Abstract
Lassa fever (LF) is a severe viral hemorrhagic fever caused by Lassa virus (LASV). The LF program at the Kenema Government Hospital (KGH) in Eastern Sierra Leone currently provides diagnostic services and clinical care for more than 500 suspected LF cases per year. Nearly two-thirds of suspected LF patients presenting to the LF Ward test negative for either LASV antigen or anti-LASV immunoglobulin M (IgM), and therefore are considered to have a non-Lassa febrile illness (NLFI). The NLFI patients in this study were generally severely ill, which accounts for their high case fatality rate of 36%. The current studies were aimed at determining possible causes of severe febrile illnesses in non-LF cases presenting to the KGH, including possible involvement of filoviruses. A seroprevalence survey employing commercial enzyme-linked immunosorbent assay tests revealed significant IgM and IgG reactivity against dengue virus, chikungunya virus, West Nile virus (WNV), Leptospira, and typhus. A polymerase chain reaction–based survey using sera from subjects with acute LF, evidence of prior LASV exposure, or NLFI revealed widespread infection with Plasmodium falciparum malaria in febrile patients. WNV RNA was detected in a subset of patients, and a 419 nt amplicon specific to filoviral L segment RNA was detected at low levels in a single patient. However, 22% of the patients presenting at the KGH between 2011 and 2014 who were included in this survey registered anti-Ebola virus (EBOV) IgG or IgM, suggesting prior exposure to this agent. The 2014 Ebola virus disease (EVD) outbreak is already the deadliest and most widely dispersed outbreak of its kind on record. Serological evidence reported here for possible human exposure to filoviruses in Sierra Leone prior to the current EVD outbreak supports genetic analysis that EBOV may have been present in West Africa for some time prior to the 2014 outbreak.
Introduction
Lassa fever (LF), a severe viral hemorrhagic fever (VHF) caused by Lassa virus (LASV), was first identified in Nigeria in 1969 (9,14,45), and subsequently identified in Sierra Leone in 1973 during an outbreak in the town of Panguma, located in the Eastern Province of Sierra Leone (29). Starting in the late 1970s and continuing through the 1980s, the Kenema Government Hospital (KGH), a 350-bed hospital located in the heart of the LF endemic zone of Sierra Leone, was the primary study site for LF research (30). The KGH LF program represented a successful partnership between the Sierra Leone Ministry of Health and Sanitation (MoHS), the United States Centers for Disease Control and Prevention, and other partners. Much of the authors' knowledge about LF and VHFs in general comes from the studies conducted at the KGH and affiliated sites during this period (18,34,35). Following the end of the civil conflict in Sierra Leone (1991–2002), a diverse group of organizations including the Sierra Leone MoHS, the World Health Organization (WHO), the United States' Office for Foreign Disaster Assistance, the United States Army Medical Research Institute of Infectious Diseases (USAMRIID), and Tulane University and its partners (the Viral Hemorrhagic Fever Consortium, VHFC) reinstated the LF clinical and laboratory research program at the KGH (4,12,30). The major aims of this effort have been to enhance the laboratory diagnostic capacity and training, to develop national and regional prevention and control strategies for LF, and to perform studies on clinical management, infection, and environmental control.
Efforts toward infection and environmental control greatly improved since the cessation of the civil conflict. Yet, the numbers of suspected cases presenting to the KGH increased (41). This is due in part to the efforts of the KGH LF outreach team, who educate healthcare providers in rural health centers to the LF threat and alert them to the renewed availability of free LF testing and treatment at the KGH. Currently, the LF program at the KGH provides diagnostic services and clinical care for more than 500 suspected LF cases per year (41) with routine laboratory testing for LF using recombinant antigen immunoassays that were implemented in 2008. Since 2008, there have been continuous improvements in LF laboratory (LFL) testing at the KGH, including developing and refining recombinant antigen enzyme-linked immunosorbent assays (ELISAs) for the detection of LASV antigens (viremia) and for LASV-specific immunoglobulin M (IgM) and immunoglobulin G (IgG) (11–13,24). A lateral flow immunoassay (LFI) was implemented in 2010 (11,12,24,41; Boisen et al. Improved Diagnosis of Lassa Fever Using ReLASV LF Immunoassays [in preparation]) for its simplicity, low cost, and widespread use in a variety of clinical and field settings, particularly in regions where electricity and other resources are not routinely available. This LFI, termed the ReLASV® Antigen Rapid Test, was developed by Corgenix, Inc., and CE marked in 2013.
In early 2014, another VHF was first documented in West Africa. Multiple cases of Ebola virus disease (EVD), a severe VHF caused by Ebola virus (EBOV), have recently been described in West Africa. These are the first reported cases of filovirus disease in the West African subregion (19,21), except for the single nonfatal case of disease caused by Taï Forest virus reported in Ivory Coast in 1994 (22) but not seen again. In the 2014 outbreak, the first EVD cases were detected in Guinea in the town of Guékédou near the Sierra Leonean and Liberian borders (Kissi Kingdom) in early February 2014. EVD subsequently spread to other regions in Guinea, including the capital city of Conakry located in the southwestern region. EVD cases were also reported in Liberia, initially in the north bordering the area of the first reported cases in Guinea and later in the capital city of Monrovia. Sierra Leone reported its first laboratory-confirmed EVD case on May 25, 2014, a case diagnosed by Augustine Goba, Director of the KGH LFL. The West African outbreak has since expanded into the most deadly and geographically dispersed EVD outbreak on record. To date (December 2, 2014), a total case count of 6,599, with 5,441 laboratory confirmed, 79 probable, and 1,079 suspected cases of EVD, have been reported in Sierra Leone, with 1,398 confirmed, probable, and suspected deaths. A total case count in Guinea, Liberia, Sierra Leone, Nigeria, Senegal, Mali, Spain, and the United States now stands at 15,935, with 5,689 deaths (46). Among patients meeting the case definition of LF, who were evaluated at the KGH Lassa Ward from 2008 to 2012, only 11% (190/1,740) were LASV antigenemic (41). Over the same time span, 65% (1,143/1,740) of suspected LF patients tested negative for either LASV antigen or anti-LASV IgM, and therefore were considered to have a non-Lassa febrile illness (NLFI). The NLFI patients had a similar range of morbidity as acute LF cases, which accounts for their high case fatality rate (CFR) of 36% observed in this prior study (41). Here, studies aimed at determining the causes of the febrile illnesses in non-LF cases presenting to the KGH are reported, including the possible involvement of filoviruses. Prior to the outbreak of EVD in West Africa, the authors had initiated development of EVD immunoassays supported by a NIAID SBIR R43 (Phase I) grant, guided by their experience with LF immunoassays. Genetic analyses of strains that have emerged in West Africa suggest that EBOV has been in Guinée Forestière (Forested Guinea) for several decades (1). The Guinean rainforest extends south into both Liberia and Sierra Leone, evoking the possibility of prior human exposure to EBOV in all three countries. Therefore, serum samples were tested from NLFI patients for the presence of non-Lassa febrile agents, including arboviruses and filoviruses, and evidence is reported for possible human exposure to EBOV in Sierra Leone prior to the current 2014 EVD outbreak.
Methods
Human subjects
The clinical research, including all human subjects tested at the KGH, was approved by the Sierra Leone Ethics and Scientific Review Committee. Suspected LF cases and close contacts of confirmed cases were eligible for enrollment in this study as outlined by the study protocol approved by Tulane University's Institutional Review Board (IRB) and following the National Institutes of Health/National Institutes of Allergy and Infectious Diseases (NIH/NIAID) guidelines governing the use of human subjects for research. All subjects in this study provided written informed consent for their case details to be published. The subjects provided explicit informed consent for their samples to be tested for the presence of other disease agents besides LASV. Human immunodeficiency virus and hepatitis virus testing was not performed due to IRB and ethics committee considerations.
Only KGH staff were involved in the administration of healthcare to patients at the KGH Lassa Ward. All medical decisions, including whether to administer ribavirin to patients, were at the sole discretion of the attending KGH Lassa Ward physician. Small blood volumes (typically 5 mL) for serum separation were collected from study subjects in vacutainer tubes by experienced phlebotomists. The blood was allowed to coagulate for 20 min at room temperature, and then the serum was separated from the coagulated blood by centrifugation (200 g for 20 min at room temperature). Aliquots of the serum fraction were used immediately for testing, and the remaining serum was stored in cryovials at −20°C for future use.
