Abstract
During development of the mammalian cerebral cortex, radial glial cells (RGCs) generate layer-specific subtypes of excitatory neurons in a defined temporal sequence, in which lower layer neurons are formed before upper layer neurons. It has been proposed that neuronal subtype fate is determined by birthdate via progressive restriction of the neurogenic potential of a common RGC progenitor. We now demonstrate that the murine cerebral cortex contains RGC sublineages with distinct fate potentials. Using in vivo genetic fate mapping and in vitro clonal analysis, we identify an RGC lineage that is intrinsically specified to generate only upper layer neurons, independently of niche and birthdate. Since upper cortical layers were expanded during primate evolution, amplification of this RGC pool may have facilitated human brain evolution.
The mammalian cerebral cortex consists of six major layers that each contain specific subtypes of neurons characterized by distinct projection patterns and gene expression profiles (1). These layer-specific classes of cortical excitatory neurons are derived from radial glial cells (RGCs) in sequential order, with neurons destined for lower layers being generated first, followed by upper layer neurons and, finally, cortical astrocytes (1). This relationship between birthdate and laminar fate of neurons in the cerebral cortex has been documented for over 50 years, although it has remained unclear whether cell fate is determined directly by birthdate (2) or if the two are linked more indirectly rather than causally (3).
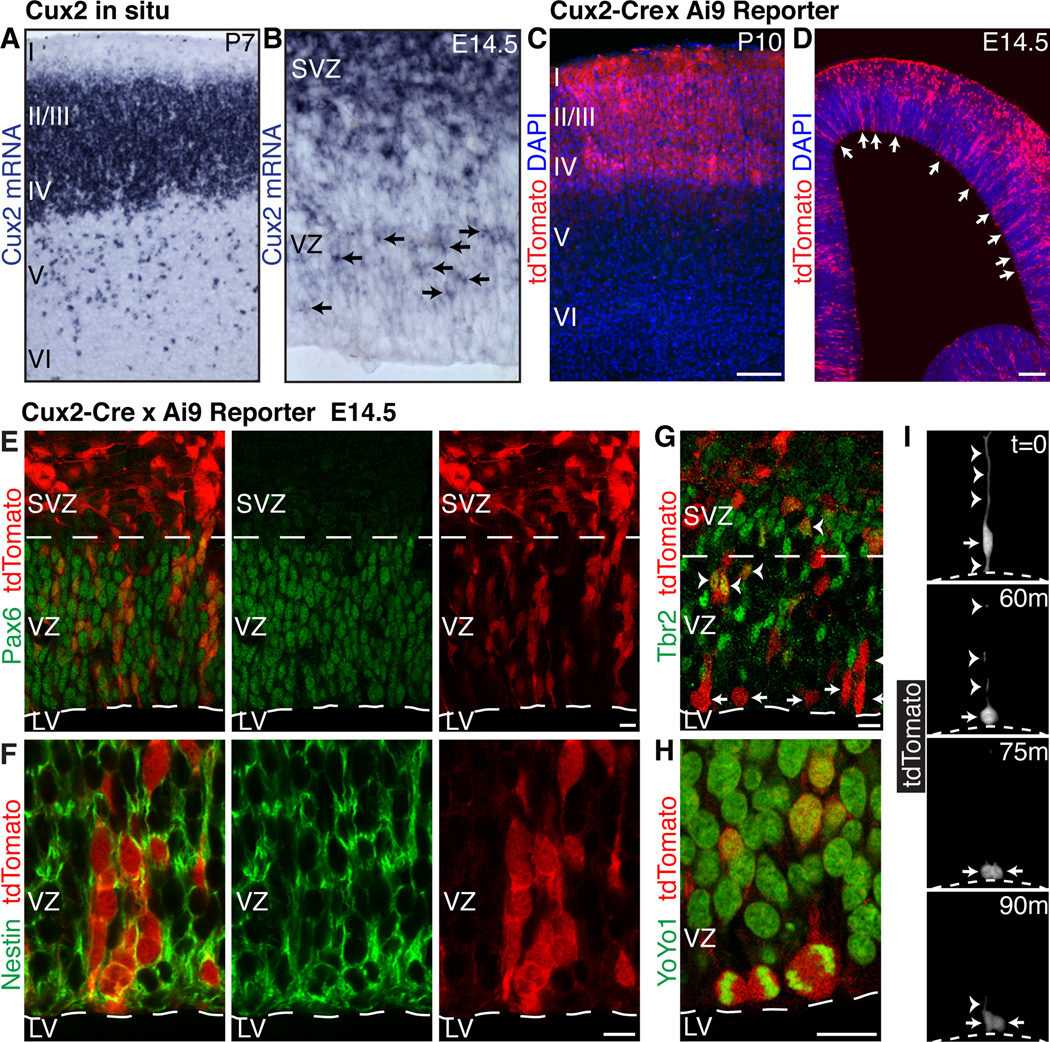
RGCs divide asymmetrically in the cortical ventricular zone to self-renew and generate neurons directly or, more commonly, indirectly via intermediate progenitor cells that divide symmetrically in the ventricular and subventricular zones (4). The transcription factor Cux2 is expressed specifically in upper layer II-IV neurons in the mature cortex (Fig. 1, A), but also in intermediate progenitors in the developing subventricular zone (5, 6), suggesting that upper vs. lower layer fate might be determined before neuronal differentiation. We found that Cux2 mRNA is also expressed in the ventricular zone in a salt-and-pepper fashion (Fig. 1, B and fig. S1, A–D), indicating that some RGCs may be committed to generate upper layer neurons. To establish the identity of ventricular zone Cux2+ cells