Abstract
In this study, we used whole genome complementation with a PAO1 cosmid library, coupled with in vitro transposon mutagenesis, to identify gene locus PA1494 as a novel inhibitor of alginate overproduction in P. aeruginosa strains possessing a wild-type mucA.
Keywords: Pseudomonas aeruginosa, alginate, biofilms, inhibitor, PA1494, muiA
Alginate overproduction is a key mechanism for the development of a chronic lung infection by P. aeruginosa in individuals with cystic fibrosis (Govan & Deretic, 1996). Alginate overproduction is achieved through increased transcription of the alginate biosynthetic operon at the algD promoter (Deretic, et al., 1987). Regulation of alginate overproduction primarily involves the alternative sigma factor AlgU (AlgT, σ22) and its cognate anti-sigma factor, MucA (Martin, et al., 1993, Martin, et al., 1993). Typically, in low alginate producing P. aeruginosa strains, AlgU is sequestered by MucA to the inner membrane (Mathee, et al., 1997, Rowen & Deretic, 2000). However, with a loss of MucA through mutations, AlgU is free to activate transcription at the algD promoter (Martin, et al., 1993, Wozniak & Ohman, 1994). Previous reports have also determined an alternative mechanism for alginate overproduction which requires AlgW, a DegS-like serine protease (Wood, et al., 2006, Qiu, et al., 2007, Cezairliyan & Sauer, 2009). AlgW is the first intramembrane protease that has been shown to degrade the periplasmic portion of MucA (Cezairliyan & Sauer, 2009). However, there are other proteases, MucP and ClpXP that are also required for alginate overproduction via the degradation of MucA (Qiu, et al., 2007, Qiu, et al., 2008). All of these proteases regulate of alginate production by degrading MucA. Additionally, it has been suggested that preventing the overproduction of alginate (Ramsey & Wozniak, 2005), and more specifically inhibiting the regulated proteolytic degradation of MucA (Damron & Goldberg, 2012), to be a potential strategy to prevent the establishment of chronic P. aeruginosa infections. In response to these suggestions, we employed whole-genome complementation coupled with in vitro transposon mutagenesis to identify a genetic locus that can inhibit alginate overproduction in P. aeruginosa strains with a wild-type mucA.
PA1494 encodes a novel inhibitor of alginate overproduction
Recently, we determined that alginate overproduction in P. aeruginosa strain PAO579 (muc-23), a derivative of PAO1, is caused by the release of AlgU from MucA via regulated intramembrane proteolysis (Withers, et al., 2012, Withers, 2013). This proteolytic cascade is initiated by the activation of AlgW through a truncated type-IV pilus (PilA108) (Withers, 2013). To identify novel inhibitors of alginate overproduction in P. aeruginosa strains with a wild-type mucA, a PAO1-derived, minimal tiling path (MTP) genomic cosmid library (Huang, et al., 2000) was conjugated into PAO579 (Govan & Fyfe, 1978). As a result, we identified that cosmid MTP87 could completely suppress alginate overproduction in PAO579 (data not shown). MTP87 covers a region of 22,757 bp from the genome of PAO1 (start: 1,618,021; end: 1,640,777). To identify the exact gene within this cosmid responsible for the multi-copy suppression of alginate overproduction, MTP87 underwent random transposon-mediated in vitro mutagenesis, and the mutated cosmids were then conjugated en masse into PAO579 and screened for alginate overproduction (Figure 1A). We observed the presence of alginate overproducing clones indicating a transposon-mediated inactivation of a specific inhibitory gene within cosmid MTP87 (Figure 1A). PCR and sequence analysis of the mutagenized MTP87 confirmed a single transposon insertion in open reading frame PA1494. Previous transcriptome analyses have shown that PA1494 is up-regulated when P. aeruginosa is exposed to azithromycin (Nalca, et al., 2006, Kai, et al., 2009) and hydrogen peroxide (Chang, et al., 2005). However, since PA1494 belongs to a class of unclassified/hypothetical genes, and its exact function is unknown, we refer to PA1494 as mucoidy inhibitor gene A, or muiA.
Figure 1.
