Abstract
The Notch pathway is integrated into numerous developmental processes and therefore is fine-tuned on many levels, including receptor production, endocytosis, and degradation. Notch is further characterized by a twofold relationship with its Delta-Serrate (DSL) ligands, as ligands from opposing cells (trans-ligands) activate Notch, whereas ligands expressed in the same cell (cis-ligands) inhibit signaling. We show that cells without both cis- and trans-ligands can mediate Notch-dependent developmental events during Drosophila oogenesis, indicating ligand-independent Notch activity occurs when the receptor is free of cis- and trans-ligands. Furthermore, cis-ligands can reduce Notch activity in endogenous and genetically induced situations of elevated trans-ligand-independent Notch signaling. We conclude that cis-expressed ligands exert their repressive effect on Notch signaling in cases of trans-ligand-independent activation, and propose a new function of cis-inhibition which buffers cells against accidental Notch activity.
DOI: http://dx.doi.org/10.7554/eLife.04415.001
Research organism: D. melanogaster
eLife digest
Many biological processes require cells to send messages to one another. Typically, this is achieved when molecules are released from one cell and make contact with companion molecules on another cell. This triggers a chemical or biological reaction in the receiving cell.
One of the most common examples of this is the Notch pathway, which is used throughout the animal kingdom and plays an important role in helping cells and embryos to develop. The Notch protein itself is a ‘receptor’ protein that is embedded in the surface of a cell, and relays signals from outside the cell to activate certain genes inside the cell. In fruit flies, two proteins called Serrate and Delta act as ‘ligands’ for Notch—by binding to Notch, they can change how this receptor works.
If Serrate or Delta are present on the outside of one cell, they can activate Notch (and hence the Notch signaling pathway) in an adjacent cell. However, if the Serrate or Delta ligands are present on the surface of the same cell as Notch they turn the receptor off, rather than activate it. Notch can also work without being activated by Serrate or Delta, but whether the ligands can inhibit this ‘ligand-independent’ Notch activation if they are on the surface of the same cell as the Notch receptor was unknown.
Palmer et al. study Notch signaling in the fruit fly equivalent of the ovary, in cells that are naturally deficient in Serrate and from which Delta was artificially removed. The Notch protein was activated when these ligands were not present. Furthermore, the developmental processes that are activated by Notch were able to proceed as normal when triggered by ligand-independent Notch signaling. In total, Palmer et al. investigated three different types of fruit fly cell, and found that ligand-independent Notch signaling can occur in all of them.
Reintroducing Delta to the same cell as Notch turns the receptor off, suggesting that ligands on the surface of the same cell as the receptor can inhibit ligand-independent Notch activity. Many genetic diseases and cancers have been linked to Notch being activated when it should not be; therefore, understanding how Notch is controlled could help guide the development of new treatments for these conditions.
Main text
Canonical Notch signaling begins when the Notch receptor receives a stimulus from a DSL-type ligand (Delta [Dl] or Serrate [Ser] in Drosophila) in an adjacent cell, which leads to γ-secretase-dependent cleavage of Notch, and translocation of the intracellular domain—NICD— into the nucleus to act as a transcriptional co-activator (de Celis, 2013). Notch may also be activated in a non-canonical, DSL-ligand independent manner (Hori et al., 2012). DSL ligands can cis-inhibit ligand-dependent Notch activation when expressed in the same cell as the receptor (Micchelli et al., 1997; Del Álamo et al., 2011). However, the possibility of a relationship between DSL-ligand independent Notch activation and cis-expressed ligands has not been explored.
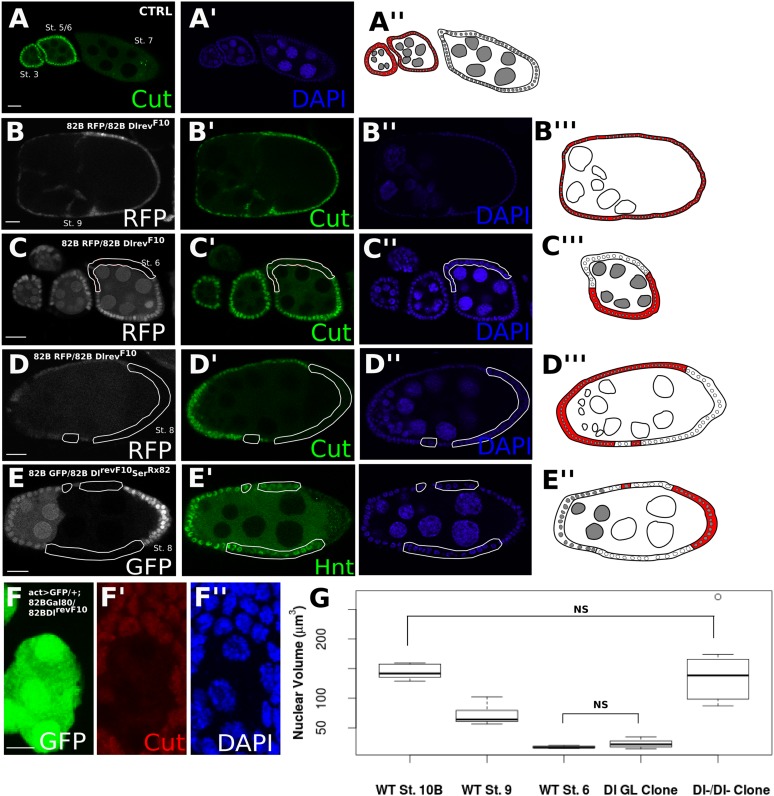
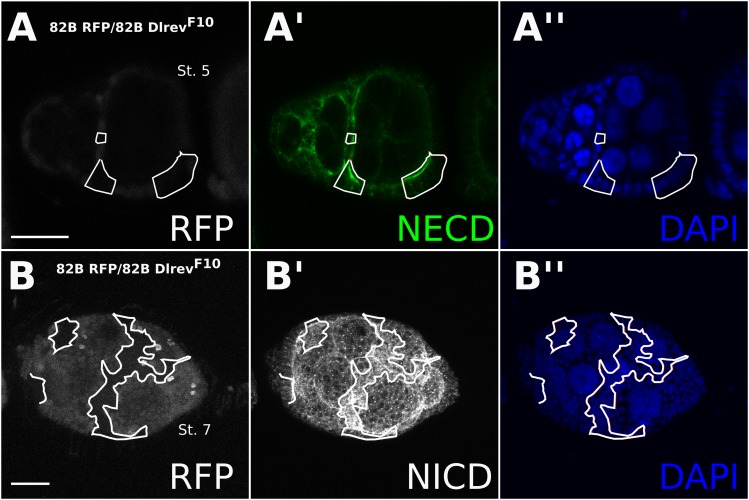
The developing Drosophila egg chamber is a convenient model for dissecting the effects of Notch ligands in cis and in trans, as Dl is the sole signaling source and the signal sending and receiving cells can be easily distinguished (Deng et al., 2001; López-Schier and St Johnston, 2001). (Figure 1—figure supplement 1 provides a brief schematic depiction of the stages of early oogenesis.) At oogenesis stage 7, Notch signaling is activated in the somatic follicle cells by a robust germline Dl upregulation, which leads to the expression of Hindsight (Hnt), downregulation of Cut, and the polyploidization of the follicle cells (Deng et al., 2001; López-Schier and St Johnston, 2001; Sun and Deng, 2005, 2007) (Figure 1A). When Dl germline mutant clones were generated (i.e., trans-activation was removed), the follicle cells failed to downregulate Cut expression, which persisted past stage 7, indicative of a failure to activate Notch (Figure 1B). In contrast, Dl follicle cell mutant clones show precocious Cut downregulation at stage 6 attributable to the relief of cis-inhibition (Poulton et al., 2011) (Figure 1C). Surprisingly, Dl mutant clones in the follicle cells bordering Dl mutant clones in the germline (i.e., a germline with no signaling source, herein referred to as Dl-/Dl- cells) show correct Hnt and Cut expression from stage 7 (Figure 1D,E, Figure 1—figure supplement 2A,B). These Dl-/Dl- clones also correctly transit into the endocycle, as their nuclear volumes are similar to wild-type follicle cells in the later stages of oogenesis after polyploidization (Figure 1F,G), whereas cells neighboring Dl-/Dl- follicle cell clones (retaining a cis-ligand but without a trans-ligand) are comparable to wild-type cells before entry to endocycle (Figure 1F,G). Removal of both cis- and trans-Dl through knockdown of Dl by RNA interference (RNAi) simultaneously in the germline and soma confirmed this finding (Figure 2—figure supplement 1A,B). Together, these observations provide evidence that follicle cells without both cis- and trans-ligand sources can still enter the endocycle stages of oogenesis. This back-up route to the endocycle is not a co-option of Ser in place of Dl, as DlRevF10SerRx82 double clones recapitulated the Dl-/Dl- phenotype (Figure 1E, Figure 1—figure supplement 2A).