and to determine their lineage potential, we employed genetic fate mapping using a mouse strain expressing Cre recombinase from the Cux2 locus (7). Crossing the Cux2-Cre driver line to reporter mouse lines led to recombination primarily in upper layer neurons in the mature cortex, with 76% of recombined cells occupying layers II-IV (Fig. 1, C and fig. S1, E–H). Only 17% and 7% of recombined cells were found in lower layers V and VI, respectively (Fig. 1, C and fig. S1, E–H), and most of these were Satb2+ excitatory neurons (74%, fig. S1, I–J), which form callosal projections similar to layer II-III neurons (8, 9). The remainder were Gad65/67+ interneurons (26%) (fig. S1, I–J) derived from the ganglionic eminences, in agreement with previous observations (6). Furthermore, in the developing cortex we identified clonal columns of recombined cells in the ventricular zone (Fig. 1, D), resembling the pattern of endogenous Cux2 mRNA expression. The majority of recombined cells in the ventricular zone expressed the RGC markers Pax6 (Fig. 1, E) and nestin (Fig. 1, F), but not the intermediate progenitor marker Tbr2 (Fig. 1, G). Recombined cells divided at the ventricular surface (Fig. 1, H and I), maintained apical and basal processes and underwent interkinetic nuclear migration (Fig. 1, I), all hallmarks of ventricular zone RGCs. These results suggest that a subset of RGCs are restricted in their lineage potential.
Fig. 1. Expression of Cux2 in a subset of RGCs suggests multiple progenitor types.
(A and B) In situ hybridization shows Cux2 mRNA at P7 (A) and E14.5 (B). Arrows indicate expression in a subset of ventricular zone cells. (C and D) Cumulative fate mapping of the Cux2 lineage in Cux2-Cre;Ai9 mice at P10 (C) and E14.5 (D). Arrows indicate recombination in a subset of ventricular zone cells. (E and F) Recombination in Pax6+ (E) and nestin+ (F) RGCs. (G) Recombination in Tbr2+ intermediate progenitors (arrowheads) and Tbr2− RGCs (arrows) in the ventricular zone. (H) Recombined mitotic cells dividing at the ventricular surface. (I) Live imaging of a slice culture from a Cux2-Cre;Ai9 embryo showing a recombined RGC (arrow) undergoing interkinetic nuclear migration and dividing at the ventricular surface (dotted line). Arrowheads, apical and basal radial processes. LV, lateral ventricle; SVZ, subventricular zone; VZ, ventricular zone. Scale bars, 100 µm (C and D) and 10 µm (E–H).
