Abstract
Countering the diabetes pandemic and consequent complications, such as nephropathy, will require better understanding of disease mechanisms and development of new diagnostic methods. Animal models can be versatile tools in studies of diabetic renal disease when model pathology is relevant to human diabetic nephropathy (DN). Diabetic models using endothelial nitric oxide synthase (eNOS) knock-out mice develop major renal lesions found in human disease. However, it is unknown whether they can also reproduce changes in urinary metabolites found in human DN. We employed Type 1 and Type 2 diabetic mouse models of DN, i.e. STZ-eNOS−/− C57BLKS and eNOS−/− C57BLKS db/db, with the goal of determining changes in urinary metabolite profile using proton nuclear magnetic resonance (NMR). Six urinary metabolites with significantly lower levels in diabetic compared to control mice have been identified. Specifically, major changes were found in metabolites from tricarboxylic acid (TCA) cycle and aromatic amino acid catabolism including 3-indoxyl sulfate, cis-aconitate, 2-oxoisocaproate, N-phenyl-acetylglycine, 4-hydroxyphenyl acetate, and hippurate. Levels of 4-hydroxyphenyl acetic acid and hippuric acid showed the strongest reverse correlation to albumin-to-creatinine ratio (ACR), which is an indicator of renal damage. Importantly, similar changes in urinary hydroxyphenyl acetate and hippurate were previously reported in human renal disease. We demonstrated that STZ-eNOS−/− C57BLKS and eNOS−/− C57BLKS db/db mouse models can recapitulate changes in urinary metabolome found in human DN and therefore can be useful new tools in metabolomic studies relevant to human pathology.
Keywords: urinary metabolites, mouse models, diabetic nephropathy, nuclear magnetic resonance
Introduction
A growing rate of diabetic kidney disease is one of the major pathologic consequences of diabetes pandemic [1]. Progression to end stage renal disease and accompanying cardiovascular complications are the primary causes of mortality in diabetic patients [2]. Introduction of new biomarkers to supplement microalbuminuria, which is currently used as the main indicator of kidney disease progression, should improve disease diagnosis and treatment. Active search for these biomarkers is underway involving genomic, transcriptomic, and proteomic technologies [3–5].
Several recent reports have demonstrated that metabolomics can be an important tool in identifying biomarkers of kidney disease and providing insights into pathogenic mechanisms [6–9]. In particular, urine metabolomics allows investigation of small biomolecules closely associated with kidney function or even originating from the kidney. In addition, urine is easier to collect and analyze compared to plasma, which is an important factor for potential clinical biomarker(s). Although metabolomic studies of diabetic kidney disease have been performed using mass spectrometry technology [6–9], proton NMR offers a number of advantages including minimal sample processing, high throughput, complete sample recovery, and direct structural identification of metabolites.
The results of several studies in animal models and human patients identified a number of urine metabolite biomarker candidates [6–8]. However, more studies utilizing different technologies are needed to identify metabolite changes common to diabetic kidney disease. In particular, finding disease-related commonalities in metabolite profiles from human patients and animal models would facilitate use of versatile model tools to help searching for clinically relevant biomarkers. In this study, we utilized robust mouse models of Type 1 and Type 2 diabetic nephropathy (DN), which most fully reproduce pathologic lesions characteristic of human DN [10–11]. We employed 1H NMR spectroscopy, which has previously been applied to studies of metabolites in diabetic plasma [12–13], to determine changes in urinary metabolite profiles in diabetic kidney disease.
We identified six urinary metabolites with significantly decreased levels in diabetic compared to control samples. These same six metabolites were showing similar changes in either Type 1 or Type 2 diabetic animal models. Levels of 4-hydroxyphenyl acetic acid and hippuric acid showed the strongest reverse correlation with the decline in renal function. Based on our findings in this study and on previous human studies [6–7], we conclude that STZ-eNOS−/− C57BLKS and eNOS−/− C57BLKS db/db mouse models can recapitulate changes in urinary metabolome found in human DN. We propose that these models can be important new experimental tools in metabolomic studies relevant to human pathology.
