Abstract
In Gram-negative bacteria, resistance to β-lactam antibacterials is largely due to β-lactamases and is a growing public health threat. One of the most concerning β-lactamases to evolve in bacteria are the Class B enzymes, the metallo-β-lactamases (MBLs). To date, penams and cephems resistant to hydrolysis by MBLs have not yet been found. As a result of this broad substrate specificity, a better understanding of the role of catalytically important amino acids in MBLs is necessary to design novel β-lactams and inhibitors. Two MBLs, the wild type IMP-1 with serine at position 262, and an engineered variant with valine at the same position (IMP-1-S262V), were previously found to exhibit very different substrate spectra. These findings compelled us to investigate the impact of a threonine at position 262 (IMP-1-S262T) on the substrate spectrum. Here, we explore MBL sequence-structure-activity relationships by predicting and experimentally validating the effect of the S262T substitution in IMP-1. Using site-directed mutagenesis, threonine was introduced at position 262, and the IMP-1-S262T enzyme, as well as the other two enzymes IMP-1 and IMP-1-S262V, were purified and kinetic constants were determined against a range of β-lactam antibacterials. Catalytic efficiencies (kcat/KM) obtained with IMP-1-S262T and minimum inhibitory concentrations (MICs) observed with bacterial cells expressing the protein were intermediate or comparable to the corresponding values with IMP-1 and IMP-1-S262V, validating the role of this residue in catalysis. Our results reveal the important role of IMP residue 262 in β-lactam turnover and support this approach to predict activities of certain novel MBL variants.
Keywords: metallo-β-lactamase, point mutation, antibiotic resistance, enzyme evolution, IMP-1 antibody
Introduction
Metallo-β-lactamases (MBLs) are bacterial resistance determinants that are becoming widely dispersed in Gram-negative bacteria. MBLs are grouped as class B β-lactamases1 (B1-B3 subclasses)2,3 and contain one or two Zn(II) ions in the active site.4,5 MBLs threaten the current status of β-lactam antibacterial chemotherapy, because they inactivate a broad range of penicillins, cephalosporins, and carbapenems via cleavage of the amide bond in the β-lactam ring.4 Currently, three major MBL families are responsible for the crisis in antimicrobial chemotherapy (NDM, VIM, and IMP family MBLs).
IMP-type MBLs belong to subclass B1,2,3 in which Zn1 is coordinated by H116, H118, and H196 (3H site) and Zn2 by D120, C221, and H263 (DCH site). IMP-type enzymes are clinically important due to their broad substrate spectra, insensitivity to currently used β-lactamase inhibitors (clavulanate, sulbactam, and tazobactam), occurrence throughout the world, and the ability of their encoding genes to spread rapidly in bacteria.5–10 In addition, IMP MBLs are a very diverse enzyme family with currently 48 variants found in clinical isolates over the course of about two decades (www.lahey.org/Studies/other.asp#table1). Many of these IMP enzymes cluster around IMP-111 and IMP-2,12 which are separated by 15% sequence divergence.8,9
An important long-term goal is to understand the evolution of substrate specificity in MBLs, so that inhibitors that restore the efficacy of β-lactam antibacterials as well as MBL-resistant β-lactams may be developed. Here, as an initial step toward this goal, we explore the role of a single amino acid substitution at the critical position 262 in the IMP-1 MBL. Residue 262 is located near the active site, proximal to the H263 Zn(II) ligand (Fig. 1). A previous study of wild types IMP-1 and IMP-6 (IMP-1-S262G), as well as variants with other amino acids at position 262, exhibited a wide array of substrate profiles toward different β-lactam antibiotics with the V262 variant being most diverse from IMP-1.13 A “domino effect” helps explain the role of residue 262 and its effect on catalytic efficiencies toward “Type I” and “Type II” substrates (Fig. 2).14 Type I substrates are defined as those containing neutral R2 groups with atoms with high electron density (e.g., nitrocefin, cephalothin, and cefotaxime), while Type II substrates contain axial methyl groups (penicillins) or positively charged R2 groups (e.g., ceftazidime and imipenem) (Fig. 2). Previously IMP-1 (S262) was shown to inactivate Type II substrates better than IMP-1-S262V (referred to as IMP-6-G262V in the original report, because IMP-6 was the starting point for that mutational study13), while the latter has a more pronounced hydrolytic preference toward Type I substrates and poorly converts Type II substrates.13 In this study, we explore the role of residue 262 and attempt to qualitatively predict catalytic efficiencies of the novel variant IMP-1-S262T. Threonine was selected as an amino acid that has features of both serine (hydroxyl group) and valine (branched) (Fig. 3). Our hypothesis is that, based on this “intermediate” structure of threonine, a variant harboring this residue at position 262 would exhibit catalytic efficiencies that are intermediate to those of the enzymes harboring serine (IMP-1) and valine (IMP-1-S262V) at this position. Validation of this hypothesis will allow us to predict the evolution of natural variants at position 262 and anticipate strategies to design effective β-lactams and inhibitors.
Figure 1.
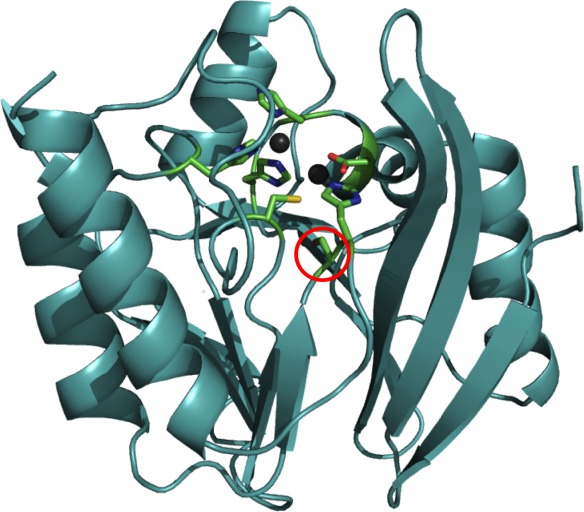
IMP-1 active site with Zn(II) ligands and residue 262. Serine 262 is highlighted with a red circle. The six remaining residues are Zn(II) ligands coordinating their respective Zn(II) ion (depicted as gray spheres). The enzyme backbone is represented as a cyan cartoon and residues as sticks with atoms colored in green (carbon), red (oxygen), blue (nitrogen), and yellow (sulfur). The top left Zn(II) ion is Zn1 in the 3H site and the bottom right one Zn2 in the DCH site. The image was created using PyMOL (www.pymol.org/) and PDB entry 1DD6 (Ref.21).
