anderson L, McKnight SL, Noah TL, Knowles M, Boucher R, Schwab U, Gilligan P, Pesci EC. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol Lett. 2002;215:41–46. doi: 10.1111/j.1574-6968.2002.tb11367.x. [DOI] [PubMed] [Google Scholar]
- Cook JMI, Harragan B (1992) In: Proceedings of the 92nd annual meeting of the american society for microbiology. Paper presented at 92nd annual meeting of the american society for microbiology, New Orleans
- Cornforth JW, James AT. Structure of a naturally occurring antagonist of dihydrostreptomycin. The Biochemical Journal. 1956;63:124–130. doi: 10.1042/bj0630124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Popat R, McNally L, Gurney J, Scott-Phillips TC, Ivens A, Diggle SP, Brown SP. Combinatorial quorum sensing allows bacteria to resolve their social and physical environment. Proc Natl Acad Sci. 2014;3(4):220–227. doi: 10.1073/pnas.1319175111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9:50–52. doi: 10.1016/s0966-842x(00)01918-1. [DOI] [PubMed] [Google Scholar]
- Daddaoua A, Fillet S, Fernández M, Udaondo Z, Krell T, Ramos JL. Genes for carbon metabolism and the ToxA virulence factor in Pseudomonas aeruginosa are regulated through molecular interactions of PtxR and PtxS. PLoS One. 2012;7:e39390. doi: 10.1371/journal.pone.0039390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Argenio DA, Wu M, Hoffman LR, Kulasekara HD, Deziel E, Smith EE, Nguyen H, Ernst RK, Larson Freeman TJ, Spencer DH, et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol. 2007;64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kievit T, Seed PC, Nezezon J, Passador L, Iglewski BH. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J Bacteriol. 1999;181:2175–2184. doi: 10.1128/jb.181.7.2175-2184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe V, Deziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology. 2009;155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- Denervaud V, TuQuoc P, Blanc D, Favre-Bonte S, Krishnapillai V, Reimmann C, Haas D, van Delden C. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J Clin Microbiol. 2004;42:554–562. doi: 10.1128/JCM.42.2.554-562.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Wu J, Tao F, Zhang LH. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem Rev. 2011;111:160–173. doi: 10.1021/cr100354f. [DOI] [PubMed] [Google Scholar]
- Denning GM, Wollenweber LA, Railsback MA, Cox CD, Stoll LL, Britigan BE. Pseudomonas pyocyanin increases interleukin-8 expression by human airway epithelial cells. Infect Immun. 1998;66:5777–5784. doi: 10.1128/iai.66.12.5777-5784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci USA. 2004;101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E, Gopalan S, Tampakaki AP, Lépine F, Padfield KE, Saucier M, Xiao G, Rahme LG. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Winzer K, Lazdunski A, Williams P, Camara M. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J Bacteriol. 2002;184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Winzer K, Chhabra SR, Worrall KE, Cámara M, Williams P. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol Microbiol. 2003;50:29–43. doi: 10.1046/j.1365-2958.2003.03672.x. [DOI] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Cámara M. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Doshi HK, Chua K, Kagda F, Tambyah PA. Multi drug resistant pseudomonas infection in open fractures post definitive fixation leading to limb loss: a report of three cases. International Journal of Case Reports and Images (IJCRI) 2011;2:1–6. [Google Scholar]
- Dubern JF, Diggle SP. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol Biosyst. 2008;4:882–888. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- Eberhard A. Inhibition and activation of bacterial luciferase synthesis. J Bacteriol. 1972;109:1101–1105. doi: 10.1128/jb.109.3.1101-1105.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Farrow JM, 3rd, Sund ZM, Ellison ML, Wade DS, Coleman JP, Pesci EC. PqsE functions independently of PqsR-Pseudomonas quinolone signal and enhances the rhl quorum-sensing system. J Bacteriol. 2008;190:7043–7051. doi: 10.1128/JB.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A, Bally M, Soscia C, Murgier M, Lazdunski A. Phosphate regulation in Pseudomonas aeruginosa: cloning of the alkaline phosphatase gene and identification of phoB-and phoR-like genes. Mol Gen Genet. 1988;212:510–513. doi: 10.1007/BF00330857. [DOI] [PubMed] [Google Scholar]
