Abstract
Neurodevelopmental disorders (NDDs) affect more than 3% of children and are attributable to single-gene mutations at more than 1000 loci. Traditional methods yield molecular diagnoses in less than one-half of children with NDD. Whole-genome sequencing (WGS) and whole-exome sequencing (WES) can enable diagnosis of NDD, but their clinical and cost-effectiveness are unknown. One hundred families with 119 children affected by NDD received diagnostic WGS and/or WES of parent-child trios, wherein the sequencing approach was guided by acuity of illness. Forty-five percent received molecular diagnoses. An accelerated sequencing modality, rapid WGS, yielded diagnoses in 73% of families with acutely ill children (11 of 15). Forty percent of families with children with nonacute NDD, followed in ambulatory care clinics (34 of 85), received diagnoses: 33 by WES and 1 by staged WES then WGS. The cost of prior negative tests in the nonacute patients was $19,100 per family, suggesting sequencing to be cost-effective at up to $7640 per family. A change in clinical care or impression of the pathophysiology was reported in 49% of newly diagnosed families. If WES or WGS had been performed at symptom onset, genomic diagnoses may have been made 77 months earlier than occurred in this study. It is suggested that initial diagnostic evaluation of children with NDD should include trio WGS or WES, with extension of accelerated sequencing modalities to high-acuity patients.
INTRODUCTION
Neurodevelopmental disorders (NDDs), including intellectual disability, global developmental delay, and autism, affect more than 3% of children. Etiologic identification of NDD often engenders a lengthy and costly differential diagnostic odyssey without return of a definitive diagnosis (1). The current etiologic evaluation of NDD is complex: primary tests include neuroimaging, karyotype, array comparative genome hybridization (array CGH) and/or single-nucleotide polymorphism arrays, and phenotype-driven metabolic, molecular, and serial gene sequencing studies. Secondary, invasive tests, such as biopsies, cerebrospinal fluid examination, and electromyography, enable diagnosis in a small percentage of additional cases. About 30% of NDDs are attributable to structural genetic variation, but more than half of patients do not receive an etiologic diagnosis (1–5). Single-gene testing for diagnosis of NDD is especially challenging because of profound locus heterogeneity and overlapping symptoms (6–10).
As predicted, the introduction of whole-genome sequencing (WGS) and whole-exome sequencing (WES) into medical practice has begun to transform the diagnosis and management of patients with genetic disease (11). Acceleration and simplification of genetic diagnosis are a result of the following: (i) multiplexed testing to interrogate nearly all genes on a physician’s differential at a cost and turnaround time approaching that of a single-gene test; (ii) the ability to analyze genes for which no other test exists; and (iii) the capacity to cast a wide net that can detect pathogenic variants in genes not yet on the clinician’s differential. The latter proves particularly powerful for diagnosing patients with rare or newly discovered genetic diseases (12) and for patients with atypical or incomplete clinical presentations (13). Furthermore, new gene and phenotype discovery has increasingly become part of the diagnostic process. The importance of molecular diagnosis is that care of such patients can then shift from interim, phenotypic-driven management to definitive treatment that is refined by genotype (11). Although early reports indicate that WES enables diagnosis of neurologic disorders (9, 14, 15), the clinical and cost-effectiveness are not known. Data are needed to guide best-practice recommendations regarding testing of probands (affected patients) alone versus trios (proband plus parents), use of WES versus WGS, and the appropriate prioritization of genomic testing in an etiologic evaluation for various clinical presentations.
Herein, we report the effectiveness of a WGS and WES sequencing program for children with NDD, featuring an accelerated sequencing modality for patients with high-acuity illness. We outline diagnostic yield and an initial analysis of the impact on time to diagnosis, cost of diagnostic testing, and subsequent clinical care.
RESULTS
Characteristics of enrolled patients
A biorepository was established at a children’s hospital in the central United States for families with one or more children suspected of having a monogenetic disease, but without a definitive diagnosis (16). Over a 33-month period, 155 families with heterogeneous clinical conditions were enrolled into the repository and analyzed by WGS or WES for diagnostic evaluation. Of these, 100 families had 119 children with NDDs and were the subjects of the analysis reported herein (Table 1). Standard WES or rapid WGS was performed on the basis of acuity of illness (16): 85 families with affected children followed in ambulatory clinics received non-expedited WES, followed by non-expedited WGS if WES was unrevealing; 15 families with infants who were symptomatic at or shortly after birth and in neonatal intensive care units (NICUs) or pediatric intensive care units (PICUs) received immediate, rapid WGS (Table 1). The mean age of the affected children in the ambulatory clinic group was about 7 years at enrollment (Table 2). Symptoms were apparent at an average of less than 1 year of age in most children (Table 2). The clinical features of each affected child were ascertained by examination of electronic health records and communication with treating clinicians and translated into Human Phenotype Ontology (HPO) terms (17). The most common features of the 119 affected children from these families were global developmental delay/intellectual disability, encephalopathy, muscular weakness, failure to thrive, microcephaly, and developmental regression (Table 1). The most common phenotype among children in the nonacute group was global developmental delay/intellectual disability (61%). Among infants enrolled from intensive care units, seizures, hypotonia, and morphological abnormalities of the central nervous system were most common. Consanguinity was noted in only four families. Our intention was to enroll and test parent-child trios; in practice, an average of 2.55 individuals per family were tested.
Table 1. Characteristics of families with NDD enrolled for acuity-guided genome- or exome-based diagnostic testing.
HPO, Human Phenotype Ontology.
| Number | ||||
|---|---|---|---|---|
| Total | Exome | Rapid genome | ||
| Families | 100 | 85 | 15 | |
| Affected children | 119 | 103 | 16 | |
| Consanguineous families | 4 | 4 | 0 | |
| NICU enrollments | 11 | 0 | 11 | |
| Clinical features by family | HPO ID(s) | |||
| Acidosis/encephalopathy | 0001941/0001298 | 11 | 9 | 2 |
| Ataxia | 0001251 | 8 | 8 | 0 |
| Autism spectrum disorder | 000729 | 10 | 10 | 0 |
| Dystonia | 0001332 | 3 | 2 | 1 |
| Global developmental delay/intellectual disability | 0001263/0001249 | 52 | 52 | 0 |
| Intrauterine growth retardation/failure to thrive | 0001511/0001508 | 27 | 23 | 4 |
| Macrocephaly | 0000256 | 9 | 8 | 1 |
| Microcephaly | 0000252 | 22 | 21 | 1 |
| Morphological abnormality of the central nervous system | 0007319 | 18 | 11 | 7 |
| Muscle weakness/severe muscular hypotonia | 0001324/0001252 | 35 | 27 | 8 |
| Neurodegeneration/developmental regression | 0002180/0002376 | 22 | 21 | 1 |
| Seizures | 0001250 | 39 | 32 | 7 |
| Visual and/or sensorineural hearing impairment | 0000505/0000407 | 17 | 15 | 2 |
Table 2. Time to diagnosis.
Average age at symptom onset, enrollment, and molecular diagnosis for children diagnosed by exome or rapid genome sequencing.
| Exome sequencing (months) |
Rapid genome sequencing (days)* |
||||
|---|---|---|---|---|---|
| Mean | Range | Mean | Median | Range | |
| Symptom onset | 6.6 | 0–90 | 8.2 | 0 | 0–90 |
| Enrollment | 83.8 | 1–252 | 43.2 | 38 | 2–154 |
| Molecular diagnosis | 95.3 | 16–262 | 107.5 | 50 | 8–521 |
Four postmortem enrollments were excluded from time-to-diagnosis calculations.
WES and WGS data
WES was performed in 16 days, to a depth of >8 gigabases (mean coverage, >80-fold; table S1). Six ambulatory patients received WGS by HiSeq X Ten after negative analysis of WES. Rapid WGS was performed in acutely ill patients, used a 50-hour protocol (16), and was to an average depth of at least 30-fold (table S1). Nucleotide variants were identified with a pipeline optimized for sensitivity to detect rare new variants, yielding 4,855,911 variants per genome and 196,280 per exome (table S1). Variants with allele frequencies <1% in a database of ~3500 individuals previously sequenced at our center, and of types that are potentially pathogenic, as defined by the American College of Medical Genetics (ACMG) (18), averaged 560 variants per exome and 835 per genome (table S1).