Clinical study
Suspected LF patients were enrolled in this clinical study after a pre-admission evaluation to determine if they presented with sufficient signs of febrile illness (Table 1) (30). Contacts of suspected LF patients were also eligible for enrollment. The suspected LF case definition was a reported or documented temperature ≥38°C for <3 weeks with absence of local inflammation and at least two major, or one major plus two minor, or at least three minor signs and symptoms of classic LF infection.
Table 1.
Positivity Rates for Suspected Lassa Cases by Three Lassa Classifications for Selected Clinical Symptoms
| Characteristic | LF (n=29)a | LPE (n=24)b | NLFI (n=24)c | Total (n=77) | p-Valued |
|---|---|---|---|---|---|
| Fatal outcome | 13/29 (45) | 4/24 (17) | 4/24 (17) | 21/77 (27) | 0.04* |
| Fever | 24/26 (92) | 21/23 (91) | 23/24 (96) | 68/73 (93) | 0.86 |
| Headache | 15/25 (60) | 13/23 (57) | 14/24 (58) | 42/72 (58) | 1.00 |
| Weakness | 12/25 (48) | 18/23 (78) | 16/24 (67) | 46/72 (64) | 0.10 |
| Dizziness | 8/25 (32) | 6/23 (26) | 14/24 (58) | 28/72 (39) | 0.06 |
| Muscle pain | 6/25 (24) | 3/23 (13) | 7/24 (29) | 16/72 (22) | 0.43 |
| Retrosternal pain | 11/25 (44) | 8/23 (35) | 11/24 (46) | 30/72 (42) | 0.73 |
| Coughing | 13/25 (52) | 13/23 (57) | 12/24 (50) | 38/72 (53) | 0.91 |
| Vomiting | 12/25 (48) | 12/23 (52) | 16/24 (67) | 40/72 (56) | 0.42 |
| Diarrhea | 7/25 (28) | 5/23 (22) | 8/24 (33) | 20/72 (28) | 0.72 |
| Bleeding | 8/25 (32) | 8/23 (35) | 6/24 (25) | 22/72 (31) | 0.81 |
| Sore throat | 6/25 (24) | 9/23 (39) | 9/24 (38) | 24/72 (33) | 0.47 |
| Abdominal pain | 7/25 (28) | 8/23 (35) | 12/24 (50) | 27/72 (38) | 0.29 |
| Runny nose | 0/25 (0) | 0/23 (0) | 1/24 (4) | 1/72 (1) | 0.65 |
| Conjunctivitis | 4/25 (16) | 2/23 (9) | 2/24 (8) | 8/72 (11) | 0.72 |
| Jaundice | 0/25 (0) | 0/23 (0) | 1/24 (4) | 1/72 (1) | 0.65 |
| Rash | 0/25 (0) | 0/23 (0) | 0/24 (0) | 0/72 (0) | — |
| Deafness | 0/25 (0) | 0/23 (0) | 1/24 (4) | 1/72 (1) | 0.65 |
| Confusion | 3/25 (12) | 1/23 (4) | 1/24 (4) | 5/72 (7) | 0.61 |
| Shortness of breath | 1/25 (4) | 0/23 (0) | 1/24 (4) | 2/72 (3) | 1.00 |
| Neck swelling | 3/25 (12) | 3/23 (13) | 1/24 (4) | 7/72 (10) | 0.61 |
Values are expressed as mean (sample size).
LF (Lassa fever) defined as PCR+(or Ag+when PCR data were unavailable).
LPE (LASV prior-exposure) defined as PCR–IgM+(or Ag–IgM+when PCR data were unavailable).
Non-Lassa febrile illness (NLFI) defined as PCR–IgM– (or Ag–IgM– when PCR data were unavailable).
p-Value for Fisher's exact test for testing for association among the three comparison groups.
—, insufficient data for calculating Fisher's exact test statistic.
p<0.05.
LASV, Lassa virus, PCR, polymerase chain reaction.
Upon enrollment, patients received an admission exam on the Lassa Ward and were screened for LF using the ReLASV® Antigen Rapid Test and ELISA assays, often within 3 h of admission. The rapid test is designed to accept either whole blood from a finger stick, plasma, or serum. Patient serum was collected and aliquoted to perform the ReLASV® Ag ELISA and ReLASV® IgM/IgG ELISAs. This serum was also used for metabolic panel and serum cytokine levels testing, and LASV reverse transcription polymerase chain reaction (RT-PCR), on-site in the LFL. Test results were used as an aid to the assessment of acute LF in conjunction with clinical signs and symptoms. Exclusion criteria included hemodynamic instability as determined by the treating physician. This study was performed in January and February 2012.
Comprehensive metabolic panel
The kinetics of 14 serum analytes were analyzed on a panel of 77 patient sera, using a Piccolo® blood chemistry analyzer (Abaxis, Inc.) with Comprehensive Metabolic Reagent Discs. Analytes characterized in these studies were sodium (Na+), potassium (K+), bicarbonate (tCO2), chloride (Cl−), calcium (Ca2+), blood urea nitrogen (BUN), creatinine (CRE), alkaline phosphatase (ALP), alanine transaminase (ALT), aspartate transaminase (AST), total bilirubin (TBIL), albumin (ALB), and total protein (TP). Data outputs were printed for each patient sample, with standard clinical values and deviations recorded.
NLFI ELISA survey
A series of NLFI patient sera (fever of unknown origin; FUOs) were selected for testing based on apparent morbidity. Symptomatic patients were identified from the Lassa Program database based on their cumulative symptoms. Patients with moderate to high symptom frequencies were selected in part based on the volume of the available sample to perform the numerous assays. A total of 77 sera were analyzed in non-LF tropical disease ELISA kits, according to the manufacturer's instructions. FUO testing included DENV Detect™ NS-1 antigen and IgM/IgG (InBios), West Nile Virus Detect™ IgM/IgG (InBios), Chikungunya IgM/IgG (Genway); Typhus IgM/IgG, (Genway), and Leptospira IgM (Core Diagnostics). Assay performance was monitored with the use of internal controls, and cutoffs were determined according to the manufacturer's instructions for individual kits.
Cytokine kinetics
Kinetics of 11 serum cytokines were analyzed by flow cytometry on a sub-panel of 53 patient sera. From the original panel of 77 sera, 24 samples did not contain enough volume to permit accurate assessment of serum cytokine levels. Serum cytokines were captured with a bead-based platform, using an eBioscience FlowCytomix Human Th1/Th2 11-plex Kit (Bender MedSystems GmbH), according to the manufacturer's instructions. Sample data were collected with an Accuri C6® benchtop cytometer (Accuri Cytometers, Inc.), and raw FACS data files were analyzed with the FlowCytomix Pro 3.0 software application (eBioscience). Data were reported in pg/mL of input serum volume.
FUO PCR survey of potential human pathogens
A panel of fever-causing agents with historical seroprevalence, sporadic emergence, or endemic status in Western Africa was chosen for a screen of viral and microbial agents present in subjects with acute LF, previous exposure to LASV, or NLFI. PCR analysis was performed on 41 of the original panel of 77 sera, due to lack of sample volume from the remaining 36 sera. The agents analyzed in the PCR survey were LASV, Zaire ebolavirus (EBOV), Marburg virus (MBGV), Crimean-Congo hemorrhagic fever virus (CCHFV), Rift Valley fever virus (RVFV), dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), chikungunya virus (CHIKV), Leptospira interrogans, Rickettsia prowasekii, Salmonella typhi, Bacillus recurrentis, Plasmodium falciparum, and Plasmodium vivax. Infectious agent or family-specific oligonucleotides and corresponding minigene amplicons were identified in the published literature (16,17,20,28,36–39,42). Inclusion criteria for PCR methodologies used in this study were previous testing on live agents, obtained from in vitro or in vivo sources, validation on nearest neighbors, and testing in controlled studies. Characteristics and sources of oligonucleotides employed in these studies are outlined in Supplementary Table S1. Nucleic acids were isolated from 100 μL of patient serum using either a Qiagen QIAmp Viral RNA Mini kit or a Genomic DNA Isolation kit (Qiagen). Oligonucleotides, buffers, and amplification conditions were as previously described in the corresponding literature references.
