Abstract
Background
Genes, RNAs, and proteins play important roles during germline development. However, the functions of non-coding RNAs (ncRNAs) on germline development remain unclear in avian species. Recent high-throughput techniques have identified several classes of ncRNAs, including micro RNAs (miRNAs), small-interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs). These ncRNAs are functionally important in the genome, however, the identification and annotation of ncRNAs in a genome is challenging. The aim of this study was to identify different types of small ncRNAs particularly piRNAs, and the role of piRNA pathway genes in the protection of chicken primordial germ cells (PGCs).
Results
At first, we performed next-generation sequencing to identify ncRNAs in chicken PGCs, and we performed ab initio predictive analysis to identify putative piRNAs in PGCs. Then, we examined the expression of three repetitive sequence-linked piRNAs and 14 genic-transcript-linked piRNAs along with their linked genes using real-time PCR. All piRNAs and their linked genes were highly expressed in PGCs. Subsequently, we knocked down two known piRNA pathway genes of chicken, PIWI-like protein 1 (CIWI) and 2 (CILI), in PGCs using siRNAs. After knockdown of CIWI and CILI, we examined their effects on the expression of six putative piRNA-linked genes and DNA double-strand breakage in PGCs. The knockdown of CIWI and CILI upregulated chicken repetitive 1 (CR1) element and RAP2B, a member of RAS oncogene family, and increased DNA double-strand breakage in PGCs.
Conclusions
Our results increase the understanding of PGC-expressed piRNAs and the role of piRNA pathway genes in the protection of germ cells.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-757) contains supplementary material, which is available to authorized users.
Keywords: Aves, Non-coding RNA, piRNA, Primordial Germ Cells
Background
Organisms that undergo sexual reproduction usually develop from the fusion of male and female gametes during the fertilization process. Germ cells are the only cells that produce functional gametes and transmit parental genetic information to the progeny. Primordial germ cells (PGCs) are the precursors of germ cells and are specified during the early days of embryonic development in all vertebrate species. The origin, migratory routes, and timing of PGC differentiation into germ cells vary among vertebrate species. In chickens, PGCs emerged initially in cleavage-stage embryos [1], and then migrated through the hypoblast layer, germinal crescent area, and blood vessels to reach the bilateral embryonic gonads by approximately embryonic day (E) 2.5 [1–3]. After reaching the gonads, PGCs undergo rapid mitotic cell division to increase their population number. Finally, PGCs differentiate into oogonia in females and prospermatogonia in males at approximately E8.0 and E13.0, respectively [4, 5]. Chicken PGCs can be easily isolated from the early embryos, and cultured long-term without losing their characteristic features [6]. Therefore, chicken PGCs act as an efficient tool for studying the early migration of germ cells, for identifying the germ cell-expressed gene functions, and for the production of transgenic birds [5, 7–10]. Recently, several reviews were emphasized the very recent progresses of PGC studies in biomedical sciences and animal biotechnology [11–14]. Early embryonic development, including the PGC lineage, is governed by the action of many genes and proteins. However, previous studies have shown that small non-coding RNAs (ncRNAs) do not efficiently govern PGCs in vertebrate species. ncRNAs are functional RNAs but lack coding sequences that can be translated into functional proteins. ncRNAs have been described as a broad class of regulatory RNA molecules whose functions continue to be characterized in a variety of model organisms and diseases [15]. The classification of ncRNAs includes highly abundant and functionally important transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small cytoplasmic RNA (scRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), micro RNAs (miRNAs), small-interfering RNAs (siRNAs), and P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) [16–18]. The sizes of most small ncRNAs range between 18 and 32 nt in length, but determining the total number of these ncRNAs in a genome remains challenging. Recently, many high-throughput technologies, including genome-wide association (GWA) studies, chromatin immunoprecipitation followed by sequencing (ChIP-seq) and RNA sequencing (RNA-seq), have been developed to examine various aspects of cellular processes, including the transcriptome, epigenome, proteome and interactome [16, 19]. Next-generation sequencing (NGS) has played an important role in genomic research and has fundamentally changed the nature of genetic experimentation [15]. NGS can be used to detect alternative splice variants using paired ends, as well as to detect relatively short reads or longer reads. In addition, NGS can detect novel classes of ncRNAs [17, 19]. In this study, we performed high-throughput NGS followed by standard annotation protocols to identify different types of small ncRNAs particularly piRNAs in chicken PGCs compared with gonadal stromal cells (GSCs) and chicken embryonic fibroblasts (CEFs), in order to verify their biofunctional activity.
Results
Next-generation sequencing of small ncRNAs
High-throughput NGS was performed in chicken PGCs using the Illumina HiSeq platform. We maintained stage X blastoderms, GSCs, and CEFs as reference samples. Standard protocols were then followed to screen the sequencing data and annotate the small ncRNAs. Raw NGS data were processed to obtain clean reads by removing low-quality reads (Q-value < 13), short read tags (<18 nt), and adaptor-ligated contaminants. After the cleaning process, the raw data contained 8,199,557 total reads in PGCs, 6,341,942 in stage X blastoderms, 9,169,772 in GSCs, and 15,180,853 in CEFs. From these total reads, there were 1,010,670 unique reads in PGCs, 487,258 in stage X blastoderms, 462,903 in GSCs, and 217,402 in CEFs (Table 1). The statistics of sequencing quality is shown in Additional file 1: Table S1. All obtained sequence reads were matched with the current release of the chicken genome in the National Center for Biotechnology Information (NCBI, Gallus gallus v.4). Among the annotated reads, the most abundant total read length was ~22 nt. However, the most abundant length of the unique reads in PGCs was ~26 nt, in contrast to the other test samples (Additional file 2: Figure S1). Among the annotated reads in PGCs, the majority of unique reads were mapped to repetitive elements (298,103), followed by exon, rRNA, intron, or tRNA sequences. In stage X blastoderms, the majority of unique reads were mapped to repetitive elements (54,461), followed by exon, rRNA, intron, or tRNA sequences. In GSCs, the majority of unique reads were mapped to exons (69,845), followed by rRNA, repetitive elements, intron, or tRNA sequences. In CEFs, the majority of unique reads were mapped to rRNAs (23,021), followed by exon, intron, tRNA, or repetitive element sequences (Figure 1). The remaining unique reads were mapped to snRNAs, snoRNAs, and scRNAs, while miRNAs were found in low frequencies in all test samples (Figure 1). We compared the RPKM (reads per kilobase per million reads) values of all unique reads to identify the upregulated ncRNAs in the test samples. Based on the 2-fold cutoff value, 14,624 (55.55%) small ncRNAs originating from repeat sequences were upregulated in PGCs. In addition, 1,281 (4.87%) rRNAs were upregulated in PGCs. In stage X blastoderms, 7,395 (26.5%) upregulated sequences were small ncRNAs originating from repeat sequences, and 7,245 (25.96%) upregulated sequences were rRNAs. In GSCs, 3,141 (63.24%) upregulated sequences were rRNAs, and 324 (6.52%) upregulated sequences were miRNAs. In CEFs, 1,261 (24.59%) upregulated sequences were rRNAs, and 742 (14.47%) upregulated sequences were miRNAs. In addition, a significant number (>25%) of unannotated sequence reads were upregulated in all test samples (Table 2).
Table 1.
Raw and processed data of next-generation sequencing
| Raw data | Clean data (Identity ≥ 90% and HSP coverage ≥ 95% to G. gallus v.4) | ||||||
|---|---|---|---|---|---|---|---|
| Sample library | Number of total reads | Total base pairs | Number of total reads | Total base pairs | Minimum base pairs | Maximum base pairs | Number of unique reads |
| PGCs | 9175177 | 449583673 | 8199557 | 193849279 | 18 | 44 | 1010670 |
| Stage X | 9243725 | 452942525 | 6341942 | 154246061 | 18 | 44 | 487258 |
| GSCs | 10314115 | 505391635 | 9169772 | 208574068 | 18 | 44 | 462903 |
| CEFs | 16705966 | 818592334 | 15180321 | 334940464 | 18 | 44 | 217402 |
Figure 1.
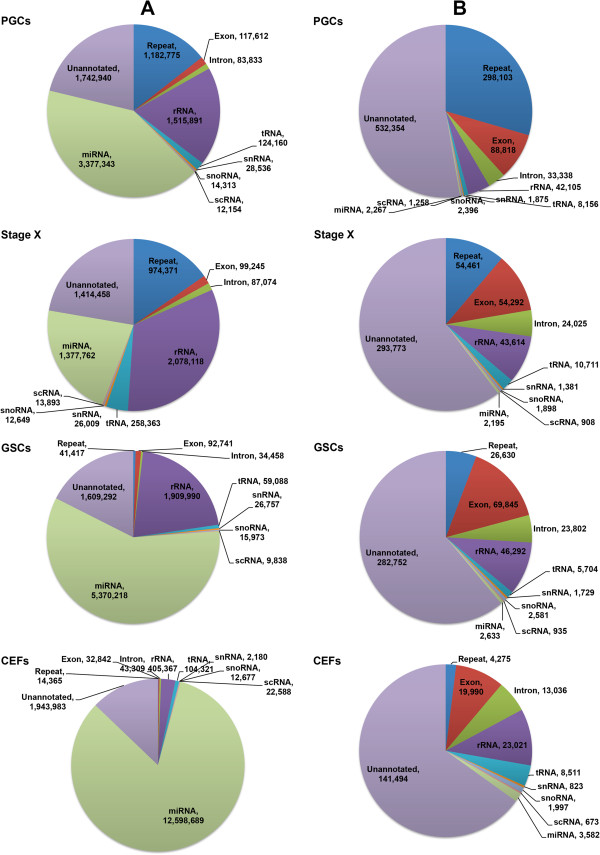
Genome distribution of total and unique reads. Genome distribution of total (A) and unique (B) reads in PGCs, stage X blastoderms, GSCs, and CEFs obtained using high-throughput next-generation sequencing following the standard annotation protocols.