Genomic diagnostic results
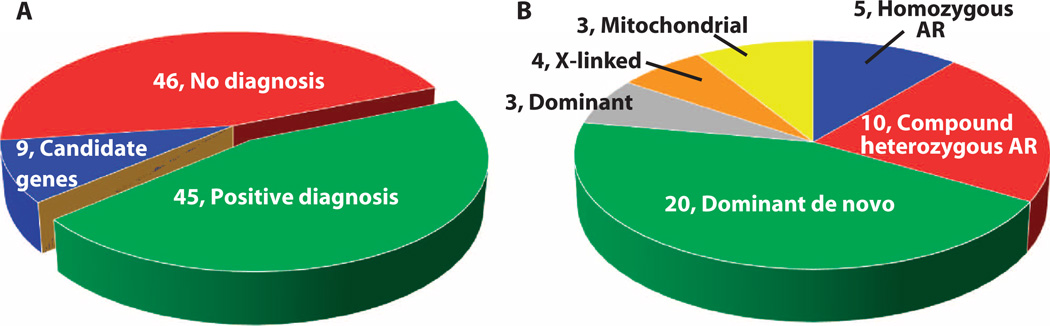
A definitive molecular diagnosis of an established genetic disorder was identified in 45 of the 100 NDD families (53 of 119 affected children) and confirmed by Sanger sequencing (Fig. 1A and Table 3). In contrast, one diagnosis was made by clinical Sanger sequencing during the 3-year study period concurrent with genomic sequencing. That patient, CMH725, had CHD7 (chromodomain helicase DNA binding protein 7)–associated CHARGE (coloboma, heart anomaly, choanal atresia, retardation, genital and ear anomalies) syndrome [Mendelian Inheritance in Man (MIM) no. 214800]. The characteristics of families receiving diagnoses by WGS and WES were explored (tables S2 and S3). Diagnoses occurred more commonly when the clinical history included failure to thrive or intrauterine growth retardation (P = 0.04) (table S3). No other clinical characteristic examined was associated with a change in rate of molecular diagnosis (table S3). The diagnostic rate differed between the acutely ill infants and nonacutely ill older patients. Seventy-three percent (11 of 15) of families with critically ill infants were diagnosed by rapid WGS. Forty percent (34 of 85) of families with children followed in ambulatory care clinics, who had been refractory to traditional diagnosis, received diagnoses: 33 by WES and 1 by WGS after negative WES. Rapid WGS in infants was performed at or near symptom onset. The nonacute, ambulatory clinic patients were older children (average age, 83.6 months) and had received a much longer period of subspecialty care and considerable prior diagnostic testing (table S4). These patients had received an average of 13.3 prior tests/panels (range, 4 to 36) with a mean cost of $19,100, whereas the acute care group had received, on average, 7 prior diagnostic tests (range, 1 to 15) with a mean cost of $9550. In patients who received diagnoses, the inheritance of causative variants was autosomal dominant in 51% (44% de novo and 7% inherited), autosomal recessive in 33% (22% compound heterozygous and 11% homozygous), X-linked in 9% (2% de novo and 7% inherited), and mitochondrial in 6.6% (4.4% de novo and 2.2% inherited) (Fig. 1B and Table 3). De novo mutations accounted for 51% (23 of 45) of diagnoses overall and 62% (23 of 37) of diagnoses in families without a prior history of NDD. Paternity was confirmed by segregation analysis of private variants in all diagnoses associated with de novo mutations in trios.
Fig. 1. Diagnoses and inheritance patterns in 100 NDD families tested by genome or exome sequencing.
(A) Diagnostic outcomes in 100 families. (B) Inheritance pattern in 45 families. AR, autosomal recessive.
Table 3. Genomic diagnoses and impact on clinical management and clinical impression of pathophysiology.
AD, autosomal dominant; AR, autosomal recessive; M, mitochondrial genome; XL, X-linked; CEDNIK, cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma; EEEI, epileptic encephalopathy early infantile;MCAHSS, multiple congenital anomalies–hypotonia–seizures syndrome; COPD, combined oxidative phosphorylation deficiency. N.A., not applicable.
| ID | Gene | MIM | Phenotype name |
Inheritance | de Novo |
Allele 1 | Allele 2 | New gene |
Atypical phenotype |
Clinical impact |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| New treatment |
Treatment discontinued |
Comorbidity evaluated |
Change in impression |
Other | ||||||||||
| 001 002 | APTX | 208920 | Ataxia, with oculomotor apraxia (22) |
AR | c.837G>A | c.837G>A | 2 | |||||||
| 006 007 | PYCR1 | 612940 | Cutis laxa type IIB (22) |
AR | c.120_121delCA | c.120_121delCA | ||||||||
| 021 | GNAS | 103580 | Pseudohypo- parathyroidism 1a |
AD | x | c.536T>C | N.A. | 1 | ||||||
| 034 | CLPB | 815750* | None | AR | c.961A>T | c.1249C>T | x | |||||||
| 036 | COQ2 | 607426 | Coenzyme Q10 deficiency 1 (58) |
AR | c.437G>A | c.1159C>T | ||||||||
| 042 | CACNA1A | 108500 | Episodic ataxia type 2 |
AD | c.574C>T | N.A. | 1 | |||||||
| 060 | TBX1 | 192430 | Velocardiofacial syndrome |
AD | c.928G>A | N.A. | 2 | 3 | 1 | |||||
| 062 | ASPM | 608716 | Primary microcephaly |
AR | c.637delA | c.637delA | ||||||||
| 067 | MT ATP6 | 256000 | Leigh syndrome (58) |
M | x | m.8993T>G | N.A. | 1 | 1 | 1 | ||||
| 079 | ASXL3 | 615485 | Bainbridge-Ropers syndrome (12) |
AD | x | c.1897_1898delCA | N.A. | 1 | 1 | |||||
| 096 | MTOR | 601231* | None (59) | AD | x | c.4448G>T | N.A. | 1 | ||||||
| 099 | IGHBMP2 | 604320 | Distal spinal muscular atrophy |
AR | c.1478C>T | c.1808G>A | ||||||||
| 102 103 | NEB | 256030 | Nemaline myopathy 2 |
AR | c.3874A>G | c.15150delT | 1 | 3 | 3 | |||||
| 146 | KIAA2022 | 300524* | XL intellectual disability |
XL | x | c.2566C>T | N.A. | x | ||||||
| 150 | COL6A1 | 158810 | Bethlem myopathy |
AD | x | c.877G>A | N.A. | |||||||
| 169 | STXBP1 | 612164 | EEEI 4 | AD | x | c.1217G>A | N.A. | |||||||
| 172 | BRAT1 | 614498 | Rigidity and multifocal seizure syndrome, lethal neonatal (16) |
AR | c.453_454ins ATCTTCTC |
c.453_454ins ATCTTCTC |
||||||||
| 190 | TRPV4 | 600175 | Spinal muscular atrophy |
AD | c.1656delC | N.A. | 2 | 1 | ||||||
| 193 | PNPLA8 | 612123* | None | AR | c.334_337delAATT | c.1975_1976delAG | x | 1 | 1 | |||||
| 194 | ARID1B | 614525 | Intellectual disability, AD 12 |
AD | x† | c.6354C>A | N.A. | 1 | 1 | |||||
| 230 | ANKRD11 | 148050 | KBG syndrome |
AD | x | c.1385_1388delCAAA | N.A. | x | 1 | 1 | ||||
| 254 255 | NDUFV1 | 252010 | Mitochondrial complex 1 deficiency (59) |
AR | c.736G>A | c.349G>A | ||||||||
| 259 | RMND1 | 614922 | COPD | AR | c.713A>G | c.1317+1G>T | x | 2 | 1 | 1 | ||||
| 301 | PIGA | 300868 | MCAHSS | XL | c.68dupG | N.A. | x | 1 | 1 | |||||
| 311 312 | PQBP1 | 309500 | Renpenning syndrome |
XL | c.459_462delAGAG | N.A. | 1 | |||||||
| 320 321 | AHCY | 613752 | Hypermethioninemia with deficiency of S-adenosylhomocysteine hydrolase |
AR | c.293C>T | c.428A>G | 1 | |||||||
| 334 335 | MECP2 | 300055 | Intellectual disability, X-linked, syndromic 13 |
XL | c.419C>T | N.A. | 1 | |||||||
| 350 | STXBP1 | 612164 | EEEI type 4 | AD | x | c.170-2 A>G | N.A. | |||||||
| 382 383 | MAGEL2 | 615547 | Prader-Willi–like syndrome |
AD | ‡ | c.1996dupC | N.A. | |||||||
| 430 | MT ND3 | 256000 | Leigh syndrome | M | x | m.10158T>C | N.A. | |||||||
| 471 | KMT2D | 147920 | Kabuki syndrome 1 |
AD | x | c.4366dupT | N.A. | |||||||
| 502 | SNAP29 | 609528 | CEDNIK syndrome |
AR | c.520+1G>T | c.520+1G>T | ||||||||
| 545 | PTPN11 | 163950 | Noonan syndrome |
AD | x | c.922A>G | N.A. | |||||||
| 564 | UPF3B | 300676 | Intellectual disability, X-linked, 14 |
AD | x | c.1091_1094delAGAG | N.A. | |||||||
| 574 | KCNB1 | 600397* | None | AD | x | c.1133T>C | N.A. | x | ||||||
| 578 | PTPN11 | 176876 | LEOPARD syndrome |
AD | x | c.1391G9003E;C | N.A. | |||||||
| 586 | MTTE | 590025 | Reversible cyclooxygenase deficiency |
M | m.14674T>C | N.A. | 2 | 2 | 1 | 2 | ||||
| 605 | TSC1 | 191100 | Tuberous sclerosis 1 |
AD | x† | c.196G>T | N.A. | x | 5 | 1 | ||||
| 629 | SCN2A | 607745 | Seizures, benign familial infantile, 3 |
AD | x | c.4877G>A | N.A. | |||||||
| 659 | KAT6B | 606170 | Genitopatellar syndrome |
AD | x† | c.3603_3606delACAA | N.A. | |||||||
| 663 | SLC25A1 | 615182 |
d-2- and l-2-OH glutaricaciduria |
AR | c.578C>G | c.82G>A | 1 | 1 | ||||||
| 672 | KCNQ2 | 613720 | EEEI type 7 | AD | x | c.913T>C | N.A. | 1 | ||||||
| 678 | GNPTAB | 252500 | Mucolipidosis II α/β | AR | c.1017_1020dupTGCA | c.1001G>A | ||||||||
| 680 | SCN2A | 613721 | EEEI type 11 | AD | x | c.2635G>A | N.A. | 1 | ||||||
| 725 | CHD7 | 214800 | CHARGE syndrome | AD | x | c.1234C>T | N.A. | |||||||
| Total | 3 | 5 | 12 | 5 | 18 | 12 | 11 | |||||||
Gene listed in MIM (Mendelian Inheritance in Man) because disease does not yet have an MIM number.
Presumed de novo.
Presumed paternal germline mosaicism.