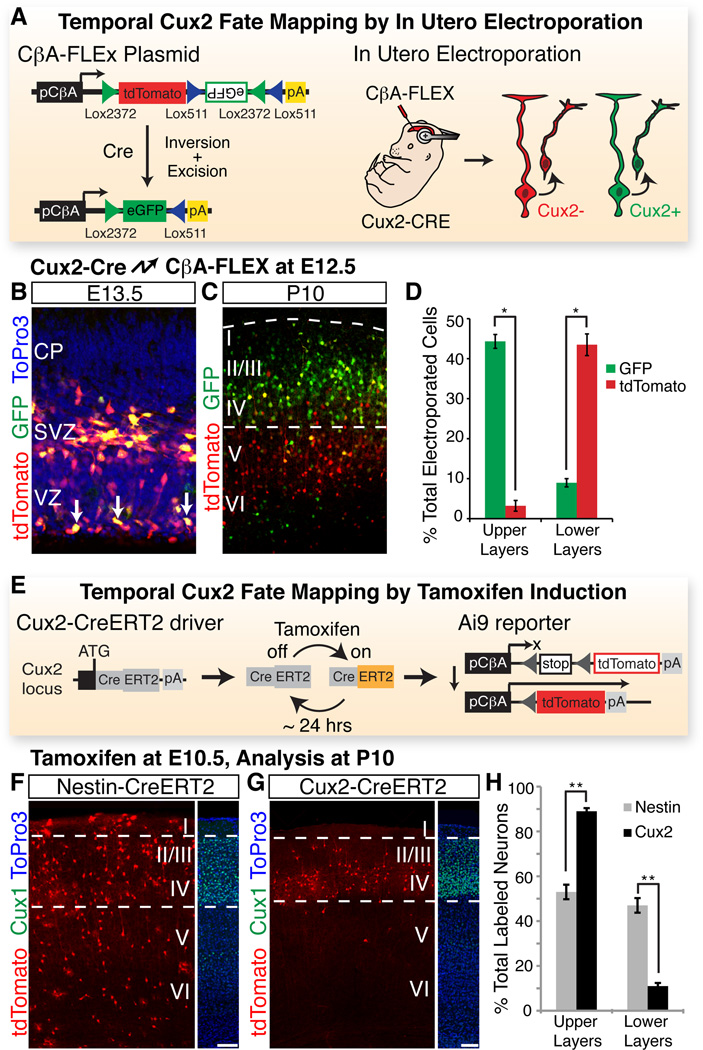
To further analyze the relationship between Cux2+ RGCs and their offspring, we introduced a Cre reporter plasmid into RGCs in Cux2-Cre embryos by in utero electroporation. In this FLEx technology based reporter plasmid (10), Cre recombination switches expression from tdTomato to GFP, thereby permitting differential fluorescent labeling of Cux2+ and Cux2− RGCs and their offspring (Fig. 2, A). Electroporation of the reporter at embryonic day (E) 12.5 and analysis at E13.5 demonstrated that a subset of electroporated RGCs had recombined the reporter and turned on GFP (Fig. 2, B), even though at this early time point the tdTomato signal remained because of protein perdurance. At postnatal day 10, 83% of neurons with recombined reporters (GFP+) had settled in upper layers II-IV (Fig. 2, C and D), whereas only 7% of the neurons with non-recombined reporters (tdTom+GFP−) were located in upper layers (Fig. 2, C and D). These results suggest that the Cux2+ progenitors constitute a lineage largely fated to become upper-layer neurons with a small contribution to lower layers, whereas the Cux2− lineage primarily generates lower layer neurons.
Fig. 2. RGCs of the Cux2 lineage are fated to generate upper layer neurons.
(A) Temporal fate mapping by in utero electroporation. Upon in utero electroporation into Cux2-Cre embryos, CβA-FLEx drives differential expression of tdTomato and eGFP in Cux2− and Cux2+ cells, respectively. (B) Electroporation of CβA-FLEx at E12.5, analysis at E13.5. Arrows identify cells in the ventricular zone that have recombined the plasmid. (C) Electroporation at E12.5, analysis at P10. (D) Percentage (± SEM) of electroporated GFP+ and tdTomato+ neurons in upper vs. lower layers at P10. *, P < 5E-4. (E) Temporal fate mapping by tamoxifen induction. Cux2- CreERT2 mice allow temporary activation of CreERT2 in the Cux2+ lineage within 6–24 hr after tamoxifen injection. CreERT2 activates the Ai9 reporter to allow permanent tdTomato labeling during this window, but not before or after. (F) E10.5 induction, P10 analysis of a Nestin- CreERT2 line, which drives recombination in Cux2+ and Cux2− RGCs. Dotted lines frame Cux1 expression in upper layers. (G) E10.5 induction, P10 analysis of the Cux2-CreERT2 line. (H) Percentage (± SEM) of recombined neurons in upper vs. lower layers for each driver. *, P < 1E-10. CP, cortical plate. Scale bars, 100 µm.