Materials and Methods
Animal studies
In this study urine samples from three groups of mice were investigated: a control group and type 1 diabetic and type 2 diabetic groups (11 mice per group). Animal experiments were performed at the AAALAC-accredited animal facilities at Vanderbilt University Medical Center according to institutional guidelines and IACUC-approved experimental protocol. Mice were housed in an approved facility and given standard chow (Lab Diet 5015; PMI Nutrition International, Richmond, IN) and water ad libitum. For type 1 diabetic model, 8 week old eNOS−/− C57BLKS were injected with low doses of STZ (50 mg/kg of BW) for 5 consecutive days to minimize potential toxic effects as previously described [11]. For the type 2 diabetic model, we used eNOS−/− C57BLKS db/db mice [10]. Both models develop robust diabetic renal disease with pathologic lesions closely approximating those found in human DN [10–11]. Diabetic mice and age-matched wild type control mice were sacrificed at 20 wks of age. Blood and spot urine were collected before sacrifice. Serum and urine samples were stored at −80°C until analysis.
Determination of blood glucose and urinary albumin excretion
Glucose levels were measured in blood collected from the tail vein using OneTouch glucometer and Ultra test strips (LifeScan, Milpitas, CA) as previously described [10–11]. Albumin and creatinine excretion was determined in spot urine collected from individually caged mice using Albuwell-M kits (Exocell Inc, Philadelphia, PA) as previously described [10–11].
NMR experiments
NMR spectra were acquired using a 14.0 T Bruker magnet equipped with a Bruker AV-III console operating at 600.13 MHz. All spectra were acquired in 3-mm NMR tubes using a Bruker 5-mm TCI cryogenically cooled NMR probe operating at 298oK. Samples were prepared as 200 μL solutions that included 100 μL of urine, 41 μL of combination of 70 mM sodium phosphate buffer, TSP, and NaN3, and 59 μL of 90%–10% H2O/D2O which served as the 2H lock solvent. TSP (3-(trimethylsilyl)propionic-2,2,3,3-d4 acid) in the buffer solution served as the zero ppm chemical shift reference.
For 1D 1H NMR, experiments were acquired using a one-dimensional nuclear Overhauser (1D-NOE) pulse sequence with presaturation solvent suppression to suppress the signal associated with water that is typically present in high concentration in mouse urine samples. The 1D-NOE experiment filters NMR signals associated with broad line widths such as those arising from proteins that might be present in urine samples and adversely affect spectral quality. Experimental conditions included: 32K data points, 13 ppm sweep width, a recycle delay of 3 seconds, a mixing time of 150 ms and 32 scans.
NMR data analysis
Principal component analysis (PCA) was performed using the AMIX program (Bruker Biospin Corp., Billerica MA). This method requires NMR data to be distributed in chemical shift bins (0.01 ppm width) while eliminating the area associated with the water solvent suppression (4.6 -5.1 ppm) and glucose (3.2-3.58, 3.62-4.108 and 5.22-5.276 ppm). Buckets with less than 5% variance were omitted prior to PCA. PCA reduces the dimensionality of the data and summarizes the similarities and differences between multiple NMR spectra using scores plots.