For patients receiving diagnoses, we sought the degree of overlap between the canonical clinical features expected for that disease and the observed clinical features in the patient. HPO terms for the clinical features in each of the 51 affected children were mapped to ~5300 MIM diseases and ~2900 genes (table S2). The Phenomizer rank of the correct diagnosis among the prioritized list of diseases matching the observed clinical features was a measure of the goodness of fit between the observed and expected presentations.(17, 19). Among the 41 affected children for whom the rank of the molecular diagnosis on the Phenomizer-derived candidate gene list was available, the median rank was 136th (range, 1st to 3103rd; table S2).
As anticipated, the time to diagnosis with 50-hour WGS was much shorter than routine WES or WGS (Table 2). Among the 11 families receiving 50-hour WGS, the fastest times to final report of a confirmed diagnosis were 6 days (n = 1), 8 days (n = 1), and 10 days (n = 2) (Table 2). Time to diagnosis was longer for recently described or previously undescribed genetic diseases and in patients whose phenotypes were atypical for the causal gene, as measured by high Phenomizer ranking or divergence from the expected disease course, such as in case CMH301 presented below.
In addition to the 45 families receiving definitive molecular diagnoses, potentially pathogenic nucleotide variants were identified in candidate disease genes in nine families. In the future, validation studies will determine whether these are indeed new disease genes. Three candidate disease genes identified during the study were subsequently validated and were included in the 45 definite diagnoses (Table 3).
Financial impact of genomic diagnoses
As a surrogate for cost-effectiveness, we determined the total cost of prior negative diagnostic testing for children who received a diagnosis. Laboratory tests, radiologic procedures, electromyograms, and nerve conduction velocity studies performed for diagnostic purposes were included (tables S4 and S5). The mean total charge for prior testing was $19,100 per family enrolled from the ambulatory care clinics (range, $3248 to $55,321; table S4). We omitted diagnostic testing at outside institutions, tests necessary for patient management (such as electroencephalograms), physician visits, phlebotomy, and other health care charges and costs. We sought to determine the cost at which, assuming a rate of diagnosis of 40% and an average charge for prior testing of $19,100 per family, WGS or WES sequencing would be cost-effective. Excluding all costs other than that of prior tests, genomic sequencing of ambulatory care patients was cost-effective at a cost of no more than $7640 per family (tables S4 and S5). Assuming WES of an average of 2.55 individuals per family, as occurred when we sought to enroll trios, it would be cost-effective as long as the cost was no more than $2996 per individual.
For 11 families enrolled from the NICU and PICU, the mean total charge of conventional diagnostic tests was $9550 (range, $3873 to $14,605; table S4). We omitted all other costs of intensive care potentially saved by earlier diagnosis, either through withdrawal of care where the prognosis rendered medical care futile or as a result of institution of an effective treatment upon diagnosis (20).
Clinical impact of genomic diagnoses
Among ambulatory care clinic patients, the mean age at symptom onset was 6.6 months (range, 0 to 90 months), enrollment was at 83.7 months (range, 1 to 252 months), and confirmed and reported diagnosis was at 95.3 months (range, 16 to 262 months) (Table 2). Among infants who received a diagnosis via rapid WGS sequencing, the median age of symptom onset was 0 day (mean, 8.2 days; range, 0 to 90 days), median age at enrollment was 38 days (range, 2 to 154 days), and median age at confirmed and reported diagnosis was 50 days (range, 8 to 521 days).
As a surrogate measure of clinical effectiveness, we assessed the short-term clinical impact of diagnoses by chart reviews and interviews with referring physicians. Diagnoses changed patient management and/ or clinical impression of the pathophysiology in 49% of the 45 families (n = 22; Table 3 and table S6). Drug or dietary treatments were started or planned in 10 children. In two, both of whom were diagnosed in infancy, there was a favorable response to the treatment. One of these, CMH663, is presented in detail below. The other, CMH680, was diagnosed with early infantile epileptic encephalopathy type 11 (MIM no. 613721) and was started on a ketogenic diet with resultant decrease in seizures. Siblings CMH001 and CMH002, with advanced ataxia with oculomotor apraxia type 1 (MIM no. 208920), were treated with oral coenzyme Q10 supplements (21, 22); however, no reversal of existing morbidity was reported. Three diagnoses enabled discontinuation of unnecessary treatments, and nine prompted evaluation for possible disease complications.
Case examples
CMH301
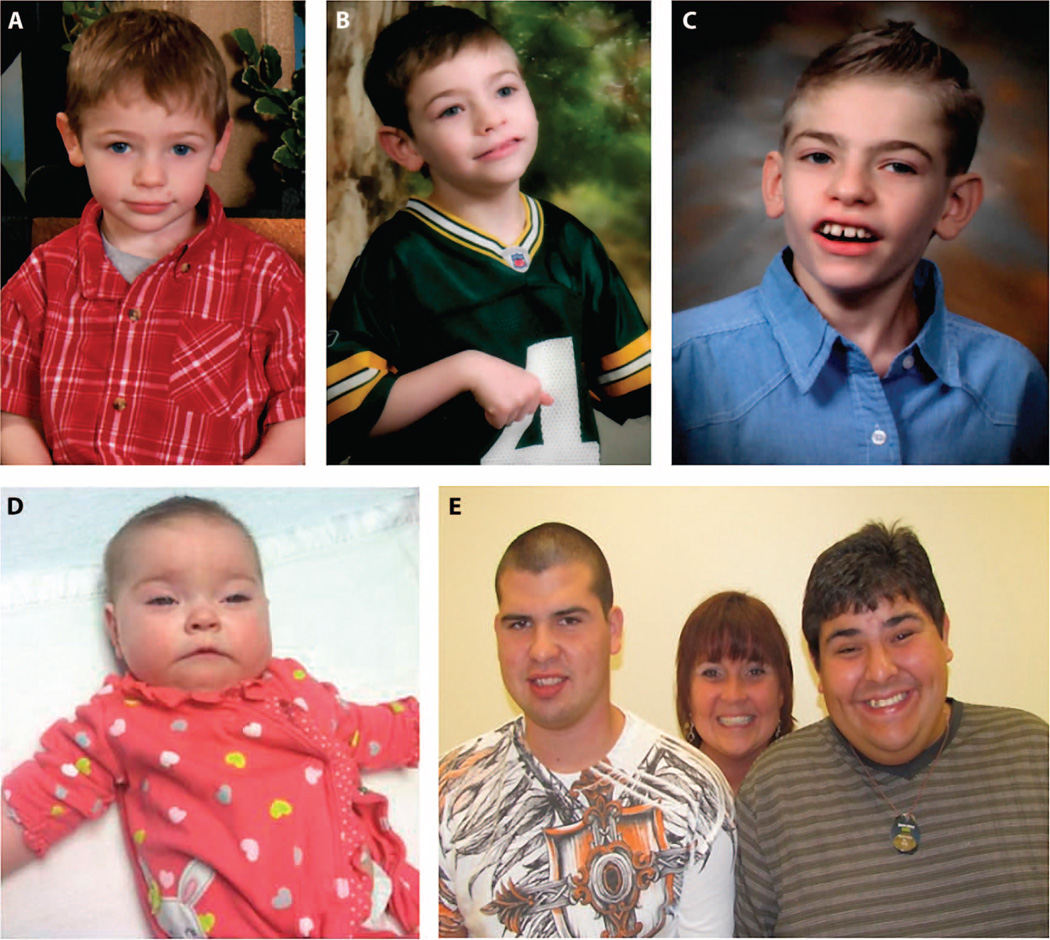
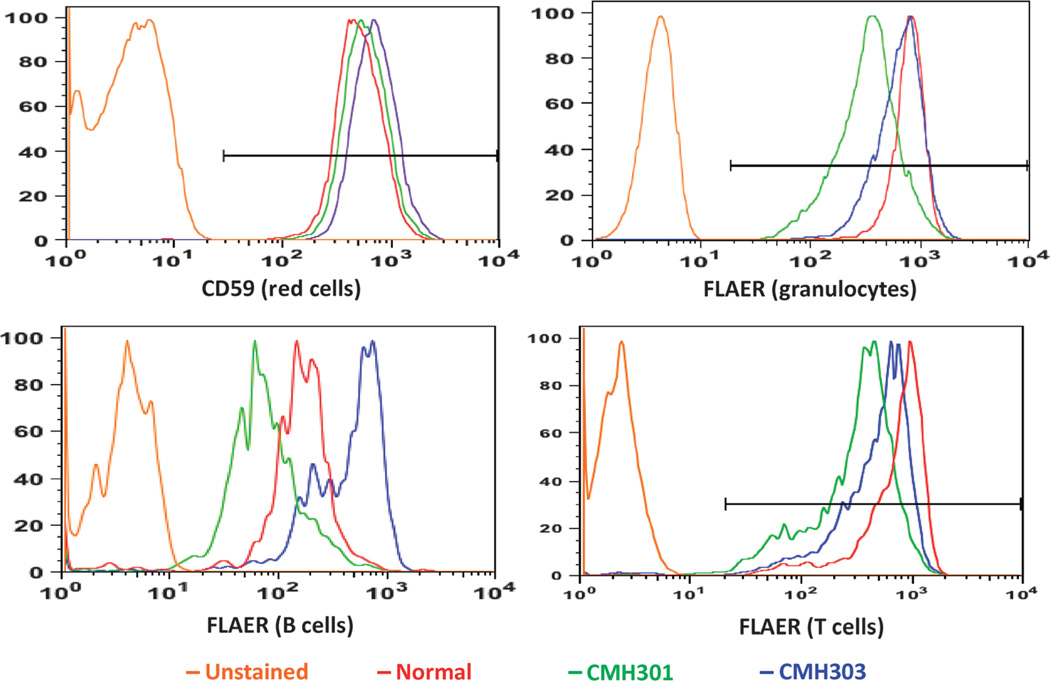
CMH301 illustrated the use of WES for diagnosis in a patient with an atypical, nonacute presentation of a recently described cause of NDD. This patient was asymptomatic until 6 months of age when he developed tonic-clonic seizures. At 1.5 years of age, he became withdrawn and developed motor stereotypies (Fig. 2A). He was diagnosed with autism spectrum disorder. Seizures occurred up to 30 times daily, despite antiepileptic treatment and a vagal nerve stimulator. At 3 years of age, he developed a tremor and unsteady gait. By age 10, he had frequent falls, loss of protective reflexes, and required a wheelchair for distances. Physical examination was notable for a long thin face, thin vermilion of the upper lip, and repetitive hand movements, including midline wringing (Fig. 2, A to C). Gait was slow and unsteady. Electroencephalogram demonstrated a left hemisphere epileptogenic focus and atypical background activity with slowing. Extensive neurologic, laboratory, and imaging evaluations were not diagnostic.WES revealed a new hemizygous variant in the class A phosphatidylinositol glycan anchor biosynthesis protein (PIGA, c.68dupG, p.Ser24LysfsX6). His unaffected mother (CMH303) was heterozygous with a random pattern (54:46) of X chromosome inactivation. PIGA has recently been associated with X-linked multiple congenital anomalies–hypotonia–seizures syndrome 2, causing death in infancy (MIM no. 300868) (23). However, Belet et al. (24) demonstrated that an early stop mutation in PIGA results in a hypomorphic protein with initiation at p.Met37. This truncated PIGA partially restores surface expression of glycosylphosphatidylinositol (GPI)–anchored proteins, consistent with the less severe phenotype in CMH301, whose variant preserves the alternative start codon. A GPI-anchored protein assay (25) confirmed decreased expression on granulocytes, T cells, and B cells, and normal erythrocyte expression (Fig. 3) consistent with the absence of hemolysis. Pyridoxine, an effective antiepileptic for at least one other GPI anchor biosynthesis disorder (26), was trialed butwas not efficacious.