Figure 1. Follicle cells without DSL ligand bordering germline cells without DSL ligand show proper Notch activation and downstream differentiation.
Illustrations legend: active Notch = white cytoplasm, inactive Notch = red cytoplasm, WT cell = grey nuclei, mutant clone = white nuclei. (A–E). Follicle cells downregulate Cut at stage 7 of oogenesis (A). DlrevF10 mutant germline cells cause late Cut expression in follicle cells (B). DlrevF10 mutant follicle cells downregulate Cut early (C). DlrevF10 follicle cell clones bordering DlrevF10 germline clones show proper Cut downregulation (D). DlrevF10SerRx82 mutant follicle cell clones bordering DlrevF10SerRx82 germline clones also show proper Hnt (E). See Figure 1—figure supplement 2 for a z-series image for 1D and 1E. These germline/follicle cell clones (D and E) show increased nuclear size comparable to wild-type (WT) follicle cells which have entered the endocycle (n = 8 for each stage/genotype) (F and G). For (G), Welch t-tests were done to assess significance between each condition. The only comparisons that were not significant were between WT stage 10B and Dl-/Dl- clones and between WT stage 6 and Dl germline clones, indicating nuclear size in germline clones alone is similar to that of cells before the endocycle, whereas Dl-/Dl- clonal nuclei are more similar in size to cells that have entered the endocycle. Scale bars represent 20 μm, except in F, where the scale bar represents 5 μm.
Figure 1—figure supplement 1. A schematic depiction of the early stages of Drosophila oogenesis.
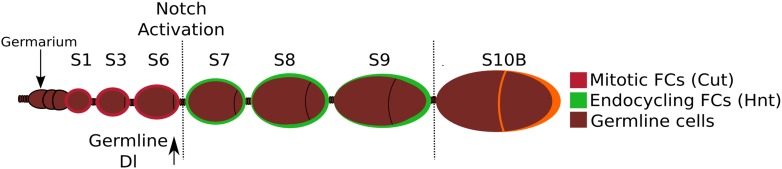
Figure 1—figure supplement 2. Z-stacked images of Dl-/Dl- clones and quantification of Cut staining in egg chamber clones.
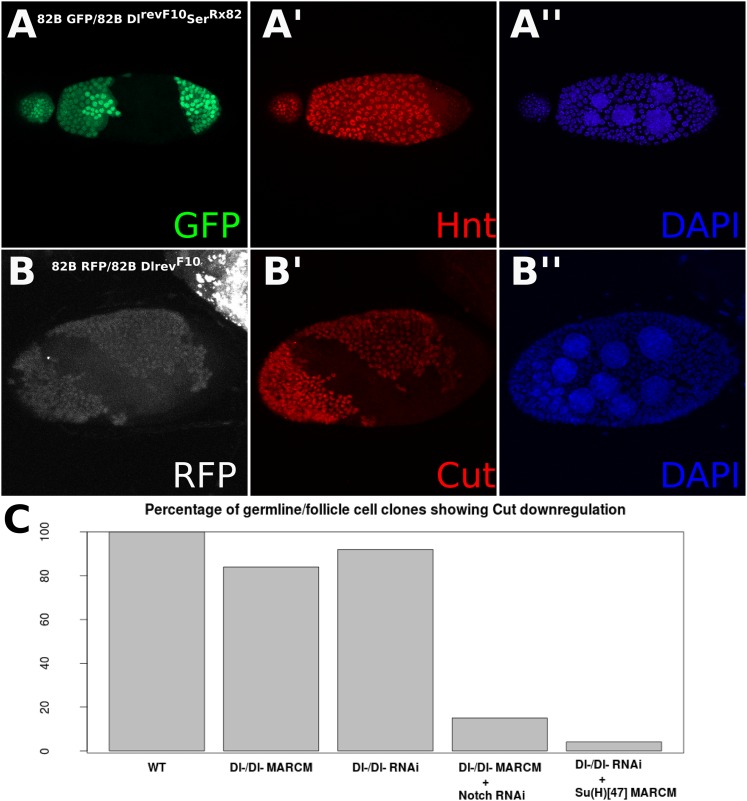
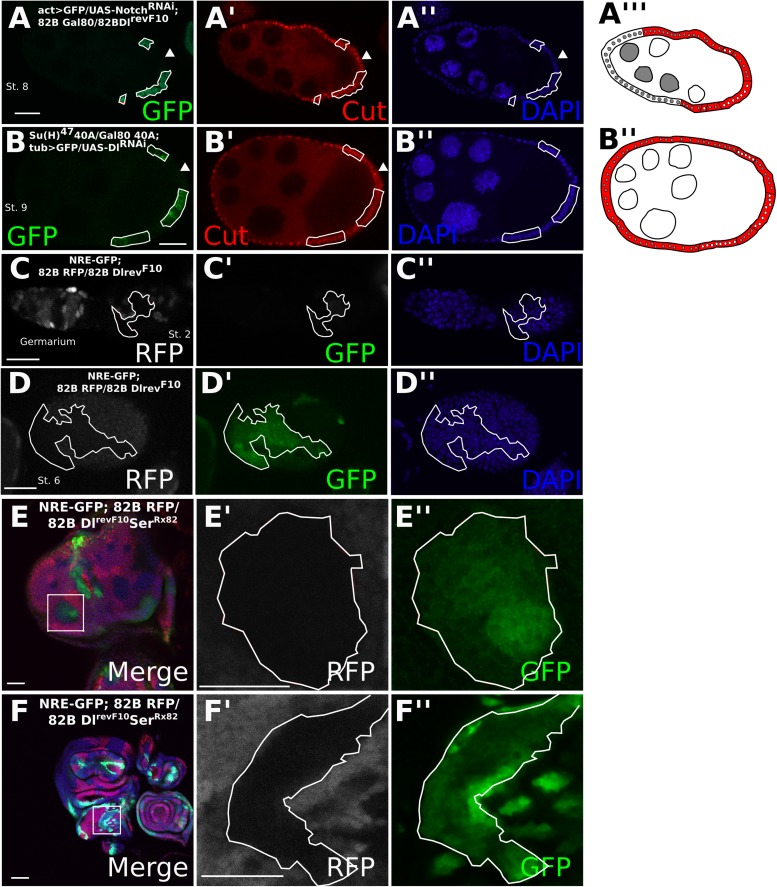
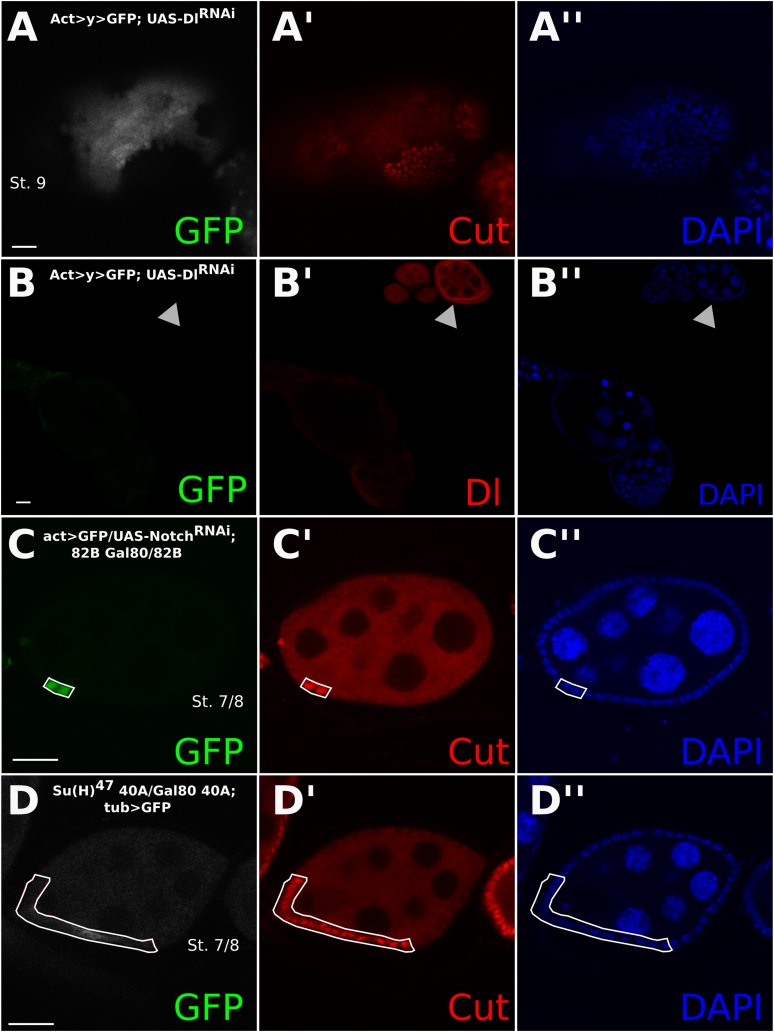
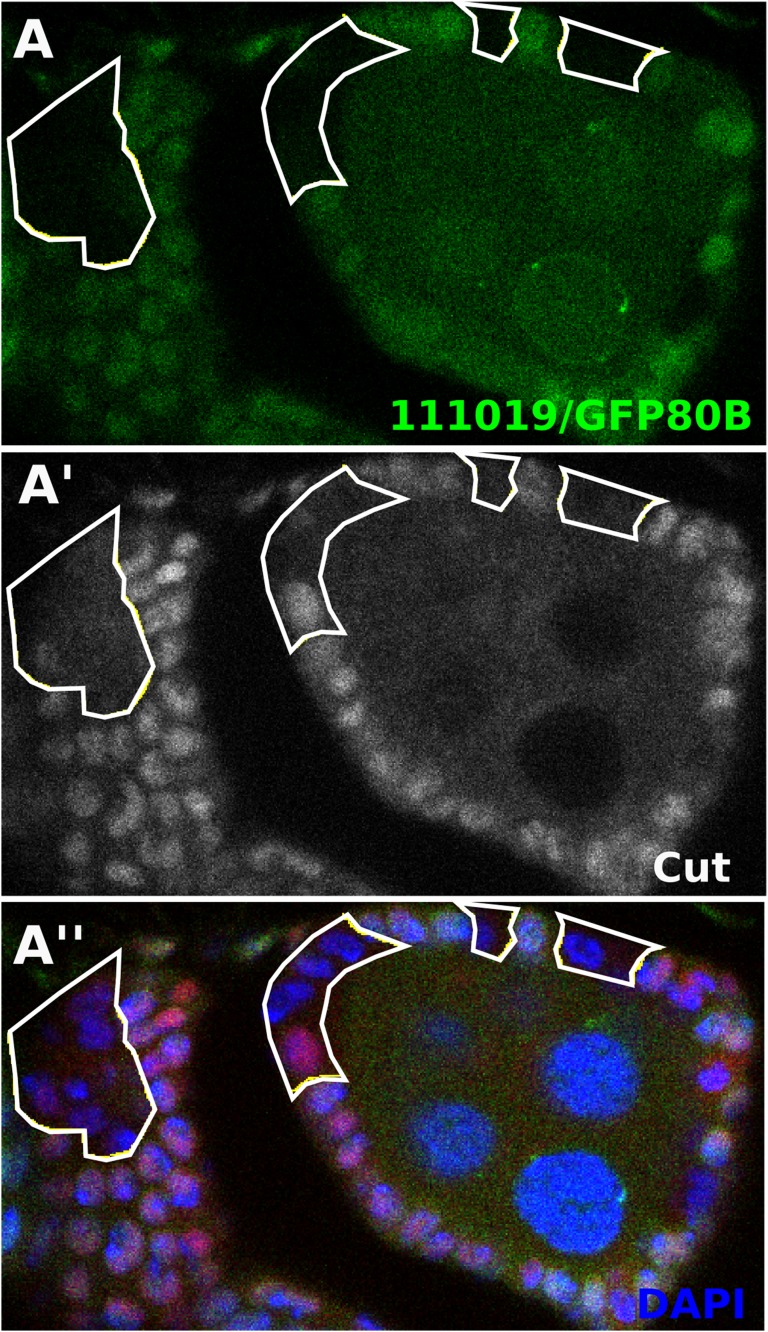