Table 2.
Unique small ncRNAs that are upregulated in PGCs, stage X blastoderms, GSCs, and CEFs
| PGCs | Stage X | GSCs | CEFs | |||||
|---|---|---|---|---|---|---|---|---|
| (PGCs vs Stage X, GSCs, CEFs) | (Stage X vs PGCs, GSCs, CEFs) | (GSCs vs PGCs, Stage X, CEFs) | (CEFs vs PGCs, Stage X, GSCs) | |||||
| Number of unique reads | % | Number of unique reads | % | Number of unique reads | % | Number of unique reads | % | |
| Repeat | 14624 | 55.55 | 7395 | 26.50 | 5 | 0.10 | 49 | 0.96 |
| Unannotated | 8394 | 31.88 | 9335 | 33.46 | 1248 | 25.13 | 2181 | 42.53 |
| rRNAs | 1281 | 4.87 | 7245 | 25.96 | 3141 | 63.24 | 1261 | 24.59 |
| Intron sense | 629 | 2.39 | 566 | 2.03 | 4 | 0.08 | 84 | 1.64 |
| tRNAs | 507 | 1.93 | 1831 | 6.56 | 37 | 0.74 | 554 | 10.80 |
| Intron antisense | 320 | 1.22 | 180 | 0.65 | 8 | 0.16 | 51 | 0.99 |
| Exon sense | 251 | 0.95 | 435 | 1.56 | 49 | 0.99 | 67 | 1.31 |
| scRNA | 92 | 0.35 | 124 | 0.44 | 20 | 0.40 | 44 | 0.86 |
| miRNAs | 96 | 0.36 | 448 | 1.61 | 324 | 6.52 | 742 | 14.47 |
| snoRNA | 75 | 0.28 | 165 | 0.59 | 96 | 1.93 | 79 | 1.54 |
| snRNA | 53 | 0.20 | 151 | 0.54 | 35 | 0.70 | 9 | 0.18 |
| Exon antisense | 6 | 0.02 | 28 | 0.10 | 0 | - | 7 | 0.14 |
| Total | 26328 | 100 | 27903 | 100 | 4967 | 100 | 5128 | 100 |
piRNAs obtained using proTRAC software
Since the most abundant base pair size (~26 nt) and annotated classification (repeat sequence origin) of unique reads in PGCs were the potential characteristic of piRNAs, we performed ab initio predictive analysis using probabilistic TRacking and Analysis of Clusters (proTRAC) software to identify putative piRNAs in chicken PGCs compared with stage X blastoderms, GSCs, and CEFs. Among all annotated and unannotated unique reads, those that met the input criteria were only accepted for analysis using proTRAC software. The proTRAC output revealed 92,373 unique piRNAs in PGCs. Among the putative piRNAs, a large number (74,337) were derived from repeat sequences. The second largest number of piRNAs (14,478) was derived from unannotated sequences. Approximately 2,827 piRNAs were derived from intronic genetic sequences, and ~ 645 piRNAs from gene exons. In addition, a small proportion of other ncRNAs were identified as piRNAs. In stage X blastoderms, proTRAC output revealed 12,124 unique piRNAs, among which 10,925 were derived from repeat sequences. In GSCs, proTRAC output revealed 4,930 unique piRNAs, among which 3,116 were derived from repeat sequences. In CEFs, proTRAC output revealed only 642 unique piRNAs, and among these piRNAs the most abundant were derived from unannotated sequences (Table 3). In order to cross-validate the putative chicken piRNAs, we performed another ab initio predictive analysis using piRNApredictor software with the sequence reads of test samples. The piRNApredictor output revealed more number of unique piRNAs in each test sample (Additional file 3: Table S2). The output results of proTRAC and piRNApredictor were then compared. In this comparison also, a large number of piRNAs were derived from repeat sequences in PGCs. The second largest number of piRNAs was derived from unannotated sequences (Additional file 4: Table S3). However, the piRNAs identified using proTRAC were only referred in the subsequent analysis in this study.
Table 3.
proTRAC output and distribution of piRNAs from next-generation sequencing
| Samples | proTRAC input | proTRAC output | Distribution of piRNAs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number of unique reads | Number of unique piRNAs | Repeat | Unannotated | Intron sense | Intron antisense | Exon sense | Exon antisense | snRNA | rRNA | miRNA | tRNA | |
| PGCs | 687544 | 92373 | 74337 | 14478 | 1961 | 866 | 524 | 121 | 50 | 22 | 13 | 1 |
| Stage X | 350113 | 12124 | 10925 | 925 | 194 | 63 | 13 | 1 | 0 | 2 | 0 | 1 |
| GSCs | 350647 | 4930 | 3116 | 1562 | 121 | 62 | 59 | 0 | 5 | 0 | 5 | 0 |
| CEFs | 159803 | 642 | 31 | 360 | 17 | 2 | 122 | 20 | 1 | 2 | 0 | 87 |
Table 4 shows the top 20 putative piRNAs originating from repeat sequences based on the RPKM values in PGCs, the majority of which were derived from the long interspersed element (LINE) or chicken repetitive 1 (CR1). Table 5 shows the top 20 putative piRNAs originating from genic sequences based on RPKM values in PGCs. We also searched for additional information on these piRNA-linked coding genes, such as their associated pathways using the Kyoto Encyclopedia of Genes and Genomes database (KEGG) [20], functional domains using protein family matrices (Pfam) [21], and molecular function gene ontologies using AmiGO, a web based application for gene ontology search [22]. Furthermore, we searched for possible functions of piRNA-linked coding genes based on earlier studies. These search results demonstrated that the majority of piRNA-linked coding genes are involved in specific pathways and are functionally related to germ cells, testis, and ovary in different vertebrates (Additional file 5: Table S4).
Table 4.
List of top 20 repeat sequences originated piRNAs that upregulated in PGCs
| piRNA seq. ID | Sequence length | Sequences | Origin | Genomic distribution | PGCs | Stage X | GSCs | CEFs | Fold change |
|---|---|---|---|---|---|---|---|---|---|
| (RPKM) | (RPKM) | (RPKM) | (RPKM) | ||||||
| ISG_3439104* | 25 | TATTTCCTAACGTCCAGCCTGAACC | Repeat seq. | LINE/CR1:0 | 1147.02 | 13.07 | 13.52 | 0 | 129.42 |
| ISG_1952422* | 27 | CCAGAACACACTTGGCCTTCCGGGCTG | Repeat seq. | LINE/CR1:0 | 928.3 | 3.78 | 17.55 | 0 | 130.55 |
| ISG_2828838 | 25 | CACTGATGGACAGGTCCTGGCTAAG | Repeat seq. | LTR/ERVL:1 | 251.4 | 2.68 | 3.92 | 0 | 114.24 |
| ISG_1920655* | 27 | ACTGAACACAGCACTCGAGGTGAGGCC | Repeat seq. | LINE/CR1:0 | 242.5 | 2.83 | 3.49 | 0 | 115.05 |
| ISG_587310 | 25 | TATTTCCTAACGTCCAGCCTGAATC | Repeat seq. | Ambi | 215.07 | 1.73 | 2.29 | 0 | 160.43 |
| ISG_434349 | 27 | TCCTTGCACAGCCACGACAGTCGCCTG | Repeat seq. | LINE/CR1:0 | 205.56 | 0.31 | 3.6 | 0 | 157.61 |
| ISG_3712010 | 25 | TACCTGTAGAACCCCTTCTTGTTGT | Repeat seq. | LINE/CR1:0 | 194.22 | 0.63 | 4.03 | 0 | 124.93 |
| ISG_347271 | 28 | TTGAACCTCATTAGGTTTTCGTGGGACC | Repeat seq. | LINE/CR1:0 | 175.2 | 2.36 | 2.83 | 0 | 101.14 |
| ISG_471296 | 27 | TGCACTCGATGCCATCGTCTGTCACTG | Repeat seq. | LINE/CR1:0 | 169.35 | 0.47 | 4.03 | 0 | 112.74 |
| ISG_2635821 | 26 | TTCCAGCGTTGTGTGATTTTAGAAGC | Repeat seq. | LINE/CR1:1 | 155.45 | 0.63 | 3.16 | 0 | 123 |
| ISG_2618644 | 26 | TGCTGACGGACTTCCCTGGGCCTGCT | Repeat seq. | LTR/ERVL:1 | 150.81 | 0.16 | 1.85 | 0 | 225 |
| ISG_3554587 | 29 | TCTGATCATCCTCTGGACTTGCTCCAAGA | Repeat seq. | LINE/CR1:0 | 149.72 | 0.63 | 2.51 | 0 | 143.16 |
| ISG_3050277 | 25 | TTTTGACTTAAAAAACGTGTGCGCC | Repeat seq. | LTR/ERVL:1 | 144.48 | 2.99 | 1.09 | 0 | 106.18 |
| ISG_1247918 | 25 | AAGAAAGACGCAGAGCTCTTGGACC | Repeat seq. | LINE/CR1:1 | 130.09 | 0.79 | 3.05 | 0 | 101.63 |
| ISG_2559548 | 26 | TTTCCATCCCTCACTGTCTCTGAGCT | Repeat seq. | LINE/CR1:0 | 120.94 | 0.16 | 2.07 | 0 | 162.79 |
| ISG_2994213 | 27 | TGTACCTGTAGAACCCCTTCTTGTTGT | Repeat seq. | LINE/CR1:0 | 119.36 | 0 | 1.74 | 0 | 205.28 |
| ISG_2004249 | 23 | TTTCCTAACGTCCAGCCTGAACC | Repeat seq. | LINE/CR1:0 | 110.95 | 0.16 | 1.64 | 0 | 185.66 |
| ISG_988698 | 26 | GATGATCAGAGGGCTGGAGCACCTCC | Repeat seq. | LINE/CR1:1 | 110.22 | 0.63 | 1.96 | 0 | 127.55 |
| ISG_1691836 | 26 | TGTACCTGTAGAACCCCTTCTTGTTG | Repeat seq. | LINE/CR1:0 | 109.73 | 0.16 | 1.64 | 0 | 183.61 |
| ISG_2499527 | 25 | AGGAATGGGCTGCCCAGAGAGGTGG | Repeat seq. | LINE/CR1:1 | 108.39 | 0 | 1.53 | 0 | 213.04 |
LINE: long interspersed element; CR1: chicken repetitive 1 element; LTR: long terminal repeat; ERVL: endogenous retroviral element; Ambi (ambiguous): denotes small RNAs overlap with more than one repeat type. *piRNAs were examined by qPCR along with CR1.