Fig. 2. Clinical features of patients CMH301, CMH663, CMH334, and CMH335.
(A to C) Patient CMH301, with multiple congenital anomalies–hypotonia–seizures syndrome 2 (PIGA, c.68dupG, p.Ser24LysfsX6) at ages 2 (A), 6 (B), and 10 years (C). (D) Infant CMH663, with compound heterozygous mutations in the mitochondrial malate/citrate transporter (SLC25A1). (E) Male patients CMH334 (left) and CMH335 (right) with X-linked Rett syndrome (MECP2, c.419C>T, p.A140V) and their mother.
Fig. 3. Expression of GPI-anchored proteins on peripheral blood cells of patient CMH301.
CMH301 was diagnosed with multiple congenital anomalies–hypotonia–seizures syndrome 2. Flow cytometric signals corresponding to CMH301 are shown by the green lines, his mother CMH303 is shown in blue, and a normal control is shown in red. Erythrocytes were stained with anti-CD59 antibodies. Granulocytes, B cells, and T cells were stained with fluorescent aerolysin (FLAER). The orange line represents an unstained normal control. The x axis is the number of cells. The y axis is fluorescence intensity, representing the abundance of protein expression on the cell surface. CMH301 has normal expression of CD59 and decreased expression of GPIanchored proteins on granulocytes, B lymphocytes, and T lymphocytes.
CMH230
CMH230 underscored the power of WES to provide a molecular diagnosis in a clinically heterogeneous, nonacute disorder. This patient was born at 37 weeks after detection of a complex congenital heart defect, growth restriction, and liver calcifications in utero. A complete atrioventricular canal defect was identified on postnatal echocardiography. Dysmorphic features included two posterior hair whorls, tall skull, short forehead, low anterior hairline, flat midface, prominent eyes, periorbital fullness, down-slanting palpebral fissures, sparse curly lashes, brows with medial flare, bluish sclerae, large protruding ears, a high nasal root, bulbous nasal tip, inverted nipples, taut skin on the lower extremities, and hypotonia. Notable were the absence of wide-spaced eyes or macrodontia. Complete repair of the atrioventricular canal was performed at 7 months of age, after which her growth improved. She was diagnosed with partial complex seizures at 15 months. By 2 years, she was able to walk independently and began to develop expressive language. Karyotype and array CGH testing were not diagnostic. The clinical findings suggested a peroxisomal disorder or congenital glycosylation defect. Very long chain fatty acids, urine oligosaccharides, and transferrin studies were not diagnostic. Two N-glycan profiles demonstrated a mild increase in monogalactosylated glycan but were not consistent with a primary congenital glycosylation defect. O-glycan profile was initially suggestive of a multiple glycosylation defect, but repeat testing was normal.
WES revealed a de novo frameshift variant in the ankyrin repeat domain 11 (ANKRD11) gene (c.1385_ 1388delCAAA, p.Thr462LysfsX47) in the proband, consistent with a diagnosis of KBG syndrome (MIM no. 148050). CMH230 did not present with the typical features of KBG, which is classically characterized by hypertelorism, macrodontia, short stature, skeletal findings, and developmental delay.
CMH663
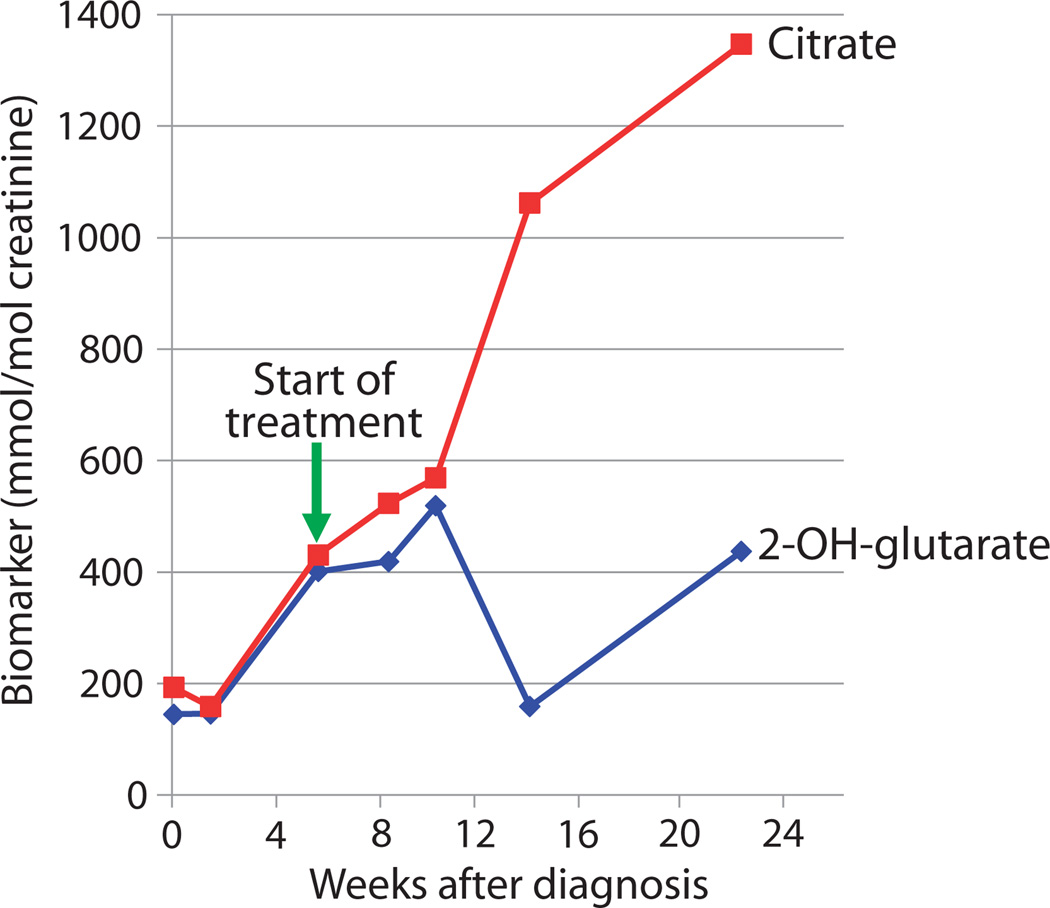
CMH663 illustrated the diagnostic use of rapid WGS in a rare cause of NDD that resulted in a change in patient management. This patient underwent evaluation at 6 months of age for delayed attainment of developmental milestones, hypotonia, mildly dysmorphic facies, and frequent episodes of respiratory distress (Fig. 2D). Extensive neurologic, laboratory, and imaging evaluations were not diagnostic. An episode of acute respiratory decompensation necessitated intubation and transfer to an intensive care unit. Electroencephalography revealed generalized slowing. Rapid WGS identified compound heterozygous missense variants in the mitochondrial malate/citrate transporter (SLC25A1, c.578C>G, p.Ser193Trp and c.82G>A, p.Ala28Thr). d-2- and l-2-hydroxyglutaric acid were elevated in plasma and urine, confirming the diagnosis of combined d-2- and l-2-hydroxyglutaric aciduria (MIM no. 615182). This disorder is associated with a poor prognosis: 8 of 13 reported patients died by 8 months of age (27). Although no standardized treatment existed, Mühlhausen et al. (28) successfully treated an affected patient with daily Na-K-citrate supplements, with subsequent decrease in biomarker concentrations and stabilization of apneic seizure-like activity that required respiratory support. CMH663 was started on oral Na-Kcitrate (1500 mg/kg per day of citrate). After 6 weeks, 2-OH-glutaric acid excretion decreased, and citric acid excretion increased (Fig. 4). Muscle tone, head control, ptosis, and alertness improved, but she subsequently developed episodes of eye twitching and upper extremity extension, correlated with left temporal and occasional right temporal spike, sharp, and slow waves suggestive of epilepsy. However, at 15 months of age, she has had no further episodes of respiratory decompensation.