Figure 2.
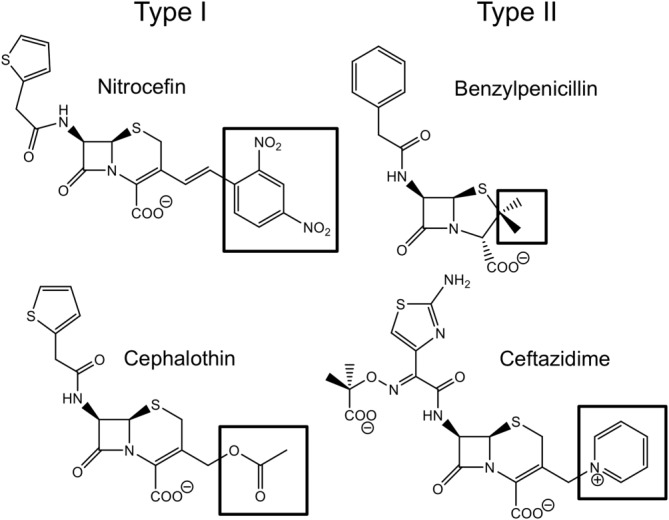
Structures of selected β-lactam substrates. Type I substrates (nitrocefin, cephalothin, and cefotaxime) possess R2 groups with atoms of high electron density (indicated by black boxes); Type II substrates (ceftazidime, benzylpenicillin, ampicillin, imipenem, and meropenem) have axial methyl groups or positively charged and/or bulky R2 groups.
Figure 3.
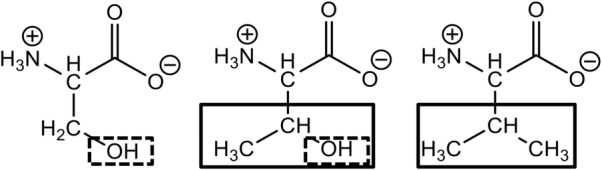
Structural characteristics of amino acids at position 262. The amino acid structures of serine (left), threonine (middle), and valine (right) are depicted in the ionized form with shared functional groups highlighted in boxes. Hydroxyl groups are highlighted in dashed boxes and branched (bulky) groups in solid boxes.
Results
Expression and biophysical characteristics
The bla genes encoding IMP-1-S262T and IMP-1-S262V were generated by polymerase chain reaction (PCR)-based site-directed mutagenesis, expressed under control of the T7 promoter in Escherichia coli cells, and gene products were purified as described previously.15 Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis and protein quantification indicated homogeneous proteins of >95% purity and overall yields of 1.1 mg and 1.3 mg per liter culture for IMP-1-S262T and IMP-1-S262V, respectively (Table 1). The percentages of protein expressed in the soluble form, determined by comparing soluble and insoluble fractions after cell lysis via SDS-PAGE, were estimated to be ∼70% and ∼50% for IMP-1-S262T and IMP-1-S262V, respectively. These values are consistent with those of IMP-1 expressed and purified in a previous study15 and used for comparison in the present study, which yielded more (∼90%) recombinant protein in the soluble fraction and more (2.0 mg) purified protein per liter culture.
Table 1.
Expression Data and Biophysical Characteristics of IMP-1, IMP-1-S262T, and IMP-1-S262V
| Property | IMP-1a | IMP-1-S262T | IMP-1-S262V |
|---|---|---|---|
| % Soluble | ∼90 | ∼70 | ∼50 |
| Yield of purified protein per liter of culture in mg | 2.0 | 1.1 | 1.3 |
| Molecular mass measured (calculated) in Da | 25,128 (25,127) | 25,125 (25,125) | 25,112 (25,113) |
| Zn(II) ions/molecule enzyme | 1.8 ± 0.2 | 1.8 ± 0.1 | 1.7 ± 0.1 |
Values reported by Liu et al.15
The molecular masses of IMP-1-S262T and IMP-1-S262V determined by electrospray ionization mass spectrometry (ESI-MS) are consistent with the theoretical masses (Table 1). Circular dichroism scans of IMP-1-S262T and IMP-1-S262V are superimposable with that of wild-type IMP-1, indicating a conserved secondary structure composed of α helices and β sheets (Fig. 4), typical for the αββα fold of the zinc metallohydrolase family of the β-lactamase fold.16,17 The differences in the amplitude of the ellipicity are most likely the result of uncertainty in the determination of protein concentration, which can be in the range of 10–15% rather than an indication of an actual change in the secondary structure. The 4-(2-pyridylazo)resorcinol (PAR) assay18 indicated both enzymes bound approximately 2 Zn(II) ions per molecule enzyme, just like IMP-1 (Table 1).
Figure 4.
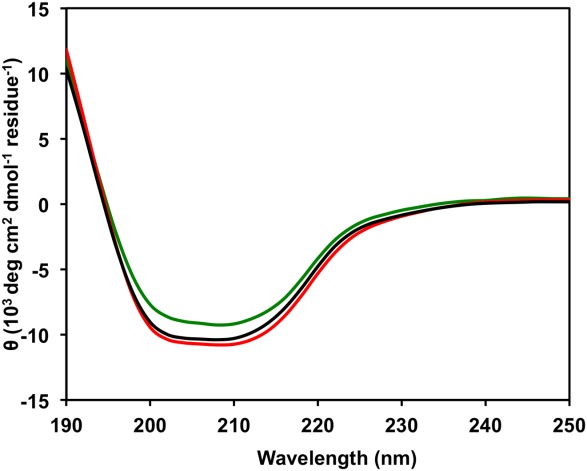
Circular dichroism scans of IMP-1 (black line), IMP-1-S262T (green line), and IMP-1-S262V (red line).