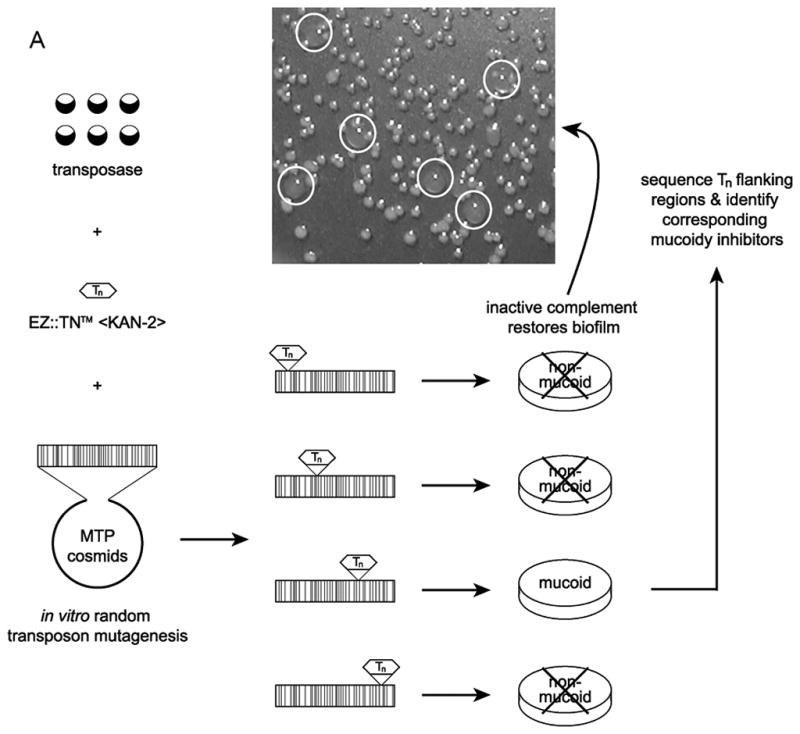
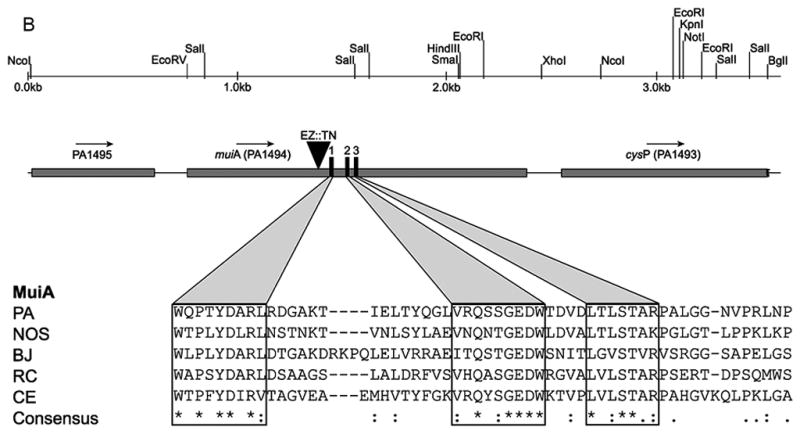
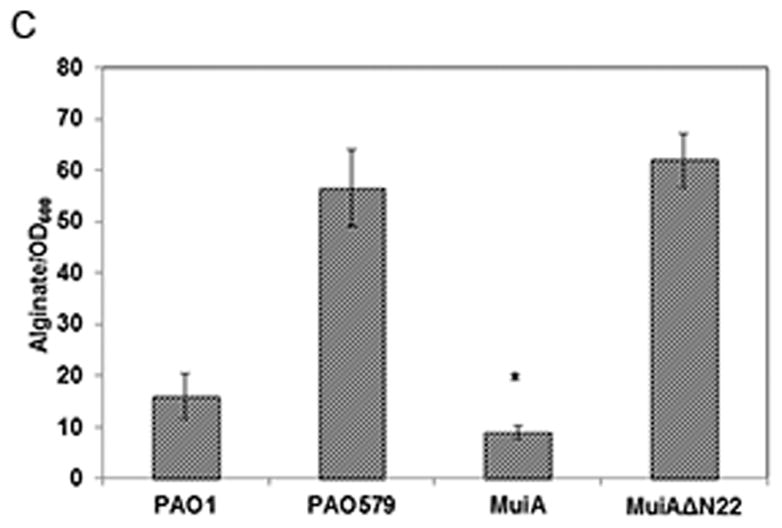
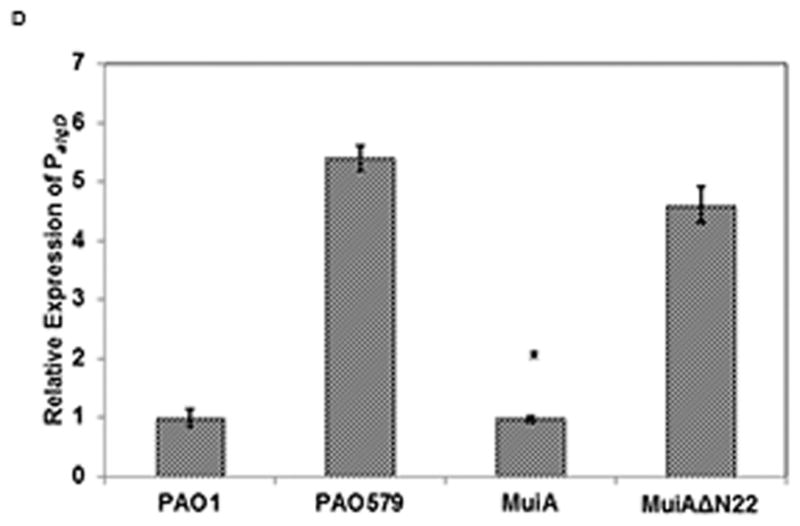
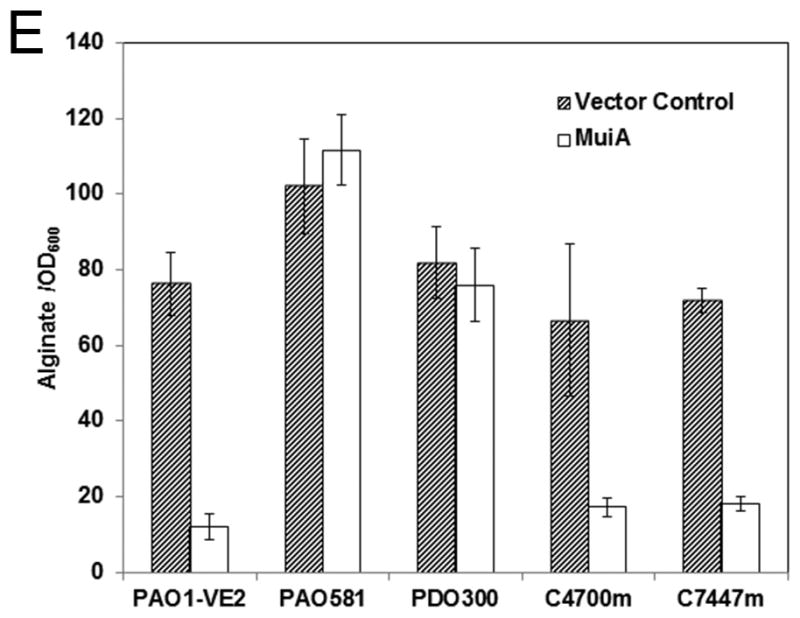
Identification and characterization of MuiA. A) MTP87 cosmid was subjected to in vitro transposon mutagenesis to generate random gene knockouts. Shown in the inset are PAO579 (muc-23) exconjugants carrying cosmid MTP87 randomly mutagenized with an EZ::TN transposon (Epicentre), selected on a PIA plate supplemented with the appropriate antibiotic, and incubated at 37°C for 48h. B) Restriction map, gene organization and Tn insertion in the muiA gene. Homology of MuiA with its orthologs. Shown are the most homologous regions (1, 2 and 3): R. capsulatus (RC; ORF1654; 534 aa), B. japonicum (BJ; CAC38742; 560 aa), Nostoc sp. (NOS; NP_484904; 545 aa), and C. elegans (CE; NP_500427; 556 aa). A single Tn insertion occurred before regions 1, 2 and 3. C) PAO1 pHERD20T, PAO579 pHERD20T (vector control), PAO579 pHERD20T-muiA (wild-type muiA) and PAO579 pHERD20T-muiAΔN22 (deletion of N terminal signal sequence) were grown on PIA plates supplemented with carbenicillin and 0.1% arabinose for 24h at 37°C then for 24h at room temperature. The alginate was collected and measured using the standard carbazole assay. The values are representative of three independent experiments. Statistical significance was determine using the Student’s t-test in comparison with PAO579 (*P<0.05). D) The β-galactosidase activity of the algD promoter was measured using PAO1 and PAO579 miniCTX-PalgD-lacZ with pHERD20T, pHERD20T-muiA or pHERD20T-muiAΔN22. All strains were incubated at 37° C on PIA plates supplemented with tetracycline, carbenicillin and 0.1% arabinose. The values for the mean and standard deviation Miller Units (One Miller Unit=1000 X (A420/−1.75 X A550/OD600 mL−1 min−1)) are shown as relative expression as compared to PAO1, and are representative of three independent experiments. Statistical significance was determine using the Student’s t-test in comparison with PAO579 (*P<0.05). E) Alginate measurements for various laboratory and clinical strains expressing pHERD20T (vector control) or pHERD20T-muiA in trans. Strains were cultured on PIA plates supplemented with 300 μg/mL of carbenicillin and 0.1% arabinose and incubated for 24h at 37°C then for 24h at room temperature. The values are representative of three independent experiments. Statistical significance was determine using the Student’s t-test in comparison with the vector control (*P<0.05).