Prior to adapting PCR protocols in the screening of FUO patient nucleic acids, each method was validated with agent-specific in vitro generated RNA transcripts, or DNA minigenes cloned in plasmid vectors (synthesized at Integrated DNA Technologies). RNA transcripts were generated by runoff in vitro transcription reactions using T7 RNA Polymerase on Sap I-linearized plasmids with cloned target amplicons. RNAs were subsequently purified through G25 Sepharose columns and were resuspended in DEPC water for analysis. Unrestricted, diluted plasmid solutions containing target DNA sequences were used directly in PCR reactions. PCR protocols were validated with target RNAs or DNAs spiked in normal human serum at 1 μg/mL, extracted with the kits indicated above, and 2 μL of nucleic acid input was used in a 50 μL PCR reaction using the manufacturer's suggested conditions, or with modifications suggested by the procedures outlined in the literature references.
Amplification of RNA targets was performed using a SuperScript III One-Step RT-PCR kit (Invitrogen). Amplification of DNA targets was performed with AmpliTaq Gold® 360 DNA Polymerase and Master Mix (Invitrogen). Amplified nucleic acids were resolved on 4% E-Gels, alongside a 50 bp ladder and relevant controls, and were digitally recorded for analysis. Patient nucleic acids samples were subsequently isolated in the same manner, and PCRs were performed under the same conditions. Plasmid DNA constructs were isolated and characterized at Tulane University, and transported to the KGH LFW on cold packs. In vitro transcription reactions and RNA purifications were performed at the KGH LFL just prior to use in PCR assays. Positive and negative RNA and DNA controls were purified freshly for use in each set of PCR reactions. Each PCR run included positive and negative control nucleic acids, and negative controls specifically designed to test for contamination of extraction and purification reagents, PCR buffers, oligonucleotides, and polymerase stocks.
ReLASV® IgG/IgM ELISA
The ReLASV® IgG/IgM ELISA Test kits utilize microwell plates coated with LASV nucleoprotein (Vybion, Inc.) (13; Boisen et al. Improved Diagnosis of Lassa Fever Using ReLASV LF Immunoassays [in preparation]). The kit includes a normal human serum control. Convalescent patient serums were used for the reference curve and positive control. Reference serum, controls, and patient serum were diluted 1:100 in sample buffer. The reference serum was additionally diluted threefold as described above. Diluted reference, controls, and samples were transferred into the microwell plate (100 μL/well) and incubated for 30 min at ambient temperature. The microwells were washed four times with 300 μL/well of phosophate buffered saline (PBS)–Tween wash solution. Peroxidase labeled Hu IgG or IgM Fc-specific caprine polyclonal reagent (Jackson ImmunoResearch Labs, Inc.) was added to the microwells (100 μL/well) and incubated at ambient temperature for 30 min. The microwells were washed four times with 300 μL/well of PBS–Tween wash solution. TMB substrate (BioFx Laboratories, Inc.) was added (100 μL/well) and incubated for 15 min before a stop solution (100 μL/well) was added. The microplates were read at 450 nm with 650 nm subtraction. Positive cutoffs were established using U.S. normal panels and LASV PCR negative contact samples (LF controls) (Boisen et al. Improved Diagnosis of Lassa Fever Using ReLASV LF Immunoassays [in preparation]).
ReEBOV™ IgM/IgG ELISA
A retrospective study was performed with banked sera samples collected between 2011 and 2014 (n=242), using a ReEBOV™ IgM/IgG ELISA platform recently developed by the VHFC. The ReEBOV™ IgM/IgG ELISA prototype utilized microwell plates coated with recombinant filovirus antigens (kindly provided by E.O. Saphire, of The Scripps Research Institute). Recombinant Zaire EBOV glycoprotein (31) and VP40 structural protein (10) were coated on high binding microwell plates and stabilized with proprietary blocking solution. The testing protocol was similar to the ReLASV® IgG/IgM ELISA described above. A positive cutoff (optical density [OD] 450 nm=0.360) was established on the 80th percentile obtained with a panel of normal sera from SL donors (n=14).
EVD convalescent serum was used as an interassay reference sample. This reference serum was diluted 1:100 followed by serial (threefold) dilutions for testing along with normal human serum control. In addition, Drs Saphire and Burton (The Scripps Research Institute) made available the EBOV GP specific human monoclonal antibody KZ52 (10,31) for use as an additional EBOV IgG reference sample. This study was performed in June 2014, shortly after the first case of EVD was diagnosed at the KGH LFL (on May 25, 2014).
Statistical analysis
Data were analyzed using SAS v9.3 (SAS Institute, Inc.). Hypotheses involving dichotomous response variables were tested using Fisher's exact test or logistic regression. For logistic regression models with multiple independent predictors, all pairwise interaction terms were included in the model. Count outcomes were modeled using Poisson regression, and distributional comparisons were carried out using the Kolmogorov–Smirnov approach. Gaussian kernel smoothing was used to smooth the categorical serostatus age distributions. Analyses were two-tailed, with a significance threshold set at p<0.05.
Results
Clinical symptoms or blood chemistries cannot distinguish LF patients from patients with NLFIs
Diagnosis of hemorrhagic fevers, such as EVD and LF, is challenged by the fact that the presenting syndrome is very nonspecific with a broad differential diagnosis. Malaria, typhoid fever, yellow fever, leptospirosis, meningococcemia, Rickettsial infections, bacterial sepsis, and many other illnesses can all be confused with VHF in the early stages of disease presentation. Even an experienced clinician may have difficulty distinguishing EVD or LF from each other in the early stages and may misdiagnose the disease on clinical grounds as a common febrile illness. To determine which pathogens may be mistaken for LF in Sierra Leone, a series of serum samples from NLFI were examined and tested for various serological and clinical parameters, and the results were compared to samples from patients with acute LF as well as evidence for previous LASV exposure.
Symptomatic patients who had provided informed consent were identified from the LF program database based on their cumulative symptoms. All subjects met the LF surveillance case definition previously discussed in Khan et al. (30). Patients with moderate to high symptom frequencies were selected in part based on the volume of the available sample to perform the numerous assays described here. Patients were subdivided into three groups depending on test results from ReLASV® rapid tests and immunoassays. Subjects who presented to the KGH Lassa Ward with LASV antigenemia were classified as having acute LF. Subjects with circulating anti-LASV IgM were considered to have had Lassa prior exposure (LPE). As discussed previously (12), LASV infection appears to interfere with class switching from a predominantly IgM to a predominantly IgG response. Thus, antigen-negative subjects with anti-LASV IgM may have been infected with LASV and become LF convalescent prior to presenting to the Lassa Ward with a NLFI. Subjects with neither LASV antigenemia nor anti-LASV IgM were considered to have a NLFI.
The LF group had a higher mortality at 45% (p=0.04) than either the subjects with LPE or NLFI (CFR in both groups: 17%; Table 1). Overall, the mortality in these cohorts was lower than the overall CFR of subjects examined in the authors' previous study that included consented subjects with known outcome presenting from 2008 to 2012 (41). The CRF in the comparable groups in that study were 69% for acute LF, 29% for LPE, and 36% for NLFI. The lower CFR in the current study is likely due in part to the lack of sufficient serum volume for the study, which biased the study to nonfatal cases compared to the overall patient cohorts.
