Abstract
The histone deacetylase inhibitor (HDACi) suberoylanilide hydroxyamic acid (SAHA), also known as vorinostat, has recently been reported to activate latent HIV-1 in patients undergoing antiretroviral therapy. It is possible that SAHA reactivation of latent viruses may involve effects on cellular transcription factors such as positive transcription elongation factor b (P-TEFb), a protein kinase whose core is composed of CDK9 and Cyclin T1. P-TEFb is recruited by the HIV-1 Tat protein to activate productive RNA polymerase II elongation of the integrated provirus. We found that SAHA treatment of isolated resting CD4+ T cells induced CDK9 Thr-186 (T-loop) phosphorylation in six of eight healthy donors and increased Cyclin T1 expression in one donor; Thr-186 phosphorylation is required for P-TEFb function. Disulfiram, another small molecule currently under evaluation in clinical trials for reactivation of latent HIV-1, was also found capable of inducing CDK9 Thr-186 phosphorylation and Cyclin T1 levels in resting CD4+ T cells from healthy donors. In a Jurkat CD4+ T cells HIV-1 latency system, disulfiram reactivated the latent provirus and induced CDK9 Thr-186 phosphorylation. Our findings suggest that small molecules capable of reactivating latent HIV-1 in resting CD4+ T cells may function in part by increasing CDK9 Thr-186 phosphorylation and perhaps Cyclin T1 expression, thereby up-regulating P-TEFb function.
HIV-1 replication is dependent upon efficient transcription of the integrated provirus by RNA polymerase II (RNAP II). Following transcription initiation from the HIV-1 LTR, RNAP II pauses due to the action of two negative elongation factors, NELF and DSIF, which associate with the RNAP II complex and inhibit elongation.1 The HIV-1 transactivator protein, Tat, recruits positive transcription elongation factor b (P-TEFb), a cellular protein kinase complex, to the TAR RNA element at the 5′ end of the nascent viral transcript. The kinase subunit of P-TEFb is CDK9 and the regulatory subunit is either Cyclin T1 or Cyclin T2. Tat binds directly to Cyclin T12 and therefore only targets Cyclin T1-containing P-TEFb. P-TEFb stimulates processive transcriptional elongation by phosphorylating the carboxyl terminal domain (CTD) of RNAP II and specific protein subunits of NELF and DSIF, thereby abrogating their inhibition of elongation. In primary resting CD4+ T cells and monocytes, P-TEFb function is down-regulated due to low expression levels of Cyclin T1 and repression of CDK9 Thr-186 (T-loop) phosphorylation, a critical posttranslational modification required for kinase activity.3–6 In metabolically active cells such as activated CD4+ T cells, the P-TEFb function is partially repressed by sequestering the kinase complex in the 7SK snRNP, which is composed of 7SK snRNA, HEXIM1/2, MEPCE, and LARP7.7,8 Additionally, P-TEFb can be found in a multiprotein complex termed the Super-Elongation Complex that is involved in stimulating transcriptional elongation.8