The muiA gene is predicted to encode a polypeptide of 551 amino acids with a predicted molecular mass of 61 kDa and an isoelectric point (pI) of 5.5. Located immediately downstream is the E. coli periplasmic sulfate-binding ortholog gene (cysP: PA1493). The muiA gene is predicted to use GTG as a start codon with a typical type-I signal sequence encoding 22 amino acids (NH2-MNRLAASPLLFAGLFASAPLLA-COOH) (Lewenza, et al., 2005), and previous proteomic analysis detected MuiA in the periplasm of PAO1. Based on MALDI-TOF/TOF survey on the proteome of the PAO1 cell envelope, the relative abundance of MuiA was in the same range as the alginate negative regulator MucB, but less than the major porin protein OprF (Imperi, et al., 2009). Additionally, we confirmed the results presented in these previous studies by detecting presence of a hemmaglutinin-tagged MuiA in the periplasm using cell fractionation and Western blot analysis (data not shown). Through BLAST analysis, we determined that MuiA is highly conserved amongst other P. aeruginosa strains; however no orthologs were identified in E. coli species, or other Pseudomonads. MuiA orthologs were found in other organisms including Rhodobacter capsulatus, Bradyrhizobium japonicum, Nostoc sp., and Caenorhabditis elegans (Figure 1B). These orthologs are all of similar size ranging from 530 to 560 amino acids in length, and are classified as conserved hypothetical proteins. An internal region of MuiA (232-274aa) displayed 3 highly conserved regions. In addition, the transposon insertion in MTP87 was located 15 bps in front of these conserved domains (Figure 1B).
Expression of muiA suppresses alginate overproduction
In order to confirm whether muiA is responsible for suppressing alginate overproduction, we used standard molecular techniques (Russell, 2001) to clone muiA into the shuttle vector pHERD20T which contains the PBAD arabinose inducible promoter (Qiu, et al., 2008). PAO1 pHERD20T, PAO579 pHERD20T (vector control) and PAO579 pHERD20T-muiA were cultured on PIA supplemented with carbenicillin and 0.1% arabinose, and the amount of alginate was measured using the standard carbazole assay (Knutson & Jeanes, 1968). When compared to the PAO1 and the vector control, there was a decrease in alginate overproduction when muiA was expressed in trans (Figure 1C). Additionally, we observed that pHERD20T-muiA can suppress mucoidy even in the absence of arabinose on PIA, suggesting that the basal expression from pHERD20T-muiA was sufficient for the suppression (data not shown). We also observe that the removal of the N-terminal signal sequence (pHERD20T-muiAΔN22) abrogated MuiA’s ability to suppress alginate overproduction in PAO579 (Figure 1C). Also, we observed that the in-frame deletion of muiA in strain PAO1 did not result in alginate overproduction, suggesting that MuiA does not play a central role in alginate regulation (data not shown). These results suggest that MuiA suppresses alginate overproduction after localization to the periplasm, and can act as a multi-copy suppressor for alginate overproduction in PAO579.
