Abstract
This article utilized “protein charge ladders”—chemical derivatives of proteins with similar structure, but systematically altered net charge—to quantify how missense mutations that cause amyotrophic lateral sclerosis (ALS) affect the net negative charge (Z) of superoxide dismutase-1 (SOD1) as a function of subcellular pH and Zn2+ stoichiometry. Capillary electrophoresis revealed that the net charge of ALS-variant SOD1 can be different in sign and in magnitude—by up to 7.4 units per dimer at lysosomal pH—than values predicted from standard pKa values of amino acids and formal oxidation states of metal ions. At pH 7.4, the G85R, D90A, and G93R substitutions diminished the net negative charge of dimeric SOD1 by up to +2.29 units more than predicted; E100K lowered net charge by less than predicted. The binding of a single Zn2+ to mutant SOD1 lowered its net charge by an additional +2.33 ± 0.01 to +3.18 ± 0.02 units, however, each protein regulated net charge when binding a second, third, or fourth Zn2+ (ΔZ < 0.44 ± 0.07 per additional Zn2+). Both metalated and apo-SOD1 regulated net charge across subcellular pH, without inverting from negative to positive at the theoretical pI. Differential scanning calorimetry, hydrogen-deuterium exchange, and inductively coupled plasma mass spectrometry confirmed that the structure, stability, and metal content of mutant proteins were not significantly affected by lysine acetylation. Measured values of net charge should be used when correlating the biophysical properties of a specific ALS-variant SOD1 protein with its observed aggregation propensity or clinical phenotype.
Keywords: Cu,Zn superoxide dismutase; amyotrophic lateral sclerosis; SOD1; amyloid; protein aggregation; motor neuron disease
Introduction
A fraction of the ∼160 mutations in the gene encoding Cu, Zn superoxide dismutase (SOD1) that are linked to amyotrophic lateral sclerosis (ALS) appear to accelerate the fibrillization of the SOD1 protein (in vivo) without significantly altering its native state structure, free energy of folding (ΔGfold), or the coordination of Cu2+ and Zn2+.1–5 These “cryptic” substitutions—which so far include D90A, E100K, D101N, and N139K2,6 —do, however, lower the negative surface potential of SOD1. Although “cryptic” mutations (some of which are used to construct transgenic mouse models of ALS7) comprise a small fraction of SOD1-ALS mutations, they suggest that perturbations in the biophysical parameters of SOD1, other than ΔGfold, can accelerate its self-assembly and result in selective death of motor neurons.
One hypothesis contends that “cryptic” substitutions accelerate self-assembly by lowering the electrostatic repulsion of negatively charged SOD1 polypeptides.1,3 This hypothesis is supported by: (i) the absence of “cryptic” mutations that increase net negative charge, and (ii) the widespread scattering of “cryptic” substitutions across 264 Å2 of the SOD1 surface.2,6 For example, less than 10% of ALS-SOD1 mutations increase formal net negative charge3,4 and those that do (such as V7E or N139D) also reduce the thermostability of holo-SOD1 and/or apo-SOD1.8 More general support for the electrostatic repulsion hypothesis can be found throughout protein science,9–11 from the century-old observation that proteins precipitate rapidly at their isoelectric point,12,13 to the measured rates of amyloid formation among non-isoelectric variants,11,14 to the engineering of “supercharged” proteins that are impervious to self-assembly.15
Because intermolecular electrostatic interactions persist through physiological buffer across long distances (the Debye radius, κ−1 = 10 Å at ε = 80, I = 0.1 M, T = 310 K), it is not immediately clear whether “cryptic” mutations would accelerate self-assembly by minimizing long-range electrostatic repulsions—that depend upon the protein’s net charge, as reported for other proteins9–11,16—or by minimizing specific, shorter-range repulsions that depend upon local patterns of charge (e.g., in-register or out-of-register patterns, per structure of the fibril17,18). Regardless of which (or whether) one scenario predominates, quantifying how ALS mutations affect the electrostatic surface potential of SOD1 in its native state will answer several important questions. Do “cryptic” amino acid substitutions diminish net charge by a single unit, or more or less than expected because of “charge regulation”?19,20 Do ALS-variant SOD1 proteins follow the same nonlinear electrostatic pathway during metal loading as the WT protein and do they regulate net charge across subcellular pH?21 Charge regulation is a process by which the pKa of a protein’s functional groups undergo cooperative increases or decreases to compensate for electrostatic perturbations within the interdependent network of ionizable residues.22
Measuring such a seemingly trivial, coarse metric as the net charge of a protein might seem unfitting for an entire research study, however, the net charge of a folded protein remains one of its most enigmatic properties to experimentally determine because few tools exist to make such a measurement (accurately).22,23 We estimate that the net charge of <0.1% of unique polypeptides in the Protein Data Bank have been accurately measured in the native state (at pH ≠ pI), including any mutant variants of these proteins. In cases where accurate measurements have been made, the measured values (denoted ZCE in this article) can differ in magnitude by up to seven-fold from predicted values that are calculated from the standard pKa of side-chains (denoted Zseq), presumably because of anomalous pKa.24,25
The isoelectric points of a few ALS-variants have been measured,26–30 however, pI is not an expression of—and does not scale with—net charge at pH ≠ pI.24 Experimentally measuring the net charge of ALS-variant SOD1 instead of predicting it is especially important because SOD1 is an electrostatically peculiar metalloenzyme. The dimeric SOD1 protein contains at least two aberrantly charged residues: each His63 residue is negatively charged when bridging Cu2+ and Zn2+ at the binuclear active site.31,32 The ionization state of His63 is dubious when bridging two Zn2+ ions at the active site (i.e., Cu0Zn4-SOD1).33 Four intrinsically disordered loops in each SOD1 dimer—spanning ∼ 120 residues in total length, including the highly charged “electrostatic loop”31–35—also undergo decreases in solvent accessibility upon metal binding. These types of metal-induced reductions in solvent accessibility could alter the pKa of ionizable residues by multiple units.36,37 The SOD1 protein is also localized at multiple subcellular sites with local pH that are above and below the theoretical pI of SOD1 (theoretical pI = 5.9 for apo-SOD1; pI = 5.3 for Cu2Zn2SOD1).21 An ALS missense mutation such as E100K might, therefore, diminish the net (negative) charge of Cu2Zn2SOD1 at cytosolic pH, but increase net (positive) charge at lysosomal pH.
In our previous study of WT SOD1, we used protein charge ladders and capillary electrophoresis (CE) (Fig. 1) to demonstrate that the actual net charge of apo-SOD1 and Cu2Zn2-SOD1 deviates from predicted values by between ∼0.5 and 11 units—and even opposite in sign—across its range of subcellular pH (this deviation accounts for the charge imparted by bound metal ions and the fixed charge associated with non-titratable metal ligands).21 In this article, we use protein charge ladders and CE to measure the net charge of four ALS-linked variants of SOD1 (including two cryptic variants), both as a function of Zn2+ stoichiometry and pH.
Figure 1.
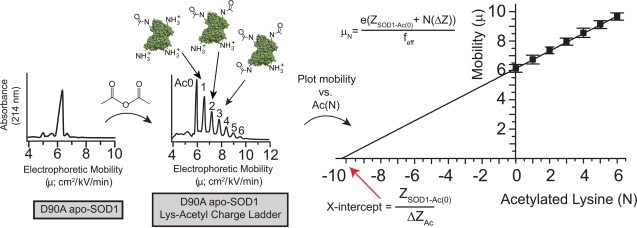
Using “protein charge ladders” and capillary electrophoresis to measure the net charge of native ALS-variant SOD1. Left plot: a capillary electropherogram of recombinant D90A apo-SOD1 at pH 7.4. Center plot: a capillary electropherogram of a “protein charge ladder” of D90A apo-SOD1 prepared by acetylating its surface Lys-ε-NH3+ (to electrostatically neutral Lys-NHCOCH3) with acetic anhydride. Right plot: a plot of the electrophoretic mobility (µ) of each rung of the ladder versus the number of acetylated lysines (N) of each rung. The x-intercept of this plot is equal to ZSOD1-Ac0/ΔZAc, that is, the net charge of unmodified protein (ZSOD1-Ac0) divided by the change in net charge imparted by each lysine acetylation (ΔZAc); feff = hydrodynamic drag; e = approximate charge of an electron. The x-intercept can be considered to represent, per se, the approximate number of Lys-NH3+ groups that would need to be added to the negatively charged protein in order to cause the protein to have an electrophoretic mobility of zero, and thus a net charge of zero.
A protein charge ladder is a set of non-isoelectric chemical derivatives of a protein generated by the acetylation of its surface Lys-ε-NH3+ (e.g., with acetic anhydride). Each Lys-acetyl derivative or “rung” differs in net charge from the next by ΔZAc ≈ 0.9 units (instead of ΔZAc = 1.0) because of charge regulation,38 but has similar shape and hydrodynamic drag. When analyzed by CE, a protein charge ladder is an internally consistent, self-calibrating electrophoretic array that can be used to determine the net charge of the zeroth rung (i.e., the unmodified protein, Fig. 1). A protein charge ladder is the only convenient tool available for accurately measuring the net charge of a folded protein.22,24
The results of our measurements demonstrate that the effective magnitude and sign of net charge of ALS-variant SOD1 proteins cannot be accurately predicted at all values of subcellular pH, even when taking into account the previously measured net charge of WT SOD1. Accurately measuring the effect of ALS mutations on the net charge of the SOD1 protein will establish a more rigorous understanding of why ALS mutations accelerate the self-assembly of SOD1 and why some mutations result in more severe clinical phenotypes than others.4,39,40
Results and Discussion
Throughout this article, and in all of our previous papers, we refer to changes in the net of proteins—changes that occur upon amino acid substitution, fluctuation in pH, or metal binding—in terms of the change in the absolute magnitude of net charge, whether it be positive or negative. For example, if metal binding changed the net charge of a protein from Z = −14 to Z = −12, we would say that metal binding reduced or diminished the net charge of the protein, that is, reduced the magnitude of net negative charge. Likewise, if increasing pH changed the net charge of a protein from Z = −12 to Z = −14, we would say that increasing pH increased the net charge of the protein. Furthermore, we would say that a protein with a net charge of Z = −3, and a separate protein with a net charge of Z = +3, each have the same magnitude of net charge. Finally, if any process abolished the net charge of either of these triply charged proteins, from Z = −3 and Z = +3, to Z = 0, we would say that both processes reduced the net charge of both proteins, that is, reduced the magnitude of charge. We use this convention because the sign of charge is intrinsically arbitrary and does not express magnitude, and the word “net” refers to magnitude, not sign.