Combinations of antiretroviral drugs can suppress active viral replication but are ineffective against transcriptionally latent proviruses that are a source of persistent HIV-1 infection, especially in memory resting CD4+ T cells. Therefore, a focus in the field is now on devising strategies to cure HIV-1 infection by flushing out the latent provirus. One such strategy has been termed “shock and kill.” This involves reactivating latent HIV-1 with small molecules and allowing the host immune system and combination antiretroviral therapy to eradicate the virus. Histone deacetylases (HDACs) are in part responsible for maintaining HIV-1 latency.9–11 A recent proof-of-concept study by Archin et al.11 provided evidence that treating HIV-infected patients with the histone deacetylase inhibitor (HDACi) suberoylanilide hydroxyamic acid (SAHA) (vorinostat) increased viral RNA expression in circulating resting CD4+ T cells. The authors concluded that inhibition of HDACs by SAHA was responsible for the increase of viral transcription.11 Vorinostat is currently being evaluated in additional clinical trials for the ability to reactivate latent HIV-1 and reduce the latent reservoir in HIV-infected individuals undergoing antiviral therapy.12
Because P-TEFb is critical for Tat-mediated HIV-1 transcription, we reasoned that SAHA treatment of resting CD4+ T cells was also likely to up-regulate P-TEFb. We isolated resting CD4+ T cells from eight healthy blood donors and treated them with 335 nM SAHA, equivalent to a single 400 mg in vivo dose that activated latent HIV-1 in patients.11 Under these conditions, resting CD4+ T cells populations are 95% pure as determined by flow cytometry.13 Cell lysates were prepared as described earlier6 at different times posttreatment and lysates were examined in immunoblots (Fig. 1). In parallel, resting cells were treated with 5 μg/ml phytohemagglutinin (PHA) as positive control as we have shown PHA induces Cyclin T1 protein expression and CDK9 Thr-186 phosphorylation.6
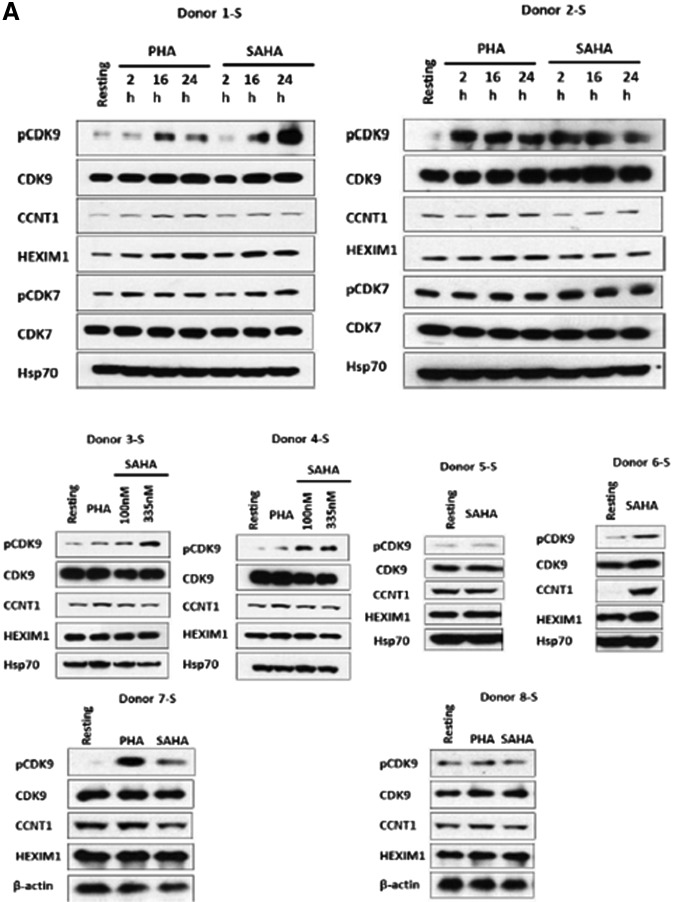
FIG. 1.
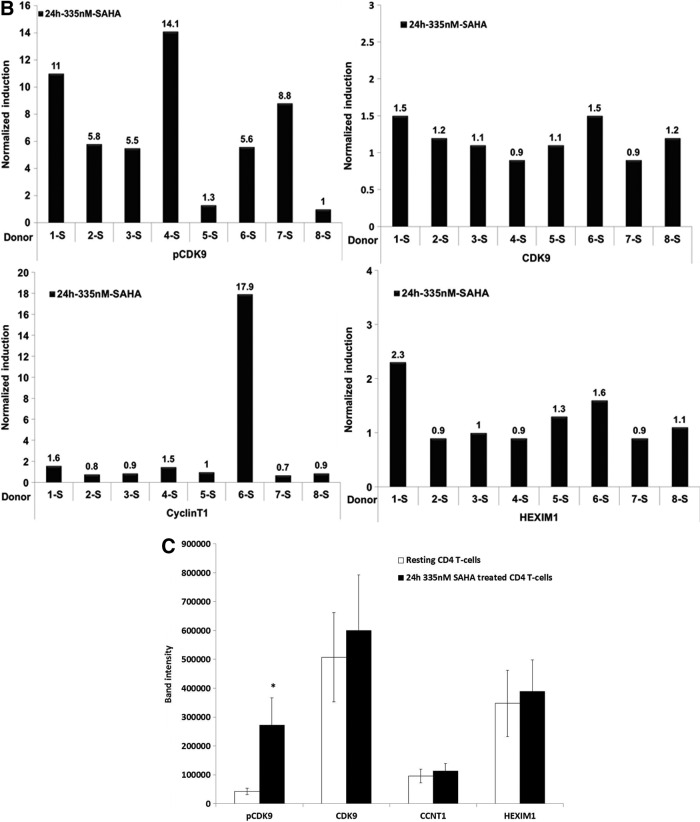
Suberoylanilide hydroxyamic acid (SAHA) induces CDK9 Thr-186 phosphorylation in resting CD4+ T cells. (A) Resting CD4+ T cells isolated by negative selection from peripheral blood of healthy individuals were treated with 335 nM SAHA or 5 μg/ml phytohemagglutinin (PHA) for the indicated times (Donors 1-S and 2-S), 100 and 335 nM SAHA for 24 h (Donors 3-S and 4-S) or 335 nM SAHA for 24 h (Donors 5-S through 8-S). Cell lysates were analyzed for the expression of indicated proteins in immunoblots. (B) Band intensities for immunoblots in (A) were quantified using Image J.23 Band intensities of either Hsp70 or β-actin were used for normalization, with the expression level of indicated proteins in untreated resting cells set at 1.0. (C) Statistical analysis (unpaired t-test) on the average normalized raw band intensities of indicated proteins for immunoblots in (A) at 24 h time point (*p<0.05).