Subjects from each of the three groups presented with similar symptoms and range of morbidity. The most frequent symptoms for all groups were fever, weakness, headache, vomiting, and coughing (Table 1). Values of clinical chemistries were also similar for the three subject groups (Table 2). LF patients had elevated levels of AST and ALT, enzymes found in the liver, cardiac and skeletal muscle, kidneys, brain, and red blood cells that can be an indicator of organ dysfunction or trauma, but the difference was not statistically significant compared to the elevations in LPE or NLFI groups. Previous studies have found AST and ALT to be significantly elevated in subjects with acute LF compared to other LPE or NLFI groups (12,24). The lack of significance in the current study appears to be due to the small sample sizes. While TP was significantly elevated in NLFI compared to LF or LPE groups, the levels of TP in LF patients was also high. Therefore, this marker has limited diagnostic or prognostic value. These results indicate that LF diagnosis should not be based on clinical symptoms or blood chemistries alone, in order not to be confounded with NLFI, which can present with similar symptoms. The use of immunodiagnostic tests provides a clearer method of differentiating LF patients from non-LF patients.
Table 2.
Clinical Chemistry Responses for Suspected Lassa Patients
| Clinical chemistry | LF (n=29)a | LPE (n=24)b | NLFI (n=24)c | Total (n=77) | p-Valued |
|---|---|---|---|---|---|
| Sodium | 137 (6) | 131 (11) | 133 (9) | 133 (26) | 0.91 |
| Potassium | 5 (6) | 5 (10) | 6 (9) | 5 (25) | 0.78 |
| tCO2 | 22 (6) | 16 (11) | 18 (9) | 18 (26) | 0.44 |
| Cl− | 92 (7) | 98 (11) | 95 (9) | 95 (27) | 0.31 |
| Glucose | 5 (7) | 3 (12) | 4 (9) | 4 (28) | 0.37 |
| Calcium | 2 (7) | 2 (11) | 2 (8) | 2 (26) | 0.50 |
| BUN | 8 (7) | 9 (12) | 6 (9) | 8 (28) | 0.58 |
| CRE | 71 (6) | 132 (12) | 100 (9) | 107 (27) | 0.59 |
| ALP | 82 (7) | 243 (12) | 147 (9) | 172 (28) | 0.25 |
| ALT | 103 (7) | 132 (12) | 75 (9) | 106 (28) | 0.67 |
| AST | 402 (7) | 156 (10) | 79 (9) | 195 (26) | 0.74 |
| TBIL | 31 (7) | 15 (11) | 19 (9) | 20 (27) | 0.77 |
| ALB | 30 (7) | 24 (12) | 31 (9) | 28 (28) | 0.19 |
| TP | 62 (7) | 60 (12) | 82 (9) | 68 (28) | 0.04* |
Values are expressed as mean (sample size).
LF defined as PCR+(or Ag+when PCR data were unavailable).
LPE defined as PCR–IgM+(or Ag–IgM+when PCR data were unavailable).
NLFI defined as PCR–IgM– (or Ag–IgM– when PCR data were unavailable).
p-Value for Kruskal–Wallis one-way analysis of variance (ANOVA) test for comparing mean values among the three comparison groups.
p<0.05.
tCO2, bicarbonate; Cl−, chloride; BUN, blood urea nitrogen; CRE, creatinine; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate transaminase; TBIL, total bilirubin; ALB, albumin; TP, total protein.
Serum cytokine levels in acute LF patients, subjects with prior exposure to LASV, and patients with NLFIs
A number of studies have shown that the levels of various cytokines and chemokines are altered during LASV infection or exposure in both humans and nonhuman primates (24,26,32,33). To explore the possibility that cytokine alterations might have diagnostic potential in distinguishing acute LF patients from subjects with prior exposure to LASV or patients with NLFIs, selected cytokines were measured using commercial assay reagents. Results were further stratified into those patients who had fatal or non-fatal outcomes. Certain cytokines showed different levels in acute LF, LPE, and NLFI (Table 3). Interferon gamma (IFN-γ) was significantly elevated in fatal LF cases of LF compared to fatal cases of LPE and NLFI. IL-1beta (IL-1β) was significantly elevated in subjects with prior LASV exposure who died compared to acute LF patients or subjects with NLFI.
Table 3.
Cytokine Laboratory Responses for Suspected Lassa Patients
| Cytokine | LF (n=25)a | LPE (n=19)b | NLFI (n=9)c | p-Valued |
|---|---|---|---|---|
| IFN-gamma | 871.16 (0.00, 2007.24) | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.0005* |
| Fatal | 1181.59 (625.21, 2547.12) | 0.00 (0.00, 1763.33) | 0.00 (0.00, 0.00) | 0.0454* |
| Nonfatal | 0.00 (0.00, 1036.89) | 0.00 (0.00, 0.00) | 0.00 (0.00, 0.00) | 0.1409 |
| IL-10 | 68.41 (0.00, 385.12) | 19.82 (7.56, 87.55) | 14.01 (0.00, 91.13) | 0.7542 |
| Fatal | 151.56 (0.00, 1232.97) | 484.85 (70.14, 1838.94) | 244.06 (91.13, 396.99) | 0.7233 |
| Nonfatal | 58.08 (0.00, 383.35) | 18.14 (0.00, 34.94) | 0.00 (0.00, 22.68) | 0.5492 |
| IL-8 | 663.00 (270.49, 1399.42) | 501.95 (125.81, 1624.92) | 133.39 (32.62, 282.90) | 0.1055 |
| Fatal | 1397.16 (821.32, 1750.38) | 1339.71 (704.54, 2073.66) | 270.02 (119.11, 420.92) | 0.1394 |
| Nonfatal | 353.29 (101.57, 522.17) | 414.65 (124.91, 1624.92) | 133.39 (16.38, 282.90) | 0.3349 |
| IL-6 | 281.01 (20.29, 928.19) | 40.56 (0.00, 1213.77) | 0.00 (0.00, 0.00) | 0.0523 |
| Fatal | 938.84 (717.56, 1174.01) | 1579.19 (646.83, 2989.08) | 2027.68 (0.00, 4055.36) | 0.7255 |
| Nonfatal | 20.29 (0.00, 108.90) | 38.40 (0.00, 102.90) | 0.00 (0.00, 0.00) | 0.1144 |
| IL-1beta | 0.00 (0.00, 0.00) | 0.00 (0.00, 103.05) | 0.00 (0.00, 0.00) | 0.0133* |
| Fatal | 0.00 (0.00, 0.00) | 70.63 (0.00, 436.28) | 0.00 (0.00, 0.00) | 0.2466 |
| Nonfatal | 0.00 (0.00, 0.00) | 0.00 (0.00, 58.79) | 0.00 (0.00, 0.00) | 0.0235* |
Values are expressed as median (interquartile range).
LF defined as PCR+(or Ag+when PCR data were unavailable).
LPE defined as PCR–IgM+(or Ag–IgM+when PCR data were unavailable).
NLFI defined as PCR–IgM– (or Ag–IgM– when PCR data were unavailable).
p-Value for Kruskal–Wallis one-way ANOVA test for comparing median values among the three comparison groups.
p<0.05.
Presence of potential febrile disease agents in subjects with acute LF, prior LASV exposure, and NLFIs
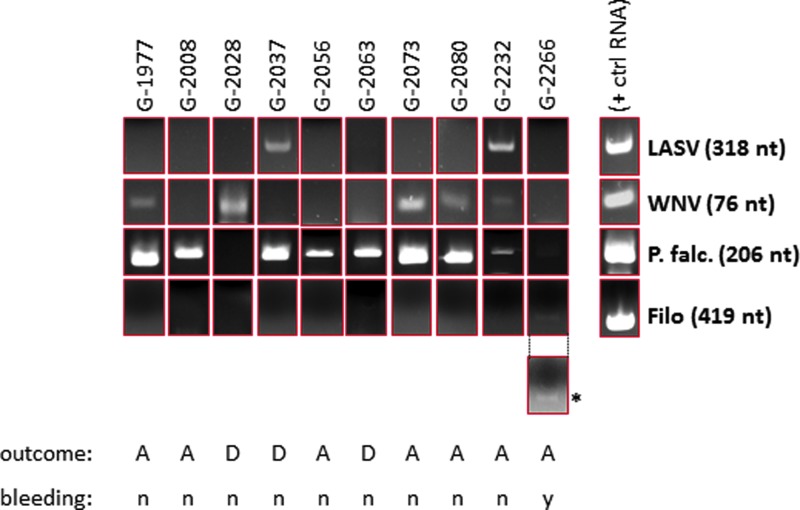
Forty-one sera were screened by RT-PCR and conventional PCR using a panel of oligonucleotides designed for specific detection of amplicons from 13 febrile agents. Nine sera tested positive for LASV (9/41, 22%), six were WNV positive (6/41, 15%), and 32 were P. falciparum positive (32/41, 78%). A single patient sample was positive for a 419 nt amplicon corresponding to a conserved filoviral L gene segment (1/41, 2.0%; Table 4 and Fig. 1). Five samples tested positive for multiple infectious agents (5/41, 12%), with three double positives for P. falciparum and WNV (3/41, 7%), one double positive for P. falciparum and LASV (1/41, 2%), and one triple positive for LASV, WNV, and P. falciparum (1/49, 2%; Table 4 and Fig. 1). The study did not identify P. vivax positive malaria (0/41, 0%), although at least one patient with this parasite has been identified over the time period studied (unpublished results). This study did not identify nucleic acids in the serum of FUO patients corresponding to YFV, DENV, CHIKV, CCHF, RFV, S. typhi STY1599, Leptospira int & RFA, R. prowasekii, and B. recurrentis amplicons.