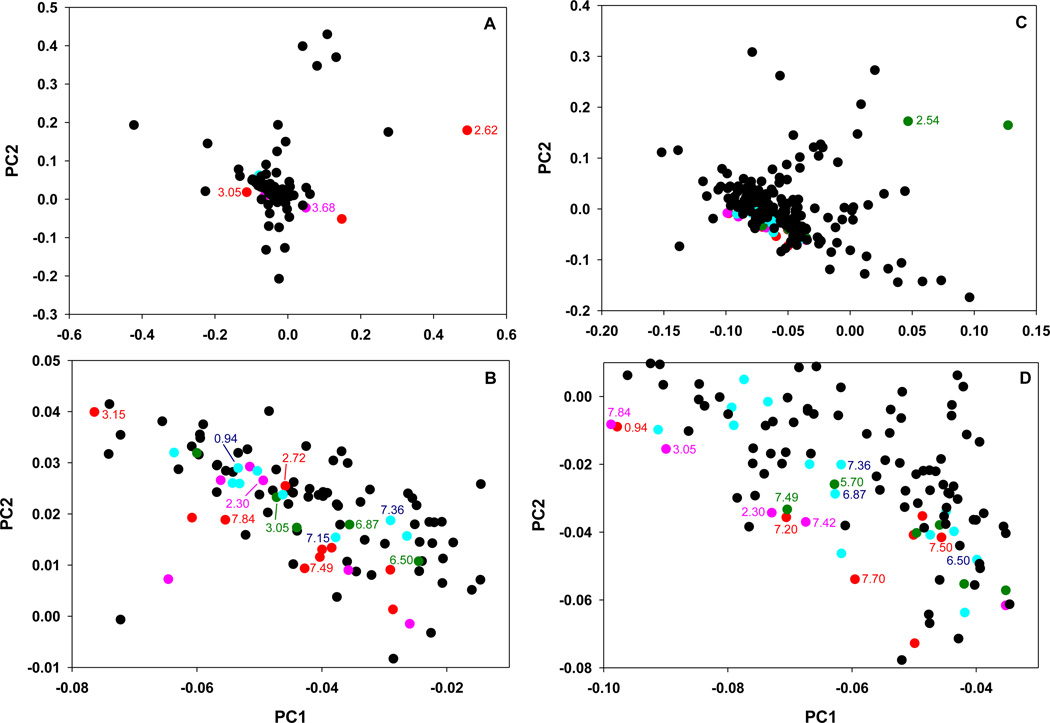
The statistical analysis of the NMR data was performed as previously published by Kennedy et. al. [14]. The P scores were calculated and automatically divided into four groups by AMIX software based on significance level. These groups were color coded within PCA loadings plots according to P score values, thus generating a heat map representation of the data (Fig. 3). Buckets that corresponded to statistically significant P scores were compared to ppm values of the peaks that were identified from 1D & 2D NMR assignments. In order for a metabolite to be selected as a candidate, the occurrence of more than one metabolite peak had to be present in the statistically significant buckets. For example, 3-indoxyl sulfate was selected since the peaks at 7.70 and 7.49 ppm were among the buckets that appeared in the significant P score ranges for both the Type 1 and Type 2 data. Candidate metabolites were quantified by measuring the sum of the areas under all the characteristic resonance peaks corresponding to a given metabolite. Statistical analysis was performed using ANOVA followed by post-hoc Tukey test. Statistical significance was determined after taking into account Bonferroni correction.
Figure 3. Loadings plot of urinary metabolomics data from mouse models of type 1 (A and B) and type 2 (C and D) diabetes.
(A and C) Heat maps are color-coded according to bucket P score values as described under Materials and Methods. For the Type 1 diabetes data, the red points corresponded to P<8.3×10−5, pink points: 8.3×10−5<P<1.6×10−4, green points: 1.6×10−4<P<2.5×10−4, blue points: 2.5×10−4<P<3.2×10−3 and black points: P>3.2×10−3. For Type 2 diabetes data, red points corresponded to P<7.5×10−5, pink points: 7.5×10−5<P<1.8×10−4, green points: 1.8×10−4<P<5.0×10−4, blue points: 5.0×10−4<P<1.0×10−3 and black points: P>1.0×10−3. The numbers indicate ppm values of the selected NMR peaks with P<3×10−3 for which metabolite assignments have been made. (B and D) Zoomed areas from the loadings plots in panels A and C, respectively.
Identification of metabolites
Metabolites were identified by comparison of acquired spectra with standard spectra using the Chenomx Profiler program (Fig. S1). Metabolite identities were further confirmed using two dimensional 1H-1H COSY and 1H-13C HQSC (see Fig. S2 and Table S1 for spectral assignments). For 2D 1H-1H COSY, experimental conditions included 2048 × 512 data matrix, 13 ppm sweep width, recycle delay of 1.5 seconds and 8 scans per increment. The data were processed using squared sinebell window function, symmetrized, and displayed in magnitude mode. Multiplicity-edited HSQC experiments were acquired using a 1024 × 256 data matrix, a J(C-H) value of 145 Hz, which resulted in a multiplicity selection delay of 34 ms, a recycle delay of 1.5 seconds and 32 scans per increment along with GARP decoupling on 13C during the acquisition time (150 ms). The data were processed using a π/2 shifted squared sine window function and displayed with CH/CH3 signals phased positive and CH2 signals phased negative.