To facilitate temporal fate-mapping of the Cux2+ lineage, we generated a mouse line in which the tamoxifen-inducible CreERT2 (11) gene is knocked into the endogenous Cux2 locus (fig. S2, A–C). By crossing Cux2-CreERT2 mice to a Cre-reporter strain and inducing recombination with a single injection of tamoxifen (Fig. 2, E), we can label the Cux2+ lineage within a narrow ~24 hour window (12, 13). Because most cortical cells are RGCs at E10.5, we injected tamoxifen at this time point to permanently label Cux2+ RGCs between E10.5 and E11.5, but not before or afterward. We performed the same experiments with Nestin-CreERT2 mice (14), which express tamoxifen-inducible Cre in all RGCs (fig. S2, D and E). While the Nestin-CreERT2 line labeled neurons that contributed to all cortical layers (Fig. 2, F and H), as well as astrocytes and oligodendrocytes (fig. S2, F and G), the Cux2-CreERT2 line labeled predominantly neurons in upper layers (Fig. 2, G and H; fig. S2, E). Thus, Cux2+ RGCs are fated to generate upper layer neurons even at the earliest stages of cortical neurogenesis. Cux2− RGCs are likely the proposed bipotent neuro-glial progenitors (15), or are perhaps further subdivided into lineage-restricted subtypes.
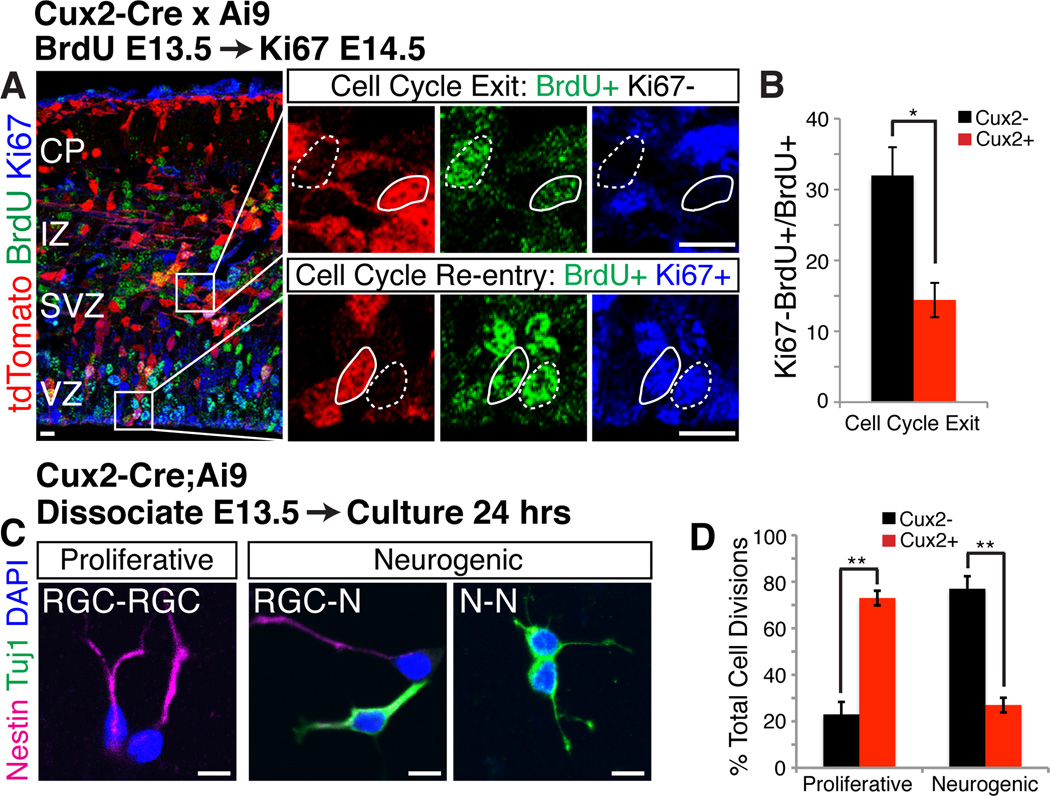
Lower layer neurons are largely born before upper layer neurons, although there is some degree of overlap in the birthdates of different neuronal subtypes (16). Since Cux2+ upper layer progenitors and Cux2− lower layer progenitors co-exist at the earliest stages of cortical development (fig. S3, A), we reasoned that during lower layer neurogenesis Cux2+ RGCs might show a greater propensity to proliferate to expand the RGC pool, while Cux2− RGCs may preferentially generate neurons. In support of this idea, the percentage of Cux2+ RGCs increased over time, whereas the percentage of Cux2− RGCs decreased (fig. S3, A). This suggests that the fraction of RGCs leaving the cell cycle at early stages of corticogenesis is lower for Cux2+ RGCs than for Cux2− progenitors. To test this hypothesis, we analyzed cell cycle exit in Cux2-Cre;Ai9 mice using a BrdU pulse to label S-phase cells at E12.5, followed by Ki67 staining at E13.5 to label cells that remained in the cell cycle (Fig. 3, B). As expected, the fraction of Cux2+ progenitors exiting the cell cycle was less than 50% that of Cux2− progenitors (Fig. 3, C).