Fig. 4. Effect of citrate supplementation on urinary citrate and 2-hydroxyglutarate in patient CMH663.
CMH663 had combined d-2- and l-2-hydroxyglutaric aciduria. CMH urinary citrate reference value for normal urine is >994 mmol/mol creatinine. CMH urinary 2-OH-glutarate reference value for normal urine is <89 mmol/mol creatinine.
CMH382 and CMH383
CMH382 and CMH383 illustrated the use of routine WGS for molecular diagnosis in patients with NDD in whom WES failed to yield a diagnosis. CMH382 was the first child born to healthy Caucasian, nonconsanguineous parents. Pregnancy was complicated by hyperemesis and preterm labor, resulting in birth at 32 weeks; size was appropriate for gestational age (AGA). She was hypotonic and lethargic after delivery. Hyperinsulinemic hypoglycemia was detected, and she spent 5 months in the NICU for respiratory and feeding support and blood sugar control. Physical examination was notable for ptosis, exotropia, high palate, smooth philtrum, inverted nipples, short upper arms with decreased elbow extension and wrist mobility, hypotonia, low muscle mass, and increased central distribution of body fat. She was diagnosed with autism spectrum disorder at age 3. Developmental quotients at ages 3 and 5 were less than 50. She required diazoxide treatment for hyperinsulinism until age 6. At age 7, she developed premature adrenarche, and an advanced bone age of 10 years was identified.
CMH383, the sibling of CMH382, was born at 34 weeks; size was AGA. Neonatal course was complicated by apnea, bradycardia, poor feeding, hyperinsulinemic hypoglycemia, and seizures. Physical examination was notable for marked hypotonia, finger contractures, and dysmorphic features similar to those of her sister’s. She had gross developmental delays and autistic features. Extensive neurologic, laboratory, and imaging evaluations were nondiagnostic. WES of both affected siblings and their unaffected parents did not reveal any shared pathogenic variants in NDD candidate genes. Subsequently, WGS was performed on CMH382 (HiSeq X Ten) and identified 156 rare, potentially pathogenic variants not disclosed by WES. Variant reanalysis revealed a new heterozygous, truncating variant in MAGE-like 2 (MAGEL2, c.1996dupC, p.Gln666 Profs*47). Further investigation revealed incomplete coverage of the MAGEL2 coding domain with WES but not with WGS (fig. S1). The variant was predicted to cause a premature stop codon at amino acid 713. Although this variant has not been reported in the literature, it is of a type expected to be pathogenic, leading to loss of protein function through either nonsense-mediated mRNA decay or production of a truncated protein.
Sanger sequencing confirmed the presence of the p.Gln666Profs*47 variant in CMH382 and her affected sibling CMH383. The variant was undetectable in DNA from the blood of either parent, suggesting gonadal mosaicism of this paternally expressed gene. MAGEL2 is a GC-rich (61%), intronless gene that maps within the Prader-Willi syndrome critical region on chromosome 15q11-q13. Truncating, de novo, paternally derived variants in MAGEL2 have recently been linked to Prader-Willi–like syndrome [Online MIM (OMIM) no. 615547] (29). Because MAGEL2 is imprinted and exhibits paternal monoallelic expression in the brain, the findings are consistent with a loss of MAGEL2 function. Although parental gonadal mosaicism is rare, this case highlighted the need to include analysis of de novo disease-causing variants in families with multiple affected siblings.
CMH334 and CMH335
Siblings CMH334 and CMH335 demonstrated that clinical heterogeneity in NDD can hinder molecular diagnosis by conventional methods and be circumvented byWES.CMH334 had a history of intellectual disability, a mixed seizure disorder with possible myoclonic epilepsy, and thrombocytopenia of unknown etiology. Scores on theWechsler Intelligence Scale for Children (Third Edition) revealed a verbal intelligence quotient (IQ) of 63, a performance IQ of 65, and a full-scale IQ of 61 (first percentile). At age 17, after a sedated dental procedure, he developed a lower extremity tremor that progressed to tremulous movements and facial twitching. A decline in school performance and development of severe anxiety led to further evaluation. Physical features included synophrys and prominent eyebrow ridges. Neurologic findings included saccadic eye movements, a resting upper extremity tremor, a perioral tremor, and tongue fasciculations (Fig. 2E). Deep tendon reflexes were brisk, but muscle tone, bulk, and strength were maintained. Speech was slow. Heel-to-toe gait was unsteady, but Romberg sign was negative. Laboratory studies suggested a possible creatine biosynthesis disorder; however, GATM (glycine amidinotransferase) and SLC6A8 (creatine transporter) sequencing was negative, and magnetic resonance spectroscopy revealed central nervous system creatine levels to be normal.
CMH335, a full brother, was also diagnosed with attention deficit hyperactivity disorder, intellectual disability, and epilepsy. Notable features included macrocephaly, bitemporal narrowing, obesity, hypotonia, intention tremor, and tongue fasciculations (Fig. 2E). At age 9, he had an episode of acute psychosis and transient loss of some cognitive skills, including inability to recognize family members. He had complete resolution of these symptoms after about 3 weeks. At age 16, he was again hospitalized for neuropsychiatric decompensation and a subacute decline in reading skills. He was found to have euthyroid thyroiditis with thyroglobulin antibodies at 2565 IU/ml (normal, <116 IU/ml), resulting in a diagnosis of Hashimoto encephalopathy. He also underwent a lengthy diagnostic evaluation, which included negative methylation studies for Prader-Willi/Angelman syndrome and an X-linked intellectual disability panel.
WES revealed a known pathogenic hemizygous variant in themethyl CpG binding protein 2 gene (MECP2, c.419C>T, p.A140V) in both boys; their asymptomatic mother was heterozygous. This variant has been previously reported as a hypomorphic allele that, unlike many MECP2 variants, is compatible with life in affected males. Such males exhibit Rett-like symptoms (MIM no. 312750); carrier females may have mild cognitive impairment or no symptoms (30).
DISCUSSION
Here, we report high rates of monogenetic disease diagnosis in children with NDDs by acuity-guided WGS or WES of trios. We also report retrospective estimates of clinical and cost-effectiveness of WGS- and WES-based diagnoses of NDD. Because NDD affects more than 3% of children, these results have broad implications for pediatric medicine.
The 45% rate of molecular diagnosis of NDD, reported herein, was modestly higher than in previous reports, in which 8 to 42% of individuals or families received diagnoses by WGS or WES (9, 14,15, 31–33). The highdiagnostic rate reported here reflected, in part, the use of rapid WGS in critically ill infants, who had little prior testing, with a resultant diagnosis rate of 73% (11 of 15 families). Nevertheless, the diagnostic yield in ambulatory patients who had received extensive prior testing (34 of 85 families, 40%) was also high in view of exclusion of readily diagnosed causes, low rate of consanguinity (4%), and inclusion criteria similar to prior studies (9, 14,15, 31, 32). Cases CMH382 and CMH383 highlighted the potential for WGS to detect variants missed by WES, particularly variants in GC-rich exons. However, a broader comparison of the diagnostic sensitivity of WGS and WES was precluded by the two distinct populations tested in this study. At present, there is no generalizable evidence for the superiority of 40-fold WGS or deep WES for diagnosis of monogenetic disorders (33–36). This may change with maturation of tools for identification of pathogenic non-exonic variants and understanding of the burden of causal chimerism and somatic mutations in genetic diseases.
Two other methodological characteristics may have contributed to the high overall diagnostic sensitivity. First, de novo mutations were the most common genetic cause of childhood NDD, accounting for 23 (51%) diagnoses (37). With the exception of curated known variants, such cases benefit from trio enrollment. Second, clinicopathologic software was used to translate individual symptoms into a comprehensive set of disease genes that was initially examined for causality (13, 16, 19). Such software helped to solve the immense interpretive problem of broad genetic and clinical heterogeneity of NDD (14, 19, 32, 38). This was exemplified in many of the cases reported (for example, CMH001, CMH002, CMH079, CMH096, CMH301, CMH334, and CMH335), where the clinical overlap with classic disease descriptions was modest, as objectively measured by the rank of the molecular diagnosis on the list of differential diagnosis derived from the clinical features with the Phenomizer tool (17, 19). A consequence is that it will be challenging to recapitulate dynamic, clinical feature–driven interpretive workflows in remote reference laboratories, where most molecular diagnostic testing is currently performed.
Broad adoption of acuity-guided allocation of WGS or WES for NDD will require prospective analyses of the incremental cost-effectiveness versus traditional testing. Decision-analytic models should include the total cost of implementation by health care systems and long-term comparisons of overall cost of care, given the chronicity of NDD (39, 40). Here, as a retrospective proxy, we identified the total charge for prior, negative diagnostic tests in families who received WES- or WES and WGS–based diagnoses. The average cost of prior testing, $19,100, appeared representative of tertiary pediatric practice in the United States (1). Assuming the observed rate of diagnosis (40%) in the ambulatory group, sequencing was found to be a cost-effective replacement diagnostic test up to $7640 per family or $2996 per individual. Although $2996 is at the lower end of the cost of clinical WES today, next-generation sequencing continues to decline in cost. Furthermore, the cost-effectiveness estimates reported herein excluded potential changes in health care cost associated with earlier diagnosis.
Two families powerfully illustrated the impact of WES on the cost and length of the NDD diagnostic odyssey. The first enrollees, CMH001 and CMH002, were sisters with progressive cerebellar atrophy (22). Before enrollment, they had 45 subspecialist visits during 7 years of progressive ataxia, and their cost of negative diagnostic studies exceeded $35,000. WES yielded a diagnosis of ataxia with oculomotor apraxia type 1. In contrast, 1 year later, siblings CMH102 and CMH103 were enrolled for WES at the first subspecialist visit. The cost of their diagnostic studies was $3248. WES yielded a diagnosis of nemaline myopathy. A third affected sibling was diagnosed by Sanger sequencing of the causative variants.