Table 4.
Positivity Rates for Suspected Lassa Cases by Three Lassa Classifications for Selected Competing Illnesses Assessed by PCR
| Competing illnesses | LF (n=10)a | LPE (n=18)b | NLFI (n=13)c | Total (n=41) | p-Valued |
|---|---|---|---|---|---|
| P. falc. malaria | 9/10 (90) | 13/18 (72) | 10/13 (77) | 32/41 (78) | 0.312 |
| P. vivax malaria | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| Filovirus | 0/10 (0) | 1/18 (6) | 0/13 (0) | 1/41 (2) | 0.641 |
| WNV | 2/10 (20) | 2/18 (11) | 2/13 (15) | 6/41 (15) | 0.803 |
| YFV | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| DENV | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| CHIKV | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| RVF | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| CCHF | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| S. typhi STY1599 | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| Lepto. Int. & RFA | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| R. prowasekii | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
| B. recurrentis | 0/10 (0) | 0/18 (0) | 0/13 (0) | 0/41 (0) | — |
Values are expressed as mean (sample size).
LF defined as PCR+(or Ag+when PCR data were unavailable).
LPE defined as PCR–IgM+(or Ag–IgM+when PCR data were unavailable).
NLFI defined as PCR–IgM– (or Ag–IgM– when PCR data were unavailable).
p-Value for Fisher's exact test for testing for association among the three comparison groups.
—, insufficient data for calculating Fisher's exact test statistic.
WNV, West Nile virus; YFV, yellow fever virus; DENV, dengue virus; CHIKV, chikungunya virus; RVF, Rift Valley fever virus; CCHF, Crimean-Congo hemorrhagic fever virus.
FIG. 1.
Representative polymerase chain reaction (PCR) panel showing single, dual, and triple infections identified by PCR screening in a subset of FUO patients. A subset of patients in the study tested positive by PCR for non-Lassa febrile illnesses (NLFIs), such as Plasmodium falciparum (G-2008, G-2056, G-2063), P. falciparum and West Nile virus (WNV; G-1977, G-2073, G-2080), or WNV alone (G-2028). One patient tested positive for Lassa virus (LASV) and P. falciparum (G-2037), and another was triple positive for LASV, WNV, and P. falciparum (G-2232). One patient in the study had a very low level of P. falciparum antigen and tested positive in a filoviral reverse transcription (RT)-PCR screen (G-2266). Positive control RNAs were generated by runoff in vitro transcription reactions from a T7 promoter immediately upstream of the corresponding amplicon. The positive control P. falciparum DNA was amplified from a cloned 206 nt 18S RNA gene fragment that generated a distinct amplicon from a P. vivax 18S RNA cDNA. Each subset of PCR reactions included a negative control RNA or DNA isolated from normal, negative serum of United States and Sierra Leone origin, as well as master PCR mixes containing oligonucleotides but without input nucleic acids. For clarity, the panel displaying the single filoviral amplicon (*) was brightness (+40%) and contrast (+40%) enhanced. The original image is shown above the enhanced panel. All other panels are shown as originally captured. The outcome (A=alive; D=deceased) and bleeding (n=no bleeding; y=bleeding) status for each patient is shown below each set of panels. Ten representative PCR screening profiles are shown. A complete profile of the PCR screening is displayed in Supplementary Table S3. FUO, fever of unknown origin. Color images available online at www.liebertpub.com/vim
Prior exposure of subjects with acute LF, prior LASV exposure, and NLFIs to additional potential febrile agents
Non-LF tropical disease ELISA kits were tested on the serum panel according to the manufacturer's instructions. IgM and IgG reactivities were recorded for LF, LPE, and NLFI patients against DENV, CHIKV, WNV, and Salmonella typhus antigens, and IgM against Leptospira without statistical significance between the classification groups (Table 5). A significant proportion of the subjects tested IgM and/or IgG positive for more than one agent (Supplementary Table S3). Only three patients in the data set did not register an IgM response to at least one agent. None of the six patient sera that tested positive in the WNV PCR scored a positive response in both the WNV IgM and IgG ELISA, suggesting active WNV infections that had not seroconverted at the time of presentation (Supplementary Table S3).
Table 5.
Positivity Rates for Suspected Lassa Cases by Three Lassa Classifications for Selected Competing Illnesses
| Seroreactivity | LF (n=29)a | LPE (n=24)b | NLFI (n=24)c | Total (n=77) | p-Valued |
|---|---|---|---|---|---|
| LASV IgG | 7/29 (24) | 12/24 (50) | 11/24 (46) | 30/77 (39) | 0.11 |
| DENV IgG | 10/29 (34) | 12/24 (50) | 13/24 (54) | 35/77 (45) | 0.35 |
| DENV IgM | 5/29 (17) | 7/24 (29) | 8/24 (33) | 20/77 (26) | 0.40 |
| CHIK IgG | 7/29 (24) | 7/24 (29) | 7/24 (29) | 21/77 (27) | 0.90 |
| CHIK IgM | 11/29 (38) | 7/24 (29) | 9/24 (38) | 27/77 (35) | 0.83 |
| Lepto IgM | 9/29 (31) | 12/24 (50) | 8/24 (33) | 29/77 (38) | 0.39 |
| WNV IgG | 10/25 (40) | 13/21 (62) | 13/21 (62) | 36/67 (54) | 0.26 |
| WNV IgM | 8/25 (32) | 5/21 (24) | 3/21 (14) | 16/67 (24) | 0.38 |
| Typhus IgG | 4/25 (16) | 5/20 (25) | 3/21 (14) | 12/66 (18) | 0.66 |
| Typhus IgM | 9/25 (36) | 7/20 (35) | 3/21 (14) | 19/66 (29) | 0.22 |
Values are expressed as mean (sample size).
LF defined as PCR+(or Ag+when PCR data were unavailable).
LPE defined as PCR–IgM+(or Ag–IgM+when PCR data were unavailable).
NLFI defined as PCR–IgM– (or Ag–IgM– when PCR data were unavailable).
p-Value for Fisher's exact test for testing for association among the three comparison groups.
LASV IgG seropositive rates were higher for LPE and NLFI (50% and 46% respectively) compared to LF (24%), suggesting these patients had prior exposure to LASV. Acute LF patients also exhibited positive IgM and IgG responses to DENV (17% and 34% respectively), CHIKV (38% and 24%), Leptospira (31%, IgM only), WNV (32% and 40%), and typhus (36% and 16%). The prevalence of antibody responses against these other agents reflects the co-circulation of these pathogens and the potential for co-infection. Despite the relatively small sample size, these results point to significant endemicity rates for these competing illnesses. However, possible cross-reactivity within arbovirus or flavivirus assays may contribute to the results reported in these studies. For example, CHIKV positivity may reflect a prior infection with O'nyong'nyong virus (ONNV), a related but separate alphavirus.