Kinetic profiles
Kinetic parameters of IMP-1 were determined previously,15 and IMP-1-S262T and IMP-1-S262V were tested against nine antibiotics: nitrocefin, cephalothin, cefotaxime, ceftazidime, benzylpenicillin, ampicillin, imipenem, meropenem, and aztreonam. All antibiotics were hydrolyzed by these MBLs except for aztreonam. The kinetic constants for IMP-1-S262V and IMP-1 hydrolysis (Table 2) are comparable to those in a previous study.13 Interestingly, when analyzing the values of IMP-1-S262T in comparison to those of IMP-1 and IMP-1-S262V, the kcat/KM values toward all but one substrate (cephalothin) are in the range between these other two proteins, as hypothesized (bolded numbers in Table 1). Even the cephalothin value is only 24% outside that range, which could be attributed to experimental uncertainty in protein determination and pipetting errors.
Table 2.
Kinetic Constants of IMP-1, IMP-1-S262T, and IMP-1-S262V with Nine β-Lactam Antibiotics
| IMP-1a | IMP-1-S262T | IMP-1-S262V | |||||||
|---|---|---|---|---|---|---|---|---|---|
| β-Lactam Antibiotic | kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) | kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) | kcat (s−1) | KM (μM) | kcat/KM (μM−1 s−1) |
| Nitrocefin | 181 ± 8 | 9 ± 1 | 20 ± 2 | 394 ± 8 | 8 ± 1 | 48 ± 5 | 562 ± 5 | 5.9 ± 0.4 | 96 ± 6 |
| Cephalothin | 50 ± 1 | 4.5 ± 0.2 | 11.1 ± 0.2 | 240 ± 20 | 9 ± 1 | 26 ± 1 | 160 ± 20 | 7.4 ± 0.7 | 21 ± 1 |
| Cefotaxime | 16.4 ± 0.4 | 5.1 ± 0.8 | 3.2 ± 0.5 | 70 ± 3 | 23 ± 2 | 4.0 ± 0.1 | 118 ± 5 | 23 ± 2 | 5.2 ± 0.3 |
| Ceftazidime | 14 ± 1 | 55 ± 7 | 0.25 ± 0.02 | 39 ± 6 | 160 ± 20 | 0.25 ± 0.01 | 2.4 ± 0.7 | 90 ± 30 | 0.028 ± 0.003 |
| Benzylpenicillin | 2000 ± 100 | 530 ± 50 | 3.8 ± 0.1 | 1000 ± 200 | 700 ± 200 | 1.4 ± 0.1 | 14 ± 1 | 400 ± 70 | 0.035 ± 0.004 |
| Ampicillin | 260 ± 30 | 210 ± 30 | 1.2 ± 0.1 | 450 ± 40 | 700 ± 100 | 0.62 ± 0.04 | 3 ± 2 | 400 ± 300 | 0.009 ± 0.003 |
| Imipenem | 105 ± 5 | 35 ± 4 | 3.0 ± 0.2 | 340 ± 30 | 150 ± 20 | 2.3 ± 0.1 | 11 ± 3 | 230 ± 60 | 0.06 ± 0.01 |
| Meropenem | 22 ± 1 | 2.9 ± 0.2 | 7.7 ± 0.4 | 57 ± 6 | 11 ± 3 | 4.6 ± 0.7 | 21.0 ± 0.3 | 34 ± 1 | 0.61 ± 0.01 |
| Aztreonam | NDb | ND | ND | ND | ND | ND | ND | ND | ND |
Experiments were performed in triplicate and values are reported as average ± standard deviation. Values for IMP-1-S262T that are within the range between IMP-1 and IMP-1-S262V are highlighted in bold.
Values reported by Liu et al.15
Not detectable.
Using equation ΔΔGenz = -RT ln((kcat/KM IMP-1-S262T)/(kcat/KM IMP-1-S262V)) to understand the energetics, this difference in kcat/KM corresponds to a negligible decrease of 0.13 kcal/mol in activation free energy.19 When analyzing kcat and KM values separately, the values for IMP-1-S262T are in the range between those of IMP-1 and IMP-1-S262V in three and four out of eight cases, respectively. For two Type I substrates, nitrocefin and cefotaxime, all three values (kcat, KM, and kcat/KM) of IMP-1-S262T lie between the values of the other two enzymes.
Susceptibility profiles
For the Type II substrates ceftazidime, benzylpenicillin, ampicillin, imipenem, and meropenem, the order of the MIC values for the three enzymes corresponds to that of the kcat/KM values (IMP-1=IMP-1-S262T>>IMP-1-S262V for ceftazidime and IMP-1>IMP-1-S262T>>IMP-1-S262V for the others). Thus, the predicted trends in catalytic efficiencies directly translate to trends in bacterial resistance levels. Actually, IMP-1-S262V does not provide E. coli DH10B cells with resistance to these Type II substrates according to Clinical and Laboratory Standards Institute standards20 (except for ceftazidime, MICs are indistinguishable from the negative controls), while IMP-1 and IMP-1-S262T do for the most part (cells expressing IMP-1-S262T are not resistant to imipenem, but intermediate). Apart from lower kcat/KM values, this can be explained with a lower expression level of IMP-1-S262V in DH10B cells as shown by western blot (below). This is also apparent for the Type I substrates cephalothin and cefotaxime, as cells expressing IMP-1-S262V show lower MIC values than IMP-1 and IMP-1-S262T, although the purified enzyme exhibits higher or comparable kcat/KM values. This observation is in agreement with a previous study,13 where purified IMP-1-S262V demonstrated high kcat/KM values toward cephalothin and cefotaxime, but cells expressing the enzyme resulted in relatively low MIC values. We attributed this difference to a lower level of functionally expressed MBL. In that respect, the previous report is consistent with the present results, although the cells (E. coli BL21 (DE3)) and expression vectors (pET30a) used were different. E. coli BL21 (DE3) cells and the pET30a expression vector were designed for overexpression of MBL in the cytoplasm and are not an ideal system for MIC assays, while E. coli DH10B cells and the pBC SK(+) expression vector used here are designed to mimic natural expression levels of MBL in the periplasm.