Another prerequisite for broad acceptance and adoption of WGS and WES for diagnosis of childhood NDD is demonstration of clinical effectiveness. The premise of genomic medicine is that early molecular diagnosis enables institution of mechanism-targeting, useful treatments before the occurrence of fixed functional deficits. Prospective clinical effectiveness studies with randomization and comparison of morbidity, quality of life, and life expectancy related to NDD have not yet been undertaken. Here, as preliminary surrogates, we retrospectively examined the time to diagnosis and changes in care upon return of new molecular diagnoses. In the ambulatory patient group, patients had been symptomatic for 77 months, on average, before enrollment. WES, if performed at symptom onset, would have had the potential to truncate the diagnostic odyssey in such cases. Time-to-diagnosis rates reported herein (WES, 11.5 months; rapid WGS, 43 days; Table 2) predict that use of rapid WGS could accelerate diagnosis by an additional 10 months. For children with progressive NDD for which treatments exist, outcomes are likely to be markedly improved by treatment institution months to years earlier than would have otherwise occurred.
Another well-established benefit of a molecular diagnosis is genetic counseling of families for recurrence risk. In the current study, there were five genetic disorder recurrences in four of the families who received diagnoses. Of equal importance, the 23 families with causative de novo variants could have been counseled earlier that, barring gonadal mosaicism, recurrence was not expected. Affected children in 49% of families receiving diagnoses by WGS or WES were reported by their physicians to have had a change in clinical management and/or clinical impression (Table 3 and table S6). A change in drug or dietary treatment either occurred or was planned in 10 families (23%), in agreement with one previous report (32). In two patients, both of whom received diagnoses in infancy, there was a favorable response to that treatment. One of these, CMH663, was presented in detail here. Given that all diagnoses were of ultrarare diseases, a recurrent finding was that the new treatment considered was supported only by case reports or studies in model systems. For example, several patients with ataxia with oculomotor apraxia type 1, which was the diagnosis for CMH001 and CMH002, had responded to oral coenzyme Q10 supplements (21). In addition to only anecdotal evidence of efficacy, the treatment of CMH001 and CMH002 with coenzyme Q10 was complicated by advanced cerebellar atrophy at time of diagnosis and the absence of pharmaceutical formulation and of pharmacokinetic, pharmacodynamic, or dosing information in children. Thus, demonstration of the clinical effectiveness of genomic medicine will require not only improved rates and timeliness of molecular diagnosis but also multidisciplinary care to identify, design, and implement candidate interventions on an N-of-1-family or N-of-1-genome basis.
NDDs exhibited a broad spectrum of monogenetic inheritance patterns and, frequently, divergence of clinical features from classical descriptions. More than 2400 genetically distinct neurologic disorders exist, underscoring the relative ineffectiveness of serial, single-gene testing (19). Furthermore, the clinical features of patients and families receiving diagnoses did not delineate a subset of NDD patients unlikely to benefit from WGS or WES. Mechanistically, the low incidence of recurrent alleles was consistent with their recent origin, as was the high rate of causative de novo mutations (32, 41, 42). Given the broad enrollment criteria used herein, it is possible that this level of genetic and clinical heterogeneity may be typical of NDD in subspecialty practice.
The evaluation of NDD patients has, historically, been constrained by the availability and cost of testing. Limited availability of tests reflects both the delay between disease gene discovery and the development of clinical diagnostic gene panels, and the adverse economics of targeted test development for ultrarare diseases. Acuity-guided WGS and WES largely circumvented these constraints. Indeed, eight of the diagnoses reported herein were in genes for which no individual clinical sequencing was available at the time of patient enrollment (ASXL3, BRAT1, CLPB, KCNB1, MTOR, PIGA, PNPLA8, and MAGEL2) (24, 43, 44).
A new candidate NDD gene or a previously undescribed presentation of a known NDD-associated gene that required additional experimental support was identified in 12 families. Three new disease-gene associations and one new phenotype were validated or reported during the study. Functional studies will need to be performed in the future for the remaining nine candidate genes, which were not included among the positive diagnoses reported here. These patients lacked causative genotypes in known disease genes and had rare, likely pathogenic changes in biologically plausible genes that exhibited appropriate familial segregation (9, 14, 32, 45). The possibility of a substantial number of new NDD genes fits with findings in other recent case series (45, 46). From a clinical standpoint, the common identification of variants of uncertain significance in candidate disease genes creates practical dilemmas that are not experienced with traditional diagnostic testing. Given the exacting principles of validation of a newdisease gene, there exists an urgent need for precompetitive sharing of the relevant pedigrees (47).
This study had several limitations. It was retrospective and lacked a control group. Clinical data were collected principally through chart review, which may have led to under- or overestimates of acute changes in management. We did not ascertain information about long-term consequences of diagnosis, such as the impact of genetic counseling. Comparisons of costs of genomic and conventional diagnostic testing excluded associated costs of testing, such as outpatient visits, and may have included tests that would nevertheless have been performed, irrespective of diagnosis. The acuity-based approach to expedited WGS and non-expedited WES was a patient care–driven approach and was not designed to facilitate direct comparisons between the two methods.
In summary, WGS and WES provided prompt diagnoses in a substantial minority of children with NDD who were undiagnosed despite extensive diagnostic evaluations. Preliminary analyses suggested that WES was less costly than continued conventional diagnostic testing of children with NDD in whom initial testing failed to yield a diagnosis. WES-based diagnoses were found to refine treatment plans in many patients with NDD. It is suggested that sequencing of genomes or exomes of trios should become an early part of the diagnostic workup of NDD and that accelerated sequencing modalities be extended to patients with high-acuity illness.
MATERIALS AND METHODS
Study design
This is a retrospective analysis of patients enrolled in a biorepository at a children’s hospital in the central United States. The repository comprised all families enrolled in a research WGS and WES program established to diagnose pediatric monogenic disorders (16). Of 155 families analyzed byWGS or WES during the first 33 months of the diagnostic program, 100 were families affected by NDD. This is a descriptive study of the 119 affected children from these families.
Study participants
Referring physicians were encouraged to nominate families for enrollment in cases with multiple affected children, consanguineous unions where both biologic parents were available for enrollment, infants receiving intensive care, or children with progressive NDD. WES was deferred when the phenotype was suggestive of genetic diseases not detectable by next-generation sequencing, such as triplet repeat disorders, or when standard cytogenetic testing or array CGH had not been obtained. Postmortem enrollment was considered for deceased probands of families receiving ongoing health care services at our institution.
NDD was characterized as central or peripheral nervous system symptoms and developmental delays or disabilities. With one exception, enrollment was from subspecialty clinics at a single, urban children’s hospital. This study was approved by the Institutional Review Board at Children’s Mercy–Kansas City. Informed written consent was obtained from adult subjects, parents of children, and children capable of assenting.
Ascertainment of clinical features in affected children
The clinical features of each affected child were ascertained by examination of electronic health records and communication with treating clinicians, translated into HPO terms (13, 16), and mapped to ~4000 monogenic diseases and ~2800 genes with the clinicopathologic correlation tools SSAGA (Symptom and Sign Associated Genome Analysis) and/or Phenomizer (17, 19) (table S2).
Exome sequencing
WES was performed in a Clinical Laboratory Improvement Amendments/College of American Pathologists–approved laboratory under a research protocol. Exome samples were prepared with either Illumina TruSeq Exome or Nextera Rapid Capture Exome kits according to the manufacturer’s protocols. (48, 49). Exon enrichment was verified by quantitative polymerase chain reaction (PCR) of four targeted loci and two nontargeted loci, both before and after enrichment (48). Samples were sequenced on Illumina HiSeq 2000 and 2500 instruments with 2 × 100 nucleotide (nt) sequences.
Genome sequencing
Genomic DNA was prepared for WGS using either Illumina TruSeq PCR-Free (rapid WGS) or TruSeq Nano (HiSeq X Ten) sample preparation according to the manufacturer’s protocols. Briefly, 500 ng of DNA was sheared with a Covaris S2 Biodisruptor, end-repaired, A-tailed, and adapter-ligated. Quantitation was carried out by real-time PCR. Libraries were sequenced by Illumina HiSeq 2500 instruments (2 × 100 nt) in rapid-run mode or by HiSeq X Ten (2 × 150 nt).
Next-generation sequencing analysis
Sequence data were generated with Illumina RTA 1.12.4.2 and CASAVA 1.8.2, aligned to the human reference NCBI (National Center for Biotechnology Information) 37 using Genomic Short-read Nucleotide Alignment Program (GSNAP) (50), and variants were detected and genotyped with the Genome Analysis Toolkit (GATK), versions 1.4 and 1.6. (51), and Alpheus v3.0 (52). Sequence analysis used FASTQ, bam, and VCF (variant call format) files. Variants were called and genotyped in WES in batches, corresponding to exome pools, using GATK 1.6 with best-practice recommendations (53). Variants were identified in WGS using GATK 1.6 without Variant Quality Score Recalibration. The largest deletion variant detected was 9992 nt, and the largest insertion was 236 nt.