Potential exposure of subjects with NLFIs to a filovirus
Phylogenic analysis of the EBOV isolates that have emerged in the West African countries of Guinea, Sierra Leone, and Liberia suggest the EBOV has been in the Guinean rain forest for several decades (1). To determine the possibility that human exposures to filoviruses may have occurred prior to the current outbreak, a serosurvey was performed on samples collected between June 2011 and March 2014 (n=242). Significant IgG seroreactivity was noted in samples collected in the years 2012–2014. A conservative cutoff O.D.450=0.360, established on the 80th percentile obtained with a panel of normal sera from SL donors (n=14), which intersected with three times the mean O.D. generated by a U.S. normal sera panel (n=13; Figs. 2 and 3A) resulted in a seropositivity rate of 22% (53/242; Fig. 3B). Only three sera from the data set reported in Figures 2 and 3 (patient IDs 2008, 2126, 2281, Supplementary Tables S2 and S3, 2012 FUO study) were also analyzed in the retrospective EBOV IgG study. Serum from the sample that tested positive in the filovirus-specific PCR in January 2012 (G-2266) was not available for testing in the EBOV IgG/IgM ELISA, preventing further characterization of this potential early case of EBOV infection without acute manifestation of EVD. The remaining serum volume was also not sufficient to permit reamplification of filoviral sequences, for cloning of selected amplicons, or for deep sequencing analysis. Patient G-2266 was a 26-year-old Sierra Leonean female who presented to the KGH LFW from another KGH ward as a suspected LF case. She presented with fever, weakness, dizziness, retrosternal pain, and bleeding at the time of presentation to the KGH LFW on January 17, 2012. Following a diagnosis of a NLFI, the patient was not admitted or followed as a suspected VHF case. The patient was LASV antigen-negative by LFI, ELISA, and PCR, and negative in P. falciparum and WNV PCR screens. She registered positive titers for LASV IgM, DENV IgG, CHIKV IgM, WNV IgM and IgG, and typhus IgM. Her serum cytokine profile was unremarkable, registering low levels of IL-10 and IL-8, at 20 pg/mL and 126 pg/mL respectively. A subset of samples collected in early 2014, prior to the identification of the EVD outbreak in Sierra Leone, was further characterized for IgM and IgG reactivity to EBOV VP40 and GP antigens independently (Supplementary Fig. S1). Generally, a correlation was not observed between IgM and IgG reactivity on both VP40 and GP antigens on the subset of sera tested. Some patients registered significant IgM reactivity against both antigens (G-3625), without corresponding measurable IgG titers (Supplementary Fig. S1A and B). Patient G-3637 registered a significant IgM titer to VP40 but not IgG, and an IgG titer to GP but not IgM. Patient G-3640 only registered an IgG titer to VP40. A series of sequential serum samples was available from some patients, drawn throughout their stay at the KGH LFW. Analysis of IgM and IgG reactivities from a given patient over time can provide valuable information on seroconversion to a given infectious agent. Four sequential serum samples were analyzed from patient G-3642, showing consistent VP40 and GP IgG throughout the testing time frame (Supplementary Fig. S1A and B). Overall, the sample size did not permit extrapolation of significant seroconversion trends, but it provided valuable insights for future studies.
FIG. 2.
Normal quantile plot of the distribution of optical density (OD) 450 nm values in Ebola virus (EBOV) immunoglobulin G (IgG) enzyme-linked immunosorbent assay (ELISA) with sera from patients with FUO collected from 2011 to March 2014 (n=242), and a set of Sierra Leone (n=14) and United States normal (n=13). Color images available online at www.liebertpub.com/vim
FIG. 3.
Distribution of OD 450 nm values in EBOV IgG combo ELISA with the EBOV GP-specific KZ52 control monoclonal antibody over a titration range (blue bars), a panel of 14 normal Sierra Leone donors (Kenema Government Hospital [KGH] LF team members, green bars), and 13 normal United States donors (brown bars). (A) Conservative cutoff OD 450 nm=0.360, established on the 80th percentile of normal sera from SL donors, which intersected three times with the mean OD generated by a United States normal sera panel, was used for analysis of the data in this study. (B) ReEBOV IgG ELISA screening of a panel of LF, NLFI, LPE, and FUO patients presenting to the KGH Lassa Ward between 2011 and March 2014 (n=242). The plot displays mean raw data obtained over a titration range with EBOV GP-specific KZ52 antibody (green bars), a negative control serum (orange bar), and a 1:100 dilution of each patient serum (blue bars). The cutoff OD 450 nm=0.360 results in a 20% positive rate for samples registering IgG antibodies specific to EBOV proteins (GP and VP40). A significant correlation between increasing IgG titers and temporal presentation of subjects at the KGH Lassa Ward could not be established (R2=0.0021; data not shown). A temporal distribution of sera analyzed, from mid-September 2011 to late April 2014, and corresponding dry and rainy seasons, and average temperatures (in °C) are graphically represented. In Sierra Leone, dry and rainy seasons correlate with specific agricultural activities: The main planting season is April–July, with harvesting occurring between September and January. Agricultural activities increase the contact between humans and animal species in fields and wooded areas. These studies did not identify an obvious correlation between seasons and the emergence of seroreactivity against EBOV antigens. Color images available online at www.liebertpub.com/vim
Discussion
All subjects included in the study met the case definition for LF. The patients were generally severely ill, irrespective of whether they were diagnosed with LF, accounting for the CFRs of 17% observed in both the LPE groups (Ag–/IgM+) and the NLFI (Ag–/IgM–) groups. Signs and symptoms alone were not sufficient to provide a clinical diagnosis of VHF. There were also no differences in clinical chemistry measurements that could be used to distinguish acute LF patients easily from other groups. Elevations of AST in LF and TP in LPE are markers that may offer clinical guidance, but they are not diagnostic. Certain cytokines showed different levels in acute LF, LPE, and NLFI. IFN-γ was significantly elevated in fatal cases of LF compared to fatal cases of LPE and NLFI. Focused research on cytokine levels in LF and other VHFs is likely to suggest possible treatment strategies. However, routine cytokine analysis is not feasible in most LF treatment centers, and the broad applicability of these findings for VHF diagnostic purposes is limited. Overall, the studies suggest that only specific diagnostic assays rather than clinical observation or other laboratory measurement can distinguish LF from other cause serious febrile illnesses in this region.
The current studies provide insight into some pathogens that may be responsible for causing NLFI in patients that present to the KGH Lassa Ward. Despite antimalarial treatment, it was found that 78% of subjects presenting with evidence of LPE or NLFI had detectable levels of P. falciparum DNA by PCR. In the LF patient group, 90% of LF patients also had PCR-detectable levels of P. falciparum in their blood. Nearly all patients were unresponsive to antimalarial therapy, and remained ill. Drug-resistant strains of malaria parasites have been circulating in West Africa, and there are concerns about the self-prescribed use of expired or inactive antimalarials in the region. Further studies of the parasite burden in LF and other febrile illness patients are needed. More studies are needed to determine whether the parasite burdens detected by PCR may be sufficient to induce VHF-like signs and symptoms that precipitated the subjects to seek medical attention.
Overall, 15% of the subjects presenting to the KGH Lassa Ward had detectable RNA in their blood that amplified with WNV-specific primers. Other studies are required to determine whether the RNA detected is specific for WNV or is a related flavivirus, and to confirm whether this important pathogen is responsible for any of the febrile disease burdens in this region of Sierra Leone. Acute infections with YFV were not detected in the cohort tested, although YFV is known to be present in Sierra Leone. A YFV vaccination program has not been implemented in the subregion, although proof of YFV vaccination is required prior to entry by all foreigners into Sierra Leone. Two cases of YFV since 2008 have been detected among patients presenting to the Lassa Ward at the KGH (unpublished data). PCR screening using oligonucleotides specific for DENV, CHIKV, RVF, CCHF, S. typhi STY1599, Lepto. Int. & RFA, R. prowasekii, and B. recurrentis did not reveal any of these or related pathogens in the panel of sera tested. Excluding the possibility that these agents are significant causes of the febrile illnesses in subjects presenting to the VHF Ward at the KGH will require the examination of larger cohorts. Moreover, it may be more appropriate to analyze other sample types. For instance, in some cases, such as S. typhi, it may be more appropriate to analyze stool samples rather than blood. PCR detects nucleic acids in patients with active infections. By contrast, serological studies detect recent to past exposures accumulated over many years. Therefore, a serological study was performed of the exposure to flaviviruses or arboviruses in the febrile illness subjects presenting to the KGH. A high level of exposures was detected in these cohorts to DENV, WNV, CHIKV, leptospirosis, and typhus. More than half of the subjects tested had anti-WNV IgG, while approximately one-fourth of the subjects tested positive for anti-WNV IgM (Supplementary Table S3). A recent study by Schoepp et al. (40) also detected the serological response to several of these same pathogens using in-house developed assays (USAMRIID), albeit at lower levels than in the authors' studies using commercially available serological assays. Cross-reactive antibodies may be present in this serosurvey. For example, CHIKV positivity may reflect a prior infection by ONNV, a related but separate alphavirus. A rapid and accurate diagnosis of flaviviruses is notoriously difficult due to the significant cross-reactivity between members of the family, namely YFV, WNV, and DENV.