Table 5.
List of top 20 genic sequences originated piRNAs that upregulated in PGCs
| piRNA seq. ID | Sequence length | Sequences | Origin | Genomic distribution | Associated gene | PGCs | Stage X | GSCs | CEFs | Fold change |
|---|---|---|---|---|---|---|---|---|---|---|
| (RPKM) | (RPKM) | (RPKM) | (RPKM) | |||||||
| ISG_2943457* | 25 | GACAGACAGAGCTGCCCCTGAGCCT | Intron_antisense | NM_001146136_intr_4 | CHIR-B5 | 55.72 | 0 | 0.87 | 0 | 191.65 |
| ISG_1259042* | 27 | AGACTGAAGATGTGCACCTGACGCCAG | Intron_sense | NM_001146141_intr_4 | CHIR-AB1 | 49.62 | 0 | 0.76 | 0 | 195.06 |
| ISG_2785619* | 26 | TTCTGCATGTTGCTCTCTGTCAGCTG | Intron_sense | NM_001030561_intr_5 | PLLP | 42.92 | 0 | 0.33 | 0 | 393.64 |
| ISG_2237691 | 25 | AGACTGAAGATCTGCACCTGACACC | Intron_sense | NM_001146136_intr_6 | CHIR-B5 | 35.6 | 0 | 0.87 | 0 | 122.45 |
| ISG_2022559* | 27 | TGTTTACTGACTGAGCTACTTTTCCCC | Intron_sense | NM_205163_intr_15 | MYO1A | 33.41 | 0 | 0.11 | 0 | 919.25 |
| ISG_2633063* | 26 | AGGGTACTGAGACATCTTGGAGACAA | Intron_antisense | NM_204716_intr_4 | SLC6A2 | 19.87 | 0 | 0.33 | 0 | 182.28 |
| ISG_1354003* | 21 | TTTCCAAGGACCAGTAGCGCT | Intron_sense | NM_205429_intr_1 | 17.5 | 18.29 | 0 | 0.55 | 0 | 100.65 |
| ISG_2087909* | 26 | TCTCAAAGGATTCCGCATCGTCGACG | Exon_antisense | NM_001030702_exon_1 | RAP2B | 11.95 | 0 | 0 | 0 | 1194.81 |
| ISG_3065006* | 28 | AAGGACCCAAATGGTAGCAGAGGCCATG | Intron_antisense | NM_205186_intr_18 | LRP8 | 9.51 | 0 | 0.11 | 0 | 261.68 |
| ISG_3356981* | 25 | AATGCTGAGAACTAAGGATGCCTCC | Intron_sense | NM_205098_intr_2 | VDR | 7.92 | 0 | 0 | 0 | 792.48 |
| ISG_3294350* | 30 | CAGGCTGTGACCCTGGAATTCCACTACACT | Intron_sense | NM_001013397_intr_72 | MYH1E | 7.44 | 0 | 0.11 | 0 | 204.65 |
| ISG_2645108* | 27 | GAGTGTGAGAAGGGCTTTGTGCAGAGC | Exon_antisense | NM_001030695_exon_5 | ZNF302 | 5.24 | 0 | 0 | 0 | 524.26 |
| ISG_2675670* | 23 | CGGTGTGGGACGAGAAGGAGAAC | Exon_sense | NM_204729_exon_2 | RGN | 4.88 | 0 | 0 | 0 | 487.68 |
| ISG_3080707* | 27 | CTGTGAGTGTGTGAGTGCGGCGGCGCG | Intron_antisense | NM_204952_intr_1 | FOXD2 | 4.27 | 0 | 0.11 | 0 | 117.42 |
| ISG_3280151* | 24 | AAGGACCTCTGAGAATTGCTTTCT | Exon_sense | NM_001030884_exon_12 | RASSF2 | 2.32 | 0 | 0 | 0 | 231.65 |
| ISG_1621199 | 27 | CAGAAGAGAAGCTGAACACAGGGTGTC | Intron_antisense | NM_001031121_intr_7 | VAMP7 | 1.95 | 0 | 0 | 0 | 195.07 |
| ISG_882291 | 28 | TTAAAGATATTTGGCTGCCTGGCTCGCC | Intron_sense | NM_001081502_intr_4 | NLGN1 | 1.95 | 0 | 0 | 0 | 195.07 |
| ISG_2363541 | 26 | AAGGACACCGAGGCTCTGCGTGCTGA | Exon_sense | NM_205525_exon_2 | APOA1 | 1.83 | 0 | 0 | 0 | 182.88 |
| ISG_2033456 | 26 | TCGGCTCGGCTCGGCTCGGCTCGGCT | Exon_antisense | NM_001006501_exon_1 | ADK | 1.1 | 0 | 0 | 0 | 109.73 |
| ISG_950928 | 27 | ATCTGCGTTTAAAGCTCTTTGCACACT | Intron_antisense | NM_001030561_intr_5 | PLLP | 1.1 | 0 | 0 | 0 | 109.73 |
CHIR-B5: immunoglobulin-like receptor CHIR-B5; CHIR-AB1: immunoglobulin-like receptor CHIR-AB1; PLLP: plasmolipin; MYO1A: myosin IA; SLC6A2: solute carrier family 6, member 2; 17.5: lectin-like protein, type II transmembrane protein (17.5); RAP2B: RAP2B, member of RAS oncogene family; LRP8: low density lipoprotein receptor-related protein 8, apolipoprotein e receptor; VDR: vitamin D3 receptor; MYH1E: myosin, heavy chain 1E, skeletal muscle; ZNF302: zinc finger protein 302; RGN: regucalcin; FOXD2: forkhead box D2; RASSF2: Ras association (RalGDS/AF-6) domain family member 2; VAMP7: vesicle-associated membrane protein 7; NLGN1: neuroligin 1; APOA1: apolipoprotein A-I; ADK: adenosine kinase. *piRNAs were examined by qPCR along with their associated genes.
Expression analysis of putative piRNAs and piRNA-linked genes
We examined the expression of three repeat sequence-linked piRNAs (ISG_3439104, ISG_1952422, ISG_1920655) that were upregulated in PGCs (RPKM value > 2) along with CR1 using quantitative real-time PCR (qPCR). The expression of the three repeat sequence-linked piRNAs and CR1 were enriched in PGCs compared with stage X blastoderms, GSCs, and CEFs. However, the expression of the piRNAs was several folds higher in PGCs than that of CR1 (Figure 2). We then performed qPCR to examine the expression of 14 genic-transcript-linked piRNAs (ISG_2943457, ISG_1259042, ISG_2785619, ISG_2022559, ISG_2633063, ISG_1354003, ISG_2087909, ISG_3065006, ISG_3356981, ISG_3294350, ISG_2645108, ISG_2675670, ISG_3080707, ISG_3280151) that were upregulated in PGCs (RPKM value > 2) along with their 14 linked genes, including immunoglobulin-like receptor CHIR-B5 (CHIR-B5), immunoglobulin-like receptor CHIR-AB1 (CHIR-AB1), plasmolipin (PLLP), myosin IA (MYO1A), solute carrier family 6, member 2 (SLC6A2), lectin-like protein, type II transmembrane protein (17.5), RAP2B, member of RAS oncogene family (RAP2B), low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (LRP8), vitamin D3 receptor (VDR), myosin, heavy chain 1E (MYH1E), zinc finger protein 302 (ZNF302), regucalcin (RGN), forkhead box D2 (FOXD2), and ras association (RalGDS/AF-6) domain family member 2 (RASSF2), respectively (Figure 3). As expected, all genic-transcript-linked piRNAs and their linked genes were highly expressed in PGCs compared with stage X blastoderms, GSCs, and CEFs.
Figure 2.