Statistical analyses
Mouse physiology data were expressed as means ± SEM; statistical analysis was performed using ANOVA followed by post-hoc Tukey test. Differences were considered statistically significant if p values were less than 0.05.
Results and Discussion
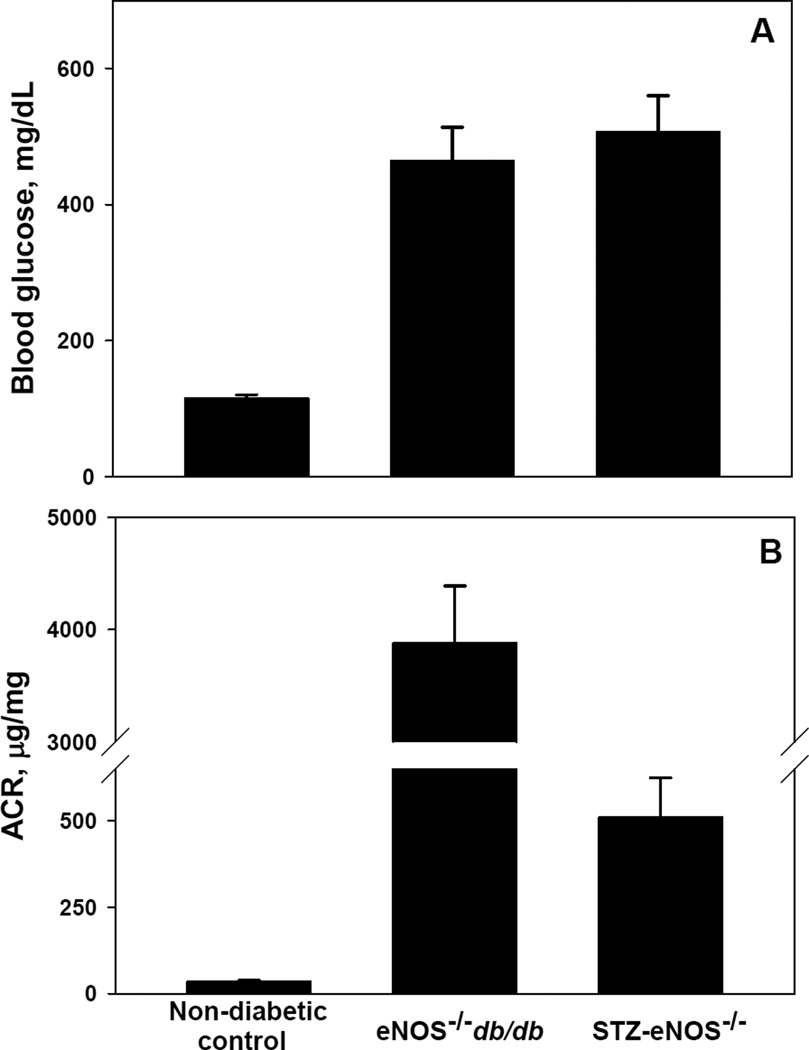
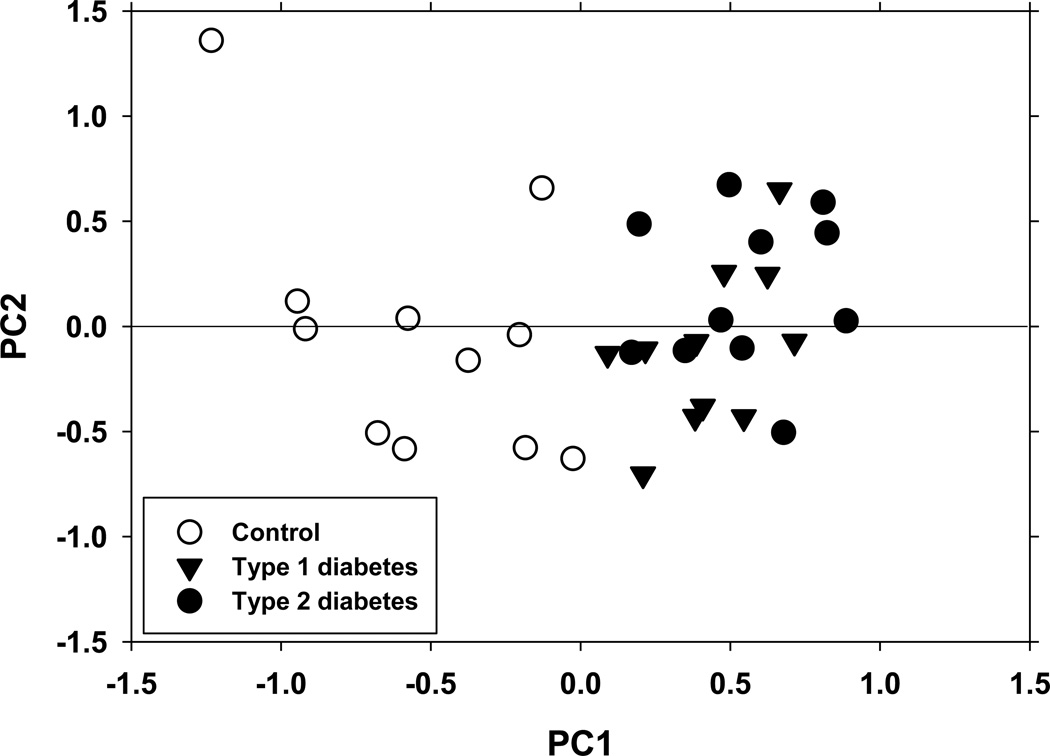
The two animal models of DN used in this study both exhibited hyperglycemia and, after about 14 weeks of diabetes, showed significantly increased urinary albumin excretion, indicative of renal damage (Fig. 1). Small molecule metabolites were analyzed in mouse urine using 1H NMR spectroscopy. Principal component (PC) analysis of the NMR spectra revealed a clear separation of the data points into non-diabetic control and diabetic clusters (Fig. 2). This separation cannot be attributed to differences in glucose concentration between control and diabetic samples as glucose proton NMR signals were removed from the spectra before the analysis. Data from Type 1 and Type 2 diabetic mouse models clustered together in the PC plot indicating similarities of NMR spectra in these two models (Fig. 2).
Figure 1. Levels of blood glucose and albumin excretion in non-diabetic C57BLKS, eNOS−/− C57BLKS db/db and STZ - eNOS−/− C57BLKS mice.
Blood glucose (A) and urinary albumin-to-creatinine ratio (B) were determined in mice at 20 weeks of age as describe under Materials and Methods. Shown are means ± SEM; (n=11).
Figure 2. Principal component analysis of the 1H NMR data from control and diabetic urine samples.
PCA scores were calculated using the data from control (open circles), diabetic type 1 (black triangles) and diabetic type 2 (black circles) urine samples as described under Materials and Methods.
In order to identify spectral features contributing to the observed data clustering, NMR spectra were divided into 0.01 ppm intervals (buckets) and signal intensities in each bucket were analyzed. The P score analysis using AMIX software identified multiple buckets with significant differences between control and diabetic samples (Fig. 3). This step provided for unbiased analysis of the spectral differences between the samples before the identities of specific metabolites were determined.