One of samples tested in the present study generated a weak PCR product for filovirus RNA. This finding is significant because the patient presented to the KGH LFW in late 2011, more than 2 years before the recent EVD outbreak was detected. A substantial portion of NLFI patients (between 6.8% and 22%, depending on the stringency of immune assay cutoff applied) presenting to the Lassa Ward and tested in this study had IgG that reacted with EBOV proteins. Prior studies have suggested that exposure to filoviruses or filovirus antigens may be relatively common in some communities in Central Africa. For example, Becquart et al. found that the prevalence of EBOV-specific IgG was 15.3% overall, increasing to 32.4% (p<0.001) in forest areas (6). No sociodemographic risk factors were found, but the antibody prevalence increased linearly up to 20 years of age. Becker et al. (5) found that 43.3% of the monkey sera and 6.9% of the human sera collected from various sites in Germany reacted positively with at least one of the three different filovirus antigens. The prevalence of EBOV antibodies in five gold-panning villages situated in northeastern Gabon was 10% (7). Other studies found a lower seroprevalence of 1% in villages in Gabon outside of recognized outbreak areas. In this latter study, only IgG antibodies were found, and no IgM antibodies were detected.
Furthermore, Schoepp et al. (40) reported that the prevalence of IgM reactivity to EBOV and MBGV was 8.6% and 3.6% respectively. Surprisingly, no evidence of IgG reactivity to filovirus proteins was noted in those studies. The samples analyzed by Schoepp et al. were collected between October 2006 and October 2008, while all of the samples analyzed in the current study were collected after mid-2011. We observed a higher level of background reactivity in our filovirus assay in older samples, potentially due to temperature fluctuations during storage that result in possible sample degradation, which may cause assay background. It is noteworthy that the samples tested by Schoepp et al. were provided by the VHFC, and thus consisted of banked samples that were collected over a 2 year period and subjected to similar storage conditions. Based on phylogenetic analyses, EBOV emergence and exposure to Sierra Leonean populations may be a relatively recent phenomenon, perhaps only a few decades old (1).
The results of the current studies support the possibility that filovirus infections occurred in Sierra Leone prior to the 2014 outbreak. An individual with EVD presenting to a health facility in this region would have a high likelihood of being diagnosed with another disease, such as malaria, on clinical grounds alone. Rare infections with EBOV could escape detection by organized health surveillance systems that are in place because symptoms are similar to diseases endemic to the region. If such cases had presented to the KGH LF program and been admitted on clinical grounds, routine testing for LF would not have revealed a filovirus infection. No outbreaks of EBOV have been detected in the Eastern Province of Sierra Leone in the recent decade despite an active surveillance program for LF, a VHF that presents with indistinguishable case presentations. Milder or asymptomatic infections with EBOV or cross-reactive but less virulent or avirulent filoviruses could account for the seroprevalences observed in this and other studies. Fruit bats and other forest animals are considered to be reservoirs for EBOV. People living in the area may be exposed to degraded or otherwise inactivated EBOV proteins by consumption of the animals or foods that have been contaminated with saliva from bats or other animals. Repeated exposure to EBOV antigens without overt infection could account for the seroprevalence recorded here.
The Manu River region of West Africa could now be endemic for two of the world's deadliest viruses. Thus, future laboratory evaluations of people presenting with signs and symptoms of VHF must receive a differential diagnosis of LF and EVD. There is no drug treatment for EVD, but supportive care can improve survival. While there is no FDA-approved treatment for LF, ribavirin can reduce mortality in acute infections, but only if used early in infection (25,27,43). Further studies of NLFIs other than LF and EVD in this population are also of interest, but a more extensive characterization than that reported here was not possible due to resource and infrastructure constraints during this project period. Distinguishing between EVD and LF infections will be a continuous challenge in West Africa going forward as the two diseases require distinct management. The EBOV-specific IgG-capture assays used in this study appear to be sensitive, specific, and robust, but more work will be needed to validate the assays fully with derivation of commercially acceptable levels of sensitivity and specificity. This will require further authentication of the assays using serum samples from EVD patients and comparison to other methods, such as IgM capture and virus neutralization. The development of a multiplexed LFI with the ability to differentiate between LASV and the filoviruses is under way, providing a panel approach for rapidly distinguishing among African hemorrhagic virus infections that present with similar symptoms.
Future studies by the VHFC to define the viruses present in febrile illness patients will make use of high-throughput technologies such as next-generation sequencing (NGS). NGS has already been used successfully as both a diagnostic tool and a means to discover novel viruses associated with human diseases (2,8,15,23,44). Applying NGS as a diagnostic tool holds the promise for a relatively unbiased view of the microbes infecting humans, as it does not require a priori knowledge of the pathogens present. It also has the potential to elucidate the spectrum of disease-causing viruses in patients with undiagnosed febrile illness, which is a common occurrence in health clinics around the world. NGS can also serve to increase the power of surveillance systems to detect infrequent zoonotic transmissions that have the potential to become pandemics. Epidemiological studies are ongoing with the aim of identifying factors that converged and potentiated the transmission and rapid dissemination of EVD throughout the Western African subregion in 2014. A potential index case for the current EVD outbreak may have been identified in Guinea (1). A recent report suggested that ecological factors may have contributed to the convergence of factors that culminated in the ongoing EVD outbreak in the Manu River region (3). Anecdotal reports of an exceptionally dry and prolonged dry season, starting with sharply drier than normal conditions at the end of the rainy season, may have had an impact in triggering the transmission of EBOV from its natural host to humans. At least one conclusion may be derived from the current situation: it is clear that geographic expansion and increased human-to-human transmission of one of the most feared infectious agents known to science is underway.
Supplementary Material
Contributor Information
Collaborators: the Viral Hemorrhagic Fever Consortium
Acknowledgments
Department of Health and Human Services NIH/NIAID Challenge and Partnership Grant Numbers AI067188 and AI082119 provided funding for the development and clinical study guidelines for these immunodiagnostics. The authors thank all the members of the VHFC for their continued support (www.vhfc.org).