In Donor 1-S, CDK9 Thr-186 phosphorylation was induced 6-fold after 16 h of SAHA treatment and 11-fold at 24 h; this compared to 4- and 3-fold inductions by PHA at these time points (Fig. 1A). In Donor 2-S, both SAHA and PHA induced CDK9 Thr-186 phosphorylation at 2 h. We also examined CDK7 T-loop phosphorylation in Donors 1-S and 2-S as CDK7 has been recently reported to phosphorylate the CDK9 Thr-186.14 A modest induction of CDK7 T-loop phosphorylation by SAHA and PHA was observed in Donor 1-S (Fig. 1A). In Donors 3-S and 4-S, resting cells were treated with 100 nM or 335 nM SAHA for 24 h. In Donor 3-S, a dose-dependent induction of CDK9 Thr-186 phosphorylation was seen, while in Donor 4-S, the induction of CDK9 Thr-186 phosphorylation was similar for both concentrations of SAHA (Fig. 1A). In Donors 5-S through 8-S, we treated resting CD4+ T cells with 335 nM SAHA for 24 h. SAHA treatment induced CDK9 Thr-186 phosphorylation in Donors 6-S and 7-S, but had little effect in Donors 5-S and 8-S (Fig. 1A). Interestingly, in Donor 6-S, SAHA induced Cyclin T1 protein expression 18-fold, while its effects were modest or not apparent for the other seven donors examined (Fig. 1A). We used flow cytometry to examine the effects of SAHA on the T cell activation markers CD25 and CD69 and observed no effect on either marker by SAHA (data not shown). We also performed flow cytometry analysis of propidium iodide-stained cells and observed no effects of SAHA on cellular proliferations (data not shown). Thus, SAHA does not activate resting CD4+ T cells under our experimental conditions.