The location of each amino acid substitution that we studied (G85R, D90A, G93R, and E100K) is shown in Supporting Information Figure S1. Although these substitutions are expected to diminish the net negative charge of SOD1 by +2 or +4 formal units (per dimer), each amino acid substitution has different effects on the structure, folding, metal affinity, and conformational stability of SOD1.2 The “cryptic” D90A and E100K substitutions occur at the surface of SOD1.6 The G85R substitution (a non-cryptic substitution within the H-bonding network that surrounds metal ligands41) weakens the affinity of SOD1 for both Cu2+ and Zn2+.30,42 The G85R substitution also reduces the thermostability of the SOD1 protein and alters its three-dimensional structure.1,41,43 The fourth protein, G93R apo-SOD1, another noncryptic variant, can coordinate metals properly but exhibits a destabilized conformation and altered structure compared to the WT apo-SOD1 protein.43,44
In this study, we only examined the electrostatic effects of Zn2+ coordination to ALS-variant SOD1. Although the dimeric SOD1 protein binds two equivalents of Cu1+/2+ and two equivalents of Zn2+ in its holo state, the protein is often isolated with more Zn2+ bound to the active site than Cu1+/2+ (e.g., Cu1Zn3-SOD1).45,46 Alterations in Zn2+ binding might modulate neurotoxicity.47,48 We also only studied SOD1 in its disulfide intact state.
Monitoring the binding of zinc ions to ALS-variant SOD1 with capillary electrophoresis
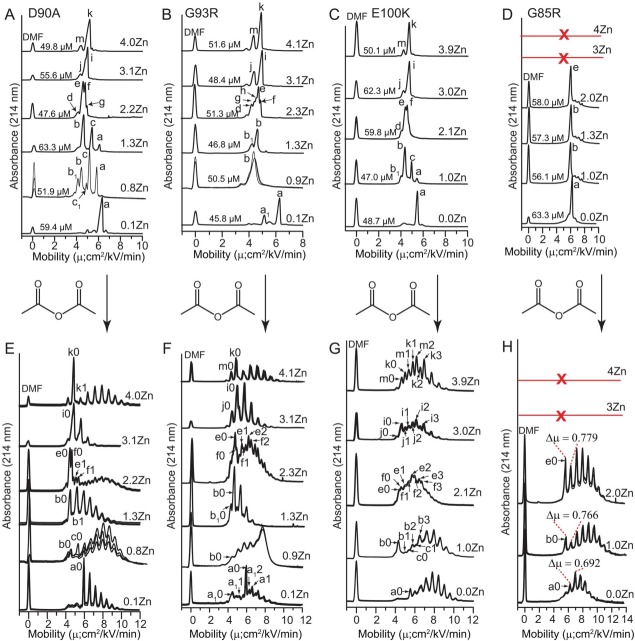
Capillary electropherograms of Zn2+ derivatives of ALS-variant SOD1 proteins are shown in Figure 2(A–D), from which lysine-acetyl “protein charge ladders” were synthesized via acetylation of each Zn2+-derivative with acetic anhydride [Fig. 2(E–H)]. Because CE separates proteins based upon their net charge and their hydrodynamic drag, each peak in the electropherograms of Figure 2(A–D) represent an SOD1 protein with a unique charge and/or a unique structure or state of oligomerization.
Figure 2.
Capillary electrophoresis of Zn2+ derivatives of ALS-variant SOD1 and Lys-acetyl protein charge ladders. (A–D) Capillary electropherograms of ALS-variant SOD1 with increasing stoichiometries of bound Zn2+. Each electropherogram is an overlay of 3 separate electropherograms from replicate experiments. Electrophoretically unique species are labeled with lower case letters and subscript numbers (a-m). The concentration of each SOD1 derivative at the time of injection is listed per dimer. The red line and “X” indicates that the G85R protein did not coordinate > 2 Zn2+/dimer. E-H) Protein charge ladders of each Zn2+ derivative of each ALS-variant SOD1 protein from panels A–D, prepared by acetylation of lysine-ε-NH3+ with acetic anhydride. Each electropherogram is an overlay of three separate electropherograms from repeated experiments. Labels such as “a0, a1, b0, b1” denote acetylation number of each electrophoretically unique species from parts A–D (i.e., “a0” = unacetylated or zeroth rung of species “a”; “b1” = singly acetylated or first rung of species “b”). Acetylation of minor species in panels A–D such as “d” in G93R were not detectable and are not labeled. Note: the E100K charge ladder of Zn3- and Zn4-SOD1 exhibited two overlapping ladders that might reflect differing reactivity of Lys residues (including K100).
At intermediate states of metalation, that is, 1–2 Zn2+, the electropherograms of each ALS-variant SOD1 protein contained more peaks (albeit minor ones) than electropherograms previously published for the WT SOD1 protein at intermediate metalation states21 [Fig. 2(A–D)]. This heterogeneity suggested that ALS-variant SOD1 proteins populated more unique structural or metalation states than WT SOD1. The most intense peak in each electropherogram was assigned to be equal to the average metal stoichiometry that was determined by ICP-MS. The heterogeneity observed in the intermediate metalation states (1Zn2+ or 2Zn2+) is interpreted in terms of subunit exchange and is discussed (along with peak assignments) in Supporting Information (Fig. S2). The terminal metalation states (e.g., 0Zn2+, or 4Zn2+) resulted in simpler electropherograms with a predominantly unimodal distribution of species. Unimodal electropherograms are expected for terminal metalation states because only one state of metalation is statistically possible.
With the exception of G85R SOD1, the binding of one to four Zn2+ to each ALS-variant SOD1 resulted in a non-linear shift in mobility. For example, the binding of a single Zn2+ to D90A, G93R and E100K apo-SOD1 caused a large reduction in mobility whereas the successive binding of the second and third stoichiometric equivalents of Zn2+ gradually increased mobility of D90A, G93R, and E100K apo-SOD1. The binding of the fourth Zn2+ cation to these three proteins did not significantly affect electrophoretic mobility. This nonlinear trend was observed previously for the WT SOD1 protein.21
The nonlinear correlation between Zn2+ stoichiometry and electrophoretic mobility reflects the fact that metal binding simultaneously alters net charge and/or decreases hydrodynamic drag nonlinearly with respect to metal stoichiometry. This nonlinear correlation between metal stoichiometry and net charge, or metal stoichiometry and hydrodynamic drag is expected in SOD1 for two reasons. First, the binding of a single Zn2+ ion affects structure more than subsequent metal ions (i.e., the so-called “one zinc effect,”35 wherein the binding of one Zn2+ to dimeric SOD1 results in the ordering of disordered loops in both subunits). Second, the binding of higher equivalents of Zn2+ (3Zn2+ and 4Zn2+) is expected to affect charge by less than the binding of 1Zn2+ or 2Zn2+ because the deprotonation of His63 will only occur—if it occurs at all—upon binding the 3rd and 4th Zn2+. Thus, two derivatives such as G85R Zn1-SOD1 and G85R Zn2-SOD1 [Fig. 2(D)] can have values of net charge that differ by multiple units, despite having nearly identical electrophoretic mobilities (that differ by <0.5 cm2/kV/min). This point is verified below by electrophoresis of protein charge ladders of each derivative.
Unlike D90A, G93R, or E100K SOD1, the G85R SOD1 protein could not coordinate > 2Zn2+ per dimer (according to ICP-MS, Supporting Information Table SI) and did not follow a semi-parabolic trend in mobility versus Zn2+ stoichiometry. Moreover, the binding of one or two Zn2+ cations to G85R did not diminish its mobility as much as observed for the other mutants [Fig. 2(D)]. An analysis of Zn2+-derivatives of G85R with ICP-MS confirmed, however, the stoichiometry of the Zn1- and Zn2-SOD1 proteins and demonstrated that the protein did coordinate up to two Zn2+ cations per dimer (Supporting Information Table SI). Analysis of Co2+ derivatives of G85R (Co1- and Co2-SOD1) with UV–vis also demonstrated that metal ions are bound at the active site of G85R SOD1 (described below).
The net electrostatic charge of ALS-variant Apo-SOD1 at pH 7.4 is lower than predicted
Protein charge ladders of each ALS-variant SOD1 protein are shown in Figure 2(E–H). The number of acetylated lysine residues was confirmed with mass spectrometry (see Supporting Information, Fig. S3). In cases where the electropherogram of the unacetylated metal derivative was characterized by a single predominant peak (e.g., apo-SOD1 or Zn4-SOD1) the acetylation of lysine residues resulted in a single charge ladder that was easily interpretable. In cases where the electropherogram of the unacetylated derivative consisted of multiple peaks, the acetylation of lysine residues resulted in multiple charge ladders (of each species) that could still be partially resolved by capillary electrophoresis and deconvoluted [Fig. 2(E–H)].
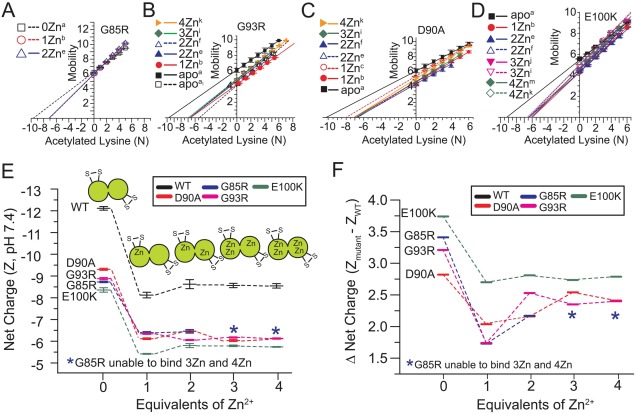
The plots of electrophoretic mobility of each rung versus its number of acetylated lysines are shown for each protein in Figure 3(A–D). The values of net charge that are derived from these plots are listed for each protein in Table 1 and are also summarized in Figure 3(E,F). At pH 7.4, each of the four ALS-variant apo-SOD1 proteins were less negatively charged (i.e., closer to electrostatic neutrality) than predicted from its amino acid sequence by a range of ΔZCE:seq = 0.64–2.29 units per dimer (Table 1).