Western blot analysis
An IMP-1-specific antibody was produced, characterized, and tested for nonspecific binding to other MBLs (will be published elsewhere). The western blot analysis indicated that IMP-1 and IMP-1-S262T were expressed at comparable levels, while the expression level of functional IMP-1-S262V was about a tenth of that (Fig. 5). Although enzyme for purification was overexpressed in the cytoplasm vs. the periplasm, a similar trend was observed here: the amount of soluble protein of IMP-1-S262V was ∼50% in this study versus ∼70% for IMP-1-S262T and ∼90% for IMP-1. Also in our previous study (cytoplasmic expression in E. coli BL21 (DE3) cells) less of IMP-1-S262V (∼10%) was found in the soluble fraction than of IMP-1 (∼20%).13 In summary, the lower expression level of functional IMP-1-S262V serves as an explanation for the relatively low MIC values for IMP-1-S262V compared with its kcat/KM values.
Figure 5.
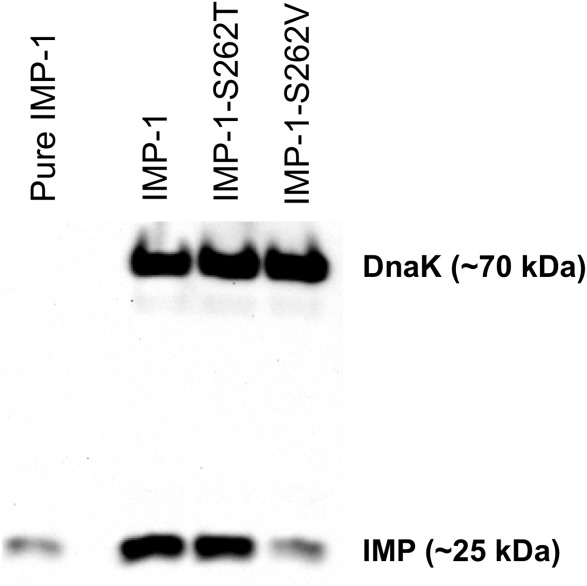
Image of a representative western blot of whole cells (E. coli DH10B) expressing IMP-1, IMP-1-S262T, and IMP-1-S262V. DnaK at ∼70 kDa served as a constitutively expressed control. The band at ∼25 kDa corresponds to the MBLs. Fifty ng of purified IMP-1 were loaded as positive control in the left lane. Expression levels of IMP-1 and IMP-1-S262T are comparable and that of IMP-1-S262V is about 10% of that.
Discussion
Catalytic efficiencies
IMP-1-S262T exhibited intermediate or comparable catalytic efficiencies to IMP-1 and IMP-1-S262V (Table 2) toward the eight tested substrates, validating our hypothesis, while aztreonam was not a substrate for any of the enzymes. IMP-1 is the most effective out of all three tested enzymes in the hydrolysis of Type II β-lactams as explained by the “domino effect”.14 IMP-1-S262V is a relatively poor enzyme for the hydrolysis of Type II substrates, possibly due to its 1-methyl-ethyl side chain (Fig. 3) that could cause unfavorable steric interactions with H263, which itself could interact unfavorably with R2 groups of Type II substrates, destabilizing its metal ligation of Zn2 (Fig. 1) and the stability of the enzyme-substrate transition state complex. This scenario has been described previously.13
Overall, the “hybrid” side chain of threonine results in catalytic efficiencies toward Type II substrates comparable to or lower than S262, but the extent depends on the substrate: for ceftazidime and carbapenems (charged and/or bulky R2 side chains) they are the same to about two thirds; for the penicillins they are about half to a third. T262 in IMP-1-S262T contains a hydroxyl group like S262 in IMP-1, which may have a favorable interaction with P68 (labeled P32 in the crystal structure21 and in our previous study14) and account for a portion of the “domino effect” with certain substrates, while the methyl group may interact unfavorably with H263, as in V262. A portion of the favorable effect of S262 in IMP-1 was missing with the corresponding residue missing the hydroxyl group (A262 in IMP-1-S262A=IMP-6-G262A), resulting in slightly lower kcat/KM values of this variant toward Type II substrates than IMP-1 (S262), but higher than IMP-6 (G262).13 The difference in kcat/KM values toward Type II substrates obtained here with IMP-1-S262T versus IMP-1-S262V is similar to but bigger than those of IMP-1 versus IMP-1-S262A, suggesting that replacing the hydroxyl group with a methyl group (T→V) has a more detrimental effect than simply removing it (S→A).
IMP-1 is very efficient in hydrolyzing Type I substrates, but there is an increase in kcat/KM values for IMP-1-S262T and IMP-1-S262V (Table 2). As suggested previously,13 increased steric interaction of the 1-hydroxyl-ethyl side chain in IMP-1-S262T and the 1-methyl-ethyl side chain in IMP-1-S262V with H263 may not be detrimental or even favorable with these substrates, as their R2 groups 2,4-dinitrostyryl (nitrocefin) and acetoxymethyl (cephalothin and cefotaxime) are kept from interacting with H263 by engaging in electrostatic interactions with neighboring residues in the binding pocket, such as N233.14
In this study meropenem behaved like a Type II substrate, although the [5-(dimethylcarbamoyl)pryrrolidin-3-yl]sulfanyl R2 group with a pKa of 7.4 (AstraZeneca, Mississauga, Ontario, Canada) could be neutral.15 However, the bulkiness of the R2 group and the axial methyl group at C1 may account for its Type II substrate-like character. Also, it is possible that this moiety is protonated due to the microenvironment in the active site of certain MBLs, but not others.