Expression of muiA decreases PalgD transcriptional activity
Previously it was reported that alginate overproduction in PAO579 was due to increased transcriptional activity at the PalgD promoter site of the alginate biosynthetic operon (Boucher, et al., 2000, Withers, 2013). To determine the effect that the expression of muiA has on PalgD activity, we used a PAO1 and PAO579 merodiploid strains (generated via miniCTX-PalgD-lacZ) that carry a chromosomal copy of the algD promoter fused with a reporter gene, lacZ. Next, we conjugated pHERD20T (vector control) pHERD20T-muiA and pHERD20T-muiAΔN22 into the PAO1 or PAO579 miniCTX-PalgD-lacZ, and cultured them on PIA plates supplemented with carbenicillin, tetracycline and 0.1% arabinose. We measured the transcriptional activity of PalgD promoter using the Miller Assay (Miller, 1972). As expected, the level of transcriptional activity in PAO579 pHERD20T was significantly higher than that in PAO1 (Figure 1D). The activity at PalgD decreased when pHERD20T-muiA was expressed in trans in PAO579 (Figure 1D). Additionally, we observed that expression of pHERD20T-muiAΔN22 did not result in a decrease in PalgD activity in PAO579 (Figure 1D). Based on these results, we concluded that expression of muiA suppresses transcriptional activity at the alginate biosynthetic operon at the algD promoter.
Expression of muiA suppresses alginate overproduction in laboratory and clinical strains with a wild-type MucA
To determine the overall robustness, and also elucidate the possible mechanism by which MuiA suppresses alginate overproduction, we conjugated pHERD20T-muiA into a variety of laboratory and clinical strains. We observed that expression of muiA in trans suppressed alginate overproduction in PAO1-VE2 (Figure 1E). PAO1-VE2 is a derivative of PAO1, and overproduces alginate due to activation of AlgW by MucE, a small envelope protein (Qiu, et al., 2007, Cezairliyan & Sauer, 2009). Similarly, expression of muiA in trans was able to suppress alginate overproduction in clinical strains C7447m and C4700m, both possessing a wild-type MucA (Figure 1E). The decrease in alginate overproduction observed in PAO1-VE2, C7447m and C4700m were all statistically significant when compared to the vector control (P<0.05). However, expression of muiA was unable to suppress alginate overproduction in the PAO1-derived, mucA25 strain, PAO581 (Figure 1E). PAO581 carries a truncated MucA25 protein which lacks the transmembrane domain of the wild-type MucA, suggesting that MucA25 is likely localized in the cytoplasm (Qiu, et al., 2008). Additionally, expression of muiA did not suppress alginate overproduction in the strain PDO300 (Figure 1E). PDO300 carries a truncated MucA22 protein. It is important to note that none of the strains examined in this study, aside from PAO579, have three tandem mutations resulting in a truncation of type IV pili. Therefore, the suppression of alginate overproduction through the expression of MuiA is not specific to those strains possessing a truncation in type IV pili. Together, these data suggests that MuiA’s ability to suppress alginate overproduction is not unique to PAO579; however it is only effective at suppressing alginate overproduction in strains with a wild-type MucA.
In summary, we coupled whole genome complementation of a PAO1 cosmid library and in vitro transposon mutagenesis to identify the genetic locus PA1494 (muiA) as a novel inhibitor of alginate overproduction in P. aeruginosa strains with a wild-type mucA. Additionally, we determined that expression of muiA in trans resulted in a decrease in alginate production, as well as transcriptional activity at the PalgD promoter. Additionally, expression of muiA in trans was only able to suppress alginate overproduction in various clinical and laboratory strains possessing a wild-type MucA.
Acknowledgments
This work was supported by the National Aeronautics and Space Administration West Virginia Space Grant Consortium (NASA WVSGC), Cystic Fibrosis Foundation (CFF-YU11G0) and NIH P20RR016477 and P20GM103434 to the West Virginia IDeA Network for Biomedical Research Excellence. We would like to thank Dr. Nathan Head for his technical assistance.
Footnotes
Authors’ Contributions
T.R.W. designed and performed experiments, analyzed data and wrote the manuscript. Y.Y. and H.D.Y. analyzed data and helped revised of the manuscript.
DISCLOSURES:
The author Hongwei D. Yu is the Chief Science Officer and Co-founder of Progenesis Technologies, LLC.