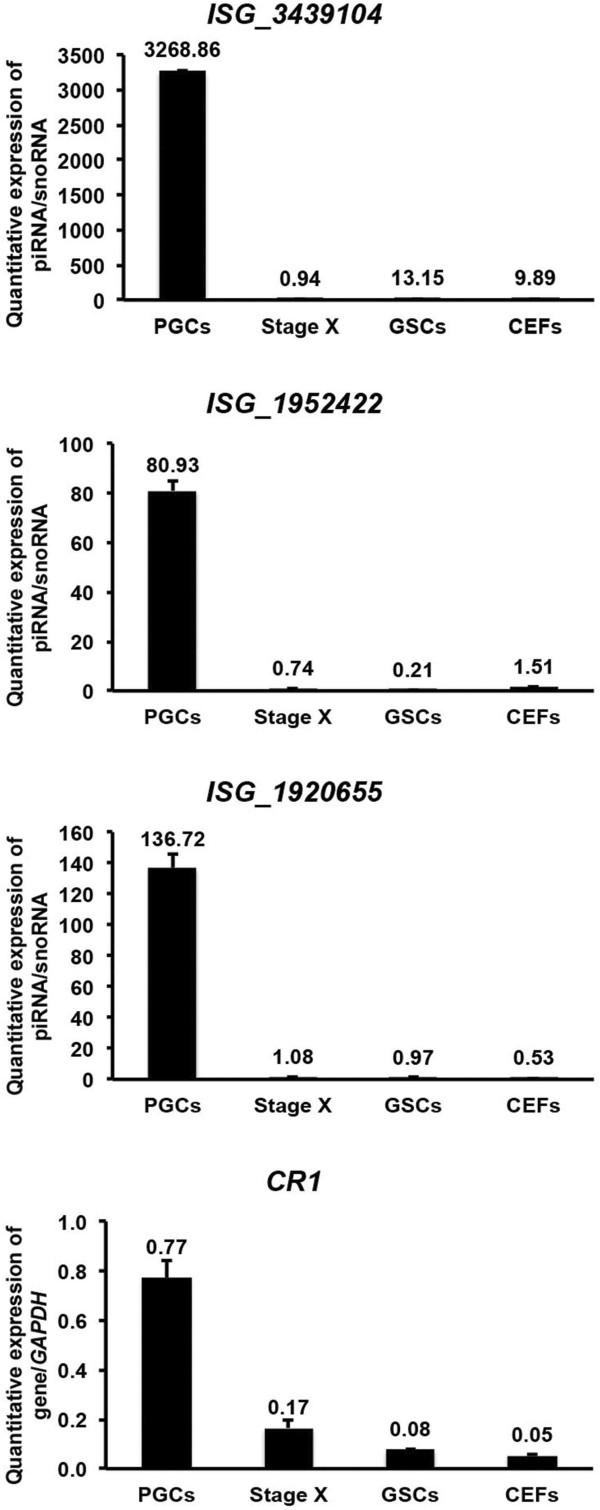
qPCR analysis of piRNAs and piRNA-linked non-coding gene. qPCR quantitative expression analysis of three repeat sequence-linked putative piRNAs that were upregulated in PGCs, along with CR1. cDNA templates used for the amplification of piRNAs and genes were prepared separately, and amplified using the appropriate piRNA- and gene-specific primers. The expression of piRNAs and CR1 were normalized against that of snoRNA and GAPDH, respectively. Bars indicate the SEM of triplicate analyses.
Figure 3.
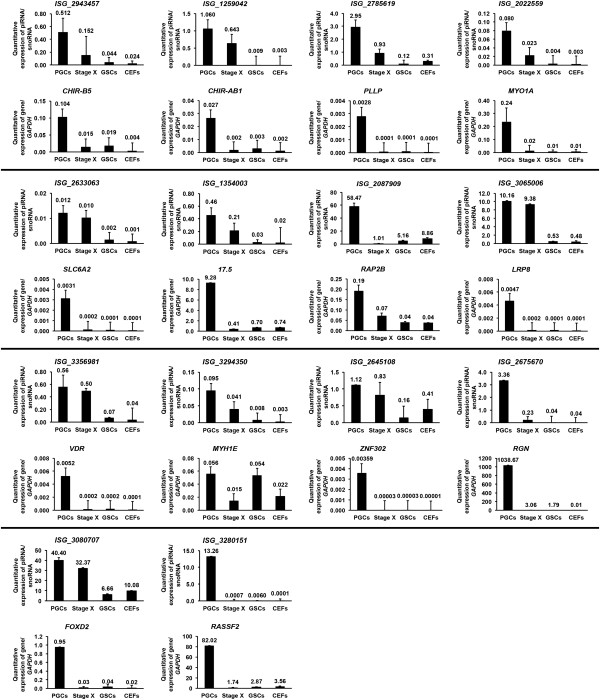
qPCR analysis of piRNAs and piRNA-linked coding genes. qPCR quantitative expression analysis of 14 genic-transcript-linked piRNAs that were upregulated in PGCs, along with their linked coding genes. cDNA templates used for the amplification of piRNAs and genes were prepared separately, and amplified with appropriate piRNA- and gene-specific primers. The expression of piRNAs and genes were normalized against that of snoRNA and GAPDH, respectively. Bars indicate the SEM of triplicate analyses.
Functional validation of piRNA-linked genes
Since the expression of the identified piRNAs and their linked genes were higher in germ cells than in other test samples, we performed an indirect functional validation of selected piRNA-linked genes by knocking down the known testis/ovary-specific piRNA pathway genes from chicken, PIWI-like protein 1 (CIWI) and 2 (CILI). We first examined the expression patterns of CIWI and CILI using qPCR and in situ hybridization. The expression of CIWI, detected by qPCR, was several folds higher in PGCs than in stage X blastoderms, GSCs, and CEFs (Figure 4A). Furthermore, the expression of CIWI was detected continually in the developing germ cells at various developmental stages of male and female gonads based on in situ localization. In the adult stage, the expression of CIWI was restricted to spermatogonia in males and oocytes in females (Additional file 6: Figure S2). Similar to CIWI, the expression of CILI was also several folds higher in PGCs detected by qPCR (Figure 4A) and localized to germ cells throughout the developmental stages based on in situ hybridization (Additional file 7: Figure S3). However, the expression of CILI was slightly stronger than that of CIWI. In the adult stage, the expression of CILI was detected in spermatogonia and spermatocytes of males and in oocytes, granulosa cells and theca cells in females (Additional file 7: Figure S3).
Figure 4.
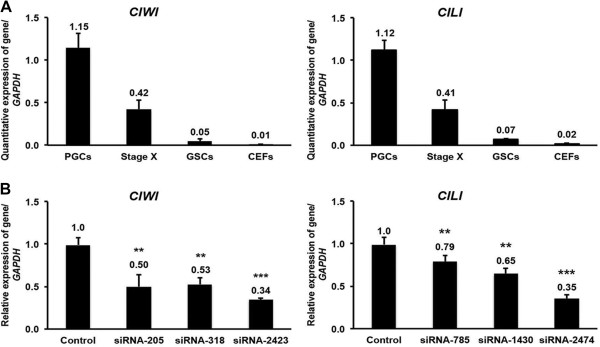
qPCR and knockdown analysis of CIWI and CILI . (A) qPCR quantitative expression analysis of the chicken piRNA pathway genes CIWI and CILI in PGCs, stage X blastoderms, GSCs, and CEFs. (B) Knockdown of CIWI and CILI in chicken PGCs. Three siRNAs each for the knockdown of CIWI and CILI were transfected into PGCs using lipofection. Approximately 48 h after transfection, knockdown efficiency was examined by qPCR amplification of CIWI and CILI. The expression of CIWI and CILI was normalized against that of GAPDH. Bars indicate the SEM of triplicate analyses. **P < 0.01 vs. control and ***P < 0.001 vs. control.
We used three siRNAs (siRNA-205, siRNA-318, and siRNA-2423) to knockdown CIWI in PGCs using RNA transfection. Approximately 48 h after transfection, all three siRNAs significantly decreased the expression of CIWI: 50% decrease by siRNA-205, 47% by siRNA-318, and 66% by siRNA-2423 (Figure 4B). Similarly, we used three other siRNAs (siRNA-785, siRNA-1430, and siRNA-2474) to knockdown CILI and found that all three significantly decreased the expression of CILI: 21% decrease by siRNA-785, 35% by siRNA-1430, and 65% by siRNA-2474 (Figure 4B). After maximum knockdown of CIWI (using siRNA-2423) and CILI (using siRNA-2474) was achieved, we examined the expression of a repeat sequence originated piRNA-linked gene (CR1) and of five genic sequences originated piRNA-linked genes (RAP2B, LRP8, VDR, ZNF302 and RGN) that play crucial role in germ cells. Compared with the control, knockdown of CIWI and CILI increased the expression of both CR1 and RAP2B by at least 2-fold in PGCs. In contrast, knockdown of CIWI and CILI decreased the expression of VDR and RGN (Figure 5).
Figure 5.