To our knowledge, this is the first analysis that predicts and explores the activity of an MBL variant across a broad spectrum of β-lactam antibacterials. Other sequence-structure-activity studies of the IMP-type β-lactamases have focused mostly on determining residues critical for activity,22–24 responsible for an altered substrate profile,25 or important for the hydrolysis of specific antibiotics.26 Interestingly, in the IMP-1 β-lactamase two residues that tolerate mutations, even though they are highly conserved, have been heretofore identified: N233,27 which has previously been assigned a role in forming an oxyanion hole,28,21 and C221,29 which is a functionally important Zn(II) ligand.21,22 Attempts have been made to predict enzyme activity of IMP variants using molecular dynamics simulations13 and computational protein design.30 These studies did not yield accurate predictions, that is, either not all variants predicted to be more active were more active, or, if they were, it only applied to some of the substrates. In the present study, we have successfully predicted activity of one variant against a broad spectrum of substrates using the available information of variants that have very closely related amino acids at the mutated position.
MIC values and western blot analysis
MIC values are not only representative of the catalytic efficiencies, but also other resistance mechanisms, such as reduced membrane permeability and efflux pumps.31 In the tested ElectroMAX E. coli DH10B cells carrying pBC SK(+) plasmids these factors that contribute to resistance were eliminated. IMP-1 conferred greater MIC values (Table 3) of all tested β-lactams except for cephalothin and ceftazidime, whose MIC values were just as high for IMP-1-S262T. The MIC values of aztreonam were equivalent and as low as those of the controls, indicating susceptibility for cells expressing the three different enzymes. Nitrocefin was not tested in the MIC assays, because it is not clinically used and quite unstable. Overall, there is a good correlation between kcat/KM values and MIC values with the exception of IMP-1-S262V, which conferred lower resistance levels than expected, in fact none at all (susceptible) for the Type II substrates. This effect can be linked to a lower expression level of functional enzyme for this variant as shown by western blot analysis. Using an IMP-1-specific polyclonal antibody, as done here, has some advantages over using tagged protein and a tag-specific antibody,27 since it cannot be excluded that a tag will have some impact on expression or other properties of recombinant protein. Therefore, it has been recommended that β-lactamases be expressed and studied without any artificially added tag.32
Table 3.
Antibiotic Susceptibility Assay of E. coli DH10B Cells Harboring pBC SK(+)blaIMP Plasmids
| MIC (µg/ml) | |||||
|---|---|---|---|---|---|
| pBC SK(+) | pBC SK(+) | pBC SK(+) | |||
| β-Lactam | blaIMP-1a | blaIMP-1-S262T | blaIMP-1-S262V | pBC SK(+)a | No Plasmida |
| Cephalothin | 512 | 512 | 128 | 16 | 16 |
| Cefotaxime | 256 | 128 | 8 | 0.125 | 0.25 |
| Ceftazidime | 256 | 256 | 1 | 0.5 | 0.5 |
| Benzylpenicillin | 256 | 128 | 32 | 32 | 32 |
| Ampicillin | 256 | 128 | 8 | 4 | 8 |
| Imipenem | 4 | 2 | 0.25 | 0.25 | 0.25 |
| Meropenem | 16 | 4 | 0.0625 | <0.0625 | 0.0625 |
| Aztreonam | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 |
MIC values were determined using the median value of six measurements.
Values for IMP-1-S262T that are within the range between IMP-1 and IMP-1-S262V are highlighted in bold.
Values reported by Liu et al.15
Conclusions
The data presented herein demonstrate readily how a single point mutation can alter substrate specificity and how sequence-structure-activity relationships can help predict hydrolytic preferences in IMP-1-S262T. Apart from one outlier (cephalothin), which can be considered within the error margin, IMP-1-S262T exhibited catalytic efficiencies in the range between those of IMP-1 and IMP-1-S262V, as predicted, and these translate into intermediate MIC values. Defining and testing hypotheses regarding the evolution of substrate specificity, as done in this report, is the ultimate test of our understanding of MBL function and its evolution under selective pressure.
We anticipate that the T262 mutant will be found in nature, as its encoding gene can be derived from the blaIMP-6 gene through a single nucleotide change and it would confer increased MICs toward ceftazidime, penicillins, and imipenem (a selective advantage in the clinical setting).25,15 However, the evolutionary advantage of expressing IMP-1-S262T versus other IMP variants will need to be defined by the clinical context in which this variant is recovered.
The approach presented here could be used to predict the phenotypes of other variants that are predicted to evolve. For instance, IMP-10 differs from IMP-1 by a V67F mutation, which leads to decreased MICs of penicillins.33 Other variants (IMP-9, IMP-31, IMP-35, and IMP-45) possess isoleucine at the same position. Therefore, an IMP-1-V67I mutant may be expected to evolve or already exist. One may expect MICs of penicillins for cells expressing this variant to be similar to IMP-1 due to isoleucine being very similar (bulky) to valine or lower due to isoleucine being larger in size than valine, just like phenylalanine in IMP-10, but to a lesser degree. We are currently investigating the IMP-1-V67I variant and have found the MIC of ampicillin to be the same as for IMP-1 and that of benzylpenicillin to be between those for IMP-1 and IMP-10, consistent with what would be expected (unpublished results). Further studies must be undertaken as novel β-lactam compounds and inhibitors of MBLs are developed, so that a deeper understanding is achieved.