Variants were annotated with the Rapid Understanding of Nucleotide variant Effect Software (RUNES v1.0) (16). RUNES incorporates data from ENSEMBL’s Variant Effect Predictor software (54), produces comparisons to NCBI Single Nucleotide Polymorphism Database, known disease variants from the Human Gene Mutation Database (55), and performs additional in silico prediction of variant consequences using RefSeq and ENSEMBL gene annotations (1, 50). RUNES categorized each variant according to ACMG recommendations for reporting sequence variation (18, 56) and with an allele frequency [minor allele frequency (MAF)] derived from Center for Pediatric Genomic Medicine’s Variant Warehouse database (16). Category 1 variants had previously been reported to be disease-causing. Category 2 variants had not previously been reported to be disease-causing but were of types that were expected to be pathogenic (loss of initiation, premature stop codon, disruption of stop codon, whole-gene deletion, frame shifting indel, and disruption of splicing). Category 3 were variants of unknown significance that were potentially disease-causing (nonsynonymous substitution, in-frame indel, disruption of polypyrimidine tract, overlap with 5′ exonic, 5′ flank, or 3′ exonic splice contexts). Category 4 were variants that were probably not causative of disease (synonymous variants that were unlikely to produce a cryptic splice site, intronic variants >20 nt from the intron/exon boundary, and variants commonly observed in unaffected individuals). Causative variants were identified primarily with Variant Integration and Knowledge INterpretation in Genomes (VIKING) software (16). Variants were filtered by limitation to ACMG categories 1 to 3 and MAF <1%. All potential monogenetic inheritance patterns including de novo, recessive, dominant, X-linked, mitochondrial, and, where possible, somatic variation were examined. Where a single likely causative variant for a recessive disorder was identified, the entire coding domain was manually inspected using the Integrated Genomics Viewer for coverage and additional variants, as were variants for that locus called in the appropriate parent that may have had low coverage in the proband (57). Expert interpretation and literature curation were performed for all likely causative variants with regard to evidence for pathogenicity. Sanger sequencing was used for clinical confirmation and reporting of all diagnostic genotypes. Additional expert consultation and functional confirmation were performed when the subject’s phenotype differed from previous mutation reports for that disease gene.
Flow cytometry
Allophycocyanin-conjugated antibodies to CD59 were obtained from Becton Dickinson. Detection of GPI-anchored protein expression on granulocytes, B cells, and T cells was performed with a fluorescent aerolysin– based assay (Protox Biotech) (25). Before staining white blood cells, whole blood was incubated in 1× red blood cell lysis buffer (Gibco). The remaining nucleated cells were identified on the basis of forward and side scatter and by staining with phycoerythrin-conjugated anti-CD3 (T cells), anti- CD15 (granulocytes), and anti-CD20 (B cells) antibodies (Becton Dickinson). Acquisition and analysis were performed by flow cytometry (FACSCalibur, Becton Dickinson) and FlowJo (Tree Star Inc). For all cell types, the isotypic control was set at 1% (25).
Supplementary Material
Acknowledgments
The authors thank L. Biesecker and the Divisions of Child Neurology, Developmental and Behavioral Sciences, Medical Genetics, Neonatology, and Rehabilitation Medicine at Children’s Mercy–Kansas City for their contribution and thank the families for their participation in this research study. A Deo lumen, ab amicis auxilium. Funding: This work was supported by the Marion Merrell Dow Foundation, Children’s Mercy–Kansas City, Patton Trust, William T. Kemper Foundation, Pat and Gil Clements Foundation, Claire Giannini Foundation, Black and Veatch, National Institute of Child Health and Human Development and National Human Genome Research Institute (grant U19HD077693), and National Center for Advancing Translational Sciences (Clinical and Translational Science Award grant TL1TR000120).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/6/265/265ra168/DC1
Fig. S1. Genome and exome coverage of MAGEL2 in patient CMH382.
Table S1. Genome and exome sequencing and variant metrics in NDD-affected children.
Table S2. Phenotype-to-gene mapping with the clinicopathologic software tool Phenomizer.
Table S3. Associations between clinical and familial characteristics and exome or genome diagnoses.
Table S4. Time to diagnosis and cost of prior etiologic clinical tests for children with NDD.
Table S5. Etiologic tests ordered before WGS/WES.
Table S6. Impact of genomic diagnosis on patient management.
Author contributions: S.E.S. and S.F.K. undertook analysis of data and wrote the manuscript; C.J.S., N.A.M., E.G.F., L.D.S., L.K.W., I.T., D.L.D., L.Z., A.M.A., S.S.N., I.A.L., B.Z., M.C., and S.H. undertook analysis of data and edited the manuscript. N.A.M., G.T., and A.N. developed the software and bioinformatic tools and pipelines; J.-B.L., B.A.H., A.T.A., N.S., J.E.P., and A.M., compiled patient information and edited the manuscript; and Z.Y., X.Y., and R.A.B. performed functional studies and helped write the manuscript.
Competing interests: A patent application has been filed for some of the software used to analyze the data reported in this manuscript. The authors declare that they have no other competing interests.
Data and materials availability: Genomic sequence data are available at the database of Genotypes and Phenotypes under accession no. phs000564.
REFERENCES AND NOTES
- 1.Shashi V, McConkie-Rosell A, Rosell B, Schoch K, Vellore K, McDonald M, Jiang YH, Xie P, Need A, Goldstein DB. The utility of the traditional medical genetics diagnostic evaluation in the context of next-generation sequencing for undiagnosed genetic disorders. Genet. Med. 2014;16:176–182. doi: 10.1038/gim.2013.99. [DOI] [PubMed] [Google Scholar]
- 2.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battaglia A, Doccini V, Bernardini L, Novelli A, Loddo S, Capalbo A, Filippi T, Carey JC. Confirmation of chromosomal microarray as a first-tier clinical diagnostic test for individuals with developmental delay, intellectual disability, autism spectrum disorders and dysmorphic features. Eur. J. Paediatr. Neurol. 2013;17:589–599. doi: 10.1016/j.ejpn.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Bartnik M, Nowakowska B, Derwińska K, Wiśniowiecka-Kowalnik B, Kędzior M, Bernaciak J, Ziemkiewicz K, Gambin T, Sykulski M, Bezniakow N, Korniszewski L, Kutkowska-Kaźmierczak A, Klapecki J, Szczałuba K, Shaw CA, Mazurczak T, Gambin A, Obersztyn E, Bocian E, Stankiewicz P. Application of array comparative genomic hybridization in 256 patients with developmental delay or intellectual disability. J. Appl. Genet. 2014;55:125–144. doi: 10.1007/s13353-013-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kashevarova AA, Nazarenko LP, Skryabin NA, Salyukova OA, Chechetkina NN, Tolmacheva EN, Sazhenova EA, Magini P, Graziano C, Romeo G, Kučinskas V, Lebedev IN. Array CGH analysis of a cohort of Russian patients with intellectual disability. Gene. 2014;536:145–150. doi: 10.1016/j.gene.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 6.Ropers HH. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 7.Vorstman JA, Ophoff RA. Genetic causes of developmental disorders. Curr. Opin. Neurol. 2013;26:128–136. doi: 10.1097/WCO.0b013e32835f1a30. [DOI] [PubMed] [Google Scholar]
- 8.Piton A, Jouan L, Rochefort D, Dobrzeniecka S, Lachapelle K, Dion PA, Gauthier J, Rouleau GA. Analysis of the effects of rare variants on splicing identifies alterations in GABAA receptor genes in autism spectrum disorder individuals. Eur. J. Hum. Genet. 2013;21:749–756. doi: 10.1038/ejhg.2012.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ligt J, Willemsen MH, van Bon BW, Kleefstra T, Yntema HG, Kroes T, Vulto-van Silfhout AT, Koolen DA, de Vries P, Gilissen C, del Rosario M, Hoischen A, Scheffer H, de Vries BB, Brunner HG, Veltman JA, Vissers LE. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 10.Lubs HA, Stevenson RE, Schwartz CE. Fragile X and X-linked intellectual disability: Four decades of discovery. Am. J. Hum. Genet. 2012;90:579–590. doi: 10.1016/j.ajhg.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green ED, Guyer MS. National Human Genome Research Institute, Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 12.Dinwiddie DL, Soden SE, Saunders CJ, Miller NA, Farrow EG, Smith LD, Kingsmore SF. De novo frameshift mutation in ASXL3 in a patient with global developmental delay, microcephaly, and craniofacial anomalies. BMC Med. Genomics. 2013;6:32. doi: 10.1186/1755-8794-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kingsmore SF, Dinwiddie DL, Miller NA, Soden SE, Saunders CJ. Adopting orphans: Comprehensive genetic testing of Mendelian diseases of childhood by next-generation sequencing. Expert Rev. Mol. Diagn. 2011;11:855–868. doi: 10.1586/erm.11.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dixon-Salazar TJ, Silhavy JL, Udpa N, Schroth J, Bielas S, Schaffer AE, Olvera J, Bafna V, Zaki MS, Abdel-Salam GH, Mansour LA, Selim L, Abdel-Hadi S, Marzouki N, Ben-Omran T, Al-Saana NA, Sonmez FM, Celep F, Azam M, Hill KJ, Collazo A, Fenstermaker AG, Novarino G, Akizu N, Garimella KV, Sougnez C, Russ C, Gabriel SB, Gleeson JG. Exome sequencing can improve diagnosis and alter patient management. Sci. Transl. Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci. Transl. Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee, ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 19.Köhler S, Doelken SC, Rath A, Aymé S, Robinson PN. Ontological phenotype standards for neurogenetics. Hum. Mutat. 2012;33:1333–1339. doi: 10.1002/humu.22112. [DOI] [PubMed] [Google Scholar]