To determine whether the entry into the endocycle in Dl-/Dl- follicle cells still requires the function of Notch, we implemented the mosaic analysis with a repressible cell marker (MARCM) system (Lee and Luo, 2001). The MARCM system enables us to create mutant clones while driving expression of a UAS transgene specifically in those clonal cells. Dl-/Dl- clones driving expression of NotchRNAi show a significantly higher proportion (p < 0.0001) of late Cut-expressing cells than the Dl-/Dl- clones alone, indicating that Notch is still required for the mitotic-to-endocycle switch (Figures 1D and 2A, Figure 1—figure supplement 2C, Figure 2—figure supplement 1C, Supplementary file 1). Likewise, MARCM clones for the null allele of Suppressor of Hairless (Drosophila Notch transcriptional effector), Su(H)47, in RNAi-induced Dl-/Dl- clones also show late Cut expression (p < 0.0001) (Figure 2B, Figure 1—figure supplement 2C, Supplementary file 1) in comparison with RNAi-induced Dl/Dl- clone controls (Figure 2—figure supplement 1A,D). A Notch activity reporter, Notch Responsive Element (NRE)-green fluorescent protein (GFP) (Stempfle et al., 2010) was also upregulated in Dl-/Dl- clones as early as stage 2, and this expression persisted beyond stage 6 (Figure 2C,D), suggesting that NRE-GFP is probably more sensitive to Notch activation than Hnt in follicle cells. Together, these results suggest that Notch activity occurs independently of canonical ligands when both cis- and trans-ligands are removed, resulting in normal downstream developmental events in the follicle cells. Consistently, DlRevF10SerRx82 double mutant clones in the wing and eye discs show a slight cell-autonomous upregulation of NRE-GFP in the clone center, which would only occur if cis-inhibition blocked a DSL-independent mode of Notch activity, as interior cells have no access to trans-ligand (Figure 2E,F). This NRE-GFP upregulation was spatially variable in the wing disc, having the highest prevalence in the notum region (25% incidence), a low incidence in the dorsal pouch (8%), whereas in the ventral pouch region it was never seen (n = 80) (Supplementary file 1), perhaps owing to the differential regulation of Notch degradation throughout the wing disc (Hori et al., 2011). As reported previously, most wing disc clones showed a higher NRE-GFP upregulation in the clone boundary where there is access to trans-ligand, indicating that the ligand-independent Notch activity observed occurs at a rather low level.