Figure 3.
Electrostatic pathway of ALS-variant SOD1 (at pH 7.4) as a function of coordinated Zn2+. (A–D) Plot of electrophoretic mobility of each rung of each ALS-variant charge ladder [derived from the protein charge ladders shown in Fig. 2(E–H)] versus the number of acetylated lysines in that rung. (E) Summary of the changes in the net charge of ALS-variant SOD1 as a function of Zn2+ stoichiometry (per dimer). The electrostatic pathway of WT SOD1 is shown for reference and was determined in our previous study.21 Values of net charge are expressed per dimer. Error bars represent standard deviation from three to five replicate experiments. (F) The effect of Zn2+ binding on the WT-variant “charge gap” (i.e., the difference in the net charge of WT and ALS-variant SOD1).
Table 1.
Comparison of Measured (ZCE) and Predicted (Zseq) Values of Net Charge of WT and ALS-variant D90A, G85R, G93R, and E100K SOD1 at pH 7.4, 20°C
| Net charge (ZCE and Zseq)a | |||||
|---|---|---|---|---|---|
| SOD1b | WTc | D90A | G85R | G93R | E100K |
| apo(a) ZCE | −12.13 ± 0.08 | −9.30 ± 0.02 | −8.71 ± 0.01 | −8.91 ± 0.04 | −8.36 ± 0.02 |
| Zseq | −13.0 | −11.0 | −11.0 | −11.0 | −9.0 |
| apo(a1) ZCE | N/A | N/A | N/A | −5.04 ± 0.02 | N/A |
| Zseq | −13.0 | −11.0 | −11.0 | −11.0 | −9.0 |
| Zn1(b) ZCE | −8.12 ± 0.23 | −6.12 ± 0.01 | −6.38 ± 0.01 | −6.25 ± 0.02 | −5.42 ± 0.01 |
| Zseq | −10.3 | −8.3 | −8.3 | −8.3 | −6.3 |
| Zn1(c) ZCE | N/A | −7.22 ± 0.03 | N/A | N/A | N/A |
| Zseq | −10.3 | −8.3 | −8.3 | −8.3 | −6.3 |
| Zn2(e) ZCE | −8.62 ± 0.30 | −6.46 ± 0.08 | −6.45 ± 0.10 | −6.09 ± 0.01 | −5.81 ± 0.11 |
| Zseq | −7.6 | −5.6 | −5.6 | −5.6 | −3.6 |
| Zn2(f) ZCE | −8.45 ± 0.27 | −6.23 ± 0.10 | N/A | −6.08 ± 0.03 | −5.85 ± 0.07 |
| Zseq | −7.6 | −5.6 | −5.6 | −5.6 | −3.6 |
| Zn3(j) ZCE | N/A | N/A | N/A | N/A | −5.48 ± 0.01 |
| Zseq | −7.9 | −5.9 | −5.9 | −5.9 | −3.9 |
| Zn3(i) ZCE | −8.55 ± 0.11 | −6.02 ± 0.02 | N/A | −6.20 ± 0.01 | −5.83 ± 0.01 |
| Zseq | −7.9 | −5.9 | −5.9 | −5.9 | −3.9 |
| Zn4(m) ZCE | N/A | N/A | N/A | N/A | −5.44 ± 0.01 |
| Zseq | −8.2 | −6.2 | −6.2 | −6.2 | −4.2 |
| Zn4(k) ZCE | −8.54 ± 0.12 | −6.12 ± 0.01 | N/A | −6.14 ± 0.01 | −5.72 ± 0.03 |
| Zseq | −8.2 | −6.2 | −6.2 | −6.2 | −4.2 |
With the exception of E100K, the measured values of net negative charge of ALS-variant apo-SOD1 proteins were lower (closer to neutrality) than predicted at pH 7.4, even when considering the previously measured net charge of WT apo-SOD1 (which is also lower than predicted from its sequence, i.e., ZCE = −12.13 per WT SOD1 dimer at pH = 7.4, compared to Zseq = −13.0).21 These data demonstrate that the nonlinear network of ionizable residues in the SOD1 protein cannot regulate its net charge in response to the G85R, D90A, and G93R amino acid substitutions. For example, the expected difference in the formal net negative charge of WT and G85R, D90A, and G93R SOD1 is ΔZWT:ALS = +2.0 units, however, the G85R substitution caused a 70% greater decrease in the net negative charge of apo-SOD1 than expected that is, ΔZWT:G85R = +3.42 ± 0.08 (Table 2).
Table 2.
“Charge Gap” Values (ΔZALS:WT) between WT and ALS-variant SOD1 at Different Metalation States as Measured by Capillary Electrophoresis at pH 7.4, 20°C
| ΔZALS:WT | ||||
|---|---|---|---|---|
| SODa | D90A | G85R | G93R | E100K |
| apoS-S | 2.82 ± 0.08 | 3.42 ± 0.08 | 3.22 ± 0.09 | 3.77 ± 0.08 |
| Zn1(b) | 2.00 ± 0.23 | 1.74 ± 0.23 | 1.87 ± 0.23 | 2.70 ± 0.23 |
| Zn2(e) | 2.16 ± 0.31 | 2.17 ± 0.32 | 2.53 ± 0.30 | 2.81 ± 0.32 |
| Zn3(i) | 2.53 ± 0.11 | N/A | 2.35 ± 0.11 | 2.72 ± 0.11 |
| Zn4(k) | 2.42 ± 0.12 | N/A | 2.40 ± 0.12 | 2.82 ± 0.12 |
The labels “a”, “e”, etc., correspond to labels of electrophoretically unique states in Figure 2.
The G93R substitution diminished the magnitude of net negative charge of apo-SOD1 by ΔZWT:G93R = +3.22 ± 0.09 (Table 2). The D90A substitution diminished net negative charge of apo-SOD1 by ΔZWT:D90A = +2.82 ± 0.08 (Table 2). A significant amount of charge regulation was exhibited by apo-SOD1 in response to the E100K substitution, which lowered net negative charge by less than the 4 units predicted: ΔZWT:E100K = +3.77 ± 0.08 (Table 2). These results are, to our knowledge, the first direct, accurate measurement of the effect of an amino acid substitution on the net charge of a folded protein at physiological pH (i.e., at pH ≠ pI). Thus, the exact cause of the deviations between predicted and measured ΔZ—which are significant and greater than experimental error—are not understood and must be the subject of a future study.
Nevertheless, we do note that a correlation existed between: (i) the difference in the predicted and measured change in net charge of apo-SOD1 upon amino acid substitution (denoted ΔΔZ, i.e., ΔZCE − ΔZseq) and (ii) the number of anionic versus cationic residues at the substitution’s locus, within a spherical radius of 10 Å from the locus (Supporting Information Fig. S4). A detailed discussion of these points can be found in Supporting Information.
We point out that in this study we only quantified the net charge of ALS-variant SOD1 proteins at relatively low ionic strength (10 mM phosphate buffer). The values of net charge that we determined might change slightly at higher ionic strength, as inferred from previous studies into the effect of ionic strength on residue pKa.49 Previous studies have shown, for example, that the pKa values of 14 different histidine residues in horse and whale myoglobin increased from an average of pKa = 6.41 at ∼ 0.2 M NaCl to pKa = 6.61 at 200 mM NaCl.49
The binding of one Zn2+ diminishes the “charge gap” between WT and ALS-variant SOD1
Upon binding a single Zn2+ cation, all four ALS-variant apo-SOD1 proteins underwent a decrease in the magnitude of net negative charge that ranged between +2.33 ± 0.01 units per dimer (for G85R) to +3.18 ± 0.02 units per dimer (for D90A). The value of net charge of metalated proteins are listed in Table 1 and shown schematically in Figure 3(E). The values of ΔZCE associated with the binding of each Zn2+ ion to each protein are shown in Table 3. These values of ΔZ0Zn→1Zn are lower for each ALS variant than the previously reported value of ΔZ0Zn→1Zn = +4.01 ± 0.20 units for WT SOD1.21
Table 3.
Charge Difference (ΔZ) Among Various Metalation States of WT and ALS-Variant SOD1 as Measured by Capillary Electrophoresis at pH 7.4, 20°C
| ΔZa | WTb | G85R | D90A | G93R | E100K |
|---|---|---|---|---|---|
| ΔZ 0(a)→1(b) Zn | 4.01 ± 0.20 | 2.33 ± 0.01 | 3.18 ± 0.02 | 2.66 ± 0.03 | 2.94 ± 0.02 |
| ΔZ 1(b)→2(e) Zn | −0.50 ± 0.27 | −0.07 ± 0.10 | −0.34 ± 0.08 | 0.16 ± 0.02 | −0.39 ± 0.11 |
| ΔZ 2(e)→3(i) Zn | 0.07 ± 0.26 | N/A | 0.44 ± 0.07 | −0.11 ± 0.01 | −0.02 ± 0.11 |
| ΔZ 3(i)→4(k) Zn | 0.01 ± 0.12 | N/A | −0.10 ± 0.02 | 0.06 ± 0.01 | 0.11 ± 0.03 |
The different magnitude of ΔZ upon binding one Zn2+ to WT apo-SOD1 compared to that of ALS-variant apo-SOD1 are outside experimental error for all four ALS variants and are considered significant. The binding of one equivalent of Zn2+ to ALS-variant apo-SOD1 protein actually diminished its “charge gap” relative to WT SOD1 [Fig. 3(F)]. The charge gap for WT and G85R apo-SOD1 is ΔZCE = +3.42 ± 0.08, and the binding of one Zn2+ ion diminished this charge gap to ΔZCE = +1.74 ± 0.23 [Fig. 3(E); Table 2]. Thus, the binding of Zn2+ partially abolishes the electrostatic differences between WT and ALS-linked SOD1, as measured by protein charge ladders and CE.