Materials and Methods
Preparation of plasmids for overexpression and minimum inhibitory concentration (MIC) assays. To mutate the blaIMP-1 gene cloned into a pET30a vector under control of the T7 promoter for cytoplasmic overexpression (pET30a-blaIMP-1, kindly provided by Merck, Sharp & Dohme Corp, Rahway, NJ) to blaIMP-1-S262T, the forward primer 5’ – G GCA AAA CTG GTT GTT CCA ACT CAC AGT GAA GTT GGA GAC GC – 3’ (bold letters indicating the mutated codon and bold underlined the mutated nucleotide) and the complementary reverse primer were designed and purchased from Integrated DNA Technologies (Coralville, IA). Polymerase chain reaction (PCR)-based site-directed mutagenesis was carried out in an Eppendorf Mastercycler Pro (Eppendorf, Hamburg, Germany) with pET30a-blaIMP-1 as a template using 30 cycles of denaturation at 95°C for 45 seconds, annealing at 51.3°C for 1 minute, and extension at 70°C for 2 minutes with additional final heating at 70°C for 10 minutes. Escherichia coli OverExpress C43 (DE3) cells (Lucigen, Middleton, WI) were transformed with DpnI (Promega Corporation, Madison, WI)-digested and deionized PCR product by electroporation according to the manufacturer’s instructions. Colonies were selected on lysogeny broth (LB) agar plates with 50 μg/ml kanamycin, and plasmids were isolated from overnight cultures using the QIAprep Spin Miniprep Kit (QIAGEN, Hilden, Germany). The correct mutation was verified by DNA sequencing (Eurofins MWG Operon, Huntsville, AL). The vector pET30a-blaIMP-1-S262V was taken from a previous study.13 The blaIMP-1 gene with its native leader sequence for periplasmic expression was excised from the pET26b vector (kindly provided by Dr. James Spencer, University of Bristol, UK) and inserted into the pBC SK(+) vector (Stratagene, Santa Clara, CA) resulting in pBC SK(+)-blaIMP-1.15 To obtain pBC SK(+)-blaIMP-1-S262T, PCR-based site-directed mutagenesis was employed using the same primers and cycling parameters as for pET30a-blaIMP-1-S262T. To obtain pBC SK(+)-blaIMP-1-S262V, pBC SK(+)-blaIMP-6 from our previous study15 served as a template and the forward primer 5’ – G GCA AAA CTG GTT GTT CCA GTT CAC AGT GAA GTT GGA GAC GC – 3’ and the complementary reverse primer were used. The cycling parameters were 16 cycles of denaturation at 96°C for 1.5 minutes, annealing at 60°C for 1 minute, and extension at 72°C for 22 minutes with additional final heating at 72°C for 10 minutes. E. coli TG1 electrocompetent cells (Lucigen, Middleton, WI) were transformed with DpnI-digested and deionized PCR product, plasmids were isolated from selected cultures, and the blaIMP mutant genes were sequenced as described above. ElectroMAX E. coli DH10B cells (Invitrogen, Grand Island, NY) were transformed with the confirmed pBC SK(+)-blaIMP mutant plasmids and used for MIC assays.
β-Lactamase expression and purification
E. coli OverExpress C43 (DE3) cells harboring pET30a-blaIMP-1, pET30a-blaIMP-1-S262T, and pET30a-blaIMP-1-S262V were grown in 1 liter of LB with 50 μg/ml kanamycin at 37°C. Protein expression was induced in the log phase by isopropyl β-D-1-thiogalactopyranoside (IPTG, Fisher Scientific, Fair Lawn, NJ) at a final concentration of 0.5 mM at 21°C for an additional four hours. Cells were then harvested by centrifugation at 3,220 g at 4°C for 45 min. Pellets were resuspended in 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS) (pH 7.0) with 100 μM ZnSO4 (MOPS buffer) and lysed by sonication (Model 120 Sonic Dismembrator; Fisher Scientific, Pittsburgh, PA) for a total of 3 min of pulsing at 70% intensity in 20-s periods interrupted by 20-s pauses. Soluble and insoluble fractions were separated by centrifugation at 38,900 g at 4°C for 35 min. The soluble fraction was syringe filtered (0.22 µm) and injected into two connected HiTrap CM FF ion-exchange columns (GE Healthcare, Piscataway, NJ) on an ÄKTA Purifier UPC 100 (GE Healthcare, Piscataway, NJ), washed with MOPS buffer until baseline absorbance at 280 nm was obtained and eluted with a 0–0.5 M NaCl gradient in MOPS buffer. Fractions containing MBL as judged by nitrocefin hydrolysis were pooled and injected into a HiPrep 26/60 Sephacryl S-100 HR column (GE Healthcare, Piscataway, NJ). Gel filtration fractions were eluted in MOPS buffer containing 100 mM NaCl and pooled for centrifugal concentration using an Amicon Ultra-15 Centrifugal Filter Unit (molecular weight cut-off of 10 kDa) (Millipore, Billerica, MA) at 3,220 g at 4°C for 20 min. Percentages of MBL in the soluble and insoluble fractions after cell lysis as well as homogeneity of purified protein (>95%) were determined via SDS-PAGE using standard protocols34 at the 25 kDa marker.
Biophysical characterization
Protein concentration was determined in triplicate using the Bio-Rad protein assay kit with bovine serum albumin (BSA) as a standard (Bio-Rad, Richmond, CA). The molecular weight of pure protein (IMP-1-S262T and IMP-1-S262V) was determined by electrospray ionization mass spectrometry (ESI-MS) at the Protein/Peptide Microanalytical Laboratory at the California Institute of Technology (Pasadena, CA). For circular dichroism (CD) experiments, all three purified enzymes were dialyzed 1:2,000 in 5 mM phosphate buffer (pH 7.00) for 24 hours and the protein concentration was determined afterwards as described above. CD scans were done using an Olis DSM 20 Spectrophotometer (Olis, Bogart, GA). Three scans from 250-190 nm were averaged and converted to molar ellipicity to analyze secondary structure. Zn(II) ion content of the MBLs was determined using the 4-(2-pyridylazo)resorcinol (PAR) (Tokyo Chemical Industry, Tokyo, Japan) assay,18 except with the use of zinc-free 50 mM MOPS buffer (pH 7.0). Zn(II) concentrations were calculated by linearly plotting detected Zn(II) released from different concentrations of MBL (determined via Bio-Rad Protein Assay) upon denaturation in comparison to a series of different Zn(II) concentrations in the same buffer as a standard.