Quantification of the immunoblots shown in Fig. 1 is presented in Fig. 1B, using either Hsp70 or β-actin as a normalization control. We performed a statistical analysis of the P-TEFb expression levels in the immunoblots for the eight donors. We found that the induction of CDK9 Thr-186 phosphorylation following SAHA treatment was statistically significant. It should be noted that in the Archin study, ex vivo SAHA treatment of isolated resting CD4+ T cells increased viral RNA expression in eight of nine patients examined for whom the basal level of HIV RNA was quantifiable.11 The variation in effects of SAHA on CDK9 Thr-186 phosphorylation and Cyclin T1 expression in our analysis of eight healthy blood donors reiterates the variability in patients' responses seen by Archin et al.11 Additionally, recent studies have shown that a small molecule termed JQ1 has variability in its ability to reactivate latent viruses in primary CD4+ T cells through its effects on P-TEFb and the bromodomain proteins Brd2 and Brd4.15–18
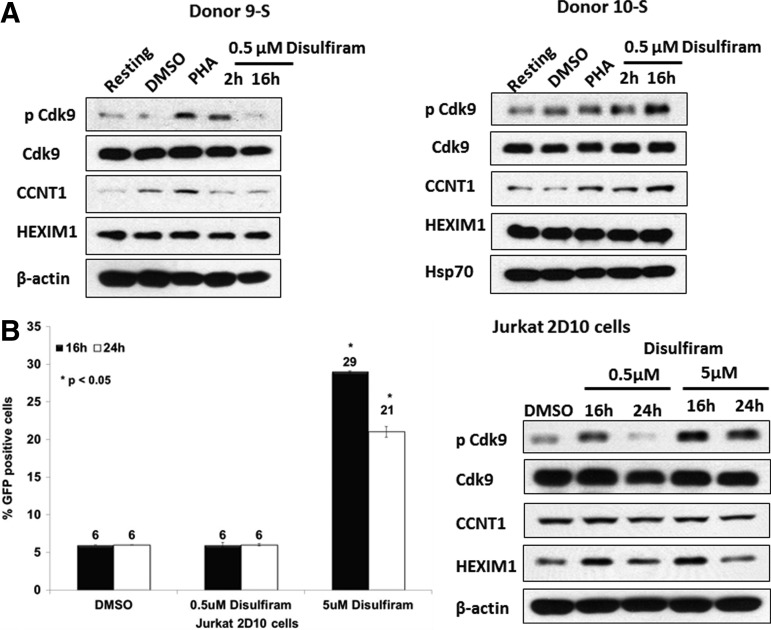
Disulfiram is an aldehyde dehydrogenase inhibitor that is used to treat alcoholism and cocaine dependence.19,20 In a study using a primary CD4+ T cell latency model, disulfiram was reported to reactivate latent HIV-1 without activating T cells.21 Disulfiram is currently being evaluated in clinical trials for the ability to reactivate latent HIV-1 and reduce the latent reservoir in HIV-infected individuals undergoing antiviral therapy.12 As with SAHA, we suspected that the induction of latent viruses in primary cells by disulfiram is likely to involve an up-regulation of P-TEFb. We therefore treated resting CD4+ T cells with an increasing dose of disulfiram for 24 h. In Donor 8-S, disulfiram did not induce either CD69 or CD25 and had no stimulatory effect on cellular proliferation as examined by propidium iodide staining (data not shown). This result is in agreement with the findings of Xing et al.21 that disulfiram does not induce T cell activation. In Donors 9-S and 10-S, we examined the effect of 0.5 μM disulfiram for 2 and 16 h on P-TEFb expression (Fig. 2A). In Donor 9-S, both PHA and disulfiram induced CDK9 Thr-186 phosphorylation at 2 h of treatment. In Donor 10-S, disulfiram treatment of resting CD4+ T cells induced CDK9 Thr-186 phosphorylation. In Donor 10-S, we also saw an induction of Cyclin T1 by disulfiram, but this induction was not seen in Donor 9-S.
FIG. 2.
Disulfiram induces positive transcription elongation factor b (P-TEFb) in primary CD4+ T cells and reactivates latent provirus in the Jurkat 2D10 cell line. (A) Resting CD4+ T cells were treated with disulfiram for the indicated times and cell lysates were examined for the expression of indicated proteins in immunoblots. (B) Duplicate Jurkat 2D10 cultures were treated with disulfiram for the indicated time points. A portion of the cells was analyzed for GFP expression by flow cytometry using BD LSR Fortessa. Cell lysates prepared from the remaining cells were analyzed in immunoblots for the expression of the indicated proteins.
We also investigated the effect of disulfiram on reactivation of latent provirus in the Jurkat 2D10 cell model system.22 Although 0.5 μM disulfiram did not reactivate latent virus, 5 μM disulfiram showed a significant reactivation of latent provirus at 16 and 24 h posttreatment (Fig. 2B). An immunoblot analysis of a portion of the disulfiram-treated cells showed an increase in CDK9 Thr-186 phosphorylation by 5 μM disulfiram, the concentration that reactivated latent virus in this system (Fig. 2B).
In summary, we have shown that SAHA and disulfiram up-regulated CDK9 Thr-186 phosphorylation in resting CD4+ T cells isolated from the majority of healthy donors examined. Additionally, these compounds induced Cyclin T1 protein levels in a minority of donors examined. This variability in response to SAHA was also seen in the Archin study,11 as well as in the studies with JQ1.18 Thus, it is likely that genetic and environmental factors play important roles in individual responses to these small molecules. Based on the data presented here, we propose that small molecules that reactivate latent HIV in resting CD4+ T cells are likely to function in part through an up-regulation of P-TEFb by increasing CDK9 Thr-186 phosphorylation and perhaps Cyclin T1 levels.