Fig. 3. During early neurogenesis, Cux2+ progenitors are mainly proliferative, while Cux2− progenitors are neurogenic.
(A) Cux2-Cre;Ai9 embryos injected with BrdU to label S-phase cells at E12.5, then immunostained at E13.5 for BrdU and the proliferation marker Ki67. High magnifications of boxed areas at right show cells that exit (BrdU+Ki67−) or re-enter (BrdU+Ki67+) the cell cycle within 24 hours. tdTomato absence (dotted outline) and presence (solid outline) identifies Cux2− and Cux2+ cells, respectively. IZ, intermediate zone. (B) Percentage (± SEM) of Cux2− vs. Cux2+ progenitors that exit the cell cycle (BrdU+Ki67−). *, P < 0.01. (C) Cortical cells from E13.5 Cux2-Cre;Ai9 embryos cultured at single-cell density for 24 hrs. Pairs of 2 nestin+ RGCs indicate proliferative divisions; neurogenic divisions produce 1 or 2 Tuj1+ neurons. Cux2− and Cux2+ cells were identified by absence/presence of tdTomato (not shown). (D) Percentage (± SEM) of proliferative and neurogenic divisions in Cux2− vs. Cux2+ cell pairs. **, P < 1E-6. Scale bars, 10 µm.
Since environmental cues such as notch-delta signaling control RGC self-renewal (17, 18), we hypothesized that notch signaling may be preferentially active in Cux2+ RGCs. However, notch signaling was not differentially regulated in Cux2+ vs. Cux2− RGCs (fig. S3, B–E), and activation of the pathway affected the two populations similarly (fig. S3, F–H). Rather, the proliferative behaviors of the two RGC subtypes appear to be at least partially intrinsic, since these differences were maintained outside their normal niche. When cells from E13.5 Cux2- Cre;Ai9 embryos were cultured in vitro at clonal density (Fig. 3, C), the majority of Cux2+ progenitors divided symmetrically to generate pairs of RGCs, whereas Cux2− progenitors preferentially underwent neurogenic divisions (Fig. 3, D). We conclude that at early stages of corticogenesis, Cux2+ RGCs fated to generate upper layer neurons preferentially proliferate to enlarge the precursor pool, while Cux2− RGCs already generate lower layer neurons.