- Frisk A, Schurr JR, Wang G, Bertucci DC, Marrero L, Hwang SH, Hassett DJ, Schurr MJ. Transcriptome analysis of Pseudomonas aeruginosa after interaction with human airway epithelial cells. Infect Immun. 2004;72:5433–5438. doi: 10.1128/IAI.72.9.5433-5438.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C. The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity. J Bacteriol. 2006;188:3169–3171. doi: 10.1128/JB.188.9.3169-3171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Iglewski BH. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambello MJ, Kaye S, Iglewski BH. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry DR, Cashel M. Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol Microbiol. 1996;19:1373–1384. doi: 10.1111/j.1365-2958.1996.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Haddad A, Jensen V, Becker T, Haussler S. The Pho regulon influences biofilm formation and type three secretion in Pseudomonas aeruginosa. Environ Microbiol Rep. 2009;1:488–494. doi: 10.1111/j.1758-2229.2009.00049.x. [DOI] [PubMed] [Google Scholar]
- Hamood AN, Griswold J, Colmer J. Characterization of elastase-deficient clinical isolates of Pseudomonas aeruginosa. Infect Immun. 1996;64:3154–3160. doi: 10.1128/iai.64.8.3154-3160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AR, Cobb E, Bodí M, Mariscal D, Vallés J, Engel JN, Rello J. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30:521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- Henry RL, Mellis CM, Petrovic L. Mucoid Pseudomonas aeruginosa is a marker of poor survival in cystic fibrosis. Pediatr Pulmonol. 1992;12:158–161. doi: 10.1002/ppul.1950120306. [DOI] [PubMed] [Google Scholar]
- Hense BA, Kuttler C, Muller J, Rothballer M, Hartmann A, Kreft JU. Does efficiency sensing unify diffusion and quorum sensing? Nature Rev Microbiol. 2007;5:230–239. doi: 10.1038/nrmicro1600. [DOI] [PubMed] [Google Scholar]
- Heurlier K, Dénervaud V, Pessi G, Reimmann C, Haas D. Negative control of quorum sensing by RpoN (σ54) in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:2227–2235. doi: 10.1128/JB.185.7.2227-2235.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y-J, Wanner BL. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol. 2010;13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JJ, Petersen A, Whiteley M, Leadbetter JR. Identification of QuiP, the product of gene PA1032, as the second acyl-homoserine lactone acylase of Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72:1190–1197. doi: 10.1128/AEM.72.2.1190-1197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackowski JT, Szepfalusi Z, Wanner DA, Seybold Z, Sielczak MW, Lauredo IT, Adams T, Abraham WM, Wanner A. Effects of P. aeruginosa-derived bacterial products on tracheal ciliary function: role of O2 radicals. Am J Physiol. 1991;260:L61–L67. doi: 10.1152/ajplung.1991.260.2.L61. [DOI] [PubMed] [Google Scholar]
- Jackson AA, Gross MJ, Daniels EF, Hampton TH, Hammond JH, Vallet-Gely I, Dove SL, Stanton BA, Hogan DA. Anr and its activation by PlcH activity in Pseudomonas aeruginosa host colonization and virulence. J Bacteriol. 2013;195:3093–3104. doi: 10.1128/JB.02169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen V, Löns D, Zaoui C, Bredenbruch F, Meissner A, Dieterich G, Münch R, Häussler S. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and-independent pathways. J Bacteriol. 2006;188:8601–8606. doi: 10.1128/JB.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Yu B, Bainton NJ, Birdsall M, Bycroft BW, Chhabra SR, Cox AJ, Golby P, Reeves PJ, Stephens S, et al. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseleau-Petit D, Vinella D, D’Ari R. Metabolic alarms and cell division in Escherichia coli. J Bacteriol. 1999;181:9–14. doi: 10.1128/jb.181.1.9-14.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude F, Kohler T, Branny P, Perron K, Mayer MP, Comte R, van Delden C. Posttranscriptional control of quorum-sensing-dependent virulence genes by DksA in Pseudomonas aeruginosa. J Bacteriol. 2003;185:3558–3566. doi: 10.1128/JB.185.12.3558-3566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M, Wiehlmann L, Huber B, Jordan D, Lauber J, Salunkhe P, Limpert AS, von Gotz F, Steinmetz I, Eberl L, et al. Global regulation of quorum sensing and virulence by VqsR in Pseudomonas aeruginosa. Microbiology. 2004;150:831–841. doi: 10.1099/mic.0.26906-0. [DOI] [PubMed] [Google Scholar]
- Kessler E, Safrin M, Olson JC, Ohman DE. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268:7503–7508. [PubMed] [Google Scholar]
- Kim EJ, Sabra W, Zeng AP. Iron deficiency leads to inhibition of oxygen transfer and enhanced formation of virulence factors in cultures of Pseudomonas aeruginosa PAO1. Microbiology. 2003;149:2627–2634. doi: 10.1099/mic.0.26276-0. [DOI] [PubMed] [Google Scholar]