All four ALS-variant proteins also underwent a significant degree of charge regulation when binding > 1 Zn2+ per dimer [Fig. 3(E)], and therefore, the binding of 2–4 Zn2+ did not lead to continued reductions in the net negative charge of SOD1 or continued reductions in the “charge gap” (Fig. 3, Tables 1 and 2). In fact, for all of the variants except for E100K, the binding of > 1 Zn2+ slightly increased the “charge gap” [Fig. 3(F), Table 2]. Therefore, the difference in the net charge between each ALS-variant protein and WT SOD1 is maximized in the apo state, minimized in the Zn1 state, and is intermediate in the Zn2–4 states (with the exception of E100K SOD1, wherein the charge gap of the Zn1 and Zn>1 states are similar). We hypothesize that the ability of SOD1 to resist reductions in its negative charge when binding multiple Zn2+ ions might be a functionally relevant property that maximizes protein solubility and maintains proper protein-protein interactions.
We point out that the linear plots of electrophoretic mobility versus the number of acetylated lysines for each metal derivative of each variant are not (and should not be) necessarily parallel or superimposable, or have identical y-intercepts, because the slope and y-intercept is a function of the hydrodynamic drag and net charge of each derivative. It is possible for two different Zn2+ derivatives of the same variant (or the same Zn2+ derivatives from two different ALS variants) to have different slopes or y-intercepts (of µ versus Ac(N)) but have similar values of net charge (i.e., similar x-intercepts) because two different proteins can have equal charge but different hydrodynamic drag. Thus, the slopes of plots of µ versus Ac(N) can provide information about the structure of the protein in question. For example, the binding of multiple Zn2+ to E100K SOD1 generally increased the slope (slope = ΔZAc / feff, feff = hydrodynamic drag) and the y-intercept (y-intercept = µAc(0) = eZAc(0) / feff). This simultaneous increase in y-intercept and slope resulted in similar x-intercepts or values of net charge between Zn2, Zn3, and Zn4 E100K (Fig. 3). We attribute this increase in slope (upon metal binding) to be caused by the decrease in hydrodynamic drag in SOD1 that likely resulted from the metal-induced ordering and tightening of the large disordered loops in SOD1 (residues ∼48–83 and 120–143).
Metal ions are bound at the active site of SOD1 and remain bound after acetylation
It is not unreasonable to suspect that the ability of SOD1 to resist continued reductions in net negative charge upon binding two, three or four Zn2+ ions is some type of experimental artifact and not, as we conclude, a “Bohr-like” effect wherein the pKa of multiple amino acid residues (or tightly bound solvent) is coupled to the binding of metal ions via conformational change (or direct deprotonation of a metal ligand). Such an artifact could be conceivably caused by the lack of metal binding (or loss of metal ions) in Zn2-SOD1, Zn3-SOD1, or Zn4-SOD1 (e.g., a charge ladder of Zn3-SOD1 is in fact a charge ladder of Zn1-SOD1). The ICP-MS analysis of metalated protein charge ladders (after removal of unbound metal ions with excessive centrifugal filtration) demonstrated, however, that Zn2+ ions remained bound to SOD1 proteins after formation of the charge ladder (Supporting Information Table SI).
Although ICP-MS provided accurate measurements of metal content, and confirmed that Zn2+ ions were bound to SOD1 before and after acetylation (Supporting Information Table SI), we wanted to ensure that Zn2+ ions were coordinating at the active site of each mutant (not somewhere on the surface, such as Cys11150). We also wanted to verify that Zn2+ remained bound to the active site after acetylation of multiple lysines. To do so, we added four equivalents of Co2+ ions to dimeric apo-SOD1 proteins in the same manner as we added Zn2+ ions and analyzed the proteins with UV–vis spectroscopy. We also acetylated these Co2+ derivatives to generate protein charge ladders. Cobalt (II) is a spectroscopically active coordination analog of the spectroscopically silent Zn2+ (d10) cation.
The UV–vis spectra of the d-d electronic excitation band (spanning 450–550 nm) in G85R Co2-SOD1 (acetylated and non-acetylated) demonstrated that a maximum of only two Co2+ ions bound to the active site, even after the addition of four Co2+ equivalents [Fig. 4(A)]. This result is consistent with ICP-MS data of Zn2+-SOD1 derivatives that showed a maximum of only two Zn2+ ions bound to G85R-SOD1 (Supporting Information Table S1).
Figure 4.
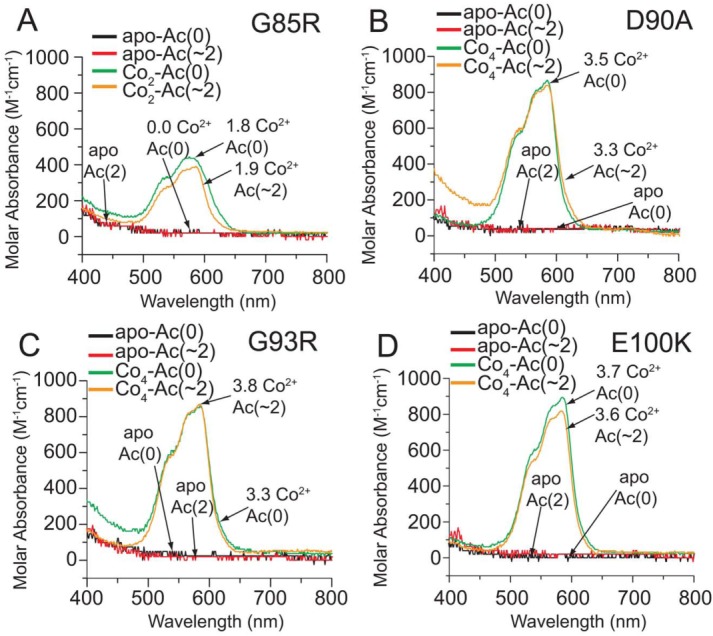
UV–vis spectra of Co2+-loaded, Lys-acetyl protein charge ladders of ALS-variant SOD1. UV–vis spectra of the Co2+ d-d electronic excitation band of (A) G85R, (B) D90A, (C) G93R, and (D) E100K SOD1 after four stoichiometric equivalents of Co2+ are added to each protein and proteins are centrifugally washed with 10 mM phosphate (pH 7.4) to remove unbound Co2+ ions. The actual stoichiometry of bound Co2+ for each derivative (as determined by ICP-MS) is listed (per dimer) next to each spectra. The acetylation number (denoted Ac(∼N)) represents the most predominant rung of the ladder according to CE (i.e., the average number of acetylated lysines in each dimer).
The intensity of the d-d electronic excitation band confirmed that all four Co2+ ions are coordinated at the active sites in the D90A, G93R, and E100K SOD1 dimers (and subsequent charge ladders). For example, the extinction coefficient for the Co2+ d-d absorption band (εd-d) in each protein (per equivalent of Co2+ ion) were: εd-d = 213 M−1 cm−1 (for G85R Co2-SOD1); 217 M−1 cm−1 (for E100K Co4-SOD1); 234 M−1 cm−1 (for G93R Co4-SOD1); and 254 M−1 cm−1 (for D90A Co4-SOD1). These εd-d values are calculated from the Co2+-replete derivatives, and do not reflect the possible differences in εd-d of Co2+ in the copper site versus Co2+ in the zinc binding site of SOD1, but are nevertheless similar to εd-d values of Co2+-replete derivatives of WT SOD1 from previous studies (e.g., 235 and 289 M−1 cm−1).21,50 Thus, the Co2+ ions—and by extrapolation, Zn2+ ions—that we detected with ICP-MS to be bound to ALS-variant SOD1 are bound at the active site and remain bound after acetylation of lysine.
Acetylated and unacetylated ALS-linked SOD1 proteins have similar thermostability and rates of amide H/D exchange
The validity of the “protein charge ladder” method rests upon the assumption that the acetylation of a few surface lysine residues on each SOD1 subunit does not significantly alter the globular structure of SOD1 (i.e., that each rung has a similar shape and hydrodynamic drag). If this assumption is wrong, then our measurements of net charge will be inaccurate. To determine if the acetylation of 2–4 lysines in each ALS-variant SOD1 subunit affected the structure of SOD1 proteins, we measured the rate of amide H/D exchange of acetylated and unacetylated apo-SOD1 with MS [Fig. 5(A)] and measured the thermostabilities with DSC (Fig. 6). Mass spectrometry allowed us to simultaneously measure the rate of amide H/D exchange of each rung under identical solution conditions and abolished the artifactual variations in measured rates that can occur in parallel experiments (because of variations in D/H back-exchange51).
Figure 5.
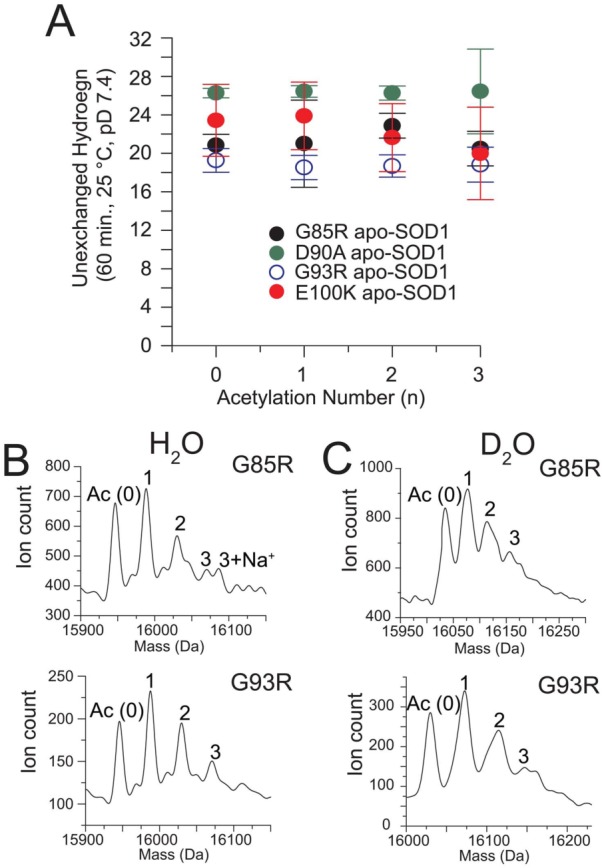
Amide H/D exchange of Lys-acetyl protein charge ladders of ALS-variant apo-SOD1 measured with ESI-MS. (A) Number of unexchanged amide hydrogens in ALS-variant apo-SOD1 (G85R, D90A, G93R, and E100K) after 60 min in D2O plotted as a function of number of acetylated lysine residues. The number of acetylated lysines measured by mass spectrometry is one-half the number observed with CE because the SOD1 dimer dissociates during ESI-MS (but not during CE). Error bars represent standard deviation of seven separate measurements. (B) Representative deconvoluted ESI-MS spectra of ALS-variant apo-SOD1 in H2O and (C) after incubation in D2O (10 mM potassium phosphate, pD 7.4) for 60 min.