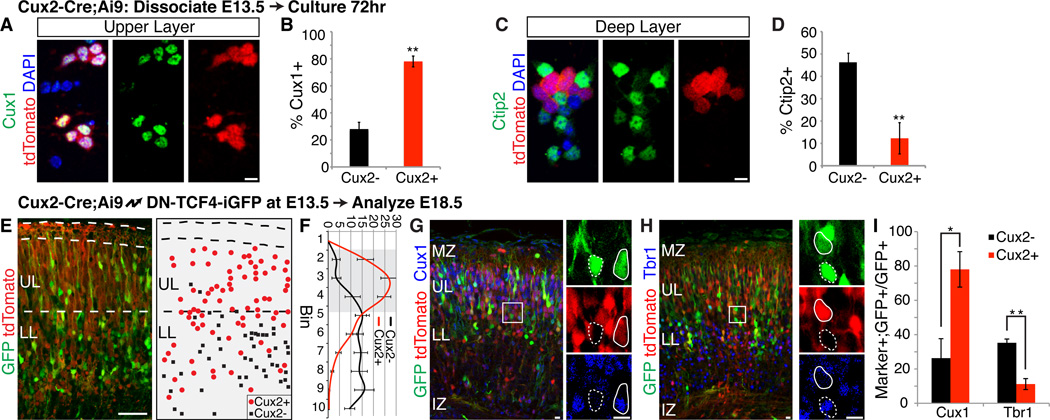
We next asked whether the generation of distinct neuronal subtypes is an intrinsic property of the two different progenitor types. Dissociated cortical cells from Cux2-Cre;Ai9 embryos were cultured until they differentiated and were stained with layer specific markers (Fig. 4, A and C). The in vivo situation was recapitulated in vitro, with ~80% of the Cux2+ progenitors generating neurons with upper layer identity (Fig. 4, B) and the majority of the Cux2− progenitors producing neurons with lower layer identity (Fig. 4, D). Thus, the neurogenic differences between the two progenitor populations are maintained outside their normal developmental niche, indicating an intrinsic aspect to their divergent fate specification. This result also suggests that birthdate may not be causative to cell fate. To test this hypothesis, we electroporated dominant-negative TCF4 into Cux2+ RGCs to force their premature cell cycle exit (19), so that they generated neurons at the expense of self renewal (fig. S4). These prematurely generated Cux2+ neurons still preferentially occupied upper layers (Fig. 4, E and F) and expressed upper layer, but not lower layer, markers (Fig. 4, G–I), indicating that their fate was not altered by the change in birthdate. Thus, Cux2+ RGCs are intrinsically specified to generate upper layer neurons, independent of niche or birthdate.
Fig. 4. Cux2+ progenitors are intrinsically specified to generate upper layer neurons, independently of niche or birthdate.
(A–D) E13.5 Cux2-Cre;Ai9 cortical cells cultured in vitro for 72 hrs and stained for Cux1 (A) or Ctip2 (C). tdTomato absence/presence identified Cux2−/ Cux2+ cells. Quantification is percentage (± SEM) of Cux2− vs. Cux2+ cells expressing Cux1 (B) or Ctip2 (D). **, P < 0.0001. Scale bars, 10 µm. (E-I) Cux2-Cre;Ai9 embryos electroporated at E13.5, analyzed at E18.5 for laminar position and molecular identity. (E) Example image shown on left; example schematic representation on right. UL, upper layers; LL, lower layers. Scale bar, 50 µm. (F) Cell position quantified by dividing the cortical plate into 10 equal-size bins. Graph shows percentage (± SEM) of electroporated Cux2− vs. Cux2+ cells in each bin. Upper layer bins are shaded. (G and H) Immunostaining for Cux1 (G) and Tbr1 (H). High magnifications of boxed areas shown at right. tdTomato absence (dotted outline) and presence (solid outline) identified Cux2− and Cux2+ cells, respectively. Scale bars, 10 µm. (I) Percentage (± SEM) of Cux2− vs. Cux2+ cells expressing Cux1 or Tbr1. *, P < 0.02; **, P < 0.005.
Our data show that a subset of RGCs are specified to generate upper layer neurons regardless of birthdate, but these progenitors are intrinsically programmed to generate neurons predominantly later than their lower layer counterparts. Thus, contrary to the prevailing model (2), our study indicates that molecular fate specification ensures proper birth order, rather than vice versa. Our data also suggest that the minor fraction of callosal projection neurons found in lower layers are derived from the same RGC pool as the major population of callosal neurons in upper layers, demonstrating a common lineage for these functionally similar neurons, irrespective of cortical layer position. Although this model applies to the broad RGC subclasses that generate intracortical vs. subcortical/subcerebral projections neurons, it remains possible that the potential of Cux2+ and Cux2− progenitors is subsequently progressively restricted to further specify neuronal subtypes within the two lineages.