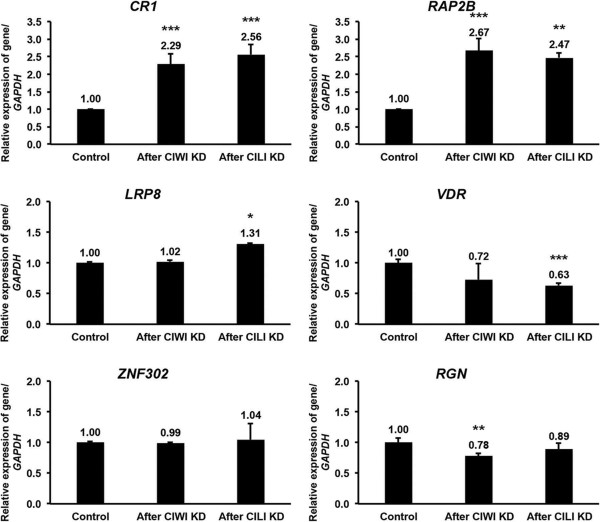
Effects of CIWI and CILI knockdown on the expression of piRNA-linked genes. Effects of chicken piRNA pathway genes CIWI and CILI knockdown on the expression of six putative piRNA-linked genes including CR1, RAP2B, LRP8, VDR, ZNF302, and RGN using qPCR. The relative expression of all genes was normalized against that of GAPDH. Bars indicate the SEM of triplicate analyses. *P < 0.05 vs. control, **P < 0.01 vs. control, and ***P < 0.001 vs. control.
Finally, we examined double-strand DNA breakage in PGCs after maximum knockdown of CIWI and CILI. Approximately 48 h after knockdown, PGCs were incubated with anti-gamma H2A.X (phospho S139) followed by incubation with phycoerythrin. Both CIWI and CILI knockdown in PGCs resulted in clear anti-gamma H2A.X staining, indicating double-strand breakage. This DNA double-strand breakage may be mediated by the increased amount of CR1 after knockdown of CIWI and CILI. Anti-gamma H2A.X staining was not detected in control PGCs (Figure 6).
Figure 6.
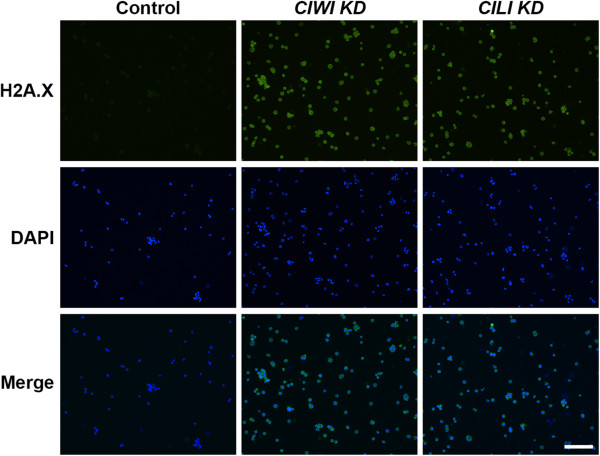
Immunocytochemical analysis of anti-gamma H2A.X expression in PGCs after knockdown of CIWI and CILI . Approximately 48 h after knockdown, PGCs fixed with paraformaldehyde were incubated with anti-gamma H2A.X followed by secondary antibody phycoerythrin to examine double-strand DNA breakage. DAPI was used for counterstaining. Bar = 100 μm.
Discussion
Protein-coding genes and proteins have been explored by several functional genomic studies in animals and plants. However, protein-coding genes account for a small proportion of the whole genome. The most exciting area in recent genomic studies has been the discovery and functional analysis of ncRNAs, which account for the majority of the genome and play critical roles in regulating protein-coding genes [23]. ncRNAs can be classified as long ncRNAs (>300 nt in length) or short ncRNAs (18-32 nt in length). Many high-throughput methods and annotation programs have been developed to identify ncRNAs. Among the high-throughput methods, the inexpensive production of large volumes of sequence data is the primary advantage of NGS over conventional methods [24]. In this study, we performed NGS to identify short ncRNAs in chicken PGCs. The data generated from high-throughput technologies can be difficult to manage [19]. Therefore, the millions of total reads obtained in this study were processed to obtain clean reads. After processing the sequences, total and unique reads were used to annotate small ncRNAs. However, a significant proportion of sequencing reads were unannotated in all test samples. Identifying ncRNA sequences and their genomic locations can be complicated, since several classes of ncRNAs are poorly conserved [15]. These unannotated reads may include novel classes of ncRNAs; therefore, their genomic conservation and potential functions on gene regulation should be explored.
In this study, annotated ncRNAs may have a specific function in an organism. For example, tRNAs and rRNAs are functionally essential for protein synthesis in all living organisms. The tRNAs of eukaryotes contain stretches of base sequences that are similar to those found in their respective rRNA, and these two RNAs share common ancestral origins rather than common functions [25]. snoRNA has diverse roles in RNA silencing, telomerase maintenance and regulation of alternative splicing. Dysregulation of snoRNAs can cause cancer in humans [26]. snRNAs transcribed from RNA polymerase II are most abundant in the nucleus of eukaryotes. snRNAs play important roles in the splicing of introns from primary genomic transcripts. Additionally, snRNAs regulate various aspects of RNA biogenesis, from transcription to polyadenylation and RNA stability [27]. miRNA is one of the best-studied classes of small RNAs. They are highly abundant and conserved among several species and involved in posttranscriptional regulation of gene expression in various tissues [16]. When we compared the RPKM values of small ncRNAs among test samples, ncRNAs originating from repeat sequences were upregulated in PGCs. In animals, siRNAs and piRNAs are mainly derived from repetitive elements (transposable elements). piRNAs are produced through the dicer-independent biogenesis pathway, which results in mature species that are longer (23-30 nt) than miRNAs and siRNAs [28]. In this report, the most abundant base pair size among the unique reads in PGCs was ~ 26 nt, a characteristic of piRNAs.
Among the classes of ncRNAs, piRNAs are important post-transcriptional regulators in germ cells. They can be classified as repetitive element sequence-derived or protein-coding genes-derived based on their genomic origins [17, 28]. Furthermore, the expression of most ncRNAs is ubiquitous, but piRNAs are expressed specifically in germ cells [17]. However, little information is available on piRNAs and their roles in germ cells of vertebrate species. In this report, we examined putative piRNAs in PGCs compared with stage X blastoderms, GSCs and CEFs. The proTRAC output showed a higher proportion of piRNAs (74,337) that were exclusively derived from different types of repeat sequences in PGCs. In addition, a significant proportion of piRNAs were derived from genomic regions and other types of ncRNAs in PGCs. Since the proTRAC software identifies piRNAs based on typical characteristics including the number of loci with T at position 1 or A at position 10 that is the so-called ping-pong signature, all identified piRNAs in chicken PGCs might be amplified in the ping-pong cycle. When we examined the expression of CR1 and three repeat sequence-linked piRNAs, piRNA expression was high in PGCs compared with CR1 expression. This result indicates that these piRNAs are already activated to control CR1 expression. Based on the expression of 14 genic-transcript-linked piRNAs along with their linked genes, the expression of most piRNAs and piRNA-linked genes were highly expressed in PGCs. This may indicate that these piRNAs are produced from their linked genes to regulate various PGC functions. Nevertheless, further investigation of the co-expression of piRNAs and their linked genes in PGCs is required. In this report, we examined pathway information, functional domains and functions of piRNA-linked genes. We found that the majority of piRNA-linked genes are functionally related to germ cells, testis, and ovary in different vertebrates. piRNAs are thought to be required for germ cell development in vertebrate species [16]. Not only piRNAs but also piRNA-linked genes showed predominant functions in the germ cells of vertebrate species [29, 30]. In addition, the expression level of specific piRNAs in stage X blastoderms was very similar to those in PGCs. These piRNAs may be localized to PGCs within stage X blastoderms, which is indicative of a post-zygotic origin [28]. Numerous genes in piRNA functional pathways have been identified, including PIWI/Argonaute family genes, tudor family genes, maelstrom homolog, and vasa homolog [31]. piRNA pathway genes play crucial roles in germline determination during meiosis, gametogenesis, and transposon silencing. These functions may involve piRNAs and may be achieved via RNA interference silencing complex (RISC) mediated epigenetic and posttranscriptional regulation [29, 32]. In Drosophila, PIWI, aubergine, or spindle-E regulates epigenetic function via heterochromatin proteins (HP1 and HP2), which participates in the formation of heterochromatin along with histone H3 lysine 9 methyltransferase [29, 33]. Silencing these genes in Drosophila caused loss of heterochromatin formation by reduction of histone methyltransferase, and delocalization of HP1 and HP2 [33]. Similarly, fission yeast lacking an RISC component argonaute, dicer, or RNA-dependent RNA polymerase caused loss of histone methyltransferase and HP1 homolog at the centromeric heterochromatin [32, 34].