Figure 6.
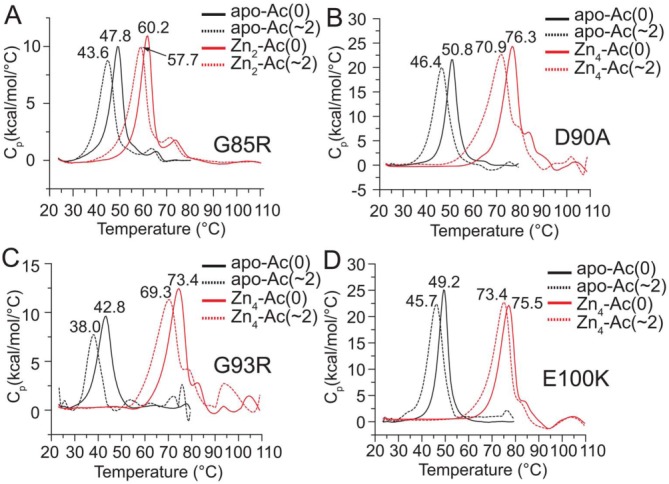
Differential scanning calorimetry of Lys-acetyl protein charge ladders of ALS-variant apo-SOD1 and Zn2+-loaded SOD1. (A) Endothermic transitions of acetylated and non-acetylated G85R apo-SOD1 and G85R Zn2-SOD1. The Tm value for each protein is listed next to each endothermic peak. The metal content was confirmed with ICP-MS. The average number of acetylated lysines in each ladder (Ac(∼N)) are listed per dimer. (B–D) Same experiments as in part A, but for D90A, G93R, and E100K apo-SOD1 and Zn4-SOD1 proteins.
All four ALS-variant apo-SOD1 proteins exhibited between ∼20 and 28 unexchanged protons per monomer, after 60 min in deuterated buffer [Fig. 5(A)]. The D90A and E100K apo-SOD1 proteins retained more unexchanged hydrogens than G93R or G85R, which is qualitatively consistent with previous measurements of H/D exchange (at 37°C)1 and correlates with the known effects of each substitution on the structure and stability of SOD1.1 The similar rates of amide H/D exchange of rungs in the charge ladder—which are resolvable with ESI-MS before deuteration [Fig. 5(B)] and after deuteration [Fig. 5(C)]—demonstrated that the acetylation of a few surface lysines in each subunit of SOD1 does not abolish the globular structure of apo-SOD1, or significantly alter its secondary or tertiary structure.
It is important to realize that the number of acetylated lysines measured by mass spectrometry [listed for each SOD1 protein in Fig. 5(A–C)] refers to the number of acetylated lysine per monomer. In contrast, the number of acetylated lysines listed for all SOD1 proteins in all other figures (i.e., electropherograms, optical spectra, thermograms, etc.) are listed per dimer and are therefore two-fold greater than the numbers of acetylated lysines shown in Figure 5. This twofold difference is due to the monomerization of dimeric SOD1 during electrospray ionization-MS, whereas the SOD1 protein was analyzed in the dimeric state during DSC, UV–vis, and CE experiments.
Differential scanning calorimetry also demonstrated that each ALS-variant SOD1 protein is properly folded, before and after lysine acetylation, and that acetylation resulted in only modest reductions in the melting transition temperature (Tm), for example, between 4 and 5°C for ALS-variant apo-SOD1 proteins and 3–5°C for Zn4-SOD1 proteins (Fig. 6). The analysis of acetylated and non-acetylated ALS-variant apo-SOD1 and Zn4-SOD1 with DSC also confirmed the presence of a large increase in Tm upon Zn2+ binding (ΔTm = ∼ 25°C). The large increase in Tm is similar to previously observed measurements for WT apo-SOD1 and Zn4-SOD1,35 and adds additional support to the contention that Zn2+ ions are bound to the active site of ALS-variant SOD1 proteins examined in this study, both before and after acetylation.
How do ALS-variant SOD1 proteins regulate net charge when binding multiple Zn2+ ions?
We hypothesize that the binding of the first Zn2+ ion to ALS-variant of SOD1 resulted in the association of an additional proton (net) and that the binding of each subsequent Zn2+ ion resulted in the dissociation of approximately two protons (net). Hence, from Zn1- to Zn4-SOD1, the SOD1 protein is expected to dissociate six protons (per dimer) in order to maintain its net negative charge. Two of these six net protons could originate from the deprotonation of two His63 to imidazolate.32 It is not known if His63 is deprotonated when it is bridging two Zn2+ at the same active site.33 Because the measured values of ALS-variant Zn4-SOD1 at pH 7.4 are similar—or more anionic in the case of E100K—than predicted values that assume each His63 to be negatively charged (Table 1), our results suggest that His63 is fully deprotonated in ALS-variant Zn4-SOD1 at pH 7.4. Nevertheless, His63 cannot account for all six released protons that are presumably dissociated from dimeric SOD1 when binding the 2nd, 3rd, and 4th Zn2+. Identifying these possible residues is beyond the scope of this article and will likely require high resolution tools such as neutron diffraction crystallography.52
It is unlikely (but nevertheless still possible) that some of these six purported protons are dissociated from positively charged histidine residues, that is, it is not likely that active-site histidines are positively charged prior to metalation and undergo deprotonation upon coordinating metals. The measured values of net charge for all four apo-SOD1 proteins suggest that the metal binding histidines in apo-SOD1 are electrostatically neutral (imidazole) prior to metal binding. For example, the calculation of Zseq for each apo-SOD1 protein at pH 7.4 assumed that all histidines are electrostatically neutral and only small, nonuniform deviations were observed between ZCE and Zseq for all four ALS-variant apo-SOD1 proteins at pH 7.4, that is, ΔZCE/seq ranged from +0.64 to −2.29 (Table 1).
ALS-linked mutations do not impair the ability of Apo-SOD1 to regulate net charge across subcellular pH
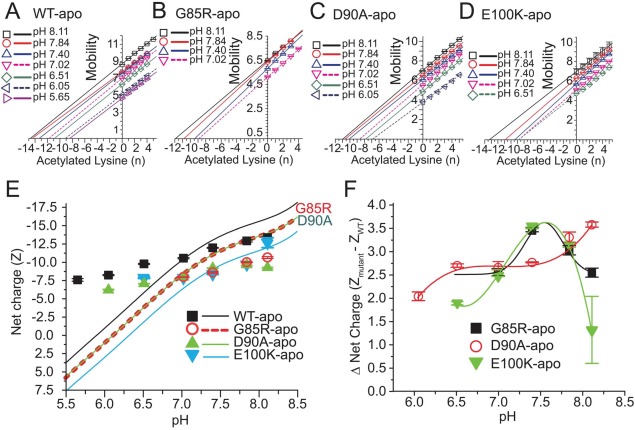
With regard to ALS, we do not know which locus or loci—cytosol, lysosome, mitochondria, or nucleus—is the primary site of SOD1 self-assembly in motor neurons and so we analyzed protein charge ladders across pH 5–8 (Supporting Information Fig. S5) and determined the net charge of ALS-variant SOD1 across pH 5–8 (Fig. 7). We could not study the unstable G93R apo-SOD1 across this entire range of pH because the charge ladders could not be resolved at low pH, presumably because the protein was absorbing onto the capillary or undergoing aggregation (charge ladders of metalated G93R SOD1 were, however, resolvable across pH 5–8). The ALS-variant D90A, G85R, and E100K apo-SOD1 proteins generally yielded resolvable protein charge ladders from pH 6.51–8.11 [Fig. S5(A–D)], from which plots of mobility versus Ac(N) were generated [Fig. 7(A–D)]. The G85R apo-SOD1 protein did not yield a resolvable charge ladder—and its net charge could not be determined—at or below pH 6.51 [Fig. S5(B)].
Figure 7.
Effect of solvent pH on net electrostatic charge of ALS-variant apo-SOD1. (A–D). Plot of electrophoretic mobility of rungs of charge ladders of ALS-variant apo-SOD1 (from Supporting Information Fig. S5) versus the number of acetylated lysines of each rung at each value of pH. (E) The measured and predicted (formal) net charge of WT and ALS-variant apo-SOD1 from pH 5.5 to 8.5. The formal values of net charge are indicated by solid and dashed lines and were calculated with the Henderson–Hasselbalch equation (see Experimental Methods). The measured values of net charge were derived from plots in part A–D. (F) Plot of the “charge gap” (i.e., the difference in net negative charge of variant and WT apo-SOD1) across pH 5.5–8.5.
In general, the net negative charge of all ALS-variant apo-SOD1 proteins that we studied did not approach electrostatic neutrality by the expected magnitude as pH was lowered from cytosolic to lysosomal pH [Fig. 7(E)]. This ability for all ALS-variant apo-SOD1 proteins to regulate net charge across pH was also observed previously for WT apo-SOD1.21 For example, the net negative charge of D90A apo-SOD1 was diminished from ZCE = −9.80 ± 0.08 at pH 8.11 to ZCE = −6.20 ± 0.01 at pH 6.05, which is smaller than the decrease that would be predicted from its sequence, that is, from Zseq = −14.2 to Zseq = +0.7 [Fig. 7(E)].
The E100K apo-SOD1 protein was, unexpectedly, more negatively charged than D90A and G85R at most values of pH across pH ∼ 6.5–8 (with the exception of pH 7.4). The E100K substitution did not generally diminish the net charge of apo-SOD1 as much as the D90A and G85R substitutions, which is a surprising result and would be impossible to predict from formal pKa values of amino acids.