- Kiratisin P, Tucker KD, Passador L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J Bacteriol. 2002;184:4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings AF, Martin LW, Sharples KJ, Roddam LF, Latham R, Reid DW, Lamont IL. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect Immun. 2013;81:2697–2704. doi: 10.1128/IAI.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosorok MR, Zeng L, West SE, Rock MJ, Splaingard ML, Laxova A, Green CG, Collins J, Farrell PM. Acceleration of lung disease in children with cystic fibrosis after Pseudomonas aeruginosa acquisition. Pediatr Pulmonol. 2001;32:277–287. doi: 10.1002/ppul.2009.abs. [DOI] [PubMed] [Google Scholar]
- Krieg DP, Helmke RJ, German VF, Mangos JA. Resistance of mucoid Pseudomonas aeruginosa to nonopsonic phagocytosis by alveolar macrophages in vitro. Infect Immun. 1988;56:3173–3179. doi: 10.1128/iai.56.12.3173-3179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laarman AJ, Bardoel BW, Ruyken M, Fernie J, Milder FJ, van Strijp JA, Rooijakkers SH. Pseudomonas aeruginosa alkaline protease blocks complement activation via the classical and lectin pathways. J Immunol. 2012;188:386–393. doi: 10.4049/jimmunol.1102162. [DOI] [PubMed] [Google Scholar]
- Latifi A, Winson MK, Foglino M, Bycroft BW, Stewart GS, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72:4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledgham F, Soscia C, Chakrabarty A, Lazdunski A, Foglino M. Global regulation in Pseudomonas aeruginosa: the regulatory protein AlgR2 (AlgQ) acts as a modulator of quorum sensing. Res Microbiol. 2003;154:207–213. doi: 10.1016/S0923-2508(03)00024-X. [DOI] [PubMed] [Google Scholar]
- Ledgham F, Ventre I, Soscia C, Foglino M, Sturgis JN, Lazdunski A. Interactions of the quorum sensing regulator QscR: interaction with itself and the other regulators of Pseudomonas aeruginosa LasR and RhlR. Mol Microbiol. 2003;48:199–210. doi: 10.1046/j.1365-2958.2003.03423.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang LH. A cell-cell communication signal integrates quorum sensing and stress response. Nature Chem Biol. 2013;9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- Lépine F, Milot S, Déziel E, He J, Rahme LG. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom. 2004;15:862–869. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Greenberg EP. Timing and localization of rhamnolipid synthesis gene expression in Pseudomonas aeruginosa biofilms. J Bacteriol. 2005;187:37–44. doi: 10.1128/JB.187.1.37-44.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LL, Malone JE, Iglewski BH. Regulation of the Pseudomonas aeruginosa quorum-sensing regulator VqsR. J Bacteriol. 2007;189:4367–4374. doi: 10.1128/JB.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightbown JW, Jackson FL. Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem J. 1956;63:130–137. doi: 10.1042/bj0630130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattmann ME, Blackwell HE. Small molecules that modulate quorum sensing and control virulence in Pseudomonas aeruginosa. J Org Chem. 2010;75:6737–6746. doi: 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa exotoxin A triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–374. doi: 10.1016/j.chom.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight SL, Iglewski BH, Pesci EC. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealson KH, Platt T, Hastings JW. Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. GeneChip® expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- Oglesby AG, Farrow JM, Lee J-H, Tomaras AP, Greenberg E, Pesci EC, Vasil ML. The influence of iron on Pseudomonas aeruginosa Physiology: a regulatory link between iron and quorum sensing. J Biol Chem. 2008;283:15558–15567. doi: 10.1074/jbc.M707840200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PW, Pier GB, Preston MJ, Goldberger O, Fitzgerald ML, Bernfield M. Syndecan-1 shedding is enhanced by LasA, a secreted virulence factor of Pseudomonas aeruginosa. J Biol Chem. 2000;275:3057–3064. doi: 10.1074/jbc.275.5.3057. [DOI] [PubMed] [Google Scholar]
- Parkins MD, Ceri H, Storey DG. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol Microbiol. 2001;40:1215–1226. doi: 10.1046/j.1365-2958.2001.02469.x. [DOI] [PubMed] [Google Scholar]
- Passador L, Cook JM, Gambello MJ, Rust L, Iglewski BH. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Passador L, Iglewski BH, Greenberg EP. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CS, Thompson JA, Xavier KB. AI-2-mediated signalling in bacteria. FEMS Microbiol Rev. 2013;37:156–181. doi: 10.1111/j.1574-6976.2012.00345.x. [DOI] [PubMed] [Google Scholar]