Kinetic assays
Steady-state kinetic parameters were determined using an SQ-2802 UV/Vis Spectrophotometer (UNICO, Dayton, NJ). To protect diluted enzymes from denaturation, a final concentration of 10 μg/mL BSA was added. Substrate and enzyme were pre-incubated separately for 5 min at 30°C in MOPS buffer before the reaction was initiated by rapid mixing. The appearance of product (nitrocefin) or disappearance of substrate (all other β-lactam antibiotics) was monitored at the specific wavelengths and converted to initial velocities using the specific extinction coefficients used previously.13,15,35 The initial velocity of each substrate was measured at 7–8 different concentrations spread out over a range chosen based on the previously published KM values of IMP-1. Three series of initial velocities were determined and analyzed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA). Nonlinear fitting of these series to the Michaelis-Menten equation under consideration of the enzyme concentration used yielded three kcat, KM, and kcat/KM values, respectively. The averages of these values ± standard deviations are reported.
Susceptibility assays
Antibiotic susceptibility was determined by minimum inhibitory concentration (MIC) assays using a broth dilution method according to the guidelines from the Clinical and Laboratory Standards Institute.36 An inoculum of 104 E. coli ElectroMAX DH10B cells harboring the blaIMP-1, blaIMP-1-S262T, and blaIMP-1-S262V genes in the pBC SK (+) plasmid were grown in Mueller-Hinton II broth (MHB) pH 7.3 at 37°C for 20 h in the presence of one of eight β-lactam antibiotics supplied in serial dilutions in a 96-well plate assay format. Cells without the plasmid and with the plasmid without the MBL gene served as controls. Six measurements were carried out, and the median was determined as the MIC value.
Western blot analysis
Western blotting was employed to analyze expression levels of wild type and artificial variants of IMP-1 under conditions comparable with those in the susceptibility assay. Totally, 5 mL cultures of E. coli DH10B cells containing pBC SK(+) plasmids harboring the various blaIMP genes in MHB containing 20 µg/mL of chloramphenicol were grown at 37°C to an OD600 of 0.8. A total of 50 µL aliquots of the cultures were pelleted and frozen overnight. Pellets were resuspended in 20 µL loading dye, separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane (Novex, Life Technologies, Carlsbad, CA) by electroblotting. Fifty ng of purified IMP-1 were used as a positive control. After blocking for 1 h with 5% nonfat dry milk, MBL presence on the blot was detected by incubation in 1 µg/mL anti-IMP-1 polyclonal antibody (Ab) (raised in rabbits injected with purified IMP-1 and purified on Protein G columns as described previously37) and 1/10,000 dilution of mouse anti-DnaK monoclonal Ab (Enzo Life Sciences) overnight at 4°C. The membrane was washed four times, 15 min each, in Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 and subsequently incubated with 1/10,000 dilution of both horeseradish peroxidase (HRP)-Protein G conjugate (Bio-Rad) and HRP-goat anti-mouse Ab conjugate (Santa Cruz Biotechnology). All Ab/protein G solutions were prepared with 5% nonfat dry milk. After four additional washes, membranes were processed for exposure using the ECL kit (GE Healthcare) and FOTO/Analyst® FX (Fotodyne).
Acknowledgments
The authors thank Dr. James Spencer (University of Bristol, UK) for the pET26b expression vector containing the IMP-1 gene with its native leader sequence and Merck, Sharp & Dohme Corp. for the pET30a expression vector containing the IMP-1 gene without the leader sequence and imipenem. We also thank Dr. H. Howard Xu and Jennifer Fleischer (California State University, Los Angeles) for assistance and training with the MIC assays and Antony Huang (California State Polytechnic University, Pomona) for assistance with the circular dichroism scans.
Glossary
- BSA
bovine serum albumin
- ESI-MS
electrospray ionization mass spectrometry
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MBL
metallo-β-lactamase
- MIC
minimum inhibitory concentration
- MOPS
3-(N-morpholino)propanesulfonic acid
- PAR
4-(2-pyridylazo)resorcinol
- PCR
polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis.