All four ALS-variant apo-SOD1 proteins regulated their net charge to the point that each protein remained negatively charged at values of pH as low as pH 6, even though each protein is expected to have a positive charge at pH < 6.5 [Fig. 7(E)]. For example, D90A apo-SOD1 is predicted to have a net charge of Zseq ≈ −1 at pH 6.0, however, protein charge ladders demonstrated that the net charge was ZCE = −6.20 ± 0.01 at pH 6.05. In the case of E100K apo-SOD1, a net charge of Zseq = 0 is predicted at pH 6.50, however, charge ladders revealed that ZCE = −7.88 ± 0.10 at pH 6.51. These data suggest that missense mutations such as E100K will diminish the net negative charge of SOD1 at all subcellular loci instead of increasing the protein’s net positive charge at lysosomal pH (pH 5) as would be predicted from their formal values of net charge. This point—which is important for rationalizing how ALS mutations electrostatically accelerate SOD1 self-assembly at different subcellular loci—would be impossible to predict from standard values of molecular charge.
The set of linear plots of mobility versus Ac(N) at different pH were not parallel for each protein, as expected and previously observed for the WT SOD1 protein [Fig. 7(A–D)].21 For example, the plots of mobility versus Ac(N) for D90A apo-SOD1 exhibited a general decrease in slope as pH was decreased from pH 8.1 to pH 6.0 [Fig. 7(C)]. This decrease suggests that the hydrodynamic drag of the D90A apo-SOD1 protein steadily increased as pH was decreased.
The “charge gap” between WT and ALS-variant Apo-SOD1 is greatest at cytosolic pH
The “charge gap” between WT and ALS-variant apo-SOD1 was not constant across pH. Generally, the gap increased from ΔZCE ≈ 2.0 at pH 6.0–6.5, to ΔZCE ≈ 3.0 at pH 7.0–7.5 [Fig. 7(F)]. The G85R and E100K apoproteins, however, exhibited decreases in the charge gap at pH > 7.5 (whereas the D90A apoprotein continued to undergo increases in the charge gap at pH > 7.5).
We do not have any definitive explanation for why the charge gap in D90A apo-SOD1 generally increased with pH. Aspartate-90 is located in the most negatively charged region of SOD1.6 It is therefore possible that Asp90 (or another nearby Asp or Glu residue) might have an abnormally high pKa that deprotonates at pH > 5 in WT SOD1 but, upon substitution of Asp90 with Ala90 might exhibit a lower, more standard pKa value. Regardless of the explanation, the electrostatic effects of ALS-linked mutations are not constant across the range of intracellular pH encountered by the SOD1 protein. The E100K and G85R substitutions will cause the largest reductions in net charge at cytosolic pH—and impart the greatest minimization of intermolecular electrostatic repulsion between apo-SOD1 proteins—and will thus maximally increase the electrostatic component of apo-SOD1’s aggregation propensity at cytosolic pH.
Charge regulation of ALS-variant Zn4-SOD1 across subcellular pH
To investigate the effect of Zn2+ binding on the ability of ALS-variant SOD1 to regulate its net negative charge across subcellular pH, we analyzed protein charge ladders of D90A, G93R and E100K Zn4-SOD1 from pH 5.5 to 8.0 with capillary electrophoresis. We were able to measure the net charge of ALS-variant Zn4-SOD1 at a lower pH than metal-free ALS-variant SOD1 because the metalated proteins yielded resolvable charge ladders at lower pH than the apo-SOD1 proteins. Protein charge ladders of Zn4-SOD1 are shown in Supporting Information Figure S6, and plots of electrophoretic mobility versus Ac(N) are shown in Figure 8(A–D). We also prepared protein charge ladders (and measured the net charge) of the WT Zn4-SOD1 protein across pH because the WT Zn4-SOD1 derivative was not included in our previous analysis (that only included analyses of WT apo-SOD1 and WT Cu2Zn2-SOD1 across subcellular pH).21
Figure 8.
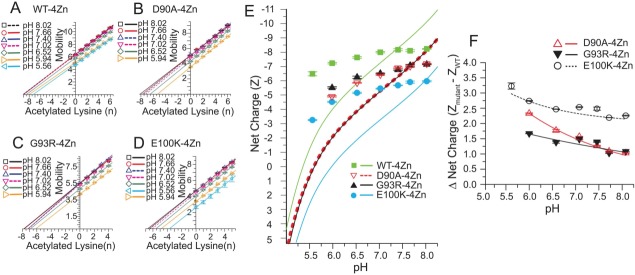
Effect of solvent pH on net electrostatic charge of WT and ALS-variant Zn4-SOD1. A–D). Plot of electrophoretic mobility of rungs of charge ladders of WT and ALS-variant Zn4-SOD1 (at different pH) against the number of acetylated lysines of each rung; plots are derived from charge ladders shown in Supporting Information Figure S6. (E) The measured and predicted (formal) net charge of WT and ALS-variant Zn4-SOD1 from pH = 5.5 to 8.0. The formal values of net charge (solid or dashed lines) were calculated as described in Materials and Methods. The measured values of net charge were derived from plots in part A–D. (F) Plot of the “charge gap” (i.e., the difference in net negative charge of variant and WT Zn4-SOD1) across pH 5.5–8.0.
The ALS-variant Zn4-SOD1 proteins regulated their net charge across pH 5.5–8.0 by a magnitude that was similar to that of the WT Zn4-SOD1 protein. The Zn2+-loaded ALS-variant proteins do not therefore undergo charge inversion from net negative to net positive as would be predicted at pH values that are below the theoretical pI (pI = 6.0 for E100K Zn4-SOD1; pI = 5.6 for G93R and D90A Zn4-SOD1). Thus, Zn2+-loaded ALS-variant SOD1 is likely to remain anionic across all of its subcellular loci. For example, the net negative charge of E100K Zn4-SOD1 was diminished from ZCE = −5.98 ± 0.04 to −3.27 ± 0.04 as pH was lowered from pH 8.02 to pH 5.56. This measured decrease in the magnitude of net negative charge is smaller than predicted across this pH range (i.e., ΔZCE = +2.71 vs. ΔZseq = +8.6). This degree of charge regulation prevented the net charge of the E100K Zn4-SOD1 protein from inverting from the predicted values of Zseq = −6 to Zseq = +2.6.
Similar degrees of charge regulation were observed for WT, G93R and D90A Zn4-SOD1 across a similar range of pH. For example, G93R Zn4-SOD1 exhibited a net charge of ZCE = −7.15 ± 0.03 at pH 8.02, which diminished in magnitude to ZCE = −5.56 ± 0.04 at pH 5.94 [Fig. 8(E)]. This fluctuation in charge of ΔZCE = +1.59 ± 0.05 is smaller than the predicted fluctuation of ΔZseq = +6.4 that is expected to occur from pH 8.0–5.9.
The “charge gap” between WT and ALS-variant Zn4-SOD1 is minimized at high (cytosolic) pH and is maximized at low (lysosomal) pH
In contrast to the “charge gap” between WT and ALS-variant apo-SOD1, which generally increased as pH was raised from pH 6–8, the “charge gap” between WT and ALS-variant Zn4-SOD1 proteins generally decreased for all three variants from pH 6–8 [Fig. 8(F)]. As pH was increased from pH 6.0–8.1, the “charge gap” of D90A Zn4-SOD1 was diminished from 2.33 ± 0.02 to 1.26 ± 0.01 and the “charge gap” of G93R Zn4-SOD1 diminished from 1.67 ± 0.01 to 1.09 ± 0.02 [Fig. 8(F)]. The “charge gap” for the E100K Zn4-SOD1 protein diminished from 3.23 ± 0.10 to 2.19 ± 0.01 across pH 5.6–8.1. The E100K Zn4-SOD1 protein possessed the smallest magnitude of net negative charge of all ALS-variant Zn4-SOD1 proteins across pH [Fig. 8(E)], whereas E100K apo-SOD1 was more similarly charged to the other ALS-variant apo-SOD1 proteins [Fig. 7(E)].
Thus, all three ALS-linked missense mutations lower the net charge of Zn4-SOD1 by a greater magnitude at lysosomal pH than at cytosolic pH. We hypothesize that the coordination of metal ions to SOD1 inhibits the protonation of residues in SOD1 and inhibits the minimization of the “charge gap” between WT and ALS-variant SOD1 at low pH.
Conclusion
This study represents the first use of protein charge ladders and capillary electrophoresis to measure how a missense mutation affects the net charge of a folded protein across physiological pH. This study represents, therefore, one of the few—to our knowledge the only—direct, accurate measurement of how a missense mutation affects the net charge of its gene product at physiological pH. The differences that we found between the predicted and effective net charge of ALS-variant SOD1 demonstrate that the combined effects of missense mutation, Zn2+ coordination, and fluctuation in pH, on the net charge of a protein must be measured and never assumed or approximated from standard pKa values of amino acid side chains and formal oxidation states of metal ions.39,40
It is almost physically impossible for our measurement of the polarity of any metal derivative of any protein in this study to be wrong (i.e., to be + instead of –). Every solution of SOD1 that we analyzed with CE contained an electrostatically neutral marker of electroosmotic flow (dimethylformamide) that functioned as a demarcation of polarity. Peaks that appear to the right of the DMF peak have a net negative charge, and peaks on the left have a net positive charge.22 All rungs of each charge ladder in this study appeared to the far right of the DMF peak (Supporting Information Fig. S6).
The deviation between the predicted and effective net charge of ALS-variant SOD1 will complicate ongoing and future attempts to rationally correlate the biophysical properties of each ALS-variant of SOD1 with its aggregation propensity and clinical phenotype (e.g., correlating net charge or ΔGfold with age of disease onset or disease duration).4,39,40 For example, researchers have found a weak linear correlation (r = 0.78) between the ΔGfold of ALS-variant apo-SOD1 proteins that are formally isoelectric to WT SOD1, and the patient survivability associated with that mutation.4 Many variants that are non-isoelectric to WT SOD1, however, lie outside this linear grouping. In most of these cases, variants with a lower magnitude of formal net charge (at pH 7.4) are associated with lower survivability (by up to 10 years) than variants with similar ΔGfold that are formally isoelectric to WT SOD1. The few ALS variants that have higher magnitudes of formal net charge (compared to WT SOD1) are associated with higher survivability (by up to 10 years) than similarly destabilized variants that are isoelectric to WT SOD1.4 Additional patient data will be required to render these correlations statistically significant because only a few ALS-linked SOD1 mutations in the current sets of data have survivability data from more than 50 patients (e.g., A4V and E100G).4 Furthermore, it is also not clear if all patients in these data sets made similar choices for palliative care (e.g., elected to have a percutaneous endoscopic gastrostomy, which can prolong life in dysphagic ALS patients by 6–8 months53).