In the following metabolite identification step the ppm bucket values with significant P scores were used to identify only those individual urinary metabolites, whose levels significantly changed in diabetic compared to control urine samples. This analysis resulted in identification of nine candidate metabolite shown in Table 1 and in Table S2. Additional statistical analysis of integrated peak areas demonstrated that out of nine candidate metabolite identified from the loadings plot, only six metabolites were significantly decreased in diabetic vs. control urines (Table 1, top six entries) and levels of only five metabolites significantly correlated with urinary ACR, a marker of renal pathology (Table 2, top five entries). The most interesting finding was that urinary levels of hydroxyphenyl acetate and hippurate were significantly decreased in diabetes vs. control (Table 1) and also showed the strongest reverse correlation with ACR in diabetic mice (Table 2, top two entries). Reduction in glomerular filtration rate reported in diabetic eNOS−/− C57BLKS db/db mice [10], is unlikely to significantly contribute to this phenomenon since several other small molecule urinary metabolites showed no correlation with ACR (Table 2). Thus, the decrease in hydroxyphenyl acetate and hippurate levels may reflect metabolic changes related to diabetes such as dysregulation of aromatic acid metabolism [15–16].
Table 1.
Levels of urinary metabolites in non-diabetic control and diabetic nephropathy model mice of Type 1 (STZ-eNOS−/−) and Type 2 (eNOS−/− db/db) diabetes
| Metabolite | Proton NMR peak position (ppm) |
Relative level in control urine |
Relative level in Type 1 diabetic urine |
P value vs. control |
Relative level in Type 2 diabetic urine |
P value vs. control |
|---|---|---|---|---|---|---|
| 3-indoxyl sulfate | 7.70, 7.49, 7.37, 7.20 | 1.48 ± 0.15 | 0.48 ± 0.06* | 4.55E-05 | 0.24 ± 0.05* | 7.78E-06 |
| N-phenyl-acetylglycine | 7.42, 7.36, 3.76, 3.68 | 3.29 ± 0.34 | 0.37 ± 0.03* | 1.07E-05 | 0.48 ± 0.10* | 8.25E-06 |
| cis-aconitate | 5.70, 3.15 | 0.81 ± 0.08 | 0.079 ± 0.009* | 1.07E-05 | 0.20 ± 0.03* | 2.19E-05 |
| 2-oxoisocaproate | 2.62, 2.09, 0.94 | 1.42 ± 0.21 | 0.24 ± 0.05* | 1.79E-04 | 0.25 ± 0.06* | 1.70E-04 |
| 4-hydroxyphenyl acetate | 6.87, 7.15 | 0.36 ± 0.06 | 0.05 ± 0.01* | 2.01E-04 | 0.04 ± 0.02* | 1.73E-04 |
| hippurate | 7.84, 7.64, 7.50, 3.95 | 1.81 ± 0.29 | 0.74 ± 0.11 | 0.017 | 0.63 ± 0.10* | 0.004 |
| 2-oxoglutarate | 3.05, 2.45 | 2.11 ± 0.47 | 0.95 ± 0.12 | 0.117 | 0.74 ± 0.17 | 0.033 |
| citrate | 2.72, 2.54 | 3.57 ± 0.62 | 2.14 ± 0.35 | 0.217 | 2.81 ± 0.39 | 0.217 |
| fumarate | 6.50 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.484 | 0.04 ± 0.01 | 0.217 |
Data presented as metabolite concentrations in urine of 20 week old mice, mean ± SEM. Relative metabolite levels represent integrated peak areas normalized to TSP standard. P values based on comparison of control vs. diabetic nephropathy groups (n=11); Bonferroni-adjusted P value cut-off was 0.0056 to adjust for 9 metabolites; asterisk indicates significant change vs. control.
Table 2.
Pearson correlation analysis of ACR vs. urinary metabolites in eNOS−/− db/db mice
| Metabolite | Pearson correlation coefficient |
P value |
|---|---|---|
| 4-hydroxyphenyl acetate | −0.727 | 0.007** |
| hippurate | −0.723 | 0.008** |
| 3-indoxyl sulfate | −0.685 | 0.014* |
| 2-oxoisocaproate | −0.621 | 0.031* |
| N-phenyl-acetylglycine | −0.599 | 0.040* |
| cis-aconitate | −0.574 | 0.051 |
| 2-oxoglutarate | −0.413 | 0.183 |
| citrate | −0.367 | 0.241 |
| fumarate | −0.098 | 0.762 |
Pearson correlation coefficient was determined for selected urinary metabolites identified in urine of type 2 diabetic mice.