- 20.Weiner J, Sharma J, Lantos J, Kilbride H. How infants die in the neonatal intensive care unit: Trends from 1999 through 2008. Arch. Pediatr. Adolesc. Med. 2011;165:630–634. doi: 10.1001/archpediatrics.2011.102. [DOI] [PubMed] [Google Scholar]
- 21.Quinzii CM, Hirano M. Primary and secondary CoQ10 deficiencies in humans. Biofactors. 2011;37:361–365. doi: 10.1002/biof.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soden SE, Saunders CJ, Dinwiddie DL, Miller NA, Atherton AM, Alnadi NA, Leeder JS, Smith LD, Kingsmore SF. A systematic approach to implementing monogenic genomic medicine. J. Genomes Exomes. 2012;1:15–24. [Google Scholar]
- 23.Johnston JJ, Gropman AL, Sapp JC, Teer JK, Martin JM, Liu CF, Yuan X, Ye Z, Cheng L, Brodsky RA, Biesecker LG. The phenotype of a germline mutation in PIGA: The gene somatically mutated in paroxysmal nocturnal hemoglobinuria. Am. J. Hum. Genet. 2012;90:295–300. doi: 10.1016/j.ajhg.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belet S, Fieremans N, Yuan X, Van Esch H, Verbeeck J, Ye Z, Cheng L, Brodsky BR, Hu H, Kalscheuer VM, Brodsky RA, Froyen G. Early frameshift mutation in PIGA identified in a large XLID family without neonatal lethality. Hum. Mutat. 2014;35:350–355. doi: 10.1002/humu.22498. [DOI] [PubMed] [Google Scholar]
- 25.Brodsky RA, Mukhina GL, Li S, Nelson KL, Chiurazzi PL, Buckley JT, Borowitz MJ. Improved detection and characterization of paroxysmal nocturnal hemoglobinuria using fluorescent aerolysin. Am. J. Clin. Pathol. 2000;114:459–466. doi: 10.1093/ajcp/114.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuki I, Takahashi Y, Okazaki S, Kawawaki H, Ehara E, Inoue N, Kinoshita T, Murakami Y. Vitamin B6–responsive epilepsy due to inherited GPI deficiency. Neurology. 2013;81:1467–1469. doi: 10.1212/WNL.0b013e3182a8411a. [DOI] [PubMed] [Google Scholar]
- 27.Nota B, Struys EA, Pop A, Jansen EE, Fernandez Ojeda MR, Kanhai WA, Kranendijk M, van Dooren SJ, Bevova MR, Sistermans EA, Nieuwint AW, Barth M, Ben-Omran T, Hoffmann GF, de Lonlay P, McDonald MT, Meberg A, Muntau AC, Nuoffer JM, Parini R, Read MH, Renneberg A, Santer R, Strahleck T, van Schaftingen E, van der Knaap MS, Jakobs C, Salomons GS. Deficiency in SLC25A1, encoding the mitochondrial citrate carrier, causes combined D-2- and L-2-hydroxyglutaric aciduria. Am. J. Hum. Genet. 2013;92:627–631. doi: 10.1016/j.ajhg.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mühlhausen C, Salomons GS, Lukacs Z, Struys EA, van der Knaap MS, Ullrich K, Santer R. Combined D2-/L2-hydroxyglutaric aciduria (SLC25A1 deficiency): Clinical course and effects of citrate treatment. J. Inherit. Metab. Dis. 2014;37:775–781. doi: 10.1007/s10545-014-9702-y. [DOI] [PubMed] [Google Scholar]
- 29.Schaaf CP, Gonzalez-Garay ML, Xia F, Potocki L, Gripp KW, Zhang B, Peters BA, McElwain MA, Drmanac R, Beaudet AL, Caskey CT, Yang Y. Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism. Nat. Genet. 2013;45:1405–1408. doi: 10.1038/ng.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudo S, Nomura Y, Segawa M, Fujita N, Nakao M, Schanen C, Tamura M. Heterogeneity in residual function of MeCP2 carrying missense mutations in the methyl CpG binding domain. J. Med. Genet. 2003;40:487–493. doi: 10.1136/jmg.40.7.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Need AC, Shashi V, Hitomi Y, Schoch K, Shianna KV, McDonald MT, Meisler MH, Goldstein DB. Clinical application of exome sequencing in undiagnosed genetic conditions. J. Med. Genet. 2012;49:353–361. doi: 10.1136/jmedgenet-2012-100819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava S, Cohen JS, Vernon H, Barañano K, McClellan R, Jamal L, Naidu S, Fatemi A. Clinical whole exome sequencing in child neurology practice. Ann. Neurol. 2014;76:473–483. doi: 10.1002/ana.24251. [DOI] [PubMed] [Google Scholar]
- 33.Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, Kwint M, Janssen IM, Hoischen A, Schenck A, Leach R, Klein R, Tearle R, Bo T, Pfundt R, Yntema HG, de Vries BB, Kleefstra T, Brunner HG, Vissers LE, Veltman JA. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 34.Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge MN, Jhangiani S, Buhay CJ, Kovar CL, Wang M, Hawes AC, Reid JG, Eng C, Muzny DM, Gibbs RA. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linderman MD, Brandt T, Edelmann L, Jabado O, Kasai Y, Kornreich R, Mahajan M, Shah H, Kasarskis A, Schadt EE. Analytical validation of whole exome and whole genome sequencing for clinical applications. BMC Med. Genomics. 2014;7:20. doi: 10.1186/1755-8794-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics. 2014;15:247. doi: 10.1186/1471-2105-15-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, Kuperman R, Witt O, Kohrman MH, Flamini JR, Wu JY, Curatolo P, de Vries PJ, Whittemore VH, Thiele EA, Ford JP, Shah G, Cauwel H, Lebwohl D, Sahmoud T, Jozwiak S. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): A multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2013;381:125–132. doi: 10.1016/S0140-6736(12)61134-9. [DOI] [PubMed] [Google Scholar]
- 38.Robinson PN. Deep phenotyping for precision medicine. Hum. Mutat. 2012;33:777–780. doi: 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- 39.Chan K, Davis J, Pai SY, Bonilla FA, Puck JM, Apkon M. A Markov model to analyze cost-effectiveness of screening for severe combined immunodeficiency (SCID) Mol. Genet. Metab. 2011;104:383–389. doi: 10.1016/j.ymgme.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan K, Lai MN, Groessl EJ, Hanchate AD, Wong JB, Clark JA, Asch SM, Gifford AL, Ho SB. Cost effectiveness of direct-acting antiviral therapy for treatment-naive patients with chronic HCV genotype 1 infection in the veterans health administration. Clin. Gastroenterol. Hepatol. 2013;11:1503–1510. doi: 10.1016/j.cgh.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 41.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, NHLBI Exome Sequencing Project. Akey JM. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blekhman R, Man O, Herrmann L, Boyko AR, Indap A, Kosiol C, Bustamante CD, Teshima KM, Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr. Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kato M, Saitsu H, Murakami Y, Kikuchi K, Watanabe S, Iai M, Miya K, Matsuura R, Takayama R, Ohba C, Nakashima M, Tsurusaki Y, Miyake N, Hamano S, Osaka H, Hayasaka K, Kinoshita T, Matsumoto N. PIGA mutations cause early-onset epileptic encephalopathies and distinctive features. Neurology. 2014;82:1587–1596. doi: 10.1212/WNL.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 44.Torkamani A, Bersell K, Jorge BS, Bjork RL, Jr, Friedman JR, Bloss CS, Cohen J, Gupta S, Naidu S, Vanoye CG, George AL, Jr, Kearney JA. De novo KCNB1 mutations in epileptic encephalopathy. Ann. Neurol. 2014;76:529–540. doi: 10.1002/ana.24263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Röpke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schröck E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: An exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 46.Musante L, Ropers HH. Genetics of recessive cognitive disorders. Trends Genet. 2014;30:32–39. doi: 10.1016/j.tig.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Kingsmore SF. A new journal and a new model for structured data dissemination for an era of genomic medicine. J. Genomes Exomes. 2012;1:1–5. [Google Scholar]
- 48.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quail MA, Otto TD, Gu Y, Harris SR, Skelly TF, McQuillan JA, Swerdlow HP, Oyola SO. Optimal enzymes for amplifying sequencing libraries. Nat. Methods. 2012;9:10–11. doi: 10.1038/nmeth.1814. [DOI] [PubMed] [Google Scholar]
- 50.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Pohl A, Malladi VS, Li CH, Learned K, Kirkup V, Hsu F, Harte RA, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Diekhans M, Cline MS, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC Genome Browser database: Extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller NA, Kingsmore SF, Farmer A, Langley RJ, Mudge J, Crow JA, Gonzalez AJ, Schilkey FD, Kim RJ, van Velkinburgh J, May GD, Black CF, Myers MK, Utsey JP, Frost NS, Sugarbaker DJ, Bueno R, Gullans SR, Baxter SM, Day SW, Retzel EF. Management of high-throughput DNA sequencing projects: Alpheus. J. Comput. Sci. Syst. Biol. 2008;1:132. doi: 10.4172/jcsb.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenson PD, Ball EV, Howells K, Phillips AD, Mort M, Cooper DN. The Human Gene Mutation Database: Providing a comprehensive central mutation database for molecular diagnostics and personalized genomics. Hum. Genomics. 2009;4:69–72. doi: 10.1186/1479-7364-4-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maddalena A, Bale S, Das S, Grody W, Richards S. ACMG Laboratory Quality Assurance Committee, Technical standards and guidelines: Molecular genetic testing for ultra-rare disorders. Genet. Med. 2005;7:571–583. doi: 10.1097/01.gim.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- 57.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dinwiddie DL, Smith LD, Miller NA, Atherton AM, Farrow EG, Strenk ME, Soden SE, Saunders CJ, Kingsmore SF. Diagnosis of mitochondrial disorders by concomitant next-generation sequencing of the exome and mitochondrial genome. Genomics. 2013;102:148–156. doi: 10.1016/j.ygeno.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith LD, Saunders CJ, Dinwiddie DL, Atherton AM, Miller NA, Soden SE, Farrow EG, Abdelmoity ATG, Kingsmore SF. Exome sequencing reveals de novo germline mutation of the mammalian target of rapamycin (MTOR) in a patient with megalencephaly and intractable seizures. J. Genomes Exomes. 2013;2:63–72. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.