We selected chicken homologs of two piRNA pathway genes, CIWI and CILI, for indirect functional validation of the putative piRNA-linked genes obtained in this study. To our knowledge, these two PIWI family members are only identified in chicken genome, and their expression pattern in PGCs was described in our recent study [30]. We used gene-specific siRNAs to knockdown CIWI and CILI. After knockdown, we examined the expression of CR1 and five piRNA-linked coding genes (RAP2B, LRP8, VDR, ZNF302 and RGN). These genes were selected based on their crucial roles in germ cells. The maximal knockdown of CIWI and CILI significantly increased the expression of CR1 and RAP2B in PGCs. piRNAs and their linked genes play a major defense role against transposable elements. The over activation of transposable elements is associated with severely impaired gametogenesis and causes DNA double-strand breakage in germ cells [35]. According to previous studies, increased CR1 expression causes DNA damage in PGCs. Therefore, the expression of RAP2B increased under these conditions. RAP2B is a member of the Ras superfamily that protects cells from DNA damage in a p53-dependent manner [36]. In contrast, knockdown of CIWI and CILI decreased the expression of VDR and RGN in PGCs, which could impair germ cell development. VDR is crucial for vitamin D3 metabolism, Ca2+ homeostasis, and gametogenesis [37–40], and RGN is crucial for Ca2+ homeostasis and gametogenesis [41, 42]. The nucleosome is composed of DNA and multiple histone protein families, H2A, H2B, H3, and H4. In mammals, the H2A family is consists of three members, H2A.1-H2A.2, H2A.Z, and H2A.X. Among these three members of H2A family, H2A.X becomes phosphorylated on residue serine 139 (the site of gamma-phosphorylation) in cells when double-stranded breaks are introduced into the DNA [43, 44]. Thus, accumulation of gamma H2A.X near break site is a rapid cellular response to the presence of DNA double-strand breakage [43]. It has been reported that the aubergine, spindle-E, and armitage mutations lead to germline-specific accumulation of gamma-H2A.X in Drosophila[45]. In addition, gamma H2A.X accumulates in normal chromosomes where the meiotic double-strand breaks are formed [46]. In this study, to further confirm the DNA double-strand breakage in PGCs, cells were subjected to H2A.X staining after knockdown of CIWI and CILI. Our results clearly indicated that PGCs undergo CR1-mediated DNA double-strand breakage after knockdown of CIWI and CILI. These results suggested that CIWI and CILI interact with certain piRNA-linked genes to protect germ cells.
Conclusions
In this study, we identified different types of ncRNAs including piRNAs in chicken PGCs. piRNAs are closely related with piRNA pathway genes, and functions specifically in germ cells. After knockdown of piRNA pathway genes CIWI and CILI, we found a transposable element-mediated DNA double-strand breakage in chicken PGCs. Several piRNA-linked genes that play crucial role in germ cells were also altered in chicken PGCs. Thus, our results significantly indicate the role of piRNA pathway genes in the protection of germ cells.
Methods
Experimental animals and animal care
The care and experimental use of White Leghorn (WL) chickens was approved (SNU-070823-5) by the Institute of Laboratory Animal Resources, Seoul National University, Korea. For fertilized eggs, chickens were maintained according to a standard management program at the University Animal Farm. The procedures for animal management, reproduction, and embryo manipulation adhered to the standard operating protocols of our laboratory.
Sample preparation
Blastodermal cells at Eyal-Giladi and Kochav (EG&K) stage X [47] were collected by gentle dissociation of stage X blastoderms (n = 30) from freshly laid eggs in phosphate buffered saline (PBS) followed by centrifugation at 1,250 rpm for 10 min. Fertilized eggs were incubated at 37.5°C under 50–60% relative humidity. PGCs and GSCs were isolated from the gonads of chicken embryos at E6.0 (mixed sex, n = ~2,000) using the magnetic-activated cell-sorter (MACS) method [48]. Gonads were dissociated by gentle pipetting in 0.25% trypsin (Invitrogen) and 0.05% ethylenediaminetetraacetic acid (EDTA). After adding 10% fetal bovine serum (FBS, Thermo Fisher Scientific) to inactivate the trypsin-EDTA and briefly centrifuging at 200 × g for 5 min, total gonadal cells were incubated with anti-stage specific embryonic antigen (SSEA-1, Santa Cruz Biotechnology) for 20 min at room temperature. Cells were washed with 1 mL MACS buffer (0.5% BSA and 2 mM EDTA in PBS), and the supernatant was completely removed by centrifugation. The pellet was mixed with 100 μL MACS buffer supplemented with 20 μL goat anti-mouse IgM microbeads for 15 min at 4°C. Cells were washed with 500 μL MACS buffer and loaded onto a MACS column (Miltenyi Biotec GmbH). After several batches of cell preparation, the PGC-enriched fraction and GSCs were separated. The survival rate of PGCs purified by MACS is shown in Additional file 8: Table S5. CEFs were collected by dissociating the embryonic body (E6.0, n = 6) in 0.25% EDTA at 37°C for 20 min. Cells were then cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific) containing 10% FBS and 1% antibiotic–antimycotic (Invitrogen) in a 5% CO2 atmosphere at 37°C. In addition, we collected the left gonads at E13.5, E15.5, and E17.5 and the testis and ovary at 1 day and 24 weeks from male and female chicken embryos or chickens (n = 3), respectively. All experiments discussed hereafter were performed at least in triplicate.
High-throughput next-generation sequencing
Approximately 5 μg total RNA from each test sample (PGCs, stage X blastoderms, GSCs, and CEFs) was used to generate high-throughput NGS data using the Illumina HiSeq (Beijing Genomics Institute, China) following standard protocols from Insilicogen, Korea. Raw NGS data were processed to obtain clean reads by removing low-quality reads (Q-value < 13), short read tags (<18 nt), and adaptor-ligated contaminants using TRIM software developed by BGI for the analysis of high-throughput sequencing. To further identify sequence reads with reliable chromosomal locations, they were mapped onto the reference genome from the NCBI (Gallus gallus v.4) using BLAST, with identity ≥ 90% and HSP coverage ≥ 95%. Among the clean reads, those with sequence lengths between 18 and 44 nt were selected as potential small ncRNAs. Basic annotations of small ncRNAs were performed following standard protocols from Insilicogen. Briefly, to identify miRNAs, sequence reads were aligned with the precursor or mature miRNAs of chickens in miRBase19 using BLASTN with an e-value cutoff of 0.01. Sequence reads from repeat regions were identified by alignment with the reference chicken repeat sequences. To identify rRNAs, tRNAs, snoRNAs, scRNAs and snRNAs, sequence reads were aligned with the reference nucleotides from GenBank records and into the Rfam database using BLASTN with an e-value of 0.01. Sequence reads from exonic or intronic regions were identified based on their genomic locations in 17,767 reference gene sets. Furthermore, a priority rule was applied to ensure that every unique small ncRNA was mapped to only one annotation: tRNAs, rRNAs, snRNAs, snoRNAs, scRNAs > miRNAs > repeats > exons > introns.
Prediction of piRNAs using proTRAC software
To obtain the putative chicken piRNAs, we performed ab initio predictive analysis using proTRAC software [49] with the sequence reads of test samples. proTRAC predicts potential piRNAs based on typical characteristics such as strand bias, the number of loci with T at position 1 or A at position 10 that is the so-called ping-pong signature, the number of loci within the typical piRNA length (26-32 nt), and the quantity of loci from infrequently mapped reads. The alignment output of sequence reads against the chicken reference genome was generated using SeqMap software [50] with the recommended ELAND3 output option, which was used as the input for proTRAC with default parameters (excluding minimum loci per cluster as 6, significance level of p ≤ 0.05 for increased hit density, and a minimum score of 1.3 for strand bias). To cross-validate the putative chicken piRNAs, we performed another ab initio predictive analysis using piRNApredictor software [51] with the sequence reads of test samples. The output results of proTRAC and piRNApredictor were then compared to control the false positive rate.
qPCR analysis of piRNAs and mRNAs
qPCR was performed to examine the expression of 3 repeat sequence-linked putative piRNAs and 14 genic-transcript-linked putative piRNAs along with their 15 linked genes in PGCs, stage X blastoderms, GSCs, and CEFs. Total RNA of the test samples was isolated using TRIzol reagent (Invitrogen). For piRNA amplification, 1 μg total RNA was reverse transcribed using a miRNA 1st-strand cDNA synthesis kit (Agilent Technologies). To elongate the piRNAs, total RNA was first treated with Escherichia coli poly-A polymerase to generate a poly-A tail at the 3′-end of each RNA molecule. Following polyadenylation, cDNAs were synthesized using the RT adaptor primer. PCR was performed using the High-Specificity miRNA qPCR Core Reagent Kit (Agilent Technologies). The PCR reaction mixture was prepared by adding 2.5 μL of 10× core PCR buffer, 2.75 μL of 50 mM MgCl2, 10 μL of 20 mM dNTPs, 1.25 μL of 20× Eva green (Biotium), 1.0 μL of 3.125 μM piRNA-specific forward primers, 1.0 μL of 3.125 μM universal reverse primer (Agilent Technologies), 0.5 μL of High-Specificity polymerase, and 2.0 μL of cDNA to a final volume of 25 μL. PCR was performed with an initial incubation at 94°C for 10 min, followed by 40 cycles at 94°C for 10 s, 60°C for 15 s, and 72°C for 20 s. The forward primer for each piRNA, and chicken snoRNA (U24, GenBank: Z48762) was designed according to the guidelines of Agilent Technologies (Additional file 9: Table S6). piRNA expression was normalized to that of chicken snoRNA, which already validated as internal control [52].
For mRNA amplification, 1 μg total RNA was reverse transcribed using the Superscript III First-Strand Synthesis System (Invitrogen). The PCR reaction mixture was prepared by adding 2 μL PCR buffer, 1.6 μL 2.5 mM dNTP, 10 pmol each forward and reverse primer, 1 μL 20× Eva green, 0.2 μL Taq DNA polymerase, and 2 μL cDNA to a final volume of 20 μL. PCR was performed with an initial incubation at 94°C for 3 min, followed by 40 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. qPCR primers for each target gene, and chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were designed using the Primer3 program (Additional file 10: Table S7). mRNA expression was normalized to that of chicken GAPDH. qPCR analysis for piRNAs and mRNAs was performed using the CFX96 real-time PCR detection system with a C1000 thermal cycler (Bio-Rad Laboratories).