Asterisks indicate significant correlation:
P<0.05 and
P<0.01 (n=11).
In order to confirm that our analyses identified all the detectable metabolites with significant changes between diabetic and control samples, we manually analyzed 1H NMR spectra using Chenomx Profiler program. Although a total of twenty-three metabolites were thus identified (Table S3), no additional metabolites with significant changes in diabetic vs. control samples were found.
It is important to note that in this study using STZ-eNOS−/− C57BLKS and eNOS−/− C57BLKS db/db mouse models major changes were detected in urinary metabolites of aromatic amino acid catabolism, similar to those found in human DN [6–7]. Moreover, similar to our results urinary levels of hippuric and hydroxyphenyl acetic acids were also significantly decreased in human diabetic renal pathology as determined in mass-spectrometry based metabolomic studies [6–7]. In contrast, in db/db mouse model, major changes were detected in serum and urine metabolites of TCA cycle [8], This finding is consistent with the fact that eNOS−/− db/db model more fully recapitulates major renal lesions found in human DN compared to the db/db model [10].
Supplementary Material
Highlights.
-
-
1H NMR spectroscopy was employed to study urinary metabolite profile in diabetic mouse models.
-
-
Mouse urinary metabolome showed major changes that are also found in human diabetic nephropathy.
-
-
These models can be new tools to study urinary biomarkers that are relevant to human disease.
Acknowledgements
This work was supported by the grants DK65138 and DK51265 from the National Institutes of Health. Mr. Cody Stothers, Mr. Josh Avance, and Mr. Deon Denson were supported by the Vanderbilt Aspirnaut program and the grant 1R25DK096999 from the National Institutes of Health. We would also like to acknowledge the grant S10RR019022 from the National Institute of Health for funding the purchase of the 600 MHz NMR spectrometer used in this study.
Abbreviations
- DN
diabetic nephropathy
- eNOS
endothelial nitric oxide synthase
- NMR
nuclear magnetic resonance
- TCA
tricarboxylic acid
- ACR
albumin-to-creatinine ratio
- STZ
streptozotocin
- NOE
nuclear Overhauser effect
- PCA
principal component analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Compliance with Ethical Standards
Authors declare that they have no conflict of interest. All institutional and national guidelines for the care and use of laboratory animals were followed. No human studies were carried out by the authors for this article.
References
- 1.Rosolowsky ET, Skupien J, Smiles AM, Niewczas M, Roshan B, Stanton R, Eckfeldt JH, Warram JH, Krolewski AS. Risk for ESRD in type 1 diabetes remains high despite renoprotection. J Am Soc Nephrol. 2011;22:545–553. doi: 10.1681/ASN.2010040354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groop PH, Thomas MC, Moran JL, Waden J, Thorn LM, Makinen VP, Rosengard-Barlund M, Saraheimo M, Hietala K, Heikkila O, Forsblom C. The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes. 2009;58:1651–1658. doi: 10.2337/db08-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma K, Lee S, Han S, Francos B, McCue P, Wassell R, Shaw MA, RamachandraRao SP. Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics. 2005;5:2648–2655. doi: 10.1002/pmic.200401288. [DOI] [PubMed] [Google Scholar]