Upper cortical layers are expanded in primates and are required for high-level associative connectivity. Defects in their function are implicated in the etiology of cognitive syndromes such as schizophrenia and autism. The subventricular zone of primates, and especially in humans, is enlarged compared to other species and contains outer subventricular RGCs that are thought to generate increased numbers of upper layer cortical neurons in the primate brain (20). Our findings suggest that an equally important evolutionary advance was the subdivision of labor amongst RGCs in the ventricular zone to generate lower and upper layer neurons.
Supplementary Material
Acknowledgments
We thank G. Fishell for Nestin-CreERT2 mice, H. Zeng for Ai9 reporter mice, P. Chambon for pCreERT2, A. Maximov for the FLEx backbone, T. Wagner, S. Courtes and P. Kazmierczak for experiments that did not appear in the final manuscript, and F. Polleux, E. Grove, N. Grillet and S. Courtes for critical comments. This work was supported by the NIH (SJF, NS060355; UM, NS046456, MH078833), Generalitat Valenciana (CGS, APOSTD/2010/064), Ministerio de Educacion (CGS, EX2009-0416; IMG, FU-2006-1238), CIRM training grant (IMG, AE), the Skaggs Institute for Chemical Biology (UM), and the Dorris Neuroscience Center (UM). SJF, IMG, CGS and UM conceived the project; SJF, CGS, IMG and AE designed, performed and analyzed experiments with assistance from SRHP and CR; SJF and UM prepared the manuscript, with revisions from CGS, IMG and AE.
Footnotes
References and Notes
- 1.Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 2.Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Curr Opin Neurobiol. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hevner RF, et al. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- 4.Kowalczyk T, et al. Intermediate neuronal progenitors (basal progenitors) produce pyramidalprojection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nieto M, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers IIIV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 6.Zimmer C, Tiveron M-C, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
- 7.Franco SJ, Martinez-Garay I, Gil-Sanz C, Harkins-Perry SR, Müller U. Reelin regulates cadherin function via Dab1/Rap1 to control neuronal migration and lamination in the neocortex. Neuron. 2011;69:482–497. doi: 10.1016/j.neuron.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcamo EA, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Britanova O, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 10.Schnütgen F, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 11.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifeninducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 13.Zervas M, Millet S, Ahn S, Joyner AL. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 14.Balordi F, Fishell G. Mosaic removal of hedgehog signaling in the adult SVZ reveals that the residual wild-type stem cells have a limited capacity for self-renewal. J Neurosci. 2007;27:14248–14259. doi: 10.1523/JNEUROSCI.4531-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa MR, Bucholz O, Schroeder T, Götz M. Late origin of glia-restricted progenitors in the developing mouse cerebral cortex. Cereb Cortex. 2009;19(Suppl 1):i135–i143. doi: 10.1093/cercor/bhp046. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi T, Goto T, Miyama S, Nowakowski RS, Caviness VS. Sequence of neuron origin and neocortical laminar fate: relation to cell cycle of origin in the developing murine cerebral wall. J Neurosci. 1999;19:10357–10371. doi: 10.1523/JNEUROSCI.19-23-10357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizutani K-I, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 18.Yoon K-J, et al. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Woodhead GJ, Mutch CA, Olson EC, Chenn A. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J Neurosci. 2006;26:12620–12630. doi: 10.1523/JNEUROSCI.3180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 22.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 23.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 25.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to CreloxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 26.Tiveron MC, Hirsch MR, Brunet JF. The expression pattern of the transcription factor Phox2 delineates synaptic pathways of the autonomic nervous system. J Neurosci. 1996;16:7649–7660. doi: 10.1523/JNEUROSCI.16-23-07649.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hand R, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Qian X, Goderie SK, Shen Q, Stern JH, Temple S. Intrinsic programs of patterned cell lineages in isolated vertebrate CNS ventricular zone cells. Development. 1998;125:3143–3152. doi: 10.1242/dev.125.16.3143. [DOI] [PubMed] [Google Scholar]
- 29.Polleux F, Ghosh A. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. 2002;2002:pl9. doi: 10.1126/stke.2002.136.pl9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.