Acknowledgments
We thank Jonathan Karn (Case Western Reserve University) for the Jurkat 2D10 cells. This work was supported by NIH Grant AI02483 to A.P.R. We thank Ying-Wooi Wan, Bioinformatics analyst, Department of Obstetrics and Gynecology, Baylor College of Medicine for statistical analysis.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Karn J. and Stoltzfus CM: Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2012;2:a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garber ME, Wei P, KewalRamani VN, Mayall TP, Herrmann CH, Rice AP, Littman DR, and Jones KA: The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev 1998;12:3512–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budhiraja S, Famiglietti M, Bosque A, Planelles V, and Rice AP: Cyclin T1 and CDK9 T-loop phosphorylation are downregulated during establishment of HIV-1 latency in primary resting memory CD4+ T cells. J Virol 2013;87:1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiang K, Sung TL, and Rice AP: Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J Virol 2012;86:3244–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong C, Kwas C, and Wu L: Transcriptional restriction of human immunodeficiency virus type 1 gene expression in undifferentiated primary monocytes. J Virol 2009;83:3518–3527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramakrishnan R, Dow EC, and Rice AP: Characterization of Cdk9 T-loop phosphorylation in resting and activated CD4(+) T lymphocytes. J Leukoc Biol 2009;86:1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dow EC, Liu H, and Rice AP: T-loop phosphorylated Cdk9 localizes to nuclear speckle domains which may serve as sites of active P-TEFb function and exchange between the Brd4 and 7SK/HEXIM1 regulatory complexes. J Cell Physiol 2010;224:84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ott M, Geyer M, and Zhou Q: The control of HIV transcription: Keeping RNA polymerase II on track. Cell Host Microbe 2011;10:426–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Archin NM, Espeseth A, Parker D, Cheema M, Hazuda D, and Margolis DM: Expression of latent HIV induced by the potent HDAC inhibitor suberoylanilide hydroxamic acid. AIDS Res Hum Retroviruses 2009;25:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras X, Schweneker M, Chen CS, McCune JM, Deeks SG, Martin J, and Peterlin BM: Suberoylanilide hydroxamic acid reactivates HIV from latently infected cells. J Biol Chem 2009;284:6782–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, et al. : Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012;487:482–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Lint C, Bouchat S, and Marcello A: HIV-1 transcription and latency: An update. Retrovirology 2013;10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budhiraja S, Ramakrishnan R, and Rice AP: Phosphatase PPM1A negatively regulates P-TEFb function in resting CD4T+ T cells and inhibits HIV-1 gene expression. Retrovirology 2012;9:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, and Fisher RP: Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 2012;19:1108–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, and Montano M: BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol 2012;92:1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehm D, Calvanese V, Dar RD, Xing S, Schroeder S, Martins L, Aull K, Li PC, Planelles V, Bradner JE, et al. : BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle 2013;12:452–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, Guo J, Wu Y, and Zhou Q: The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res 2013;41:277–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu J, Gaiha GD, John SP, Pertel T, Chin CR, Gao G, Qu H, Walker BD, Elledge SJ, and Brass AL: Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep 2012;2:807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faiman MD, Jensen JC, and Lacoursiere RB: Elimination kinetics of disulfiram in alcoholics after single and repeated doses. Clin Pharmacol Ther 1984;36:520–526 [DOI] [PubMed] [Google Scholar]
- 20.McCance-Katz EF, Kosten TR, and Jatlow P: Disulfiram effects on acute cocaine administration. Drug Alcohol Depend 1998;52:27–39 [DOI] [PubMed] [Google Scholar]
- 21.Xing S, Bullen CK, Shroff NS, Shan L, Yang HC, Manucci JL, Bhat S, Zhang H, Margolick JB, Quinn TC, et al. : Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Virol 2011;85:6060–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, and Karn J: Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol 2008;82:12291–12303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abramoff MD, Magelhaes PJ, and Ram SJ: Image processing with ImageJ. Biophotonics Int 2004;11:36–42 [Google Scholar]