- Pesci EC, Pearson JP, Seed PC, Iglewski BH. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G, Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol. 2000;182:6940–6949. doi: 10.1128/jb.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi G, Williams F, Hindle Z, Heurlier K, Holden MT, Cámara M, Haas D, Williams P. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin HR, Butler SM, Wohl MEB, Geller DE, Colin AA, Schidlow DV, Johnson CA, Konstan MW, Regelmann WE. Pulmonary exacerbations in cystic fibrosis. Pediatr Pulmonol. 2004;37:400–406. doi: 10.1002/ppul.20023. [DOI] [PubMed] [Google Scholar]
- Rahme LG, Tan M-W, Le L, Wong SM, Tompkins RG, Calderwood SB, Ausubel FM. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Ausubel FM, Cao H, Drenkard E, Goumnerov BC, Lau GW, Mahajan-Miklos S, Plotnikova J, Tan MW, Tsongalis J, et al. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA. 2000;97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G, Schuster M, Greenberg EP, Bertani I, Grasso M, Venturi V, Zennaro E, Leoni L. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2007;66:1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- Redfield RJ. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 2002;10:365–370. doi: 10.1016/s0966-842x(02)02400-9. [DOI] [PubMed] [Google Scholar]
- Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- Roy-Burman A, Savel RH, Racine S, Swanson BL, Revadigar NS, Fujimoto J, Sawa T, Frank DW, Wiener-Kronish JP. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis. 2001;183:1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- Ryall B, Davies JC, Wilson R, Shoemark A, Williams HD. Pseudomonas aeruginosa, cyanide accumulation and lung function in CF and non-CF bronchiectasis patients. Eur Respir J. 2008;32:740–747. doi: 10.1183/09031936.00159607. [DOI] [PubMed] [Google Scholar]
- Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaber JA, Carty NL, McDonald NA, Graham ED, Cheluvappa R, Griswold JA, Hamood AN. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J Med Microbiol. 2004;53:841–853. doi: 10.1099/jmm.0.45617-0. [DOI] [PubMed] [Google Scholar]
- Schafhauser J, Lepine F, McKay G, Ahlgren HG, Khakimova M, Nguyen D. The stringent response modulates 4-hydroxy-2-alkylquinoline biosynthesis and quorum-sensing hierarchy in Pseudomonas aeruginosa. J Bacteriol. 2014;196:1641–1650. doi: 10.1128/JB.01086-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer JW, Boulette ML, Whiteley M. More than a signal: non-signaling properties of quorum sensing molecules. Trends Microbiol. 2009;17:189–195. doi: 10.1016/j.tim.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Schuster M, Greenberg EP. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Schuster M, Greenberg EP. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics. 2007;8:287. doi: 10.1186/1471-2164-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci USA. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet Q, Zhang LH. Anti-activator QslA defines the quorum sensing threshold and response in Pseudomonas aeruginosa. Mol Microbiol. 2011;80:951–965. doi: 10.1111/j.1365-2958.2011.07622.x. [DOI] [PubMed] [Google Scholar]
- Siehnel R, Traxler B, An DD, Parsek MR, Schaefer AL, Singh PK. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2010;107:7916–7921. doi: 10.1073/pnas.0908511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sio CF, Otten LG, Cool RH, Diggle SP, Braun PG, Bos R, Daykin M, Camara M, Williams P, Quax WJ. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun. 2006;74:1673–1682. doi: 10.1128/IAI.74.3.1673-1682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D’Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA. 2006;103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson LP. Cyanide as a metabolic inhibitor. In: Vennesland EECB, Knowles CJ, Westley J, Wissing F, editors. Cyanide in biology. London: Academic Press; 1981. pp. 11–28. [Google Scholar]
- Stewart GS, Williams P. lux genes and the applications of bacterial bioluminescence. Journal of general microbiology. 1992;138:1289–1300. doi: 10.1099/00221287-138-7-1289. [DOI] [PubMed] [Google Scholar]
- Strempel N, Neidig A, Nusser M, Geffers R, Vieillard J, Lesouhaitier O, Brenner-Weiss G, Overhage J. Human host defense peptide LL-37 stimulates virulence factor production and adaptive resistance in Pseudomonas aeruginosa. PLoS One. 2013;8:e82240. doi: 10.1371/journal.pone.0082240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitil AL, Cashel M, Zyskind JW. Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J Biol Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- Tan TT. “Future” threat of gram-negative resistance in Singapore. Ann Acad Med Singap. 2008;37:884–890. [PubMed] [Google Scholar]