Figure 2. Cis-ligand represses ligand-independent Notch activity in the follicle cells and imaginal discs.
DlrevF10 mutant MARCM germline/follicle cell clones co-expressing NotchRNAi show prolonged Cut expression (A). Su(H)47 MARCM mutant germline/follicle cell clones co-expressing DlRNAi show failure to enter the endocycle (B). Germline clones are shown by late Cut expression in wild-type follicle cells (A, B, see arrowheads). See Figure 2—figure supplement 1A for control DlRNAi-induced germline follicle cell clones. Notch Responsive Element-green fluorescent protein (NRE-GFP) is upregulated beginning from stage 2 (C) and through later stages (D) in DlRevF10 germline and follicle cell clones. NRE-GFP is also upregulated cell-autonomously in DlRevF10SerRx82 mutant clones in eye (E) and wing (F) imaginal discs. Scale bars represent 20 μm.
Figure 2—figure supplement 1. Control experiments relating to Figure 2.
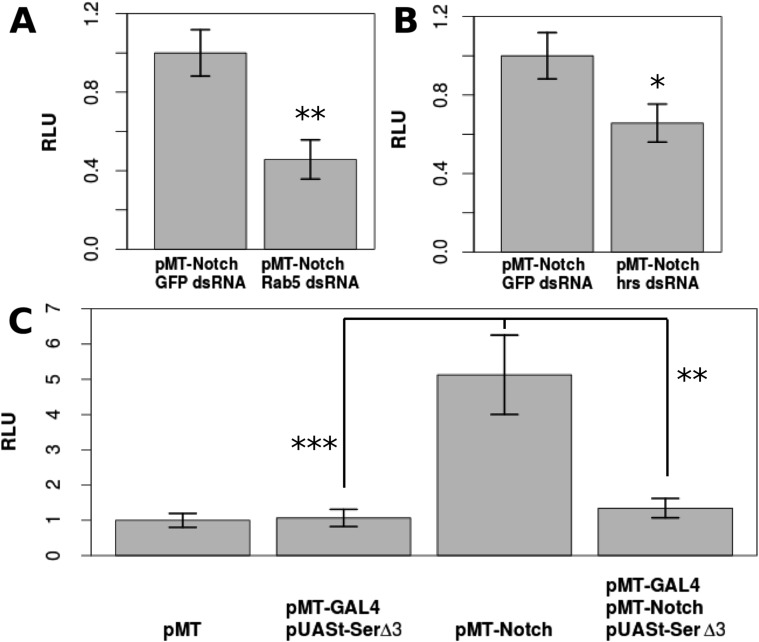
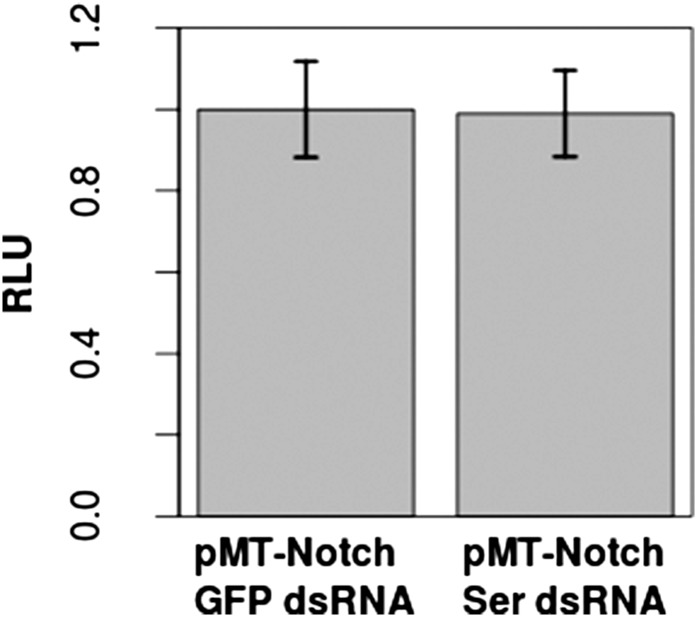
Drosophila S2 cells are reported to have no Dl expression and a very low level of Ser expression, which had no effect on Notch signaling (Fehon et al., 1990; Graveley et al., 2011) (Figure 3—figure supplement 1), and have been used as a model to study ligand-independent Notch activity (Hori et al., 2011). Upon transfection with pMT-NFL, a CuSO4-inducible full-length Notch construct, Notch activation was increased by a factor of 5.13 compared with the control cells, as indicated by a NRE-firefly luciferase reporter gene (p < 0.0001) (Figure 3C). Notch activation in S2 cells is at least partially dependent on endosomal trafficking, as double-stranded (ds) RNA against early endosome component, Rab5, or multivesicular body sorting protein, hrs, reduced the levels of Notch activation (Figure 3A,B). This is consistent with the in vivo studies indicating that ligand-independent Notch activation relies heavily on receptor trafficking (Hori et al., 2012) (Rab5 p = 0.00623, hrs p = 0.0159), and our observation that Notch accumulates in Dl-/Dl- clones (Figure 3—figure supplement 2). A requirement for trafficking is consistent with the results of others who have demonstrated aberrant Notch activation in follicle cell mutants for trafficking components (Wilkin et al., 2004; Vaccari et al., 2008; Schneider et al., 2013), such as tsg101 mutant clones, which show early Notch activation in the follicle cells (Figure 3—figure supplement 3). Furthermore, co-transfecting pMT-NFL with pMT-GAL4 and pUASt-Serdel3, a form of Ser that cannot activate Notch, but only cis-inhibit, (Fleming et al., 2013) almost entirely abolished the Notch activation detected when NFL was transfected alone (p = 0.0048) (Figure 3C). These results suggest that if Notch is expressed in a cell free of cis- and trans-ligands, DSL ligand-independent activity will occur and that cis-inhibition is extremely efficient in preventing this ‘accidental’ Notch activity as it travels through the endosomal pathway en route to degradation.
Figure 3. DSL-ligand-independent Notch activity in S2 cells is buffered by cis-ligand.