References
- Boucher JC, Schurr MJ, Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- Cezairliyan BO, Sauer RT. Control of Pseudomonas aeruginosa AlgW protease cleavage of MucA by peptide signals and MucB. Mol Microbiol. 2009;72:368–379. doi: 10.1111/j.1365-2958.2009.06654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Small DA, Toghrol F, Bentley WE. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics. 2005;6:115. doi: 10.1186/1471-2164-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damron FH, Goldberg JB. Proteolytic regulation of alginate overproduction in Pseudomonas aeruginosa. Mol Microbiol. 2012;84:595–607. doi: 10.1111/j.1365-2958.2012.08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Gill JF, Chakrabarty AM. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan JR, Fyfe JA. Mucoid Pseudomonas aeruginosa and cystic fibrosis: resistance of the mucoid from to carbenicillin, flucloxacillin and tobramycin and the isolation of mucoid variants in vitro. J Antimicrob Chemother. 1978;4:233–240. doi: 10.1093/jac/4.3.233. [DOI] [PubMed] [Google Scholar]
- Govan JR, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Whitchurch CB, Croft L, Beatson SA, Mattick JS. A minimal tiling path cosmid library for functional analysis of the Pseudomonas aeruginosa PAO1 genome. Microb Comp Genomics. 2000;5:189–203. doi: 10.1089/omi.1.2000.5.189. [DOI] [PubMed] [Google Scholar]
- Imperi F, Ciccosanti F, Perdomo AB, et al. Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen. Proteomics. 2009;9:1901–1915. doi: 10.1002/pmic.200800618. [DOI] [PubMed] [Google Scholar]
- Kai T, Tateda K, Kimura S, et al. A low concentration of azithromycin inhibits the mRNA expression of N-acyl homoserine lactone synthesis enzymes, upstream of lasI or rhlI, in Pseudomonas aeruginosa. Pulm Pharmacol Ther. 2009;22:483–486. doi: 10.1016/j.pupt.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Knutson CA, Jeanes A. A new modification of the carbazole reaction: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- Lewenza S, Gardy JL, Brinkman FS, Hancock RE. Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen. Genome Res. 2005;15:321–329. doi: 10.1101/gr.3257305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Holloway BW, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DW, Schurr MJ, Mudd MH, Govan JR, Holloway BW, Deretic V. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc Natl Acad Sci U S A. 1993;90:8377–8381. doi: 10.1073/pnas.90.18.8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathee K, McPherson CJ, Ohman DE. Posttranslational control of the algT (algU)-encoded sigma22 for expression of the alginate regulon in Pseudomonas aeruginosa and localization of its antagonist proteins MucA and MucB (AlgN) J Bacteriol. 1997;179:3711–3720. doi: 10.1128/jb.179.11.3711-3720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. beta-galactosidase assay. In: Miller JH, editor. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor; New York: 1972. pp. 352–355. [Google Scholar]
- Nalca Y, Jansch L, Bredenbruch F, Geffers R, Buer J, Haussler S. Quorum-sensing antagonistic activities of azithromycin in Pseudomonas aeruginosa PAO1: a global approach. Antimicrob Agents Chemother. 2006;50:1680–1688. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Rowen DW, Yu HD. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2007;104:8107–8112. doi: 10.1073/pnas.0702660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Eisinger VM, Head NE, Pier GB, Yu HD. ClpXP proteases positively regulate alginate overexpression and mucoid conversion in Pseudomonas aeruginosa. Microbiology. 2008;154:2119–2130. doi: 10.1099/mic.0.2008/017368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. PBAD-based shuttle vectors for functional analysis of toxic and highly-regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol. 2008;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey DM, Wozniak DJ. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol Microbiol. 2005;56:309–322. doi: 10.1111/j.1365-2958.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- Rowen DW, Deretic V. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol. 2000;36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- Russell JSaDW. Molecular Cloning A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Withers TR, Johnson SL, Yu HD. Draft Genome Sequence for Pseudomonas aeruginosa Strain PAO579, a Mucoid Derivative of PAO381. J Bacteriol. 2012;194:6617. doi: 10.1128/JB.01406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers TR, Damron FH, Yin, Yeshi Yu, Hongwei D. Truncation of type-IV pilin induces mucoidy in Pseudomonas aeruginosa strain PAO579. Microbiology Open. 2013;2:459–470. doi: 10.1002/mbo3.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LF, Leech AJ, Ohman DE. Cell wall-inhibitory antibiotics activate the alginate biosynthesis operon in Pseudomonas aeruginosa: Roles of sigma (AlgT) and the AlgW and Prc proteases. Mol Microbiol. 2006;62:412–426. doi: 10.1111/j.1365-2958.2006.05390.x. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Ohman DE. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]