- 4.Iyengar SK, Abboud HE, Goddard KA, Saad MF, Adler SG, Arar NH, Bowden DW, Duggirala R, Elston RC, Hanson RL, Ipp E, Kao WH, Kimmel PL, Klag MJ, Knowler WC, Meoni LA, Nelson RG, Nicholas SB, Pahl MV, Parekh RS, Quade SR, Rich SS, Rotter JI, Scavini M, Schelling JR, Sedor JR, Sehgal AR, Shah VO, Smith MW, Taylor KD, Winkler CA, Zager PG, Freedman BI. Genome-wide scans for diabetic nephropathy and albuminuria in multiethnic populations: the family investigation of nephropathy and diabetes (FIND) Diabetes. 2007;56:1577–1585. doi: 10.2337/db06-1154. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez ML, Distefano JK. The role of non-coding RNAs in diabetic nephropathy: potential applications as biomarkers for disease development and progression. Diabetes Res Clin Pract. 2013;99:1–11. doi: 10.1016/j.diabres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, Saito R, Pu M, Sharma S, You YH, Wang L, Diamond-Stanic M, Lindenmeyer MT, Forsblom C, Wu W, Ix JH, Ideker T, Kopp JB, Nigam SK, Cohen CD, Groop PH, Barshop BA, Natarajan L, Nyhan WL, Naviaux RK. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24:1901–1912. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Kloet FM, Tempels FW, Ismail N, van der Heijden R, Kasper PT, Rojas-Cherto M, van Doorn R, Spijksma G, Koek M, van der Greef J, Makinen VP, Forsblom C, Holthofer H, Groop PH, Reijmers TH, Hankemeier T. Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study) Metabolomics. 2012;8:109–119. doi: 10.1007/s11306-011-0291-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Wang X, Aa J, Qin W, Zha W, Ge Y, Liu L, Zheng T, Cao B, Shi J, Zhao C, Yu X, Wang G, Liu Z. GC/TOFMS analysis of metabolites in serum and urine reveals metabolic perturbation of TCA cycle in db/db mice involved in diabetic nephropathy. Am J Physiol Renal Physiol. 2013;304:F1317–F1324. doi: 10.1152/ajprenal.00536.2012. [DOI] [PubMed] [Google Scholar]
- 9.Abbiss H, Maker GL, Gummer J, Sharman MJ, Phillips JK, Boyce M, Trengove RD. Development of a non-targeted metabolomics method to investigate urine in a rat model of polycystic kidney disease. Nephrology (Carlton) 2012;17:104–110. doi: 10.1111/j.1440-1797.2011.01532.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhao HJ, Wang S, Cheng H, Zhang MZ, Takahashi T, Fogo AB, Breyer MD, Harris RC. Endothelial nitric oxide synthase deficiency produces accelerated nephropathy in diabetic mice. J Am Soc Nephrol. 2006;17:2664–2669. doi: 10.1681/ASN.2006070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanetsuna Y, Takahashi K, Nagata M, Gannon MA, Breyer MD, Harris RC, Takahashi T. Deficiency of endothelial nitric-oxide synthase confers susceptibility to diabetic nephropathy in nephropathy-resistant inbred mice. Am J Pathol. 2007;170:1473–1484. doi: 10.2353/ajpath.2007.060481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maher AD, Crockford D, Toft H, Malmodin D, Faber JH, McCarthy MI, Barrett A, Allen M, Walker M, Holmes E, Lindon JC, Nicholson JK. Optimization of human plasma 1H NMR spectroscopic data processing for high-throughput metabolic phenotyping studies and detection of insulin resistance related to type 2 diabetes. Anal Chem. 2008;80:7354–7362. doi: 10.1021/ac801053g. [DOI] [PubMed] [Google Scholar]
- 13.Makinen VP, Soininen P, Forsblom C, Parkkonen M, Ingman P, Kaski K, Groop PH, Ala-Korpela M. 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Mol Syst Biol. 2008;4:167. doi: 10.1038/msb4100205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodpaster AM, Romick-Rosendale LE, Kennedy MA. Statistical significance analysis of nuclear magnetic resonance-based metabonomics data. Anal Biochem. 2010;401:134–143. doi: 10.1016/j.ab.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Crandall EA, Fernstrom JD. Effect of experimental diabetes on the levels of aromatic and branched-chain amino acids in rat blood and brain. Diabetes. 1983;32:222–230. doi: 10.2337/diab.32.3.222. [DOI] [PubMed] [Google Scholar]
- 16.Gibson CJ. Diurnal alterations in retinal tyrosine level and dopamine turnover in diabetic rats. Brain Res. 1988;454:60–66. doi: 10.1016/0006-8993(88)90803-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.