Note Added in Proofs
Tragically, two co-authors—Mohamed Fullah and Sheik Humarr Khan—who contributed greatly to public health and VHF research efforts in Sierra Leone, contracted EVD and lost their battle with the disease before this manuscript could be published. A third author—Sidiki Safa—also succumbed to a non-EVD-related illness. We wish to honor their memory.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Baize S, Pannetier D, Oestereich L, et al. . Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 2014;371:1418–1425 [DOI] [PubMed] [Google Scholar]
- 2.Barzon L, Lavezzo E, Militello V, et al. . Applications of next-generation sequencing technologies to diagnostic virology. Int J Mol Sci 2011;12:7861–7884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bausch DG, and Schwarz L. Outbreak of Ebola virus disease in Guinea: where ecology meets economy. PLoS Negl Trop Dis 2014;8:e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bausch DG, Sesay SS, and Oshin B. On the front lines of Lassa fever. Emerg Infect Dis 2004;10:1889–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker S, Feldmann H, Will C, et al. . Evidence for occurrence of filovirus antibodies in humans and imported monkeys: do subclinical filovirus infections occur worldwide? Med Microbiol Immunol 1992;181:43–55 [DOI] [PubMed] [Google Scholar]
- 6.Becquart P, Wauquier N, Mahlakoiv T, et al. . High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLoS One 2010;5:e9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertherat E, Renaut A, Nabias R, et al. . Leptospirosis and Ebola virus infection in five gold-panning villages in northeastern Gabon. Am J Trop Med Hyg 1999;60:610–615 [DOI] [PubMed] [Google Scholar]
- 8.Bodewes R, Kik MJ, Raj VS, et al. . Detection of novel divergent arenaviruses in boid snakes with inclusion body disease in the Netherlands. J Gen Virol 2013;94:1206–1210 [DOI] [PubMed] [Google Scholar]
- 9.Bond N, Schieffelin JS, Moses LM, et al. . A historical look at the first reported cases of Lassa fever: IgG antibodies 40 years after acute infection. Am J Trop Med Hyg 2012;88:241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bornholdt ZA, Noda T, Abelson DM, et al. . Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell 2013;154:763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Branco LM, Boisen ML, Andersen KG, et al. . Lassa hemorrhagic fever in a late term pregnancy from northern Sierra Leone with a positive maternal outcome: case report. Virol J 2011;8:404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branco LM, Grove JN, Boisen ML, et al. . Emerging trends in Lassa fever: redefining the role of immunoglobulin M and inflammation in diagnosing acute infection. Virol J 2011;8:478., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branco LM, Matschiner A, Fair JN, et al. . Bacterial-based systems for expression and purification of recombinant Lassa virus proteins of immunological relevance. Virol J 2008;5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckley SM, Casals J. Lassa fever, a new virus disease of man from West Africa. 3. Isolation and characterization of the virus. Am J Trop Med Hyg 1970;19:680–691 [DOI] [PubMed] [Google Scholar]
- 15.Capobianchi MR, Giombine E, and Rozera G. Next-generation sequencing technology in clinical virology. Clin Microbiol Infect 2013;19:15–22 [DOI] [PubMed] [Google Scholar]
- 16.Carletti F, Bordi L, Chiappini R, et al. . Rapid detection and quantification of Chikungunya virus by a one-step reverse transcription polymerase chain reaction real-time assay. Am J Trop Med Hyg 2007;77:521–524 [PubMed] [Google Scholar]
- 17.da Silva NS, da Silva-Nunes M, Malafronte RS, et al. . Epidemiology and control of frontier malaria in Brazil: lessons from community-based studies in rural Amazonia. Trans R Soc Trop Med Hyg 2010;104:343–350 [DOI] [PubMed] [Google Scholar]
- 18.Demby AH, Chamberlain J, Brown DW, et al. . Early diagnosis of Lassa fever by reverse transcription-PCR. J Clin Microbiol 1994;32:2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon MG, and Schafer IJ. Ebola viral disease outbreak—West Africa, 2014. MMWR Morb Mortal Wkly Rep 2014;63:548–551 [PMC free article] [PubMed] [Google Scholar]
- 20.Drosten C, Gottig S, Schilling S, et al. . Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol 2002;40:2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldmann H. Ebola—a growing threat? N Engl J Med 2014;371:1375–1378 [DOI] [PubMed] [Google Scholar]
- 22.Formenty P, Boesch C, Wyers M, et al. . Ebola virus outbreak among wild chimpanzees living in a rain forest of Cote d'Ivoire. J Infect Dis 1999;179:S120–126 [DOI] [PubMed] [Google Scholar]
- 23.Gire SK, Goba A, Andersen KG, et al. . Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014;345:1369–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grove JN, Branco LM, Boisen ML, et al. . Capacity building permitting comprehensive monitoring of a severe case of Lassa hemorrhagic fever in Sierra Leone with a positive outcome: case report. Virol J 2011;8:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunther S, Asper M, Roser C, Luna LK, Drosten C, Becker-Ziaja B, Borowski P, Chen HM, Hosmane RS. Application of real-time PCR for testing antiviral compounds against Lassa virus, SARS coronavirus and Ebola virus in vitro. Antiviral Res 2004;63:209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hensley LE, Smith MA, Geisbert JB, et al. . Pathogenesis of Lassa fever in cynomolgus macaques. Virol J 2011;8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huggins JW. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Rev Infect Dis 1989;11:S750–761 [DOI] [PubMed] [Google Scholar]
- 28.Jiang J, Temenak JJ, and Richards AL. Real-time PCR duplex assay for Rickettsia prowazekii and Borrelia recurrentis. Ann N Y Acad Sci 2003;990:302–310 [DOI] [PubMed] [Google Scholar]
- 29.Keane E, and Gilles HM. Lassa fever in Panguma Hospital, Sierra Leone, 1973–6. Br Med J 1977;1:1399–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan SH, Goba A, Chu M, et al. . New opportunities for field research on the pathogenesis and treatment of Lassa fever. Antiviral Res 2008;78:103–115 [DOI] [PubMed] [Google Scholar]
- 31.Lee JE, Fusco ML, Hessell AJ, et al. . Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008;454:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahanty S, Bausch DG, Thomas RL, et al. . Low levels of interleukin-8 and interferon-inducible protein-10 in serum are associated with fatal infections in acute Lassa fever. J Infect Dis 2001;183:1713–1721 [DOI] [PubMed] [Google Scholar]
- 33.Malhotra S, Yen JY, Honko AN, et al. . Transcriptional profiling of the circulating immune response to lassa virus in an aerosol model of exposure. PLoS Negl Trop Dis 2013;7:e2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick JB, Webb PA, Krebs JW, et al. . A prospective study of the epidemiology and ecology of Lassa fever. J Infect Dis 1987;155:437–444 [DOI] [PubMed] [Google Scholar]
- 35.Monath TP, Maher M, Casals J, et al. . Lassa fever in the Eastern Province of Sierra Leone, 1970–1972. II. Clinical observations and virological studies on selected hospital cases. Am J Trop Med Hyg 1974;23:1140–1149 [DOI] [PubMed] [Google Scholar]
- 36.Mourya DT, Yadav PD, Shete AM, et al. . Detection, isolation and confirmation of Crimean-Congo hemorrhagic fever virus in human, ticks and animals in Ahmadabad, India, 2010–2011. PLoS Negl Trop Dis 2012;6:e1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olschlager S, Lelke M, Emmerich P, et al. . Improved detection of Lassa virus by reverse transcription-PCR targeting the 5′ region of S RNA. J Clin Microbiol 2010;48:2009–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SH, Kim HJ, Cho WH, et al. . Identification of Salmonella enterica subspecies I, Salmonella enterica serovars Typhimurium, Enteritidis and Typhi using multiplex PCR. FEMS Microbiol Lett 2009;301:137–146 [DOI] [PubMed] [Google Scholar]
- 39.Reitstetter RE. Development of species-specific PCR primer sets for the detection of Leptospira. FEMS Microbiol Lett 2006;264:31–39 [DOI] [PubMed] [Google Scholar]
- 40.Schoepp RJ, Rossi CA, Khan SH, et al. . Undiagnosed acute viral febrile illnesses, Sierra Leone. Emerg Infect Dis 2014;20:1176–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaffer JG, Grant DS, Schieffelin JS, et al. . Lassa fever in post-conflict Sierra Leone. PLoS Negl Trop Dis 2014;8:e2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirato K, Miyoshi H, Kariwa H, et al. . Detection of West Nile virus and Japanese encephalitis virus using real-time PCR with a probe common to both viruses. J Virol Methods 2005;126:119–125 [DOI] [PubMed] [Google Scholar]
- 43.Snell NJ. Ribavirin—current status of a broad spectrum antiviral agent. Expert Opin Pharmacother 2001;2:1317–1324 [DOI] [PubMed] [Google Scholar]
- 44.Wang J, Selleck P, Yu M, et al. . Novel phlebovirus with zoonotic potential isolate from ticks, Australia. Emerg Inf Dis 2014;20:1040–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watts GM. Lily Lyman Pinneo. Lancet 2013;380:1552 [Google Scholar]
- 46.WHO: Ebola Response Roadmap Situation Report November 26, 2014. Available at http://apps.who.int/iris/bitstream/10665/144498/1/roadmapsitrep_26Nov2014_eng.pdf?ua=1 (accessed December2, 2014)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.