Trafficking is important for Notch activation in S2 cells, as treatment with Rab5 dsRNA (A) or hrs dsRNA (B) significantly decreases the amount of Notch activated in S2 cells as shown by Notch-responsive luciferase activity (NRE-firefly) in relative light units (RLU). Transfecting only pMT-NotchFL into S2 cells causes a 5.13-fold increase in Notch activation, which is almost entirely reduced (1.34-fold from the negative control) by co-transfection of pMT-GAL4 and pUASt-Serdel3 (C). Each experiment was carried out with two technical replicates and three biological replicates. Means of the technical replicates were used to carry out a paired t-test (n = 3) for each comparison. Error bars represent standard deviation (SD).
Figure 3—figure supplement 1. Addition of Ser dsRNA had no effect on the Notch activation in S2 cells in comparison with cells treated with control green fluorescent protein (GFP) dsRNA, indicating that the small amount of Ser expression is either not translated or does not significantly contribute to Notch activation upon transfection with pMT-NFL.
Figure 3—figure supplement 2. Notch accumulates in Dl-/Dl- clones.
Figure 3—figure supplement 3. Follicle cells mutant for ESCRT component tsg101 show early Notch activity in the follicle cells (Vaccari et al., 2008).
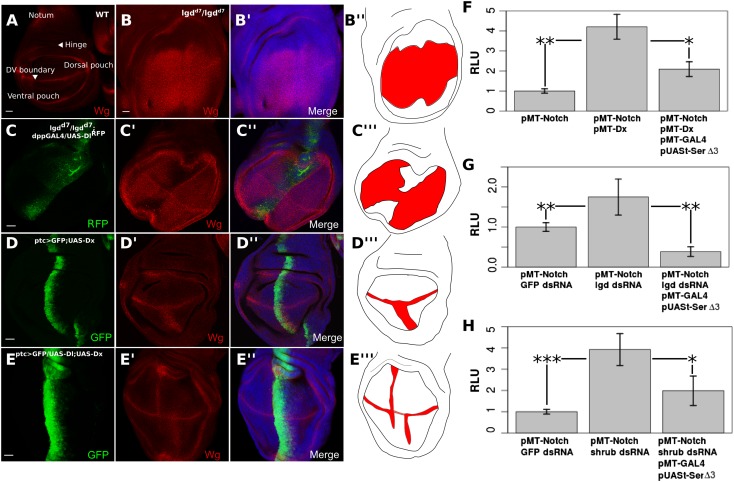
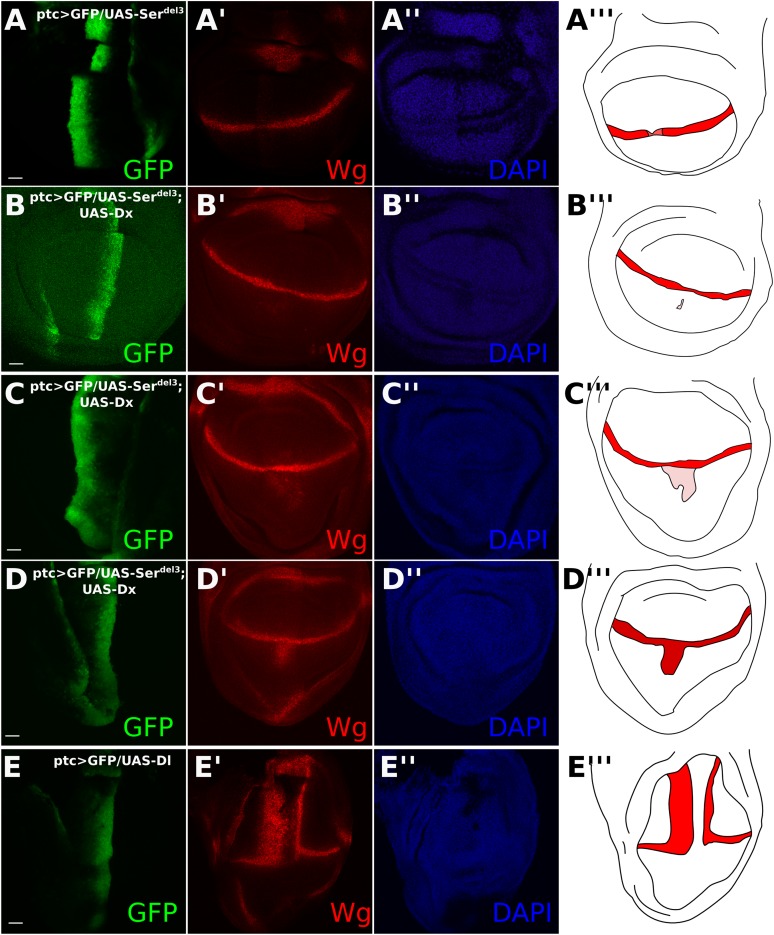
We next explored whether cis-inhibition can also block ligand-independent Notch activity induced in aberrant genetic backgrounds. The Notch target, Wingless (Wg) is normally expressed along the dorsoventral boundary of the wing disc (Figure 4A). Lethal giant disc (lgd) homozygous mutant (lgdd7) larvae display overgrown imaginal discs and ubiquitous ligand-independent Notch activation in the wing pouch region, as shown by upregulation of Wg (Figure 4B). Notch activation in lgd mutant cells is caused by a defect in Notch trafficking and degradation, as the receptor is aberrantly transported to the limiting membrane of the lysosome which facilitates production of NICD (Childress et al., 2006; Gallagher and Knoblich, 2006; Jaekel and Klein, 2006; Schneider et al., 2013). Using dpp-GAL4 to misexpress UAS-Dl along the anterior–posterior axis of the wing disc in lgdd7 homozygous larvae, Wg expression was considerably reduced along the dpp expression domain, indicating that cis-inhibition can block the ligand-independent Notch activity observed in this situation (Figure 4C). Overexpression of Deltex (Dx), an E3 ubiquitin ligase that stimulates Notch monoubiquitination and promotes its trafficking to the lysosomal limiting membrane, has also been shown to induce ligand-independent Notch activation specifically in the ventral wing pouch region (Matsuno et al., 2002; Hori et al., 2004; Wilkin et al., 2008; Schneider et al., 2013) (Figure 4D). We used patched (ptc)-GAL4 to drive expression of UAS-Dx with either UAS-Dl or UAS-Serdel3, whose ectopic expression leads to a reduction of Wg staining along the dorsoventral boundary (Micchelli et al., 1997; Fleming et al., 2013) (controls in Figure 4—figure supplement 1A,E). Co-expression of Dx and Dl led to a decrease in Wg expression in the ventral ptc domain as compared with expression of Dx alone (Figure 4E). When UAS-Dx and UAS-Serdel3 were co-expressed, there was a small but noticeable, albeit variable, decrease in Dx-induced Notch activation (Figure 4—figure supplement 1B–D). This incomplete reduction was probably due to the previously noted, slightly compromised, cis-inhibitory potential of UAS-Serdel3 (Fleming et al., 2013) (Figure 4—figure supplement 1A). Taken together, these results provide evidence that cis-ligand has a negative effect on the raised levels of DSL-ligand independent Notch activation incurred in genetically abnormal cells.
Figure 4. Notch ligand buffers against genetically induced DSL-independent activation.