- Thompson LS, Webb JS, Rice SA, Kjelleberg S. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2003;220:187–195. doi: 10.1016/S0378-1097(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Toder DS, Gambello MJ, Iglewski BH. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- Van Delden C, Iglewski BH. Cell-to-cell signaling and Pseudomonas aeruginosa infections. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Delden C, Pesci EC, Pearson JP, Iglewski BH. Starvation selection restores elastase and rhamnolipid production in a Pseudomonas aeruginosa quorum-sensing mutant. Infect Immun. 1998;66:4499–4502. doi: 10.1128/iai.66.9.4499-4502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Delden C, Comte R, Bally AM. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventre I, Ledgham F, Prima V, Lazdunski A, Foglino M, Sturgis JN. Dimerization of the quorum sensing regulator RhlR: development of a method using EGFP fluorescence anisotropy. Mol Microbiol. 2003;48:187–198. doi: 10.1046/j.1365-2958.2003.03422.x. [DOI] [PubMed] [Google Scholar]
- von Bodman SB, Willey JM, Diggle SP. Cell–cell communication in bacteria: united we stand. J Bacteriol. 2008;190:4377–4391. doi: 10.1128/JB.00486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade DS, Calfee MW, Rocha ER, Ling EA, Engstrom E, Coleman JP, Pesci EC. Regulation of Pseudomonas quinolone signal synthesis in Pseudomonas aeruginosa. J Bacteriol. 2005;187:4372–4380. doi: 10.1128/JB.187.13.4372-4380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westblade LF, Ilag LL, Powell AK, Kolb A, Robinson CV, Busby SJ. Studies of the Escherichia coli Rsd-sigma70 complex. J Mol Biol. 2004;335:685–692. doi: 10.1016/j.jmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Beatson SA, Comolli JC, Jakobsen T, Sargent JL, Bertrand JJ, West J, Klausen M, Waite LL, Kang PJ, et al. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol Microbiol. 2005;55:1357–1378. doi: 10.1111/j.1365-2958.2005.04479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev. 2001;25:365–404. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Greenberg EP. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J Bacteriol. 2001;183:5529–5534. doi: 10.1128/JB.183.19.5529-5534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Lee KM, Greenberg EP. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Parsek MR, Greenberg EP. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P, Bainton NJ, Swift S, Chhabra SR, Winson MK, Stewart GS, Salmond GP, Bycroft BW. Small molecule-mediated density-dependent control of gene expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol Lett. 1992;100:161–167. doi: 10.1111/j.1574-6968.1992.tb14035.x. [DOI] [PubMed] [Google Scholar]
- Winson MK, Camara M, Latifi A, Foglino M, Chhabra SR, Daykin M, Bally M, Chapon V, Salmond GP, Bycroft BW, et al. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:9427–9431. doi: 10.1073/pnas.92.20.9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K, Falconer C, Garber NC, Diggle SP, Camara M, Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J Bacteriol. 2000;182:6401–6411. doi: 10.1128/jb.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolz C, Hohloch K, Ocaktan A, Poole K, Evans RW, Rochel N, Albrecht-Gary AM, Abdallah MA, Doring G. Iron release from transferrin by pyoverdin and elastase from Pseudomonas aeruginosa. Infect Immun. 1994;62:4021–4027. doi: 10.1128/iai.62.9.4021-4027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Estrada O, Zaborina O, Bains M, Shen L, Kohler JE, Patel N, Musch MW, Chang EB, Fu YX, et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309:774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- Xiao G, Deziel E, He J, Lepine F, Lesic B, Castonguay MH, Milot S, Tampakaki AP, Stachel SE, Rahme LG. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- Xiao G, He J, Rahme LG. Mutation analysis of the Pseudomonas aeruginosa mvfR and pqsABCDE gene promoters demonstrates complex quorum-sensing circuitry. Microbiology. 2006;152:1679–1686. doi: 10.1099/mic.0.28605-0. [DOI] [PubMed] [Google Scholar]
- Yanagihara K, Tomono K, Kaneko Y, Miyazaki Y, Tsukamoto K, Hirakata Y, Mukae H, Kadota J, Murata I, Kohno S. Role of elastase in a mouse model of chronic respiratory Pseudomonas aeruginosa infection that mimics diffuse panbronchiolitis. Journal of medical microbiology. 2003;52:531–535. doi: 10.1099/jmm.0.05154-0. [DOI] [PubMed] [Google Scholar]
- Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, et al. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA. 2009;106:6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborina O, Lepine F, Xiao G, Valuckaite V, Chen Y, Li T, Ciancio M, Zaborin A, Petrof EO, Turner JR, et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007;3:e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Murphy PJ, Kerr A, Tate ME. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]