In the current study, we only examined the electrostatic effects of formally non-isoelectric ALS-linked amino acid substitutions. When considering the strong correlation between the structural environment of an ionizable amino acid residue and its pKa (a point that has been recognized for over 50 years54) it is reasonable to hypothesize that a destabilizing isoelectric ALS-linked mutation such as A4V—which alters the structural environment of at least 20 residues in the apo-SOD1 dimer55,56 and raises pI values during native isoelectric focusing28 by up to ΔpI = +0.25—might lower the net charge of SOD1 by a degree that will increase its propensity to self-assemble into toxic oligomers.
Materials and Methods
Purification of ALS-variant SOD1
Recombinant ALS-variant SOD1 proteins (D90A, G85R, G93R, and E100K) were expressed in Saccharomyces cerevisiae and purified as previously described.21 Protein concentrations were determined by UV–vis spectroscopy using λmax = 280 nm and ε = 10,800 cm−1 M−1. Mass spectrometry was used to confirm the absence of tryptophan oxidation or any other post-translational modification that might alter the optical absorbance of SOD1 at 280 nm.
Demetalation and remetalation of ALS-variant SOD1
Metal ions (Cu2+, Zn2+) were removed from recombinant ALS-variant SOD1 by dialysis as previously described.21 All glassware and plastic utensils were soaked in 20 mM EDTA and rinsed with ultra-pure, metal-free MilliQ water before any contact with SOD1 proteins. Zinc (II) ions and Co2+ ions were then titrated back into demetalated SOD1 proteins by the slow and stepwise addition of micro-liter aliquots of ZnSO4 (5 mM) or CoCl2 (5 mM) into solutions of apo-SOD1 protein (∼100 µM SOD1) in 100 mM sodium acetate buffer, pH 5.5, as previously described.21 To remove any metal ions that remained unbound after each metal titration was completed, SOD1 protein solutions underwent at least three cycles of centrifugal filtration (i.e., a 10–20 fold dilution into 10 mM potassium phosphate, pH 7.4, followed by a 10–20 fold centrifugal concentration). The stoichiometry of Zn2+ and Co2+ bound to SOD1 was determined with an Elan 9000 Inductively-Coupled Plasma Mass Spectrometer (ICP-MS) before and after acetylation, as previously described.21
Preparation of Lys-NHCOCH3 charge ladders of ALS-variant SOD1
Lysine-ε-NH3+ protein charge ladders of ALS-variant apo-SOD1 and charge ladders of Zn2+ derivatives and Co2+ derivatives of ALS-variant SOD1 were synthesized with acetic anhydride as previously described for the WT SOD1 protein.21 Capillary electrophoresis and electrospray ionization mass spectrometry (ESI-MS) were used to determine the degree of acetylation after solutions of protein were washed (via centrifugal filtration) and transferred into a potassium phosphate buffer (10 mM, pH 7.4). Only the lowest rungs of each ladder (i.e., typically first 5–7 rungs) were generated (instead of a full ladder consisting of 23 rungs) because this number of rungs is sufficient to generate a linear plot of electrophoretic mobility versus the number of acetylated lysines.
Capillary electrophoresis
A Beckman P/ACE CE system equipped with a bare, fused silica capillary (surrounded by a liquid-cooled jacket) was used for all CE experiments. Electrophoresis was performed at 20°C (to prevent Joule heating) using a running buffer of 10 mM potassium phosphate, at various pHs ranging from pH ∼ 5–8. A small amount of dimethylformamide (DMF) was added to each solution of SOD1 before electrophoresis as an electrostatically neutral marker of electroosmotic flow (final [DMF] ≈ 1 µM during CE of sample).
Differential scanning calorimetry and amide hydrogen/deuterium exchange
To ensure that the acetylation of lysine in SOD1 did not significantly affect the structure or conformational stability of SOD1, we analyzed acetylated and non-acetylated SOD1 proteins with differential scanning calorimetry (DSC) and amide hydrogen/deuterium (H/D) exchange, as previously described for the WT SOD1 protein.21
Calculation of formal values of net charge from amino acid sequences
The formal values of net charge of SOD1 proteins were calculated from their respective amino acid sequences using the Henderson–Hasselbalch equation, following the same assumptions as previously described.21 Briefly, for calculations involving apo-SOD1 proteins, we used standard pKa values of free amino acids21; N-terminal acetylation was accounted for each subunit by subtracting a positive unit of charge. The net charge of Zn4-SOD1 proteins was similarly calculated with four additional assumptions: (i) all metal coordinating residues were treated as non-titratable across pH 5–8 (which is consistent with the ability of SOD1 to remain enzymatically active across this pH range50); (ii) each of the His63 residues were treated as anionic (imidazolate) and non-titratable whereas all other metal-coordinating histidines were treated as electrostatically neutral; (iii) each Asp83 residue (a metal binding residue) was treated as anionic and non-titratable; and (iv) two units of positive charge were added for each of the four Zn2+ ions.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
References
- Rodriguez JA, Shaw BF, Durazo A, Sohn SH, Doucette PA, Nersissian AM, Faull KF, Eggers DK, Tiwari A, Hayward LJ, Valentine JS. Destabilization of apoprotein is insufficient to explain Cu,Zn-superoxide dismutase-linked ALS pathogenesis. Proc Natl Acad Sci USA. 2005;102:10516–10521. doi: 10.1073/pnas.0502515102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw BF, Valentine JS. How do ALS-associated mutations in superoxide dismutase 1 promote aggregation of the protein? Trends Biochem Sci. 2007;32:78–85. doi: 10.1016/j.tibs.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Sandelin E, Nordlund A, Andersen PM, Marklund SS, Oliveberg M. Amyotrophic lateral sclerosis-associated copper/zinc superoxide dismutase mutations preferentially reduce the repulsive charge of the proteins. J Biol Chem. 2007;282:21230–21236. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- Bystrom R, Andersen PM, Grobner G, Oliveberg M. SOD1 mutations targeting surface hydrogen bonds promote amyotrophic lateral sclerosis without reducing apo-state stability. J Biol Chem. 2010;285:19544–19552. doi: 10.1074/jbc.M109.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassall KA, Stubbs HR, Primmer HA, Tong MS, Sullivan SM, Sobering R, Srinivasan S, Briere LA, Dunn SD, Colon W, Meiering EM. Decreased stability and increased formation of soluble aggregates by immature superoxide dismutase do not account for disease severity in ALS. Proc Natl Acad Sci USA. 2011;108:2210–2215. doi: 10.1073/pnas.0913021108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw BF, Moustakas DT, Whitelegge JP, Faull KF. Taking charge of proteins from neurodegeneration to industrial biotechnology. Adv Protein Chem Struct Biol. 2010;79:127–164. doi: 10.1016/S1876-1623(10)79004-0. [DOI] [PubMed] [Google Scholar]
- Jonsson PA, Graffmo KS, Brannstrom T, Nilsson P, Andersen PM, Marklund SL. Motor neuron disease in mice expressing the wild type-like D90A mutant superoxide dismutase-1. J Neuropathol Exp Neurol. 2006;65:1126–1136. doi: 10.1097/01.jnen.0000248545.36046.3c. [DOI] [PubMed] [Google Scholar]
- Shi Y, Rhodes NR, Abdolvahabi A, Kohn TP, Cook NP, Marti AA, Shaw BF. Deamidation of asparagine to aspartate destabilizes Cu, Zn superoxide dismutase, accelerates fibrillization and mirrors ALS-linked mutations. J Am Chem Soc. 2013;135:15897–15908. doi: 10.1021/ja407801x. [DOI] [PubMed] [Google Scholar]
- Buell AK, Hung P, Salvatella X, Welland ME, Dobson CM, Knowles TP. Electrostatic effects in filamentous protein aggregation. Biophys J. 2013;104:1116–1126. doi: 10.1016/j.bpj.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- Osborne TB. The basic character of the protein molecule and the reactions of edestin with definite quantities of acids and alkalies. J Am Chem Soc. 1902;24:39–78. [Google Scholar]
- Csonka FA, Murphy JC, Jones DB. The iso-rlectric points of barious proteins. J Am Chem Soc. 1926;48:763–768. [Google Scholar]
- Chiti F, Calamai M, Taddei N, Stefani M, Ramponi G, Dobson CM. Studies of the aggregation of mutant proteins in vitro provide insights into the genetics of amyloid diseases. 2002;99:16419–16426. doi: 10.1073/pnas.212527999. Proc Natl Acad Sci USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Phillips KJ, Liu DR. Supercharging proteins can impart unusual resilience. J Am Chem Soc. 2007;129:10110–10112. doi: 10.1021/ja071641y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittschmitt JP, Scholtz JM. The role of protein stability, solubility, and net charge in amyloid fibril formation. Protein Sci. 2003;12:2374–2378. doi: 10.1110/ps.03152903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganowsky A, Liu C, Sawaya MR, Whitelegge JP, Park J, Zhao M, Pensalfini A, Soriaga AB, Landau M, Teng PK, Cascio D, Glabe C, Eisenberg D. Atomic view of a toxic amyloid small oligomer. Science. 2012;335:1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova MI, Sievers SA, Guenther EL, Johnson LM, Winkler DD, Galaleldeen A, Sawaya MR, Hart PJ, Eisenberg DS. Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. 2014;111:197–201. doi: 10.1073/pnas.1320786110. Proc Natl Acad Sci USA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso da Silva FL, Bostrom M, Persson C. Effect of charge regulation and ion-dipole interactions on the selectivity of protein-nanoparticle binding. Langmuir. 2014;30:4078–4083. doi: 10.1021/la500027f. [DOI] [PubMed] [Google Scholar]