Wing discs were stained with Wg antibody and illustrations are colored red where Wg is expressed (A–E). A wing disc with regions of interest is labeled and WT Wg staining shown (A). lgdd7/lgdd7 wing discs show ubiquitous Wg expression in the wing pouch as a result of DSL-ligand-independent Notch activity (B). Misexpression of UAS-Dl in lgdd7/lgdd7 discs causes a reduction in Wg staining along the anteroposterior boundary of the pouch (C). ptcGAL4 drives UAS-Dx causing ectopic Notch activity in the ventral wing pouch (D). Co-expression of Dx with Dl reduces Wg staining in the ptc domain (E), although, as in lgdd7/lgdd7 discs, the reduction is not complete towards the dorsoventral boundary. Cis-ligand also decreases Notch activation caused by genetic defects in S2 cells (F–H). Co-transfection with pMT-NFL and pMT-Dx caused a significant increase in Notch luciferase reporter expression, and adding Serdel3 significantly reduced this Dx-induced activation (F). Cells treated with lgd dsRNA (G) or ESCRT-III component, shrub, dsRNA (H) also caused significant increases in Notch reporter activity, either of which could be blocked by addition of Serdel3. For each of the S2 cell experiments, means were taken for technical duplicates and used for a paired t-test for three biological replicates. Error bars represent SD. Scale bars represent 20 μm.
Figure 4—figure supplement 1. Co-expression of UAS-Dx and UAS-Serdel3 has a variable effect on DSL-independent Notch activation.
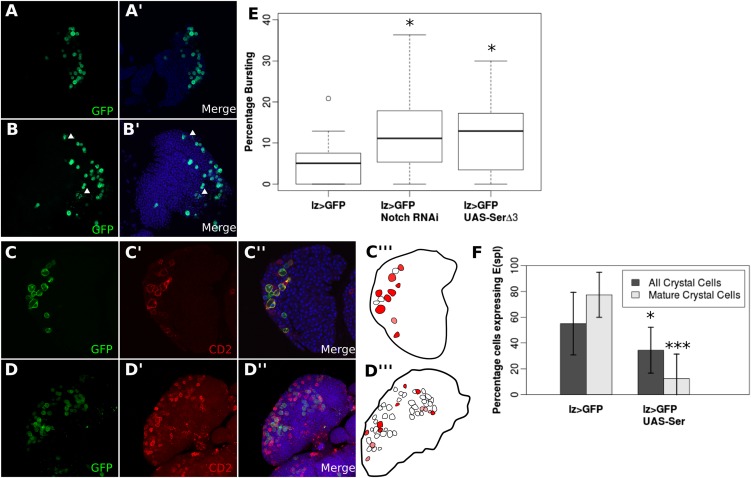
Figure 4—figure supplement 2. Endogenous DSL-independent Notch activity in crystal cells is reduced by cis-inhibition.
Figure 4—figure supplement 3. Reduced Notch reporter activity in crystal cells was not caused by indirect effects on early ligand-dependent Notch signaling in prohaemocytes.
To quantify this effect, we co-transfected pMT-Dx with pMT-NFL, causing an increase by a factor of 4.21 (p = 0.0021) in the Notch activation compared with transfecting pMT-NFL alone (Figure 4F). Transfection of pMT-NFL, pMT-Dx, pMT-GAL4, and pUASt-Serdel3 significantly (p = 0.0194) reduced the level of Notch activation (Figure 4F). We next treated cells with dsRNA for either lgd or shrub (a component of the ESCRT-III complex). Lgd dsRNA induced an increase in Notch activation by a factor of 1.73 compared with GFP dsRNA-treated cells (p = 0.00286) (Figure 4G). Likewise, shrub dsRNA caused a 3.93-fold increase (p < 0.0001) in Notch activation in S2 cells (Figure 4H) (Thompson et al., 2005). Expression of Serdel3 in both situations led to a significant decrease in the amount of Notch activated in comparison with Notch-expressing cells treated with control dsRNA (lgd p = 0.0093, shrub p = 0.0257) (Figure 4G,H).
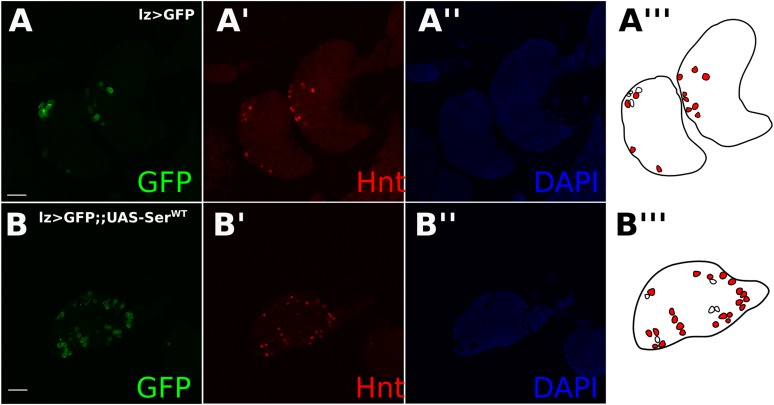
To explore whether cis-acting ligands might block endogenous raised levels of ligand-independent Notch activation, in addition to the raised levels induced by genetic defects, we examined the effect of increased ligand expression in crystal cells in the larval lymph gland, which have recently been shown to have ligand-independent Notch activation (Mukherjee et al., 2011). Notch activity in crystal cells promotes cell survival, and decreased Notch activity leads to a ‘bursting’ phenotype (Mukherjee et al., 2011) (Figure 4—figure supplement 2B,E). Evidence for this bursting phenotype is provided by the disorganization of membrane-associated GFP (Mukherjee et al., 2011). Using Lozenge (Lz)-GAL4, a crystal cell lineage-specific driver (Terriente-Felix et al., 2013) to misexpress UAS-NotchRNAi or UAS-Serdel3 led to a significantly higher proportion of cells showed the ‘bursting’ phenotype than wild-type crystal cells (NotchRNAi p = 0.0434, Serdel3 p = 0.0286) (Figure 4—figure supplement 2A,B,E). Furthermore, overexpression of UAS-SerWT led to a significant decrease of the Notch reporter E(spl):mβ-CD2expression in mature crystal cells (Figure 4—figure supplement 2C,D,F). Reduced Notch reporter activity was not caused by indirect effects on early ligand-dependent Notch signaling in prohaemocytes, as Hnt, a Notch target in differentiating crystal cells, (Terriente-Felix et al., 2013) was unaffected by ligand misexpression (Figure 4—figure supplement 3A,B). These observations indicate that increased ligand expression in crystal cells decreases cell survival by blocking Notch ligand-independent activation, and therefore the buffering role of cis-expressed ligand can be extended to endogenous cases of DSL-independent Notch activity.
In this study, we show that cells devoid of DSL ligands activate Notch sufficiently to stimulate reporter activity, and in the ovarian follicle cells the level of activation is above the threshold required to mediate normal Notch-induced downstream developmental events. During development, this type of noncanonical Notch activity is normally prevented by cis-expressed DSL ligands in numerous tissues. Cis-inhibition can also attenuate DSL-ligand independent Notch activity both in endogenous and genetically induced situations. Mechanistically, this could be explained if DSL ligands sequestered Notch at the membrane, made Notch more sensitive to degradation, or increased the stability of the heterodimer as it travels through the endosomal pathway. As we and others (Fiuza et al., 2010) have shown that increasing or decreasing ligand has variable effects on receptor distribution among tissues, and given that we observe a consistent effect among tissues on Notch activation upon cis-ligand removal, we prefer the stability hypothesis. Fiuza et al. (2010) show that ligand affects Notch stability during Notch activation by EDTA, giving support to the stability hypothesis as the most parsimonious explanation (Fiuza et al., 2010). It is suggested that retaining a pool of translated Notch receptor keeps the pathway in a condition capable of almost instant activation (Sprinzak et al., 2010). Therefore, we propose that a role of cis-ligands might be to keep the Notch pathway in a state of readiness by buffering against unintentional stochastic Notch activity resulting from normal processing through the endosomes. Endogenously, this may aid the ability of a cell to mediate future Notch-dependent developmental events that have strict temporal regulation.