Expression analysis of CIWIand CILI
The expression patterns of two known piRNA pathway genes from chicken, CIWI and CILI, were examined using qPCR and in situ hybridization. For qPCR, cDNA samples from PGCs, stage X blastoderms, GSCs, and CEFs were amplified using the appropriate CIWI and CILI primers (Additional file 10: Table S7), as described above. For in situ hybridization, cDNA from PGCs was amplified using primers targeting CIWI (F: 5′-CCT GAT GGT GTA GGA GAT GGA; R: 5′-CAA GGA AAG CCA GTT TAT GGG) [GenBank: NM_001098852] and CILI (F: 5′-TGA GCC CCG ACA TCC ACA G; R: 5′-TTC TTG GGC AGG CAG TGG TT) [GenBank: JN248386]. The CIWI and CILI PCR products were cloned into the pGEM-T plasmid vector (Promega) and transformed into E. coli strain DH5α. After we verified the cloned sequence, the recombinant plasmid containing CIWI and CILI was amplified using T7- and SP6-specific primers and was subjected to cRNA probe preparation using a digoxigenin RNA labeling kit (Roche Diagnostics). Localization of CIWI and CILI during limited stages of germ cell development (E13.5, E15.5, E17.5, 1 day, and 24 weeks) in male and female chickens was examined as described previously [53]. The mRNA signal was visualized as a brown color, and images were captured under a Zeiss Axiophot light microscope (Carl Zeiss).
Knockdown of CIWIand CILIand their effects on putative piRNA-linked genes
Gonadal PGCs were cultured briefly for mass production as described previously [8]. siRNAs targeting CIWI and CILI were designed using the RNAi designer tool (Invitrogen), which includes a sequence similarity search of the input sequence using the BLAST program. Three siRNA sequences, siRNA-205 (sense: 5′-AGA CAC UAG GAU UAC AGA U; antisense: 5′-AUC UGU AAU CCU AGU GUC U), siRNA-318 (sense: 5′-CAC GUU AGA GAA UCA AAA A; antisense: 5′-UUU UUG AUU CUC UAA CGU G), and siRNA-2423 (sense: 5′-ACU GAA ACC AGA UCA UGU A; antisense: 5′-UAC AUG AUC UGG UUU CAG U), against CIWI were synthesized with incorporation of a 5′-fluorescein isothiocynate (FITC) modification. In addition, three siRNA sequences, siRNA-785 (sense: 5′-GAA UUU GGU GGC UCU GCU G; antisense: 5′-CAG CAG AGC CAC CAA AUU C), siRNA-1430 (sense: 5′- AUG CUU CGA CAC CUU CGA G; antisense: 5′-CUC GAA GGU GUC GAA GCA U), and siRNA-2474 (sense: 5′-AUG CUU CGA CAC CUU CGA G; antisense: 5′-CUC GAA GGU GUC GAA GCA U), against CILI were synthesized. Commercially available control siRNA (sense: 5′-CCU ACG CCA CCA AUU UCG U; antisense: 5′-GGA UGC GGU GGU UAA AGC A), which was not a scrambled sequence of the test siRNAs, was purchased from Bioneer Corporation (Daejeon, Korea). To knockdown the genes, siRNAs were transfected into PGCs (500 pmol siRNA in lipofectamine + Opti-MEM per 1 × 105 cells) as described previously [52]. After transfection for 48 h, FITC expression was confirmed in PGCs, and total RNA was extracted using TRIzol reagent. The efficiency of CIWI and CILI knockdown, and their effects on the expression of six candidate putative piRNA-linked genes including CR1, RAP2B, LRP8, VDR, ZNF302, and RGN were measured using qPCR.
Immunocytochemical analysis
Immunocytochemistry was performed to examine DNA double-strand breakage in CIWI and CILI knockdown PGCs. Approximately 48 h after knockdown, PGCs were fixed in 3.7% paraformaldehyde and incubated with 1:200 diluted rabbit polyclonal to gamma H2A.X (phospho S139) antibodies (Abcam, ab11174) overnight at 4°C. After washing with PBS, PGCs were incubated with secondary antibody labeled with phycoerythrin (anti-rabbit IgG, Santa Cruz Biotechnology) for 1 h at room temperature. Cells were finally mounted with Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI, Vector Laboratories), and analyzed under a fluorescence microscope (Nikon Corporation).
Statistical analysis
Statistical analysis in gene knockdown experiments was performed using the Student’s t test of the SAS software (SAS Institute). Significant differences between control and treatments were analyzed using the general linear model (PROC-GLM) of the SAS software. Statistical significance was ranked as *P < 0.05, **P < 0.01, or ***P < 0.001.
Supporting data
The raw and processed data of this project have been deposited to the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE60400.
Electronic supplementary material
Additional file 1: Table S1: The statistics of sequencing quality. (PDF 41 KB)
Additional file 2: Figure S1: Size distribution of total and unique reads. Size distribution of total (A) and unique (B) reads in PGCs, stage X blastoderms, GSCs, and CEFs obtained using high-throughput next-generation sequencing following the standard annotation protocols. (TIFF 2 MB)
Additional file 3: Table S2: piRNApredictor output and distribution of piRNAs from next-generation sequencing. (PDF 42 KB)
Additional file 4: Table S3: Comparison of proTRAC and piRNApredictor outputs. (PDF 42 KB)
Additional file 5: Table S4: Pathways and functional information of genic sequences originated piRNA-associated genes. (PDF 149 KB)
Additional file 6: Figure S2: mRNA localization of CIWI. mRNA localization of chicken piRNA pathway gene CIWI during limited stages of germ cell development in male and female chickens. S: sense control. Bar = 200 μm (1st and 3rd columns) and 50 μm (2nd and 4th columns). (TIFF 8 MB)
Additional file 7: Figure S3: mRNA localization of CILI. mRNA localization of chicken piRNA pathway gene CILI during limited stages of germ cell development in male and female chickens. S: sense control. Bar = 200 μm (1st and 3rd columns) and 50 μm (2nd and 4th columns). (TIFF 9 MB)
Additional file 8: Table S5: Survival rate of PGCs purified by MACS. (PDF 39 KB)
Additional file 9: Table S6: qPCR primers used for the amplification of piRNAs. (PDF 41 KB)
Additional file 10: Table S7: qPCR primers used for the amplification of genes. (PDF 78 KB)
Acknowledgements
This work was supported by the Next-Generation BioGreen 21 Program [PJ008142] from the Rural Development Administration, Republic of Korea.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DR, SIL and JYH conceived and designed the study. DR, SIL, HJL, YMK and YAS performed the experiments. DR, SIL, TSP, MJ, SJN, HJ and JYH analyzed the data. DR, SIL, TSP and JYH wrote the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Deivendran Rengaraj, Email: deivendran@snu.ac.kr.
Sang In Lee, Email: at314@snu.ac.kr.
Tae Sub Park, Email: taesubpark@snu.ac.kr.
Hong Jo Lee, Email: cszjjang@naver.com.
Young Min Kim, Email: ypoc01@hanmail.net.
Yoon Ah Sohn, Email: letitbe6@snu.ac.kr.
Myunghee Jung, Email: mhjung@insilicogen.com.
Seung-Jae Noh, Email: sjnoh@insilicogen.com.
Hojin Jung, Email: hjjung@insilicogen.com.
Jae Yong Han, Email: jaehan@snu.ac.kr.