References
- Ambler RP. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frere JM. Standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother. 2001;45:660–663. doi: 10.1128/AAC.45.3.660-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garau G, Garcia-Saez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frere JM, Dideberg O. Update of the standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother. 2004;48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder MW, Spencer J, Vila AJ. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc Chem Res. 2006;39:721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- Bebrone C. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol. 2007;74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Pitout JD, Nordmann P. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2007;2:501–512. doi: 10.2217/17460913.2.5.501. [DOI] [PubMed] [Google Scholar]
- Oelschlaeger P, Ai N, Duprez KT, Welsh WJ, Toney JH. Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J Med Chem. 2010;53:3013–3027. doi: 10.1021/jm9012938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- Gutkind GO, Di Conza J, Power P, Radice M. β-Lactamase-mediated resistance: a biochemical, epidemiological and genetic overview. Curr Pharm Des. 2013;19:164–208. [PubMed] [Google Scholar]
- Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. Molecular characterization of an enterobacterial metallo β-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother. 1994;38:71–78. doi: 10.1128/aac.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio ML, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, Amicosante G, Rossolini GM. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother. 2000;44:1229–1235. doi: 10.1128/aac.44.5.1229-1235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger P, Mayo SL, Pleiss J. Impact of remote mutations on metallo-β-lactamase substrate specificity: implications for the evolution of antibiotic resistance. Protein Sci. 2005;14:765–774. doi: 10.1110/ps.041093405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger P, Schmid RD, Pleiss J. Modeling domino effects in enzymes: molecular basis of the substrate specificity of the bacterial metallo-β-lactamases IMP-1 and IMP-6. Biochemistry. 2003;42:8945–8956. doi: 10.1021/bi0300332. [DOI] [PubMed] [Google Scholar]
- Liu EM, Pegg KM, Oelschlaeger P. The sequence-activity relationship between metallo-β-lactamases IMP-1, IMP-6, and IMP-25 suggests an evolutionary adaptation to meropenem exposure. Antimicrob Agents Chemother. 2012;56:6403–6406. doi: 10.1128/AAC.01440-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiyasu H, Osaka K, Ishino Y, Toh H. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 2001;503:1–6. doi: 10.1016/s0014-5793(01)02686-2. [DOI] [PubMed] [Google Scholar]
- Hunt JB, Neece SH, Ginsburg A. The use of 4-(2-pyridylazo)resorcinol in studies of zinc release from Escherichia coli aspartate transcarbamoylase. Anal Biochem. 1985;146:150–157. doi: 10.1016/0003-2697(85)90409-9. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace KM, Taracila M, Wallace CJ, Hujer KM, Bethel CR, Hornick JM, Bonomo RA. Elucidating the role of Trp105 in the KPC-2 β-lactamase. Protein Sci. 2010;19:1714–1727. doi: 10.1002/pro.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. 2012. pp. M100–S22. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere JM, Payne DJ, Bateson JH, Abdel-Meguid SS. Crystal structure of the IMP-1 metallo β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry. 2000;39:4288–4298. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- Haruta S, Yamaguchi H, Yamamoto ET, Eriguchi Y, Nukaga M, O’Hara K, Sawai T. Functional analysis of the active site of a metallo-β-lactamase proliferating in Japan. Antimicrob Agents Chemother. 2000;44:2304–2309. doi: 10.1128/aac.44.9.2304-2309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta S, Yamamoto ET, Eriguchi Y, Sawai T. Characterization of the active-site residues asparagine 167 and lysine 161 of the IMP-1 metallo β-lactamase. FEMS Microbiol Lett. 2001;197:85–89. doi: 10.1111/j.1574-6968.2001.tb10587.x. [DOI] [PubMed] [Google Scholar]
- Materon IC, Palzkill T. Identification of residues critical for metallo-β-lactamase function by codon randomization and selection. Protein Sci. 2001;10:2556–2565. doi: 10.1110/ps.40884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyobe S, Kusadokoro H, Ozaki J, Matsumura N, Minami S, Haruta S, Sawai T, O’Hara K. Amino acid substitutions in a variant of IMP-1 metallo-β-lactamase. Antimicrob Agents Chemother. 2000;44:2023–2027. doi: 10.1128/aac.44.8.2023-2027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materon IC, Beharry Z, Huang W, Perez C, Palzkill T. Analysis of the context dependent sequence requirements of active site residues in the metallo-β-lactamase IMP-1. J Mol Biol. 2004;344:653–663. doi: 10.1016/j.jmb.2004.09.074. [DOI] [PubMed] [Google Scholar]
- Brown NG, Horton LB, Huang W, Vongpunsawad S, Palzkill T. Analysis of the functional contributions of Asn233 in metallo-β-lactamase IMP-1. Antimicrob Agents Chemother. 2011;55:5696–5702. doi: 10.1128/AAC.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Fast W, Benkovic SJ. On the mechanism of the metallo-β-lactamase from Bacteroides fragilis. Biochemistry. 1999;38:10013–10023. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]
- Horton LB, Shanker S, Mikulski R, Brown NG, Phillips KJ, Lykissa E, Venkataram Prasad BV, Palzkill T. Mutagenesis of zinc ligand residue Cys221 reveals plasticity in the IMP-1 metallo-β-lactamase active site. Antimicrob Agents Chemother. 2012;56:5667–5677. doi: 10.1128/AAC.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelschlaeger P, Mayo SL. Hydroxyl groups in the ββ sandwich of metallo-β-lactamases favor enzyme activity: a computational protein design study. J Mol Biol. 2005;350:395–401. doi: 10.1016/j.jmb.2005.04.044. [DOI] [PubMed] [Google Scholar]
- Frere JM. Quantitative relationship between sensitivity to β-lactam antibiotics and β-lactamase production in gram-negative bacteria–I. Steady-state treatment. Biochem Pharmacol. 1989;38:1415–1426. doi: 10.1016/0006-2952(89)90180-9. [DOI] [PubMed] [Google Scholar]
- Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyobe S, Kusadokoro H, Takahashi A, Yomoda S, Okubo T, Nakamura A, O’Hara K. Detection of a variant metallo-β-lactamase, IMP-10, from two unrelated strains of Pseudomonas aeruginosa and an Alcaligenes xylosoxidans strain. Antimicrob Agents Chemother. 2002;46:2014–2016. doi: 10.1128/AAC.46.6.2014-2016.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Pegg KM, Liu EM, Lacuran AE, Oelschlaeger P. Biochemical characterization of IMP-30, a metallo-β-lactamase with enhanced activity toward ceftazidime. Antimicrob Agents Chemother. 2013;57:5122–5126. doi: 10.1128/AAC.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. 2012. pp. M07–A8. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard–Ninth Edition. Clinical and Laboratory Standards Institute, Wayne, PA.
- Hujer AM, Page MG, Helfand MS, Yeiser B, Bonomo RA. Development of a sensitive and specific enzyme-linked immunosorbent assay for detecting and quantifying CMY-2 and SHV β-lactamases. J Clin Microbiol. 2002;40:1947–1957. doi: 10.1128/JCM.40.6.1947-1957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