Materials and methods
Drosophila stocks and generation of clones
The following fly stocks were used for Drosophila crosses. hs-flp122;;FRT82B RFP (Poulton et al., 2011), FRT82B DlRevF10 (Haenlin et al., 1990), FRT82B DlRevF10SerRx82 (BDSC #6300), hs-FLP122; act-GAL4 UAS-GFP;FRT82B Gal80, UAS-NotchRNAi (VDRC #1112—no expression in germline cells), UAS-DeltaRNAi (BDSC #34322—able to express in germline cells); hsFLP GFPstau; act > y+ > GAL4, UAS-GFP, hs-flp122; Gal80 FRT40A; tubGAL4 UASGFP, Su(H)47FRT40A (Morel and Schweisguth, 2000), NRE-EGFP (BDSC #30727; Stempfle et al., 2010), ubx-FLP;;FRT82B RFP, patched-GAL4 UAS-GFP (Hinz et al., 1994), UAS-DlMyc (a gift from Marc Muskavitch), tsg101111019 from Kyoto stock center, UAS-SerWT (BDSC #5815), UAS-Serdel3−tom (a gift from Robert J Fleming) (Graveley et al., 2011), UAS-Deltex (a gift from Martin Baron), lgdd740A (BDSC #25087), dppGAL4 (BDSC #7007), lz-GAL4 UAS-GFP (BDSC #6314). To create FRT82B, DlRevF10 germline/follicle cell clones by the FLP/FRT or MARCM methods (Golic and Lindquist, 1989; Lee and Luo, 2001) (e.g., Figures 1B,D–F, 2A,C–D, Figure 1—figure supplement 2A–B), crossed flies were subjected to a 2 hr heat shock at 37°C for two consecutive days while in the mid-pupal to late-pupal stages. Flies were sorted three days after eclosion, and then kept for an extra three days at 25° before an additional 1-hr heat shock and incubation at 29°C with yeast paste for two more days before dissection. FLP-out-induced DlRNAi germline/follicle cell clones (e.g., Figure 2B, Figure 2—figure supplement 1A,B) were produced by two consecutive 50-min heat shocks, followed by incubation at 25°C for a week and then transfer to yeasted vials in the 29°C incubator for dissection two days later. Evidence for MARCM and FLP-out-induced germline clones was provided by small nuclei and late Cut expression, as the UASt-GFP transgene does not reliably express in the germline. Follicle cell clones alone were produced by two 50-min heat shocks, followed by two days’ incubation at 29°C (e.g., Figure 1C and Figure 2—figure supplement 1C–D). Imaginal disc FLP-FRT-induced mutant clones were produced either by a ubx-FLP or a 1-hr heat shock with hs-FLP122 two days after egg laying. All other crosses were kept at 25°C unless otherwise noted. In lymph gland studies, Grubbs' test was used to identify significant (p < 0.05) outliers, which were omitted from further analyses.
Immunostaining
Ovaries, imaginal discs, or lymph glands were dissected in phosphate-buffered saline (PBS), fixed in 10% formaldehyde, washed three times in PBS + Triton-X (PBT), and then blocked for at least 1 hr in PBT with goat serum. Tissues were then either stained overnight with mouse anti-Cut (DSHB 2B10, 1:30), mouse anti-Hindsight (DSHB 1G9, 1:15), mouse anti-NICD (DSHB C179C6, 1:15), mouse anti-NECD (DSHB C4582H, 1:15), mouse anti-Wingless (DSHB 4D4, 1:20), mouse anti-Dl (DSHB C594.9B, 1:15), rabbit anti-βGal (MP Biomedical, Santa Ana, CA. SKU #08559761), or rabbit anti-GFP (abcam, Cambridge, UK. ab290—NRE-GFP was co-stained with this antibody to increase reporter sensitivity) primary antibodies. Tissues were mounted on slides after PBT washes and secondary antibody incubation. 4',6-Diamidino-2-phenylindole (DAPI) was used to stain nuclei. Samples were then analyzed with a Zeiss 510 or Leica SP2 confocal microscope and after analysis with the Image J software. Nuclear volume quantification was done with the Volumest plug-in for ImageJ.
S2 cell transfection and RNA interference
S2 cells were grown under standard conditions and passaged once every three days in serum-free Gibco media (Invitrogen, Waltham, MA) supplemented with antibiotics. In preparation for transfection 106 cells per milliliter were seeded into either 24-well plates or 96-well plates for experiments with or without dsRNA treatment, respectively. Transfections were carried out with Qiagen Effectene (Qiagen, Netherlands) transfection reagent according to the manufacturer's instruction. Plasmids used for transfection were pMT-NotchFL (a gift from Renjie Jiao), pMT-GAL4 (DGRC #1042), pUASt-Serdel3 (a gift from Robert J Fleming), pMT-Deltex (a gift from Spyros Artavanis-Tsakonas), NRE-firefly luciferase (a gift from Sarah Bray), or Renilla luciferase (a gift from Sarah Bray). Aliquots (75 ng for 24-well plates or 50 ng for 96-well plates) of each non-luciferase plasmid were added and, where applicable, 10 ng of each luciferase plasmid. DNA concentration between transfections was kept constant with an empty vector. For experiments without dsRNA treatment, CuSO4 was added to a concentration of 500 µM 24 hr after transfection, and cells were assayed 24 hr later. dsRNA was transcribed in vitro using the RiboMAX large-scale RNA production system-T7 kit (Promega, Madison, WI). The following primers were used to amplify genomic DNA taken from a single male fly from the NRE-GFP stock:
Rab5
Forward: GAATTAATACGACTCACTATAGGGCAGGGGACGAATTTCATTTG
Reverse: GAATTAATACGACTCACTATAGGGAAAACCCTGCGCTTTCTTCT
Hrs
Forward: GAATTAATACGACTCACTATAGGGAATCGCCAACAATCAAGTCC
Reverse: GAATTAATACGACTCACTATAGGGCGTGCAGCACTACTTTCCAA
Lgd
Forward: GAATTAATACGACTCACTATAGGGAGATGCCTCTGAGGAACCCGTCCAG
Reverse: GAATTAATACGACTCACTATAGGGAGAGTGTGGGTTCTGGGGCAGCAGT
Shrub
Forward: GAATTAATACGACTCACTATAGGGACTTTTATGCAGGGACGTGG
Reverse: GAATTAATACGACTCACTATAGGGTCCCTCGCTTCGAACTAAAA
Serrate
Forward: GAATTAATACGACTCACTATAGGGTCTCACCAACCAACCAATCA
Reverse: GAATTAATACGACTCACTATAGGGCACAATATAGAGCGCGACGA
GFP
Forward: GAATTAATACGACTCACTATAGGGAGCTGGACGGCGACGTAAAC
Reverse: GAATTAATACGACTCACTATAGGGATGGGGGTGTTCTGCTGGTAG
Cells were treated with dsRNA at a concentration of 50 nM, and then transfected shortly after. CuSO4 was added to a concentration of 500 µM later that day. Cells were incubatedfor five days, with an additional treatment of dsRNA on the fourth day.
Luciferase assay
Cells were transfected with plasmids of interest together with an NRE-driving firefly luciferase expression and a constitutively activated Renilla luciferase to control for transfection efficiency. Luciferase measures were inspected with the Dual-Luciferase Assay Kit (Promega) in 96-well luminometer plates. Each transfection was performed in duplicate and repeated several times. Student's t test was used to test for statistical significance.