References
- 1.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127(12):2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 2.Ginsburg M, Eyal-Giladi H. Temporal and spatial aspects of the gradual migration of primordial germ cells from the epiblast into the germinal crescent in the avian embryo. J Embryol Exp Morphol. 1986;95:53–71. [PubMed] [Google Scholar]
- 3.Ginsburg M, Eyal-Giladi H. Primordial germ cells of the young chick blastoderm originate from the central zone of the area pellucida irrespective of the embryo-forming process. Development. 1987;101(2):209–219. doi: 10.1242/dev.101.2.209. [DOI] [PubMed] [Google Scholar]
- 4.Aramaki S, Sato F, Kato T, Soh T, Kato Y, Hattori MA. Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res. 2007;330(1):45–52. doi: 10.1007/s00441-007-0435-1. [DOI] [PubMed] [Google Scholar]
- 5.Nakamura Y, Yamamoto Y, Usui F, Mushika T, Ono T, Setioko AR, Takeda K, Nirasawa K, Kagami H, Tagami T. Migration and proliferation of primordial germ cells in the early chicken embryo. Poult Sci. 2007;86(10):2182–2193. doi: 10.1093/ps/86.10.2182. [DOI] [PubMed] [Google Scholar]
- 6.Choi JW, Kim S, Kim TM, Kim YM, Seo HW, Park TS, Jeong JW, Song G, Han JY. Basic fibroblast growth factor activates MEK/ERK cell signaling pathway and stimulates the proliferation of chicken primordial germ cells. PloS One. 2010;5(9):e12968. doi: 10.1371/journal.pone.0012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim JN, Park TS, Park SH, Park KJ, Kim TM, Lee SK, Lim JM, Han JY. Migration and proliferation of intact and genetically modified primordial germ cells and the generation of a transgenic chicken. Biol Reprod. 2010;82(2):257–262. doi: 10.1095/biolreprod.109.079723. [DOI] [PubMed] [Google Scholar]
- 8.Park TS, Han JY. piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc Natl Acad Sci U S A. 2012;109(24):9337–9341. doi: 10.1073/pnas.1203823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glover JD, Taylor L, Sherman A, Zeiger-Poli C, Sang HM, McGrew MJ. A novel piggyBac transposon inducible expression system identifies a role for AKT signalling in primordial germ cell migration. PloS One. 2013;8(11):e77222. doi: 10.1371/journal.pone.0077222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naeemipour M, Dehghani H, Bassami M, Bahrami A. Expression dynamics of pluripotency genes in chicken primordial germ cells before and after colonization of the genital ridges. Mol Reprod Dev. 2013;80(10):849–861. doi: 10.1002/mrd.22216. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura Y, Kagami H, Tagami T. Development, differentiation and manipulation of chicken germ cells. Dev Growth Differ. 2013;55(1):20–40. doi: 10.1111/dgd.12026. [DOI] [PubMed] [Google Scholar]
- 12.Nishijima K, Iijima S. Transgenic chickens. Dev Growth Differ. 2013;55(1):207–216. doi: 10.1111/dgd.12032. [DOI] [PubMed] [Google Scholar]
- 13.Park TS, Kang KS, Han JY. Current genomic editing approaches in avian transgenesis. Gen Comp Endocrinol. 2013;190:144–148. doi: 10.1016/j.ygcen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 14.Intarapat S, Stern CD. Chick stem cells: current progress and future prospects. Stem Cell Res. 2013;11(3):1378–1392. doi: 10.1016/j.scr.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 16.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92(5):255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Jung CH, Hansen MA, Makunin IV, Korbie DJ, Mattick JS. Identification of novel non-coding RNAs using profiles of short sequence reads from next generation sequencing data. BMC Genomics. 2010;11:77. doi: 10.1186/1471-2164-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasold M, Langenberger D, Binder H, Stadler PF, Hoffmann S. DARIO: a ncRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2011;39:W112–W117. doi: 10.1093/nar/gkr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11(7):476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub, Web Presence Working Group AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T, Wang S, Wu R, Zhou X, Zhu D, Zhang Y. Identification of long non-protein coding RNAs in chicken skeletal muscle using next generation sequencing. Genomics. 2012;99(5):292–298. doi: 10.1016/j.ygeno.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 25.Bloch DP, McArthur B, Widdowson R, Spector D, Guimaraes RC, Smith J. tRNA-rRNA sequence homologies: evidence for a common evolutionary origin? J Mol Evol. 1983;19(6):420–428. doi: 10.1007/BF02102317. [DOI] [PubMed] [Google Scholar]
- 26.Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem. 2013;54:53–77. doi: 10.1042/bse0540053. [DOI] [PubMed] [Google Scholar]
- 27.Valadkhan S, Gunawardane LS. Role of small nuclear RNAs in eukaryotic gene expression. Essays Biochem. 2013;54:79–90. doi: 10.1042/bse0540079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shao P, Liao JY, Guan DG, Yang JH, Zheng LL, Jing Q, Zhou H, Qu LH. Drastic expression change of transposon-derived piRNA-like RNAs and microRNAs in early stages of chicken embryos implies a role in gastrulation. RNA Biol. 2012;9(2):212–227. doi: 10.4161/rna.18489. [DOI] [PubMed] [Google Scholar]
- 29.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim TH, Yun TW, Rengaraj D, Lee SI, Lim SM, Seo HW, Park TS, Han JY. Conserved functional characteristics of the PIWI family members in chicken germ cell lineage. Theriogenology. 2012;78(9):1948–1959. doi: 10.1016/j.theriogenology.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Lim SL, Tsend-Ayush E, Kortschak RD, Jacob R, Ricciardelli C, Oehler MK, Grutzner F. Conservation and expression of PIWI-interacting RNA pathway genes in male and female adult gonad of amniotes. Biol Reprod. 2013;89(6):136. doi: 10.1095/biolreprod.113.111211. [DOI] [PubMed] [Google Scholar]
- 32.Allshire R. Molecular biology. RNAi and heterochromatin--a hushed-up affair. Science. 2002;297(5588):1818–1819. doi: 10.1126/science.1075874. [DOI] [PubMed] [Google Scholar]
- 33.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Chikka MR, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 34.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 35.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, He Y, Lee KH, Dubois W, Li Z, Wu X, Kovalchuk A, Zhang W, Huang J. Rap2b, a novel p53 target, regulates p53-mediated pro-survival function. Cell Cycle. 2013;12(8):1279–1291. doi: 10.4161/cc.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dornas RA, Oliveira AG, Kalapothakis E, Hess RA, Mahecha GA, Oliveira CA. Distribution of vitamin D3 receptor in the epididymal region of roosters (Gallus domesticus) is cell and segment specific. Gen Comp Endocrinol. 2007;150(3):414–418. doi: 10.1016/j.ygcen.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Craig TA, Sommer S, Sussman CR, Grande JP, Kumar R. Expression and regulation of the vitamin D receptor in the zebrafish, Danio rerio. J Bone Miner Res. 2008;23(9):1486–1496. doi: 10.1359/jbmr.080403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerchbaum E, Obermayer-Pietsch B. Vitamin D and fertility: a systematic review. Eur J Endocrinol. 2012;166(5):765–778. doi: 10.1530/EJE-11-0984. [DOI] [PubMed] [Google Scholar]
- 40.Smolikova K, Mlynarcikova A, Scsukova S. Effect of 1alpha,25-dihydroxyvitamin D3 on progesterone secretion by porcine ovarian granulosa cells. Endocr Regul. 2013;47(3):123–131. doi: 10.4149/endo_2013_03_123. [DOI] [PubMed] [Google Scholar]
- 41.Bruggeman V, Van den Bergh G, Clerens S, Dumez L, Onagbesan O, Arckens L, Decuypere E. Effect of a single in ovo injection of 2,3,7,8-tetrachlorodibenzo-p-dioxin on protein expression in liver and ovary of the one-day-old chick analyzed by fluorescent two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 2006;6(8):2576–2585. doi: 10.1002/pmic.200500237. [DOI] [PubMed] [Google Scholar]
- 42.Laurentino SS, Correia S, Cavaco JE, Oliveira PF, de Sousa M, Barros A, Socorro S. Regucalcin, a calcium-binding protein with a role in male reproduction? Mol Hum Reprod. 2012;18(4):161–170. doi: 10.1093/molehr/gar075. [DOI] [PubMed] [Google Scholar]
- 43.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 44.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12(2):162–169. doi: 10.1016/S0959-437X(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 45.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12(1):45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Jang JK, Sherizen DE, Bhagat R, Manheim EA, McKim KS. Relationship of DNA double-strand breaks to synapsis in Drosophila. J Cell Sci. 2003;116(Pt 15):3069–3077. doi: 10.1242/jcs.00614. [DOI] [PubMed] [Google Scholar]
- 47.Eyal-Giladi H, Kochav S. From cleavage to primitive streak formation: a complementary normal table and a new look at the first stages of the development of the chick. I. General morphology. Dev Biol. 1976;49(2):321–337. doi: 10.1016/0012-1606(76)90178-0. [DOI] [PubMed] [Google Scholar]
- 48.Han JY, Park TS, Kim JN, Kim MA, Lim D, Lim JM, Kim H. Gene expression profiling of chicken primordial germ cell ESTs. BMC Genomics. 2006;7:220. doi: 10.1186/1471-2164-7-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenkranz D, Zischler H. proTRAC-a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformatics. 2012;13:5. doi: 10.1186/1471-2105-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24(20):2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Wang X, Kang L. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics. 2011;27(6):771–776. doi: 10.1093/bioinformatics/btr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee SI, Lee BR, Hwang YS, Lee HC, Rengaraj D, Song G, Park TS, Han JY. MicroRNA-mediated posttranscriptional regulation is required for maintaining undifferentiated properties of blastoderm and primordial germ cells in chickens. Proc Natl Acad Sci U S A. 2011;108(26):10426–10431. doi: 10.1073/pnas.1106141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rengaraj D, Lee SI, Yoo M, Kim TH, Song G, Han JY. Expression and knockdown analysis of glucose phosphate isomerase in chicken primordial germ cells. Biol Reprod. 2012;87(3):57. doi: 10.1095/biolreprod.112.101345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1: The statistics of sequencing quality. (PDF 41 KB)
Additional file 2: Figure S1: Size distribution of total and unique reads. Size distribution of total (A) and unique (B) reads in PGCs, stage X blastoderms, GSCs, and CEFs obtained using high-throughput next-generation sequencing following the standard annotation protocols. (TIFF 2 MB)
Additional file 3: Table S2: piRNApredictor output and distribution of piRNAs from next-generation sequencing. (PDF 42 KB)
Additional file 4: Table S3: Comparison of proTRAC and piRNApredictor outputs. (PDF 42 KB)
Additional file 5: Table S4: Pathways and functional information of genic sequences originated piRNA-associated genes. (PDF 149 KB)
Additional file 6: Figure S2: mRNA localization of CIWI. mRNA localization of chicken piRNA pathway gene CIWI during limited stages of germ cell development in male and female chickens. S: sense control. Bar = 200 μm (1st and 3rd columns) and 50 μm (2nd and 4th columns). (TIFF 8 MB)
Additional file 7: Figure S3: mRNA localization of CILI. mRNA localization of chicken piRNA pathway gene CILI during limited stages of germ cell development in male and female chickens. S: sense control. Bar = 200 μm (1st and 3rd columns) and 50 μm (2nd and 4th columns). (TIFF 9 MB)
Additional file 8: Table S5: Survival rate of PGCs purified by MACS. (PDF 39 KB)
Additional file 9: Table S6: qPCR primers used for the amplification of piRNAs. (PDF 41 KB)
Additional file 10: Table S7: qPCR primers used for the amplification of genes. (PDF 78 KB)