- Kurut A, Henriques J, Forsman J, Skepo M, Lund M. Role of histidine for charge regulation of unstructured peptides at interfaces and in bulk. Proteins. 2014;82:657–667. doi: 10.1002/prot.24445. [DOI] [PubMed] [Google Scholar]
- Shi Y, Mowery RA, Shaw BF. Effect of metal loading and subcellular pH on net charge of superoxide dismutase-1. J Mol Biol. 2013;425:4388–4404. doi: 10.1016/j.jmb.2013.07.018. [DOI] [PubMed] [Google Scholar]
- Gitlin I, Carbeck JD, Whitesides GM. Why are proteins charged? Networks of charge-charge interactions in proteins measured by charge ladders and capillary electrophoresis. Angew Chem Int Ed Engl. 2006;45:3022–3060. doi: 10.1002/anie.200502530. [DOI] [PubMed] [Google Scholar]
- Winzor DJ. Determination of the net charge (valence) of a protein: a fundamental but elusive parameter. Anal Biochem. 2004;325:1–20. doi: 10.1016/j.ab.2003.09.035. [DOI] [PubMed] [Google Scholar]
- Colton IJ, Anderson JR, Gao JM, Chapman RG, Isaacs L, Whitesides GM. Formation of protein charge ladders by acylation of amino groups on proteins. J Am Chem Soc. 1997;119:12701–12709. [Google Scholar]
- Shaw BF, Schneider GF, Bilgicer B, Kaufman GK, Neveu JM, Lane WS, Whitelegge JP, Whitesides GM. Lysine acetylation can generate highly charged enzymes with increased resistance toward irreversible inactivation. Protein Sci. 2008;17:1446–1455. doi: 10.1110/ps.035154.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso M, Massignan T, Samengo G, Cheroni C, De Biasi S, Salmona M, Bendotti C, Bonetto V. Insoluble mutant SOD1 is partly oligoubiquitinated in amyotrophic lateral sclerosis mice. J Biol Chem. 2006;281:33325–33335. doi: 10.1074/jbc.M603489200. [DOI] [PubMed] [Google Scholar]
- Choi J, Rees HD, Weintraub ST, Levey AI, Chin LS, Li L. Oxidative modifications and aggregation of Cu,Zn-superoxide dismutase associated with Alzheimer and Parkinson diseases. J Biol Chem. 2005;280:11648–11655. doi: 10.1074/jbc.M414327200. [DOI] [PubMed] [Google Scholar]
- Chevreux S, Llorens I, Solari PL, Roudeau S, Deves G, Carmona A, Testemale D, Hazemann JL, Ortega R. Coupling of native IEF and extended X-ray absorption fine structure to characterize zinc-binding sites from pI isoforms of SOD1 and A4V pathogenic mutant. Electrophoresis. 2012;33:1276–1281. doi: 10.1002/elps.201100596. [DOI] [PubMed] [Google Scholar]
- Wenisch E, Vorauer K, Jungbauer A, Katinger H, Righetti PG. Purification of human recombinant superoxide dismutase by isoelectric focusing in a multicompartment electrolyzer with zwitterionic membranes. Electrophoresis. 1994;15:647–653. doi: 10.1002/elps.1150150191. [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc Natl Acad Sci USA. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Ann Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Shin DS, Getzoff ED, Tainer JA. The structural biochemistry of the superoxide dismutases. Biochim Biophys Acta. 2010;1804:245–−262. doi: 10.1016/j.bbapap.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strange RW, Antonyuk SV, Hough MA, Doucette PA, Valentine JS, Hasnain SS. Variable metallation of human superoxide dismutase: atomic resolution crystal structures of Cu-Zn, Zn-Zn and as-isolated wild-type enzymes. J Mol Biol. 2006;356:1152–1162. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Solution structure of Apo Cu,Zn superoxide dismutase: role of metal ions in protein folding. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- Potter SZ, Zhu H, Shaw BF, Rodriguez JA, Doucette PA, Sohn SH, Durazo A, Faull KF, Gralla EB, Nersissian AM, Valentine JS. Binding of a single zinc ion to one subunit of copper-zinc superoxide dismutase apoprotein substantially influences the structure and stability of the entire homodimeric protein. J Am Chem Soc. 2007;129:4575–4583. doi: 10.1021/ja066690+. [DOI] [PubMed] [Google Scholar]
- Isom DG, Castaneda CA, Cannon BR, Velu PD, Garcia-Moreno EB. Charges in the hydrophobic interior of proteins. Proc Natl Acad Sci USA. 2010;107:16096–16100. doi: 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno B. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci USA. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon MK, Zydney AL. Determination of effective protein charge by capillary electrophoresis: effects of charge regulation in the analysis of charge ladders. Anal Chem. 2000;72:5714–5717. doi: 10.1021/ac000752b. [DOI] [PubMed] [Google Scholar]
- Prudencio M, Hart PJ, Borchelt DR, Andersen PM. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum Mol Genet. 2009;18:3217–3226. doi: 10.1093/hmg/ddp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Johnson JL, Agar NY, Agar JN. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Antonyuk SV, Seetharaman SV, Whitson LJ, Taylor AB, Holloway SP, Strange RW, Doucette PA, Valentine JS, Tiwari A, Hayward LJ, Padua S, Cohlberg JA, Hasnain SS, Hart PJ. Structures of the G85R variant of SOD1 in familial amyotrophic lateral sclerosis. J Biol Chem. 2008;283:16169–16177. doi: 10.1074/jbc.M801522200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward LJ, Rodriguez JA, Kim JW, Tiwari A, Goto JJ, Cabelli DE, Valentine JS, Brown RH., Jr Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J Biol Chem. 2002;277:15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- Rodriguez JA, Valentine JS, Eggers DK, Roe JA, Tiwari A, Brown RH, Jr, Hayward LJ. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J Biol Chem. 2002;277:15932–15937. doi: 10.1074/jbc.M112088200. [DOI] [PubMed] [Google Scholar]
- Shipp EL, Cantini F, Bertini I, Valentine JS, Banci L. Dynamic properties of the G93A mutant of copper-zinc superoxide dismutase as detected by NMR spectroscopy: implications for the pathology of familial amyotrophic lateral sclerosis. Biochemistry. 2003;42:1890–1899. doi: 10.1021/bi026704y. [DOI] [PubMed] [Google Scholar]
- Lelie HL, Liba A, Bourassa MW, Chattopadhyay M, Chan PK, Gralla EB, Miller LM, Borchelt DR, Valentine JS, Whitelegge JP. Copper and zinc metallation status of copper-zinc superoxide dismutase from amyotrophic lateral sclerosis transgenic mice. J Biol Chem. 2011;286:2795–2806. doi: 10.1074/jbc.M110.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinartaitė L, Saraboji K, Nordlund A, Logan DT, Oliveberg M. Folding catalysis by transient coordination of Zn2+ to the Cu ligands of the ALS-associated enzyme Cu/Zn superoxide dismutase 1. J Am Chem Soc. 2010;132:13495–13504. doi: 10.1021/ja1057136. [DOI] [PubMed] [Google Scholar]
- Johansson AS, Vestling M, Zetterstrom P, Lang L, Leinartaite L, Karlstrom M, Danielsson J, Marklund SL, Oliveberg M. Cytotoxicity of superoxide dismutase 1 in cultured cells is linked to Zn2+ chelation. PLoS One. 2012;7:e36104. doi: 10.1371/journal.pone.0036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahadorani S, Mukai ST, Rabie J, Beckman JS, Phillips JP, Hilliker AJ. Expression of zinc-deficient human superoxide dismutase in Drosophila neurons produces a locomotor defect linked to mitochondrial dysfunction. Neurobiol Aging. 2013;34:2322–2330. doi: 10.1016/j.neurobiolaging.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YH, Fitch CA, Bhattacharya S, Sarkisian CJ, Lecomte JT, Garcia-Moreno EB. Salt effects on ionization equilibria of histidines in myoglobin. Biophys J. 2000;79:1637–1654. doi: 10.1016/S0006-3495(00)76414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhu H, Eggers DK, Nersissian AM, Faull KF, Goto JJ, Ai J, Sanders-Loehr J, Gralla EB, Valentine JS. Copper(2+) binding to the surface residue cysteine 111 of His46Arg human copper-zinc superoxide dismutase, a familial amyotrophic lateral sclerosis mutant. Biochemistry. 2000;39:8125–8132. doi: 10.1021/bi000846f. [DOI] [PubMed] [Google Scholar]
- Venable JD, Okach L, Agarwalla S, Brock A. Subzero temperature chromatography for reduced back-exchange and improved dynamic range in amide hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2012;84:9601–9608. doi: 10.1021/ac302488h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalevsky AY, Chatake T, Shibayama N, Park SY, Ishikawa T, Mustyakimov M, Fisher Z, Langan P, Morimoto Y. Direct determination of protonation states of histidine residues in a 2 Å neutron structure of deoxy-human normal adult hemoglobin and implications for the Bohr effect. J Mol Biol. 2010;398:276–291. doi: 10.1016/j.jmb.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Spataro R, Ficano L, Piccoli F, La Bella V. Percutaneous endoscopic gastrostomy in amyotrophic lateral sclerosis: effect on survival. J Neurol Sci. 2011;304:44–48. doi: 10.1016/j.jns.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Tanford C, Taggert VG. Ionization linked changes in protein conformation. II. The N to R transition in beta-lactoglobulin. J Am Chem Soc. 1961;83:1634–1638. [Google Scholar]
- Durazo A, Shaw BF, Chattopadhyay M, Faull KF, Nersissian AM, Valentine JS, Whitelegge JP. Metal-free superoxide dismutase-1 and three different amyotrophic lateral sclerosis variants share a similar partially unfolded beta-barrel at physiological temperature. J Biol Chem. 2009;284:34382–34389. doi: 10.1074/jbc.M109.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw BF, Durazo A, Nersissian AM, Whitelegge JP, Faull KF, Valentine JS. Local unfolding in a destabilized, pathogenic variant of superoxide dismutase 1 observed with H/D exchange and mass spectrometry. J Biol Chem. 2006;281:18167–18176. doi: 10.1074/jbc.M600623200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.