Acknowledgements
Dongyu Jia discovered the phenotype of Hindsight, and Cut expression in Dl-/Dl- (Dl germline/follicle cell) clones, and developed the project as DSL ligand- independent mitotic cycle/endocycle switch initially. We thank Marc Muskavitch, Martin Baron, Robert Fleming, Renjie Jiao, Spyros Artavanis-Tsakonas, Sarah Bray, the Bloomington Drosophila Stock Center, the Vienna Drosophila RNAi Center, the TRiP at Harvard Medical School, and the Developmental Studies Hybridoma Bank for providing us with stocks and reagents. We also thank Yi-Chun Huang for technical assistance, and Gary Struhl, John Poulton, Pang-Kuo Lo, Gengqiang Xie, Jen Kennedy, Steven Lenhert, and Gabriel Calvin for helpful comments and suggestions while preparing the manuscript. W-MD is supported by NIH grants R01GM072562 and NSF IOS-1052333.
Funding Statement
The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Science Foundation IOS-1052333 to Wu-Min Deng.
National Institutes of Health R01GM072562 to Wu-Min Deng.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
WHP, Conception and design, Acquisition of data, Analysis and interpretation of data, Drafting or revising the article.
DJ, Conception and design, Acquisition of data.
W-MD, Conception and design, Analysis and interpretation of data, Drafting or revising the article, Contributed unpublished essential data or reagents.
Additional files
Supplementary clonal data file. Excel worksheet containing clonal data from the egg chamber and the wing disc.
References
- Childress JL, Acar M, Tao C, Halder G. Lethal giant discs, a novel c2-domain protein, restricts notch activation during endocytosis. Current Biology. 2006;16:2228–2233. doi: 10.1016/j.cub.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF. Understanding the determinants of notch interactions with its ligands. Science Signaling. 2013;6:pe19. doi: 10.1126/scisignal.2004079. [DOI] [PubMed] [Google Scholar]
- Del Álamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in notch signalling. Current Biology. 2011;21:R40–R47. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Deng WM, Althauser C, Ruohola-Baker H. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development. 2001;128:4737–4746. doi: 10.1242/dev.128.23.4737. [DOI] [PubMed] [Google Scholar]
- Fehon RG, Kooh PJ, Rebay I, Regan CL, Xu T, Muskavitch MA, Artavanis-Tsakonas S. Molecular interactions between the protein products of the neurogenic loci Notch and Delta, two EGF-homologous genes in Drosophila. Cell. 1990;61:523–534. doi: 10.1016/0092-8674(90)90534-L. [DOI] [PubMed] [Google Scholar]
- Fiuza UM, Klein T, Martinez Arias A, Hayward P. Mechanisms of ligand-mediated inhibition in Notch signaling activity in Drosophila. Developmental Dynamics. 2010;239:798–805. doi: 10.1002/dvdy.22207. [DOI] [PubMed] [Google Scholar]
- Fleming RJ, Hori K, Sen A, Filloramo GV, Langer JM, Obar RA, Artavanis-Tsakonas S, Maharaj-Best AC. An extracellular region of Serrate is essential for ligand-induced cis-inhibition of Notch signaling. Development. 2013;140:2039–2049. doi: 10.1242/dev.087916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher CM, Knoblich JA. The conserved c2 domain protein lethal (2) giant discs regulates protein trafficking in Drosophila. Developmental Cell. 2006;11:641–653. doi: 10.1016/j.devcel.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenlin M, Kramatschek B, Campos-Ortega JA. The pattern of transcription of the neurogenic gene Delta of Drosophila melanogaster. Development. 1990;110:905–914. doi: 10.1242/dev.110.3.905. [DOI] [PubMed] [Google Scholar]
- Hinz U, Giebel B, Campos-Ortega J. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- Hori K, Fostier M, Ito M, Fuwa TJ, Go MJ, Okano H, Baron M, Matsuno K. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development. 2004;131:5527–5537. doi: 10.1242/dev.01448. [DOI] [PubMed] [Google Scholar]
- Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. The Journal of Cell Biology. 2011;195:1005–1015. doi: 10.1083/jcb.201104146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Sen A, Kirchhausen T, Artavanis-Tsakonas S. Regulation of ligand-independent Notch signal through intracellular trafficking. Communicative & Integrative Biology. 2012;5:374–376. doi: 10.4161/cib.19995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaekel R, Klein T. The Drosophila notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Developmental Cell. 2006;11:655–669. doi: 10.1016/j.devcel.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in Neurosciences. 2001;24:251–254. doi: 10.1016/S0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- López-Schier H, St Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes & Development. 2001;15:1393–1405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuno K, Ito M, Hori K, Miyashita F, Suzuki S, Kishi N, Artavanis-Tsakonas S, Okano H. Involvement of a proline-rich motif and RING-H2 finger of Deltex in the regulation of Notch signaling. Development. 2002;129:1049–1059. doi: 10.1242/dev.129.4.1049. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124:1485–1495. doi: 10.1242/dev.124.8.1485. [DOI] [PubMed] [Google Scholar]
- Morel V, Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes & Development. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- Mukherjee T, Kim WS, Mandal L, Banerjee U. Interaction between Notch and Hif-α in development and survival of Drosophila blood cells. Science. 2011;332:1210–1213. doi: 10.1126/science.1199643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton JS, Huang YC, Smith L, Sun J, Leake N, Schleede J, Stevens LM, Deng WM. The microRNA pathway regulates the temporal pattern of Notch signaling in Drosophila follicle cells. Development. 2011;138:1737–1745. doi: 10.1242/dev.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Troost T, Grawe F, Martinez-Arias A, Klein T. Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome. Journal of Cell Science. 2013;126:645–656. doi: 10.1242/jcs.116590. [DOI] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, Garcia-Ojalvo J, Elowitz MB. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempfle D, Kanwar R, Loewer A, Fortini ME, Merdes G. In vivo reconstitution of gamma-secretase in Drosophila results in substrate specificity. Molecular and Cellular Biology. 2010;30:3165–3175. doi: 10.1128/MCB.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng WM. Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells. Development. 2005;132:4299–4308. doi: 10.1242/dev.02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Deng WM. Hindsight mediates the role of notch in suppressing hedgehog signaling and cell proliferation. Developmental Cell. 2007;12:431–442. doi: 10.1016/j.devcel.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terriente-Felix A, Li J, Collins S, Mulligan A, Reekie I, Bernard F, Krejci A, Bray S. Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development. 2013;140:926–937. doi: 10.1242/dev.086785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Mathieu J, Sung HH, Loeser E, Rørth P, Cohen SM. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Developmental Cell. 2005;9:711–720. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Vaccari T, Lu H, Kanwar R, Fortini ME, Bilder D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. The Journal of Cell Biology. 2008;180:755–762. doi: 10.1083/jcb.200708127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkin MB, Carbery AM, Fostier M, Aslam H, Mazaleyrat SL, Higgs J, Myat A, Evans DA, Cornell M, Baron M. Regulation of notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Current Biology. 2004;14:2237–2244. doi: 10.1016/j.cub.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Wilkin M, Tongngok P, Gensch N, Clemence S, Motoki M, Yamada K, Hori K, Taniguchi-Kanai M, Franklin E, Matsuno K, Baron M. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of Notch in the endosomal trafficking pathway. Developmental Cell. 2008;15:762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]